Abstract
Musashi1 (MSI1) is an RNA-binding protein that plays critical roles in nervous-system development and stem-cell self-renewal. Here, we examined its role in the progression of glioma. Short hairpin RNA (shRNA)-based MSI1-knock down (KD) in glioblastoma and medulloblastoma cells resulted in a significantly lower number of self renewing colony on day 30 (a 65% reduction), compared with non-silencing shRNA-treated control cells, indicative of an inhibitory effect of MSI1-KD on tumor cell growth and survival. Immunocytochemical staining of the MSI1-KD glioblastoma cells indicated that they ectopically expressed metaphase markers. In addition, a 2.2-fold increase in the number of MSI1-KD cells in the G2/M phase was observed. Thus, MSI1-KD caused the prolongation of mitosis and reduced the cell survival, although the expression of activated Caspase-3 was unaltered. We further showed that MSI1-KD glioblastoma cells xenografted into the brains of NOD/SCID mice formed tumors that were 96.6% smaller, as measured by a bioluminescence imaging system (BLI), than non-KD cells, and the host survival was longer (49.3±6.1 days vs. 33.6±3.6 days; P<0.01). These findings and other cell biological analyses suggested that the reduction of MSI1 in glioma cells prolonged the cell cycle by inducing the accumulation of Cyclin B1. Furthermore, MSI1-KD reduced the activities of the Notch and PI3 kinase-Akt signaling pathways, through the up-regulation of Numb and PTEN, respectively. Exposure of glioma cells to chemical inhibitors of these pathways reduced the number of spheres and living cells, as did MSI1-KD. These results suggest that MSI1 increases the growth and/or survival of certain types of glioma cells by promoting the activation of both Notch and PI3 kinase/Akt signaling.
Introduction
In recent years, substantial research has elucidated the involvement of cancer stem cells in brain tumors [1]. The concept of cancer stem cells originated from observations that not all cancer cells are equal [2]. Brain tumor tissue, for example, which includes both rapidly proliferating cells and post-mitotic differentiated cells, is hypothesized to be generated from the progeny of cancer stem/progenitor cells. Although cancer stem cells are still hypothetical, they are considered a new clinical target for cancer treatments. Thus, many researchers have been trying to distinguish and identify cell surface markers for cancer stem cells, although the validity of these markers is still extensively debated. In addition, cytoplasmic factors in proliferative cell fractions have been investigated as selective markers of cancer stem cells. Among these, the RNA-binding protein Musashi1 (MSI1) has been identified as a candidate marker of cancer stem cells for glioblastoma [3], [4].
The Musashi (Msi) family of RNA-binding proteins is evolutionarily conserved [5], [6]. Msi was originally identified in Drosophila melanogaster as a regulator of asymmetric cell division [7], [8], and its mouse counterpart is expressed in stem and progenitor cells in various tissues [6]. Two members of this family, MSI1 and Musashi2 (MSI2), have been identified in mammals [5], [9], [10]. MSI1 is a marker of neural stem progenitor cells (NS/PCs), and functions in stem-cell maintenance in the nervous system and other tissues [11], [12]. We previously demonstrated that MSI1 activates the Notch signaling pathway by translational repression of the mRNA for the Numb protein [13], a negative regulator of the Notch-signaling pathway [14], [15]. The mechanism of MSI1-mediated translational repression was recently clarified in detail: MSI1 competitively inhibits the interaction between eukaryotic translation initiation factor 4G (eIF4G) and poly(A)-binding protein (PABP), thereby blocking translation [16]. Through translational regulation, MSI1 supports the maintenance of the renewal ability of NS/PCs [6]. In addition, it was recently shown that an MSI family molecule suppresses the translation of Numb in chronic myelogenous leukemia (CML) cells [17]. MSI1 is expressed in NS/PCs in the developing embryonic and mature adult brain [11], [12]. In addition, many studies have shown that MSI1 is up-regulated in tumors such as medulloblastoma [18], [19], glioma [3], [4], astrocytoma [20], retinoblastoma [21], and colorectal adenoma [22]. Correlations have been established between the expression level of MSI1 and the grade of malignancy, the cells' proliferative activity [4], and their immaturity, in human glioma [3].
In the present study, to gain insight into the mechanism by which MSI1 contributes to tumor-cell generation or maintenance, we analyzed MSI1's functions in malignant glioblastoma. Furthermore, the contribution of MSI1 to cancer-cell progression and its usefulness as a potential target for cancer therapy have not been examined in animal models in vivo. A few reports have been published on the role of MSI1-knockdown (KD) in suppressing tumor-cell proliferation in a medulloblastoma cell line [23] and in adenocarcinoma xenografts in nude mice [24]. However, there are no studies clarifying the detailed role of MSI in the tumor growth of glioblastoma, which is the most malignant form of glioma. Hence, here we examined the role of MSI1 in the abnormal growth of glioma cells. Our results show that MSI1 enhance the growth and/or survival of glioma cells by inducing the Notch and PI3K/Akt signaling pathways, through post-transcriptional regulatory mechanisms.
Results
MSI1 expression in glioblastoma cell lines and low-passage cells obtained from glioblastoma specimens
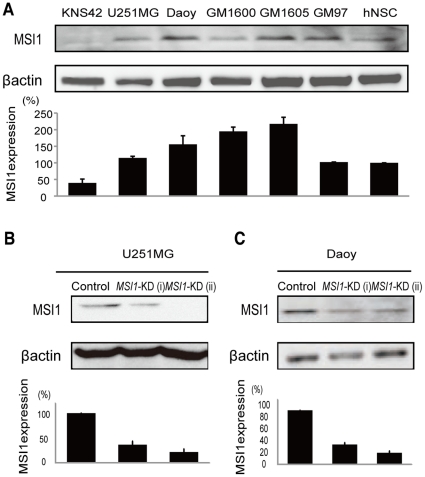
We examined various glioma cells for MSI1 protein expression by immunoblotting, and found it in human glioblastoma cell lines (KNS42 and U251MG), low-passage glioblastoma cell lines obtained from patients (GM1600, GM1605, and GM97), and a human medulloblastoma cell line (Daoy) (Fig. 1A; compare with the level in hNSCs). Each cell line and the primary cells from glioblastoma patients expressed the MSI1 protein at different levels, even though there were no differences in the patients' poor prognosis. The MSI1 protein expression was greater in cancer cells than in hNSCs, except for the KNS42 glioblastoma line. Lentivirus-mediated transduction of two separate anti-MSI1 shRNAs (MSI1-KD (i) and MSI1-KD (ii)) selectively depleted the production of MSI1 protein in the U251MG line (Fig. 1B), to 43% and 27%, respectively, and in the Daoy cells (Fig. 1C), to 36% and 21%, respectively, compared to the level in control cells transduced with non-silencing shRNA. The shRNA target sequences of MSI1 KD (i) and MSI1 KD (ii) are shown in materials and methods.
Figure 1. MSI1 expression in glioblastoma cell lines, medulloblastoma cell lines, and low-passage cells from glioblastoma patients.
(A) Immunoblots showing the MSI1 expression in human glioblastoma cell lines (U251MG, and KNS42), low-passage glioblastoma cells obtained from patients GM97, GM1600, GM1605, and a human medulloblastoma cell line (Daoy). (B and C) The selective depletion of MSI1 protein by treatment with shRNAs in U251MG cells (B) and Daoy cells (C). β-actin was used as a loading control in all the experiments.
