Abstract
The secreted Mycobacterium tuberculosis (Mtb) heme binding protein Rv0203 has been shown to play a role in Mtb heme uptake. In this work we use spectroscopic (absorption, electron paramagnetic resonance and magnetic circular dichrosim) methods to further characterize the heme coordination environments of His-tagged and native protein forms, Rv0203-His and Rv0203-notag, respectively. Rv0203-His binds the heme molecule through bis-His coordination and is low spin in both ferric and ferrous oxidation states. Rv0203-notag is high spin in both oxidation states and shares spectroscopic similarity with pentacoordinate oxygen ligated heme proteins. Mutagenesis experiments identified that residues Tyr59, His63 and His89 are required for Rv0203-notag to efficiently bind heme, reinforcing the hypothesis based on our previous structural and mutagenesis studies of Rv0203-His. While Tyr59, His63 and His89 are required for heme binding to Rv0203-notag, comparison of the absorption spectra of the Rv0203-notag mutants suggest the heme-ligand may be the hydroxyl group of Tyr59, although an exogenous hydroxide cannot be ruled out. Additionally, we measured the heme affinities of Rv0203-His and Rv0203-notag using stopped flow techniques. The rates for heme binding to Rv0203-His and Rv0203-notag are similar, 115 (μM s)-1 and 133 (μM s)-1, respectively. However, the heme off-rates differ quite dramatically, whereby Rv0203-His gives biphasic dissociation kinetics with fast and slow rates of 0.0019 s-1 and 0.0002 s-1, respectively, and Rv0203-notag has a single off-rate of 0.082 s-1. The spectral and heme binding affinity differences between Rv0203-His and Rv0203-notag suggest that the His-tag interferes with heme binding. Furthermore, these results imply that the His-tag has the ability to stabilize heme binding as well as alter heme ligand coordination of Rv0203 by providing an unnatural histidine ligand. Moreover, the heme affinity of Rv0203-notag is comparable to that of other heme transport proteins implying that Rv0203 may act as an extracellular heme transporter.
Mycobacterium tuberculosis (Mtb) is the causative agent of tuberculosis (TB), a disease responsible for approximately 1.5 million deaths per year (1). During its infection, Mtb has to maintain iron homeostasis while residing in its human host. Mtb has therefore developed a sophisticated siderophore mediated iron uptake pathway, which can scavenge non-heme-iron from a variety of sources including the human iron-transport protein transferrin, and subsequently transfers scavenged iron to membrane bound mycobactin (2-4) or the iron transport system, IrtA/B (5). This pathway, however, is metabolically costly since mycobactin biosynthesis requires at least eleven different enzymes (6). Moreover, the human innate immune system features a siderophore binding protein, siderocalin, which sequesters siderophores and has been shown to inhibit Mtb replication in macrophages by disrupting the bacterium's siderophore-mediated iron scavenging abilities (7).
An alternative form of iron uptake is via the ubiquitous heme group which contains iron. Over the last two decades heme acquisition pathways have been discovered in a growing number of bacteria. The ability to acquire heme has been demonstrated to enhance bacterial virulence in two species, Staphylococcus aureus (8) and Escherichia coli (9). There are several mechanisms of heme acquisition; the ligand can be acquired by cell surface associated heme and hemoprotein receptors (10-13), secreted high affinity heme scavengers (hemophores) (14-17), and in conjunction with hemoprotein specific proteases (18-20). Recently, studies have shown that mycobacteria may utilize heme as an alternative iron source (21-23). Experiments using a siderophore knockout of Mtb (MtbΔmbtB) showed that heme can sustain bacterial growth in vitro (22), whereas it was observed that the attenuated M. bovis strain, Bacille Calmette-Guérin siderophore knockout mutant (BCGΔmbtB), grew in a mouse-model suggesting that this mutant utilized heme as an iron source in vivo (21). A region of the Mtb genome encoding seven mycobacteria-specific proteins (Rv0201c to Rv0207c) has been identified as essential for Mtb heme uptake (22). A secreted protein, Rv0203, is encoded in this genomic region. Rv0203 has low sequence homology to all other non-mycobacterial proteins in the NCBI database and no assigned function until this point. The role Rv0203 in heme uptake was investigated by double knockout experiments (MtbΔmbtBΔRv0203), which demonstrated that Rv0203 is required for efficient growth of MtbΔmbtB in heme containing media, although Rv0203 is not essential for survival under these in vitro conditions (22). Furthermore, Rv0203 is the only Mtb culture filtrate (secreted) protein that binds to heme-agarose and it has been shown that the mature, His-tagged form of recombinant Rv0203 (Rv0203-His with no signal peptide) binds heme (22).
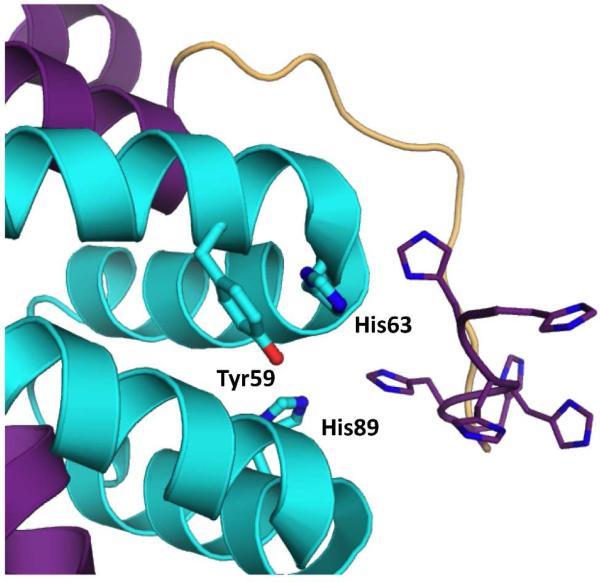
The apo-crystal structure of Rv0203-His has been solved. It is entirely α-helical and features a novel fold (22). Analysis of the apo-Rv0203-His structure revealed a similar heme binding motif to that of Serratia marcescens HasA (24) (a well-characterized hemophore) whereby His89 is 7.8 Å apart from Tyr59, which is hydrogen-bonded to His63 (Figure 1), leading to the hypothesis that Tyr59, His63 and His89 may represent the Rv0203 heme binding site. This hypothesis was reinforced through the observation that the Y59A-Rv0203-His mutant does not bind heme, although both the H63A- and H89A-Rv0203-His mutants retain similar heme binding properties as WT-Rv0203-His (22).
Figure 1.
Crystal structure of apo-Rv0203 (pdb code: 3MAY), including a manually built in N-terminal His-tag. The residues MHHHHHHGSVPRGAS were not observed in the electon density map of Rv0203 and were built using COOT (61) solely to illustrate how the His-tag may come into proximity with the proposed heme binding site. The polypeptide chains A and B are in represented in cartoon and colored in purple and cyan, respectively. The proposed active site residues, Tyr59, His63 and His89 are in stick representation and the His residues from the His-tag are represented as lines.
In this work, we have produced a tagless version of Rv0203 (Rv0203-notag) and provide evidence that the His-tag of Rv0203-His interferes with heme binding and acts as an unnatural ligand to the heme-iron. While Rv0203-notag retains the ability to bind heme, it displays differences in both heme coordination and heme binding affinity compared to Rv0203-His. Mutational analysis indicates that the heme binding site of Rv0203-notag consists of Tyr59, His63 and His89, which are the same residues as determined for Rv0203-His (22). However, the UV-visible (UV-vis) absorption and magnetic circular dichroism (MCD) spectra indicate the coordination structure of the heme-iron in Rv0203-His is bis-His, whereas Rv0203-notag binds heme through an oxygen-ligand. The heme binding affinities of both Rv0203-His and Rv0203-notag were measured using stopped flow techniques, which showed that the affinity of Rv0203-notag to heme is approximately two orders of magnitude lower than that of Rv0203-His. The heme binding affinity of Rv0203-notag is similar to that of several heme-trafficking proteins but lower than that of the well-characterized hemophore HasA (25), suggesting that Rv0203 does not scavenge heme by an affinity driven mechanism.
MATERIALS AND METHODS
Cloning, expression and purification of Rv0203
The cloning, expression and purification protocols for Rv0203-His and Mtb proteins have been described previously (26). Briefly for Rv0203-notag, the truncated gene encoding mature Rv0203 (residues 37-136) was cloned into pET30a (Stratagene) using NcoI and HindIII restriction enzyme sites to produce a fusion protein with an N-terminal enterokinase-cleavable His-tag. After enterokinase cleavage, three amino acid (AMA) residues represent the only non-native N-terminal residues of Rv0203-notag. For Rv0203-His (N-terminal His-tagged fusion protein), the truncated gene encoding residues 37-136 was cloned into pQE30 (Qiagen), described previously (22). The translated protein sequence contains 15 non-native amino acid residues at the N-terminus (MRGSHHHHHHGSVPR.).
Plasmids expressing Rv0203-notag and Rv0203-His were transformed into BL21-Gold (DE3) and XL1 Blue cells, respectively. Cells were grown at 37°C in LB medium containing 30 μg/mL kanamycin or 50 μg/mL ampicillin for Rv0203-notag and Rv0203-His, respectively. Protein expression was induced when cells reached an OD of approximately 0.8 by the addition of 1 mM isopropyl β-D-1-thiogalactopyranoside. The cells were harvested after 4 h by centrifugation at 5100 RPM for 20 min and resuspended in 50 mM Tris pH 7.4, 350 mM NaCl.
The cells were lysed by sonication after addition of egg hen lysozyme and phenylmethylsulfonyl fluoride. The cell lysate was centrifuged at 14000 RPM for 30 min and the supernatant collected. After addition of 400 μL Proteoblock protease inhibitor cocktail (Fermentas), the supernatant was loaded onto a Ni2+-charged HisTrap column (GE Healthcare) and eluted with a linear imidazole gradient. Both Rv0203 histagged constructs (Rv0203-notag prior to enterokinase treatment and Rv0203-His) eluted between 100 mM and 200 mM imidazole. Fractions containing Rv0203 histagged proteins were identified by SDS-PAGE, pooled and concentrated using a Centricon centrifugal concentrator (Millipore). Final purification of Rv0203-His was achieved by running the protein over an S75 gel filtration column (GE Healthcare) equilibrated with 50 mM Tris pH 7.4, 150 mM NaCl, which yielded nearly 100% pure protein. To produce Rv0203-notag, Rv0203 with a cleavable histag was further purified over an S75 gel filtration column and buffer exchanged into cleavage buffer (20 mM Tris, pH 7.4, 50 mM NaCl, 2 mM CaCl2). Fractions containing the protein of interest were identified by SDS-PAGE, pooled and diluted to approximately 1 mg/mL of protein. Cleavage of the His-tag was accomplished by adding 10 activity units of recombinant enterokinase (Novagen) to the protein. The cleavage reaction was monitored by SDS-PAGE and run until completion. Recombinant enterokinase was removed using a Recombinant Enterokinase Capture Kit (Novagen). The cleaved His-tag was separated from Rv0203-notag by adding NaCl to a 1 M concentration and loading the protein onto a Ni2+-charged HiTrap column (GE Healthcare). The flow-through and protein which eluted at 10 mM imidazole was collected. The final purification step of Rv0203-notag over an S75 gel filtration column yielded nearly 100% pure protein. All experiments were performed in 50 mM Tris pH 7.4, 150 mM NaCl unless otherwise stated.
