Abstract
Background
The Ampelopsis clade (Ampelopsis and its close allies) of the grape family Vitaceae contains ca. 43 species disjunctly distributed in Asia, Europe, North America, South America, Africa, and Australia, and is a rare example to study both the Northern and the Southern Hemisphere intercontinental disjunctions. We reconstruct the temporal and spatial diversification of the Ampelopsis clade to explore the evolutionary processes that have resulted in their intercontinental disjunctions in six continents.
Results
The Bayesian molecular clock dating and the likelihood ancestral area analyses suggest that the Ampelopsis clade most likely originated in North America with its crown group dated at 41.2 Ma (95% HPD 23.4 - 61.0 Ma) in the middle Eocene. Two independent Laurasian migrations into Eurasia are inferred to have occurred in the early Miocene via the North Atlantic land bridges. The ancestor of the Southern Hemisphere lineage migrated from North America to South America in the early Oligocene. The Gondwanan-like pattern of intercontinental disjunction is best explained by two long-distance dispersals: once from South America to Africa estimated at 30.5 Ma (95% HPD 16.9 - 45.9 Ma), and the other from South America to Australia dated to 19.2 Ma (95% HPD 6.7 - 22.3 Ma).
Conclusions
The global disjunctions in the Ampelopsis clade are best explained by a diversification model of North American origin, two Laurasian migrations, one migration into South America, and two post-Gondwanan long-distance dispersals. These findings highlight the importance of both vicariance and long distance dispersal in shaping intercontinental disjunctions of flowering plants.
Background
Understanding the underlying mechanisms for the evolution of wide-ranging disjunct patterns has long been a major focus of biogeography [1-5]. Taxa disjunct at the global level involving both Northern and Southern Hemisphere are particularly informative because their histories may have general implications for other groups. Biogeographic history in the Northern Hemisphere is complicated, but has usually been explained by the widespread distribution of the Boreotropical flora in the Eocene and followed by appearance of more temperate forest elements during the mid-Tertiary extirpations of thermophilic elements in response to climatic cooling episodes of the late Eocene and the Plio-Pleistocene [6-11]. The Southern Hemisphere is often interpreted to show a vicariance pattern attributed to the sequential breakup of Gondwanan landmasses [12]. Recent studies on Nothofagus have demonstrated the relevance of long distance dispersal rather than vicariance in shaping Gondwanan distributional patterns [13-16].
Commonly three main routes for the migration of taxa between the Northern and the Southern Hemisphere have been recognized (Figure 1). The first is the opening of biotic exchanges between North and South America at various times in the Tertiary [1,17]. The second hypothetical migration route is the trans-Tethyan dispersal between Europe and Africa [18]. The third is less common concerning the possible route between Asia and Australia in the Miocene and later [18,19]. These three routes can be viewed as alternative hypotheses for the ex situ origin of elements of global diversity.
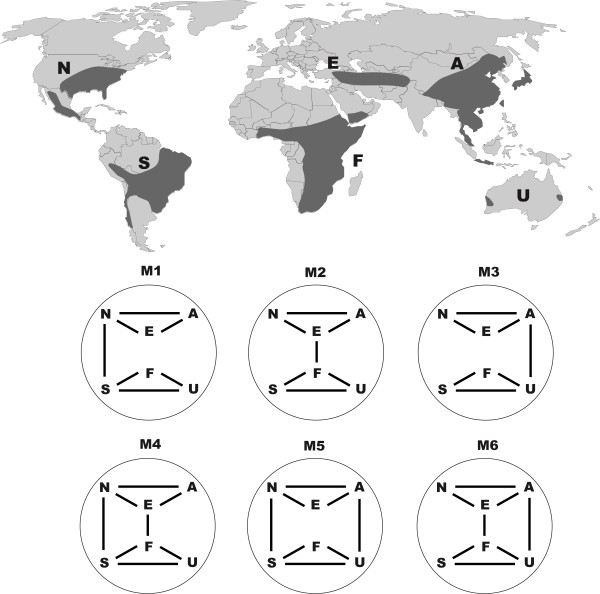
Figure 1.
Delimitation of the six areas for taxa in the Ampelopsis clade and the schematic models used in the Lagrange analyses. N = North America; A = eastern Asia; E = Europe to central Asia; F = Africa; S = South America; and U = Australia.
There are many examples of global distribution between the Southern and the Northern Hemisphere, particularly in pantropical families (e.g., Rubiaceae, Annonaceae, Lauraceae) or in many large cosmopolitan genera, such as Ranunculus [20], Senecio [21], and Lobelia [22,23]. Nevertheless, all of them have continuous distribution from the tropics to the temperate zones. Molecular studies usually show a very complex disjunct history of large taxa with relatively low resolution [24,25]. There are also some taxa with relatively few species that exhibit an intercontinental disjunction involving both the Northern and the Southern Hemisphere. However, the disjunction in such taxa usually involves only one or two northern or southern landmasses. Examples include Caltha (Ranunculaceae) with a global disjunction but absent from Africa [26] and Thamnosma (Rutaceae) disjunct between North America and Africa [27]. Of great interest, we have recently found a truly global disjunct pattern in a small group including Ampelopsis Michx. and its relative taxa from the grape family, Vitaceae [28-30].
Vitaceae is a well-known group of flowering plants having a largely pantropical distribution in Asia, Africa, Australia, the neotropics, and the Pacific islands, with only a few genera in temperate regions [31-33]. As currently circumscribed, Ampelopsis is one of the few genera mostly restricted to the north temperate zone. It has approximately 25 species disjunctly distributed in Eurasia (c. 22 spp.) and North and Central America (3 spp.). Recent phylogenetic analyses based on plastid or nuclear sequences revealed that there are at least two disjunctions between the New and the Old World in the genus [28,29]. More interestingly, both chloroplast and nuclear data clearly suggested that the African Rhoicissus Planch. and the South American Cissus striata Ruiz & Pav. complex form a clade nested within the paraphyletic Ampelopsis [28,29]. Rhoicissus consists of about 12 species endemic to tropical and southern Africa. The Cissus striata complex contains four species from South America [29,34]. Furthermore, the Australian genus Clematicissus Planch. seemed to be closely related to the Cissus striata - Rhoicissus clade based on plastid data [30,35]. There are only two Clematicissus species known from Australia: C. opaca from Australia's eastern region and C. angustissima from the west coast [30]. Therefore, the Northern Hemisphere Ampelopsis and its three close relatives (Rhoicissus, Clematicissus and the Cissus striata complex) from the Southern Hemisphere (hereafter referred to as the Ampelopsis clade) demonstrate an unusual global intercontinental disjunct pattern involving six continents (Figure 1). Yet morphologically, the Ampelopsis clade seems to be heterogeneous, such as in having 4-7-merous flowers and fleshy to dry fruits [29].
The Ampelopsis clade offers a good opportunity to explore the origin and evolution of the global intercontinental disjunct pattern in flowering plants, especially concerning both Northern and Southern Hemisphere intercontinental disjunctions. A hierarchical global distribution was predicted by our previous studies, but with limited sampling [28,29]. We herein employ phylogenetic, molecular dating, and biogeographic methods to reconstruct the evolutionary history of the Ampelopsis clade based on a comprehensive sampling scheme using four plastid regions (trnL-F, rps16, psbA-trnH, and atpB-rbcL).
