Abstract
DNAzymes are catalytically active DNA molecules that use metal cofactors for their enzymatic functions. While a growing number of DNAzymes with diverse functions and metal selectivities have been reported, the relationships between metal ion selectivity, conserved sequences and structures responsible for selectivity remain to be elucidated. To address this issue, we report biochemical assays of a family of previously reported in vitro selected DNAzymes. This family includes the clone 11 DNAzyme, which was isolated by positive and negative selection, and the clone 18 DNAzyme, which was isolated by positive selection alone. The clone 11 DNAzyme has a higher selectivity for Co2+ over Pb2+ compared with clone 18. The reasons for this difference are explored here through phylogenetic comparison, mutational analysis and stepwise truncation. A novel DNAzyme truncation method incorporated a nick in the middle of the DNAzyme to allow for truncation close to the nicked site while preserving peripheral sequences at both ends of the DNAzyme. The results demonstrate that peripheral sequences within the substrate binding arms, most notably the stem loop, loop II, are sufficient to restore its selectivity for Co2+ over Pb2+ to levels observed in clone 11. A comparison of these sequences’ secondary structures and Co2+ selectivities suggested that metastable structures affect metal ion selectivity. The Co2+ selectivity of the clone 11 DNAzyme showed that the metal ion binding and selectivities of small, in vitro selected DNAzymes may be more complex than previously appreciated, and that clone 11 may be more similar to larger ribozymes than to other small DNAzymes in its structural complexity and behavior. These factors should be taken into account when metal-ion selectivity is required in rationally designed DNAzymes and DNAzyme-based biosensors.
Keywords: biotechnology, cobalt, DNAzymes, metal ions, metastable structure, nucleic acids
Introduction
The discovery of the first catalytic DNA molecule (also called a DNAzyme or deoxyribozyme) settled a fundamental question: whether DNA, from a four-letter alphabet of building blocks lacking the 2’-hydroxyl present in RNA, could form sufficiently complex secondary and tertiary structures to achieve DNA catalysis. Since 1994, when the first DNAzyme was isolated through in vitro selection,[1] many DNAzymes with significant rate enhancements over uncatalyzed reactions, high substrate selectivities, and diverse catalytic functions have been isolated; they have also been shown to have potential as pharmaceutical drugs, sensors, and logic gate mathematical regulators.[2] In contrast to the significant advances in isolating and applying DNAzymes, the understanding of the structure–function relationships of these DNAzymes is progressing much more slowly. Understanding these relationships will enrich our knowledge of chemical biology and nucleic acid biochemistry, and will, in turn, produce more customizable DNAzymes for practical applications.
DNAzymes have been shown to selectively recruit metal ions to perform diverse functions similar to those performed by protein and RNA enzymes. For example, the 8–17 RNA-cleaving DNAzyme[3–5] is more than 100 times more selective for Pb2+ than for any other metal ion. In addition, a number of DNAzymes with high selectivities for Co2+,[6, 7] Cu2+,[8] Hg2+,[9, 10] Mg2+,[4] Mn2+,[7] Pb2+,[1] Zn2+,[5,11] and porphyrins[12] have been reported. Finally, a recently selected DNAzyme has selectivity for UO22+ that is a million-fold higher than for other metal ions.[13, 14] These high metal ion selectivities have established DNAzymes as a new class of efficient metal ion sensors,[9, 13, 15] with detection limits as low as 45 pm or 11 ppt.[13]
In contrast to metalloprotein enzymes, and to a certain degree even ribozymes,[16] relatively little is known about the factors that determine DNAzymes’ metal-ion selectivities. This is because no three-dimensional structure of a DNAzyme in an active conformation has yet been reported. To address this issue, studies on the interactions between DNA and inorganic metal complexes,[17] and between nucleotides and metal ions[18] have provided insights into ligand preferences and ligand geometries in larger nucleic acid strands. Metal ions are essential for the folding and optimal activity of almost all reported DNAzymes, and divalent metal ions have been implicated as direct participants in catalysis. Metal ions have also been reported to affect the tertiary structures and mechanisms of DNAzymes and ribozymes.[19–21] An improved understanding of the process by which metalloenzymes selectively bind metal ions will be invaluable to engineer DNAzymes with high activity and selectivity for use as biosensors.
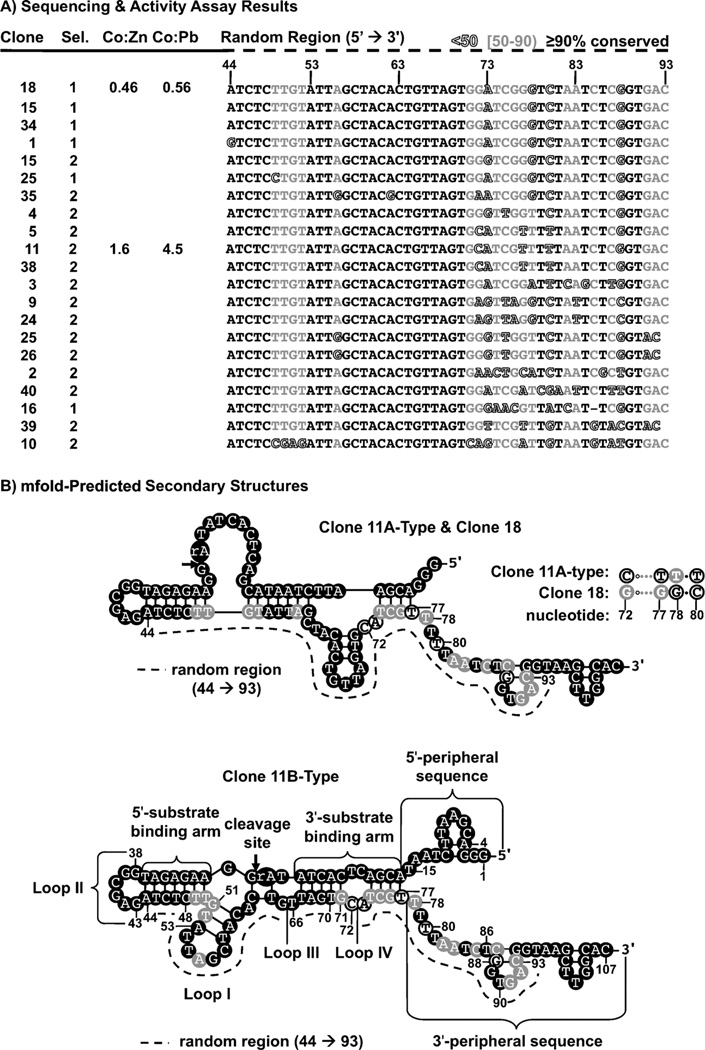
A primary example of the challenge in understanding metal ion selectivity is finding metalloenzymes that can differentiate Co2+ from Zn2+. Not only do these metal ions have identical charges, they also have nearly identical ionic radii and ligand donor set preferences.[22] Therefore, designing a molecule with high selectivity for Co2+ over Zn2+ is very difficult. In fact, although metalloproteins are known to bind metal ions with high selectivity, most Zn2+-binding proteins can bind Co2+ with almost 100% activity. The problem of selectivity is further compounded for RNA-cleaving DNAzymes because of the background hydrolytic activity of Zn2+ and Co2+.[23] To find molecules that differentiate between these two metal ions, we previously performed in vitro selection centered on phosphodiester cleavage. We used a negative-selection approach to obtain DNAzymes more selective for Co2+ over Zn2+ and Pb2+.[6] Two alternative selection methods were carried out to isolate Co2+-selective sequences. Selection 1 resulted in a DNAzyme population that was active in the presence of Co2+, but was also active in the presence of Zn2+ and Pb2+. To address this limitation, selection 2 incorporated several rounds of negative selection to increase the Co2+ selectivity by removing DNAzymes active in the presence of Zn2+ and Pb2+. This negative selection approach produced a population with increased selectivity for Co2+ over both Zn2+ and Pb2+. Several of the sequences obtained in these selections are shown in Figure 1A. The clone 11 DNAzyme, isolated during selection 2, was the most selective for Co2+ (Co:Zn 1.6, Co:Pb 4.5) and had the highest activity (kobs 0.18 min−1) of all of the DNAzymes isolated by either selection method. Clone 11’s sequence was similar to that of clone 18, a DNAzyme isolated by Selection 1. By using mfold to predict the secondary structures,[24] clone 11 was found to have two putative secondary structures: 11A and 11B (Figure 1B). While the secondary structures of 11A and clone 18 were identical, clone 18 had poor Co2+ selectivity and only moderate activity (kobs 0.044 min−1). Interestingly, a comparison of clones 11 and 18 showed that only four different nucleotides, C72, T77, T78 and T80 (Figure 1B), decreased the Co2+ selectivity nearly eightfold (Table 1). It was hypothesized, then, that clone 11’s enhanced selectivity and activity resulted from the 11B secondary structure.
