Abstract
Subunit 7a of mouse cytochrome c oxidase (Cox) displays a contractile muscle-specific isoform, Cox7a1, that is the major cardiac form. To gain insight into the role of this isoform, we have produced a new knockout mouse line that lacks Cox7a1. We show that homozygous and heterozygous Cox7a1 knockout mice, although viable, have reduced Cox activity and develop a dilated cardiomyopathy at 6 weeks of age. Surprisingly, the cardiomyopathy improves and stabilizes by 6 months of age. Cox7a1 knockout mice incorporate more of the “liver-type” isoform Cox7a2 into the cardiac Cox holoenzyme and, also surprisingly, have higher tissue ATP levels.
Keywords: Mitochondria, dilated cardiomyopathy, oxidative phosphorylation, electron transport chain, gene knockout
1. Introduction
Mitochondrial1 genetic disease can be divided into those due to mutations in the mitochondrial genome and those associated with mutations in nuclear-encoded genes for mitochondrial proteins. A number of mitochondrial genome mutations are occasionally associated with cardiomyopathy, e.g. MELAS (Mitochondrial Encephalopathy Lactic Acidosis and Stroke-like episodes (Deschauer et al., 2001)), sometimes as the most prominent pathological feature (Silvestri et al., 2001). However, isolated cardiomyopathy is almost unknown with these maternally inherited disorders. On the other hand, mutations in many genes have been associated with dominantly inherited dilated cardiomyopathy and mouse models have been developed for a large number of them. Most of these involve mutations in structural proteins of the cardiac myofibrillary contractile structure, as summarized in Table 1. Other dilated cardiomyopathy genes affect ionic regulation of cardiac contractility: sodium-channel gates type V, alpha subunit, more frequently associated with long QT syndrome (SCN5A (Olson et al, 2005), Scn5A (Papadatos et al., 2002), phospholamban (PLN (Schmitt et al., 2003), and Pln (Luo et al., 1994)). Surprisingly, there have been only a small number of studies linking isolated dilated cardiomyopathy to mutations in nuclear-encoded genes for mitochondrial proteins. Examples for such mouse models are knockout of mitochondrial heat shock protein Hsp40 (Hayashi et al., 2006), heart-specific knockout of mitochondrial thioredoxin reductase TrxR2 (Conrad et al., 2004), and knockout of mitochondrial creatine kinase (Nahrendorf et al., 2005). Maternally inherited pure cardiomyopathy is rarely observed (Casali et al., 1999).
Table 1.
Human and mouse cardiomyopathy genes involved in the cardiomyofibrillary contractile structure.
| Protein | Human gene/Clinical study | Reference | Mouse gene/Mouse model | Reference |
|---|---|---|---|---|
| Cardiac myosin binding protein 3 | MYBPC3 | 1 | ||
| Cysteine and glycine rich protein 3 | CSRP3 | 2 | Csrp3 | 12 |
| Desmin | DES | 3 | Des | 13 |
| Lamin A/C | LMNA | 4 | Lmna | 14 |
| LIM-Domain binding 3 | LDB3 | 5 | Ldb3 | 15 |
| Myosin light chain 2 | MYL2 | 6 | MYL2 transgene | 16 |
| Myosin heavy chain 7 | MYH7 | 7 | Myh7 | 17 |
| Sarcoglycan delta | SGCD | 8 | Sgcd | 18 |
| Titin | TTN | 9 | Ttn | 19 |
| Tropomyosin | TPM1 | 10 | Tpm1 | 20 |
| Troponin T2 | TNNT2 | 11 | Tnnt2 | 21 |
References: 1, Dhandapany et al, 2009; 2, Knoll et al., 2002; 3, Dalakas et al., 2000; 4, Fatkin et al., 1999; 5, Vatta et al., 2003; 6, Poetter et al., 1996; 7, Kamisago et al., 2000; 8, Tsubata et al., 2000; 9, Satoh et al., 1999; 10, Olson et al., 2001; 11, Thierfelder et al., 1994; 12, Arber et al., 1997; 13, Li et al., 1996; 14, Mounkes et al., 2005; 15, Zheng et al., 2009; 16, Szczesna-Cordary et al., 2005; 17, Geisterfer-Lowrance et al., 1996; 18, Coral-Vazquez et al., 1999; 19, Radke et al., 2007; 20, Muthuchamy et al., 1999; 21, Tardiff et al., 1998.
Mitochondrial energy generation is essential for the functional health of cardiac tissue and is provided by the oxidative phosphorylation (OxPhos) complexes located in the mitochondrial inner membrane. Cytochrome c oxidase (Cox), the terminal protein complex of the mitochondrial electron transport chain (ETC), consists of 13 proteins, 3 encoded by the mitochondrial genome and 10 encoded by the nuclear genome. Cox is the proposed rate-limiting enzyme of the ETC in intact cells (Villani et al., 2003; Acin-Perez et al., 2003), and it contains at least five subunits with tissue-specific isoforms, uniquely among the electron transport complexes, suggesting a regulatory role in modulating energy metabolism. These are Cox4i1/Cox4i2; Cox6a1/Cox6a2; Cox6b1/Cox6b2; Cox7a1/Cox7a2; Cox8a/Cox8b/Cox8c. The Cox7a isoforms are part of the subunit 6a/7a/8 trio where the more restricted member is expressed primarily in heart and skeletal muscle (Kadenbach et al., 1987; Lomax and Grossman, 1989; Lenka et al., 1998).
We present here a new knockout mouse line that lacks the “heart-type” isoform of cytochrome c oxidase subunit 7a (Cox7a1; Cox7ah in previous terminology), which is expressed in heart and skeletal muscle in mammals. It might be anticipated that mild mutations in the heart-type isoforms for subunits 6a (Cox6a2), 7a (Cox7a1), or 8 (Cox8b) would be better tolerated than Cox mutations in genes without isoforms and could survive to show cardiac myopathy. We find indeed that Cox7a1 null mice are viable and fertile, producing increased amounts of the Cox7a2 isoform. Cox7a1 null mice demonstrate dilated cardiomyopathy in heterozygotes and homozygotes and, unusually, the cardiomyopathy tends to improve with age. We present here the initial characterization of Cox7a1 null animals, which includes the surprising discovery that knockout animals show higher ATP levels than do wild-type.
2. Materials and Methods
2.1 Chemicals
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise.