Site directed mutagenesis
The Y59A-, Y59H-, Y59F-, H63A- and H89A-Rv0203-notag single mutants were made by in vitro site directed mutagenesis using the Quick Change site-directed mutagenesis kit (Stratagene). The H63A/H89A-Rv0203-notag double mutant was produced by introducing the H63A mutation into the plasmid containing H89A-Rv0203-notag. The purification procedure for the mutant forms of Rv0203-notag is identical to that of wild type protein.
Heme solution preparation and reconstitution of apo-proteins
Approximately 4 mg of heme were dissolved in 0.4 mL of ice cold 0.1 M NaOH and vortexed periodically. After 30 min, 0.4 mL of 1 M Tris, pH 7.4 was added to the solution. The solution was subsequently centrifuged for 10 min at 4° C at 13000 RPM. The supernatant was then diluted with 50 mM Tris, pH 7.4, 150 mM NaCl and centrifuged again at 5100 RPM for 10 min to remove any heme aggregates. Final concentrations were determined using ε385 = 58.44 (mM cm)-1 (27). Heme solutions were used within 12 h.
Apo-protein was reconstituted by slowly adding 1.25-fold excess heme to the apo-protein and subsequently nutating the protein at 4° C overnight. Excess heme was removed from Rv0203-His and Rv0203-notag by passing the samples over a G-25 desalting column (GE Healthcare). The heme contents of the holo-proteins were measured using the pyridine hemochromagen assay (28) and the protein concentration was measured by the modified Lowry assay (Pierce).
To make unsaturated Rv0203-notag for the ferrous Rv0203-notag UV-vis absorption spectrum and for the off-rate experiment with unsaturated Rv0203-notag, apo-Rv0203-notag was reconstituted with a heme to protein ratio of 1:10 and 1:3.3, respectively.
Ferrous hemoproteins were made by anaerobic reduction with sodium dithionite. Between 1-2 mM acqueous sodium dithionite was added to degassed ferric protein from a stock solution of 200 mM sodium dithionite, dissolved in 50 mM Tris, pH approximately 9.0, 150 mM NaCl. The ferrous CO adducts of Rv0203-notag and Rv0203-His were prepared by saturating the reduced holo-protein solutions with CO gas.
Spectroscopic measurements
All absorption spectra were recorded at room temperature on a Beckman DU800 spectrophotometer using cells with 1 cm pathlength, except for heme titration measurements, which were recorded on a Varian Cary 3E dual beam spectrophotometer.
MCD spectra were measured with a magnetic field strength of 1.41 T by using a JASCO J815 spectropolarimeter. This instrument was equipped with a JASCO MCD-1B electromagnet and interfaced with a Silicon Solutions PC through a JASCO IF-815-2 interface unit. All spectral measurements were performed using 0.2- or 1.0-cm cuvette at 4 °C. Data acquisition and manipulation has been described previously (29). UV-visible absorption spectra were recorded before and after the MCD measurements to verify sample integrity
Electron paramagnetic resonance (EPR) spectra were taken on a Bruker EMX500 spectrometer fitted with a Bruker ER041x microwave bridge and an Oxford Instruments continuous flow liquid He cryostat. The temperature was maintained at 4.5 K and monitored using an Oxford ITC503S temperature control unit. The magnetic field frequency was calibrated using a 2,2-diphenyl-1-picrylhydrazyl (DPPH) standard (g=2.0036).
Far-UV circular dichroism (CD) spectra were recorded at 4°C on a Jasco J-715 spectropolarimeter using 0.1 cm pathlength cells. The bandwidth was set to 0.5 nm, the response time was equal to 2 s and the wavelength step was equal to 0.2 nm per increment.
Heme titration
Small amounts of a concentrated heme solution (approximately 250 μM) in 50 mM Tris pH 7.4, 150 mM NaCl were titrated into cuvettes containing either protein or buffer. Spectra were recorded after a 5 min incubation period at room temperature.
Preparation of apo-H64Y/V68F-Mb
A dilute solution (approximately 50 μM) of H64Y/V68F-Mb (30) was acidified to pH 3.0 on ice and heme was extracted using two equal volume washes of ice cold methyl-ethyl-ketone. Apo-H64Y/V68F-Mb was then extensively dialyzed at 4°C against 50 mM Tris, pH 7.4, 150 mM NaCl to remove methyl-ethyl-ketone and the protein concentration measured using ε280=17 (mM cm)-1.
Off-rate measurement (apo-Myoglobin-assay)
To measure heme dissociation, holo-Rv0203-notag and a tenfold excess of H64Y/V68F-Mb were mixed in a SX.18MV stopped flow spectrophotometer (Applied Photophysics) by rapid, equal volume mixing at room temperature. The reaction was monitored at 410 nm or 600 nm until the reaction had completed. The rate of heme dissociation from holo-Rv0203-His was measured in a conventional spectrophotometer by mixing the holo-protein with tenfold excess apo-H64Y/V68F-Mb and monitoring the reaction at 600 nm for up to three hours. The resulting time-courses for both Rv0203-notag and Rv0203-His were fitted to single and double exponential functions using Graphpad Prism.
For the off-rate measurements of the various Rv0203-notag mutants, the holo-protein to apo-H64Y/V68F-Mb concentration ratio was 1:10 (protein to protein). Since it is not possible to fully heme-load the mutant proteins, especially Y59A- and H63A/H89A-Rv0203-notag, the heme contents of each mutant is different. To permit visual comparison between mutants, the data were normalized between 0 and 100, such that 100 equals the respective total differential absorption over the course of the experiment for each mutant. As discussed in the results section, the percentage of heme loading is not expected to affect the off-rate as long as apo-H64Y/V68F-Mb is in excess.
On-rate measurement
Stocks of ferrous CO-heme were made by anaerobically reducing fresh heme solutions at various concentrations with 2 mM sodium dithionite and then bubbling CO through the ferrous heme solutions. The heme concentrations of ferrous CO-heme stock were verified using ε407=147 (mM cm)-1 (31). Holo-protein samples were degassed, reduced with 2 mM sodium dithionite and saturated with CO.
Stopped flow measurements were carried out in an anaerobic environment by equal volume mixing in an Applied Photophysics SX.18MV device at room temperature. The loading syringes were assembled in an anaerobic glove box and fitted to the vertical position of a three-way stopcock. A wash syringe containing CO saturated buffer and 2 mM dithionite was attached to the horizontal position of the stopcock and used to purge the stopped flow system before each experiment.
Initially, the reactions of either 2 μM apo-Rv0203-notag or apo-Rv0203-His with 1 μM ferrous CO-heme was measured by time-resolved rapid scan spectrophotometry to find the wavelength of maximal absorbance change, which is 427 nm for both proteins. This method was also used to determine the values of kobs for wild-type (WT)-Rv0203-notag and the various mutants. On-rates, kon, for Rv0203-notag and Rv0203-His were determined by mixing various concentrations of apo-protein with 0.5 μM ferrous CO-heme and monitoring the reactions at 427 nm in photomultiplier mode. The experimental data was monophasic and was fitted to single exponential curves using Graphpad Prism. At concentrations greater than 3 μM apo-protein, a large portion of the reaction occurs within the 2 ms instrument deadtime. It should be noted that the values in the kobs measurement differ from those in the on-rate experiment due to the different instrument settings, diode array mode and photomultiplier mode, respectively. Diode array mode records fewer datapoints in the first ms of the reaction and therefore produces a smaller value for kobs.
RESULTS
Heme reconstitution and heme titration of Rv0203
Recombinant Rv0203-His was expressed as fusion protein with a non-cleavable N-terminal His-tag. A new construct was engineered to produce Rv0203 with an N-terminal enterokinase cleavable His-tag to produce Rv0203-notag. After enterokinase cleavage three non-native amino acid residues remain on the N-terminus of Rv0203-notag (AMA). Both proteins are present as predominantly apo-proteins with no endogenous heme content (<1%) after purification by Ni2+-affinity followed by gel filtration chromatography. Previously, it has been shown that Rv0203-His binds heme in a 1:1 heme to protein ratio (22). A heme titration experiment with Rv0203-notag indicates that the binding stoichiometry is also 1:1 (Figure 2). To reconstitute the apo-proteins, heme was added to both Rv0203-His and Rv0203-notag to a 1.25-fold molar excess and unbound heme removed over a desalting column, which produces rust colored holo-Rv0203-His and green colored holo-Rv0203-notag.
Figure 2.
Heme titration experiments with 5 μM apo-Rv0203-notag indicating 1:1 heme to protein stoichiometry. The curve was generated by measuring the difference spectrum between the protein sample and heme-only sample to find the wavelength of maximal absorbance difference, which was 412 nm. The saturation concentration is reached when the differential absorbance remains constant.
The oxidation state of both reconstituted holo-proteins was determined by adding the oxidizing agent potassium ferricyanide to the holo-proteins and observing the change in the absorption spectrum between 450 nm and 700 nm, a region sensitive to the oxidation state of the iron. Addition of ferricyanide did not alter the absorption spectra of either Rv0203-notag or Rv0203-His, indicating that the aerobically reconstituted proteins are both present in their ferric states (Figure S1).
Heme binding does not change the Rv0203-notag secondary structure and oligomeric state
To ensure that no large secondary structure changes occurred upon heme binding, the far-UV circular dichroism (CD) spectra of holo and apo-Rv0203-notag were recorded, as shown in Figure 3. The spectra of holo and apo-Rv0203-notag are identical and predominantly α-helical, thus ruling out significant secondary structure perturbation upon heme binding. These results reflect what had previously been reported for Rv0203-His (22). The oligomeric states of holo-Rv0203-notag and apo-Rv0203-notag were determined by analytical gel filtration chromatography and both apo- and holo-Rv0203-notag elute as a dimer (Figure S2).
Figure 3.