Effect of MSI1-KD on glioma cell colony formation
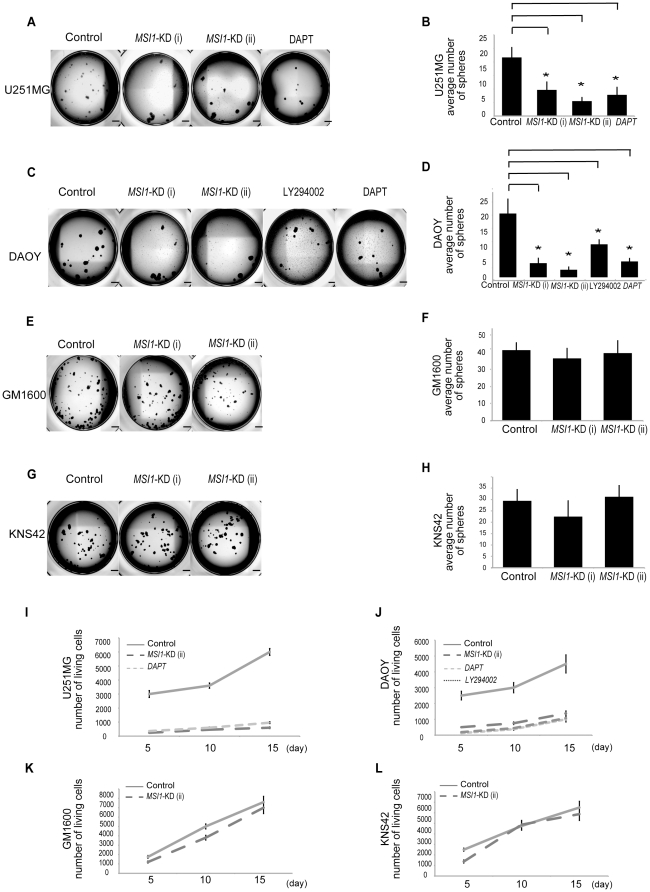
To evaluate the effect of MSI1-KD on tumor cell colony formation in vitro, we performed methylcellulose gel colony-forming assays in the presence of EGF and FGF2. Under these conditions, the cells form spherical colonies. The number of colony (at a starting density of 2000 cells per well and counting colonies larger than 100 µm), which indicated progenitor cell proliferation, showed a reduction of 65%, 84%, and 58% in the spheres generated from MSI1-KD U251MG, Daoy, and GM1605 cells, respectively. By contrast, in GM1600 (Fig. 2E,F) and KNS42 (Fig. 2.G,H) cells, which have lower endogenous MSI1 expression, the number of colonies was not reduced by MSI1 shRNA compared to control shRNA. Next, primary spheres were dissociated into single cells and further cultured in methylcellulose-containing medium, resulting in the reformation of spheres (secondary spheres). These secondary spheres were used in all experiments. MSI1-KD reduced the number of spheres compared to control shRNA in U251MG (Fig. 2A) and Daoy cells (Fig. 2C). The spheres were then dissociated and the total cell number was counted. On day 15, the total cell number in MSI1-KD spheres was reduced by 83%, 55% and 39% of control in U251MG (Fig. 2I), Daoy (Fig. 2J) and GM1605 (Fig. S3) cells, respectively; however no decrease in cell number was observed in GM1600 (Fig. 2K) and KNS42 (Fig. 2L) cells, suggesting that MSI1 does not contribute to proliferation in these cells. Sphere number and cell survival were decreased in U251MG cells exposed to DAPT (γ-secretase inhibitor) and in Daoy cells exposed to LY294002 (PI3 kinase inhibitor) and DAPT. These results suggest that MSI1 regulates both the Notch pathway and the PI3 kinase/Akt pathway in glioma cells. This is consistent with the observation that cell proliferation was repressed in MSI1-KD spheres compared to control spheres, and suggests that MSI1-KD may disorder the cell cycle in U251MG and Daoy.
Figure 2. Colony-forming cell assay and effects of MSI1-KD.
(A,B,C and D) MSI1 shRNA decreased the number of colony-forming cells. (A) Representative images of the secondary colonies formed by the control shRNA-treated and MSI1-KD U251MG cells. (Bar = 500 µm). The number of spheres was reduced in U251 cells exposed to 10 µM DAPT. (B) On Day 30, secondary spheres larger than 100 µm in diameter were counted. U251MG glioblastoma cells transduced with MSI1 shRNA and cells treated with 10 µM DAPT (γ-secretase inhibitor) generated significantly fewer spheroid cell colonies than cells transduced with control shRNA. Error bars represent SEM, *P<0.01(C) Representative images of the secondary colonies obtained in cells transduced with control shRNA and in MSI1-KD PTEN-intact Daoy cells exposed to 10 µM LY294002 (PI3 kinase inhibitor) and DAPT. (D) On Day 30, secondary spheres larger than 100 µm in diameter were counted. PTEN-intact Daoy cells transduced with MSI1 shRNA and cells exposed to LY294002 and DAPT generated significantly fewer spheroid colonies than cells transduced with control shRNA. Error bars represent SEM, *P<0.01(E and G). Representative images of the secondary colonies formed by cells expressing low endogenous MSI1. GM1600 (Fig. 2E) and KNS42 (Fig. 2G) transduced with control shRNA and MSI1 shRNA (F and H). On day 30, secondary spheres larger than 100 µm in diameter were counted. There was no significant difference between control shRNA and MSI1 shRNA in GM1600 (Fig. 2F) or KNS42 (Fig. 2H). (I, J, and K) MSI1-KD inhibits colony formation. Proliferation was assessed in glioblastoma and medulloblastoma cell lines (cell density of 2×103 cells/well) were assessed for cell proliferation by cell counting. The total cell count in the MSI1-KD groups on day 15 was reduced by 83%, and 55% in the (I) U251MG and (J) Daoy cells, respectively, as compared to the control groups.
MSI1-KD inhibits the growth of glioblastoma xenografts
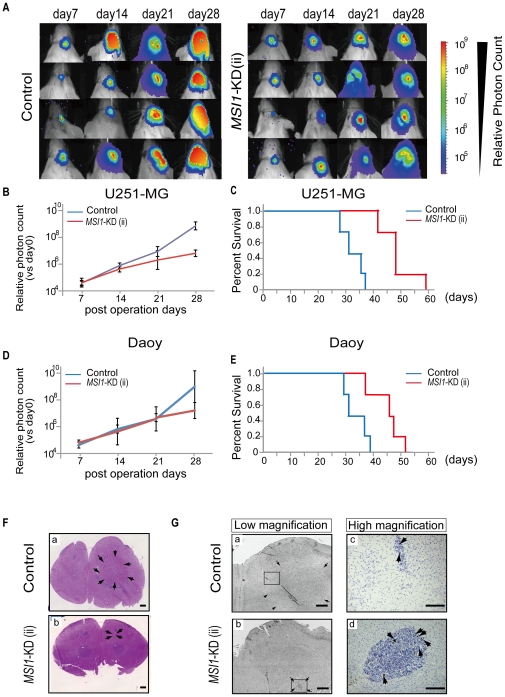
To assess the effect of MSI1-KD on tumor growth, U251MG or Daoy cells, treated with MSI1-KD (ii) or control shRNA, were transplanted into the right striatum of NOD-SCID mice. The transplanted cells were transfected with a bicistronic reporter gene encoding a luminescent protein (CBRluc) and a fluorescent protein (Venus) separated by an IRES. The tumor growth was monitored in live animals by bioluminescence imaging (BLI) using the IVIS system® (Caliper Life Sciences, Hopkinton, MA), with which the grafted cells could be identified by their fluorescent Venus and bioluminescent Luciferase signals. The results of the experiments using U251MG and Daoy cells are shown in Fig. 2, respectively. Treatment of U251MG cells with MSI1-KD resulted in a 96.6% reduction in BLI on Day 28 compared to the control shRNA group (Fig. 2A and B), and the host survival time in the MSI1-KD group (49.3±6.1 days) was significantly longer than that in the shRNA control group (33.6±3.6 days; P<0.01) (Fig. 2C). In Daoy cells, MSI1-KD cells resulted in a 97.3% reduction in BLI on Day 28 compared to the control shRNA group, the survival time of the MSI1-KD group (45.5±5.4 days) was significantly longer than that of the shRNA control group (34.0±4.1 days; P<0.01)(Fig. 2D and E). The successful transplantation of the control U251MG cells (Fig. 2F-a) and of MSI1-KD U251MG cells (Fig. 2F-b) into the mouse brain was confirmed by hematoxylin and eosin staining. However, unexpectedly, the number of positive cells for phosphohistone H3 (Fig. 2G-b, d) in the MSI1-KD-cell-transplanted mice was much larger than the number in the controls (Fig. 2G-a, c), indicating that the peri-metaphase (M-phase) cell-cycle regulation might be abnormal in the MSI1-KD cells. Taken together, these results suggest that the downregulation of MSI1 in the tumors led to a reduction in tumor tissue mass and to disordered cell proliferation and cell-cycle regulation.