Figure 1.
The artificial phylogenetic analysis and secondary structures of the clone 11 and clone 18 DNAzymes. A) The aligned sequences from selections 1 and 2 that were used for artificial phylogenetic analysis. Sequences are arranged according to similarity. Shading indicates the degree of nucleotide conservation: positions that were highly (>90%) conserved are shown in black, positions that were moderately (50–90%) conserved are shown in grey, and nonconserved (<50%) positions are outlined. Positions 71–83 were highly variable. Nucleotide positions are based on the full-length clone 11 sequence. B) The sequence variability of each sequence overlaid with their mfold-predicted secondary structures. Cleavage sites are indicated by arrows. Two secondary structures were predicted for clone 11: the 11A-type and the 11B-type secondary structure. Clone 18 differs from clone 11 by only four nucleotides (72, 77, 78 and 80), and the single secondary structure predicted for clone 18 is identical to that of the 11A-type secondary structure.
Table 1.
The kinetic data and Co2+ selectivity of clones 11 and 18 and the 11B trans-cleaving DNAzymes. Kinetic data was obtained at pH 7.0 with metal ions (500 µm) and NaCl (500 mm).
| Construct | kobs [min−1] | Co:Zn | Co:Pb | ||
|---|---|---|---|---|---|
| Co2+ | Zn2+ | Pb2+ | |||
| Clone 11 | 0.18 | 0.11 | 0.040 | 1.6 | 4.5 |
| Clone 18 | 0.044 | 0.096 | 0.078 | 0.46 | 0.56 |
| 11B | 0.088 | 0.15 | 0.079 | 0.59 | 1.1 |
To elucidate the relationship between Co2+ selectivity and DNAzyme sequence and structure in these systems, phylogenetic comparison, mutational analysis and stepwise truncation were performed. We found that peripheral sequences elements enhanced the activity and Co2+ selectivity of clone 11, and that metastable structures might also play a role.
Results
Artificial phylogenetic analysis
To trace selectivity differences between clone 11 and clone 18 DNAzymes, artificial phylogenetic analysis was performed for the sequences obtained from selections 1 and 2.[6] We sought to test the premise that sequences showing similar metal selectivities would also show similar structural characteristics. A similar approach had been used in the early RNA secondary-structure predictions (phylogenetic analysis and the hypothesis that RNAs with similar functions but different species of origin have similar structures).[25]
The artificial phylogenetic analysis used six sequences from selection 1 and 15 sequences from selection 2 (Figure 1A). These sequences were chosen based on the similarities between clone 11 and clone 18. Within the region randomized for selection, a highly conserved region from positions 53 to 70 was apparent, while nucleotides 71–86 showed considerable variability. By superimposing the sequence alignment in Figure 1A onto the active secondary structure of the Co2+-selective Clone 11 DNAzyme, we showed that the highly conserved region between 53 to 70 helps form the 5’ substrate binding arm, stem loop I, and the 3’ substrate-binding arm (Figure 1 B). The more variable region (positions 71–86) coincides with the corresponding terminal 3’ substrate-binding arm that contains loop IV and the adjacent 3’-single-stranded region. Surprisingly, the four positions that distinguish clone 11 from 18 (positions 72, 77, 78, and 80) show the highest variability. The variability at these positions was unexpected, because sequence or structural motifs that increase selectivity were expected to be highly conserved. The sequence variability at positions 72 and 73 suggests that loop IV helps increase Co2+ selectivity. The short helical region distal to loop IV contains T77, which may also play a role in Co2+ selectivity. While T78 and T80 are part of a region predicted to be unstructured, they may participate in tertiary interactions affecting Co2+ selectivity.
Stepwise truncation of peripheral sequence elements
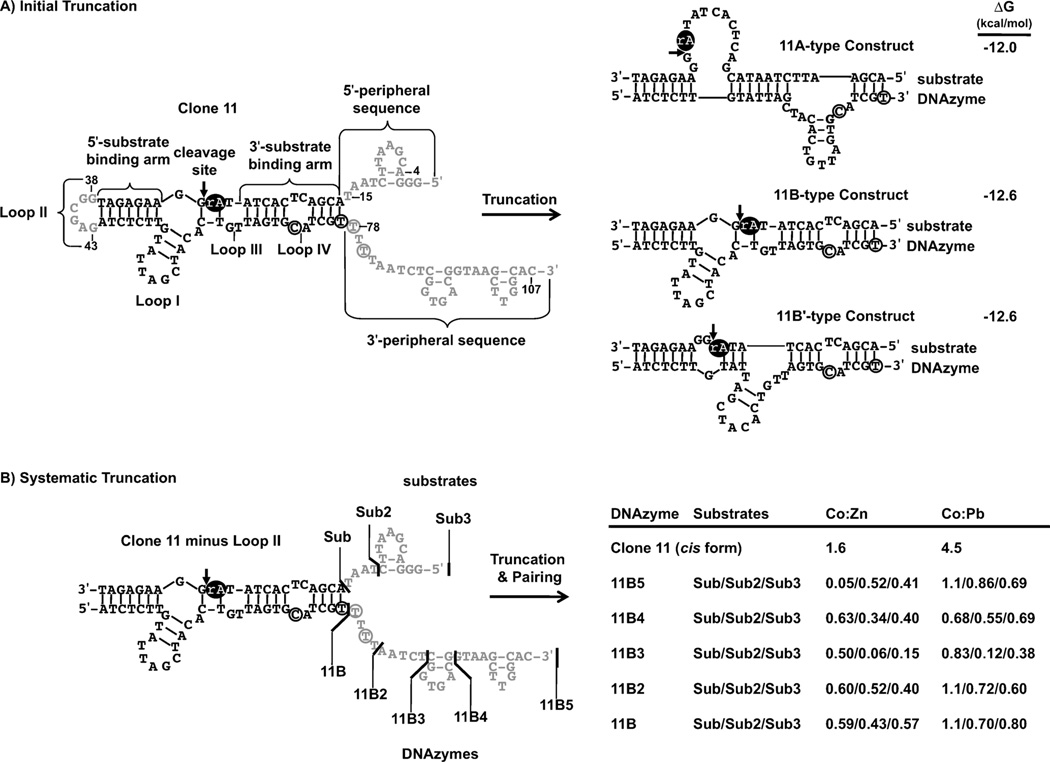
Next, the effect of peripheral sequences on Co2+ selectivity was investigated by truncating the clone 11 DNAzyme. After in vitro selection, constructs have been routinely truncated to contain only mfold-predicted secondary structures of interest. The cis-cleaving clone 11 DNAzyme was truncated to the 11B trans-cleaving construct (11B-trans) based on the 11B-type active secondary structure (Figure 2A). Interestingly, truncation of the 5’ and 3’ peripheral sequences resulted in another predicted secondary structure (11B’, Figure 2A) that resembled neither that of clone 11 nor that of clone 18. 11B’ retained the base pairing within both substrate binding arms but had disruptions at stem loop I and at the base pairs at the cleavage site. Co:Pb selectivity of the truncated 11B trans construct was four times less than that of clone 11 (Table 1), and its Co:Zn selectivity was three times lower (Table 1). The distal 5’ and 3’ sequence elements (nucleotides 1–15 and 78–107) that are adjacent to the 3’ substrate binding arm can be thought of as “peripheral”, but might also be integral to selectivity: the decrease in Co2+ selectivity might have been due to the deletion of portions of these regions.
Figure 2.
Truncation of the clone 11 cis-cleaving DNAzyme. The portions of clone 11 that were removed are shown in grey and the ribonucleotide is shown as a black oval. A) The clone 11 cis-cleaving DNAzyme was initially truncated to the 11B-trans cleaving DNAzyme (11B-trans) by deleting loop II and all peripheral sequences. Mfold predicted three different secondary structures for 11B-trans: 11A-type, 11B-type, and 11B’ (shown here with corresponding ΔG values). The 11B and 11B’ structures differ only at positions 51–67, and 11B’ has a single-stranded region immediately following the cleavage site. Only 5–6 base pairs differ between 11B and 11B’. B) The clone 11 cis-cleaving DNAzyme was then modified by deleting loop II and systematically truncating peripheral sequences by small increments. This produced three substrate variants (Sub3, Sub2 and Sub) and five DNAzyme variants (11B5, 11B4, 11B3, 11B2 and 11B). These sequences were paired in every possible permutation and their metal-dependent activities were measured. Co:Zn and Co:Pb selectivity indices for each permutation are reported for the results using Co2+, Zn2+, and Pb2+ (500 µm). The full results are listed in Table 2.