Methods
Taxon sampling, DNA sequencing, and phylogenetic analyses
We sample 28 of the 43 species (65%) of the Ampelopsis clade including all three North American Ampelopsis species, 15 of the 22 Eurasian Ampelopsis, five of the 12 Rhoicissus species from Africa, three of the four species from the Cissus striata complex from South America, and the two Australian Clematicissus species (Additional file 1, Table S1). The sampling covers the entire extant geographic range of Ampelopsis and its close relatives from both the Northern and Southern Hemisphere. But we still have some missing taxa in our sampling scheme, such as Rhoicissus with only 41% species sampled. However, each group is supposed to be monophyletic based on morphological, biogeographic, and molecular evidence [28-30,34,36]. The missing taxa in our sampling should have little effect in the present study that focused on phylogenetic relationships among genera and biogeographic evolution at the intercontinental level. In order to place our analyses of the Ampelopsis clade in a broad framework on the family level, we sampled 66 additional taxa from the other major groups in Vitaceae (i.e., the Vitis - Parthenocissus - Ampelocissus clade, the core Cissus clade, and the Cyphostemma - Caryatia - Tetrastigma clade) plus three Leea species of Leeaceae based on previous investigations [28,29].
Total DNAs were extracted from silica gel dried leaves using the Dneasy Plant Mini Kit (QIAGEN, Crawley, UK). Amplification and sequencing followed Soejima and Wen (2006) for trnL-F, rps16, and atpB-rbcL, and Meng et al. [37] for psbA-trnH. DNA sequences were assembled using Sequencher v4.1.4 (Gene Codes Corp., Ann Arbor, Michigan, USA). Sequence alignment was initially performed using MUSCLE 3.8.31 [38] in the multiple alignment routine followed by manual adjustment in Se-Al v2.0a11 (http://tree.bio.ed.ac.uk/software/seal/). The chloroplast genome is generally considered as one unit without recombination although there have been reports of recombination in the chloroplast genome [39]. Therefore, we combined all the plastid data (trnL-F, rps16, psbA-trnH, and atpB-rbcL) in our analysis. The combined plastid data were analyzed using Bayesian inference as implemented in MrBayes 3.1.2 [40]. The best-fit model of nucleotide substitution (GTR + I + G) was determined by MrModelTest 2.3 [41] using the Akaike Information Criterion (AIC). Variation of gaps in our sequences is not complicate. A total of 31 binary characters were coded for gaps according to Simmons and Ochoterena [42] and separated into independent partition in all analyses. Bayesian tree topology and posterior probabilities (PP) were determined from two independent runs of four incrementally heated chains. Runs were performed for 5 million generations with sampling of trees every 500th generation. When the log-likelihood scores were found to have stabilized, a consensus tree was calculated after omitting the first 10% of trees as burn-in.
Divergence time estimation
For molecular dating analyses, the strict molecular clock model was rejected from our dataset based on a likelihood ratio test performed in PAUP* [43]. Therefore, we estimated node ages within the Ampelopsis group using a Bayesian relaxed clock model as implemented in BEAST v1.6.1 [44]. We largely followed the dating strategies in Nie et al. (2010), which analyzed diversification in Parthenocissus of Vitaceae. After optimal operator adjustment as suggested by the output diagnostics from several preliminary BEAST runs, two final independent runs (each 50 million generations) were performed on a cluster of Mac XServes used for analysis of biological data at the Smithsonian Institution (http://topazweb.si.edu). Tracer version 1.5 was used to check for convergence between the runs [44]. After discarding the first 10% samples as burn-in, the trees and parameter estimates from the two runs were combined using LogCombiner 1.6.1 [44]. Results were considered reliable once the effective sampling size (ESS) for all parameters exceeded 200 as suggested by the program manual [45]. The samples from the posterior were summarized on the maximum clade credibility tree using the program TreeAnnotator 1.6.1 [44] with posterior probability limit of 0.5 and mean node heights summarized.
Fossil seeds of Vitaceae can be differentiated to the generic level [46,47]. The oldest best preserved seed fossil of the family is from the late Paleocene of the Beicegel Creek locality in North Dakota. This fossil is undoubtedly assigned to Ampelocissus s.l. (as A. parvisemina Chen & Manchester) and is easily distinguished from all other vitaceous genera by its long, parallel ventral infolds and a centrally positioned oval chalazal scar [46]. Since the Ampelocissus s.l. is not monophyletic with Vitis nested within it [28], the A. parvisemina fossil thus may represent an early member of the Ampelocissus clade retaining some characters shared with its common ancestor to Vitis [46]. The stem age of the Ampelocissus - Vitis clade was thus fixed at 58.5 ± 5.0 million years ago (Ma).
For the root age of Vitaceae, Nie et al. (2010) fixed the split between Vitaceae and Leea as 85 ± 4 based on the estimated age of 78-92 Ma by Wikström et al. (2001). Recently, Bell et al. (2010) reported an estimate ranging from 65 (45 - 81) to 48 (21 - 79) Ma for the crown age Vitis - Leea clade, which is roughly consistent with the earliest fossil evidences of Vitaceae in the Palaeocene [46]. However, their results may have underestimated for Vitaceae because the oldest fossil of A. parvisemina is undoubtedly assigned to the Ampelocissus s.l. within Vitaceae and the family is predicted to have a Cretaceous history in view of its basal position in the rosids and the presence of Cretaceous rosid fossils [48]. The time estimates of angiosperms by Magallón and Castillo (2009) also suggested a pre-Tertiary origin as 90.65 (90.47 - 90.84) to 90.82 (90.64 - 91) Ma for Vitaceae. The inferences from Magallón and Castillo (2009) and Wikström et al. (2001) are close, although the later was criticized for the nonparametric rate smoothing method and for calibrating the tree using only a single calibration point. Therefore, we used the estimate from Magallón and Castillo (2009) and set the normal prior distribution of 90.7 ± 1.0 Ma for the stem age of the family. A low standard error was used because of the narrow 95% confidence from Magallón and Castillo (2009).
Ancestral area reconstruction
Several methods have been recently proposed that take into account of genetic branch lengths, phylogenetic uncertainty, and branch length uncertainty for reconstructing distributional change through evolutionary time, using either maximum likelihood [49] or Bayesian inference [50]. The ancestral area of the Ampelopsis clade was reconstructed with the likelihood analysis using the program Lagrange version 20110117 [49,51]. Unlike the DIVA method [52], this likelihood approach incorporates an explicit dispersal-extinction-cladogenesis (DEC) model of dispersal routes available at historical intervals correlating stochastic events with lineage persistence [49]. The likelihood analysis is prone to estimate wide ancestral ranges for early-branching lineages [53-55]. In our case, ancestral ranges were assumed to include no more than two areas since all extant species in the Ampelopsis clade are restricted into only one area. Moreover, spatial and temporal constraints (e.g., area distances, continent connections, dates of geological origin) may be imposed in the DEC model estimation, providing a more accurate estimation of the ancestral ranges and hypothesis testing of different geographic scenarios. We did not conduct the Bayesian calculation of ancestral geographic distributions with standard continuous-time Markov chains (CTMCs), because geologic information (e.g., the presence and dissolution of land bridges and island chains) is not explicitly incorporated into the analyses.
We herein used the likelihood method to test a null model and six alternative biogeographic scenarios (Figure 1) based on the hypothesized dispersal or migration routes between the Northern and the Southern Hemisphere. Six areas were delimited by continental divisions and the extant distributions of the Ampelopsis clade: 1) N - North America including Central America; 2) S - South America; 3) F - Africa; 4) A - eastern Asia; 5) E - Europe to central Asia; and 6) U - Australia (Figure 1). The unconstrained null model (M0) assumes that spatial and temporal distribution has no effect on biogeographic patterns of evolution and allows geographic ranges to include any possible combination of continents and permits direct dispersal between any area pairs. The M1 model favors a migration route from North to South America (N - S) with the biogeographic connections between Europe and Africa (E - F) and eastern Asia and Australia (A - U) excluded from our analyses. Similarly, the M2 model considers the connection between E - F and did not allow other possibilities. The migration route between Asia and Australia (M3) seems less likely, but we still considered it in our analyses as a comparison. We also test models that allow two connections between the Northern and Southern hemispheres (M4 - M6 in Figure 1). Following Ree et al. (2005), the results between models were assessed by directly comparing their log-likelihood scores. The conventional cut-off value over two log-likelihood units was considered statistically significant, and models with lower likelihood score were rejected [56,57].