Treatment with MSI1-KD induces mitosis and G2/M arrest in spheres of glioblastoma cells
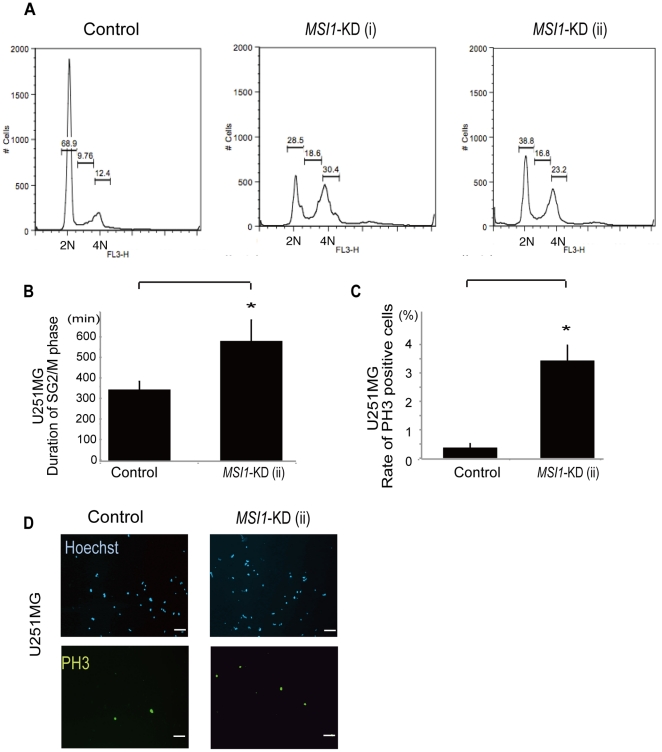
The decrease in the MSI1 protein level in brain tumor cells led to a reduction in the transplanted tumor mass in the host mouse brain (Fig. 2) and a repression of cell proliferation (Fig. 3). We therefore surmised that MSI1 regulates both the cell-cycle and cell survival. To analyze the effects of MSI1-KD on the cell cycle, MSI1-KD U251MG cells were analyzed for cell-cycle distribution by flow-cytometry. Compared to control cells, a higher number of MSI1-KD U251MG cells accumulated in the G2/M phase, and significantly fewer cells in the G0–G1 and S1 phases were detected. These results clearly demonstrate that the knockdown of MSI1 in U251MG cells resulted in changes in the cell-cycle phase distribution (Fig. 4A).
Figure 3. Xenografts expressing MSI1-KD showed reduced BLI and longer survival.
Briefly, 1×104 human glioblastoma cells were transplanted into the right striatum of NOD/SCID mice (n = 4), and tumor growth was monitored in live animals. The MSI1-KD group exhibited better survival than the control group (n = 4, *P<0.05). (A) Representative BLI images of mice receiving implants of MSI1 shRNA lentivirus-transfected U251MG cells on day 28. (B&D) Relative photon count of BLI compared with that day0 was shown. The tumor average photon count in MSI1-KD group was inferior to that in control groups in U251MG cells (Fig. 3B) and in Daoy cells (Fig. 3D) (C) Kaplan-Meier survival curves for the control and MSI1-KD mice in U251MG cells (*P<0.05). The survival time in the MSI1-KD group (49.3±6.1 days) was significantly longer than that in the shRNA control group in U251-MG cells (33.6±3.6 days; P<0.01). (E) Kaplan-Meier survival curves for the control and MSI1-KD mice are shown in Daoy cells (*P<0.05). The survival time of the MSI1-KD group (45.5±5.4 days) was significantly longer than that of the shRNA control group in Daoy cells (34.0±4.1 days; P<0.01). (F) Mouse brain transplanted with control cells (F-a) and MSI1-KD cells (F-b) was stained for hematoxylin and eosin (Bar = 1 mm). (G) Brain transplanted with control cells [G-(a, c)] and MSI1-KD cells [G-(b, d)] was immunostained for PH3 [(a, b) Bar = 800 µm (c, d) Bar = 200 µm]. In G-(a, b), surrounding arrows show the transplanted tumor area. G-(c, d) indicated the high magnification images of the squared region of G-(a, b), respectively.
Figure 4. Knockdown of MSI1 induces G2/M arrest.
MSI1-KD cells were analyzed by fluorescence-activated cell sorting. A representative cell-cycle profile for each treatment is indicated. 2N represents the G0–G1 phase and 4N represents the G2/M phase. An increased number of MSI1 KD-positive cells accumulated in the G2/M phase. (B) Time-lapse microscopy using a fluorescent ubiquitination-based cell-cycle indicator (Fucci) probe system showed that MSI1-KD cells retained the green fluorescence for a much longer time (4 days). Error bars represent SEM, *P<0.01.The spheres were dissociated, and the dissociated cells were fixed to coverslips and stained for PH3 in U251MG cells (C and D). Cell counts are plotted as bar graphs. An average of 25 high-power fields was counted. Error bars represent SEM, *P<0.01. (D) Representative images of control cells and MSI1-KD cells stained for PH3 and Hoechst. There were more PH3-positive MSI1-KD cells than control cells. (Bar = 500 µm).
To further analyze the cell-cycle status of MSI1-KD U251MG cells, we used a ubiquitination-based fluorescent cell-cycle indicator (Fucci) probe-based system, in which transfected cells show green fluorescence during the G2/M phase of the cell cycle [25]. The MSI1-KD cells retained the green fluorescence for a much longer time (4 days), as observed by time-lapse microscopy (Fig. 4B), than the control U251MG cells, suggesting a clear relationship between MSI1 and the cell cycle.
The cell colonies were then dissociated, fixed, and immunostained for phosphohistone H3 (PH3) to assess the cells' mitotic status. Both MSI1-KD U251MG cells and control U251MG cells stained positive for PH3, a marker for mitosis. Compared to the control cells, however, the MSI1-KD-transduced U251MG cells (Fig. 4C and 4D) showed a significant, 10-fold increase in PH3-positive cells. In GM1605 cells, the MSI1-KD-transduced cells demonstrated an increase of 2.8-fold (Fig. S1). Taken together, these results indicate that MSI1 is involved in cell-cycle control, especially at the M-phase, in brain tumor cells.
MSI1-KD increases Caspase-3-independent cell death through a mechanism other than cellular senescence or apoptosis
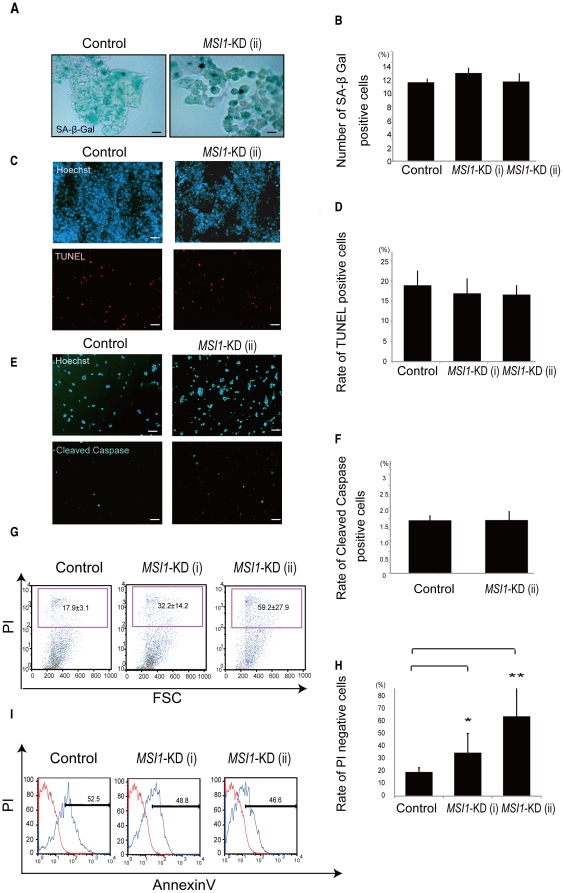
Apoptosis, senescence, or the alteration of cell survival was possible reason for the cell-number reduction in the MSI1-KD tumor cell colonies. To determine whether any of these processes was involved, we performed immunocytochemical experiments. To evaluate the state of senescence, the MSI1-KD cells were stained for senescence-activated β-Galactosidase (SA-β-Gal) [26]. There was no significant difference between the control and MSI1-KD groups on day 30 (Fig. 5A and B). Next, the MSI1-KD cells were dissociated, fixed, and immunostained for TUNEL and anti-activated Caspase-3 to examine apoptotic targets. There was no significant difference in the number of TUNEL- (Fig. 5C and 5D) or activated Caspase-3-positive cells (Fig. 5E and 5F) between the MSI1-KD U251MG cells and control cells. Similar results were obtained using MSI-KD GM1605 cells (Fig. S2). Further flow-cytometric population analysis showed that the number of annexin V positive cells in the MSI1-KD group were the same cells compared with control group (Fig. 5I), inducing the conclusion that there were no difference in apoptotic cells between MSI1-KD U251MG cells and control cells. However, flow-cytometric population analysis revealed that the number of cells that incorporated propidium-iodide (PI) was higher for the MSI1-KD cells compared with control cells (Fig. 5G and 5H), indicating that there were more dead MSI1-KD U251MG cells than control cells. Therefore, MSI1-KD appeared to induce cell death simultaneously with the occurrence of cell-cycle dysregulation. However, no increase in cleaved-Caspase-3-positive cells and in TUNEL positive cells were observed in the MSI1-KD tumor cells. Collectively, these results suggested that the MSI1-KD-induced reduction in colony-forming cells in the glioblastoma cell line occurs through cell death, but via a pathway other than apoptosis, possibly necrosis.
Figure 5. MSI1-KD cells may undergo non-apoptotic cell death.