As the large-scale truncation described above decreased the Co2+ selectivity, a systematic truncation approach was adopted to find which regions were responsible for the decreased selectivity. Short peripheral sequence elements were systematically deleted from the enzyme and substrate strands of clone 11 (Figure 2 B). This produced a series of trans-cleaving enzyme and substrate strands of various lengths, and every possible combination of enzyme and substrate strand was constructed and assayed for Co2+ selectivity. All truncated constructs were most active in the presence of Zn2+ and were less selective for Co2+ than clone 11 (Table 2). Of the Co2+, Zn2+, and Pb2+ selectivities, the Co2+-dependent activity typically showed the greatest decrease: Co:Zn selectivities decreased as much as 32-fold and Co:Pb selectivities decreased nearly 40-fold. These changes in Co2+ selectivity were surprising for the sequences that retained C72, T77, T78, and T80 (the four nucleotides that distinguish clone 11 and clone 18). Deleting only Loop II from clone 11 produced a full-length, trans-cleaving clone 11 variant that also had reduced Co2+ selectivity. There are several possible explanations for this. First, the mfold-predicted structures could be incorrect. However, the introduction of point mutations (data not shown) and the activity of the trans-cleaving constructs designed by mfold support the mfold-derived clone 11 structures. Another possibility is that interstrand secondary structure forms between the enzyme and substrates of the Loop II deletion constructs. Alternatively, the trans-cleaving configuration might inhibit formation of a selective Co2+ binding pocket, as clone 11 was selected as a cis-cleaving DNAzyme. The results of the stepwise truncation prompted alternative truncation strategies, to investigate the role that loop II and other peripheral sequences play in determining the Co2+ selectivity of clone 11.
Table 2.
The Co2+ selectivity over Zn2+ and Pb2+ is shown for each step-wise truncation of clone 11. Pseudo-first-order rate constants for substrate cleavage by truncated Clone 11 constructs were determined. The enzyme strand is denoted by “11B” followed by the substrate strand. All permutations of enzyme and substrate strands were assayed. Note that 11B/Sub is the 11B-trans construct. All data were obtained from assays with Co2+, Zn2+ or Pb2+ (500 µm).
| Construct | kobs [min−1] | Co:Zn | Co:Pb | ||
|---|---|---|---|---|---|
| Co2+ | Zn2+ | Pb2+ | |||
| 11B5/Sub3 | 0.045 | 0.11 | 0.065 | 0.41 | 0.69 |
| 11B5/Sub2 | 0.057 | 0.11 | 0.066 | 0.52 | 0.86 |
| 11B5/Sub | 0.025 | 0.53 | 0.022 | 0.05 | 1.1 |
| 11B4/Sub3 | 0.043 | 0.10 | 0.062 | 0.40 | 0.69 |
| 11B4/Sub2 | 0.048 | 0.14 | 0.087 | 0.34 | 0.55 |
| 11B4/Sub | 0.075 | 0.12 | 0.11 | 0.63 | 0.68 |
| 11B3/Sub3 | 0.015 | 0.099 | 0.039 | 0.15 | 0.38 |
| 11B3/Sub2 | 0.014 | 0.24 | 0.12 | 0.06 | 0.12 |
| 11B3/Sub | 0.070 | 0.14 | 0.084 | 0.50 | 0.83 |
| 11B2/Sub3 | 0.018 | 0.045 | 0.030 | 0.40 | 0.60 |
| 11B2/Sub2 | 0.094 | 0.18 | 0.13 | 0.52 | 0.72 |
| 11B2/Sub | 0.12 | 0.20 | 0.11 | 0.60 | 1.1 |
| 11B/Sub3 | 0.008 | 0.014 | 0.010 | 0.57 | 0.80 |
| 11B/Sub2 | 0.069 | 0.16 | 0.099 | 0.43 | 0.70 |
| 11B/Sub | 0.088 | 0.15 | 0.079 | 0.59 | 1.1 |
Alternative truncation strategies
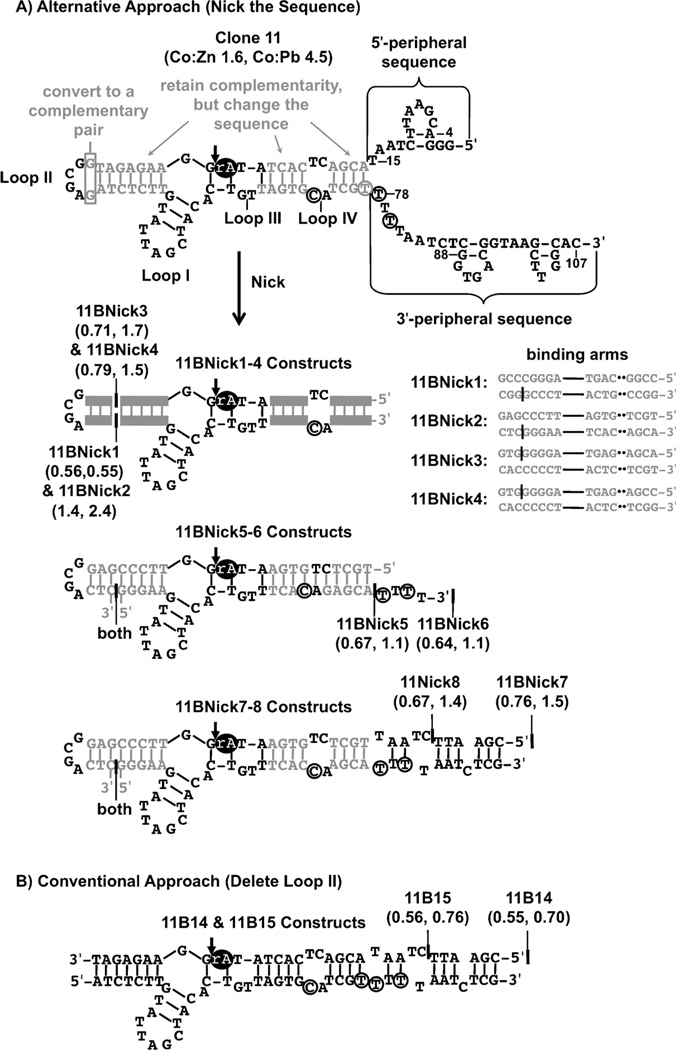
Investigating the influence of peripheral sequences on Co2+ selectivity required sequences with loop II intact; therefore, alternatives to conventional DNAzyme truncation strategies[4, 5, 26] were developed. Cis-cleaving constructs with peripheral sequence truncations had poor Co2+ selectivities and were difficult to synthesize because of their length (>100 nucleotides) and embedded ribonucleotide. Instead, novel trans-cleaving constructs (Figure 3A) were designed to provide a platform preserving loop II while testing the peripheral sequence elements’ contributions to Co2+ selectivity. This platform placed a nick in the 5’-substrate binding arm and incorporated loop II as a terminal stem loop on either the enzyme or substrate strand. As shown in Figure 3A, “nicked” constructs were designed on the basis of mfold analysis, to choose sequences that 1) preserved the 11B-type secondary structure of clone 11, 2) retained the primary sequence of the conserved loop 1 region (nucleotides 51–66), and 3) retained substrate binding arm base-pairing. Criterion 2 was based on the results of the artificial phylogenetic analysis. In addition, preliminary mutational analysis showed that helical regions of the substrate binding arms were mutation tolerant and that loops I and III were mutation intolerant (data not shown). Mutations in the substrate binding arms preserved substrate recognition in the 5’-substrate binding arm and reduced the self-complementarity of the enzyme and substrate strands. During the design process, the 5’-substrate binding arm was also lengthened by one base pair, converting the six-base loop II into a tetraloop. The additional base pair and tetraloop allowed the helical region of the terminal stem loop to participate in substrate recognition in the 5’-substrate binding arm. The sequences of constructs designed for the truncation study are shown in Figure 3A.
Figure 3.
Truncation of the clone 11 cis-cleaving DNAzyme by conventional and alternative truncation approaches. The portions of clone 11 that were changed during truncation are shown in grey. The ribonucleotide is shown as a black oval. A) An alternative truncation approach to the conventional method (shown in Figure 2B) preserved the secondary structure of 11B, retained many conserved nucleotides from clone 11, and preserved the substrate binding arm base pairs. The nucleotides within clone 11’s binding arms were modified, but its base pairing interactions were preserved. Then the 5’ substrate-binding arm was nicked at one of two positions. Thus the two resulting truncated constructs retained loop II. In order to preserve the base pairing in the 5’-substrate binding arm, two nucleotides in loop II were base-paired, converting loop II to a tetraloop. The peripheral sequences were then systematically truncated. In 11BNick5 and 11BNick6, two additional point mutations were introduced in loop IV to increase the substrate binding arm complementarity. The mfold-predicted secondary structures of each of the constructs obtained by the alternative approach (11BNick1–8) are shown. 11BNick1–4 have secondary structures that differ only in the substrate binding arm sequences, as shown. Their Co:Zn and Co:Pb selectivity indices are shown in parentheses. B) The truncation of clone 11 by the conventional approach described in Figure 2B produced two additional constructs: 11B14 and 11B15. Modifications included 1) deleting loop II and 2) systematically truncating the peripheral sequences.
