Abstract
Background
Plants adopt different reproductive strategies as an adaptation to growth in a range of climates. In Arabidopsis thaliana FRIGIDA (FRI) confers a vernalization requirement and thus winter annual habit by increasing the expression of the MADS box transcriptional repressor FLOWERING LOCUS C (FLC). Variation at FRI plays a major role in A. thaliana life history strategy, as independent loss-of-function alleles that result in a rapid-cycling habit in different accessions, appear to have evolved many times. The aim of this study was to identify and characterize orthologues of FRI in Brassica oleracea.
Results
We describe the characterization of FRI from Brassica oleracea and identify the two B. oleracea FRI orthologues (BolC.FRI.a and BolC.FRI.b). These show extensive amino acid conservation in the central and C-terminal regions to FRI from other Brassicaceae, including A. thaliana, but have a diverged N-terminus. The genes map to two of the three regions of B. oleracea chromosomes syntenic to part of A. thaliana chromosome 5 suggesting that one of the FRI copies has been lost since the ancient triplication event that formed the B. oleracea genome. This genomic position is not syntenic with FRI in A. thaliana and comparative analysis revealed a recombination event within the A. thaliana FRI promoter. This relocated A. thaliana FRI to chromosome 4, very close to the nucleolar organizer region, leaving a fragment of FRI in the syntenic location on A. thaliana chromosome 5. Our data show this rearrangement occurred after the divergence from A. lyrata. We explored the allelic variation at BolC.FRI.a within cultivated B. oleracea germplasm and identified two major alleles, which appear equally functional both to each other and A. thaliana FRI, when expressed as fusions in A. thaliana.
Conclusions
We identify the two Brassica oleracea FRI genes, one of which we show through A. thaliana complementation experiments is functional, and show their genomic location is not syntenic with A. thaliana FRI due to an ancient recombination event. This has complicated previous association analyses of FRI with variation in life history strategy in the Brassica genus.
Keywords: FRIGIDA, Flowering time, vernalization, synteny, Brassica oleracea, Arabidopsis thaliana
Background
The switch to reproductive development is a fundamental process in the plant life cycle. The molecular mechanisms underlying this developmental transition have been extensively studied in Arabidopsis thaliana. An integrated network of environmentally responsive genetic pathways converge on a common set of targets to quantitatively regulate the genes required to switch the apical meristem from a vegetative to a floral state [1-3]. One important environmental cue is prolonged cold, which accelerates flowering in a process termed vernalization and aligns pollination and seed set with the favourable conditions of spring. Variation in requirement for vernalization exists in many plant species and this influences life history strategy with plants requiring vernalization adopting a perennial, biennial or winter annual habit in contrast to summer annuals, which flower in the first growing season. This is in contrast to other species that are more reliant on photoperiodic signals or endogenous cues e.g. rice [4]. The significant fitness consequences of flowering time variation, demonstrated in annual [5,6] and perennial plants [7], have most likely contributed to the evolution of the extensive variability in flowering time control. Flowering also influences the pattern of growth throughout the seasons and affects many agronomic characters including the quantity and quality of crop production. This is particularly apparent in cultivated brassicas, where variation in the flowering process has been selected to produce a diverse array of economically important morphological forms.
A major determinant in the variation of vernalization requirement in A. thaliana is allelic variation at FRIGIDA (FRI) [8-11]. FRI represses flowering by promoting the expression of the floral repressor FLOWERING LOCUS C (FLC) [12,13]. Vernalization acts antagonistically to FRI and accelerates flowering by down-regulating FLC. A number of rapid-cycling variants of A. thaliana that do not need vernalization were found to have arisen through loss of function of FRI, an evolutionary step that has occurred multiple times [8,9,11,14]. Parallel evolution through allelic variation at a common target has been found in other organisms [15]. It was therefore interesting to ask whether a similar evolutionary step has occurred in other plant species. Many other species do show variation in vernalization requirement and it is an important agronomic trait in many major crops. For example, in B. oleracea (horticultural brassicas) vernalization-requiring biennials are represented by cabbage and Brussels sprouts, with summer annual crops including some calabrese and cauliflower cultivars. Orthologues of FRI have been identified in A. lyrata [16], Capsella species [17] and the halophyte Thellungiella halophila [18] within the Brassicaceae, and more broadly in Medicago truncatula, Lotus japonicus, Vitis vinifera [19], Populus balsamifera [20] and Oryza sativa [21]. To date natural variation in vernalization requirement has been associated with FRI polymorphism in A. lyrata [8] and allelic variation in one orthologue in Brassica napus (BnaA.FRI.a) has been associated with flowering time variation [22].
We are interested in understanding the molecular basis of variation in flowering time and vernalization requirement in horticultural brassicas. Genetic information from A. thaliana can generally be applied to Brassica species because of their evolutionary relatedness. The Arabidopsis and Brassica genera are in the same family (Brassicaceae) with B. oleracea thought to have arisen from a triplication of an ancestral genome similar to that of Arabidopsis [23-26]. Genetic information on the control of flowering in Arabidopsis can be applied to Brassica species because of the colinearity of the Arabidopsis and Brassica genomes [23,27,28]. This has been used to infer candidate genes that might account for QTL underlying flowering time and other variation [22,29,30]; however, in some instances it can be misleading [31]. Here, we identify the two FRI genes in the B. oleracea genome and map their genomic locations. We also explored allelic variation at one of the FRI loci in cultivated B. oleracea germplasm. These new data will provide the necessary information to elucidate how general a role FRI plays in life history variation in the Brassicaceae.
Results and discussion
Two FRI genes are present in the Brassica oleracea genome
The BoFRI genes were isolated from the JBo BAC library of the B. oleracea Chinese kale genotype A12DHd [32] through hybridization with an A. thaliana FRI genomic clone. From seven positive BAC clones two that showed distinct FRI hybridization patterns (JBo72I23 and JBo88G16, Figure 1a, b) were selected for sub-cloning. Analysis of these confirmed they carried different Brassica paralogues designated BolC.FRI.a and BolC.FRI.b [33] and referred to hereafter as BoFRIa and BoFRIb [GenBank JN191450 and JN191449]. As in other species, BoFRIa and BoFRIb contain three exons encoding predicted open reading frames (ORFs) of 594 and 585 residues respectively (Figure 1c). BoFRIa contains two coiled-coil domains, typically involved in protein oligomerisation (as predicted by COILS http://www.ch.embnet.org/software/COILS_form.html[34]), very similar to the predicted structure of the A. thaliana FRI (AtFRI) [8,35]. In contrast, BoFRIb is predicted to contain only one coiled-coil domain in the C-terminal region as was found to be the case for two of the four FRI identified in B. napus [22].
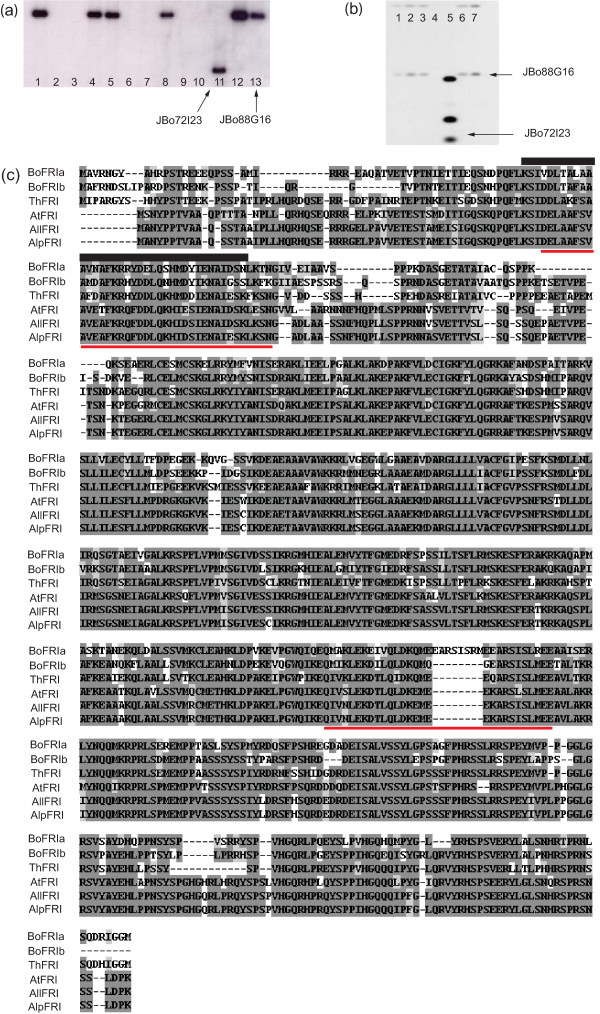
Figure 1.