(A and B) SA-β-Gal staining was performed to assess the senescence in controls and MSI1-KD cells. (A) Representative images of control cells and MSI1-KD cells stained for SA-β-Gal (Bar = 200 µm). (C, D, E, and F) Colonies were dissociated, and the cells were fixed to coverslips and stained for TUNEL in U251 MG cells (C and D), and for cleaved Caspase-3 in U251MG cells (E and F). Cell counts are plotted as bar graphs. An average of 25 high power fields was counted. (G and H) The number of PI-positive cells (indicating dead cells) was greater in the MSI1-KD cell population than in the control cell population. Error bars represent SEM. *P<0.01.**P<0.01. (I) The number of annexinV-positive in the MSI1-KD cells (indicating apoptotic cells) was not different from that in the control cell population.
MSI1-KD alters the expression of several cell cycle-related markers and PTEN-PI3 kinase/Akt pathway-related molecules in some glioma cells
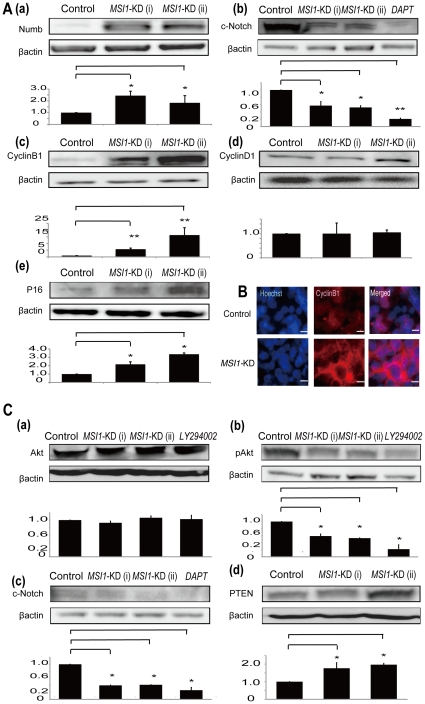
To clarify the events occurring in the MSI1-KD cells, we analyzed whether there were differences in the expression levels of various proteins between MSI1-KD cells and control shRNA-transduced U251MG (Fig. 6A) and Daoy cells (Fig. 6C). MSI1 activates Notch signaling thorough the MSI1-Numb-Notch pathway [6], [27], and the excessive translational repression of Numb by the constitutive overexpression of MSI promotes the development and progression of CML blast crisis [17]. Therefore, we examined the MSI1-KD mediated alteration of the Numb protein level and Notch activity, as represented by the cleaved-Notch level in glioma cells. Furthermore, because we found that MSI1 reduction was accompanied by cell-cycle prolongation, as shown in Figs. 3 and 4, and apparently regulated the cell-cycle, especially at the M-phase, we examined the changes in cell-cycle-related markers caused by MSI1-KD. Cyclin B1 is involved in mitosis, and Cyclin D1 is involved in the G1/S transition; PTEN are suppressors of Akt pathways. An increased expression of the p16 gene as organisms age reduces the proliferation of stem cells [28]. Since our results shows MSI1 was involved in glioma survival, we surmised MSI1 was related to the regulation of PTEN-PI3 kinase/Akt pathway. Our results showed that MSI1-KD induced the up-regulation of Numb, Cyclin B1, p16, and reduced the level of cleaved Notch in U251MG cells. Furgthermore, MSI1-KD up-regulated PTEN, and down-regulated the level of cleaved Notch and Akt phosphorylation at theonine 308 (Phospho-Akt (Thr308)) in PTEN-intact Daoy cells (Fig. 6C). There was no change in the expression of Cyclin D1 compared to control U251MG cells (Fig. 6A) and no change in the expression of Akt in Daoy cells (Fig. 6C). Cyclin B1 was more strongly expressed in MSI1-KD cells than in control cells by immunocytochemistry (Fig. 6B). In the MSI1 KD cells, Cyclin B1 was located in the cytoplasm, indicating the cells were in early mitosis. In M-phase, the nucleus has split in two and is not entirely stained by Cyclin B1 (Fig. 6B). Taken together, these results show that a decrease in MSI1 led to decreases in both the Notch pathway in U251MG cells and the PI3 kinase-Akt signaling activity in Daoy cells. Furthermore, Cyclin B1 was up-regulated, which is consistent with the prolongation of metaphase observed upon MSI1-KD.
Figure 6. MSI1-KD changed the expression level of various genes.
(A) Immunoblots showing the expression of various cell-cycle markers and downstream genes of MSI1 in control and MSI1-KD in U251MG cell lines. MSI1-KD resulted in the up-regulation of Numb, Cyclin B1, and P16, the downregulation of cleaved Notch, and no change in CyclinD1. (B) Immunocytochemistry showed that Cyclin B1 was potently expressed in MSI1-KD cells compared with control cells. (Bar = 200 µm). (C) In PTEN-intact Daoy cells, the immunoblot analysis showed that MSI1-KD led to the up-regulation of PTEN, and down-regulation of cleaved Notch and p-Akt with no change in total Akt level. Error bars represent SEM (*P<0.01).
Discussion
Here, by knocking down MSI1 in glioma cells, we found that MSI1 may promote cell proliferation and survival, and its loss can be detrimental. MSI1-KD in medulloblastoma Daoy cells induces apoptosis and mitotic catastrophe, suggesting a potential mechanism for its role in tumorigenesis [23]. MSI1-KD in adenocarcinoma cells results in tumor growth arrest in xenografts, reduced cancer cell proliferation, and increased apoptosis [24]. MSI1 acts as a key determinant of the mammary lineage through its inability to coordinate cell-cycle entry and activate the Notch and Wnt pathways [24], [27]. In human breast cancer patients, MSI1 is a negative prognostic indicator of patient survival, and is indicative of tumor cells with stem cell-like characteristics [29]. These previous reports and our current findings showed that MSI1-KD reduces tumor-cell survival and tumor-xenograft growth, and support its possible identification as a novel target for glioblastoma therapy.
In this study, using an in vitro approach, we showed that MSI1 is highly expressed in various glioblastoma cell lines (Fig. 1). MSI1-KD treated cells generated a reduced number of glioma spheres (Fig. 2) and stained positive for M-phase markers (Fig. 4). In addition, a tumor-growth assay in vivo using NOD-SCID mice demonstrated that xenografts expressing MSI1-KD resulted in a smaller tumor size (as seen by a marked decrease in BLI) and longer mouse survival compared with their counterparts treated with control shRNA-expressing xenografts (Fig. 3). These results were consistent with previous studies indicating that high MSI1 expression is associated with a poor prognosis in glioma [3], [4].
Furthermore, in glioblastoma cells, we found that MSI1-KD led to decreased Notch signaling activity, as represented by the level of cleaved Notch, and to the accumulation of Numb. Our group previously reported that MSI1 represses Numb translation and consequently augments the Notch signaling activity [27]. Other groups have shown that MSI1 up-regulates the Notch signaling activity in medulloblastoma [18], colon cancer [24], mammary progenitor cells [29], and endometrial carcinoma [30]. The nuclear translocation of Notch1, which results in the activation of its downstream pathway, may contribute to the induction of tumorigenesis by increasing proliferation and inhibiting apoptosis [2], [31].
A recent report showed that the MSI2-mediated downregulation of Numb significantly impairs the development and propagation of CML blast crisis in vitro and in vivo [17]. Indeed, disorganization of the Notch signaling balance by breakdown of the Musashi-Numb axis is directly linked to the malignancy in the CML blast crisis [32]. Consequently, modulation of the Musashi-Numb-Notch pathway influences the tumorigenicity of various tumors including glioblastoma, as described in this study. MSI1-KD probably leads the glioblastoma cells to mitotic catastrophe, resulting in cell death by the accumulation of Numb protein and subsequent inhibition of the Notch signaling pathway. (Fig. 7)
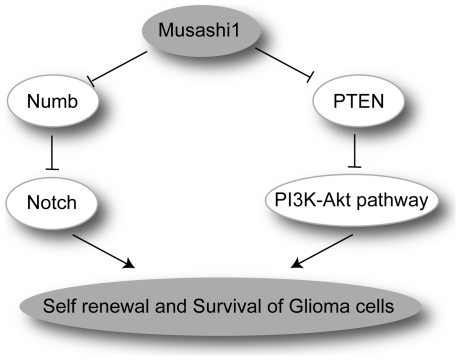
Figure 7. Current model for the involvement of Musashi-1 in glioma development.
The elevated level of the RNA-binding protein Musashi-1 leads to the down-regulation of Numb and PTEN (Imai et al., unpublished), resulting in increased activity of the Notch and PI3K/Akt-pathways, respectively. These events could lead to the enhanced self-renewal and survival of glioma cells and subsequent glioma development. In addition to these mechanisms, the downregulation of Cyclin B1 expression and prolongation of the cell cycle could be involved in Musashi1-mediated glioma development.