Constructs 11BNick1–4, which were generated by this method, differ only in their substrate binding arms; they contain no peripheral sequences and are predicted to form the 11B-type secondary structure. In addition, 11BNick1, 11BNick2, and 11BNick 4 were predicted to form the 11B’-type structure. Two additional constructs, 11BNick5 and 11BNick6, were designed to incorporate base pairs in place of loop IV. 11BNick6 also has peripheral sequences (nucleotides 77–81) appended. Each nicked construct containing loop II was tested for Co2+ selectivity (Table 3). 11BNick1 (Co:Zn 0.56±0.15, Co:Pb 0.55±0.16), 11BNick5 (Co:Zn 0.67±0.11, Co:Pb 1.1±0.3), and 11BNick6 (Co:Zn 0.64±0.03, Co:Pb 1.1±0.3) showed no improvement in Co2+ selectivity. Constructs 11BNick3 and 11BNick4 showed an increased preference for Co2+ over Pb2+ (Co:Pb 11BNick3 1.7- fold, 11BNick4 1.5-fold). In contrast, 11BNick2 showed an increased Co2+ selectivity over Zn2+ (Co:Zn 1.4±0.3) and Pb2+ (Co:Pb 2.4±0.5). 11BNicks2’s selectivity for Co2+ over Zn2+ and Pb2+ was found to increase further as the metal-ion concentration was decreased to 50 µm (Co:Zn 1.6±0.2, Co:Pb 3.2±0.7, Table 4).
Table 3.
The Co2+ selectivity over Zn2+ or Pb2+ for constructs 11BNick1–6. The effect of the alternative truncation platform involving a nick in the 5’-substrate binding arm is examined. Metal ion selectivities and pseudo-first-order rate constants for each construct are shown.
| Construct | kobs [min−1] | Co:Zn | Co:Pb | ||
|---|---|---|---|---|---|
| Co2+ | Zn2+ | Pb2+ | |||
| 11B | 0.088±0.006 | 0.15±0.01 | 0.079±0.016 | 0.59±0.06 | 1.1±0.2 |
| 11BNick1 | 0.042±0.004 | 0.075±0.019 | 0.076±0.021 | 0.56±0.15 | 0.55±0.16 |
| 11BNick2 | 0.010±0.002 | 0.0073±0.0008 | 0.0041±0.0004 | 1.4±0.3 | 2.4±0.5 |
| 11BNick3 | 0.027±0.004 | 0.038±0.010 | 0.016±0.002 | 0.71±0.22 | 1.7±0.3 |
| 11BNick4 | 0.015±0.002 | 0.019±0.003 | 0.010±0.002 | 0.79±0.14 | 1.5±0.3 |
| 11BNick5 | 0.18±0.01 | 0.27±0.04 | 0.16±0.04 | 0.67±0.11 | 1.1±0.3 |
| 11BNick6 | 0.18±0.01 | 0.28±0.01 | 0.16±0.05 | 0.64±0.03 | 1.1±0.3 |
Table 4.
The Co2+ selectivity of nicked constructs containing peripheral sequence elements. The increase in Co2+ selectivity attributable to the presence of loop II and peripheral sequence elements is examined. Pseudo-first-order rate constants and metal selectivities are shown for the given metal ion concentration.
| Construct | kobs [min−1] | Co:Zn | Co:Pb | |||
|---|---|---|---|---|---|---|
| Co2+ | Zn2+ | Pb2+ | ||||
| 11B | 500 | 0.088±0.006 | 0.15±0.01 | 0.079±0.016 | 0.59±0.06 | 1.1±0.2 |
| 50 | 0.022±0.002 | 0.025±0.002 | 0.021±0.001 | 0.88±0.09 | 1.1±0.1 | |
| 11BNick2 | 500 | 0.010±0.002 | 0.0073±0.0008 | 0.0041±0.0004 | 1.4±0.3 | 2.4±0.5 |
| 50 | 0.0016±0.0001 | 0.0010±0.0001 | 0.0005±0.0001 | 1.6±0.2 | 3.2±0.7 | |
| 11BNick7 | 500 | 0.13±0.01 | 0.17±0.04 | 0.086±0.022 | 0.76±0.20 | 1.5±0.4 |
| 50 | 0.034±0.001 | 0.037±0.003 | 0.012±0.003 | 0.92±0.09 | 2.8±0.7 | |
| 11BNick8 | 500 | 0.094±0.008 | 0.14±0.01 | 0.065±0.008 | 0.67±0.08 | 1.4±0.2 |
| 50 | 0.022±0.001 | 0.029±0.002 | 0.010±0.004 | 0.76±0.07 | 2.2±0.8 | |
| 11B14 | 500 | 0.077±0.001 | 0.14±0.02 | 0.11±0.03 | 0.55±0.06 | 0.70±0.18 |
| 50 | 0.022±0.003 | 0.026±0.003 | 0.012±0.001 | 0.85±0.15 | 1.8±0.3 | |
| 11B15 | 500 | 0.056±0.007 | 0.10±0.01 | 0.074±0.014 | 0.56±0.10 | 0.76±0.17 |
| 50 | 0.013±0.002 | 0.015±0.001 | 0.0071±0.0004 | 0.87±0.15 | 1.8±0.4 |
Although the near threefold increase in 11BNick2’s Co:Zn and Co:Pb selectivity is modest, the reproducible effect supports a role for loop II in Co2+ selectivity. The conversion of loop II to a tetraloop and the absence of additional peripheral sequence elements in these constructs may have limited the magnitude of the increased Co2+ selectivity. The increased selectivity of these constructs, however, demonstrated the utility of this alternative truncation platform in investigating the Co2+ selectivity of the clone 11 system. Also, because most DNAzyme configurations incorporate a small catalytic core flanked by substrate binding arms, further refinement of the alternative truncation platform may lead to a general method for retaining the selectivity and activity of RNA-cleaving DNAzymes.
Loop II is sufficient to increase Co2+ selectivity
To further investigate the role of peripheral sequence elements in Co2+ selectivity, additional constructs were designed based on the alternative truncation platform. The 11BNick2 construct was selected for further study because of its improved Co2+ selectivity over both Zn2+ and Pb2+, as shown above. Constructs based on 11BNick2 contained both loop II and peripheral sequences elements such as T78 and T80 (Figure 3 A). A unique secondary structure element, not predicted in the full length clone 11 cis- or trans-sequences or sequence variants, was predicted for construct 11BNick7 in peripheral sequence regions (nucleotides 4–15, 78–88; Figure 3A).
These peripheral sequence elements’ contributions to Co2+ selectivity was investigated in the 11BNick7 construct (Table 4). 11BNick7 and 11BNick8 showed reproducible but insignificant improvements in Co2+ selectivity (Co:Pb 1.3–1.4-fold) over the 11B construct. No improvement was observed in the Co:Zn selectivity. At lower concentrations of metal ions, however, the Co:Pb selectivity was improved for 11BNick7 (Co:Pb 1.5±0.4) and 11BNick8 (Co:Pb 2.2±0.8), with levels similar to those observed in the 11BNick2 construct. This level corresponds to an increase in Co2+ selectivity of up to 2.9-fold over Pb2+, compared with the 11Btrans system. No increase in Co2+ selectivity was observed for 11B-trans at the same concentration of metal ions, suggesting that the effect is specific to the constructs using the 11BNick2 construct from the alternative truncation platform.
To investigate whether the peripheral sequences forming loop II were alone sufficient to increase Co2+ selectivity, two additional constructs, 11B14 and 11B15, were designed using the conventional truncation approach (Figure 3 B). Aside from the absence of loop II, construct 11B14 is identical to 11BNick7, and 11B15 is identical to 11BNick8. The Co:Zn selectivity for both 11B14 (Co:Zn 0.55±0.06) and 11B15 (Co:Zn 0.56±0.10) showed no increase over the 11B-trans construct (Table 4). Both constructs mirrored the improvement in the Co:Pb selectivity seen with 11BNick7 and 11BNick8 at metal ion concentrations of 50 µm. The 11BNick 7 (Co:Pb 2.8±0.7) construct, however, showed a more significant increase over 11B14 (Co:Pb 1.8±0.3) or 11B15 (Co:Pb 1.8±0.4). While constructs containing the 3’ and 5’ peripheral sequences showed increases in Co:Pb selectivity, the 11BNick2 construct utilizing the alternative truncation platform and loop II showed over a threefold increase in Co:Pb selectivity exceeding gains in all other constructs.
Exploring possible metastable structures
The investigation described above was based primarily on the most stable secondary structure predicted by mfold, and truncation was based on the 11B secondary structure. However, systematic truncation to preserve the most stable secondary structure mostly resulted in a loss of metal ion selectivity. We concluded that some metastable structures predicted by mfold might play a role. Three distinct secondary structures—11A, 11B and 11B’—were predicted for DNAzyme constructs based on clone 11 (Figure 2A). The third structure, 11B’, was identified during the design of constructs for the truncation study. 11B and 11B’ differ by the secondary structure adopted within the highly conserved region (nucleotides 51–67). Since this region is highly conserved, adopts two different secondary structures, and interfaces with the cleavage site, the structure within this region could be functionally important.