Cloning BoFRIa and BoFRIb. (a) Southern analysis of A12 BACs identified by colony hybridisation and probed with AtFRI. Lanes 1, 4, 5, 8, 11, 12 and 13 contain clones that show homology to FRIGIDA. BACs in lanes 2, 3, 6, 7, and 10 do not cross hybridise (b) HindIII digest of six BACs probed with conserved region from exons 2 and 3 of BoFRIa. Lane 5 contains JBo72I23 from which BoFRIa was sequenced. Lane 7 contains JBo88G16. Lanes 1, 2, 3 and 6 contain four further BACs showing the same hybridization pattern as JBo88G16. Note the intensity of the hybrization is indicative of the sequence divergence between BoFRIa and BoFRIb (See 1c) (c) Comparison of the protein sequences of BoFRIa and BoFRIb with other members of the FRI sub-family. From top to bottom they are Brassica oleracea BoFRIa, Brassica oleracea BoFRIb, Thelliungiella halophila ThFRI, Arabidopsis thaliana AtFRI Arabidopsis lyrata ssp lyrata AllFRI and Arabidopsis lyrata ssp petraea AlpFRI. The N-terminal domain containing the conserved region of 37 amino acids (indicated by solid bar) that defines copies of FRIGIDA from other members of the FRI superfamily [17]. The coiled-coil domains are indicated by the red lines.
AtFRI is the original member of a family of seven proteins in A. thaliana which, apart from the two predicted coiled-coil domains, show no homology with any other proteins and whose function has yet to be determined. Recent analysis of the FRI protein family [19] identified a conserved core central domain. Outside of this domain significant variation is observed that allows the FRI family to be subdivided into five distinct groups. AtFRI and its orthologues in other species are defined by a conserved region of 37 amino acids in the N-terminal region of the protein. The BoFRI proteins we describe here contain this conserved 37 amino acid region reinforcing the view they are FRI orthologues; however, the amino acids either side of this region show lower homology to AtFRI (Figure 1c). This region includes much of the first predicted coiled-coil in BoFRIa. Variations in this domain in BoFRIb result in the loss of a predicted coiled-coil, emphasising a possible functional significance for the amino acid polymorphisms in this region. A similar degree of divergence from AtFRI is found in the N-terminal region of an orthologue of FRI isolated from the halophyte T. halophila and in four orthologues of FRI identified in B. napus [18,22]. By contrast, there is extensive amino acid conservation between BoFRIa, BoFRIb and AtFRI in the central and C-terminal regions (Figure 1c). Transgenic analysis of the functional domains of AtFRI in A. thaliana where the N or C terminus was deleted revealed that the N-terminal region was less important for function [19], perhaps explaining the high degree of divergence observed.
The BoFRI genes map to regions that are non-syntenic with A. thaliana FRI
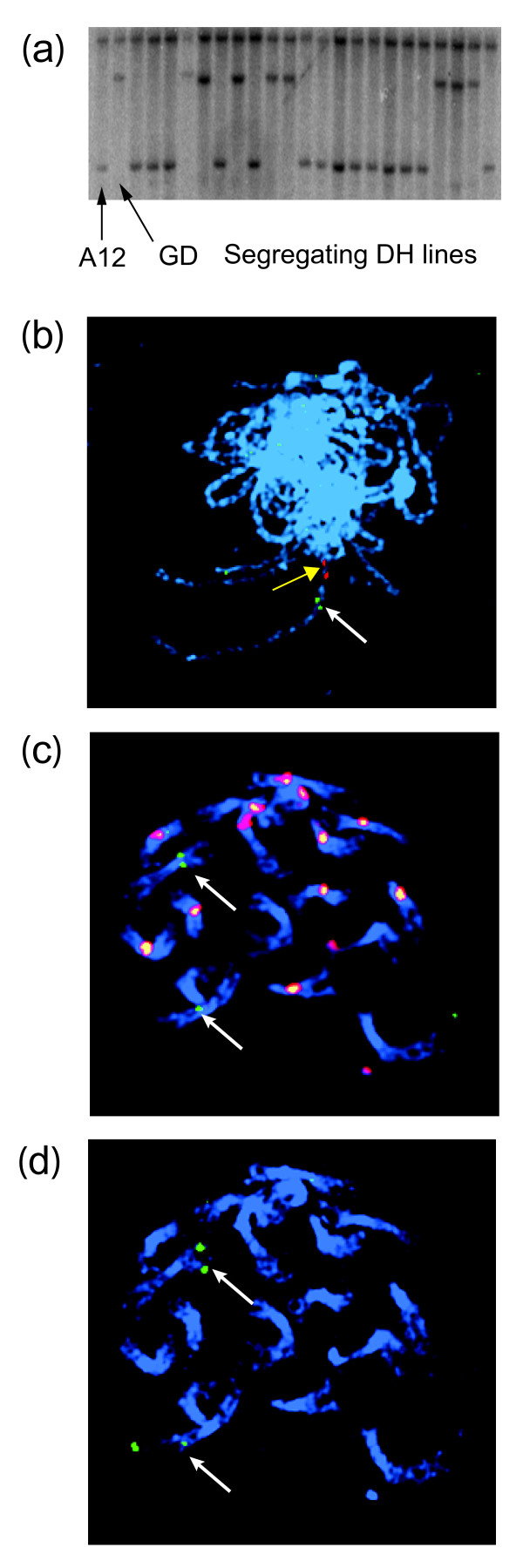
A genomic fragment including exon 2, intron 2 and exon 3 of BoFRIa (and showing a high level of conservation in BoFRIb and AtFRI) was hybridized to mapping filters from two B. oleracea mapping populations: Chinese kale × calabrese (var. alboglabra x var. italica; A12DHdxGDDH33, [36]; Figure 2a) and cauliflower × Brussels sprout (var. botrytis x var. gemmifera; N × G [37]). RFLPs for one of the two BoFRI loci segregated in the A12DHdxGDDH33 mapping population that allowed this locus to be mapped to 39.5 cM on linkage group C3 of the B. oleracea genome. The locus mapping to C3 was identified as BoFRIa by fluorescence in situ hybridization (FISH) with BAC JBo72I23, from which BoFRIa was originally sequenced (Figure 2b). JBo88G16 was located on the short arm of chromosome C9 by FISH (Figure 2c, d). Therefore, the second locus, BoFRIb, was on linkage group C9. Two further BACs showing the same restriction pattern as JBo88G16 (Figure 1b) hybridized to the same location on C9 (data not shown). These results confirm that the B. oleracea genome contains two orthologues.
Figure 2.
Mapping BoFRIa and BoFRIb. (a) A12DHdxGDDH33 mapping population probed with conserved region from exons 2 and 3 of BoFRIa. Two loci are identified; one monomorphic (upper band) and a second segregating with the two parental alleles (lower two bands). (b) Meiotic pachytene spread with JBo72I23 (BoFRIa, green, white arrow) hybridizing to C3 between the telomere and JBo62M08 (red, yellow arrow). (c) and (d) JBo88G16 (BoFRIb) hybridizes to the short arm of C9: (c) mitotic metaphase with JBo88G16, (green, arrows), BoB061G14 and 45S rDNA (red), (d) reprobe of (c) with JBo32J18 (green, arrows), a marker for C9.
Comparative analysis of the Brassica and A. thaliana genomes has shown that the chromosomal regions of C3 and C9 to which the two BoFRI loci have been mapped are syntenic to a region of A. thaliana chromosome 5 and not to the top of A. thaliana chromosome 4, where AtFRI (At4g00650) is located [23]. This region of chromosome 5 includes a number of genes known to be involved in the control of flowering including FLC, FY and CONSTANS (CO). Several QTL studies have found loci for flowering time variation mapping to this genomic region in a number of Brassica populations including B. oleracea [38-41], B. rapa [42-46]; B. nigra [47,48] and B. napus [29,49]. The mapping we have undertaken reveals the proximity of BoFRI not only to BoFLC, but also BoFY and BoCO; other flowering time genes that have been mapped previously. The sequences of BoFRIa and BoFRIb further allow us to identify which of the four orthologues of AtFRI recently identified in B. napus [22] are the two C genome copies. The four copies of FRI were designated BnaA.FRI.a, and BnaX.FRIb-d. Comparison of the amino acid sequences of these proteins with BoFRIa and BoFRIb suggest that BnaX.FRI.d is the orthologue of BoFRIa and the C genome homoeologue of BnaA.FRI.a. This conclusion is further supported by the fact that BnaA.FRI.a was mapped to a region of A3 homoeologous to the region of C3 where we have mapped BoFRIa. Comparison of the amino acid sequence of BnaX.FRI.c shows it to be identical to that of BoFRIb. BnaX.FRI.c appears most similar to BnaX.FRI.b and is therefore likely to be the A genome homoeologue of BoFRIb in B. napus.