The reduction of MSI1 in glioblastoma cells caused other abnormalities in cell-cycle regulation. MSI1-KD increased the number of PH3-positive cells (Fig. 4C, Fig. S1), and the cell-cycle analysis using PI revealed that the number of MSI1-KD cells in the G2/M phase increased (Fig. 4A). Time-lapse assays using Fucci demonstrated that MSI1-KD sphere cells stayed longer in the S/G2/M phase of the cell cycle than cells in control spheres (Fig. 4B). There was a significant decrease in the number of MSI1-KD cells accumulating in the G0–G1 phase, supporting the previous finding that MSI1 promotes G1/S transition through the activation of both the Notch and Wnt signaling pathways [29]. Immunoblot analysis revealed that MSI1-KD in glioblastoma cells resulted in the accumulation of Cyclin B1 due to prolongation of the M-phase (Fig. 6). Interestingly, MacNicol's group found that Msi1 activates the cytoplasmic polyadenylation of c-Mos, Cyclin B1, and Cyclin B4 mRNA in Xenopus oocytes [33], [34]. In the Xenopus oocyte, cytoplasmic polyadenylation is thought to be linked with translation activation [35]. The c-Mos gene is related to meiotic cell-cycle progression. However, in our glioblastoma cells, Cyclin B1 was probably appropriately down-regulated by MSI1 through an indirect or direct pathway. Why MSI1 has different functions in various contexts is an interesting question for future study.
There was no difference in the expression of the apoptotic marker activated Caspase-3 (Fig. 5E, 5F, and Fig. S2) or in TUNEL staining (Fig. 5C and 5D) in MSI1-KD-expressing cells, and the number of annexinV-positive cells was the same as the shRNA-transduced control cells (Fig. 5I), but the number of PI-positive dead cells increased (Fig. 5G and 5H). In the present study, the decrease in MSI1 in glioblastoma cells led to cell growth retardation and a defect in non-apoptotic cell survival, accompanied by a cell-cycle abnormality, which resulted in an ectopic accumulation of Cyclin B1. These findings showed that the decrease in cell growth was caused by prolongation of the cell cycle, especially by a prolonged M-phase, and an increase in cell death by a Caspase-3-independent mechanism. Previously, inappropriate increases in nuclear Cyclin B1 have been found in mitotic catastrophe in colorectal adenocarcinomas, nasopharyngeal carcinoma, and colon cancer [36], [37]. It has been suggested that an elevated level of Cyclin B1 or premature activation and nuclear entry of CDK1/Cyclin B1 may be sufficient to induce mitosis before the completion of DNA replication, thus causing cell death during mitosis [38]. An inhibition of Notch signaling results in prolonged G2-M cell-cycle arrest before the induction of apoptosis in Kaposi's sarcoma, and leads to mitotic catastrophe, in which the destruction of nuclear structures is observed [39].
These phenomena correspond to mitotic catastrophe. The initial step in mitotic catastrophe is evidence of DNA damage in a cell that is undergoing mitosis [39]. A hallmark of mitotic catastrophe is the entry of cells into mitosis despite the presence of damaged DNA, resulting in the activation of cell death [40], [41]. These facts led us to hypothesize that Notch inhibition in glioblastoma cells results in mitotic catastrophe, which is finally resolved in prolongation of the cell cycle and delayed cell growth. Therefore, MSI1-KD probably leads the glioblastoma cells to undergo mitotic catastrophe, resulting in cell death caused by the accumulations of Numb and Cyclin B1 proteins and by the inhibition of Notch.
It is known that glioblastomas often contain mutations and deletions in the tumor suppressor gene PTEN, whose alteration affects the phosphatidyl-inositol-3 kinase (PI3K)/Akt pathway [42], [43]. In MSI1-KD Daoy cells, the expression of PTEN was increased (Fig. 6C). Recently, in an MSI1-RNA co-immunoprecipitation experiment, we found that PTEN mRNA is a putative downstream target of MSI1 in mouse neural stem cells (Imai et al., unpublished results). In the present study we observed an increase in the PTEN protein level, a decrease in the phosphorylation level of Akt (at Thr308), and no alteration in the Akt protein level in MSI1-KD cells. Accordingly, it is possible that PTEN is down-regulated by MSI1 in glioma cells. The number of spheres and living cells was reduced in PTEN-intact Daoy cells upon exposure to LY294002 (PI3 kinase inhibitor) and DAPT (γ-secretase inhibitor), a phenotype also observed upon MSI1-KD (Fig. 2).
We hypothesize that competence for cell survival may be regulated by the MSI1-PTEN-PI3 kinase/Akt pathway in glioma cells. PTEN negatively regulates neural stem cell self-renewal by modulating G0–G1 cell-cycle entry [44]. Notably, in hematopoietic stem cells, deletion of the Pten gene results in the generation of leukemic stem cells but the depletion of normal hematopoietic stem cells [45]. Taking these reports and our data together, our current model is that MSI1 plays a role in glioma development by enhancing the self-renewal and survival of glioma cells through the MSI1-Numb-Notch pathway and the MSI1-PTEN-PI3 kinase-Akt pathway (Fig. 7).
Although the MSI1-binding targets in HEK293 have been described [46], they are not known in glioblastoma cells, and our future research will be directed at identifying the downstream pathway of MSI1 through microarray analysis and proteomic profiling in glioblastoma cells. Knowledge about the downstream target genes of MSI1 will shed light on the mechanism by which these proteins promote cancer cell progression. Based on our in vitro and in vivo findings, we propose that tumor stem cells treated with MSI1-KD lose their self-renewal ability and undergo a non-apoptotic programmed cell death, which may explain the difference in tumor growth between the control and MSI1-KD groups. However, results showed that the level of MSI1 expression did not always correlate with malignancy: considering the fact that gliomas are highly variable in genetic changes/mutations and cellular origin, it is conceivable that both MSI1-dependent and -independent mechanisms are involved in the regulation of malignancy in glioma.
Materials and Methods
This study was approved by our local institutional ethical review board,“KEIO university ethical committee”. The establishment of glioblastoma cell lines GM97, GM1600, and GM1605 was specifically approved by the institutional review board of the university of California at Los Angels. This study was performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki. All the animal experiments were conducted according to the Guidelines for the Care and Use of Laboratory Animals of the Keio University School of Medicine. The KEIO permit number is #2634 and the UCLA approval number is: UCLA IRB APPROVED PROTOCOL: #10-000344. Written informed consent was obtained from all participants involved in the study.
Cell culture
Human glioblastoma cell lines U251MG, KNS42, KNS81, KNS60, and SF126 were purchased from Japanese Collection of Research Bioresources (JCRB). Other glioma cell lines, U87MG and T98G were purchased from ATCC. GM97, GM1600, and GM1605 were established from surgically resected specimens from glioblastoma patients at the University of California Los Angeles, in accordance with protocols approved by the University of California Los Angeles Institutional Review Board, as previously described [29], [47]. Briefly, after the tumor was histologically diagnosed as glioblastoma by a board-certified neuropathologist, it was finely minced with a scalpel and resuspended in complete Iscove's modified 20% Dulbecco's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Omega), 2 mM glutamine, 5 µg/mL each of insulin and transferrin, 5 ng/mL selenium (ITS Culture Supplement; Collaborative Biosciences, Bedford, MA). The GM97, GM1605, and GM1600 cell lines were passaged fewer than 10 times. The T98G, KNS42, KNS81, KNS60, SF126, GM97, GM1605, and GM1600 cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich Chemical Co., St. Louis, MO). The U251MG cell line was maintained in modified Eagle's medium (MEM) (Sigma-Aldrich). Sphere formation was performed as previously described [48]. Cells were cultured in serum-free DMEM HAM/F12 medium (Sigma-Aldrich Chemical Co., St. Louis, MO) supplemented with 1×B27 (Invitrogen), 20 ng/mL epidermal growth factor (EGF; Peprotech Invitrogen), and 20 ng/mL fibroblast growth factor-2 (FGF-2; Peprotech Invitrogen) for immunostaining and western blot experiments. The cultures were incubated at 37°C in a 5% CO2 cell culture incubator (Sanyo). Glioblastoma and medulloblastoma cells grown as sphere cultures were collected by gentle centrifugation (800×g, 5 min) and trypsinized with 0.05% TrypLESelect (Invitrogen Corp., Carlsbad, CA) for 5 min by pipetting. Cells were washed twice with phosphate buffered saline (PBS), counted, and seeded at a density of 2000 cells per 200 µL into 96-well ultralow-attachment plates (Corning).
Immunoblotting
Immunoblot analysis was performed as described previously [16]. Total cell extract (about 30 µg) was fractionated by 10% SDS-PAGE. Proteins were detected using the appropriate primary antibodies and horseradish peroxidase-linked secondary antibodies, and visualized by chemiluminescence (Amersham Biosciences).