To investigate the effects of these metastable structures in the clone 11 system, correlations between Co2+ selectivity and predicted secondary structure were examined (Table 5). A cursory look at the predicted secondary structures of the constructs used in this study revealed three different scenarios. Predicted secondary structures for clone 11-related sequences fell into several categories: 11A only, 11B along with 11A or 11B’, or 11B only. Clone 18 is an example of the first scenario, forming only the 11A-type structures and showing poor Co2+ selectivity (Co:Zn 0.46, Co:Pb 0.56). Constructs truncated based on the 11A-type structure were inactive. Considering the second situation, sequences forming the 11B- and 11B’-type structures also had decreased Co:Zn selectivity (0.06–0.79) relative to Clone 11. In addition, 11B2/Sub2 forms the 11A and 11B structures and shows low Co2+ selectivity (Co:Zn 0.41, Co:Pb 0.69). No increase in Co:Zn selectivity was observed for the 11BNick1, 11BNick3, and 11BNick4 constructs (Co:Zn 0.56–0.77), which form the 11B- and 11B’-type structures. These three sequences were predicted to form the 11B and 11B’ structures. While 11BNick3 did show a mild improvement in Co:Pb selectivity, the Co:Zn selectivity was still limited. The 11B14 and 11B15 constructs were of particular interest because each contained additional peripheral sequence elements and were predicted to form all three secondary structures. Improvements in the Co:Pb selectivity among 11BNick7, 11BNick8, 11B14, and 11B15 (1.6–2.5-fold) did not appear to correlate with their predicted secondary structures. 11BNick2 was predicted to form only the 11B-type structure. It also contained loop II and utilized the alternative truncation platform. This construct showed the largest improvement in Co:Pb selectivity (2.9-fold), restoring the Co:Pb selectivity to near clone 11 levels.
Table 5.
Effect of metastable structures on selectivity. The predicted metastable structures for selected constructs are compared with the observed metal selectivity. Constructs predicted to form only the 11B structure show higher Co2+ selectivity. All metal selectivity values were determined at 50 µm metal concentration unless otherwise stated.
| Construct | Predicted structure | Co:Zn | Co:Pb |
|---|---|---|---|
| Clone 11 | 11A/11B | 1.6[a] | 4.5[a] |
| Clone 18 | 11A | 0.46[a] | 0.56[a] |
| 11B/Sub | 11B/11B’ | 0.59±0.06[a] | 1.1±0.2[a] |
| 0.88±0.091 | 1.1±0.1 | ||
| 11B5/Sub3 | 11A/11B | 0.41[a] | 0.69[a] |
| 11B5/Sub | 11B/11B’ | 0.05[a] | 1.1[a] |
| 11B3/Sub2 | 11B/11B’ | 0.06[a] | 0.12[a] |
| 11BNick1 | 11B/11B’ | 0.56±0.15 | 0.55±0.16 |
| 11BNick2 | 11B | 1.4±0.3[a] | 2.4±0.5[a] |
| 1.6±0.2 | 3.2±0.7 | ||
| 11BNick3 | 11B/11B’ | 0.71±0.22 | 1.7±0.3 |
| 11BNick4 | 11B/11B’ | 0.79±0.14 | 1.5±0.3 |
| 11BNick5 | 11B | 0.67±0.11 | 1.1±0.3 |
| 11BNick6 | 11B | 0.64±0.03 | 1.1±0.3 |
| 11BNick7 | 11B | 0.76±0.20[a] | 1.5±0.4[a] |
| 0.92±0.09 | 2.8±0.7 | ||
| 11BNick8 | 11B | 0.67±0.08[a] | 1.4±0.2[a] |
| 0.76±0.07 | 2.2±0.8 | ||
| 11B14 | 11A/11B/11B’ | 0.55±0.06[a] | 0.70±0.18[a] |
| 0.85±0.15 | 1.8±0.27 | ||
| 11B15 | 11A/11B/11B’ | 0.56±0.10[a] | 0.76±0.17[a] |
| 0.87±0.15 | 1.8±0.4 |
Values obtained at 500 µm.
Discussion
Role of peripheral sequence elements in determining metal-ion selectivity
In this study, we carried out biochemical assays of a previously selected Co2+-selective DNAzyme[6] to identify sequence elements responsible for its subtle Co2+ selectivity. This selectivity is characterized by the selectivity index, the ratio of a DNAzyme’s phosphodiester transfer activity in the presence of Co2+ to its activity in the presence of the next-most competing ions, Zn2+ or Pb2+. Even though the changes in the selectivity indices were relatively small, the differences in the rates of the reactions in the presence of different metal ions exceeded the error measurements. Such a small metal selectivity index reflects the fact that Co2+ and Zn2+ are highly similar metal ions with the same ionic radii and similar preferences in ligand donor sets. Even metalloproteins that are known to have extremely high selectivity for other metal ions display a much smaller selectivity index between Co2+ and Zn2+, as Co2+ in almost all zinc proteins can be substituted for Zn2+ and retain ~100% activity.
The most important finding from this study is that truncating peripheral sequences in clone 11 tended to reduce Co2+ selectivity, even though the phosphodiester transfer activity remained. Truncating peripheral sequences is common practice in DNAzyme research. It often results in a smaller DNAzyme with similar activity to the originally selection product. We show here that when subtle differences in the metal-ion selectivity of highly similar metal ions such as Co2+ and Zn2+ are concerned, the role played by peripheral sequences cannot be ignored. In the clone 11 system, loop II partially restored the Co2+ selectivity over Pb2+ to levels observed in the cis-cleaving clone 11 DNAzyme.
Peripheral sequence elements have been found to modulate function of other DNAzymes and ribozymes. For example, the P5abc peripheral element facilitates the proper folding of the P4–P6 domain in the Tetrahymena thermophila group I intron.[27] A construct of the Schistosoma mansoni hammerhead ribozyme incorporating peripheral sequences showed an increase in activity.[28, 29] Folding studies have shown that other ribozymes also utilize peripheral sequences for proper folding and catalytic activity. In the case of DNAzyme systems, the activity of the X-motif DNAzyme relies on peripheral sequence elements for activity.[30] A group of recently isolated, transitionmetal-dependent DNAzymes also utilizes peripheral sequences to modulate catalytic activity[31, 32] and to stabilize the 8–17 motif within a larger DNAzyme structure.[32]
Sequence elements important for enhanced Co2+ selectivity
We found that the peripheral sequence forming loop II in the alternative truncation platform is important for increased Co2+ selectivity. Restoring the Co2+ selectivity of 11BNick2 to levels similar to those observed in clone 11 also led to insights into the structure-function relationships in clone 11. First, the similarities between the ionic radii and coordination geometry of Co2+ and Zn2+ make the twofold increase in the Co:Zn selectivity of 11BNick2 particularly significant. The peripheral sequence element loop II may assist in forming tertiary structural features that facilitate the three-dimensional arrangement of ligand donor elements contributing to clone 11’s Co:Zn selectivity. Second, the restoration of the Co2+ selectivity over Pb2+ remains equally significant because of the high background rate of Pb2+-mediated phosphodiester hydrolysis. Additional peripheral sequences beyond loop II may be required to produce a structure that further increases the selectivity over Zn2+ and Pb2+ to levels observed in clone 11. The tertiary structure formed may involve loop II along with the conserved region (positions 51–70) or the 3’ substrate binding arm to provide a three-dimensional arrangement of functional groups leading to the preferential binding of Co2+.
In the clone 11 DNAzyme system, the results from “nicked” constructs lend credence to the idea that peripheral sequences increase Co2+ selectivity. For example, converting loop II to a tetraloop or introducing mutations into the substrate-binding arms may have contributed to differential Co2+ selectivity. These possibilities, however, are unlikely to be the sole reasons for the increased Co:Zn selectivity. Loop II alone was sufficient to produce modest but reproducible increases in selectivity over those observed in clone 11. It is also important to note that increased Co2+ selectivity over both Zn2+ and Pb2+ was observed in a construct that contains only two of the four nucleotides that distinguish clone 11 from clone 18. This raises the possibility that nucleotides 72 and 77 contribute to a three-dimensional structure that preferentially binds Co2+ either through direct Co2+ binding or through a tertiary fold that uses these nucleotides to form a Co2+-selective metalbinding pocket. The modest increase in Co2+ selectivity over Zn2+ also suggests that secondary or tertiary elements present in the full-length clone 11 DNAzyme are only approximated in the truncated “nicked” platform. These results support the possibility that peripheral sequence elements contribute to a tertiary structure with sufficient complexity to arrange DNAzyme ligand donor groups in a Co2+-selective metal-binding pocket.