2.2 Generation of cytochrome c oxidase subunit 7a isoform 1 (heart-type) knockout mice
The Cox7a1 gene is located on chromosome 7 (see NCBI sequence NT_039413.6) and contains three exons (for mRNA sequence see NCBI reference sequence NM_009944.3), which were replaced by homologous recombination with the Neo cassette as part of the pPNT vector (Tybulewicz et al., 1991). Upstream (5’) and downstream (3’) genomic DNA of the Cox7a1 gene was amplified with the Expand Long Template PCR System (Roche, Indianapolis, IN, USA) in conjunction with buffer 3 according to the manufacturer's instructions (all PCRs were 50 μL reactions and nucleotide, primer, and template DNA concentrations were used as recommended). Since the 5’-genomic sequence was not available at the time we generated the knockout mice, 5’-genomic DNA was first amplified by ‘one way PCR’ using Cox7a1 exon I-specific outer primer P1 (5’-CTGGAAGAGCTTCTGCTTCTCTGCCAC-3’) and nested primer P2 (5’-TTCTAAGTGGCTTCTGGTAGATGAGC-3’) together with primers QT, Qinner, and Qouter as described (Hüttemann et al., 2007). PCR fragments were cloned into the pGEM-T Easy vector (Promega, Madison, WI, USA) and sequenced, allowing the design of upstream 5’-forward primers. To generate the 5’-arm for homologous recombination a plasmid clone was used as template DNA (100 ng) with Eco RI restriction site-containing forward primer P3 (5’-TTTTTTGAATTCCCCCGCCCCTC-3’) and reverse primer P2 (touch-up PCR with 2 min initial denaturation at 93 °C; 5 cycles: 30 sec, 93 °C; 30 sec, 50 °C; 2 min, 69 °C; 25 cycles: 30 sec, 93 °C; 30 sec, 60 °C; 2 min, 69 °C), following restriction digestion with EcoR I and Bgl II, the latter of which cleaves internally, resulting in a 1381 bp fragment, which was cloned into the EcoR I/BamH I sites of the pPNT vector (since the Bgl II site of the fragment and the BamH I site of the vector were used for cloning, which contain matching sticky ends, these sites were lost in the targeting vector and the recombinant). To generate the 3’-arm for homologous recombination a 6139 bp fragment was generated with an outer PCR (2 min initial denaturation at 93 °C; 30 cycles: 30 sec, 93 °C; 30 sec, 60 °C; 10 min, 69 °C; 300 ng of total genomic DNA were used as template DNA) using primers P4 forward (5’-GTGACACCAAGAAGCTTGGAGGAC-3’) and P5 reverse (5’-GTCTCTGACCTCTGACCTTCAACC-3’), followed by a nested PCR with 1 μL of a 1:20 dilution of the outer PCR as template and restriction site-containing nested primers P6 forward with a Xho I site (5’-AAACCCGGCTCGAGCCTGTGTGTCTCTG-3’) and P7 reverse with a Not I site (5’-GGAGCATGGCGGCCGCCATGGATGTATCCT-3’) (touch-up PCR with 2 min initial denaturation at 93 °C; 5 cycles: 30 sec, 93 °C; 30 sec, 52 °C; 10 min, 69 °C; 25 cycles: 30 sec, 93 °C; 30 sec, 65 °C; 10 min, 69 °C). The product with the expected size was obtained as judged by agarose gel electrophoresis, and the DNA was digested with Xho I and Not I and cloned into the corresponding restriction sites of the pPNT vector. Correct orientation and sequence were confirmed by sequencing. Finally, vector DNA, which contained the upstream and downstream sequences of the Cox7a1 gene flanking the Neo cassette in the pPNT vector, was linearized with Not I, resulting in a 14,868-bp fragment, and electroporation of R1 mouse embryonic stem cells was performed as a paid service at the Transgenic Animal Model Core (TAMC), University of Michigan (Ann Arbor, MI, USA). Recombinant clones that had undergone homologous recombination in the correct locus were identified using a genotyping protocol (see next section). Positively identified clones were microinjected into C57BL/6J blastocysts at the TAMC. Chimeric animals with a high contribution from the ES cell clone and low contribution from the host embryo as judged by coat color contribution (embryonic stem cells derived from 129 mice produce agouti fur) were selected. Animals were genotyped (see below) and breeding was continued for more than 8 generations using C57BL/6J mice.
2.3. Cox7a1 genotyping
Genomic DNA was isolated from punched ear tissues or mouse tails with the Wizard Genomic DNA Purification Kit (Promega). PCR genotyping was performed using the Expand Long Template PCR System (Roche) in conjunction with buffer 3 according to the manufacturer's instructions (all PCRs were 25 μL reactions with 100 ng template DNA, and nucleotide and primer concentrations were used as recommended). Two separate PCRs were performed to identify the wild-type and the recombinant alleles using primer pairs P8/P9 and P8/P10, respectively: forward primer P8 (5’-CGCCCATTTCACATTCTCAGCACTGGAG-3’), wild-type reverse primer P9 located in Cox7a1 exon I (5’-AAGAGCTTCTGCTTCTCTGCCAC-3’), recombinant-specific reverse primer P10 located in the Neo gene (5’-ACGGTATCGCCGCTCCCGATTCGCAG-3’). PCR conditions were 2 min initial denaturation at 93 °C; 13 cycles: 18 sec, 93 °C; 40 sec, 54 °C; 2 min, 69 °C; 22 cycles: 18 sec, 93 °C; 35 sec, 55 °C; 2 min, 69 °C plus 4 sec per each new cycle. PCR products were loaded and separated on a 1% agarose gel containing ethidium bromide, and wild-type and recombinant PCR products were observed at about 0.7 and 1 kb, respectively.
2.4. Isolation of heart mitochondria
Mice were sacrificed by cervical dislocation and heart mitochondria were isolated aCoxrding to Sayen et al. (Sayen et al., 2003) with minor changes. Briefly, to increase yield two mouse hearts of the same genotype were combined for each isolation and rapidly minced in ice-cold MSE buffer (220 mM mannitol, 70 mM succrose, 2 mM EGTA, 5 mM MOPS (pH 7.4), 2 mM taurine) supplemented with 0.2% fatty acid-free bovine serum albumin. Heart tissue was homogenized with a polytron-type tissue grinder at 11,000 rpm for 2.5 sec, followed by 2 strokes at 500 rpm with a loose Potter-Elvehjem tissue grinder. The homogenate was centrifuged at 500xg for 5 min at 4 °C and the supernatant was collected. The mitochondria were pelleted by centrifugation twice at 3,000×g for 5 min at 4 °C. The final pellet was rinsed and resuspended in 50 μL MSE buffer. Protein concentration was determined using the Bio-Rad Dc protein determination kit (Bio-Rad, Hercules, CA, USA).
2.5. Enzyme activity assays via spectrophotometer
Cytochrome c oxidase (Cox) and citrate synthase (CS) activities were determined in tissue homogenates at 30 °C. For Cox activity measurements, homogenized tissue samples described above for mitochondrial isolation were solubilized with 0.1% (w/v) dodecyl maltoside (DDM). The Cox activity was analyzed in 40 mM phosphate buffer (pH 7.2), the reaction was started by addition of 30 μM reduced cytochrome c, and changes of absorbance at 550 nm were recorded; enzyme activity was calculated using an extinction coefficient of ε = 19.6 mM−1 cm−1 and expressed as nmol oxidized cytochrome c /min/mg protein.
Citrate synthase (CS) activity was analyzed as described (Chowdhury et al., 2000). Briefly, 0.05 mg of homogenized tissue was solubilized with 0.1% of DDM in media containing 100 mM Tris–Cl (pH 8.1), 0.1 mM dithionitrobenzoic acid, and 50 μM acetyl-CoA. The reaction was started with 0.5 mM oxaloacetic acid and changes of absorbance at 412 nm were recorded for 1 min. Enzyme activity was calculated from the absorbance data using an extinction coefficient of ε = 13.6 mM−1 cm−1.