Results
The total length of the aligned data matrix is 3933 bp. The Bayesian consensus tree is highly congruent with the maximum clade credibility tree obtained from BEAST and the later is shown in Figure 2 with PP support values > 0.50. Our results support the monophyly of the Ampelopsis clade with three major lineages resolved within the Ampelopsis clade (Figure 2). Two distinct lineages (hereafter named as North I and II) correspond to the two sections of Ampelopsis [58]. North I includes all species of section Leeaceifoliae with pinnately to bipinnately compound leaves. North II consists of taxa of section Ampelopsis with simple or palmately-divided or palmately-compound leaves. The Southern Hemisphere taxa (the African Rhoicissus, the South American Cissus striata complex, and the Australian Clematicissus) form a clade (the South group in Figure 2).
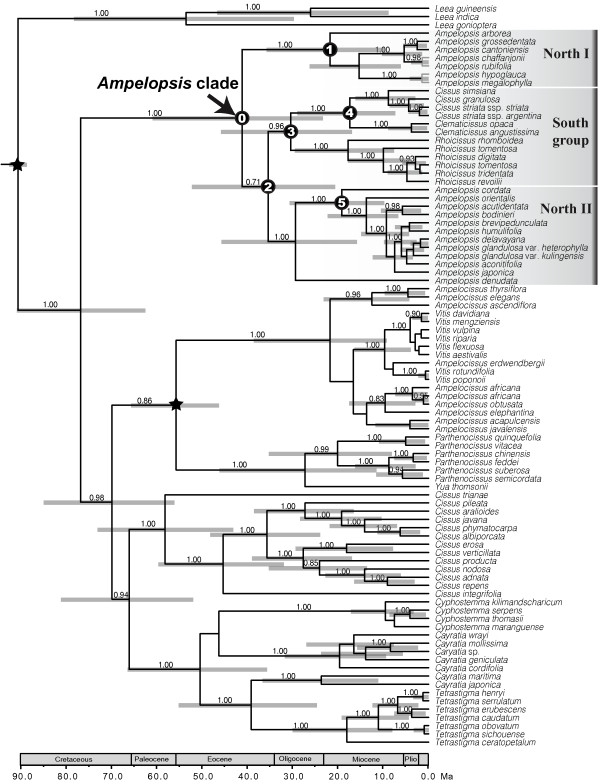
Figure 2.
Maximum clade credibility tree inferred with BEAST, with the 95% highest posterior density indicated by gray bars. Nodes of interests were marked as 0 to 5 as in Table 1; and calibrations are indicated with black stars. Values above branches represent Bayesian posterior probabilities.
A total of 100 million generations (2 runs of 50 million generations each) are necessary to reach sufficient ESS. The Ampelopsis clade is estimated to have diverged from its close relatives in Vitaceae at 41.2 (23.4 - 61.0) Ma in the Eocene. The Bayesian estimates also suggest that all the other major clades of Vitaceae (e.g., Vitis - Parthenocissus - Ampelocissus clade, core Cissus clade, and Cyphostemma - Cayratia - Tetrastigma) had already diversified in the Eocene (Figure 2). Ages of major groups within the Ampelopsis clade obtained in our study are summarized in Table 1.
Table 1.
Results of molecular dating and ancestral range reconstruction for major nodes within the Ampelopsis clade.
| -lnL | D | E | Node 0: Crown Ampelopsis clade |
Node 1: Crown North I |
Node 2: Stem South group |
Node 3: Crown South group |
Node 4: Crown Cissus striata complex - Clematicissus |
Node 5: Disjunct |
|
|---|---|---|---|---|---|---|---|---|---|
| Molecular dating with BEAST (Ma) |
41.2 (23.4-61.0) | 21.8(6.2-26.3) | 35.5(20.7-52.3) | 30.5(16.9-45.9) | 17.4(7.4-29.0) | 19.2(6.7-22.3) | |||
| M0 | 32.42 | 0.003401 | 0.004318 | N|NF (0.25) | A|N (0.81) | F|N (0.32) | S|F (0.22), U|F (0.21) |
U|S (0.63) | N|A (0.47) |
| M1 | 30.19 | 0.007085 | 0.003586 | N|NS (0.61) | A|N (0.85) | S|N (0.92) | S|F (0.54) | U|S (0.79) |
N|A (0.46), N|E (0.36) |
| M2 | 34.47 | 0.00986 | 0.008992 | A|E (0.25), N|E (0.20) |
A|N (0.51) | F|E (0.55) | F|F (0.39) | U|S (0.32) | N|E (0.51) |
| M3 | 32.94 | 0.007539 | 0.005598 | A|AU (0.60) | A|N (0.68) | U|A (0.90) | U|F (0.42), U|U (0.41) |
U|S (0.77) | N|A (0.72) |
| M4 | 30.65 | 0.006486 | 0.003784 | N|NS (0.61) | A|N (0.84) | S|N (0.91) | S|F (0.51) | U|S (0.78) | N|A (0.46), N|E (0.35) |
| M5 | 30.89 | 0.006096 | 0.003555 | N|NS (0.58) | A|N (0.84) | S|N (0.86) | S|F (0.51) | U|S (0.79) | N|A (0.48), N|E (0.35) |
| M6 | 33.3 | 0.007028 | 0.005918 | A|AU (0.55) | A|N (0.66) | U|A (0.80) | U|U (0.40), U|F (0.37) |
U|S (0.73) | N|A (0.65) |
The nodes of interest are shown in Figure 2, and the likelihood scores (-lnL), and estimates of dispersal (D) and extinction (E) rates (events per million years) are given. M0 is a null model without constraints and M1-M6 are alternative models in the Lagrange analyses. Only the highest relative probability is shown. Bold text represents the model with a significantly better likelihood when compared with the other models tested (more than two log-likelihoods better)
Patterns of temporal and spatial distribution of the Ampelopsis clade are inferred using the maximum likelihood DEC method. We compare seven models (i.e., a null and six alternatives) for the six areas (Figure 1) and the effects of different models on likelihood reconstructions are shown in Table 1. Analyses based on M1, M4, and M5 typically have lower likelihood scores than other models and produced nearly identical results (Table 1). For example, all of them suggested that the ancestral range split at the stem South lineage is between North and South America (Node 2 in Table 1). Our results also suggest that the three models (M1, M4, and M5) are significantly different from the others (M2, M3, and M6) with scores over two log-likelihood units. The M1 model is suggested as the best one with the highest likelihood score, and this model is shown in Figure 3.