Far-UV CD spectra of apo-Rv0203-notag compared with holo-Rv0203-notag. The protein concentration was 10.75 μM for apo-Rv0203-notag and 7.5 μM for holo-Rv0203-notag in 5 mM phosphate buffer, pH 7.4. Each spectrum represents the average of two scans.
Absorption and MCD spectra of holo-Rv0203-His
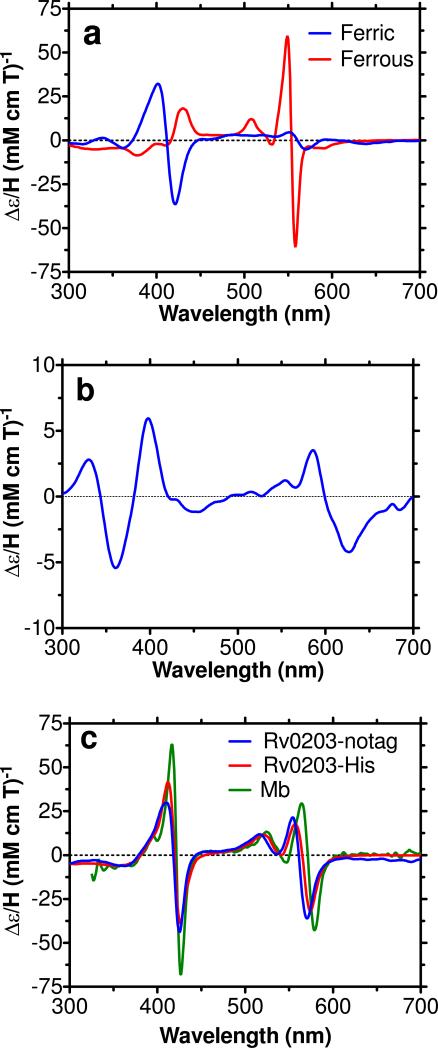
The absorption spectra of ferric, ferrous and the ferrous carbonyl adduct of holo-Rv0203-His are shown in Figure 4a. Ferric holo-Rv0203-His has a Soret peak at 412 nm and a visible maximum at 533 nm with a shoulder at 560 nm, indicating a low spin (S=1/2), six-coordinate ferric complex, which has similar spectral features as hexacoordinate bis-His ligated heme-proteins (Table 1). The ferric absorption spectrum also has a small feature at 350 nm, indicative of a minor amount high-spin heme. Upon anaerobic reduction of holo-Rv0203-His with sodium dithionite, a red-shift in the Soret peak to 426 nm is apparent, as is the emergence of two distinct peaks at 530 nm and 559 nm (Figure 4a), which indicates that the heme-iron retains its low spin six-coordinate structure in the ferrous state (Figure 4a). Inspection of ferric and ferrous Rv0203-His MCD (Figure 5a) spectra reveal similarities with bis-His heme coordinated proteins including cytochrome b5 (Table 2) and H93G-Mb (bis-imidizole) (40), indicating that the most likely coordination mode for holo-Rv0203-His is bis-His.
Figure 4.
UV-vis absorption spectra of (a) holo-Rv0203-His and (b) holo-Rv0203-notag in 50 mM Tris pH 7.4, 150 mM NaCl. The ferric holo-proteins are prepared as described in the methods. The ferrous derivatives were produced by anaerobically adding 1 mM sodium dithionite to either ferric holo-Rv0203-notag or holo-Rv0203-His. The ferrous CO adducts were produced by bubbling CO gas through the ferrous holo-proteins. The extinction coefficients of the holo-proteins are based on the heme contents.
Table 1.
Comparison of the absorption features of ferric holo-Rv0203-notag and holo-Rv0203-His with five- and six-coordinate heme-complexes from the literature. Extinction coefficients and peaks marked with * were estimated by visual inspection.
| Protein (ref.) | Ligands | Soret/nm, (ε/(mM cm)-1) | Visible/nm |
|---|---|---|---|
| Rv0203-His (this work) | His, His | 412 (78) | 533, 560 |
| Rv0203-notag (this work) | O | 392 (60) | 569, 596 |
| Cytochrome b5 (32) | His, His | 412 (116) | 531, 559 |
| Neuroglobin (33) | His, His | 413 | 533, 562 |
| HasA (34) | His, Tyr | 406 | 497, 537, 568, 618 |
| H25Y-hHO-1 (35) | Tyr | 393 | 500*, 612 |
| Dap1p (36) | Tyr | 398 | 500, 530, 620 |
| H93Y-swMb (37) | Tyr | 402 (100) | 480, 524, 598 |
| H93G-Mb (alkaline) (38) | OH- | 403 (104) | 482, 600 |
| H93G-Mb (BME) (39) | S- | 391 (65*) | 510, 618 |
| Heme (pH 7.4) | OH- | 385 (58) | 611 |
Figure 5.
MCD spectra of (a) ferric and ferrous holo-Rv0203-His and (b) ferric holo-Rv0203-notag and (c) the ferrous CO adducts of holo-Rv0203-notag and holo-Rv0203-His in 50 mM Tris pH 7.4, 150 mM NaCl compared to that of myoglobin (Mb) in 100 mM potassium phosphate buffer, pH 7 at 4 °C.
Table 2.
MCD features of ferric and ferrous holo-Rv0203-His compared to six-coordinate, bis-His-ligated cytochrome b5 in both oxidation states.
| Protein (ref.) | Ligands | Ox. State | Soret/nm (Δε/H/(mM cm T)-1) | Visible/nm (Δε/H/(mM cm T)-1) |
|---|---|---|---|---|
| Rv0203-His (this work) | His, His | Fe(III) | 402 (32.2), 421 (-36.4) | 551 (4.6), 570 (-5.1) |
| Rv0203-His (this work) | His, His | Fe(II) | 430 (18.2) | 549 (59.1), 558 (-60.8) |
| Cytochrome b5 (32) | His, His | Fe(III) | 405 (68.4), 418 (-98.0) | 553 (9.0), 571 (-14.5) |
| Cytochrome b5 (32) | His, His | Fe(II) | 425 (15.0) | 552 (160.1), 561 (-171.0) |
Absorption and MCD spectra of holo-Rv0203-notag
The UV-vis absorption spectrum of ferric holo-Rv0203-notag is shown in Figure 4b and features a relatively weak and broad Soret peak with a maximum at 392 nm and two broad visible peaks around 569 nm and 596 nm. The location of the Soret peak below 400 nm and the charge transfer band at 596 nm indicate the heme-iron is high spin (S=5/2). Investigation of the literature did not reveal a known high-spin heme-protein of great spectral similarity with Rv0203-notag, which suggests Rv0203-notag has a unique heme-coordination environment (Table 1). The closest spectroscopic resemblance is found between ferric holo-Rv0203-notag and pentacoordinate heme proteins with anionic ligands, which are known to feature a relatively weak Soret peak at wavelengths below 400 nm and a charge-transfer band near 600 nm.
Reduction of holo-Rv0203-notag by sodium dithionite produces a red-shifted Soret peak and merged α- and β-bands. However, the Soret peak is broad and resembles that of partially acid denatured deoxymyoglobin (41), indicating that heme has partially dissociated from holo-Rv0203-notag (Figure S3). To characterize heme-bound ferrous Rv0203-notag, an unsaturated form of holo-Rv0203-notag was produced that was more refractory to heme loss, whereby the heme to protein ratio in the sample is 1:10. After reduction of unsaturated holo-Rv0203-notag, the Soret peak maximum shifts to 423 nm, and a single peak emerges at 557 nm (Figure 4b), indicating a high spin five coordinate heme-iron structure. The observation that excess Rv0203-notag is required to bind ferrous heme suggests that Rv0203-notag binds heme less tightly in the ferrous oxidation state compared to the ferric state.
Further characterization of ferric holo-Rv0203-notag was undertaken using MCD spectroscopy. The overall MCD intensity of ferric holo-Rv0203-notag is relatively weak. Of particular interest, the location of the absorbance maximum at 398 nm and two minima at 527 nm and 627 nm as well as the spectral shape in the visible region (Figure 5b) share some commonality with the MCD spectra of five-coordinate oxygen–bound heme molecules such as the myoglobin cavity mutant H93Y-Mb that has a tyrosine ligand (38), and methoxide ligated Fe-protoporphyrin IX dimethyl ester (Fe-PPIXDME) (38) (Table 3).
Table 3.
MCD features of ferric holo-Rv0203-notag compared to selected five-coordinate, oxygen ligated heme molecules.
| Protein (ref.) | Ligand | Ox. State | Soret/nm (Δε/H/(mM cm T)-1) | Visible/nm (Δε/H/(mM cm T)-1) |
|---|---|---|---|---|
| Rv0203-notag (this work) | O | Fe(III) | 398 (6.0) | 527 (0), 627 (-4.2) |
| H93Y-Mb (pH 5.1-10.5) (38) | Tyr | Fe(III) | 398 (13.2) | 528 (-1.34), 621 (-10.9) |
| H93G-Mb (pH 10) (38) | OH- | Fe(III) | 397 (13.9) | 534 (-1.94), 621 (-5.21) |
| Fe-PPIXDME + CH3O- (38) | CH3O- | Fe(III) | 398 (13.7) | 530 (-2.28), 618 (12.8) |
Ferrous carbonyl adducts of holo-Rv0203-notag and holo-Rv0203-His
The absorption spectra of the ferrous CO holo-protein adducts of Rv0203-notag and Rv0203-His are similar (Figure 4). They both feature an intense Soret peak near 420 nm, which is similar to His-Fe-CO ligated proteins, including ferrous CO hHb (42) and the ferrous CO imidazole adduct of H93G-Mb (40), but different from Tyr-Fe-CO ligated proteins such as HasA (43), which have a Soret peak at a lower wavelength. This suggests both carbonyl adducts of Rv0203-His and Rv0203-notag may contain a His residue as their endogenous ligand.
The MCD properties of the ferrous CO adducts of both holo-Rv0203-His and holo-Rv0203-notag are very similar to those of His-ligated myoglobin (Figure 5c) and imidazole-coordinated H93G-Mb (40), confirming His-Fe-CO ligation under these conditions. Given the spectroscopic similarities between ferrous CO Rv0203-notag and ferrous CO Rv0203-His it is likely that under these experimental conditions their coordination environments are equivalent.