2.6. 2D-gel electrophoresis and Western blot analysis
Blue native (BN)-PAGE (Schagger, 2001) was used for the separation of native mitochondrial OxPhos complexes as described (Klement et al., 1995). Isolated mitochondria were solubilized using 4 g of DDM/g of protein for 15 min on ice in a buffer containing 1.75 M aminocaproic acid, 2 mM EDTA, and 75 mM Bis-Tris (pH 7.0). Samples were centrifuged for 20 min at 20,000xg and Serva Blue G dye was added to the collected supernatant (0.1 g/g DDM). Protein aliquots of the samples were separated on 5-13% (w/v) polyacrylamide gradient minigels. To analyze the small Cox subunits, the first-dimension band of Cox was dissected, incubated for 1h in denaturing buffer (1% (w/v) SDS, 1% (v/v) mercaptoethanol), and separated in the second dimension on a high resolution Kadenbach gel (Tris-glycine SDS-PAGE) containing urea (Kadenbach et al., 1983). The gel was silver stained (Silver staining kit, Bio-Rad).
For standard Western analysis Tricine SDS-PAGE (Schagger and Von Jagow, 1987) was performed on 10% (w/v) polyacrylamide slab minigels using samples denatured for 30 min at 30 °C in sample lysis buffer (2% (v/v) mercaptoethanol, 4% (w/v) SDS, 10mM Tris-Cl and 10% (v/v) glycerol). Proteins were transferred onto PVDF membranes (0.2 μm, Bio-Rad, Hercules, CA) using semi-dry electrotransfer at 0.7 mA/cm2 for 1h. The membranes were blocked in 5% nonfat dry milk (NFDM) in PBS containing Tween 20 (0.1% v/v) (PBST) for 2h, washed with PBST for 10 min, and incubated with primary antibodies. The primary monoclonal antibodies (MitoSciences, Eugene, OR, USA) were diluted in PBST as follows: anti-F1a subunit of ATP synthase (1:5,000), anti-core1 subunit of complex III (1:5,000), anti-Cox subunit I (1:5,000), anti-Cox subunit 4 (1:1,000), anti-SDH 70 kD subunit (1:5000), and anti-NDUFB6 subunit of complex I (1:3,000). We also used monoclonal anti-GAPDH (1:1,000) as cytosolic marker (G8795, Sigma-Aldrich) and polyclonal anti-MnSOD (1:3,000) as oxidative stress marker (ab13534, Abcam, Cambridge, MA, USA). The membranes were washed three times in PBST for 10 min and incubated with secondary antibody (1:10,000 in PBST) for 1h. Anti-F1a, anti-core1, anti-Cox I, anti-Cox4, anti-NDUFB6 and anti-GAPDH were incubated with anti-mouse IgG conjugated with horseradish peroxidase (HRP) and anti-MnSOD with anti-rabbit IgG conjugated with HRP (GE Healthcare, Piscataway, NJ, USA) secondary antibody. The membranes were washed three times in PBST, the signal was detected by the chemiluminescence method (ECL+ kit, GE Healthcare), and band intensities were quantified using the program ImageQuant version 5.1 (Molecular Dynamics, Sunnyvale, CA, USA).
2.7. Polarographic measurements
Cox activity was determined in a closed 200 μL chamber equipped with a micro Clark-type oxygen electrode (Oxygraph system, Hansatech, Norfolk, UK) in 10 mM K-HEPES (pH 7.4), 40 mM KCl, 1% Tween 20, 2 μM oligomycin, 1 mM PMSF, 10 mM KF, and 2 mM EGTA. Tissue samples were sonicated and measurements were performed at 25 °C in the presence of 20 mM ascorbate and increasing amounts of cow heart Cyc from 0-30 μM, and in the presence of 5 mM ADP, an allosteric activator of Cox, or 5 mM ATP, an allosteric inhibitor of Cox, after incubation with an ATP regenerating system, as described (Lee, Salomon, Ficarro, Mathes, Lottspeich, Grossman, Huttemann, 2005). Enzymatic activity is defined as oxygen consumed [μM]/(min·protein [mg]). Protein concentration was determined with the Dc protein assay kit (Bio-Rad).
2.8. ATP assay
The bioluminescent method in conjunction with the boiling method was used to determine ATP levels in heart tissue samples as described (Lee et al., 2005).
2.9. Isolation of mRNA and quantitative PCR
Total RNA was extracted from snap frozen cardiac tissue from 19 week old male mice of each genotype using TRIzol reagent (Invitrogen Corporation, Carlsbad, CA, USA) according to the manufacturer's protocol. DNase was applied to the RNA samples according to the manufacturer's protocol for the RQ1 RNase-Free DNase kit (Promega Corporation). cDNA for each genotype was produced from 2 μg of each mRNA sample according to the ProtoScript AMV First Strand cDNA Synthesis Kit protocol (New England BioLabs, Ipswich, MA, USA) using oligo d(T) primers. Primers were purchased from Integrated DNA Technologies (Coralville, IA, USA) and were analyzed using PrimerQuest and OligoAnalyzer 3.1 (Integrated DNA Technologies). Primers were tested for efficiency with a 1:2 series dilution of wild-type cDNA ranging from 250 ng/μL to 1.0 ng/μL in 25 μL reactions (12.5 μL Brilliant II Sybr Green Master Mix (Stratagene, La Jolla, CA, USA), 5 μL H2O, 100 nM forward and reverse primers, and 2.5 μL template) on a Stratagene MX3000P qPCR machine. Denaturation at 95 °C for 10 min was followed by 40 cycles of amplification, which included denaturation at 95 °C for 30 sec and annealing at 60 °C for 1 min. Threshold cycle (CT) values were determined by a software-calculated threshold for each primer used (Stratagene). Following amplification, a melting curve was performed by raising the temperature from 55 °C to 95 °C and measuring every 0.5 °C interval for fluorescence change. All primer pairs were shown to have reaction efficiencies between 90 and 110% as well as a single specific product, inferred by observation of one peak in melting curve analysis.