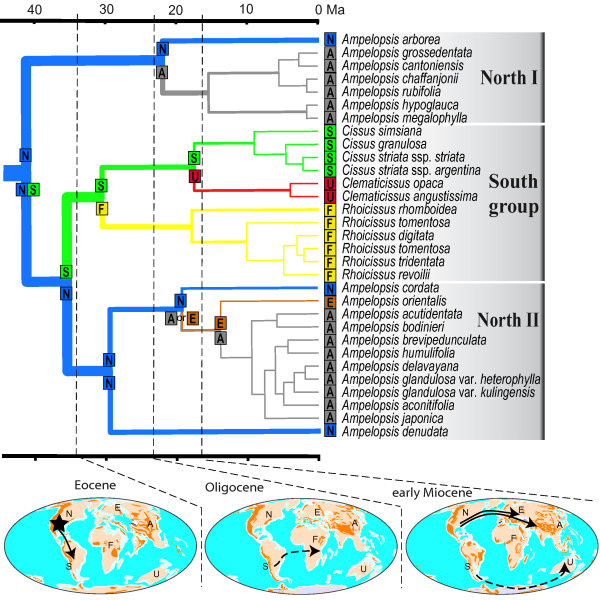
Figure 3.
Biogeographic scenario for the global disjunction of the Ampelopsis clade based on molecular dating and the best M1 model with the highest likelihood score in the ancestral range analyses. Long distance dispersal is indicated as dash lines and migration as solid lines. The ancestral area of the Ampelopsis clade is shown with black stars on the maps. The tree branches and ranges on the tree are coded as follows: blue = North America (N); grey = eastern Asia (A); orange = Europe and central Asia (E); green = South America (S); yellow = Africa (F); and red = Australia (U).
Discussion
North American origin
The Ampelopsis clade is composed of two distinct Laurasian lineages each disjunct between the Old and the New World and one South group with a Gondwana-like intercontinental disjunction: (Africa (Australia, and South America)) (Figure 3). The most likely model M1 in the DEC analyses suggested that the Ampelopsis clade had an early diversification in North America with a geographic split between N (North America) and NS (North and South America) (Table 1; Figure 3). The fact that most fossil records of the family including the oldest record in the Paleocene are found from North America [46] is consistent with this "out of North America" hypothesis. Although South America is inferred as part of the ancestral areas (Table 1), it seems less likely to be the ancestral area of the Ampelopsis clade than North America because there are very few fossils known before the Eocene of South America [47]. Furthermore, phylogenetic results also contradict the possibility of South American origin because the South American group is well embedded within the Ampelopsis clade (Figure 2). Asia also seems less likely than North America to be the ancestral area of the Ampelopsis clade, in spite of its highest extant species richness of the lineage. No seed records with ages younger than Oligocene are known from Asia [47].
The ancestral area for a taxon is usually expected to be correlated with high extant species richness. For example, eastern Asia usually has a higher level of species diversity and endemism, and has been suggested to be the ancestral area for many eastern Asian - eastern North American disjunct groups [59-63]. Donoghue and Smith (2004) found a predominance of directionality from Asia to the New World. Of the 29 lineages they analyzed with an eastern Asian and eastern North American disjunction, only one lineage showed directionality from eastern North America to eastern Asia. However, Wen et al. (2010) reported many more lineages with North American origins and subsequent migrations into eastern Asia, with 29 of the total 98 examined (30%) lineages migrated/dispersed from the New World to the Old World. It seems that eastern Asia has been over-emphasized as an ancestral area for Laurasian taxa due to its retention of the greater number of species [62,64]. North America is supported to have played an important role in the early evolution of the two Ampelopsis lineages in spite of the lower species diversity today in North America compared with eastern Asia. The lower species richness in North America is often explained by the hypothesis that both North America and eastern Asia were occupied by Boreotropical elements in the early Tertiary but North America suffered more severe extinctions with global cooling beginning in the late Eocene or Oligocene [10,63]. The high level of species diversity and endemism in Asia can also be attributed to secondary diversification due to habitat heterogeneity as well as a lower rate of extinctions in the late Tertiary [60,61].
Diversification pattern in the Southern Hemisphere
The likelihood analyses using Lagrange based on M1, M4, and M5 suggest that the most likely route between the Northern and the Southern Hemisphere is from North to South America, although more options of connections are permitted in M4 (between Europe and Africa) and M5 (between eastern Asia and Australia) (Table 1 and Figure 3). Ampelopsis may have dispersed into South America via scattered continental and/or volcanic islands that connected North and South America at various times in the Tertiary [1,17], such as via the proto Greater Antilles (ca. 50 Ma) or via GAARlandia that existed around 33 - 35 Ma [17,65]. The separation of the southern lineage from its Laurasian ancestor at 35.5 (20.7 - 52.3) Ma in the late Eocene broadly coincides with a possible biological connection between North and South America around the Eocene-Oligocene boundary [66]. Fossil seeds found from the Eocene of South America are closely related to those of the Central American Ampelocissus, indicating the possible floristic connection between North and South America at that time [46].
Another possible migration route is that the Laurasian ancestors of the Ampelopsis clade reached Africa using the Boreotropical connection via the North Atlantic and Europe in the late Eocene to early Oligocene. However, this hypothesis is rejected by the Lagrange analysis (see M2 in Table 1). The model M4 with two possible connections between the Northern and Southern Hemisphere (N - S and E - F, Figure 1) did not support the European-African (E - F) route. Although the separation of the southern lineage from its Laurasian ancestor in the early Oligocene broadly coincides with the disruption of the Boreotropical flora around the Eocene-Oligocene boundary [66], both the shallow seas that separate Africa from Eurasia and the dry belt in northern Africa were barriers to biotic exchange between the two continents in the early to mid Tertiary [1,67,68]. The third hypothesis (the M3 model, Table 1) is that Ampelopsis entered the Southern Hemisphere via the Asian - Australian connection. This model seems quite unlikely based on our analyses (Table 1). The model M5 that permits two connections between Northern and Southern Hemisphere (N - S, A - U, Figure 1) also prefers the connection between North and South America (N - S) rather than the Asian - Australian connection (A - U). The availability of biotic interchange between Australia and Asia beginning at the Miocene [18] is too recent to support this scenario.
The divergence time 30.5 (95% HPD: 16.9 - 45.9) Ma in the early Oligocene was estimated for the first split between South America and Africa (node 3 in Figure 2 and Table 1). This time is well after the last possible connection of Africa and other southern landmasses at around 96-105 Ma [18,19]. We thus argue that long distance dispersal (LDD) is the most plausible mechanism for their southern intercontinental disjunction. Vitaceae taxa are usually dispersed by animals, especially birds [32,69-71]. All taxa in the Ampelopsis clade except the Australian Clematicissus angustissima bear fleshy berries that may have facilitated LDD. In particular, LDD has been recently accepted by a number of studies as the driving force for plant disjunctions in the Southern Hemisphere, especially for those with relatively recent divergence times [5,20,72-74]. Biogeographic studies on Vochysiaceae [75] suggested a LDD from South America to Africa in the Oligocene. Givnish et al. [76] showed that the single African genus Maschocephalus of Rapateaceae is of recent origin in the late Miocene and reached Africa from South America via LDD. Dispersals between Africa and South America have also been suggested for a number of well-studied taxa, such as in Melastomataceae [77], and Simaroubaceae [73].
Our results support that the southern lineage of Ampelopsis arrived in Australia from South America in the early Miocene (node 4 in Figure 2). Migration between Australia and South America may be alternatively explained by a trans-Antarctic exchange [12]. This Antarctic route existed during the late Cretaceous-early Tertiary and was interrupted only in the late Eocene (30-35 Ma) when the South Tasman Sea opened up between Australia and eastern Antarctica [19,78]. This route is supported by evidence from several plant groups, such as Annonaceae [79], and Sapotaceae [80]. Yet the split between the South American Cissus striata complex and the Australian Clematicissus in the early Miocene is too young to be explained by an Antarctic migration. LDD is the most plausible explanation for this disjunction. Finally, the disjunction of the two Clematicissus species in eastern and western Australia [30,81] may represent a relict distribution and their divergence time in the Pliocene is consistent with the aridification in central Australia at that time [82,83].