Electron paramagnetic resonance (EPR) spectroscopy of Rv0203-notag and Rv0203-His
Ferric holo-Rv0203-His displayed a mixture of spin 5/2 and spin 1/2 populations. The dominant species are two low spin 1/2 populations which display overlapping features at g = 2.8, 2.4, 2.2 and 2.0 (Figure 6a). The high spin species has two features at g⊥ = 5.8 and g∥ = 2.0, but it represents only a minor amount of the total heme-iron. Furthermore, the high spin population correlates with the small amount of high spin heme detected in the absorption spectrum, which additionally suggests it corresponds to only a small percentage of the total heme species. Thus, ferric holo-Rv0203-His is predominately low spin which is in good agreement with its absorption spectrum (Figure 4a).
Figure 6.
X-band EPR spectra of (a) holo-Rv0203-His (440 μM) and (b). holo-Rv0203-notag (450 μM). Both proteins were in 100 mM phosphate buffer at pH 7.4. Spectra were recorded at 4.5 K, 9.39 GHz microwave frequency, 0.02 mW microwave power for holo-Rv0203-notag and 10 mW for holo-Rv0203-His, 10 G modulation amplitude at 100 kHz modulation frequency. The feature at g=2.1 in (b) represents a Cu(II) impurity, which was also present in a buffer-only sample.
The EPR spectrum of ferric holo-Rv0203-notag is typical of a high spin heme-protein with two g values at g⊥ = 6.0 and g∥ = 2.0 (Figure 6b). These EPR features are in agreement with the high spin assignment based on the absorption spectrum (Figure 4b).
Y59A- and H63A/H89A-Rv0203-notag mutants lose the ability to bind heme
Mutagenesis experiments on Rv0203-His had demonstrated that Tyr59 is necessary for heme binding (22). The Y59A-, Y59H- and Y59F-Rv0203-notag mutants were produced to investigate whether this residue plays a similarly important heme binding role in Rv0203-notag.
The heme binding properties of the various mutants were compared by using a heme reconstitution experiment in which each mutant was incubated with a 1.25-fold molar excess heme, unbound heme removed over a desalting column and the respective heme and protein concentrations determined. Using this method, WT-Rv0203-notag is about 80% heme loaded. Y59A-Rv0203-notag, on the other hand, is unable to efficiently bind heme as only 14% of the protein is complexed with heme after desalting (Figure 7a).
Figure 7.
(a) Amount of heme loading onto WT-Rv0203-notag compared to Rv0203-notag mutants after heme reconstitution and removal of free heme over a G-25 desalting column. Protein concentrations were determined by the modified Lowry method (Pierce) and heme concentrations using the pyridine hemochromagen assay (28). The results represent the average of at least three independent experiments. (b) Absorption spectra of WT-Rv0203-notag compared to H63A/H89A-Rv0203-notag and Y59A-Rv0203-notag. The heme concentration is 10 μM per sample. (c) Absorption spectra of Rv0203-notag compared to the Y59F-, Y59H-, H63A- and H89A-Rv0203-notag mutants. All samples contain approximately 10 μM heme.
The Y59H-Rv0203-notag and Y59F-Rv0203-notag mutants are 42% and 61% heme bound, respectively, which suggests the amount of bound heme to Rv0203-notag correlates with the hydrophobicity and amount of conjugation of the sidechain of residue 59. Additionally, the difference in heme retention between WT-Rv0203-notag and the Y59F-Rv0203-notag mutant indicates the hydroxide residue contributes to heme binding, either in the form of ligation to the heme-iron, or indirectly through H-bonding interactions.
Residues His63 and His89 are in close proximity to Tyr59, whereby His63 hydrogen bonds to Tyr59, and His89 is approximately 8 Å from Tyr59. Both residues are proposed to be part of the heme binding pocket for Rv0203-His (22). Therefore, the H63A- and H89A-Rv0203-notag single mutants as well as the H63A/H89A-Rv0203-notag double mutant were produced. Both H63A-Rv0203-notag and H89A-Rv0203-notag are similarly compromised in their heme binding ability, with H63A-Rv0203-notag being 48% and H89A-Rv0203-notag being 41% heme loaded (Figure 7a). The H63A/H89A-Rv0203-notag double mutant is severly deficient in its heme binding ability and contains 29% holo-protein (Figure 7a), indicating that together His63 and His89 are required for efficient heme incorporation.
Absorption spectra of Y59A-Rv0203-notag and H63A/H89A-Rv0203-notag are indicative of weak, non-specific binding
The Y59A-Rv0203-notag mutant displays no clear peaks in its ferric absorption spectrum between 450 nm and 700 nm and contains a larger Soret peak shoulder at 350 nm than WT-Rv0203-notag, which is indicative of nonspecific binding or of weak heme-protein interactions within the heme binding pocket (Figure 7b). In contrast, the ferric absorption spectra of the Y59H- and Y59F-Rv0203-notag mutants are nearly identical to that of WT-Rv0203-notag, suggesting the electronic environment of the heme molecule remains unchanged (Figure 7c). The similarities of the absorption spectra between Y59F-Rv0203-notag and WT-Rv0203-notag introduce the possibility that the phenolic oxygen is not coordinated to the heme-iron, but rather that a hydroxide molecule occupies the fifth coordination site. Given the similarities between five-coordinate hydroxide ligated heme compounds compared to phenolate ligated heme, neither possibility can be excluded based on these data (Table 1).
The absorption spectrum of ferric H63A/H89A-Rv0203-notag is seen in Figure 6b. It displays similarly broad features as Y59A-Rv0203-notag, which points towards non-specific heme binding. The single mutants H63A- and H89A-Rv0203-notag on the other hand are electronically identical compared to WT-Rv0203, suggesting a very similar coordination structure (Figure 7c). Given these results it is likely that both His63 and His89 interact with the heme molecule through noncovalent interactions but do not coordinate the heme-iron.
Rv0203-notag heme binding kinetics demonstrates both a fast heme on- and off-rate
Rv0203-notag and Rv0203-His both bind heme too tightly to measure their heme binding affinities by direct titration. Therefore, the on-rate, kon, was measured by stopped flow mixing, and monitoring the change in absorbance upon heme binding as a function of time. The off-rate, k-heme, was determined using the myoglobin assay (30). Heme binding to Rv0203 can be described by scheme 1:
| (Scheme 1) |
where Rv0203-Heme represents a weakly bound intermediate and Holo-Rv0203 is the final heme-bound protein.
When the rate k-heme is sufficiently slow and when protein is in excess, the observed pseudo first order rate constant, kobs, is given by the hyperbolic equation 1:
| (Eq. 1) |
where Kd = k-1/k1. When kcoord is not rate limiting, which is the case at low protein concentrations, the reaction is bimolecular and the apparent second order association-rate constant, kon, is given by kcoord/Kd. Under these conditions, the measured rate, kobs, is linearly dependent on protein concentration (31).
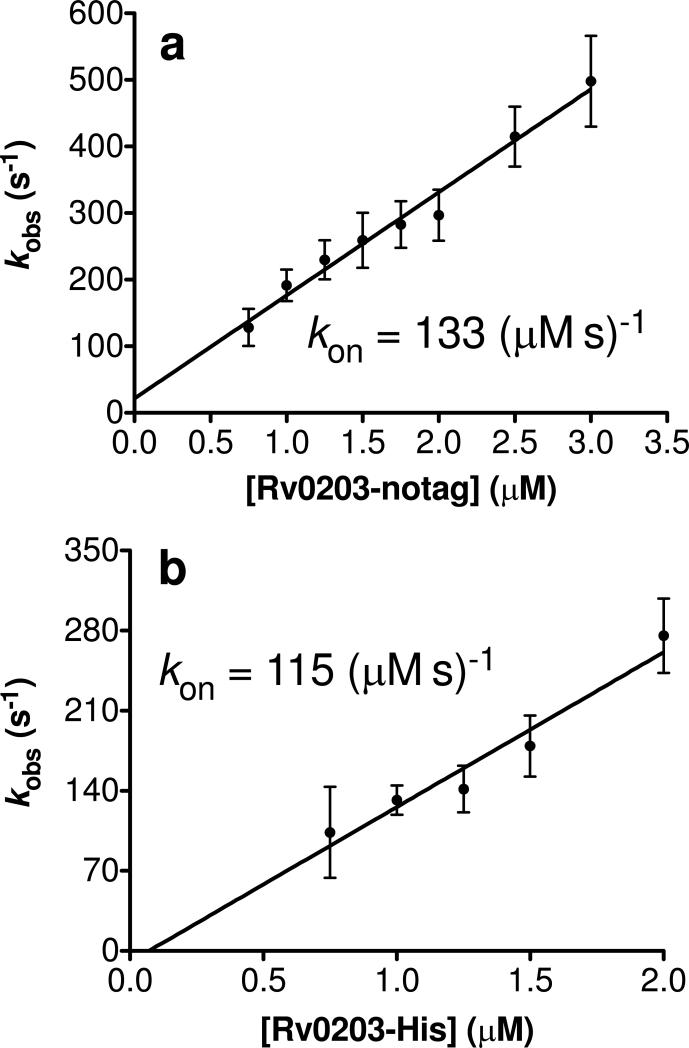
To prevent heme aggregation, which would cause the reaction to become multiphasic, we measured binding of the ferrous heme carbonyl-adduct to Rv0203-notag. The oxidation state of the heme group in this expeirment is not believed to affect the result since monomeric ferric cyanoheme binds with similar rates as ferrous CO-heme (31, 44). Ferrous CO-heme binding to Rv0203-notag was measured at increasing protein concentrations and the data was fitted to single exponential functions to obtain values for kobs (Figure S4). The linear slope of kobs vs. the concentration of apo-protein (Figure 8a) yielded the second order rate constant kon = 133 ± 19 (μM s)-1 for Rv0203-notag, which is comparable to that of several heme binding proteins (Table 4).
Figure 8.
(a) Plot of kobs of ferrous CO-heme binding vs. protein concentration to obtain the apparent second order rate constant for heme binding to Rv0203-notag and (b) Rv0203-His.
Table 4.