Antibodies used in immunoblot analysis
The following primary antibodies were used for immunoblots: rat anti-MSI1 14H1 [49] or anti-MSI1 14H1 biotinylated (1∶500), rabbit anti-PTEN (1∶500) (Cell Signaling Technology), rabbit anti-p16 (1∶500) (Cell Signaling Technology), mouse anti-p21 (1∶500) (Cell Signaling Technology), rabbit anti phosphorylation at Threonine 308 of Akt (1∶250) (Cell Signaling Technology), rabbit Akt (1∶500) (Cell Signaling Technology), and mouse anti-β-Actin (1∶1000) (Sigma-Aldrich Corp MO). For detection, horseradish peroxidase (HRP)-conjugated anti-rabbit IgG (Jackson ImmunoResearch) (1∶1000), HRP-conjugated anti-rat IgG (Jackson ImmunoResearch) (1∶1000), HRP-conjugated anti-mouse IgG (Jackson ImmunoResearch) (1∶1000), and HRP-conjugated anti-goat IgG (Jackson ImmunoResearch) (1∶1000) were used.
Colony-forming assay
Primary spheres were collected, incubated in 0.25% trypsin-EDTA for 5 min at 37°C, and triturated until single-cell suspension was obtained. The cells were spun at 800 g for 5 min at 4°C and resuspended in the aforementioned sphere culture medium. The cells were cultured in the above medium with 0.8% methylcellulose (Nacalai Tesque 22224-55) as previously described [50]. The colony-forming cell assay was performed by seeding 3×103 tumor cells/well into 96-well ultralow-attachment plates (Corning) in 200 µL medium. Every four days, 50 µL of the fresh growth medium was added. The number of colonies was counted after 30 days, once the spheres were greater than 100 µm in diameter. The colony (sphere) was observed using fluorescence microscopy (BIOREVO BZ-9000; Keyence). U251MG and DAOY cells were exposed to 10 µM LY294002 (PI3 kinase inhibitor, Promeg,WI) and 10 µM DAPT (γ-secretase inhibitor, Sigma-Aldrich Corp., Saint Louis, MO) for 16 days. The culture medium was replaced every 4 days. Sphere formation and cell numbers were assessed. The proteins were collected 24 hours after the last addition of LY294002 and DAPT. For immunocytochemistry, the spheres were collected, incubated in 0.25% trypsin-EDTA for 5 min at 37°C, and triturated until a single-cell suspension was plated in the aforementioned sphere culture medium without serum on poly-D-lysine/laminin (Sigma P7405/Invitrogen 23017-015)-coated 24-well chamber slides (Iwaki 5732-008) and cultured for 2 hours. The samples were observed with a universal fluorescence microscope (Axioplan 2 imaging; Carl Zeiss).
Flow cytometer-assisted cell sorting
For cell-cycle analysis, dissociated cells (1×106 cells) were incubated in 1 ml of hypotonic propidium iodide (PI) solution (1 µg/ml PI, 0.1% citric acid, 0.2% NP-40, 10 µg/ml RNaseA) [51] for 30 min at 48°C, followed by a 15-min incubation at 37.8°C to digest the RNA. The cells (1×107cells) were suspended in 100 µlHanks' balanced salt solution (HBSS) and incubated on ice for 30 min with fluorescein isothiocyanate (FITC)-conjugated Annexin V (BD Biosciences) to detect apoptotic cells. The cells were washed, resuspended in HBSS containing 1 µg/ml PI, and analyzed by FACS Caliber (BD Biosciences). The effects of cell death were assessed by Annexin V-FITC flow cytometry as previously described [52]. Briefly, the spheres of U251-MG cells were harvested and stained with Annexin V-FITC according to the instructions of the manufacturer. Cell samples were analyzed on a FACS and apoptotic fractions were determined.
RNA interference experiment
MSI1 expression was reduced using a pLKO lentiviral vector (Sigma) expressing targeting and non-targeting sequences. Concentrated virus harboring the MSI1 shRNA or a non-silencing shRNA as a negative control was prepared was prepared in 80 µl of PBS. , 20 µl of virus solution was mixed with culture medium to transduce U251MG, Daoy, GM1605, and GM1600 cells. Infected cells were selected by incubation with 2.5 µg/mL puromycin (Sigma-Aldrich) for 10 days. The cells were then harvested and lysed. The efficacy and specificity of the shRNAs were evaluated by immunoblot analysis (Fig. 1).
Virus production
HEK293FT cells (Invitrogen) were transfected with the lentivirus constructs pCMV-VSV-G-RSV-Rev and pCAG-HIVgp by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions [53]. After incubation for 72 hours, the virus-containing supernatant was collected and centrifuged at 25000 rpm for 90 min at 4°C. Pelleted virus particles were dissolved in 80 ml of PBS.
RNAi target sequence
MSI1-targeting RNAi
5′-CCGGCAAGATGGTGACTCGAACGAACTCGAGTTCGTTCGAGTCACCATCTTGTTTTTG-3′ in the 3′-UTR of MSI1 mRNA 370 (shown as MSI1-KD(i)) and 5′-CCGGCACGTTTGAGAGTGAGGACATCTCGAGATGTCCTCACTCTCAAACGTGTTTTTG-3′ in the 3′-UTR (untranslated region) of MSI1 mRNA 529 (shown as MSI1-KD(ii)) (Oligo ID # V2HS_280120; Open Biosystems, Huntsville, AL).
Non-targeting control vector 5′-CAACAAGATGAAGAGCACCAA-3′ (MISSION Non-Target shRNA Control Vector code# SHC002 Sigma)
Immunohistochemistry
Animals were anesthetized and transcardially perfused with 4% PFA. The whole brain was removed and postfixed for 8 hr in 4% PFA, soaked overnight in 15% followed by 30% sucrose, and embedded in OCT compound. Coronal sections 14-µm thick were made with a Cryostat (Leica, Wetzlar, Germany) and processed for immunohistochemical analysis.
The tissues were fixed with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) (pH 7.4), followed by cryoprotection with 20% sucrose. Frozen 14-µm-thick tissue sections were prepared using a cryostat (CM3000, Leica). The slides were washed 3 times in PBS, blocked for 1 hour in a buffer containing 10% goat serum in PBS, and incubated overnight at 4°C with rat anti-MSI1 antibody 14H1 [49]. The slides were washed 3 times with PBS, and the antigen was visualized with ABC Vectastain and DAB as substrates (Vector Labs, Burlingame, CA). The slides were counterstained with Harris-modified hematoxylin (Thermo-Fisher, Pittsburgh, PA) and mounted in Permount. The sections were incubated with primary antibodies in TNB blocking buffer (PerkinElmer) at 4°C overnight, and then with fluorescent dye-conjugated secondary antibodies at room temperature for 1 hr. The images were observed by fluorescence microscopy (Axioplan2 Imaging, Carl Zeiss and BZ9000, Keyence Osaka, Japan).
Antibodies used in immunocytochemistry
For immunocytochemistry, rat anti-Cyclin B1 (1∶500) (Santa Cruz Biotechnology) was used in addition to antibodies used in immunoblot analysis shown above. The secondary antibodies were as follows: Alexa488 conjugated goat anti-mouse IgG (Invitrogen) (1∶1000), Alexa488-conjugated goat anti-rabbit IgG (Invitrogen) (1∶1000), Alexa488-conjugated goat anti-rat IgG (Invitrogen) (1∶1000), Alexa488-conjugated donkey anti-rabbit IgG (Invitrogen) (1∶1000), Alexa488-conjugated donkey anti-rat IgG (Invitrogen) (1∶1000), Alexa555-conjugated goat anti-rat IgG (Invitrogen) (1∶1000), Alexa555-conjugated goat anti-rabbit IgG (Invitrogen) (1∶1000), Alexa555-conjugated donkey anti-goat IgG (Invitrogen) (1∶1000), anti-Hoechst(Sigma-Aldrich) (1∶1000), and Alexa568-conjugated donkey anti-sheep IgG (Molecular Probes) (1∶1000).
Xenograft transplantation into mouse brain
All the animal experiments were conducted according to the Guidelines for the Care and Use of Laboratory Animals of the Keio University School of Medicine. Mice were anesthetized and the implantation of partially dissociated human glioma spheres (1×104 cells in 3 µl of PBS) into the right striatum was performed stereotactically (2 mm lateral and 1 mm anterior to bregma; depth 3 mm from dura). We used immunodeficient mice (NOD/SCID mice; Charles River, Tokyo, Japan) to avoid rejection of the xenografts. The survival after glioblastoma cell transplantation was evaluated for up to 2 months.