A recombination event specific to the A. thaliana lineage has relocated the FRIGIDA gene to the top of chromosome 4
In A. thaliana, the AtFRI locus is located at the top of chromosome 4. However, it has previously been reported that the orthologue of FRI in A. lyrata maps to linkage group 8 [50,51]. This linkage group is orthologous to the lower arm of A. thaliana chromosome 5 [50,52,53]. Interestingly, an annotated gene model in this region of A. thaliana chromosome 5 (At5g51090) shows a high degree of homology to AtFRI, containing parts of intron 1 and exon 3 but lacking other parts of the coding region, thus it may be a pseudogene [50]. Genevestigator data suggest At5g51090 is expressed at very low levels, supporting this hypothesis [54]. Downstream, in the opposite orientation, is At5g51100, encoding an iron superoxide dismutase and the BoFRIa BAC clone contains 3' sequence showing homology to exons 3-9 of this A. thaliana chromosome 5 gene (Figure 3a).
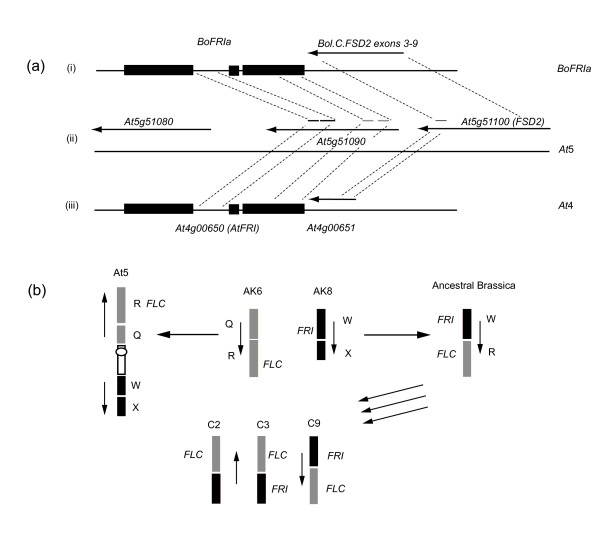
Figure 3.
BoFRIa and BoFRIb map to regions of the B. oleracea genome that are non-syntenic with A. thaliana. (a) Comparison of BoFRIa genomic clone with annotated regions of A. thaliana chromosomes 4 and 5 represented in the 5' to 3' orientation. Solid black rectangles correspond to the three exons that make up FRI. Arrows indicate the orientation of other genes. (i) BoFRIa, genomic clone. (ii) Region of A. thaliana chromosome 5 containing an iron superoxide dismutase (FSD2, At5g51100) and At51090 showing homology to FRI intron 1 and exon 3. (iii) Region of A. thaliana chromosome 4 containing AtFRI and At4g400651 showing homology to exon 9 of FSD2. (b) Proposed derivation of A. thaliana and B. oleracea linkage groups from ancestral karyotype (AK6 and AK8) as suggested in [53] showing the position of FRI and FLC. The rearrangement of ancestral blocks W and R results in orthologues of FRI and FLC being brought into close proximity in the ancestral Brassica genome. This region is present on chromosomes C2, C3 and C9 of B. oleracea and is located in regions of the B. oleracea genome showing synteny to A. thaliana chromosome 5. BoFRIa is located on C3 and BoFRIb on C9. The copy of FRI on C2 has been lost.
Synteny has been studied extensively in the Brassicaceae genomes due to its potential for gene identification and marker development. Arabidopsis and Brassica are thought to have diverged about 43 Mya with a triplication of an ancestral genome (similar to that of Arabidopsis) occurring approximately 23 Mya and giving rise to modern day diploid Brassica genomes [55]. A. thaliana and A. lyrata are thought to have diverged around 13 Mya, with a reduction in chromosome number, from the ancestral karyotype of n = 8 (as represented in A. lyrata) to the derived state in A. thaliana of n = 5 [24,55,56]. The ancestral karyotype of the Brassicaceae is proposed to be eight chromosomes composed of 24 conserved chromosomal blocks [57]. These blocks can be rearranged to model the genome structure of A. thaliana, A. lyrata and the modern day diploid Brassicas [24]. Thus the genomic composition of the nine chromosomes (C1-C9) of B. oleracea and ten chromosomes (A1-A10) of B. rapa can be related to both the ancestral karyotype and the A. thaliana genome.
The ancestral genomic blocks QR and WX from chromosomes 6 and 8 respectively of the ancestral karyotype, and today represented by A. lyrata [24], have been recombined in the ancestral Brassica genome prior to triplication, leading to the block WR being represented three times in the B. rapa genome on A2, A3 and A10 [25,26]. The paralogous regions of B. oleracea are on C2, C3 and C9 (Figure 3b). This rearrangement brings orthologues of FLC (block R) and FRI (block W) together on these chromosomes. Thus in A. lyrata and B. oleracea, FRI maps quite closely to VIN3 (also in block W and required for vernalization [58], as well as its major target FLC (block R; Figure 3b). BoFRIa mapping to C3 and BoFRIb to C9 thus represent two of the three syntenic regions. The third paralogue of FRI appears to have been lost from C2 during B. oleracea evolution; such gene loss is not uncommon [59]. This is in contrast to the current location of FRI at top of chromosome 4 in A. thaliana that shows homology to block O from chromosome 6 of the ancestral karyotype.
The data we present here suggest that the chromosomal rearrangements that occurred during the evolution of the ancestral Brassicaceae genome into A. thaliana included a recombination/rearrangement event that relocated a genomic region containing AtFRI to a position near the distal end of the short arm of chromosome 4, close to the nucleolar organiser region, leaving a non-functional remnant in the genomic position on chromosome 5 that is syntenic with FRI in the other Brassicaceae (A. lyrata, B. oleracea; [25,52]).
Two common alleles exist for BoFRIa in diverse genotypes of B. oleracea
The original sequences of BoFRIa and BoFRIb were obtained from A12DHd, one of the parents of the mapping population used in [38,39]. These studies mapped QTL for flowering time on C3 in the region where we have mapped BoFRIa. We therefore sequenced 650 bp from exon 1 of BoFRIa from the other parent of this population, GDDH33 (data not shown). The GDDH33 sequence showed two amino acid substitutions (A118V and Q125E) compared to A12DHd. Thus the parents of this population are carrying different alleles of BoFRIa and it is possible that variation at BoFRIa is contributing to variation in flowering time in this population. Single amino acid substitutions have also been identified in alleles of BnaA.FRI.a sequenced from the parents of the Tapidor × Ningyou7 (TN) mapping population from B. napus and mapped to a region underlying a QTL for flowering time variation [22].
We sequenced BoFRIa and BoFRIb for two additional genotypes of B. oleracea italica. The A12DHd reference sequence is derived from a BAC clone of a Chinese kale, B. oleracea alboglabra, which flowers after 8 weeks [38] and can be considered a rapid-cycling type, not requiring vernalization. We therefore selected two additional genotypes of broccoli (Brassica oleracea italica); E1, which has a facultative vernalization response, flowering earlier following a period of cold, but which matures in October/November (Autumn) of the year of planting and E8 which has an obligate vernalization requirement and matures in April/May (Spring) of the following year. BoFRIb is highly conserved between the three genotypes with only 5 amino acid substitutions (D6G, K20Q, Q372K and R532W in E1 and N105T in E8). The sequencing of BoFRIa in these genotypes identified a polymorphic region in exon 1 that included two deletions of seven and three amino acids in E8 relative to E1, either side of the conserved block of 37 amino acids that defines the FRI proteins, (Figure 1c). Thirteen non-synonymous and 12 synonymous substitutions differentiate the A12DHd and E1 BoFRIa alleles from the E8 allele. We therefore designated the E1 and E8 BoFRIa alleles as BoFRIa-1 and BoFRIa-4 respectively [GenBank JN191393, JN191392].