Kinetic parameters for heme binding to selected heme-proteins.
| Protein (ref.) | kon (μM s)-1 | k-heme (s-1) | Ka (M-1) | Method |
|---|---|---|---|---|
| Rv0203-notag (this work) | 133 ± 19 | Single Exp: 8.2 × 10-2 ± 1.6 × 10-2 | 1.6 × 109 (single exp.) | Stopped flow |
| Double Exp: kf (22%): 7.7 × 10-1 ± 5.7 × 10-2 ks: 5 × 10-2 ± 1 × 10-2 | ||||
| Rv0203-His (this work) | 115 ± 30 |
kf (31%): 1.9 × 10-3 ± 6 × 10-4 ks: 2 × 10-4 ± 1 × 10-4 |
5.8 × 1011 M-1 (using slow phase for k-heme) | Stopped flow |
| Mb (31) | 70 | 8.4 × 10-7 | 8×1013 | Stopped flow |
| H93G-Mb (31) | 70 | 1.2 × 10-2 | 6 × 109 | Stopped flow |
| BSA (31) | ~50 | 1.1 × 10-2 | 4 × 109 | Stopped flow |
| IsdA (46) | 100 | 2.6 × 10-4 | 3.8 × 1011 | Stopped flow |
| IsdB(47) | kf: 2.8 × 10-2, ks: 4.2 × 10-3 | > 2.6 × 106 | Stopped flow, Fluorescence titration | |
| Shp (48) | 1.6 | 3 × 10-4 | 5.3 × 109 | Stopped flow |
| HtsA (48) | 80 | 2.6 × 10-3 | 3.1 × 1010 | Stopped flow |
| HasA (15) (25) | 16 | 3 × 10-4 (calculated from k-heme = kon/Ka) | 5.3 × 1010 | Stopped flow, ITC |
| PhuS (49) | 0.18 (calculated from kon=Ka*k-heme) | 3.6 × 10-2 | 5.0 × 106 | Fluorescence titration |
| HmuT (50) | 3.4 × 109 (1st heme molecule) | ITC |
The off-rate was measured using the apo-H64Y/V68F-myoglobin assay whereby the apo-myoglobin mutant acts as a high affinity heme scavenger (30). The rate of the reaction depends solely on the unimolecular rate of heme release, k-heme. Heme release from ferric holo-Rv0203-notag was recorded over time (Figure S5a) and the timecourse fitted to both a single and a double exponential function (Figure 9a and Figure S6). Both fitting choices produced statistically good results (R2 > 98%), although the double exponential fit produced slighty more normal residuals (Figure S7). Fitting a single exponential function to the data produces a rate constant of k-heme of 0.082 ± 0.016 s-1, whereas double exponential fitting yields a fast phase, kf, equal to 0.77 ± 0.57 s-1 and a slow phase, ks, equal to 0.05 ± 0.01 s-1. There is insufficient experimental evidence to favor one model over the other. The fast and slow phases of heme release and the amplitudes of the fast and slow phases in the biphasic model do not change when the experiment is conducted with holo-Rv0203-notag that has been reconstituted with a heme to protein ratio of 0.33. This indicates that the heme off-rate is not dependent upon the amount of heme loading. Further, this result suggests that it is unlikely that two different heme binding sites exist, which have two different heme binding affinities. Instead, the biphasic kinetics are likely due to different conformations of Rv0203-notag, which resembles the results seen in the individual NEAT1, NEAT3 and NEAT4 domains of the heme-protein IsdX2 from Bacillus anthracis, which each bind one heme per domain (45). Using the value of k-heme obtained from the single exponential fit we were able to calculate the affinity constant, Ka =1.6 × 109 M-1, for heme to Rv0203-notag.
Figure 9.
Time courses of heme transfer from (a) holo-Rv0203-notag measured at 410 nm and (b) holo-Rv0203-His, measured at 600 nm, to a 10-fold excess amount of apo-H64Y/V68F-Mb The exponential fitting results for holo-Rv0203-notag at 600 nm is experimentally indistinguishable to that at 410 nm and shown in Figure S6. The experiments were run six times and a representative change in absorbance against time is shown.
Heme binding is slightly slowed for all Rv0203-notag mutants
To compare the relative rates of heme binding between WT-Rv0203-notag and the Rv0203-notag mutants, 1 μM ferrous CO-heme was rapidly mixed with 2 μM apo-Rv0203-notag and the observed rate constant, kobs, determined by single exponential fitting of the experimental binding curve (Table 5). Since the value of kobs is expected to differ depending on the respective on-rate, kon, kobs serves as an estimate for comparing heme binding rates between mutants. The values for kobs are slightly slower compared to wild type for all Rv0203-notag mutants, in line with the evidence that each mutant is less capable to bind heme compared to WT-Rv0203-notag (Figure 7a). Besides binding heme slower, the reaction amplitudes for both Y59H-, and Y59A-Rv0203 are approximately 40% - 50% lower than for WT-Rv0203-notag (data not shown), which suggests ferrous CO-heme incorporation is incomplete under the experimental conditions. Ferrous CO-heme binding to H63A/H89A-Rv0203-notag was not measured since the ferrous CO absorption spectrum of this mutant indicates nearly all heme is present as free ferrous CO-heme.
Table 5.
Off-rates and kobs for the Rv0203-notag mutants compared to WT-Rv0203-notag.
| Rv0203-notag mutant | Single Exp. k-heme (s-1) | Double Exp. kf and ks (s-1) | kobs (s-1) |
|---|---|---|---|
| Y59F | 0.094 ± 0.02 | kf: 1.2 ± 0.5 (15% ± 5) ks: 0.07 ± 0.0 | 87 ± 17 |
| Y59H | 0.1 ± 0.03 | kf: 0.95 (30%), ks: 0.06 | 82 ± 37 |
| Y59A | 0.367 ± 0.069 | kf: 2.15 ± 0.78 (36% ± 15) ks: 0.07 ± 0.04 | 82 ± 24 |
| H63A | 0.0896 ± 0.028 | kf: 0.3 ± 0.14 (30% ± 2) ks: 0.04 ± 0.01 | 109 ± 8 |
| H89A | 0.114 ± 0.037 | kf: 0.92 ± 0.27 (27% ± 0) ks: 0.08 ± 0.02 | 80 ± 9 |
| H63A/H89A | 0.234 ± 0.072 | kf: 2.01 ± 1.15 (28 ± 9) ks: 0.08 ± 0.056 | n/a |
| WT | 0.082 ± 0.16 | kf: 0.77 ± 0.57 (22% ± 6) ks: 0.05 ± 0.01 | 126 ± 13 |
| H64Y/V58F-Mb + Ferric Heme | 1.8 (ferric heme binding) | kf: 4.5 (42%) ks: 0.13 (ferric heme binding) |
Heme release is fast for H63A/H89A-Rv0203-notag and Y59A-Rv0203-notag
The kinetic parameter determining the heme binding affinity is the off-rate k-heme. Therefore, the heme off-rate of each Rv0203-notag mutant was measured using the apo-H64Y/V68F-Mb assay. As seen in Figure 10a, heme release from of Y59A- and H63A/H89A-Rv0203-notag occurs faster than for WT-Rv0203-notag. A double exponential fit describes the data better for Y59A- and H63A/H89A-Rv0203-notag since the single exponential fitting results are unable to model both the rapid heme release in the early parts of the reaction and the later slower stage of heme release (Figure 10a). The rate constants describing, k-heme, for Y59A-, H63A/H89A-Rv0203-notag using either single or double exponential fitting are faster for both mutants compared to wild type protein. In fact, when using the double exponential fitting results, the rate of heme release from the Y59A- and H63A/H89A Rv0203-notag mutants is about half as fast as ferric free heme binding to apo-H64Y/V68F-Mb under the same conditions (Table 5). This indicates that the faster dissociation kinetics is likely due to heme having dissociated from these mutants prior to the experiment or due to nonspecifically bound heme, which yields similarly rapid heme off-rates (31).
Figure 10.
(a) Time courses of heme dissociation from Y59A-Rv0203-notag (red), H63A/H89A-Rv0203-notag (green) compared to WT-Rv0203-notag (blue) and single exponential fitting results. (b) Time course for heme dissociation from H63A- (green), H89A- (red), and Y59F-Rv0203-notag (brown) compared to WT-Rv0203-notag (blue) and single exponential fitting curves. The data was normalized to account for the different amounts of heme per sample, as explained in the methods section.
The rates of heme release from the H63A-, H89A-, Y59H- and Y59F-Rv0203-notag mutants are quite similar to that of WT protein (Figure 10b). As with WT-Rv0203-notag, both single and double exponential functions fit the data and the results for both fitting choices are given in Table 5. Interestingly, using a double exponential fit, the fast phase of k-heme for H63A-Rv0203-notag is slower than that of WT-Rv0203-notag. However, the difference falls within the experimental error for the fast phase of heme release of WT-Rv0203-notag and therefore is unlikely to be relevent.
Rv0203-His has a higher heme binding affinity than Rv0203-notag
The heme binding affinity of Rv0203-His was measured analogously to Rv0203-notag. The on-rate is kon=115 ± 30 (μM s)-1, which is similar to that of Rv0203-notag (Figure 8b). Unlike Rv0203-notag, fitting the heme dissociation kinetics requires a biphasic fit, as judged by the residuals (Figure S8). Using a biphasic model, the fast phase of k-heme is equal to 0.0019 ± 0.0006 s-1 and the slow phase is equal to 0.0002 ± 0.0001 s-1 (Figures. 9b and S5b). The Ka for Rv0203-His, using the slow phase for k-heme is equal to 5.8 × 1011 M-1, thus indicating the heme binding affinity is approximately two orders of magnitude higher for Rv0203-His compared to that of Rv0203-notag (Table 4).