Role of secondary structure in metal selectivity
Comparing structural features present in clone 11 with those of other DNAzyme/ribozyme systems provided further insights. The poor conservation of an element essential for selectivity in clone 11 is mirrored in the Sc.ai5γ group II self-splicing intron, which shows poor conservation of the sequence required for substrate recognition.[33] A Mg2+-dependent secondary structure rearrangement at this substrate recognition site in the group II intron is proposed to facilitate substrate binding.[34] In other ribozyme systems, secondary structural motifs contribute to metal binding and tertiary structure formation. Loop–loop interactions mediate tertiary contacts necessary for the optimal activity of many ribozymes as well as the ribosome.[20, 28, 35, 36] A single-stranded loop in the hammerhead ribozyme stabilizes the tertiary fold and facilitates catalytic core formation.[37] Loop regions have also been implicated in G-quartet formation and stability,[38] as observed in a recently isolated kinase DNAzyme that uses the loop region of a G-quartet to form a distal tertiary contact required for activity and G-quartet formation.[39] Helical junctions, also present in clone 11, facilitate the assembly of the tertiary structure in many large and small ribozymes,[36, 40, 41] the RNA component of telomerase, and most notably in the recently isolated nuclease[4, 5, 42] and ligase[43] DNAzymes. The triple-helix junction in the 8–17 system comprises the catalytic core and serves as a hinge point for tertiary folding.[44] Crystal structures of the ribosome[20] and the L1 ligase ribozyme[45] both demonstrate the formation of tertiary contacts and the organization of divalent metal ion binding pockets mediated by helical junctions. Further studies on the clone 11 system are likely to confirm that secondary structure motifs perform similar functions: forming a tertiary structure that contributes to the DNAzyme’s ability to bind Co2+ with high selectivity.
Role of metastable structures in determining metal selectivity
Our results also suggest that metastable structures predicted by mfold may affect metal selectivity. Metastable structures are involved in the folding and function of many other DNA and RNA systems. Differential folding and metal ion-dependent secondary and tertiary structure rearrangements in many ribozymes are likely to proceed through metastable structures.[23, 46–48] For example, metastable structures contribute to the folding of the hepatitis delta virus[48] and the P4–P6 domain of the group I intron,[47] as well as mediating splicing activity of the thymidylate synthase group I intron.[47] The group II intron has also demonstrated differential folds based on the identity of the metal-ion cofactor.[49] Riboswitches have also been shown to undergo metal-ion-dependent structural transitions that affect cofactor binding and activation.[50] In the case of DNAzymes, in vitro selected transition-metal-ion-dependent systems show multiple structures for similar sequences.[31] In addition, allosteric DNAzymes undergo analyte-dependent structural changes.[51] Other effects of metastable structures are seen in the transcription and activity of viral and mRNA sequences.[40, 47, 52] Finally, a number of conditions, including monovalent[53] and divalent metal ion concentration,[21, 54] temperature,[51] pH,[53] and peripheral sequence elements[27, 55] affect the folding behavior and prevalence of metastable structures.
The influence of metastable structures on the metal selectivity of clone 11 is supported by its Co2+ selectivity and multiple predicted secondary structures, as well as the precedent of metastable structures observed in other DNA/RNA systems. Results from truncation studies show that the largest decreases in Co:Zn and Co:Pb selectivity (Table 5) correlate with constructs showing multiple predicted secondary structures. Interestingly, two of these predicted secondary structures, 11B and 11B’, lie within a sequence region that is highly conserved and intolerant to point mutations. The correlation of Co2+ selectivity with multiple predicted secondary structures suggests the possibility of interconversion between the 11B and 11B’ structures. A structural rearrangement may have the functional role of repositioning the single riboadenosine, which is predicted to be base-paired in the 11B-type structure, to allow for the incoming nucleophile’s in-line attack on the scissile phosphate. Competing metastable structures may also help explain the persistence of Pb2+- and Zn2+-dependent activity after multiple rounds of negative selection, as observed with the persistence of inactive sequences following negative selection.[56] Finally, peripheral sequences and Co2+ metal ions may help stabilize the 11B-type structure, an effect mediated by both peripheral sequences and metals in other DNAzymes/ribozymes[27, 32, 55] and only approximated in truncated constructs in the clone 11 system. A complex but synergistic relationship may exist between metal-ion cofactors and peripheral sequence elements that contribute to Co2+ selectivity in clone 11.
Conclusions
Clone 11 requires both its primary sequence and the peripheral sequence element loop II to be selective for Co2+ over Zn2+ and Pb2+. Loop II’s structural features and nucleotides 72 and 77 likely contribute tertiary contacts that either stabilize or form a Co2+ selective binding pocket. This study provides a foundation to further investigate the relationship between DNAzyme structure and analyte selectivity. The success of the “nicked” strategy in reintroducing peripheral sequence elements and in restoring Co2+ selectivity suggests the potential of this approach as a general method for truncating and studying DNAzymes/ribozymes when the traditional truncation method fails to work. As the effects of peripheral sequences and secondary structure on cofactor selectivity are better understood, increasing or altering cofactor selectivity may be possible through rational design and in vitro selection based on established motifs. With its increased selectivity at low metal-ion concentrations, clone 11 is a good candidate for the analyte-responsive element of a biosensor. Finally, insights about the potential contributions of secondary structures to a DNAzyme’s overall function may be important in future DNAzyme/ribozyme-based biotechnology applications that require high activity and high analyte selectivity.
Experimental Section
Materials
NaCl, CoCl2, ZnCl2 and Pb(CH3COO)2 were purchased from Alfa Aesar at puratronic grade (99.998% or greater purity, metals basis). HPLC-purified DNA oligonucleotides were purchased from Integrated DNA Technologies. Additional DNA-RNA chimeric oligomers were purchased from TriLink Biotechnologies and were purified by the company unless otherwise indicated. HEPES was purchased from Sigma–Aldrich. All buffers were treated with Chelex 100 (Sigma–Aldrich) to remove divalent metal ions. The radiolabeling of DNA-chimeric substrates was carried out using redivue [γ-32P]ATP (Amersham Biosciences) and T4 polynucleotide kinase (Invitrogen).
Artificial phylogenetic analysis and design of clone 11 constructs
The sequences used in the alignment, including clone 11 and 18, were derived from previously described in vitro selection experiments.[6] Sequence alignments of sequences from selections 1 and 2 were constructed using the MultAlin folding program.[57] Highly conserved sequences have greater than 90% consensus, moderately conserved sequences have between 50% and 90% consensus, and nonconserved sequences have less than 50% consensus.
The design of constructs for the Co2+ selectivity studies was based on the sequence and structure of clone 11, clone 18 or the 11B trans-cleaving construct (11B-trans), as predicted by the mfold DNA folding algorithm.[24] Oligonucleotides were purchased from Integrated DNA Technologies. Mutations were introduced at key locations as determined by factors such as the alignment of selection 1 and 2 sequences, previously tested truncations, attempts to minimize self-complementarity among enzyme and substrate strands, or attempts to stabilize or destabilize secondary structures predicted for trans-cleaving clone 11 constructs. The 11B-trans construct was designed by truncating nucleotides 1–15, 38–43, and 78–107. Alternative truncation strategies were developed to investigate the Co2+ selectivity. Truncated constructs were designed that retained nucleotides 38–43 while truncating positions 1–15 and 78–107 to various degrees. An additional strategy was developed that placed a single nick at one of two locations within the 5’-substrate binding arm of clone 11. Mutations were made that retained the predicted secondary structure and activity of clone 11 and stabilized the formation of the nicked helix. Additional truncated “nicked” constructs were designed that contained nucleotides 1–15 and 78–107 to various degrees (clone 11 numbering). The sequences of all constructs tested are shown in Figure 3A.
Kinetic assays
The kinetics of the cleavage of cis- and trans-constructs at a single riboadenosine was monitored by a radioactive assay. Preparation of 32P-radiolabeled DNA substrates for assays was carried out as follows: the DNA substrate or cis-cleaving construct (20 pmol), [γ-32P]ATP (Amersham, 0.3 mm), and T4 polynucleotide kinase (1.25 UµL−1) were heated at 37 °C for 45 min. in a reaction mixture that contained Tris·HCl (70 mm, pH 7.6), KCl (0.1m), MgCl2 (10 mm), and 2-mercaptoethanol (1 mm). The labeled product was then desalted using a Sep-Pak Plus C-18 cartridge, flash frozen, and lyophilized. DNA samples were prepared at twice the final concentration in HEPES buffer (50 mm, pH 7.0) with NaCl (500 mm). The NaCl concentration was chosen because the Co2+-dependent activity begins to plateau at 300 mm NaCl (Bruesehoff and Lu, unpublished data). Reactions were performed under single turnover conditions using a DNAzyme (1 µm) and its 32P-radiolabeled substrate (30 nm), where the concentrations listed are the final concentrations. Samples were annealed by heating to 95°C for 3 min., then cooling to ambient temperature over 15 min. Each reaction was initiated by adding an equal volume of CoCl2, ZnCl2, or Pb(CH3COO)2 to the DNAzyme solution. Aliquots (5 µL) were removed periodically and transferred to stop buffer (10 µL) containing urea (8m) and EDTA (50 mm). Samples were then separated on a 20% polyacrylamide gel and exposed to a storage phosphor screen (Molecular Dynamics). Gels were analyzed by scanning the storage phosphor screen on a Storm 840 phosphorimager (Molecular Dynamics). The cleavage efficiency was calculated at time t using the following equation: y=100*[Ic/(Iu+Ic)], where y is the percent cleaved product, Ic is the intensity of the cleaved substrate and Iu is the intensity of the uncleaved substrate. Pseudo-firstorder rate constants were determined by fitting an equation of the form y=yo+a (1−e−kt) to the data using SigmaPlot 8.0, where y is the percent cleaved product as a function of time t, yo is the background product at time t=0, a is the fraction of the pool cleaved at time t = ∞, and k is the observed rate constant.