Laurasian migrations
The DEC reconstruction suggests a North American - eastern Asian split (N|A) for the North I disjunction and a North American - eastern Asian or North American - Europe split (N|A or N|E) for the disjunction in the North II lineage (Table 1; Figure 3). The two Northern Hemisphere disjunctions may have involved the North Atlantic land bridges or the Bering land bridge from North America to eastern Asia [10,84]. We prefer to use the North Atlantic route because it is well supported in sect. Ampelopsis (North II in Figure 3). The southern North American Ampelopsis denudata diverged first, followed by the southeastern US A. cordata. The western Asian/southern European A. orientalis is then sister to the large Asian clade. The relative position of these areas in the cladogram is congruent with the migration of the lineage from North America to Europe across the North Atlantic land bridges, and the lineage then reached Asia subsequently (Figure 3).
The Bayesian molecular clock dating with fossil calibration suggests an early Miocene split of the two disjunct groups in Ampelopsis (nodes 1 and 5 in Figure 2). The divergence times are also consistent with the possibility of the North Atlantic migration route. Based on paleogeological, zoological, and botanical fossil evidence, Tiffney (1985b) argued for the importance of the North Atlantic land bridges to tropical or warm temperate species in the early Eocene to middle Miocene. A continuous belt of Boreotropical elements covered much of the southern part of North America, southern Eurasia, and northwestern Africa in the Eocene [9,66]. At that time, plant migrations through direct land connection or across limited water gaps were possible through the North Atlantic land bridges. Significant cooling during the Oligocene resulted in southward retreats and the extirpation of some lineages comprising this flora [64,85,86]. A gradual warming period occurred into the early Miocene, resulting in the expansion of some evergreen and thermophilic lineages in Europe and North America [87,88]. Dispersal of Boreotropical or warm temperate thermophilic elements, such as Ampelopsis, is therefore considered likely across the North Atlantic land bridges during this period. There are very few extant Vitaceae species in Europe, but many vitaceous seeds were reported in the early Tertiary of Europe [47,89]. Together, these lines of evidence strongly suggest that the Ampelopsis clade may have used the corridors via the North Atlantic land bridges as a pathway to reach Eurasia in the early Miocene.
Conclusions
Our results suggest a complex history of diversification in the Ampelopsis clade to explain the global disjunctions that includes a North American origin, two Laurasian migrations, one migration into South America, and two post-Gondwanan LDDs. These findings may have general implications for the origin and diversification of plants with global disjunctions. Asia, Africa, or South America has often been suggested as the ancestral area for many intercontinental disjunct groups [4,90,91]. Evidence from the Ampelopsis clade suggests that North America may have played an important role in the origin of some modern flowering plants in spite of its often lower species diversity when compared with other areas, such as eastern Asia [92] or Africa [93]. Recent biogeographic analyses of several other groups appear to provide additional examples of North American origins (e.g., Phryma [94]; Simaroubaceae [73]). This study also highlights the importance of the North and South American route in the global diversification between the Northern and the Southern Hemisphere [95]. This route apparently played an important role in the wide distribution of many pantropical plants in the early Tertiary, such as Annonaceae [96,97], Malpighiaceae [91], and Rubiaceae [98].
Recent biogeographic analyses have underscored the relative importance of LDD to intercontinental disjunctions in the Southern Hemisphere than traditionally assumed [3,13]. The fit between area cladograms and the history of tectonic fragmentation might have been overstated [5]. If we accepted the results of our calibration analyses as absolute, rather than minimum ages, then the Southern Hemisphere clade is too young to have been achieved by the Gondwanan breakup. The Gondwana-like disjunction in the Ampelopsis clade was reconstructed to have a North American origin with an initial migration into South America and then dispersed from South America into Africa and Australia independently via LDD. A similar example from Lycium (85 spp., Solanaceae) was suggested a New World origin of Lycium with recent dispersal from the Americas to Africa, and then to eastern Asia [99-101]. Dispersal has been hypothesized to be the dominant pattern in this genus that has red, fleshy, bird-dispersed fruits [99]. On the other hand, however, the Laurasian lineages in Ampelopsis clade favor a vicariance migration pattern from North America via the North Atlantic land bridges to Eurasia. Our results thus support both the Laurasian migrations and the post-Gondwanan LDD to explain the global disjunction of the Ampelopsis clade.
Authors' contributions
JW, ZLN, HS, and SRM conceived the ideas; JW, YM, QL and ZLN collected the materials; ZLN and YM analyzed the data; and ZLN and JW led the writing. All authors read and approved the final submission.
Supplementary Material
Table S1. Voucher information and GenBank accession numbers of the Ampelopsis clade and representative taxa in Vitaceae. Abbreviations of herbaria are as follows: KUN, Kunming Institute of Botany, Chinese Academy of Sciences; and US, the United States National Herbarium. Accession numbers beginning with JQ indicate sequences generated for this study and the others were obtained from GenBank. A dash means sequences missing.
Contributor Information
Ze-Long Nie, Email: niezl@mail.kib.ac.cn.
Hang Sun, Email: hsun@mail.kib.ac.cn.
Steven R Manchester, Email: stevsteven@flmnh.ufl.edu.
Ying Meng, Email: mengying@mail.kib.ac.cn.
Quentin Luke, Email: quentin.luke@swiftkenya.com.
Jun Wen, Email: wenj@si.edu.
Acknowledgements
This study was supported by grants from the National Science Foundation (DEB 0743474 to SR Manchester and J Wen), the Natural Science Foundation of China (NSFC 31061160184 to H Sun and 31129001 to J Wen), the NSFC-Yunnan Joint project (U1136601 to H Sun), and the John D and Catherine T MacArthur Foundation (to J Wen, R Ree, and G Mueller). Laboratory work was conducted in and partially supported by the Laboratory of Analytical Biology of the National Museum of Natural History, the Smithsonian Institution. Fieldwork in North America was supported by the Small Grants Program of the National Museum of Natural History, the Smithsonian Institution. Michael Nee, Maurizio Rossetto, Ihsan Al-Shehbaz, and Daigui Zhang kindly helped with the sample collection.