DISCUSSION
The His-tag of Rv0203-His interferes with heme binding
Previous mutational analysis of Rv0203-His identified residue Tyr59 to be essential for heme binding, whereas H63A and H89A single mutations did not abolish heme binding and displayed similar ferric absorption spectra as WT-Rv0203-His (Figure S9) (22). In this current study, the combined results from absorption, EPR and MCD spectra of Rv0203-His suggest that the heme-iron is low spin and coordinated through two His residues (Tables 1 and 2). This assignment is supported by the Rv0203-His absorption spectrum that does not contain a charge transfer band typical of tyrosine ligation near or above 600 nm (Table 1), and the MCD features of Rv0203-His that are dissimilar to those of hydroxide or Tyr coordinated heme-proteins (36, 38) (Table 3), but similar to bis-His ligated heme proteins (Table 2). The EPR spectrum of Rv0203-notag suggests the heme-iron is high spin and the absorption and MCD spectra of Rv0203-notag indicate the predominant axial ligand to the heme-iron is anionic oxygen. Thus, we have conflicting biophysical results for the heme-iron ligation between Rv0203-His and Rv0203-notag. The biophysical evidence leads one to speculate that the second His ligand to holo-Rv0203-His is donated by the His-tag. From the structure of apo-Rv0203-His, His63 and His89 are located in a solvent accessible cleft approximately 16 Å from the N-terminus of an adjacent subunit, whereby the flexible 15 amino acid N-terminal tag (including the hexa-His-tag) could feasibly bridge this gap to ligate the heme-iron (Figure 1). To date, there is only one well-documented case of His-tag interference with heme binding. Within the cytochrome c maturation protein, CcmE, heme is pentacoordinated through His130 (51, 52), whereas in the presence of a His-tag, heme binding is bis-His hexacoordinated, suggesting one His is provided by the tag (53, 54). The heme binding sites of both Rv0203-His and CcmE are both surface exposed, which suggests that His-tag interference can occur when the heme group is solvent accessible in conjunction with the heme group being in close proximity to the His-tag,
Analysis of heme coordination for Rv0203-notag and Rv0203-His
The absorption spectra of both ferric and ferrous Rv0203-notag (Figure 4b) and the ferric Rv0203-notag EPR spectrum are indicative of high spin heme molecules (Figure 6b). The relatively blue-shifted position of the Soret peak in the absorption spectrum could indicate coordination through an anionic oxygen or cysteine ligand. The MCD spectrum of Rv0203-notag (Figure 5b) most closely resembles oxygen ligated heme molecues. The possibility of Cys ligation can be excluded based on two observations. First, there are only two cysteines residues within Rv0203 and from the crystal structure of apo-Rv0203, they form a disulfide bond (22). Second, the ferric MCD spectra of Cys ligated heme proteins display strong negative features in the Soret region (39), which is not observed in the MCD spectrum for holo-Rv0203-notag. Since the nature of the axial ligand was not evident from the spectrosocopic evidence alone, a mutational analysis within the proposed heme binding site was carried out. The Y59A-Rv0203-notag mutant provides the strongest evidence that this residue is required for heme binding. This mutant does not bind heme efficiently, features a rapid rate for k-heme, similar to nonspecifically bound or free heme, and has a poorly resolved absorption spectrum (Figures 7 and 10). This result mirrors what was observed in Rv0203-His (22). Furthermore, the presence of a Tyr residue in the heme binding site would resemble that of many heme transporters and hemophores, including HasA (24), PhuT (55) and the NEAT domain family of heme-proteins (12, 56, 57). This propensity for Tyr is intriguing since very few heme-proteins contain Tyr ligands other than heme-transport type proteins. It is possible that the commonality of Tyr ligands in bacterial heme transport proteins is due to its preference for ferric metal, which is the oxidation state of heme within the most abundant host heme source, met-hemoglobin (11). The experimental evidence indictates that the most important characteristic of residue 59 is its ability to form π-π stacking interactions with the heme group. The Y59F-Rv0203-notag mutant is relatively uncompromised in its ability to bind heme, has an similar off-rate, k-heme, compared to wild type protein and displays little spectral differences compared to ferric wild type Rv0203-notag, all of which suggests the availability of a phenol ring at position 59 is critical for heme binding (Figures 7 and 10).
Despite the evidence supporting oxygen ligation, the exact coordination structure of WT-Rv0203-notag cannot be conclusively inferred from this data. On the one hand, the heme reconstitution assay indicates the hydroxyl group of Tyr59 is required for complete heme incorporation into Rv0203-notag, which could indicate the hydroxyl group coordinates to the metal. On the other hand, the similar ferric absorption spectra of WT-Rv0203-notag and Y59F-Rv0203-notag indicate the oxygen ligand may be in the form of an exogenous hydroxide molecule. We cannot distinguish either possibility since an exogenous hydroxide molecule can substitute for the tryosinate hydroxyl group in the Y59F-Rv0203-notag mutant and the absorption spectra of Tyr and OH- ligated heme-groups are known to be very similar. If hydroxide is the sole heme ligand, as suggested for Y59F-Rv0203-notag, it would resemble the ligation mode of the myoglobin mutant H93G-Mb (38). In the case of H93G-Mb, heme is bound to the protein through non-covalent interactions between the heme group and the residues surrounding the heme binding pocket (31).
The Rv0203-notag residues His63 and His89 have been shown to be critical for tight heme binding. For H63A-, H89A- and H63A/H89A-Rv0203-notag mutants the off-rates increase, while the values for kobs decrease slightly for both single mutants (Table 5). Furthermore, the H63A/H89A-Rv0203-notag double mutant displays a poorly resolved ferric absorption spectrum. Therefore, these residues likely constitute, with Tyr59, the heme binding pocket. It is, however, very unlikely that His63 and His89 bind to the heme-iron since the MCD spectrum of Rv0203-notag does not resemble those of high spin His-coordinated heme-proteins, which lack the derivative shaped feature in the visible region seen in Rv0203-notag (40, 58) (Tables 2 and 3). Moreover, the H63A/H89A-Rv0203-notag double mutant, although unable to bind heme well, retains more heme than the Y59A-Rv0203-notag single mutant, which suggests neither His-residue is as critical in heme binding as is Tyr59. Instead, His63 and His89 likely interact non-covalently with the heme group. As such, the residues His63 and His89 may fulfil roles similar to the His and Tyr residues surrounding the heme binding site of NEAT domain heme binding proteins. While all NEAT domain proteins coordinate heme through a conserved Tyr residue, additional His and Tyr residues are present to form electrostatic and π-π stacking interactions with the heme group (12, 56, 57).
The roles of His63 and His89 in heme binding, are different between Rv0203-His and Rv0203-notag. Rv0203-His is hexacoordinate, where one His ligand is donated by either His63 or His89 and the other by the His-tag. In Rv0203-His, the heme binding pocket therefore contains coordination redundancy. Such is also the case for the periplasmic heme transporter PhuS from Pseudomonas aeruginosa (59). PhuS presents two His residues, His209 and His212, to one face of the heme molecule, with His209 being the physiological ligand. A H209A mutant results in recovery of heme binding by His212. The PhuS heme binding site further contains His210, which does not coordinate to the heme-iron but its presence is required for complete heme loading onto PhuS as it forms hydrogen bond interactions with both possible heme ligands. In Rv0203-His, either His63 or His89 may assume the role of PhuS His209, whereby the H63A and H89A single mutations of Rv0203-His do not abolish heme binding since either residue can coordinate the heme molecule in the absence of the other.
The carbonyl adducts for both Rv0203-His and Rv0203-notag ligate heme through an endogenous His residue. This indicates that for Rv0203-notag a ligand switch from an oxygen ligand to a nitrogen ligand must occur either during reduction or due to CO binding trans of the fifth ligand. A ligand switch, in favor of His upon reduction has been reported when ferric heme has a oxygen ligand, as in the myoglobin mutant H93Y-Mb (37) and Chlamydomonas hemoglobin (60). The endogenous His coordination of the carbonyl adduct further supports the hypothesis that the heme binding site consists of Tyr59, His63 and His89.
Implication of kobs and k-heme differences across Rv0203-notag mutants for the heme binding mechanism
The differences in heme loading between WT-Rv0203-notag and the H63A-Rv0203-notag mutant is most likely due to different on-rates since their respective off-rates are nearly identical. Similarly, the Y59F-, Y59H- and H89A-Rv0203-notag mutants have slightly slower values for kobs and feature a small increase in k-heme, which suggests both on- and off-rates cause the reduced heme binding capabilities. In the case of the Y59A- and H63A/H89A-Rv0203-notag both the kobs and to a greater extent the off-rate are affected for both mutants, implying that these mutants are compromised in both their heme binding and heme retention capabilities (Figure 10 and Table 5). The significantly higher values of k-heme for Y59A- and H63A/H89A-Rv0203-notag indicate these mutants either contain loosely coordinated heme in the heme binding pocket or, alternatively, heme is nonspecifically bound. Considering the poorly resolved ferric absorption spectra (Figure 7b), the later possibility is more likely. It is unclear why heme release from H63A/H89A- and Y59A-Rv0203-notag mutants appear to be biphasic but it is reasonable to postulate that more than one site where heme binds nonspecifically exists. The larger differences in heme off-rates compared to on-rates between Rv0203-notag heme binding site mutants is in line with obervations from mutational analyses within heme binding sites of other heme binding proteins (15, 31).
The lack of large differences between Rv0203-notag mutants in the rates of kobs can be explained by the fact that heme binding is a two-step mechanism (scheme 1) where, under the experimental conditions, the formation of the Rv0203-Heme intermediate is rate limiting. Since the overall heme binding pocket structure is unlikely to be very different between mutants, the rate kobs is not expected to vary depending on the mutant. The second step in binding, the formation of Holo-Rv0203, involves binding pocket rearrangements (31) and may be more variable between the Rv0203-notag mutants. However, we could not determine the rate governing the second heme-binding step, kcoord, since we were unable to measure heme binding when the amount of apo-protein is in large excess compared to the heme concentration, which is the required condition for kcoord measurement.
Heme binding affinities and functional implications
The on-rate of both Rv0203-notag (133 μM-1 s-1) and Rv0203-His (115 μM-1 s-1) is similar to that of several heme-proteins including myoglobin (31), the heme transporter IsdA (46) and the heme receptor HtsA (48) (Table 4). Rv0203-notag features a fast rate of heme release (0.082 s-1), similar to that of BSA and H93G-Mb (31) and the heme transport domain of IsdB, IsdB-N2 (47). Due to the relatively fast off-rate, the Ka for Rv0203-notag (1.6 × 109 M-1) is lower than that of the hemophore HasA (25). It is comparable to that of IsdB-N2 (47), the myoglobin cavity mutant H93G-Mb, BSA (31), the Streptococcus pyogenes cell surface heme transporter Shp (48) and the Yersinia pestis periplasmic heme transporter HmuT (50) (Table 4). The heme binding affinitiy of Rv0203-His and Rv0203-notag differ; whereby the respective Ka for Rv0203-His is 5.8 × 1011 M-1 (using the slow phase for k-heme) (Table 4). The higher affinity of Rv0203-His could be due to the second axial ligand donated by the His-tag representing a higher kinetic barrier for heme release or due to additional non-covalent interactions between the His-tag and the heme group.
The lower heme binding constant of Rv0203-notag compared to the known hemophore HasA (Table 4) suggests that Rv0203-notag is not able to acquire heme from hemoglobin based on an affinity gradient. Instead, its Ka falls into the range of several heme transport proteins such as the heme periplasmic trafficking proteins PhuS (49) and HmuT (50) which bind heme with affinities of 5.0 × 106 M-1 and 3.4 × 109 M-1, respectively, and BSA, an important heme-trafficking protein in the bloodstream, which binds free heme with a Ka of 4 × 109 M-1 (31). Taken together, the fast off-rate and the similarities in heme binding affinity with BSA and other heme transport proteins may indicate that Rv0203-notag functions as a heme transporter. We previously proposed that the transmembrane proteins, MmpL3 and MmpL11, which have been shown to play a role in heme uptake (22), are heme-receptors and may interact with heme bound Rv0203. Both proteins contain predicted extracellular domains, which bind heme and may facilitate heme transport across the mycobacterial membrane.