SA-β-gal staining
Senescence-associated β-galactosidase (SA-β-gal) staining was performed as described previously [26]. Briefly, the control and MSI1-KD spheres were washed in PBS, dissociated, fixed with 2% formamide-0.2% glutaraldehyde in PBS for 5 min at room temperature, and incubated overnight at 37°C in fresh X-Gal solution (1 mg/ml X-Gal, 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 150 mM NaCl, and 2 mM MgCl2 in 40 mM citric acid/sodium phosphate buffer, pH 6.0). The next day, the cells were rinsed with PBS, and the percentages of SA-β-gal-positive (blue) cells were determined after scoring 100 cells for each sample using a bright-field microscope.
Statistical analysis
Statistical analyses were performed using Student's t-test and Dunnett's test. Student's t-test was used to compare the control shRNA groups with the MSI1-KD groups in the colony-formation assay, immunocytochemistry, and the cell number assay. Values are presented as the mean ± SEM. Significance was accepted at P<0.05
TUNEL staining
The cells were stained with Hoechst 33258 and then treated with the ApopTag
Red In Situ Apoptosis Detection Kit (Intergen Co. Ltd, NY, USA). The number of positive cells was counted by an IN Cell Analyzer 2000 (GE Healthcare Bioscience).
Bioluminescence Imaging (BLI)
To monitor the tumor growth in live animals, we used the IVIS system® (Caliper Life Sciences, Hopkinton, MA). The cells used for grafting were transduced with a lentivirus containing the click beetle red Luciferase (CBRluc) coding sequence and a Venus bicistronic reporter gene connected by an internal ribosomal entry site (IRES) (EF1α-CBRluc-IRES-Venus). Specifically, the lentivirus was produced with pCSII-EFp-CBR Luc-IRES2-Venus [13]. A Xenogen-IVIS 100-cooled CCD optical macroscopic imaging system was used for the bioluminescence imaging (BLI). All the images were analyzed with the Igor (WaveMetrics, Lake Oswego, OR) and Living Image software (Caliper Life Sciences), and the optical signal intensity was expressed as photon counts, in units of photons per second. The results were displayed as a pseudocolor photon count image superimposed on a gray-scale anatomic image. To quantify the measured luminescence, we defined a specific region of interest (ROI) that covered the area at and around the implanted cells. We used the same ROI for all the animals at all time points to ensure uniform data collection.
Time-lapse imaging assay
Cells were transfected with fucci2;mAG-hGem(1/110), which labels nuclei in the S/G2/M phases [25]. The virus-infected (colored) cells were sorted by flow cytometry. Cells stably expressing the S/G2/M marker were grown in 96-well dishes in serum-free DMEM HAM/F12 (Sigma-Aldrich) supplemented with 1×B27 (Invitrogen), 20 ng/mL epidermal growth factor (EGF; Peprotech Invitrogen), and 20 ng/mL fibroblast growth factor-2 (FGF-2; Peprotech Invitrogen), at 37°C with 5% CO2. The cells were imaged for 96 hours in an incubator (Tokai Hit) on the microscope stage. Fifty-one images in the z-axis, both bright-field and single-color (green), were captured at 20-min intervals. An inverted microscope (IX-71, Olympus) was fitted with a Nipkow disc scanning confocal unit (CSU10, Yokogawa Electric Corp.), EM-CCD camera (iXON BV-887, Andor), filter wheel, and z motor (Mac5000, Ludl Electronic Products). As our imaging device has an attached auto xy stage (Sigma Koki), several spheres can be monitored in one assay. Device control and image analysis were performed using MetaMorph software (Universal Imaging).
Self renewing sphere formation was observed by time-lapse imaging microscopy.
Supporting Information
The spheres were dissociated, and the dissociated cells were fixed to coverslips and stained for PH3 in GM1605 cells.
(TIF)
Colonies were dissociated, and the cells were fixed to coverslips and stained for for cleaved Caspase-3 in GM1605 cells.
(TIF)
Low passage cells from glioblastoma patients (cell density of 2×103 cells/well) were assessed for cell proliferation by cell counting. The total cell count in the MSI1-KD-treated groups on day 15 was reduced by 39% in the GM1605 cells, as compared to the control groups.
(TIF)
Acknowledgments
We thank Dr. Keisuke Kuida for critical reading of the manuscript. We are grateful to members of the Okano laboratory, Keio University for helpful advice and discussions, especially S. Shibata, S. Kawase, H. Kawahara, and N. Onishi for technical support and helpful discussions, and T. Harada for tender animal care. We thank Mr. Suzuki for helping with the FACS experiment. Dr. Hirotaka James Okano kindly supplied the EF1α-promoter-CBRLuc-IRES-Venus construct.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by three grants, a Grant-in-Aid for Scientific Research from the Japan Science and Technology Corporation, Ministry of Education, Science, Sports, Culture and Technology, Japan (MEXT) to HO, TI, and JM, Grant-in-Aid from the Sumitomo foundation to TI, and a Grant-in-Aid for the Global COE Program at Keio University. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 2.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 3.Kanemura Y, Mori K, Sakakibara S, Fujikawa H, Hayashi H, et al. Musashi1, an evolutionarily conserved neural RNA-binding protein, is a versatile marker of human glioma cells in determining their cellular origin, malignancy, and proliferative activity. Differentiation. 2001;68:141–152. doi: 10.1046/j.1432-0436.2001.680208.x. [DOI] [PubMed] [Google Scholar]
- 4.Toda M, Iizuka Y, Yu W, Imai T, Ikeda E, et al. Expression of the neural RNA-binding protein Musashi1 in human gliomas. Glia. 2001;34:1–7. doi: 10.1002/glia.1034. [DOI] [PubMed] [Google Scholar]
- 5.Okano H, Imai T, Okabe M. Musashi: a translational regulator of cell fate. J Cell Sci. 2002;115:1355–1359. doi: 10.1242/jcs.115.7.1355. [DOI] [PubMed] [Google Scholar]
- 6.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, et al. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 7.Okabe M, Imai T, Kurusu M, Hiromi Y, Okano H. Translational repression determines a neuronal potential in Drosophila asymmetric cell division. Nature. 2001;411:94–98. doi: 10.1038/35075094. [DOI] [PubMed] [Google Scholar]
- 8.Nakamura M, Okano H, Blendy JA, Montell C. Musashi, a neural RNA-binding protein required for Drosophila adult external sensory organ development. Neuron. 1994;13:67–81. doi: 10.1016/0896-6273(94)90460-x. [DOI] [PubMed] [Google Scholar]
- 9.Sakakibara S, Nakamura Y, Satoh H, Okano H. Rna-binding protein Musashi2: developmentally regulated expression in neural precursor cells and subpopulations of neurons in mammalian CNS. J Neurosci. 2001;21:8091–8107. doi: 10.1523/JNEUROSCI.21-20-08091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sakakibara S, Imai T, Hamaguchi K, Okabe M, Aruga J, et al. Mouse-Musashi-1, a neural RNA-binding protein highly enriched in the mammalian CNS stem cell. Dev Biol. 1996;176:230–242. doi: 10.1006/dbio.1996.0130. [DOI] [PubMed] [Google Scholar]
- 11.Sakakibara S, Okano H. Expression of neural RNA-binding proteins in the postnatal CNS: implications of their roles in neuronal and glial cell development. J Neurosci. 1997;17:8300–8312. doi: 10.1523/JNEUROSCI.17-21-08300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawase S, Imai T, Miyauchi-Hara C, Yaguchi K, Nishimoto Y, et al. Identification of a Novel Intronic Enhancer Responsible for the Transcriptional Regulation of Musashi1 in Neural Stem/Progenitor Cells. Mol Brain. 2010;4:14. doi: 10.1186/1756-6606-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masuda H, Maruyama T, Hiratsu E, Yamane J, Iwanami A, et al. Noninvasive and real-time assessment of reconstructed functional human endometrium in NOD/SCID/gamma c(null) immunodeficient mice. Proc Natl Acad Sci U S A. 2007;104:1925–1930. doi: 10.1073/pnas.0604310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spana EP, Doe CQ. Numb antagonizes Notch signaling to specify sibling neuron cell fates. Neuron. 1996;17:21–26. doi: 10.1016/s0896-6273(00)80277-9. [DOI] [PubMed] [Google Scholar]
- 15.Shen Q, Zhong W, Jan YN, Temple S. Asymmetric Numb distribution is critical for asymmetric cell division of mouse cerebral cortical stem cells and neuroblasts. Development. 2002;129:4843–4853. doi: 10.1242/dev.129.20.4843. [DOI] [PubMed] [Google Scholar]