Transcript levels of genes of interest from the heterozygous and knockout Cox7a1 genotypes were compared to the Gapdh (Mus musculus glyceraldehyde 3-phosphate dehydrogenase) gene as the reference housekeeping gene in the wild-type genotype as internal standard. Gapdh was shown to be an appropriate reference gene by the 2-ΔCT method (Livak, Schmittgen, 2001), and was the most stable according to geNorm version 3.5 (Vandesompele et al. 2002) out of Gapdh, B2m, Rpl13, and Actb. Primers were: Gapdh forward 5’-ACATCAAGAAGGTGGTGAAGCAGG-3’ and reverse 5’-ATCGAAGGTGGAAGAGTGGGAGTT-3’; Cox7a1 (heart-type) forward 5’-AAAACCGTGTGGCAGAGAAG-3’ and reverse 5-CCAGCCCAAGCAGTATAAGC-3’; Cox7a2 (liver-type) forward 5’-GCTATGGCTGCATTTCCCAAGAAG-3’ and reverse 5’-ACTCATCAGATTCCTGGTCCATCG-3’; cytochrome c somatic (Cycs) forward 5’-AGCCACGCTTTACCCTTCGTTCTT-3’ and reverse 5-CCACTATCACTCATTTCCCTGCCATTCTC-3’; core 1 of complex III (Mt-Co1) forward 5’-AGCAGAATTAGGTCAACCAGGTGC-3’ and reverse 5’-TGGGACAAGTCAGTTTCCAAAGCC-3’; MnSOD (Sod2) forward 5’-GATGTTACAACTCAGGTCGCTCTTC-3’ and reverse 5’-CAGCAACTCTCCTTTGGGTTCTC-3’; complex I Ndufb6 forward 5’-GTCTCTTCGCTGTTTCTCATGTGC-3’ and reverse 5’-TATCCTGGGCTTCGAGCTAACAATGG-3’; Tfam forward 5’-CAGATGGCTGAAGTTGGACGAAGTG-3’ and reverse 5’-GGCTTTGAGACCTAACTGGTTTCTTG-3’. Twenty μL quantitative PCR reactions (10 μL 2X Power SYBR® Master Mix (Applied Biosystems, Foster City, CA, USA), 6.8 μL H2O, 300 nM primers, 2.0 μL cDNA template) were run on the Applied Biosystems StepOnePlus™ Real-Time PCR System (Applied Biosystems) using the standard run protocol. The primer efficiencies on the StepOnePlus were observed to be comparable to those on the Stratagene MX3000P. Each gene of interest was run on all three genotypes in triplicate with a positive control and a no template negative control. Relative levels of transcripts were calculated using the 2-ΔΔCT method (Livak, Schmittgen, 2001), using CT data generated by the StepOnePlus analysis software. All PCR data are presented as mean ± standard deviation.
DNA isolation for comparing the relative amounts of mitochondrial and nuclear DNA was carried out on the cell homogenate fractions left over after RNA isolation, using the TRIzol method according to the manufacturer's instructions (Invitrogen). Relative amounts of mtDNA were determined with primers mMitoF1: CTAGAAACCCCGAAACCAAA and mMitoR1: CCAGCTATCACCAAGCTCGT and compared with endogenous levels of mB2M (NT_039207) using primers mB2MF1: ATGGGAAGCCGAACATACTG and mB2MR1: CAGTCTCAGTGGGGGTGAAT.
2.10. Echocardiography
Cardiac function in mice was determined by echocardiography. Animals were scanned while under 2% isoflurane anesthesia with a 30 MHz frequency probe (Vevo 770, Visual Sonics, Toronto, Ontario, Canada). Image acquisition and measurements and calculations used VisualSonics software (version 2.3). Hemodynamic parameters including heart rate, cardiac output, left ventricular mass, left atrium dimension, ejection fraction, and fractional shortening were acquired in a paraternal long-axis view. Ventricular wall thickness (septal and posterior wall) and internal dimensions were measured using the leading edge technique as recommended by the American Society for Echocardiography (Gardin et al., 2002). For determination of diastolic function, the ratio of Doppler mitral inflow E wave velocity to tissue Doppler left ventricular lateral wall e’ velocity was calculated for each animal. Female wild type mice were used as controls for echo imaging.
2.11. Light microscopy
Routine histology was performed on fresh, 4% paraformaldehyde fixed tissue. Sections were cut at 5 μm and stained with trichrome to especially study the amounts of collagen in the heart.
2.12. Electron microscopy
Cardiac tissue was incubated overnight in Karnovsky's fixative and this was then replaced with Karnovsky's storage buffer. Several 1-mm3 pieces of heart wall were transferred to 70% ethanol and embedded in plastic resin. One-micrometer-thick sections were prepared and stained with toluidine blue. The thin sections were stained with osmium tetroxide and uranylic acid and examined with a Zeiss 910 electron microscope (Seattle, WA, USA). Random pictures of each section were taken at 5000× to count the number of mitochondria and at 1000× to count the number of dark and light fibers. The fine structure of the mitochondria was examined at 31,000×.
3. Results
3.1. Generation of knockout mice
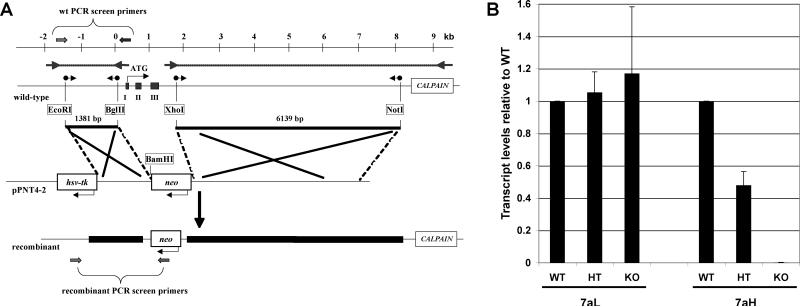
Chimeric animals were obtained after electroporation of our construct (Fig. 1A) into mouse R1 embryonic stem cells. We have replaced exon I, which contains the start ATG, and exons II and III, with the neo cassette. Cox7a1 transcripts could no longer be detected by PCR or qPCR (Fig. 1B) in the knockout. About half the wild-type amount of Cox7a1 was detected in heterozygotes. Cox7a2 transcript levels show an upward trend in heterozygotes and knockouts (Fig. 1B), although not reaching significance. At the protein level, however, a clear increase in Cox7a2 was seen in hearts of knockout animals (see section 3.4 and Fig 4).
Fig. 1. Cox7a1 knockout mouse.
A) Schematic representation of the knockout strategy. The PCR-based amplification of the 3′- and 5′-Cox7a1 regions (outer PCR: arrows with dotted lines) was followed by a nested PCR with primers containing the indicated restriction sites (sphere-tailed arrows). Two fragments are generated, the 800-bp promoter region to the end of intron I (1381-bp fragment), and the 6139-bp region spanning from intron III to the calpain gene. These fragments were cloned into the respective sites of the pPNT vector (pPNT7a1), leading to a replacement of exons I to III by the neo cassette in the recombinant. B) Relative transcript level of Cox7a isoforms in each genotype. Determination by qPCR is relative to Gapdh.
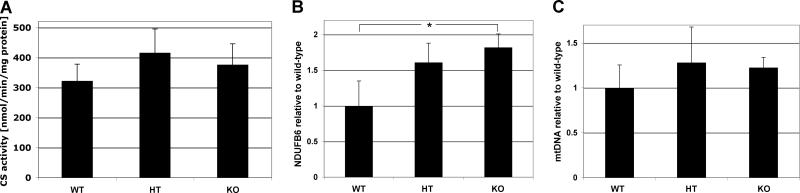
Fig. 4. Mitochondrial components trend upwards in heterozygote and null mice.
A) Citrate synthase (CS) activity was measured as described in Materials and Methods. There was a trend in which CS activity was 29% and 17% increased in the heterozygotes and the Cox7a1 knockouts, respectively. However, differences were not statistically significant (p=0.13 WT-HT; p=0.27, WT-KO) (n=4). B) Ndufb6 gene expression was measured by qPCR as described. *, p<0.05. C) MtDNA content of Cox7a1+/+, Cox7a1+/- and Cox7a1-/- was determined by qPCR as described in Materials and Methods, relative to a single copy nuclear gene (mouse β-2 microglobulin) using total DNA.