CONCLUSIONS
The results of this work suggest that the heme binding site of Rv0203 consists of Tyr59, His63 and His89. Furthermore, Tyr59 appears to play a key role in heme-iron ligation during in vitro heme reconstitution of both Rv0203-His and Rv0203-notag. The different heme coordination structures between holo-Rv0203-notag and holo-Rv0203-His raise the question of how the His-tag creates bis-His heme ligation when holo-Rv0203-notag is most likely oxygen-ligated. The Y59A-Rv0203-His mutant does not bind heme (22). Therefore, it is likely that heme binding in Rv0203-His is initiated through action of residue Tyr59 to form a 5-coordinate heme structure in a mechanism similar to heme binding in Rv0203-notag. Once the heme group is located in the heme binding pocket of Rv0203-His, the His-tag may bind to the open face of the heme-molecule, thus changing the structural and electronic properties of the heme group and inducing a trans-ligand switch from an oxygen to either His63 or His89. The observation that either His63 or His89 can bind heme in Rv0203-His and that both residues are important for full heme incorporation in Rv0203-notag leads to the hypothesis that these histidine residues could play a major role during heme transfer from a host hemoprotein or to the mycobacterial heme-receptor. Rv0203 is a mycobacterial-specific, secreted heme binding protein involved in mycobacterial heme uptake (22). This work offers the first insights into heme binding properties of Rv0203 and sets the stage for the development of inhibitors against this new pathway of heme-iron uptake in Mtb.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Masanori Sono and Shengfang Sun (University of South Carolina) and Pat Farmer (University of California, Irvine) for useful discussions. We would also like to thank Drs. Tom Poulos and Nick Chim for critical reading of the manuscript and Dr. Nick Chim, Guneet Singh and Evgeny Fadeev for assistance with the experiments. John Olsen kindly provided the clone to produce the H64Y/V68F-myoglobin mutant. We also thank the reviewers of this paper for their helpful and insightful comments.
This work has been supported by a grant from the National and Californian American Lung Association (RG-78755-N to C.W.G.) and the National Institutes of Health (AI081161 to C.W.G. and GM26730 to J.H.D.)
ABBREVIATIONS
- BME
beta-mercaptoethanol
- BSA
bovine serum albumin
- EPR
electron paramagnetic resonance
- Far-UV CD
far-ultra violet circular dichroism
- Fe-PPIXDME
Fe-protoporphyrin IX dimethylester
- hHb
human hemoglobin
- hHO
human heme oxygenase
- hMb
human myoglobin
- HRP
horse radish peroxidase
- ITC
isothermal titration calorimetry
- Mb
myoglobin
- MCD
magnetic circular dichroism
- NCBI
National Center for Biotechnology Information
- OD
Opitcal density
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- swMb
sperm whale myoglobin
Footnotes
SUPPORTING INFORMATION AVAILABLE. UV-vis absorption spectra of ferricyanide treated holo-Rv0203-notag and holo-Rv0203-His and the size exclusion chromotography trace for holo- and apo-Rv0203-notag. Ferrous holo-Rv0203-notag in a 1:1 complex with heme. Time courses of ferrous CO-heme binding to apo-Rv0203-notag with single exponential fitting results. Time resolved absoption specta of heme dissociation of ferric holo-Rv0203-notag and ferric holo-Rv0203-His. Experimental data and single exponential fitting the apo-myoglobin experiment for Rv0203-notag measured at 600 nm and the residual plots for both single and double exponential fitting of the apo-myoglobin experiment of Rv0203-notag and Rv0203-His. Ferric absorption spectra of holo-H63A-, H89A and WT-Rv0203-His. This information is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.World Health Organization 2011/2012 Tuberculosis Global Facts. 2011.
- 2.Gobin J, Horwitz MA. Exochelins of Mycobacterium tuberculosis remove iron from human iron-binding proteins and donate iron to mycobactins in the M. tuberculosis cell wall. J. Exp. Med. 1996;183:1527–1532. doi: 10.1084/jem.183.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ratledge C. Iron, mycobacteria and tuberculosis. Tuberculosis (Edinb) 2004;84:110–130. doi: 10.1016/j.tube.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Snow GA. Mycobactins: iron-chelating growth factors from mycobacteria. Bacteriol. Rev. 1970;34:99–125. doi: 10.1128/br.34.2.99-125.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez GM, Smith I. Identification of an ABC transporter required for iron acquisition and virulence in Mycobacterium tuberculosis. J. Bacteriol. 2006;188:424–430. doi: 10.1128/JB.188.2.424-430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodriguez GM. Control of iron metabolism in Mycobacterium tuberculosis. Trends Microbiol. 2006;14:320–327. doi: 10.1016/j.tim.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Johnson EE, Srikanth CV, Sandgren A, Harrington L, Trebicka E, Wang L, Borregaard N, Murray M, Cherayil BJ. Siderocalin inhibits the intracellular replication of Mycobacterium tuberculosis in macrophages. FEMS Immunol. Med. Microbiol. 2010;58:138–145. doi: 10.1111/j.1574-695X.2009.00622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626–1628. doi: 10.1126/science.1099930. [DOI] [PubMed] [Google Scholar]
- 9.Hagan EC, Mobley HL. Haem acquisition is facilitated by a novel receptor Hma and required by uropathogenic Escherichia coli for kidney infection. Mol. Microbiol. 2009;71:79–91. doi: 10.1111/j.1365-2958.2008.06509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mazmanian SK, Skaar EP, Gaspar AH, Humayun M, Gornicki P, Jelenska J, Joachmiak A, Missiakas DM, Schneewind O. Passage of heme-iron across the envelope of Staphylococcus aureus. Science. 2003;299:906–909. doi: 10.1126/science.1081147. [DOI] [PubMed] [Google Scholar]
- 11.Zhu H, Xie G, Liu M, Olson JS, Fabian M, Dooley DM, Lei B. Pathway for heme uptake from human methemoglobin by the iron-regulated surface determinants system of Staphylococcus aureus. J. Biol. Chem. 2008;283:18450–18460. doi: 10.1074/jbc.M801466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grigg JC, Ukpabi G, Gaudin CF, Murphy ME. Structural biology of heme binding in the Staphylococcus aureus Isd system. J. Inorg. Biochem. 2010;104:341–348. doi: 10.1016/j.jinorgbio.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 13.Honsa ES, Maresso AW. Mechanisms of iron import in anthrax. Biometals. 2011;24:533–545. doi: 10.1007/s10534-011-9413-x. [DOI] [PubMed] [Google Scholar]
- 14.Izadi N, Henry Y, Haladjian J, Goldberg ME, Wandersman C, Delepierre M, Lecroisey A. Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry. 1997;36:7050–7057. doi: 10.1021/bi962577s. [DOI] [PubMed] [Google Scholar]
- 15.Yukl ET, Jepkorir G, Alontaga AY, Pautsch L, Rodriguez JC, Rivera M, Moenne-Loccoz P. Kinetic and Spectroscopic Studies of Hemin Acquisition in the Hemophore HasAp from Pseudomonas aeruginosa. Biochemistry. 2010;49:6646–6654. doi: 10.1021/bi100692f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cescau S, Cwerman H, Letoffe S, Delepelaire P, Wandersman C, Biville F. Heme acquisition by hemophores. Biometals. 2007;20:603–613. doi: 10.1007/s10534-006-9050-y. [DOI] [PubMed] [Google Scholar]
- 17.Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu. Rev. Microbiol. 2004;58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 18.Lewis JP, Dawson JA, Hannis JC, Muddiman D, Macrina FL. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 1999;181:4905–4913. doi: 10.1128/jb.181.16.4905-4913.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Otto BR, van Dooren SJ, Nuijens JH, Luirink J, Oudega B. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J. Exp. Med. 1998;188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yongqing T, Potempa J, Pike RN, Wijeyewickrema LC. The lysine-specific gingipain of Porphyromonas gingivalis : importance to pathogenicity and potential strategies for inhibition. Adv. Exp. Med. Biol. 2011;712:15–29. doi: 10.1007/978-1-4419-8414-2_2. [DOI] [PubMed] [Google Scholar]
- 21.Tullius MV, Harth G, Maslesa-Galic S, Dillon BJ, Horwitz MA. A Replication-Limited Recombinant Mycobacterium bovis BCG vaccine against tuberculosis designed for human immunodeficiency virus-positive persons is safer and more efficacious than BCG. Infect. Immun. 2008;76:5200–5214. doi: 10.1128/IAI.00434-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tullius MV, Harmston CA, Owens CP, Chim N, Morse RP, McMath LM, Iniguez A, Kimmey JM, Sawaya MR, Whitelegge JP, Horwitz MA, Goulding CW. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5051–5056. doi: 10.1073/pnas.1009516108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones CM, Niederweis M. Mycobacterium tuberculosis can utilize heme as an iron source. J. Bacteriol. 2011;193:1767–1770. doi: 10.1128/JB.01312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnoux P, Haser R, Izadi N, Lecroisey A, Delepierre M, Wandersman C, Czjzek M. The crystal structure of HasA, a hemophore secreted by Serratia marcescens. Nat. Struct. Biol. 1999;6:516–520. doi: 10.1038/9281. [DOI] [PubMed] [Google Scholar]
- 25.Deniau C, Gilli R, Izadi-Pruneyre N, Létoffé S, Delepierre M, Wandersman C, Briand C, Lecroisey A. Thermodynamics of Heme Binding to the HasASM Hemophore: Effect of Mutations at Three Key Residues for Heme Uptake. Biochemistry. 2003;42:10627–10633. doi: 10.1021/bi030015k. [DOI] [PubMed] [Google Scholar]