Acknowledgements
This work has been supported by the National Institutes of Health (ES016865), the Office of Science (BER), the U.S. Department of Energy, Grant No. DE-FG02-08ER64568, and the U.S. National Science Foundation, Grant No. CTS-0120978. We thank Dr. D.K. Garner for reviewing this manuscript.
References
- 1.Breaker RR, Joyce GF. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 2.a) Sen D, Geyer CR. Curr. Opin. Chem. Biol. 1998;2:680–687. doi: 10.1016/s1367-5931(98)80103-8. [DOI] [PubMed] [Google Scholar]; b) Breaker RR. Science. 2000;290:2095–2096. doi: 10.1126/science.290.5499.2095. [DOI] [PubMed] [Google Scholar]; c) Lu Y. Chem. Eur. J. 2002;8:4588–4596. doi: 10.1002/1521-3765(20021018)8:20<4588::AID-CHEM4588>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]; d) Peracchi A. Chem Bio Chem. 2005;6:1316–1322. doi: 10.1002/cbic.200500098. [DOI] [PubMed] [Google Scholar]; e) Dass CR, Choong PFM, Khachigian LM. Mol. Cancer Ther. 2008;7:243–251. doi: 10.1158/1535-7163.MCT-07-0510. [DOI] [PubMed] [Google Scholar]; f) Silverman SK. Chem. Commun. 2008:3467–3485. doi: 10.1039/b807292m. [DOI] [PubMed] [Google Scholar]; g) Liu J, Cao Z, Lu Y. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]; h) Schlosser K, Li Y. Chem. Biol. 2009;16:311–322. doi: 10.1016/j.chembiol.2009.01.008. [DOI] [PubMed] [Google Scholar]; i) Zhang X-B, Kong R-M, Lu Y. Annu. Rev. Anal. Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.a) Faulhammer D, Famulok M. Angew. Chem. 1996;108:2984–2988. Angew. Chem. Int. Ed. Engl.1996, 35, 2837 –2841; [Google Scholar]; b) Peracchi A. J. Biol. Chem. 2000;275:11693–11697. doi: 10.1074/jbc.275.16.11693. [DOI] [PubMed] [Google Scholar]; c) Brown AK, Li J, Pavot CM-B, Lu Y. Biochemistry. 2003;42:7152–7161. doi: 10.1021/bi027332w. [DOI] [PubMed] [Google Scholar]; d) Cruz RPG, Withers JB, Li Y. Chem. Biol. 2004;11:57–67. doi: 10.1016/j.chembiol.2003.12.012. [DOI] [PubMed] [Google Scholar]; e) Schlosser K, Gu J, Sule L, Li Y. Nucleic Acids Res. 2008;36:1472–1481. doi: 10.1093/nar/gkm1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Santoro SW, Joyce GF. Proc. Natl. Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Zheng W, Kwon AH, Lu Y. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruesehoff PJ, Li J, Augustine AJ, III, Lu Y. Comb. Chem. High Throughput Screening. 2002;5:327–335. doi: 10.2174/1386207023330264. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z, Mei SHJ, Brennan JD, Li Y. J. Am. Chem. Soc. 2003;125:7539–7545. doi: 10.1021/ja035208+. [DOI] [PubMed] [Google Scholar]
- 8.Carmi N, Balkhi HR, Breaker RR. Proc. Natl. Acad. Sci. USA. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu J, Lu Y. Angew. Chem. 2007;119:7731–7734. Angew. Chem. Int. Ed.2007, 46, 7587 –7590. [Google Scholar]
- 10.Hollenstein M, Hipolito C, Lam C, Dietrich D, Perrin DM. Angew. Chem. 2008;120:4418–4422. doi: 10.1002/anie.200800960. Angew. Chem. Int. Ed.2008, 47, 4346 –4350. [DOI] [PubMed] [Google Scholar]
- 11.a) Santoro SW, Joyce GF, Sakthivel K, Gramatikova S, Barbas CF., III J. Am. Chem. Soc. 2000;122:2433–2439. doi: 10.1021/ja993688s. [DOI] [PubMed] [Google Scholar]; b) Hoadley KA, Purtha WE, Wolf AC, Flynn-Charlebois A, Silverman SK. Biochemistry. 2005;44:9217–9231. doi: 10.1021/bi050146g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Sen D. Biochemistry. 1997;36:5589–5599. doi: 10.1021/bi962694n. [DOI] [PubMed] [Google Scholar]
- 13.Liu J, Brown AK, Meng X, Cropek DM, Istok JD, Watson DB, Lu Y. Proc. Natl. Acad. Sci. USA. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brown AK, Liu J, He Y, Lu Y. Chem Bio Chem. 2009;10:486–492. doi: 10.1002/cbic.200800632. [DOI] [PubMed] [Google Scholar]
- 15.a) Li J, Lu Y. J. Am. Chem. Soc. 2000;122:10466–10467. [Google Scholar]; b) Liu J, Lu Y. J. Am. Chem. Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]; c) Lu Y, Liu J, Li J, Bruesehoff PJ, Pavot CM-B, Brown AK. Biosens. Bioelectron. 2003;18:529–540. doi: 10.1016/s0956-5663(03)00013-7. [DOI] [PubMed] [Google Scholar]; d) Achenbach JC, Chiuman W, Cruz RPG, Li Y. Curr. Pharm. Biotechnol. 2004;5:321–336. doi: 10.2174/1389201043376751. [DOI] [PubMed] [Google Scholar]; e) Chiuman W, Li Y. Nucleic Acids Res. 2007;35:401–405. doi: 10.1093/nar/gkl1056. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Liu J, Lu Y. J. Am. Chem. Soc. 2007;129:9838–9839. doi: 10.1021/ja0717358. [DOI] [PubMed] [Google Scholar]; g) Shen Y, Mackey G, Rupcich N, Gloster D, Chiuman W, Li Y, Brennan JD. Anal. Chem. 2007;79:3494–3503. doi: 10.1021/ac070235u. [DOI] [PubMed] [Google Scholar]; h) Zhao W, Chiuman W, Lam JC, Brook MA, Li Y. Chem. Commun. 2007:3729–3731. doi: 10.1039/b705335e. [DOI] [PubMed] [Google Scholar]; i) Lee JH, Wang Z, Liu J, Lu Y. J. Am. Chem. Soc. 2008;130:14217–14226. doi: 10.1021/ja803607z. [DOI] [PMC free article] [PubMed] [Google Scholar]; j) Zhao W, Lam JC, Chiuman W, Brook MA, Li Y. Small. 2008;4:810–816. doi: 10.1002/smll.200700757. [DOI] [PubMed] [Google Scholar]
- 16.a) DeRose VJ. Curr. Opin. Struct. Biol. 2003;13:317–324. doi: 10.1016/s0959-440x(03)00077-0. [DOI] [PubMed] [Google Scholar]; b) Sigel RKO, Pyle AM. Chem. Rev. 2007;107:97–113. doi: 10.1021/cr0502605. [DOI] [PubMed] [Google Scholar]
- 17.a) Boerner LJK, Zaleski JM. Curr. Opin. Chem. Biol. 2005;9:135–144. doi: 10.1016/j.cbpa.2005.02.010. [DOI] [PubMed] [Google Scholar]; b) Jiang Q, Xiao N, Shi P, Zhu Y, Guo Z. Coord. Chem. Rev. 2007;251:1951–1972. [Google Scholar]; c) Zeglis BM, Boland JA, Barton JK. Biochemistry. 2009;48:839–849. doi: 10.1021/bi801885w. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Zeglis BM, Pierre VC, Kaiser JT, Barton JK. Biochemistry. 2009;48:4247–4253. doi: 10.1021/bi900194e. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Dong X, Wang X, Lin M, Sun H, Yang X, Guo Z. Inorg. Chem. 2010;49:2541–2549. doi: 10.1021/ic100001x. [DOI] [PubMed] [Google Scholar]
- 18.a) Lippert B. Chem. Biodiversity. 2008;5:1455–1474. doi: 10.1002/cbdv.200890135. [DOI] [PubMed] [Google Scholar]; b) Mucha A, Knobloch B, Jezowska-Bojczuk M, Kozlowski H, Sigel RK. Dalton Trans. 2008:5368–5377. doi: 10.1039/b805911j. [DOI] [PubMed] [Google Scholar]; c) Nakano S, Hirayama H, Sugimoto N. Nucleic Acids Symp. Ser; 2009. pp. 229–230. [DOI] [PubMed] [Google Scholar]; d) Sigel RKO, Sigel H. Acc. Chem. Res. 2010;43:974–984. doi: 10.1021/ar900197y. [DOI] [PubMed] [Google Scholar]; e) Khutia A, Sanz Miguel PJ, Lippert B. Chem. Eur. J. 2011;17:4195–4204. doi: 10.1002/chem.201002722. [DOI] [PubMed] [Google Scholar]