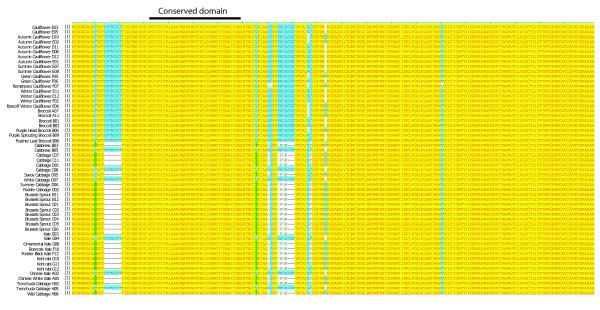
We focused our subsequent analysis on BoFRIa as this showed most polymorphism and extended our analysis to include 55 genotypes from the cultivated B. oleracea Diversity Foundation Set developed at the University of Warwick (BolDFS, King et al. http://www.brassica.info/resource/plants/diversity_sets.php; [60] Table 1). A 650 bp region of BoFRIa covering the exon 1 polymorphic region containing the two deletions was sequenced (Table 1, GenBank JN191394-191448). We identified six BoFRIa alleles within this subset of 55 genotypes from the BolDFS. These can be divided into two groups; BoFRIa 1-3 and BoFRIa 4-6 where alleles in the second group include the seven amino acid and three amino acid deletions. The BoFRIa-1 and BoFRIa-4 alleles were the most common within the 55 genotypes studied. In addition the two deletions identified in BoFRIa-4 always co-occurred and were found at high frequency together with a small number of non-synonymous nucleotide polymorphisms. The two deletions, which had been found in the late-flowering broccoli, were over-represented in B. oleracea vegetable types such as Brussels sprouts and kohl rabi with a winter annual or biennial habit, usually grown for consumption of their vegetative rather than floral forms (Table 1, Figure 4). Interestingly BnaX.FRI.d from the B. napus winter variety Express [22], which we have identified here as the C genome homologue of BoFRIa in B. napus has both of the deletions identified in the BoFRI 4-6 class of alleles that are overrepresented in brassica vegetable types with a winter annual or biennial habit. On closer examination BnaX.FRI.d was found to have the same amino acid sequence as BoFRIa-5 and is also present in the European winter type and Chinese semi-winter type parental lines of the TN mapping population [22].
Table 1.
Amino acid polymorphisms in BoFRIa from cultivated genotypes of Brassica oleracea
| Line | Accession name | Crop type | Crop group | Ref | origin | cult/landrace/DH | 32 | 36-42 | 97 | 102 | 103 | 106 | 107 | 108 | 109 | 110-112 | 118 | 119 | 125 | 172 | BoFRIa Allele |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SIR5a | Siria DH line 5a | Cauliflower | Cauliflower | E03 | N/A | DH | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU002838 | ALGROMAJO NO 2 | Cauliflower | Cauliflower | E05 | NL | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004832 | TOSCANO | Autumn cauliflower | Cauliflower | D10 | ITA | cultivar | T | V | V | S | P | K | D | A | V | C | E | F | 2 | ||
| HRIGRU005458 | DI JESI | Autumn cauliflower | Cauliflower | E02 | ITA | cultivar | T | V | V | S | P | K | D | A | V | C | E | F | 2 | ||
| HRIGRU006254 | TASMAN | Autumn cauliflower | Cauliflower | D11 | AUS | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004239 | CANBERRA | Autumn cauliflower | Cauliflower | D08 | AUS | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU008558 | GIGANTE DI NAPOLI NATALINO * | Autumn cauliflower | Cauliflower | D12 | ITA | N/A | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU006797 | SOFIA | Autumn cauliflower | Cauliflower | E01 | SP | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004814 | BIANCO NAPOLETANE NATALINO | Summer cauliflower | Cauliflower | E07 | ITA | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004991 | ALL THE YEAR ROUND | Summer cauliflower | Cauliflower | E08 | UK | N/A | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004847 | VERDE DI MACERATA | Green caulilfower | Cauliflower | F05 | ITA | N/A | T | V | V | S | P | K | D | A | V | C | E | F | 2 | ||
| HRIGRU004850 | DI ALBENGA | Green caulilfower | Cauliflower | F06 | ITA | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004861 | ROMANESCO NATALINO | Romanesco cauliflower | Cauliflower | F07 | ITA | cultivar | T | V | A | P | P | K | D | A | V | C | E | V | 3 | ||
| HRIGRU002891 | ST MALO HALF HATIF | Winter cauliflower | Cauliflower | E11 | FRA | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004492 | WINTER ROSCOFF | Winter cauliflower | Cauliflower | E12 | IRE | landrace | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU006230 | LATE QUEEN | Winter cauliflower | Cauliflower | F02 | IND | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| ROS152b | Roscoff type F1 DJ1356 DH line 152b | Roscoff winter cauliflower | Cauliflower | E06 | N/A | DH | T | V | V | S | P | K | D | A | V | C | E | F | 2 | ||
| Cor12b | Corvette DH line 12b | broccoli | Broccoli | A07 | N/A | DH | T | V | V | S | P | K | D | A | V | C | E | F | 2 | ||
| CAL 18b | DH line Royal Sluis F1 RS71343 (DJ6546) | broccoli | Broccoli | A11 | N/A | DH | T | V | V | S | P | K | D | A | V | C | E | F | 2 | ||
| HRIGRU002398 | PICOLINI DI PALERMO | broccoli | Broccoli | B01 | ITA | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU011802 | MUGNULI | broccoli? | Broccoli | B02 | ITA | landrace | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU005276 | CIMA VIOLETTA NATALINO | Purple head broccoli | Broccoli | B08 | ITA | cultivar | T | V | V | S | P | K | D | A | V | C | E | F | 2 | ||
| HRIGRU003543 | PURPLE SPROUTING LATE IMPROVED | Sprouting broccoli | Broccoli | B09 | UK | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU005416 | CAVOLO CAVOLINA RIZZA | Feather leaf broccoli | Broccoli | B06 | ITA | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004705 | RAMOSO CALABRESE PRECOCE | Calabrese | Broccoli | B03 | ITA | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU005425 | CAVOLO BROCCOLO NATALINO | Calabrese | Broccoli | B05 | ITA | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| BOH 85c | Bohmerwaldkohl DH line 85c | Cabbage | Cabbage | C07 | N/A | DH | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU005652 | SHETLAND CABBAGE | Cabbage | Cabbage | C11 | UK | N/A | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU007833 | LARGE BLOOD RED | Cabbage | Cabbage | D01 | IND | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HA 84a | Hawke DH line Ha84a | Cabbage | Cabbage | C08 | N/A | DH | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004773 | CAVOLO VERZA SAN GIOVANNI | Savoy cabbage | Cabbage | D05 | ITA | cultivar | T | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 5 |
| HRIGRU011490 | COUVE REPOLHO BACALAN | White cabbage | Cabbage | D07 | PORT | landrace | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU004771 | CAVOLO CAPPUCCIO MEDIO NAPOLETANE | Summer cababge | Cabbage | D06 | ITA | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU002574 | CATTLE (EARLY DRUMHEAD) | Fodder cabbage | Cabbage | D02 | UK | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| AC582 | DH ex. Nym | Brussels sprout | Brussels sprout | B11 | N/A | DH | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU000342 | EVESHAM GIANT | Brussels sprout | Brussels sprout | B12 | UK | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU000605 | WILHELMSBURGER | Brussels sprout | Brussels sprout | C01 | DEN | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU002227 | SANDA ROEM VAN CASTRICUM | Brussels sprout | Brussels sprout | C02 | UK | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU002787 | GROENENBOOM LATE SELECTION | Brussels sprout | Brussels sprout | C03 | NL | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU005086 | OLD BEDFORDSHIRE STOCK | Brussels sprout | Brussels sprout | C04 | UK | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU006212 | CAVOLO DI BRUXELLES MEZZO NANO | Brussels sprout | Brussels sprout | C05 | ITA | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU008226 | LOCAL SELECTION | Brussels sprout | Brussels sprout | C06 | BHUTAN | landrace | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| CGN14111 | Butzo | Kale | Kale | G03 | N/A | N/A | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU006226 | GIANT JERSEY KALE | Kale | Kale | G04 | UK | cultivar | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU009846 | RED ON GREEN | Ornamental kale | Kale | G08 | JPN | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU003598 | WESTLAND WINTER VERDURA | Borecole kale | Kale | F10 | UK | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU006210 | CAVOLO NERO DI TOSCANA O * | Fodder black kale | Kale | F12 | ITA | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU011183 | PURPLE VIENNA | Kohl rabi | Kohl rabi | G10 | USA | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU008267 | WHITE VIENNA | Kohl rabi | Kohl rabi | G11 | IS | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU005443 | CAVOLO FORTE | Purple kohl rabi | Kohl rabi | G12 | ITA | cultivar | S | DEL | I | A | S | S | P | N | K | DEL | A | C | E | F | 6 |
| HRIGRU007543 | CHINESE KALE | Chinese kale | Alboglabra | A02 | CHINA | landrace | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| Senna (GK95186) | Senna | Chinese white kale | Alboglabra | A05 | N/A | DH | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU009490 | COUVE CORTE | Tronchuda cabbage | Tronchuda cabbage | H02 | PORT | landrace | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 |
| HRIGRU009574 | COUVE PENCA DE GONDOMAR | Tronchuda cabbage | Tronchuda cabbage | H05 | PORT | landrace | T | V | V | S | P | K | D | A | A | C | Q | F | 1 | ||
| HRIGRU007796 | Wild cabbage | Wild cabbage | H08 | UK | N/A | S | DEL | I | A | S | S | P | N | K | DEL | A | Y | E | F | 4 | |
Details of amino acid polymorphisms in the 55 B. oleracea genotypes from the BolDFS genotypes [60] for the first 650 bp of BoFRIa. The number of genotypes in a BoFRIa allele type within each vegetable type is also listed
Figure 4.