References
- Raven PH, Axelrod DI. Angiosperm biogeography and past continental movements. Annals of the Missouri Botanical Garden. 1974;61(3):539–673. doi: 10.2307/2395021. [DOI] [Google Scholar]
- Raven PH. Plant species disjunctions: a summary. Annals of the Missouri Botanical Garden. 1972;59(2):234–246. doi: 10.2307/2394756. [DOI] [Google Scholar]
- Upchurch P. Gondwanan break-up: legacies of a lost world? Trends in Ecology & Evolution. 2008;23(4):229–236. doi: 10.1016/j.tree.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Donoghue MJ, Bell CD, Li JH. Phylogenetic patterns in Northern Hemisphere plant geography. International Journal of Plant Sciences. 2001;162:S41–S52. doi: 10.1086/323278. [DOI] [Google Scholar]
- de Queiroz A. The resurrection of oceanic dispersal in historical biogeography. Trends in Ecology & Evolution. 2005;20(2):68–73. doi: 10.1016/j.tree.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Graham A. In: Floristics and Paleofloristics of Asia and Eastern North America. Graham A, editor. Amsterdam: Elsevier; 1972. Outline of the origin and historical recognition of floristic affinities between Asia and eastern North America; pp. 1–18. [Google Scholar]
- Milne RI, Abbott RJ. The origin and evolution of tertiary relict floras. Advances in Botanical Research. 2002;38:281–314. [Google Scholar]
- Tiffney BH. Perspectives on the origin of the floristic similarity between eastern Asia and eastern North America. Journal of the Arnold Arboretum. 1985;66(1):73–94. [Google Scholar]
- Tiffney BH. The Eocene North Atlantic land bridge: its importance in tertiary and modern phytogeography of the Northern hemisphere. Journal of the Arnold Arboretum. 1985;66(2):243–273. [Google Scholar]
- Wen J. Evolution of eastern Asian and eastern North American disjunct distributions in flowering plants. Annual Review of Ecology and Systematics. 1999;30:421–455. doi: 10.1146/annurev.ecolsys.30.1.421. [DOI] [Google Scholar]
- Sanmartín I, Enghoff H, Ronquist F. Patterns of animal dispersal, vicariance and diversification in the Holarctic. Biological Journal of the Linnean Society. 2001;73(4):345–390. [Google Scholar]
- Sanmartín I, Ronquist F. Southern Hemisphere biogeography inferred by event-based models: Plant versus animal patterns. Systematic Biology. 2004;53(2):216–243. doi: 10.1080/10635150490423430. [DOI] [PubMed] [Google Scholar]
- Knapp M, Stockler K, Havell D, Delsuc F, Sebastiani F, Lockhart PJ. Relaxed molecular clock provides evidence for long-distance dispersal of Nothofagus (southern beech) PLos Biology. 2005;3(1):38–43. doi: 10.1371/journal.pbio.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook LG, Crisp MD. Not so ancient: the extant crown group of Nothofagus represents a post-Gondwanan radiation. Proceedings of the Royal Society B-Biological Sciences. 2005;272(1580):2535–2544. doi: 10.1098/rspb.2005.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder HP, Crisp MD. Nothofagus and Pacific biogeography. Cladistics. 1995;11(1):5–32. doi: 10.1111/j.1096-0031.1995.tb00002.x. [DOI] [PubMed] [Google Scholar]
- Swenson U, Hill RS, McLoughlin S. Biogeography of Nothofagus supports the sequence of Gondwana break-up. Taxon. 2001;50(4):1025–1041. doi: 10.2307/1224719. [DOI] [Google Scholar]
- Iturralde-Vinent MA, MacPhee RDE. Paleogeography of the Caribbean region: Implications for cenozoic biogeography. Bulletin of the American Museum of Natural History. 1999. pp. 1–95.
- Morley RJ. Interplate dispersal paths for megathermal angiosperms. Perspectives in Plant Ecology, Evolution and Systematics. 2003;6(1-2):5–20. doi: 10.1078/1433-8319-00039. [DOI] [Google Scholar]
- McLoughlin S. The breakup history of Gondwana and its impact on pre-Cenozoic floristic provincialism. Australian Journal of Botany. 2001;49(3):271–300. doi: 10.1071/BT00023. [DOI] [Google Scholar]
- Emadzade K, Gehrke B, Peter Linder H, Hörandl E. The biogeographical history of the cosmopolitan genus Ranunculus L. (Ranunculaceae) in the temperate to meridional zones. Molecular Phylogenetics and Evolution. 2011;58(1):4–21. doi: 10.1016/j.ympev.2010.11.002. [DOI] [PubMed] [Google Scholar]
- Pelser PB, Nordenstam B, Kadereit JW, Watson LE. An ITS phylogeny of tribe Senecioneae (Asteraceae) and a new delimitation of Senecio L. Taxon. 2007;56:1077–1104. doi: 10.2307/25065905. [DOI] [Google Scholar]
- Antonelli A. Higher level phylogeny and evolutionary trends in Campanulaceae subfam. Lobelioideae: Molecular signal overshadows morphology. Molecular Phylogenetics and Evolution. 2008;46(1):1–18. doi: 10.1016/j.ympev.2007.06.015. [DOI] [PubMed] [Google Scholar]
- Antonelli A. Have giant lobelias evolved several times independently? Life form shifts and historical biogeography of the cosmopolitan and highly diverse subfamily Lobelioideae (Campanulaceae) BMC Biology. 2009;7:82. doi: 10.1186/1741-7007-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettpelz E, Hoot SB, Samuel R, Ehrendorfer F. Multiple origins of Southern Hemisphere Anemone (Ranunculaceae) based on plastid and nuclear sequence data. Plant Systematics and Evolution. 2002;231(1):143–151. doi: 10.1007/s006060200016. [DOI] [Google Scholar]
- Meyer KM, Hoot SB, Arroyo MTK. Phylogenetic affinities of South American Anemone (Ranunculaceae), including the endemic segregate genera, Barneoudia and Oreithales. International Journal of Plant Sciences. 2010;171(3):323–331. doi: 10.1086/650153. [DOI] [Google Scholar]
- Schuettpelz E, Hoot SB. Phylogeny and biogeography of Caltha (Ranunculaceae) based on chloroplast and nuclear DNA sequences. American Journal of Botany. 2004;91(2):247–253. doi: 10.3732/ajb.91.2.247. [DOI] [PubMed] [Google Scholar]
- Thiv M, van der Niet T, Rutschmann F, Thulin M, Brune T, Linder HP. Old-New World and trans-African disjunctions of Thamnosma (Rutaceae): Intercontinental long-distance dispersal and local differentiation in the succulent biome. American Journal of Botany. 2010;98(1):76–87. doi: 10.3732/ajb.1000339. [DOI] [PubMed] [Google Scholar]
- Soejima A, Wen J. Phylogenetic analysis of the grape family (Vitaceae) based on three chloroplast markers. American Journal of Botany. 2006;93(2):278–287. doi: 10.3732/ajb.93.2.278. [DOI] [PubMed] [Google Scholar]
- Wen J, Nie Z-L, Soejima A, Meng Y. Phylogeny of Vitaceae based on the nuclear GAI1 gene sequences. Canadian Journal of Botany. 2007;85(8):731–745. doi: 10.1139/B07-071. [DOI] [Google Scholar]
- Rossetto M, Crayn DM, Jackes BR, Porter C. An updated estimate of intergeneric phylogenetic relationships in the Australian Vitaceae. Canadian Journal of Botany. 2007;85:722–730. doi: 10.1139/B07-022. [DOI] [Google Scholar]
- Chen ZD, Ren H, Wen J. In: Flora of China. Wu Z-Y, Hong D-Y, Raven PH, editor. Vol. 12. Beijing and St. Louis, Mo.: Science Press and Missouri Botanical Garden Press; 2007. Vitaceae. [Google Scholar]
- Wen J. In: The families and genera of vascular plants, volume 9. Kubitzki K, editor. Vol. 9. Berlin: Springer-Verlag; 2007. Vitaceae; pp. 466–478. [Google Scholar]
- Li CL. Vitaceae, Flora Reipublicae Popularis Sinica. 2. Vol. 48. Beijing: Science Press; 1998. [Google Scholar]
- Lombardi JA. Systematics of Vitaceae in South America. Canadian Journal of Botany. 2007;85:712–721. doi: 10.1139/B07-021. [DOI] [Google Scholar]
- Rossetto M, Jackes BR, Scott KD, Henry RJ. Is the genus Cissus (Vitaceae) monophyletic? Evidence from plastid and nuclear ribosomal DNA. Systematic Botany. 2002;27(3):522–533. [Google Scholar]
- Gerrath JM, Wilson T, Posluszny U. Morphological and anatomical development in the Vitaceae. VII. Floral development in Rhoicissus digitata with respect to other genera in the family. Canadian Journal of Botany. 2004;82:198–206. doi: 10.1139/b03-120. [DOI] [Google Scholar]
- Meng Y, Wen J, Nie Z-L, Sun H, Yang YP. Phylogeny and biogeographic diversification of Maianthemum (Ruscaceae: Polygonatae) Molecular Phylogenetic and Evolution. 2008;49(2):424–434. doi: 10.1016/j.ympev.2008.07.017. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein DB, Palmer JD, Thompson WF. Structural evolution and flip-flop recombination of chloroplast DNA in the fern genus Osmunda. Current Genetics. 1986;10(11):835–841. doi: 10.1007/BF00418530. [DOI] [Google Scholar]
- Huelsenbeck JP, Ronquist R. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Nylander JAA. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University. 2004.