- 19.a) Li Y, Breaker RR. Curr. Opin. Struct. Biol. 1999;9:315–323. doi: 10.1016/S0959-440X(99)80042-6. [DOI] [PubMed] [Google Scholar]; b) Liu Y, Sen D. J. Mol. Biol. 2010;395:234–241. doi: 10.1016/j.jmb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 20.Klein DJ, Moore PB, Steitz TA. RNA. 2004;10:1366–1379. doi: 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pyle A. J. Biol. Inorg. Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 22.Cotton FA, Wilkinson G, Murillo CA, Bochmann M. Advanced Inorganic Chemistry. 6th ed. New York: Wiley; 1999. [Google Scholar]
- 23.Erat MC, KO R. J. Biol. Inorg. Chem. 2008;13:1025–1036. doi: 10.1007/s00775-008-0390-7. [DOI] [PubMed] [Google Scholar]
- 24.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fox GE, Woese CR. Nature. 1975;256:505–507. doi: 10.1038/256505a0. [DOI] [PubMed] [Google Scholar]
- 26.a) Bruesehoff PJ. Dissertation/thesis. Champaign (USA): University of Illinois at Urbana; 2003. [Google Scholar]; b) Mei SHJ, Liu Z, Brennan JD, Li Y. J. Am. Chem. Soc. 2003;125:412–420. doi: 10.1021/ja0281232. [DOI] [PubMed] [Google Scholar]
- 27.Engelhardt MA, Doherty EA, Knitt DS, Doudna JA, Herschlag D. Biochemistry. 2000;39:2639–2651. doi: 10.1021/bi992313g. [DOI] [PubMed] [Google Scholar]
- 28.Osborne EM, Schaak JE, Derose VJ. RNA. 2005;11:187–196. doi: 10.1261/rna.7950605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roychowdhury-Saha M, Burke DH. RNA. 2006;12:1846–1852. doi: 10.1261/rna.128906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazarev D, Puskarz I, Breaker RR. RNA. 2003;9:688–697. doi: 10.1261/rna.2600503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiuman W, Li Y. J. Mol. Biol. 2006;357:748–754. doi: 10.1016/j.jmb.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 32.Schlosser K, Lam JCF, Li Y. Nucleic Acids Res. 2006;34:2445–2454. doi: 10.1093/nar/gkl276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruschel D, Sigel RK. J. Inorg. Biochem. 2008;102:2147–2154. doi: 10.1016/j.jinorgbio.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 34.Erat MC, Kovacs H, Sigel RKO. J. Inorg. Biochem. 2010;104:611–613. doi: 10.1016/j.jinorgbio.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 35.a) Pley HW, Flaherty KM, McKay DB. Nature. 1994;372:111–113. doi: 10.1038/372111a0. [DOI] [PubMed] [Google Scholar]; b) Wilson TJ, Lilley DMJ. RNA. 2002;8:587–600. doi: 10.1017/s1355838202020514. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Saksmerprome V, Roychowdhury-Saha M, Jayasena S, Khvorova A, Burke DH. RNA. 2004;10:1916–1924. doi: 10.1261/rna.7159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilley DMJ. RNA. 2004;10:151–158. doi: 10.1261/rna.5217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baird NJ, Westhof E, Qin H, Pan T, Sosnick TR. J. Mol. Biol. 2005;352:712–722. doi: 10.1016/j.jmb.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 38.Fujimoto T, Miyoshi D, Tateishi-Karimata H, Sugimoto N. Nucleic Acids Symp. Ser; 2009. pp. 237–238. [DOI] [PubMed] [Google Scholar]
- 39.McManus SA, Li Y. J. Mol. Biol. 2008;375:960–968. doi: 10.1016/j.jmb.2007.10.080. [DOI] [PubMed] [Google Scholar]
- 40.Krasilnikov AS, Xiao Y, Pan T, Mondragón A. Science. 2004;306:104–107. doi: 10.1126/science.1101489. [DOI] [PubMed] [Google Scholar]
- 41.Zappulla DC, Cech TR. Proc. Natl. Acad. Sci. USA. 2004;101:10024–10029. doi: 10.1073/pnas.0403641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiuman W, Li Y. Chem. Biol. 2006;13:1061–1069. doi: 10.1016/j.chembiol.2006.08.009. [DOI] [PubMed] [Google Scholar]; Shen Y, Chiuman W, Brennan JD, Li Y. Chem Bio Chem. 2006;7:1343–1348. doi: 10.1002/cbic.200600195. [DOI] [PubMed] [Google Scholar]
- 43.Coppins RL, Silverman SK. J. Am. Chem. Soc. 2005;127:2900–2907. doi: 10.1021/ja044881b. [DOI] [PubMed] [Google Scholar]
- 44.Liu J, Lu Y. J. Am. Chem. Soc. 2002;124:15208–15216. doi: 10.1021/ja027647z. [DOI] [PubMed] [Google Scholar]
- 45.Robertson MP, Scott WG. Science. 2007;315:1549–1553. doi: 10.1126/science.1136231. [DOI] [PubMed] [Google Scholar]
- 46.Andersen AA, Collins RA. Proc. Natl. Acad. Sci. USA. 2001;98:7730–7775. doi: 10.1073/pnas.141039198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nagel JHA, Pleij CWA. Biochimie. 2002;84:913–923. doi: 10.1016/s0300-9084(02)01448-7. [DOI] [PubMed] [Google Scholar]
- 48.Brown TS, Chadalavada DM, Bevilacqua PC. J. Mol. Biol. 2004;341:695–712. doi: 10.1016/j.jmb.2004.05.071. [DOI] [PubMed] [Google Scholar]
- 49.Steiner M, Rueda D, Sigel RKO. Angew. Chem. 2009;121:9920–9924. doi: 10.1002/anie.200903809. Angew. Chem. Int. Ed.2009, 48, 9739 – 9742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamauchi T, Miyoshi D, Kubodera T, Nishimura A, Nakai S, Sugimoto N. FEBS Lett. 2005;579:2583–2588. doi: 10.1016/j.febslet.2005.03.074. [DOI] [PubMed] [Google Scholar]
- 51.Zivarts M, Liu Y, Breaker RR. Nucleic Acids Res. 2005;33:622–631. doi: 10.1093/nar/gki182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suo Z, Johnson KA. Biochemistry. 1997;36:14778–11485. doi: 10.1021/bi971963m. [DOI] [PubMed] [Google Scholar]; Paiva AM, Sheardy RD. J. Am. Chem. Soc. 2005;127:5581–5585. doi: 10.1021/ja043783n. [DOI] [PubMed] [Google Scholar]
- 53.Inoue M, Miyoshi D, Sugimoto N. Nucleic Acids Symp. Ser; 2005. pp. 243–244. [DOI] [PubMed] [Google Scholar]
- 54.a) Pyle AM. Science. 1993;261:709–714. doi: 10.1126/science.7688142. [DOI] [PubMed] [Google Scholar]; b) Miyoshi D, Nakao A, Toda T, Sugimoto N. FEBS Lett. 2001;496:128–133. doi: 10.1016/s0014-5793(01)02416-4. [DOI] [PubMed] [Google Scholar]; c) Li W, Miyoshi D, Nakano S, Sugimoto N. Biochemistry. 2003;42:11736–11744. doi: 10.1021/bi034168j. [DOI] [PubMed] [Google Scholar]
- 55.Michel F, Westhof E. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 56.a) Soukup GA, Breaker RR. Curr. Opin. Struct. Biol. 2000;10:318–325. doi: 10.1016/s0959-440x(00)00090-7. [DOI] [PubMed] [Google Scholar]; b) Roth A, Breaker RR. Sioud M. Methods in Molecular Biology, Vol. 252: Ribozymes and siRNA protocols. Totowa: Humana; 2004. pp. 145–164. [DOI] [PubMed] [Google Scholar]
- 57.Corpet F. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]