Comparison of first 160 amino acids of BoFRIa from 55 BolDFS genotypes. Protein comparison of BoFRIa from cultivated genotypes of B. oleracea listed by crop type. The plate co-ordinates refer to those listed in Table 1. The conserved region of 37 amino acids that defines FRI from other members of the FRI superfamily is delineated by the horizontal black bar.
Functional analysis of BoFRIa alleles in A. thaliana
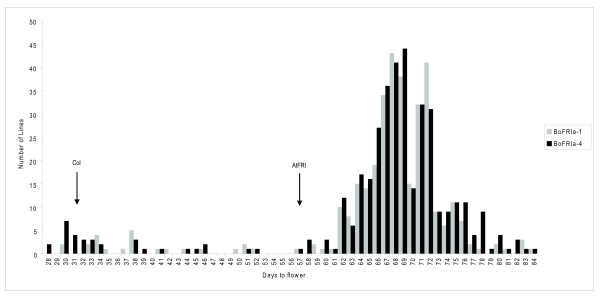
To ascertain if the two most common BoFRIa alleles conferred any functional differences we undertook transformation experiments. The coding and 3'UTR sequences from the BoFRIa-1 and BoFRIa-4 alleles were used to replace the AtFRI coding and 3' UTR sequences in an A. thaliana genomic clone. By retaining common regulatory sequences in the 5' region from the AtFRI gene we hoped to normalise expression and thus focus on the structural differences between the two Brassica proteins. These constructs were transformed into the rapid-cycling A. thaliana accession Columbia (Col-0). Col-0 carries a loss-of-function mutation within AtFRI, but has a functional FLC so these experiments would determine if BoFRIa could complement the fri mutation in Col-0, and induce late flowering. Both BoFRIa alleles complemented the loss-of-function mutation with > 100 primary (T1) transformed plants containing each of the BoFRIa alleles flowering very late compared to Col-0 plants and surprisingly also later than Col-0 transformed with a functional AtFRI (Figure 5).
Figure 5.
Distribution of flowering times in T1 transformants carrying BoFRIa-1 and BoFRIa-4 alleles. Histogram of the flowering time of T1 lines transformed with BoFRIa-1 and BoFRIa-4 measured as days to flower. The flowering time of wild type Col-0 plants and Col-0 transformed with AtFRI are indicated.
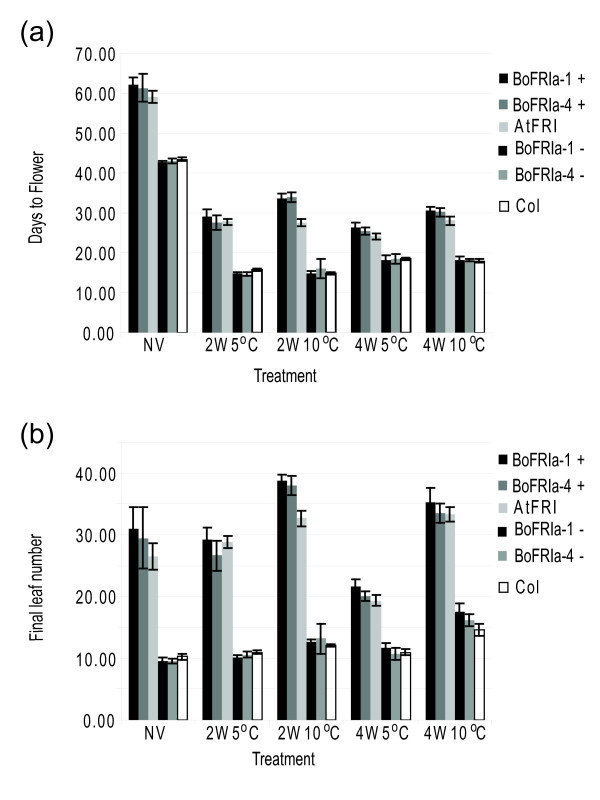
To investigate the functionality of BoFRIa-1 and BoFRIa-4 alleles under different environmental conditions five transformants carrying each allele were analysed in the next (T2) generation. Flowering time was analysed as days-to-flower and total leaf number in plants that had no vernalization, or had experienced two or four weeks vernalization, at either 5°C or 10°C (Figure 6). In all treatments, except two weeks at 10°C (2W10°C), plants with either BoFRIa allele flowered as late as those carrying AtFRI. At 2W10°C plants carrying either of the BoFRIa alleles flowered later than AtFRI. Figure 6 also shows that plants undergoing a vernalization treatment at 10°C compared to 5°C continue to grow and initiate leaves at a faster rate. Thus, when considering total leaf number as a measure of flowering time it appears that only 4W5°C was an effective vernalization treatment (Figure 6).
Figure 6.
Functional analysis of the two most common BoFRIa alleles. Average flowering time of T2 families transformed with the two major BoFRIa alleles BoFRIa-1 and BoFRIa-4 and compared to Col-0 transformed with the AtFRI allele and Col-0 wild type. (a) Flowering time measured as days-to-flower. The error bars show 95% confidence intervals. (b) Flowering time measured as final leaf number. The error bars show 95% confidence intervals. Segregating progeny with and without the transgene are indicated by + and - respectively.
Expression of the coding sequences of the two BoFRIa alleles under the AtFRI 5' regulatory sequences showed that both alleles can produce functionally equivalent proteins that may, under some environmental conditions, be even stronger with respect to flowering time effects than the endogenous A. thaliana protein (Figure 6). In contrast, two A. lyrata FRI alleles conferred a quantitative difference in flowering time by both association and transgenic studies [16]. The maintenance of both A. lyrata alleles at intermediate frequencies in natural populations suggests they are differentially selected in different environments. If the BoFRIa alleles do underlie flowering time QTL then there must be expression differences between the two genes to account for the difference in flowering time. Both these genes could be expressed in a very different pattern to AtFRI as the rearrangement that moved it to chromosome 4 resulted in completely different 5' sequences less than 1 kb upstream of the transcription start site and places it in a very different chromatin context since it is now 200-300 kb downstream of the heterochromatic nucleolar organizer region NOR4 [61].
Conclusions
Knowledge of B. oleracea FRI gene number, functionality and map position now puts us in a strong position to undertake an extensive investigation into the contribution of allelic variation at FRI to flowering, vernalization and life history behaviours. Differences in life history between A. lyrata and A. thaliana such as outcrossing versus selfing and a perennial compared to annual habit may result in a requirement for some level of FRI functionality in A. lyrata that is optional in A. thaliana [16]. B. oleracea, like A. lyrata, is a largely outcrossing species and some wild B. oleracea, thought to be the progenitor of the modern crop plants, have been reported to keep flowering for up to 20 years [62]. Our analysis of BoFRIa suggests that only a small number of functional BoFRIa alleles are captured within the cultivated B. oleracea germplasm. To date we have found no evidence for loss-of-function mutations that are frequent in AtFRI. Further analysis of the 5' and 3' regulatory regions of BoFRI is now underway. The proximity of BoFRI to BoFLC, BoFY and BoCO opens up new questions of how this may influence flowering behaviour. It will be particularly important to be in a position to select specific alleles in breeding programmes to allow us to enhance robustness against increasing climate variability.