3.2. Visual and microscopic characterization of Cox7a1 knockout
The knockout mice look normal, are born in the expected Mendelian ratios, and seem normally active in their cages. There have been 4 spontaneous deaths among about 75 mice (at about 60, 80, 100, and 160 days), which is much higher than the usual incidence of spontaneous death in our mouse colonies but we do not have a database for comparison. The fertility of the mice is comparable (litter size 3.9 ± 1.45; n=10) to the litter size of the C57BL/6J controls in our colony (4.0 ± 1.51; n=8).
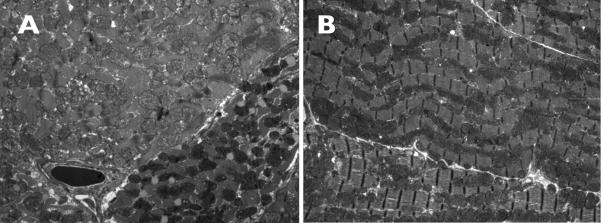
Light microscopy did not reveal any changes in the cardiac tissue. The amount of collagen surrounding vessels was similar in the Cox7a1-/- and the wild types. When examined in the electron microscope (Fig. 2), the surface area and the number of mitochondria per section at constant magnification did not differ between the Cox7a1-/- and control hearts (209.8 ± 36.8, 242.2 ± 70.2; p=0.38, n=5 each). There was no difference in the size of the mitochondria between the two genotypes. However, there was a difference noted in the number of “dark” mitochondria between the two groups: 8.5% dark mitochondria in the wild types and 9.8% in the Cox7a1-/- (Fig. 2). A given fiber is entirely composed of one or the other, i.e., there is no mixing in a given fiber. Dark mitochondria are dense but otherwise unremarkable. Their cristae look the same as those of light mitochondria and they do not have superimposed inclusions. There is no evidence of myocyte degeneration, abnormal glycogen storage, or any other changes to suggest a metabolic problem.
Fig 2. Electron microscope images of cardiac sections.
Tissues from (A) Cox7a1+/+ and (B) Cox7a1-/- mouse hearts were examined in thin sections after staining with osmium tetroxide and uranylic acid as described in Methods. Darker mitochondria (white arrowhead) of unknown etiology were present in the -/- hearts in greater numbers than the more usual structures seen in wild-type sections (white arrow).
3.3. Biochemistry
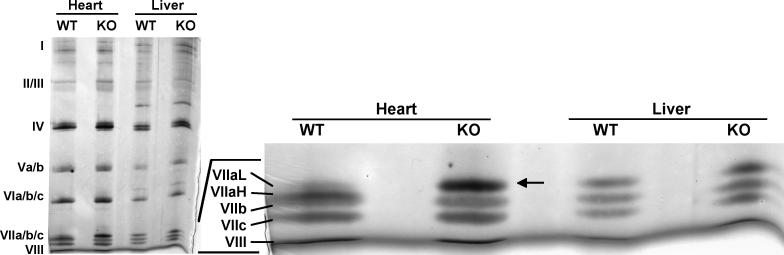
We employed blue native polyacrylamide gel electrophoresis (BN-PAGE) for detailed analysis of the impact of the Cox7a1 knockout on the content and subunit composition of heart Cox. This technique allows for separation of intact multisubunit protein complexes, and thus is ideally suited for study of OxPhos complexes. Initial analysis of the in-gel stained proteins revealed no significant difference between the content of Cox holoenzyme, as well as of other OxPhos complexes, in heart mitochondria of wild-type and knockout mice (data not shown). For determination of Cox subunit composition, we dissected the Cox bands from the first dimension of a native gel and resolved them under denaturing conditions by SDS-PAGE to separate individual subunits. An 18% acrylamide gel containing urea was used to allow for separation of the two Cox7a isoforms, which differ by only 0.3 kDa and normally comigrate. Proteins were visualized by silver staining (Fig. 3). As expected, in heart from wild-type both 7a1 (major portion) and 7a2 (residual amount) were detected. The 7a1 isoform migrated as an apparent double-band with the 7b subunit; the proper migration of 7a2 was confirmed by using liver Cox as an internal control. In contrast, the 7a1 band was completely absent in the Cox from knockouts. Interestingly, the isoform loss was compensated by upregulation of 7a2, which was approximately 5-fold increased compared to wild-type.
Fig. 3. Lack of Cox7a1 (Cox7aH) in the Cox7a1 knockouts is complemented by increased Cox7a2 (Cox7aL) isoform incorporation into the Cox holoenzyme.
For 2D-gel electrophoresis, heart and liver mitochondria were isolated from Cox7a1 knockout (KO) and wild-type mice (WT) from two pooled hearts and a liver each. OxPhos complexes were solubilized using dodecyl maltoside and run on BisTris 5-13% blue native PAGE in the first dimension. Bands corresponding to Cox were dissected and resolved by 6 M urea/18% SDS-PAGE (Lee et al., 2005), optimized for the separation of smaller Cox subunits 7 (VII) a, b, c, and 8 (VIII). Cox subunits were visualized by silver staining. In the absence of Cox7a1 in the knockouts, significantly increased protein levels of the liver-type isoform were observed (arrow).
In addition to the qualitative determination of Cox content by BN-PAGE above, we investigated the amount of mitochondrial proteins by SDS/PAGE and Western blotting. Samples of heart homogenates were evaluated for enzyme activity, probed with antibodies to representative subunits of OxPhos complexes, or examined for transcript level by qPCR (see Materials and Methods). A trend to increased amounts in the heterozygote and null were commonly seen, reaching significance in some cases. Results are shown here for citrate synthase activity (Fig. 4A) as an indicator of mitochondrial mass and transcript levels of Ndufb6 (Fig. 4B). A similar trend was seen for amounts of mtDNA per nuclear genome (Fig. 4C).