- Simmons MP, Ochoterena H. Gaps as characters in sequence-based phylogenetic analyses. Systematic Biology. 2000;49(2):369–381. [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), version 4.0b10. Sunderland, Massachusetts: Sinauer Associates; 2003. [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Rawlence N, Rambaut A. A rough guide to BEAST 1.4. 2007. http://code.google.com/p/beast-mcmc/
- Chen I, Manchester SR. Seed morphology of modern and fossil Ampelocissus (Vitaceae) and implications for phytogeography. American Journal of Botany. 2007;94(9):1534–1553. doi: 10.3732/ajb.94.9.1534. [DOI] [PubMed] [Google Scholar]
- Chen I. PhD thesis. Gainesville: University of Florida; 2009. History of Vitaceae inferred from morphology-based phylogeny and the fossil record of seeds. [Google Scholar]
- Wang H, Moore MJ, Soltis PS, Bell CD, Brockington SF, Alexandre R, Davis CC, Latvis M, Manchester SR, Soltis DE. Rosid radiation and the rapid rise of angiosperm-dominated forests. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3853–3858. doi: 10.1073/pnas.0813376106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Systematic Biology. 2008;57(1):4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009;5(9):e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ree RH, Moore BR, Webb CO, Donoghue MJ. A likelihood framework for inferring the evolution of geographic range on phylogenetic trees. Evolution. 2005;59(11):2299–2311. [PubMed] [Google Scholar]
- Ronquist F. Dispersal-vicariance analysis: A new approach to the quantification of historical biogeography. Systematic Biology. 1997;46:195–203. doi: 10.1093/sysbio/46.1.195. [DOI] [Google Scholar]
- Lamm KS, Redelings BD. Reconstructing ancestral ranges in historical biogeography: properties and prospects. Journal of Systematics and Evolution. 2009;47(5):369–382. doi: 10.1111/j.1759-6831.2009.00042.x. [DOI] [Google Scholar]
- Ree RH, Sanmartin I. Prospects and challenges for parametric models in historical biogeographical inference. Journal of Biogeography. 2009;36(7):1211–1220. doi: 10.1111/j.1365-2699.2008.02068.x. [DOI] [Google Scholar]
- Santos JC, Coloma LA, Summers K, Caldwell JP, Ree R, Cannatella DC. Amazonian amphibian diversity is primarily derived from late Miocene Andean lineages. PLOS Biology. 2009;7(3):e1000056. doi: 10.1371/journal.pbio.1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M. The maximum likelihood approach to reconstructing ancestral character states of discrete characters on phylogenies. Systematic Biology. 1999;48:612–622. doi: 10.1080/106351599260184. [DOI] [Google Scholar]
- Edwards AWF. Likelihood. Baltimore: Johns Hopkins University Press; 1992. [Google Scholar]
- Galet P. Recherches sur les methods d'identification et de classification des Vitacées temperées. II Thèse, présentée a la Faculté des Sciences. Université de Montpellier; 1967. [Google Scholar]
- Latham RE, Ricklefs RE. In: Species Diversity in Ecological Communities: Historical and Geographical Perspectives. Ricklefs RE, Schluter D, editor. Chicago: Univ. Chicago Press; 1993. Continental comparisons of temperate-zone tree species diversity; pp. 294–314. [Google Scholar]
- Xiang Q-Y, Zhang WH, Ricklefs RE, Qian H, Chen ZD, Wen J, Li JH. Regional differences in rates of plant speciation and molecular evolution: A comparison between eastern Asia and eastern North America. Evolution. 2004;58(10):2175–2184. doi: 10.1111/j.0014-3820.2004.tb01596.x. [DOI] [PubMed] [Google Scholar]
- Wen J Ickert-Bond S Nie Z-L Li R Long M, Gu H, Zhou Z. Timing and modes of evolution of eastern Asian - North American biogeographic disjunctions in seed plants Darwin's Heritage Today: Proceedings of the Darwin 2010 Beijing International Conference 2010Beijing: Higher Education Press; 252–269.22330132 [Google Scholar]
- Donoghue MJ, Smith SA. Patterns in the assembly of temperate forests around the Northern Hemisphere. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2004;359(1450):1633–1644. doi: 10.1098/rstb.2004.1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne RI. Northern hemisphere plant disjunctions: a window on tertiary land bridges and climate change? Annals of Botany. 2006;98(3):465–472. doi: 10.1093/aob/mcl148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchester SR. Biogeographical relationships of North American Tertiary floras. Annals of the Missouri Botanical Garden. 1999;86(2):472–522. doi: 10.2307/2666183. [DOI] [Google Scholar]
- Iturralde-Vinent MA. Meso-Cenozoic Caribbean paleogeography: Implications for the historical biogeography of the region. International Geology Review. 2006;48(9):791–827. doi: 10.2747/0020-6814.48.9.791. [DOI] [Google Scholar]
- Wolfe JA. Some aspects of plant geography of the Northern Hemisphere during the late Cretaceous and Tertiary. Annals of the Missouri Botanical Garden. 1975;62(2):264–279. doi: 10.2307/2395198. [DOI] [Google Scholar]
- Dewey JF, Pitman WCI, Ryan WBF, Bonnin J. Plate Tectonics and the Evolution of the Alpine System. Geological Society of America Bulletin. 1973;84(10):3137–3180. doi: 10.1130/0016-7606(1973)84<3137:PTATEO>2.0.CO;2. [DOI] [Google Scholar]
- Cox CB. Plate tectonics, seaways and climate in the historical biogeography of mammals. Memórias do Instituto Oswaldo Cruz. 2000;95(4):509–516. doi: 10.1590/s0074-02762000000400012. [DOI] [PubMed] [Google Scholar]
- Tiffney BH, Barghoorn ES. Fruits and seeds of Brandon lignite. 1. Vitaceae. Review of Palaeobotany and Palynology. 1976;22:169–191. doi: 10.1016/0034-6667(76)90001-4. [DOI] [Google Scholar]
- McAtee WL. Virginia creeper as a winter food for birds. Auk. 1906;23:346–347. [Google Scholar]
- Ridley HN. In: The dispersal of plants throughout the world. Kent: L. Reeve, editor. 1930. [Google Scholar]
- Givnish TJ, Renner SS. Tropical intercontinental disjunctions: Gondwana breakup, immigration from the boreotropics, and transoceanic dispersal. International Journal of Plant Sciences. 2004;165(4):S1–S6. doi: 10.1086/424022. [DOI] [Google Scholar]
- Clayton JW, Soltis PS, Soltis DE. Recent long-distance dispersal overshadows ancient biogeographical patterns in a pantropical angiosperm family (Simaroubaceae, Sapindales) Systematic Biology. 2009;58(4):395–410. doi: 10.1093/sysbio/syp041. [DOI] [PubMed] [Google Scholar]
- Emadzade K, Hörandl E. Northern Hemisphere origin, transoceanic dispersal, and diversification of Ranunculeae DC. (Ranunculaceae) in the Cenozoic. Journal of Biogeography. 2011;38(3):517–530. doi: 10.1111/j.1365-2699.2010.02404.x. [DOI] [Google Scholar]
- Sytsma KJ, Litt A, Zjhra ML, Pires JC, Nepokroeff M, Conti E, Walker J, Wilson PG. Clades, clocks, and continents: Historical and biogeographical analysis of Myrtaceae, Vochysiaceae, and relatives in the Southern Hemisphere. International Journal of Plant Sciences. 2004;165(4):S85–S105. doi: 10.1086/421066. [DOI] [Google Scholar]