Methods
Cloning BoFRI genes
The JBo BAC library was hybridised with the AtFRI genomic clone, originally from the accession Stockholm [8] and seven BACs identified, six having identical restriction patterns and one different. Purified genomic DNA was prepared (Qiagen Maxi Prep Kit) from two of these BACs (72I23 and 88G16) and used to generate shotgun libraries (TOPOr Shotgun Kit) of 1-2 kb fragments, in the pCRR4Blunt-TOPOR vector, giving 6-fold coverage. Colonies from these libraries were gridded onto nylon membrane (HyBond-N+) and hybridised to three probes generated from AtFRI (the 5' region, exon 1, and the 3' end of exon 3 and 3' UTR). BAC sub-clones were identified with each of these probes. Sequence analysis confirmed that the two BACs carried different Brassica paralogues.
Mapping BoFRI loci in B. oleracea mapping populations
Genetic mapping
Mapping filters of the A12DHdxGDDH33 mapping population were produced and hybridized with a conserved BoFRI probe as described in [36,63]. A 900 bp conserved region from exons 2 and 3 from BoFRIb was amplified from A12DHd genomic DNA with primers J2NG_F1 (5' AAGTATCAAGCGTGGAAAGCA 3') and J2NG_R1 (5' GTTACGAGGAGACCTGTGATT 3') and used to probe both the A12DHdxGDDH33 and NxG mapping filters (supplied by Graham Teakle, WHRI). Linkage analysis to map the BoFRIa locus was performed using Joinmap 3.0 [64] with the mapping data provided at BrassicaDB http://brassica.bbsrc.ac.uk/BrassicaDB/.
Fluorescence in situ hybridisation (FISH)
FISH was performed on chromosome spreads from the A12DHd genotype of B. oleracea using methods described in [65]. The chromosomes are now named according to their corresponding linkage group. JBo72I23 was applied to meiotic pachytene spreads together with JBo62M08, a BAC which is associated with the RFLP marker pN22 on C3 at 42 cM and previously assigned to chromosome C3 by FISH. JBo88G16 (BoFRIb) was applied to mitotic metaphase spreads together with BAC BoB061G14, which hybridizes to pericentromeric heterochromatin of six pairs of chromosomes, and a 45S rDNA probe from clone pTa71 [66], EMBLX07841. The chromosome pair to which JBo88G16 hybridized lacked signals from the other probes and had morphology suggestive of C9. Therefore, slides were reprobed with JBo32J18, a BAC associated with BoFLC1 which has been mapped to a region between pN47E4NM (87 cM) and pN3E1 (103 cM) on C9 [31,67] and confirmed to be on C9 by FISH (unpublished). Two further BACs showing the same restriction pattern as JBo88G16 were applied separately with JBo88G16 to pachytene spreads.
Sequencing BoFRIa in BolDFS
The B. oleracea diversity foundation set (BolDFS) is a core collection of lines that represent the genetic variation across the morphologically diverse crops of this species http://www.brassica.info/resource/plants/diversity_sets.php. DNA was isolated using the DNeasy 96 Plant Kit (Qiagen) and amplified using the GenomiPhi whole genome amplification kit (GE healthcare). A 650 bp fragment of BoFRIa was amplified from genomiphied DNA of 55 genotypes of the BolDFS by PCR with primers YWFRI_F (5'CGCACATCGTCCATCAACAAG 3') and FRIJ1_R2 (5'ATCCTTCACCCACCAGCCT 3') using AMPLITAQ GOLD TAQ DNA Polymerase (Life Technologies Ltd (Invitrogen Division)). Sequence analysis was conducted using AlignX in Vector NTI (Invitrogen).
Functional analysis of BoFRIa alleles
Plasmid pFRIg (in pBluescript-KS+, Stratagene) was mutagenised to introduce a BamHI site immediately 5' of the ATG (plasmid pFRIg-B). Digestion of pFRIg-B with BamHI plus ClaI allowed removal of the AtFRI coding sequences, leaving the 5' region of AtFRI. A 4.3 kb fragment containing BoFRIa was isolated from genomic DNA of lines E1 and E8 by PCR with primers BoFRI1_Bam_ATG (5'CTTCCGCGGATCCCATGGCCGTCCGTAAC3') and BoFRI1_R2_ClaI (5'CAGAGATCGATCTCGAGAAAGGTAGCTGTTT 3'), using PfuUltra II Fusion HS DNA Polymerase (Agilent Technologies) and sequenced. PCR products were digested with BamHI plus ClaI and the purified fragments ligated into BamHI plus ClaI-digested pFRIg-B to give final constructs containing the 5'UTR of AtFRI with the coding and 3' UTR sequences of BoFRIa-1 (in BoFRIa-1) or BoFRIa-4 (in BoFRIa-4). pFRIg-B was used as the A. thaliana FRI control. The final constructs were ligated into binary vector pSLJ755I6 (a gift from Prof. Jonathan Jones, http://www.tsl.ac.uk/research/jonathan-jones/plasmids.htm), on an EcoRI plus XhoI fragment (pFRIg-B) or EcoRI plus ClaI fragments (from BoFRIa-1 and BoFRIa-4). The constructs were transferred into Agrobacterium by triparental mating [68] and transformed into A. thaliana accession Col-0 by a floral dipping method (modified from [69]). T1 transformants were isolated by selection for Basta™ resistance. T2 seed were collected and flowering time determined by days-to-flower excluding the period of vernalization treatment and final leaf number at flowering.
Plant growth
T2 A. thaliana seeds were sown on 'Arabidopsis mix' (Scotts® Levington F2 8.75 l bags,100 l of grit, 200 g of Imidasect® 5 gr.) in plastic pots (7 cm × 7 cm) and stratified in a vernalization chamber at 5°C with an 8 h photoperiod and constant humidity for 3 days. Pots were moved to a naturally lit long day glasshouse for 7 days in May 2010 to allow germination and pre-growth. Seedlings not receiving a vernalization treatment remained in the glasshouse; seedlings to be vernalization treated were transferred back to a vernalization room or controlled environment cabinet (Snijder Economic Deluxe) for a treatment of either two or four weeks at 5°C or 10°C. After vernalization, 20 plants per line were transplanted into trays with 40 cells of 2 cm × 2 cm and returned to the glasshouse. Trays were moved regularly to random positions to prevent any positional effects on plant growth. Flowering time was recorded as either total leaf number (rosette leaves plus cauline leaves at flowering) or bolting time; bolting time was scored as the number of days-to-flowering determined when the inflorescence stem was 3 cm tall.
Abbreviations
QTL: Quantitative trait loci; BAC: Bacterial artificial chromosome; ORF: open reading frame; FISH: fluorescence in situ hybridization.
Authors' contributions
JI and CD conceived and designed the experiments, supervised the work and wrote the paper. CL, YZ and JI analyzed the BoFRIa alleles. ES and JI analyzed the BolDFS and GT contributed DNA from the BolDFS. EH performed the FISH experiments. All authors approved and read the final manuscript.
Contributor Information
Judith A Irwin, Email: judith.irwin@jic.ac.uk.
Clare Lister, Email: clare.lister@jic.ac.uk.
Eleni Soumpourou, Email: eleni.soumpourou@jic.ac.uk.
Yanwen Zhang, Email: yw_09@yahoo.com.
Elaine C Howell, Email: e.c.howell@bham.ac.uk.
Graham Teakle, Email: graham.teakle@warwick.ac.uk.
Caroline Dean, Email: caroline.dean@jic.ac.uk.
Acknowledgements
We thank David Laurie, Lars Østergaard and Martin Trick for critical reading of the manuscript, Theresa Townsend for help with initial BAC identification, David Turner (JIC genome lab), for BAC library construction and initial sequencing of BoFRIa and BoFRIb, John Walshaw for advice on coiled-coil analysis and Sue Kennedy at Elsoms Seeds Ltd for seeds of E1 and E8. We also thank Andreas Mueller for early access to the B. napus FRI sequences following electronic publication. This work was funded by Defra grant HH3708SFV, Defra feasibility Horticulture LINK grant HL0186, and BBSRC Strategic Grant to The John Innes Centre. The B. oleracea Diversity Foundation Set was produced at the University of Warwick with funding from Defra projects IF0157 and HH3723XS.