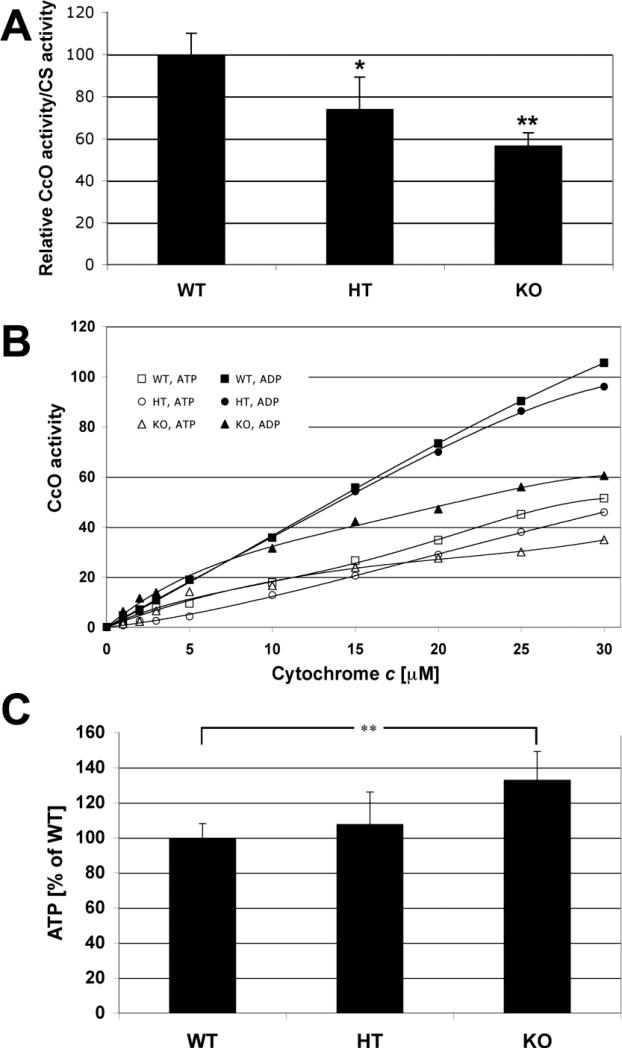
Cox activity was reduced in heterozygote (HT) and null (KO) animals. Measured spectrophotometrically relative to citrate synthase activity, and in the absence of allosteric nucleotides, Cox activity was reduced 26 and 53%, respectively, in HT and KO mice (Fig. 5A). Cox was also analyzed polarographically in heart tissue homogenates with added adenine nucleotides and increasing amounts of cytochrome c. Heart tissue was harvested from 20-week males from the same litter. At maximal turnover, Cox activity in the presence of allosteric inhibitor ATP was decreased by 9% and 32% in the heterozygotes and knockouts, respectively (Fig. 5B, open symbols). In the presence of allosteric activator ADP, heterozygotes and knockout mice showed a 7% and 42% decrease in Cox activity (Fig. 5B, closed symbols). Surprisingly, ATP levels were significantly higher in knockout hearts compared to wild-type (p < 0.01) and appeared intermediate in amount when examined in heterozygotes (Fig. 5C).
Fig. 5. Cox activity and ATP levels in Cox7a1 heterozygous and knockout mice.
A) Cox activity is decreased in hearts of heterozygous and knockout mice. Cox activity of heart homogenates was determined with the spectrophotometric method and standardized to citrate synthase activity. Wild-type (WT) was set to 100%. Heterozygotes (HT) and Cox7a1 knockout mice had 26% and 53% reduced Cox activities, respectively. Note that measurements were performed in the absence of allosteric Cox activity modulators ADP and ATP (n=4; *, p<0.5; **, p<0.01). B) Cox activity is decreased in heart tissue of Cox7a1 knockout mice. Cox activity was determined in heart tissue homogenates of wild-type (WT, squares), heterozygote, (HT, circles), and Cox7a1 knockout mice (KO, triangles) in the presence of allosteric Cox inhibitor ATP (open symbols) and allosteric activator ADP (closed symbols) with the polarographic method by increasing the amount of substrate cytochrome c. Cox activity is defined as consumed O2 [nmol]/min/protein [mg]. Shown are representative measurements (n = 4 each; standard deviation < 5% at maximal turnover). C) ATP levels are increased in the Cox7a1 knockout mice. ATP concentrations of heart median cross-sections were determined with the bioluminescence method. ATP levels are 7% and 33% increased in the heterozygotes (HT) and Cox7a1 knockouts (KO) in comparison to the wild-types (WT), respectively. ATP at 100% is equivalent to 170 μg ATP/mg of solubilized protein (n = 6 animals in each group, measured in triplicates each; **, p < 0.01).
3.4. Echocardiography
Heart weights determined by echocardiography increased significantly, about 15-20% in the knockout at 4-6 months compared to wild type at 4 months of age (Table 2). This weight increase was present by 6–9 weeks: for example, wild type male hearts at 9 weeks showed ECHO-determined left ventricular weights of 87.8 ± 15.6 mg whereas Cox7a1-/- hearts at 6 weeks weighed 115.7 ± 10.9 mg (mean ± SD; p < 0.05).
Table 2.
Comparison of echocardiographic findings in CcO7a1 null (KO) mice at indicated weeks of age and wild-type at 4 weeks.
| WT | KO | |
|---|---|---|
| Age (months) | 4 | 4-6 |
| Number | 4 | 9 |
| FS %* | 27.7 ± 3.1 | 17.7 ± 2.1 |
| EF %* | 54.0 ± 5.0 | 36.9 ± 3.8 |
| LVPWd (mm) | 0.8 ± 0.1 | 0.76 ± 0.04 |
| LV mass (mg)* | 98.4 ± 9.0 | 121.8 ± 7.6 |
| SV (mcl) | 32.7 ± 2.6 | 36.1 ± 3.8 |
| HR (bpm)* | 364.8 ± 15.2 | 329.0 ± 9.7 |
| CO (ml/min) | 11.9 ± 0.7 | 11.8 ± 1.2 |
| LV E/e’* | 4.2 ± 0.2 | 5.0 ± 0.3 |
p < 0.01 WT vs KO; 2-tailed t-test).
FS, fractional shortening; EF, ejection fraction; LVPWd, left ventricular posterior wall dimensions; LV, left ventricle; SV, stroke volume; HR, heart rate; CO, volume of blood ejected into aorta/min; LV E/e’, index of left atrial pressure.
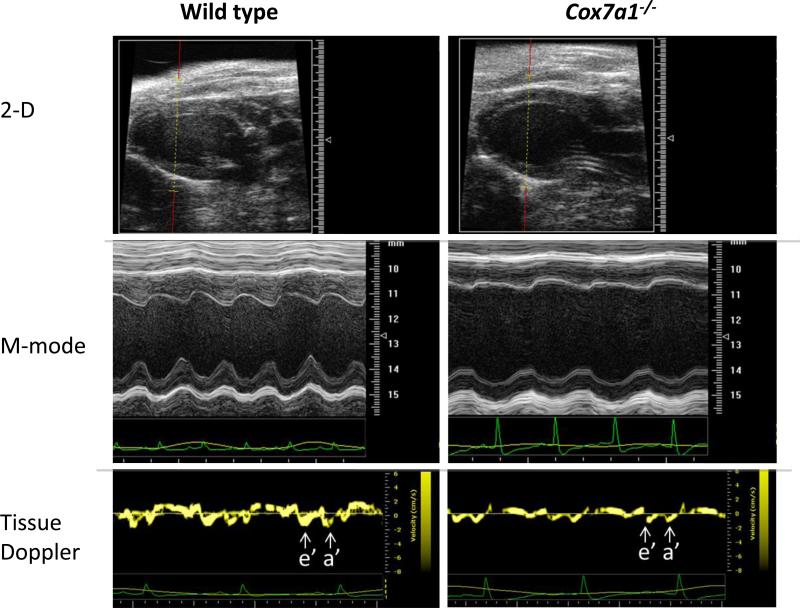
Cardiac echocardiography demonstrated dilated cardiomyopathy and reduced systolic and diastolic function in Cox7a1 KO mice (Fig. 6). At one month of age Cox7a1 KO mice exhibited a 15-20% enlargement of the left ventricular (LV) cavity dimension, which was accompanied by a 1.5-fold higher calculated left ventricular mass (Fig. 6A). LV diastolic dysfunction, as assessed by LV tissue Doppler, was not associated with significant enlargement of the left atrium (Table 2, Fig. 6B). Mice heterozygous for Cox7a1 also displayed dilated cardiomyopathy that was similar in severity to Cox7a1 KO mice (Table 2).