- 16.Kawahara H, Imai T, Imataka H, Tsujimoto M, Matsumoto K, et al. Neural RNA-binding protein Musashi1 inhibits translation initiation by competing with eIF4G for PABP. J Cell Biol. 2008;181:639–653. doi: 10.1083/jcb.200708004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito T, Kwon HY, Zimdahl B, Congdon KL, Blum J, et al. Regulation of myeloid leukaemia by the cell-fate determinant Musashi. Nature. 2010;466:765–768. doi: 10.1038/nature09171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yokota N, Mainprize TG, Taylor MD, Kohata T, Loreto M, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
- 19.Nakano A, Kanemura Y, Mori K, Kodama E, Yamamoto A, et al. Expression of the Neural RNA-binding protein Musashi1 in pediatric brain tumors. Pediatr Neurosurg. 2007;43:279–284. doi: 10.1159/000103307. [DOI] [PubMed] [Google Scholar]
- 20.Ma YH, Mentlein R, Knerlich F, Kruse ML, Mehdorn HM, et al. Expression of stem cell markers in human astrocytomas of different WHO grades. J Neurooncol. 2008;86:31–45. doi: 10.1007/s11060-007-9439-7. [DOI] [PubMed] [Google Scholar]
- 21.Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- 22.Schulenburg A, Cech P, Herbacek I, Marian B, Wrba F, et al. CD44-positive colorectal adenoma cells express the potential stem cell markers musashi antigen (msi1) and ephrin B2 receptor (EphB2). J Pathol. 2007;213:152–160. doi: 10.1002/path.2220. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez-Diaz PC, Burton TL, Burns SC, Hung JY, Penalva LO. Musashi1 modulates cell proliferation genes in the medulloblastoma cell line Daoy. BMC Cancer. 2008;8:280. doi: 10.1186/1471-2407-8-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sureban SM, May R, George RJ, Dieckgraefe BK, McLeod HL, et al. Knockdown of RNA binding protein musashi-1 leads to tumor regression in vivo. Gastroenterology. 2008;134:1448–1458. doi: 10.1053/j.gastro.2008.02.057. [DOI] [PubMed] [Google Scholar]
- 25.Sakaue-Sawano A, Kurokawa H, Morimura T, Hanyu A, Hama H, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132:487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 26.Dimri GP, Lee X, Basile G, Acosta M, Scott G, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Imai T, Tokunaga A, Yoshida T, Hashimoto M, Mikoshiba K, et al. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol. 2001;21:3888–3900. doi: 10.1128/MCB.21.12.3888-3900.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 29.Wang XY, Yin Y, Yuan H, Sakamaki T, Okano H, et al. Musashi1 modulates mammary progenitor cell expansion through proliferin-mediated activation of the Wnt and Notch pathways. Mol Cell Biol. 2008;28:3589–3599. doi: 10.1128/MCB.00040-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gotte M, Greve B, Kelsch R, Muller-Uthoff H, Weiss K, et al. The adult stem cell marker musashi-1 modulates endometrial carcinoma cell cycle progression and apoptosis via notch-1 and p21(WAF1/CIP1). Int J Cancer. 2011;129:2042–2049. doi: 10.1002/ijc.25856. [DOI] [PubMed] [Google Scholar]
- 31.Schwanbeck R, Martini S, Bernoth K, Just U. The Notch signaling pathway: Molecular basis of cell context dependency. Eur J Cell Biol. 2011;90:572–581. doi: 10.1016/j.ejcb.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Nishimoto Y, Okano H. New insight into cancer therapeutics: induction of differentiation by regulating the Musashi/Numb/Notch pathway. Cell Res. 20:1083–1085. doi: 10.1038/cr.2010.122. [DOI] [PubMed] [Google Scholar]
- 33.MacNicol MC, Cragle CE, MacNicol AM. Context-dependent regulation of Musashi-mediated mRNA translation and cell cycle regulation. Cell Cycle. 2011;10:39–44. doi: 10.4161/cc.10.1.14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richter JD. CPEB: a life in translation. Trends Biochem Sci. 2007;32:279–285. doi: 10.1016/j.tibs.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 36.Tsao YP, Huang SJ, Chang JL, Hsieh JT, Pong RC, et al. Adenovirus-mediated p21((WAF1/SDII/CIP1)) gene transfer induces apoptosis of human cervical cancer cell lines. J Virol. 1999;73:4983–4990. doi: 10.1128/jvi.73.6.4983-4990.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bhatt K, Zhou L, Mi QS, Huang S, She JX, et al. MicroRNA-34a is induced via p53 during cisplatin nephrotoxicity and contributes to cell survival. Mol Med. 2010;16:409–416. doi: 10.2119/molmed.2010.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heald R, McLoughlin M, McKeon F. Human wee1 maintains mitotic timing by protecting the nucleus from cytoplasmically activated Cdc2 kinase. Cell. 1993;74:463–474. doi: 10.1016/0092-8674(93)80048-j. [DOI] [PubMed] [Google Scholar]
- 39.Curry CL, Reed LL, Broude E, Golde TE, Miele L, et al. Notch inhibition in Kaposi's sarcoma tumor cells leads to mitotic catastrophe through nuclear factor-kappaB signaling. Mol Cancer Ther. 2007;6:1983–1992. doi: 10.1158/1535-7163.MCT-07-0093. [DOI] [PubMed] [Google Scholar]
- 40.Ueno M, Katayama K, Yamauchi H, Nakayama H, Doi K. Cell cycle and cell death regulation of neural progenitor cells in the 5-azacytidine (5AzC)-treated developing fetal brain. Exp Neurol. 2006;198:154–166. doi: 10.1016/j.expneurol.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 41.Castedo M, Perfettini JL, Roumier T, Yakushijin K, Horne D, et al. The cell cycle checkpoint kinase Chk2 is a negative regulator of mitotic catastrophe. Oncogene. 2004;23:4353–4361. doi: 10.1038/sj.onc.1207573. [DOI] [PubMed] [Google Scholar]
- 42.Mischel PS, Cloughesy TF. Targeted molecular therapy of GBM. Brain Pathol. 2003;13:52–61. doi: 10.1111/j.1750-3639.2003.tb00006.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwanami A, Cloughesy TF, Mischel PS. Striking the balance between PTEN and PDK1: it all depends on the cell context. Genes Dev. 2009;23:1699–1704. doi: 10.1101/gad.1832909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Groszer M, Erickson R, Scripture-Adams DD, Dougherty JD, Le Belle J, et al. PTEN negatively regulates neural stem cell self-renewal by modulating G0–G1 cell cycle entry. Proc Natl Acad Sci U S A. 2006;103:111–116. doi: 10.1073/pnas.0509939103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yilmaz OH, Valdez R, Theisen BK, Guo W, Ferguson DO, et al. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 46.de Sousa Abreu R, Sanchez-Diaz PC, Vogel C, Burns SC, Ko D, et al. Genomic analyses of musashi1 downstream targets show a strong association with cancer-related processes. J Biol Chem. 2009;284:12125–12135. doi: 10.1074/jbc.M809605200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu KV, Zhu S, Cvrljevic A, Huang TT, Sarkaria S, et al. Fyn and SRC are effectors of oncogenic epidermal growth factor receptor signaling in glioblastoma patients. Cancer Res. 2009;69:6889–6898. doi: 10.1158/0008-5472.CAN-09-0347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaneko Y, Sakakibara S, Imai T, Suzuki A, Nakamura Y, et al. Musashi1: an evolutionally conserved marker for CNS progenitor cells including neural stem cells. Dev Neurosci. 2000;22:139–153. doi: 10.1159/000017435. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida S, Shimmura S, Nagoshi N, Fukuda K, Matsuzaki Y, et al. Isolation of multipotent neural crest-derived stem cells from the adult mouse cornea. Stem Cells. 2006;24:2714–2722. doi: 10.1634/stemcells.2006-0156. [DOI] [PubMed] [Google Scholar]
- 51.Deitch AD, Law H, deVere White R. A stable propidium iodide staining procedure for flow cytometry. J Histochem Cytochem. 1982;30:967–972. doi: 10.1177/30.9.6182188. [DOI] [PubMed] [Google Scholar]
- 52.Li Y, Lal B, Kwon S, Fan X, Saldanha U, et al. The scatter factor/hepatocyte growth factor: c-met pathway in human embryonal central nervous system tumor malignancy. Cancer Res. 2005;65:9355–9362. doi: 10.1158/0008-5472.CAN-05-1946. [DOI] [PubMed] [Google Scholar]
- 53.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma IM. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The spheres were dissociated, and the dissociated cells were fixed to coverslips and stained for PH3 in GM1605 cells.
(TIF)
Colonies were dissociated, and the cells were fixed to coverslips and stained for for cleaved Caspase-3 in GM1605 cells.
(TIF)
Low passage cells from glioblastoma patients (cell density of 2×103 cells/well) were assessed for cell proliferation by cell counting. The total cell count in the MSI1-KD-treated groups on day 15 was reduced by 39% in the GM1605 cells, as compared to the control groups.
(TIF)