References
- Simpson GG, Dean C. Flowering - Arabidopsis, the Rosetta stone of flowering time? Science. 2002;296:285–289. doi: 10.1126/science.296.5566.285. [DOI] [PubMed] [Google Scholar]
- Baurle I, Dean C. The timing of developmental transitions in plants. Cell. 2006;125:655–664. doi: 10.1016/j.cell.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61:1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- Hayama R, Yokol S, Tamaki S, Yano M, Shimamoto K. Adaptation of photoperiodic control pathways produced short-day flowering in rice. Nature. 2003;422:719–722. doi: 10.1038/nature01549. [DOI] [PubMed] [Google Scholar]
- Franks SJ, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci (USA) 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczek AM, Roe JL, Knapp MC, Cooper MD, Lopez-Gallego C, Martin LJ, Muir CD, Sim S, Walker A, Anderson J, Egan JF, Moyers BT, Petipas R, Giakountis A, Charbit E, Coupland G, Welch SM, Schmitt J. Effects of Genetic Perturbation on Seasonal Life History Plasticity. Science. 2009;323:930–934. doi: 10.1126/science.1165826. [DOI] [PubMed] [Google Scholar]
- Sandring S, Riihimaki MA, Savolainen O, Agren J. Selection on flowering time and floral display in an alpine and lowland population of Arabidopsis lyrata. J Evolution Biol. 2007;20:558–567. doi: 10.1111/j.1420-9101.2006.01260.x. [DOI] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science. 2000;290:344–347. doi: 10.1126/science.290.5490.344. [DOI] [PubMed] [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C. Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol. 2003;132:1107–1114. doi: 10.1104/pp.103.021212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempe J, Balasubramanian S, Sureshkumar S, Singh A, Schmid M, Weigel D. Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genetics. 2005;1(1):e6. doi: 10.1371/journal.pgen.0010006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Aranzana MJ, Lister C, Baxter C, Nicholls C, Nordborg M, Dean C. Role of FRIGIDA and FLOWERING LOCUS C in determining variation in flowering time of Arabidopsis. Plant Physiol. 2005;138:1163–1173. doi: 10.1104/pp.105.061309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: The central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci (USA) 2000;97:3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell. 2001;13:935–941. doi: 10.1105/tpc.13.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Corre V, Roux F, Reboud X. DNA polymorphism at the FRIGIDA gene in Arabidopsis thaliana: extensive nonsynonymous variation is consistent with local selection for flowering time. Mol Biol Evol. 2002;19:1261–1271. doi: 10.1093/oxfordjournals.molbev.a004187. [DOI] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra S, Villarreal G Jr, Dickson M, Grimwood J, Schmutz J, Myres RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Kuittinen H, Niittyvuopio A, Rinne P, Savolainen O. Natural variation in Arabidopsis lyrata vernalization requirement conferred by a FRIGIDA indel polymorphism. Mol Biol Evol. 2008;25:319–329. doi: 10.1093/molbev/msm257. [DOI] [PubMed] [Google Scholar]
- Slotte T, Huang H, Lascoux M, Ceplitis A. Polyploid speciation did not confer instant reproductive isolation in Capsella (Brassicaceae) Mol Biol Evol. 2008;25:1472–1481. doi: 10.1093/molbev/msn092. [DOI] [PubMed] [Google Scholar]
- Fang Q, Liu J, Xu Z, Song R. Cloning and characterization of a flowering time gene from Thellungiella halophila. Acta Bioch Bioph Sin. 2008;40:747–753. [PubMed] [Google Scholar]
- Risk JM, Laurie RE, Macknight RC, Day CL. FRIGIDA and related proteins have a conserved central domain and family specific N- and C- terminal regions that are functionally important. Plant Mol Biol. 2010;73:493–505. doi: 10.1007/s11103-010-9635-2. [DOI] [PubMed] [Google Scholar]
- Keller SR, Levsen N, Ingvarsson PK, Olson MS, Tiffin P. Local selection across a latitudinal gradient shapes nucleotide diversity in Balsam Poplar, Populus balsamifera L. Genetics. 2011;188:941–952. doi: 10.1534/genetics.111.128041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P. et al. A draft sequence of the rice genome (Oryza sativa L. ssp japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- Wang N, Qian W, Suppanz I, Wei L, Mao B, Long Y, Meng J, Müller AE, Jung C. Flowering time variation in oilseed rape (Brassica napus L.) is associated with allelic variation in the FRIGIDA homologue BnaA.FRI.a. J Exp Bot. 2011;62:5641–5658. doi: 10.1093/jxb/err249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin IAP, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ. Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics. 2005;171:765–781. doi: 10.1534/genetics.105.042093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Trick M, Kwon SJ, Choi SR, Fraser F, Soumpourou E, Drou N, Wang Z, Lee SY, Yang TJ, Mun JH. Complexity of genome evolution by segmental rearrangement in Brassica rapa revealed by sequence-level analysis. BMC Genomics. 2009;10:539. doi: 10.1186/1471-2164-10-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun JH, Kwon SJ, Kim JA, Jin M, Kim JS, Lim MH, Lee SI, Hong JK, Park TH, Lee SC, Kim BJ, Seo MS, Baek S, Lee MJ, Shin JY, Hahn JH, Hwang YJ, Lim KB, Park JY, Lee J, Yang TJ, Yu HJ, Choi IY, Choi BS, Choi SR, Ramchairy N, Lim YP, Fraser F, Drou N, Soumpourou E. et al. Sequence and structure of Brassica rapa chromosome A3. Genome Biol. 2010;11:R94. doi: 10.1186/gb-2010-11-9-r94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Kondo M, Fujimura M, Nunome T, Fukuoka H, Hirai M, Matsumoto S. Simple sequence repeat-based comparative genomics between Brassica rapa and Arabidopsis thaliana: the genetic origin of clubroot resistance. Genetics. 2006;173:309–319. doi: 10.1534/genetics.104.038968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Li G, Yang B, Qiu D, Farnham M, Quiros C. High-density Brassica oleracea linkage map: identification of useful new linkages. Theor Appl Genet. 2007;115:277–287. doi: 10.1007/s00122-007-0568-3. [DOI] [PubMed] [Google Scholar]
- Long Y, Shi J, Qiu D, Li R, Zhang C, Wang J, Hou J, Zhao J, Shi L, Park BS. Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genome wide alignment with Arabidopsis. Genetics. 2007;177:2433. doi: 10.1534/genetics.107.080705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smooker AM, Wells R, Morgan C, Beaudoin F, Cho K, Fraser F, Bancroft I. The identification and mapping of candidate genes and QTL involved in the fatty acid desaturation pathway in Brassica napus. Theor App Genet. 2011;122:1075–1090. doi: 10.1007/s00122-010-1512-5. [DOI] [PubMed] [Google Scholar]
- Razi H, Howell EC, Newbury HJ, Kearsey MJ. Does sequence polymorphism of FLC paralogues underlie flowering time QTL in Brassica oleracea? Theor App Genet. 2008;116:179–192. doi: 10.1007/s00122-007-0657-3. [DOI] [PubMed] [Google Scholar]
- O'Neill CM, Bancroft I. Comparative physical mapping of segments of the genome of Brassica oleracea var. alboglabra that are homoeologous to sequenced regions of chromosomes 4 and 5 of Arabidopsis thaliana. Plant J. 2000;23:233–243. doi: 10.1046/j.1365-313x.2000.00781.x. [DOI] [PubMed] [Google Scholar]
- Østergaard L, King GK. Standardized gene nomenclature for the Brassica genus. Plant Methods. 2008;4:10–13. doi: 10.1186/1746-4811-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting Coiled Coils from Protein Sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Michaels SD, Bezerra IC, Amasino RM. FRIGIDA-related genes are required for the winter-annual habit in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:3281–3285. doi: 10.1073/pnas.0306778101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohuon EJR, Keith DJ, Parkin IAP, Sharpe AG, Lydiate DJ. Alignment of the conserved C genomes of Brassica oleracea and Brassica napus. Theor Appl Genet. 1996;93:833–839. doi: 10.1007/BF00224083. [DOI] [PubMed] [Google Scholar]
- Sebastian RL, Kearsey MJ, King GJ. Identification of quantitative trait loci controlling developmental characteristics of Brassica oleracea L. Theor Appl Genet. 2002;104:601–609. doi: 10.1007/s001220100743. [DOI] [PubMed] [Google Scholar]