Fig 6. Echo analysis of wild-type and Cox7a1-/- mice.
Two-dimensional and M-mode imaging of the left ventricle in the long-axis view demonstrates LV chamber enlargement and impaired contractility for Cox7a1-/- mice. Spectral Tissue Doppler imaging of the LV lateral wall demonstrates diminished e’ wave velocities and a’ wave for Cox7a1-/- mice, consistent with LV diastolic dysfunction.
Serial echocardiography demonstrated that impaired cardiac systolic and diastolic function tended to improve with age for Cox7a1 null mice (not shown). However, LV ejection fraction and tissue Doppler measurements suggested impairment in all Cox7a1 KO mouse hearts examined by echocardiography. Ejection fraction impairment was also seen in heterozygotes.
4. Discussion
We have shown that homozygous and heterozygous Cox7a1 knockout mice develop a dilated cardiomyopathy at 6 weeks of age, although one that improves by 6 months of age and thereafter stabilizes. In the future this will be further assessed by cardiac monitoring, with and without exercise stress. Since heterozygotes also showed cardiomyopathy, one would expect humans with Cox7A1 mutations to have a dominantly inherited cardiomyopathy. Although the null mice seem to have an increased rate of sporadic death, we show that they are able to survive knockout of “heart/skeletal muscle-type” gene Cox7a1, in part by producing more of the “liver-type” isoform Cox7a2 (Fig. 3), and in part by producing more ATP (Fig. 5C). We note that these mice have not been subjected to any exogenous stress or cardiovascular fitness regimens.
Two features of these mice are surprising and will need to be explored in future work. The first is the production of higher ATP levels in a mouse that is missing the dominant cardiac isoform of Cox subunit 7a (with appropriate overall reduction of Cox activity (Figs. 5A, B)). The second surprising feature is the that KO mice develop cardiomyopathy despite their production of more ATP than wild-type. Energy restriction is known to underlie the development of cardiomyopathy (Huss and Kelly, 2005; Marin-Garcia and Goldenthal, 2008; Burelle et al., 2010). Increased ATP production can result from the relatively modest contribution of an increased number of mitochondria. We indeed observe a trend in these unstressed samples of increased mitochondrial material in heterozygous and knockout animals for most components examined. These include activity of citrate synthase (Fig. 4A), transcripts such as for Ndufb6 (Fig. 4B), MnSOD, core 1, somatic cytochrome c, and Tfam, ROS production (not shown), and relative amount of mtDNA (Fig. 4C). By analogy with ragged red fiber disease, a marked proliferation of mitochondria observed in muscle fibers carrying certain mtDNA mutations (Sarnat and Marin-Garcia, 2005), we suggest that a response to reduced energy production per mitochondrion can elicit mitochondrial proliferation to provide increased net energy production without high membrane potential, with dilation being a space accommodation and therefore an adaptive response. A recent study documented increased ATP production during a novel response to stress of mitochondrial hyperfusion (Tondera et al., 2009). In addition to increased mitochondrial mass as an explanation for increased tissue ATP levels one might speculate that some of the metabolic flux, which is mainly driven by fatty acids in normal heart, is shifted to glycolysis. The development of cardiomyopathy may result from damage accompanying ATP production under stress conditions.
There is one other study that analyzed a mouse line lacking a Cox subunit isoform gene, i.e., heart/skeletal muscle-specific subunit 6a22 (Radford et al., 2002). Cox activity was reduced to 23% compared to wild-type animals. In contrast to our finding of a replacement of 7a1 with 7a2 in the knockouts, the Cox6a2 knockout mice did not show increased levels of isoform 6a1, likely explaining the more severe decrease in Cox activity due to incomplete assembly. Interestingly, Cox6a2 knockout animals did not develop dilated cardiomyopathy, and echocardiograms showed normal left ventricular function (Radford et al., 2002). Using isolated heart preparations the authors found that hearts lacking Cox6a2 could not be subjected to standard perfusion conditions and these had to be omitted. However, using heterozygous animals instead, they showed similar cardiac performance for most parameters they analyzed except for stroke work at high left atrial pressure, which was reduced in hearts derived from Cox6a2+/- animals (Radford et al., 2002).
An interesting disease model utilized a targeted muscle knockout of Cox10, which encodes the protoheme:heme-O-farnesyl transferase required for biosynthesis of heme a, to recapitulate a mitochondrial disease (Diaz et al., 2005). These mice had <5% Cox activity in their muscle at 2.5 months but differed only slightly from wild-type in maximum force development and fatigability, suggesting a significant ability for glycolysis to support energetic load, at least in the short term. Cox10 knockouts die by 5-7 months of age. By contrast, the Cox7a1 knockout mice develop a cardiomyopathy over early times and then show an improvement. This may reflect the more benign nature of this knockout, possibly resulting from the compensating increase of Cox7a2. Taken together, these results may point to a limitation of glycolysis to support energetic load over a longer term.
The heart-type mRNAs of subunits 6a, 7a, and 8 are transcribed in contractile muscle only, whereas the liver-type isoforms are transcribed to various degrees in all tissues including heart and skeletal muscle (Preiss et al., 1994). The expression of the liver-type mRNAs of Cox, including Cox6a1 and Cox7a2, is regulated post-transcriptionally by binding of glutamate dehydrogenase to the 3’-untranslated region of the mRNAs (Preiss et al., 1994; Schillace et al., 1994; Preiss et al., 1995). Since the 3’-mRNA sequence regions required for glutamate dehydrogenase binding do not show significant homology among subunits 6a, 7a, and 8 (Preiss and Lightowlers, 1993), it is possible that post-transcriptional regulation affects subunits 6a and 7a differently, perhaps allowing more 7a2 protein to be translated and incorporated into the holoenzyme.
Loss of Cox7a1 leads to a subunit switch in Cox, resulting in a non-canonical “hybrid” enzyme containing two heart isoforms (6a and 8) and a liver isoform of subunit 7a. Our model, therefore, represents a valuable tool for study of the role of tissue-specific isoforms of Cox since this combination does not normally occur. Mammalian Cox functions as a dimer (Tsukihara et al., 1995). It is not clear in the wild-type holoenzyme whether the Cox dimer contains both 7a isoforms of the same tissue type or whether a mixed dimer (hybrid) can be formed. An interesting possibility would be regulatory control by forming the dimer mixture.
The apparent stabilization in older mice of the dilated cardiomyopathy may be related to the increased Cox7a2 expression in the knockouts. This is possible because both rodent (Kuhn-Nentwig and Kadenbach, 1985b; Kuhn-Nentwig and Kadenbach, 1985a) and human (Van Kuilenburg et al., 1992; Bonne et al., 1993) heart contain liver- and heart-type isoforms as the minor and major fraction, respectively. Interestingly, cow, sheep, dog, and rabbit heart only expresses the heart-type isoform (Linder et al., 1995). However, despite the increased Cox7a2 expression in the knockouts, the systolic and diastolic function of all Cox7a1 null mice is impaired. It is interesting to consider that the mildly reduced ejection fraction and impaired diastolic function observed in these mice could provide a clinically relevant model for examining idiopathic dilated cardiomyopathy in humans.