- 26.Goulding CW, Perry LJ. Protein production in Escherichia coli for structural studies by X-ray crystallography. J. Struct. Biol. 2003;142:133–143. doi: 10.1016/s1047-8477(03)00044-3. [DOI] [PubMed] [Google Scholar]
- 27.Dawson JH, Elliot DC, Elliot WH, Jones KM. Data for Biochemical Research. Oxford University Press; Oxford, England: 1986. [Google Scholar]
- 28.Bartsch RG. Cytochromes: Bacterial. Methods in Enzymology. 1971;23:344–363. [Google Scholar]
- 29.Huff AM, Chang C, Cooper D, Smith K, Dawson JH. Imidazole- and alkylamine-ligated iron(II,III) chlorin complexes as models for histidine and lysine coordination to iron in dihydroporphyrin-containing proteins: characterization with magnetic circular dichroism spectroscopy. Inorg. Chem. 1993;32:1460–1466. [Google Scholar]
- 30.Hargrove MS, Singleton EW, Quillin ML, Ortiz LA, Phillips GN, Jr., Olson JS, Mathews AJ. His64(E7)-->Tyr apomyoglobin as a reagent for measuring rates of hemin dissociation. J. Biol. Chem. 1994;269:4207–4214. doi: 10.2210/pdb1mgn/pdb. [DOI] [PubMed] [Google Scholar]
- 31.Hargrove MS, Barrick D, Olson JS. The Association Rate Constant for Heme Binding to Globin Is Independent of Protein Structure. Biochemistry. 1996;35:11293–11299. doi: 10.1021/bi960371l. [DOI] [PubMed] [Google Scholar]
- 32.Cheek J, Mandelman D, Poulos TL, Dawson JH. A study of the K(+)-site mutant of ascorbate peroxidase: mutations of protein residues on the proximal side of the heme cause changes in iron ligation on the distal side. J. Biol. Inorg. Chem. 1999;4:64–72. doi: 10.1007/s007750050290. [DOI] [PubMed] [Google Scholar]
- 33.Uno T, Ryu D, Tsutsumi H, Tomisugi Y, Ishikawa Y, Wilkinson AJ, Sato H, Hayashi T. Residues in the distal heme pocket of neuroglobin. Implications for the multiple ligand binding steps. J. Biol. Chem. 2004;279:5886–5893. doi: 10.1074/jbc.M311748200. [DOI] [PubMed] [Google Scholar]
- 34.Caillet-Saguy C, Turano P, Piccioli M, Lukat-Rodgers GS, Czjzek M, Guigliarelli B, Izadi-Pruneyre N, Rodgers KR, Delepierre M, Lecroisey A. Deciphering the structural role of histidine 83 for heme binding in hemophore HasA. J. Biol. Chem. 2008;283:5960–5970. doi: 10.1074/jbc.M703795200. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Moenne-Loccoz P, Hildebrand DP, Wilks A, Loehr TM, Mauk AG, Ortiz de Montellano PR. Replacement of the Proximal Histidine Iron Ligand by a Cysteine or Tyrosine Converts Heme Oxygenase to an Oxidase. Biochemistry. 1999;38:3733–3743. doi: 10.1021/bi982707s. [DOI] [PubMed] [Google Scholar]
- 36.Ghosh K, Thompson AM, Goldbeck RA, Shi X, Whitman S, Oh E, Zhiwu Z, Vulpe C, Holman TR. Spectroscopic and Biochemical Characterization of Heme Binding to Yeast Dap1p and Mouse PGRMC1p. Biochemistry. 2005;44:16729–16736. doi: 10.1021/bi0511585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Egeberg KD, Springer BA, Martinis SA, Sligar SG, Morikis D, Champion PM. Alteration of sperm whale myoglobin heme axial ligation by site-directed mutagenesis. Biochemistry. 1990;29:9783–9791. doi: 10.1021/bi00494a004. [DOI] [PubMed] [Google Scholar]
- 38.Pond AE, Roach MP, Sono M, Rux AH, Franzen S, Hu R, Thomas MR, Wilks A, Dou Y, Ikeda-Saito M, Ortiz de Montellano PR, Woodruff WH, Boxer SG, Dawson JH. Assignment of the Heme Axial Ligand(s) for the Ferric Myoglobin (H93G) and Heme Oxygenase (H25A) Cavity Mutants as Oxygen Donors Using Magnetic Circular Dichroism. Biochemistry. 1999;38:7601–7608. doi: 10.1021/bi9825448. [DOI] [PubMed] [Google Scholar]
- 39.Qin J, Perera R, Lovelace LL, Dawson JH, Lebioda L. Structures of Thiolate- and Carboxylate-Ligated Ferric H93G Myoglobin: Models for Cytochrome P450 and for Oxyanion-Bound Heme Proteins. Biochemistry. 2006;45:3170–3177. doi: 10.1021/bi052171s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pond AE, Roach MP, Thomas MR, Boxer SG, Dawson JH. The H93G myoglobin cavity mutant as a versatile template for modeling heme proteins: ferrous, ferric, and ferryl mixed-ligand complexes with imidazole in the cavity. Inorg. Chem. 2000;39:6061–6066. doi: 10.1021/ic0007198. [DOI] [PubMed] [Google Scholar]
- 41.Tang Q, Kalsbeck WA, Olson JS, Bocian DF. Disruption of the heme iron-proximal histidine bond requires unfolding of deoxymyoglobin. Biochemistry. 1998;37:7047–7056. doi: 10.1021/bi9729413. [DOI] [PubMed] [Google Scholar]
- 42.Antonini E, Brunori M. Hemoglobin and Myoglobin in their Reactions with Ligands. North-Holland Publ.; Amsterdam: 1971. [Google Scholar]
- 43.Lukat-Rodgers GS, Rodgers KR, Caillet-Saguy C, Izadi-Pruneyre N, Lecroisey A. Novel Heme Ligand Displacement by CO in the Soluble Hemophore HasA and Its Proximal Ligand Mutants: Implications for Heme Uptake and Release. Biochemistry. 2008;47:2087–2098. doi: 10.1021/bi7019518. [DOI] [PubMed] [Google Scholar]
- 44.Kawamura-Konishi Y, Kihara H, Suzuki H. Reconstitution of myoglobin from apoprotein and heme, monitored by stopped-flow absorption, fluorescence and circular dichroism. Eur. J. Biochem. 1988;170:589–595. doi: 10.1111/j.1432-1033.1988.tb13738.x. [DOI] [PubMed] [Google Scholar]
- 45.Honsa ES, Fabian M, Cardenas AM, Olson JS, Maresso AW. The five near-iron transporter (NEAT) domain anthrax hemophore, IsdX2, scavenges heme from hemoglobin and transfers heme to the surface protein IsdC. J. Biol. Chem. 2011;286:33652–33660. doi: 10.1074/jbc.M111.241687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu M, Tanaka WN, Zhu H, Xie G, Dooley DM, Lei B. Direct hemin transfer from IsdA to IsdC in the iron-regulated surface determinant (Isd) heme acquisition system of Staphylococcus aureus. J. Biol. Chem. 2008;283:6668–6676. doi: 10.1074/jbc.M708372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gaudin CF, Grigg JC, Arrieta AL, Murphy ME. Unique Heme-Iron Coordination by the Hemoglobin Receptor IsdB of Staphylococcus aureus. Biochemistry. 2011;50:5443–5452. doi: 10.1021/bi200369p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nygaard TK, Blouin GC, Liu M, Fukumura M, Olson JS, Fabian M, Dooley DM, Lei B. The mechanism of direct heme transfer from the streptococcal cell surface protein Shp to HtsA of the HtsABC transporter. J. Biol. Chem. 2006;281:20761–20771. doi: 10.1074/jbc.M601832200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhakta MN, Wilks A. The mechanism of heme transfer from the cytoplasmic heme binding protein PhuS to the delta-regioselective heme oxygenase of Pseudomonas aeruginosa. Biochemistry. 2006;45:11642–11649. doi: 10.1021/bi060980l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mattle D, Zeltina A, Woo JS, Goetz BA, Locher KP. Two stacked heme molecules in the binding pocket of the periplasmic heme-binding protein HmuT from Yersinia pestis. J. Mol. Biol. 2010;404:220–231. doi: 10.1016/j.jmb.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Arnesano F, Banci L, Barker PD, Bertini I, Rosato A, Su XC, Viezzoli MS. Solution structure and characterization of the heme chaperone CcmE. Biochemistry. 2002;41:13587–13594. doi: 10.1021/bi026362w. [DOI] [PubMed] [Google Scholar]
- 52.Enggist E, Thony-Meyer L, Guntert P, Pervushin K. NMR structure of the heme chaperone CcmE reveals a novel functional motif. Structure. 2002;10:1551–1557. doi: 10.1016/s0969-2126(02)00885-7. [DOI] [PubMed] [Google Scholar]
- 53.Daltrop O, Stevens JM, Higham CW, Ferguson SJ. The CcmE protein of the c-type cytochrome biogenesis system: unusual in vitro heme incorporation into apo-CcmE and transfer from holo-CcmE to apocytochrome. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9703–9708. doi: 10.1073/pnas.152120699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stevens JM, Daltrop O, Higham CW, Ferguson SJ. Interaction of heme with variants of the heme chaperone CcmE carrying active site mutations and a cleavable N-terminal His tag. J. Biol. Chem. 2003;278:20500–20506. doi: 10.1074/jbc.M212925200. [DOI] [PubMed] [Google Scholar]
- 55.Ho WW, Li H, Eakanunkul S, Tong Y, Wilks A, Guo M, Poulos TL. Holo- and apo-bound structures of bacterial periplasmic heme-binding proteins. J. Biol. Chem. 2007;282:35796–35802. doi: 10.1074/jbc.M706761200. [DOI] [PubMed] [Google Scholar]
- 56.Grigg JC, Vermeiren CL, Heinrichs DE, Murphy ME. Haem recognition by a Staphylococcus aureus NEAT domain. Mol. Microbiol. 2007;63:139–149. doi: 10.1111/j.1365-2958.2006.05502.x. [DOI] [PubMed] [Google Scholar]
- 57.Sharp KH, Schneider S, Cockayne A, Paoli M. Crystal structure of the heme-IsdC complex, the central conduit of the Isd iron/heme uptake system in Staphylococcus aureus. J. Biol. Chem. 2007;282:10625–10631. doi: 10.1074/jbc.M700234200. [DOI] [PubMed] [Google Scholar]
- 58.Ikeda-Saito M, Hori H, Andersson LA, Prince RC, Pickering IJ, George GN, Sanders CR, Lutz RS, McKelvey EJ, Mattera R. Coordination structure of the ferric heme iron in engineered distal histidine myoglobin mutants. J. Biol. Chem. 1992;267:22843–22852. [PubMed] [Google Scholar]
- 59.Block DR, Lukat-Rodgers GS, Rodgers KR, Wilks A, Bhakta MN, Lansky IB. Identification of two heme-binding sites in the cytoplasmic heme-trafficking protein PhuS from Pseudomonas aeruginosa and their relevance to function. Biochemistry. 2007;46:14391–14402. doi: 10.1021/bi701509n. [DOI] [PubMed] [Google Scholar]
- 60.Couture M, Das TK, Lee HC, Peisach J, Rousseau DL, Wittenberg BA, Wittenberg JB, Guertin M. Chlamydomonas chloroplast ferrous hemoglobin. Heme pocket structure and reactions with ligands. J. Biol. Chem. 1999;274:6898–6910. doi: 10.1074/jbc.274.11.6898. [DOI] [PubMed] [Google Scholar]
- 61.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.