- Bohuon EJR, Ramsay LD, Craft JA, Arthur AE, Marshall DF, Lydiate DJ, Kearsey MJ. The association of flowering time quantitative trait loci with duplicated regions and candidate loci in Brassica oleracea. Genetics. 1998;150:393–401. doi: 10.1093/genetics/150.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AM, Howell EC, Kearsey MJ. More QTL for flowering time revealed by substitution lines in Brassica oleracea. Heredity. 1999;83:586–596. doi: 10.1038/sj.hdy.6886050. [DOI] [PubMed] [Google Scholar]
- Okazaki K, Sakamoto K, Kikuchi R, Saito A, Togashi E, Kuginuki Y, Matsumoto S, Hirai M. Mapping and characterization of FLC homologs and QTL analysis of flowering time in Brassica oleracea. Theor Appl Genet. 2007;114:595–608. doi: 10.1007/s00122-006-0460-6. [DOI] [PubMed] [Google Scholar]
- Uptmoor R, Schrag T, Stützel H, Esch E. Crop model based QTL analysis across environments and QTL based estimation of time to floral induction and flowering in Brassica oleracea. Mol Breeding. 2008;21:205–216. doi: 10.1007/s11032-007-9121-y. [DOI] [Google Scholar]
- Schranz ME, Quijada P, Sung SB, Lukens L, Amasino R, Osborn TC. Characterization and effects of the replicated flowering time gene FLC in Brassica rapa. Genetics. 2002;162:1457–1468. doi: 10.1093/genetics/162.3.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka M, Tamura K, Hayashi M, Fujimori Y, Ohkawa Y, Kuginuki Y, Harada K. Mapping of QTLs for bolting time in Brassica rapa (syn. campestris) under different environmental conditions. Breeding Sci. 2005;55:127–133. doi: 10.1270/jsbbs.55.127. [DOI] [Google Scholar]
- Lou P, Zhao JJ, Kim JS, Shen SX, Del Carpio DP, Song XF, Jin MN, Vreugdenhil D, Wang XW, Koornneef M, Bonnema G. Quantitative trait loci for flowering time and morphological traits in multiple populations of Brassica rapa. J Exp Bot. 2007;58:4005–4016. doi: 10.1093/jxb/erm255. [DOI] [PubMed] [Google Scholar]
- Yang X, Yu YJ, Zhang FL, Zou ZR, Zhao XY, Zhang DS, Xu JB. Linkage Map Construction and Quantitative Trait Loci Analysis for Bolting Based on a Double Haploid Population of Brassica rapa. J Integrative Plant Biol. 2007;49:664–671. doi: 10.1111/j.1744-7909.2007.00447.x. [DOI] [Google Scholar]
- Zhao J, Kulkarni V, Liu N, Del Carpio DP, Bucher J, Bonnema G. BrFLC2 (FLOWERING LOCUS C) as a candidate gene for a vernalization response QTL in Brassica rapa. J Exp Bot. 2010;61:1817–1825. doi: 10.1093/jxb/erq048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson T, Shavorskaya O, Lagercrantz U. Multiple flowering time QTLs within several Brassica species could be the result of duplicated copies of one ancestral gene. Genome. 2001;44:856–864. [PubMed] [Google Scholar]
- Osterberg MK, Shavorskaya O, Lascoux M, Lagercrantz U. Naturally occurring indel variation in the Brassica nigra COL1 gene is associated with variation in flowering time. Genetics. 2002;161:299–306. doi: 10.1093/genetics/161.1.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires JC, Zhao JW, Schranz ME, Leon EJ, Quijada PA, Lukens LN, Osborn TC. Flowering time divergence and genomic rearrangements in resynthesized Brassica polyploids (Brassicaceae) Biol J Linn Soc. 2004;82:675–688. doi: 10.1111/j.1095-8312.2004.00350.x. [DOI] [Google Scholar]
- Kuittinen H, de Haan AA, Vogl C, Oikarinen S, Leppala J, Koch M, Mitchell-Olds T, Langley CH, Savolainen O. Comparing the linkage maps of the close relatives Arabidopsis lyrata and A. thaliana. Genetics. 2004;168:1575–1584. doi: 10.1534/genetics.103.022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaulieu J, Belzile F, Jean M. Linkage maps for Arabidopsis lyrata subsp. lyrata and Arabidopsis lyrata subsp. petraea combining anonymous and Arabidopsis thaliana-derived markers. Genome. 2007;50:142–150. doi: 10.1139/g06-144. [DOI] [PubMed] [Google Scholar]
- Yogeeswaran K, Frary A, York TL, Amenta A, Lesser AH, Nasrallah JB, Tanksley SD, Nasrallah ME. Comparative genome analyses of Arabidopsis spp.: inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 2005;15:505–515. doi: 10.1101/gr.3436305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu TT, Pattyn P, Bakker EG, Cao J, Cheng JF, Clark RM, Fahlgren N, Fawcett JA, Grimwood J, Gundlach H, Haberer G, Hollister JD, Ossowski S, Ottilar RP, Salamov AA, Schneeberger K, Spannagl M, Wang X, Yang L, Nasrallah ME, Bergelson J, Carrington JC, Gaut BS, Schmutz J, Mayer KFX, Van De Peer Y, Grigoriev IV, Nordborg M, Weigel D, Guo YL. The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet. 2011;43:476–481. doi: 10.1038/ng.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Wdimayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics. 2008. p. 420747.q. [DOI] [PMC free article] [PubMed]
- Beilstein MA, Nagalingum NS, Clements MD, Manchester SR, Mathews S. Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc Natl Acad Sci (USA) 2010;107:18724–18728. doi: 10.1073/pnas.0909766107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MA, Matschinger M. Evolution and genetic differentiation among relatives of Arabidopsis thaliana. Proc Natl Acad Sci (USA) 2007;104:6272–6277. doi: 10.1073/pnas.0701338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak MA, Berr A, Pecinka A, Schmidt R, McBreen K, Schubert I. Mechanisms of chromsome number reduction in Arabidopsis thaliana and related Brassicaceae species. Proc Natl Acad Sci (USA) 2006;103:5224–5229. doi: 10.1073/pnas.0510791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung SB, Amasino RM. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature. 2004;427:159–164. doi: 10.1038/nature02195. [DOI] [PubMed] [Google Scholar]
- Town CD, Cheung F, Maiti R, Crabtree J, Haas BJ, Wortman JR, Hine EE, Althoff R, Arbogast TS, Tallon LJ, Vigouroux M, Trick M, Bancroft I. Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell. 2006;18:1348–1359. doi: 10.1105/tpc.106.041665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allender CJ, Allainguillaume J, Lynn J, King GJ. Simple sequence repeats reveal uneven distribution of genetic diversity in chloroplast genomes of Brassica oleracea L. and (n = 9) wild relatives. Theor Appl Genet. 2007;114:609–618. doi: 10.1007/s00122-006-0461-5. [DOI] [PubMed] [Google Scholar]
- Schmidt R, West J, Cnops G, Love K, Balestrazzi A, Dean C. Detailed description of four YAC contigs representing 17 Mb of chromosome 4 of Arabidopsis thaliana ecotype Columbia. Plant J. 1996;9:755–765. doi: 10.1046/j.1365-313X.1996.9050755.x. [DOI] [PubMed] [Google Scholar]
- Raybould AF, Mogg RJ, Clarke RT, Gliddon CJ, Gray AJ. Variation and population structure at microsatellite and isozyme loci in wild cabbage (Brassica oleracea L.) in Dorset (UK) Genet Resour Crop Ev. 1999;46:351–360. doi: 10.1023/A:1008658630440. [DOI] [Google Scholar]
- Sharpe AG, Parkin IAP, Keith DJ, Lydiate DJ. Frequent non-reciprocal translocations in the amphidiploid genome of oilseed rape (Brassica napus) Genome. 1995;38:1112–1121. doi: 10.1139/g95-148. [DOI] [PubMed] [Google Scholar]
- van Ooijen JW, Voorrips RE. JoinMap version 3.0: Software for the calculation of genetic linkage map. Wageningen: Plant Research International; 2001. [Google Scholar]
- Howell EC, Barker GC, Jones GH, Kearsey MJ, King GJ, Kop EP, Ryder CD, Teakle GR, Vicente JG, Armstrong SA. Integration of the cytogenetic and genetic linkage maps of Brassica oleracea. Genetics. 2002;161:1225–1234. doi: 10.1093/genetics/161.3.1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach WL, Bedbrook JR. Cloning and characterisation of ribosomal genes from wheat and barley. Nucleic Acids Res. 1979;7:1869–1885. doi: 10.1093/nar/7.7.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salathia NS. Regulation of Biological clocks i Brassica olerace and Arabidopsis thalian. University of Warwick: PhD Dissertation; 2003. [Google Scholar]
- Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci (USA) 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelleteir G. In planta Agrobacterium mediated gene transfer by infltration of adult Arabidopsis thaliana plants. C R Acad Sci Paris Life Sci. 1993;316:1194–1199. [Google Scholar]