- Givnish TJ, Millam KC, Evans TM, Hall JC, Pires JC, Berry PE, Sytsma KJ. Ancient vicariance or recent long-distance dispersal? Inferences about phylogeny and South American-African disjunctions in Rapateaceae and Bromeliaceae based on ndhF sequence data. International Journal of Plant Sciences. 2004;165(4 suppl):s35–s54. [Google Scholar]
- Renner SS. Multiple Miocene Melastomataceae dispersal between Madagascar, Africa and India. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2004;359(1450):1485–1494. doi: 10.1098/rstb.2004.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawver LA, Gahagan LM. Evolution of Cenozoic seaways in the circum-Antarctic region. Palaeogeography, Palaeoclimatology, Palaeoecology. 2003;198(1-2):11–37. doi: 10.1016/S0031-0182(03)00392-4. [DOI] [Google Scholar]
- Richardson JE, Chatrou LW, Mols JB, Erkens RHJ, Pirie MD. Historical biogeography of two cosmopolitan families of flowering plants: Annonaceae and Rhamnaceae. Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2004;359(1450):1495–1508. doi: 10.1098/rstb.2004.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartish IV, Antonelli A, Richardson JE, Swenson U. Vicariance or long-distance dispersal: historical biogeography of the pantropical subfamily Chrysophylloideae (Sapotaceae) Journal of Biogeography. 2011;38(1):177–190. doi: 10.1111/j.1365-2699.2010.02389.x. [DOI] [Google Scholar]
- Jackes BR, Rossetto M. A new combination in Clematicissus Planch. (Vitaceae) Telopea. 2006;11(3):390–391. [Google Scholar]
- Crisp M, Cook L, Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Philosophical Transactions of the Royal Society of London Series B-Biological Sciences. 2004;359(1450):1551–1571. doi: 10.1098/rstb.2004.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RS. Origins of the southeastern Australian vegetation. Philosophical Transactions of the Royal Society B: Biological Sciences. 2004;359:1537–1549. doi: 10.1098/rstb.2004.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J. Evolution of eastern Asian-Eastern North American biogeographic disjunctions: A few additional issues. International Journal of Plant Sciences. 2001;162:S117–S122. doi: 10.1086/322940. [DOI] [Google Scholar]
- Wolfe JA. In: Eocene-Oligocene Biotic and Climatic Evolution. Prothero DR, Berggren WA, editor. Princeton: Princeton University Press; 1992. Climatic, floristic, and vegetational changes near the Eocene/Oligocene boundary in North America; pp. 421–436. [Google Scholar]
- Berggren WA, Prothero DP. In: Eocene-Oligocene Climatic and Biotic Evolution. Prothero DP, Berggren WA, editor. Princeton: Princeton University Press; 1992. Eocene-Oligocene climatic and biotic evolution: an overview; pp. 1–28. [Google Scholar]
- Mai DH. Tertiäre Vegetationsgeschichte Europas. New York: Fischer, Jena, Stuttgart; 1995. [Google Scholar]
- Graham A. Late Cretaceous and Cenozoic History of North American Vegetation. Oxford: Oxford University Press; 1999. [Google Scholar]
- Mai DH. Neue Früchte und Samen aus paläozänen Ablagerungen Mitteleuropas. Feddes Repertorium. 1987;98:197–229. [Google Scholar]
- Muellner AN, Savolainen V, Samuel R, Chase MW. The mahogany family "out-of-Africa": divergence time estimation, global biogeographic patterns inferred from plastid rbcL DNA sequences, extant, and fossil distribution of diversity. Molecular Phylogenetics and Evolution. 2006;40(1):236–250. doi: 10.1016/j.ympev.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Davis CC, Bell CD, Mathews S, Donoghue MJ. Laurasian migration explains Gondwanan disjunctions: evidence from Malpighiaceae. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(10):6833–6837. doi: 10.1073/pnas.102175899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian H. Floristic relationships between eastern Asia and North America: Test of Gray's hypothesis. American Naturalist. 2002;160(3):317–332. doi: 10.1086/341523. [DOI] [PubMed] [Google Scholar]
- Lavin M, Thulin M, Labat JN, Pennington RT. Africa, the odd man out: Molecular biogeography of dalbergioid legumes (Fabaceae) suggests otherwise. Systematic Botany. 2000;25(3):449–467. doi: 10.2307/2666689. [DOI] [Google Scholar]
- Nie Z-L, Sun H, Beardsley PM, Olmstead RG, Wen J. Evolution of biogeographic disjunction between eastern Asia and eastern North America in Phryma (Phrymaceae) American Journal of Botany. 2006;93(9):1343–1356. doi: 10.3732/ajb.93.9.1343. [DOI] [PubMed] [Google Scholar]
- Wen J, Ickert-Bond SM. Evolution of the Madrean-Tethyan disjunctions and the North and South American amphitropical disjunctions in plants. Journal of Systematics and Evolution. 2009;47(5):331–348. doi: 10.1111/j.1759-6831.2009.00054.x. [DOI] [Google Scholar]
- Doyle JA, Sauquet H, Scharaschkin T, Le Thomas A. Phylogeny, molecular and fossil dating, and biogeographic history of Annonaceae and Myristicaceae (Magnoliales) International Journal of Plant Sciences. 2004;165(suppl 4):s35–s67. [Google Scholar]
- Erkens RHJ, Maas JW, Couvreur TLP. From Africa via Europe to South America: migrational route of a species-rich genus of Neotropical lowland rain forest trees (Guatteria, Annonaceae) Journal of Biogeography. 2009;36(12):2338–2352. doi: 10.1111/j.1365-2699.2009.02162.x. [DOI] [Google Scholar]
- Antonelli A, Nylander JAA, Persson C, Sanmartin I. Tracing the impact of the Andean uplift on Neotropical plant evolution. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(24):9749–9754. doi: 10.1073/pnas.0811421106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin RA, Miller JS. Relationships within tribe Lycieae (Solanaceae): paraphyly of Lycium and multiple origins of gender dimorphism. American Journal of Botany. 2005;92(12):2044–2053. doi: 10.3732/ajb.92.12.2044. [DOI] [PubMed] [Google Scholar]
- Miller JS, Kamath A, Damashek J, Levin RA. Out of America to Africa or Asia: Inference of dispersal histories using nuclear and plastid DNA and the S-RNase self-incompatibility locus. Molecular Biology and Evolution. 2011;28(1):793–801. doi: 10.1093/molbev/msq253. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yokoyama J, Ohashi H. Phylogeny and biogeography of the genus Lycium (Solanaceae): inferences from chloroplast DNA sequences. Molecular Phylogenetics and Evolution. 2001;19:246–258. doi: 10.1006/mpev.2001.0921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Voucher information and GenBank accession numbers of the Ampelopsis clade and representative taxa in Vitaceae. Abbreviations of herbaria are as follows: KUN, Kunming Institute of Botany, Chinese Academy of Sciences; and US, the United States National Herbarium. Accession numbers beginning with JQ indicate sequences generated for this study and the others were obtained from GenBank. A dash means sequences missing.