Supplementary Material
Highlights.
Nuclear Cox subunit Cox7a1 knockout leads to dilated cardiomyopathy
Non-tissue specific subunit Cox7a2 increases to partially compensate
Knockout shows reduced Cox activity but increased ATP production
Acknowledgements
We thank Joanna Poulton (Oxford) for helpful discussions, Silke Suer for help with mtDNA qPCR, and Afshan Malik (Kings College London) for the mtDNA and mB2M PCR primer sequences.
Sources of funding
This work was supported by a grant supplement from the National Institutes of Health (GM48517; LIG), the Center for Molecular Medicine and Genetics, Wayne State University, Detroit (MH), and a grant from the National Institutes of Health (GM089900; MH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Non-standard Abbreviations and Acronyms
Cox, cytochrome c oxidase; OxPhos, oxidative phosphorylation; ETC, electron transport chain
Note that designation of the “heart/muscle” isoform for subunits 6a and 7a is reversed, e.g., 6a2 and 7a1 are the heart-type isoforms.
References
- Acin-Perez R, Bayona-Bafaluy MP, Bueno M, Machicado C, Fernandez-Silva P, Perez-Martos A, Montoya J, Lopez-Perez MJ, Sancho J, Enriquez JA. An intragenic suppressor in the cytochrome c oxidase I gene of mouse mitochondrial DNA. Hum. Mol. Genet. 2003;12(3):329–339. doi: 10.1093/hmg/ddg021. [DOI] [PubMed] [Google Scholar]
- Arber S, Hunter JJ, Ross J, Jr., Hongo M, Sansig G, Borg J, Perriard J-C, Chien KR, Caroni P. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88(3):393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- Bonne G, Seibel P, Possekel S, Marsac C, Kadenbach B. Expression of human cytochrome-c oxidase subunits during fetal development. Eur. J. Biochem. 1993;217(3):1099–1107. doi: 10.1111/j.1432-1033.1993.tb18342.x. [DOI] [PubMed] [Google Scholar]
- Burelle Y, Khairallah M, Ascah A, Allen BG, Deschepper CF, Petrof BJ, Des Rosiers C. Alterations in mitochondrial function as a harbinger of cardiomyopathy: Lessons from the dystrophic heart. J. Mol. Cell. Cardiol. 2010;48(2):310–321. doi: 10.1016/j.yjmcc.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casali C, Damati G, Bernucci P, Debiase L, Autore C, Santorelli FM, Coviello D, Gallo P. Maternally inherited cardiomyopathy: Clinical and molecular characterization of a large kindred harboring the A4300G point mutation in mitochondrial deoxyribonucleic acid. J. Amer. Coll. Cardiol. 1999;33(6):1584–1589. doi: 10.1016/s0735-1097(99)00079-0. [DOI] [PubMed] [Google Scholar]
- Chowdhury SK, Drahota Z, Floryk D, Calda P, Houstek J. Activities of mitochondrial oxidative phosphorylation enzymes in cultured amniocytes. Clin. Chim. Acta. 2000;298(1-2):157–173. doi: 10.1016/s0009-8981(00)00300-4. [DOI] [PubMed] [Google Scholar]
- Conrad M, Jakupoglu C, Moreno SG, Lippl S, Banjac A, Schneider M, Beck H, Hatzopoulos AK, Just U, Sinowatz F, Schmahl W, Chien KR, Wurst W, Bornkamm GW, Brielmeier M. Essential role for mitochondrial thioredoxin reductase in hematopoiesis, heart development, and heart function. Mol. Cell. Biol. 2004;24(21):9414–9423. doi: 10.1128/MCB.24.21.9414-9423.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coral-Vazquez R, Cohn RD, Moore SA, Hill JA, Weiss RM, Davisson RL, Straub V, Barresi R, Bansal D, Hrstka RF, Williamson R, Campbell KP. Disruption of the sarcoglycan-sarcospan complex in vascular smooth muscle: A novel mechanism for cardiomyopathy and muscular dystrophy. Cell. 1999;98(4):465–474. doi: 10.1016/s0092-8674(00)81975-3. [DOI] [PubMed] [Google Scholar]
- Dalakas MC, Park K-Y, Semino-Mora C, Lee HS, Sivakumar K, Goldfarb LG. Desmin myopathy, a skeletal myopathy with cardiomyopathy caused by mutations in the desmin gene. New Eng. J. Med. 2000;342(11):770–780. doi: 10.1056/NEJM200003163421104. [DOI] [PubMed] [Google Scholar]
- Deschauer M, Muller T, Wieser T, Schulte-Mattler W, Kornhuber M, Zierz S. Hearing impairment is common in various phenotypes of the mitochondrial DNA A3243G mutation. Arch. Neurol. 2001;58(11):1885–1888. doi: 10.1001/archneur.58.11.1885. [DOI] [PubMed] [Google Scholar]
- Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat. Genet. 2009;41(2):187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz F, Thomas CK, Garcia S, Hernandez D, Moraes CT. Mice lacking cox10 in skeletal muscle recapitulate the phenotype of progressive mitochondrial myopathies associated with cytochrome c oxidase deficiency. Hum. Mol. Genet. 2005;14(18):2737–2748. doi: 10.1093/hmg/ddi307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatkin D, Macrae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ, Jr., Spudich S, De Girolami U, Seidman JG, Seidman CE. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. New Eng. J. Med. 1999;341(23):1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- Gardin JM, Adams DB, Douglas PS, Feigenbaum H, Forst DH, Fraser AG, Grayburn PA, Katz AS, Keller AM, Kerber RE, Khandheria BK, Klein AL, Lang RM, Pierard LA, Quinones MA, Schnittger I. Recommendations for a standardized report for adult transthoracic echocardiography: A report from the american society of echocardiography's nomenclature and standards committee and task force for a standardized echocardiography report. J. Am. Soc. Echocardiogr. 2002;15(3):275–290. doi: 10.1067/mje.2002.121536. [DOI] [PubMed] [Google Scholar]
- Geisterfer-Lowrance AA, Christe M, Conner DA, Ingwall JS, Schoen F, Seidman CE, Seidman JG. A mouse model of familial hypertrophic cardiomyopathy. Science. 1996;272(5262):731–734. doi: 10.1126/science.272.5262.731. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Imanaka-Yoshida K, Yoshida T, Wood M, Fearns C, Tatake RJ, Lee JD. A crucial role of mitochondrial HSP40 in preventing dilated cardiomyopathy. Nat. Med. 2006;12(1):128–132. doi: 10.1038/nm1327. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kelly DP. Mitochondrial energy metabolism in heart failure: A question of balance. J. Clin. Invest. 2005;115(3):547–555. doi: 10.1172/JCI200524405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttemann M, Lee I, Liu J, Grossman LI. Transcription of mammalian cytochrome c oxidase subunit IV-2 is controlled by a novel conserved oxygen responsive element. FEBS J. 2007;274(21):5737–5748. doi: 10.1111/j.1742-4658.2007.06093.x. [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Jarausch J, Hartmann R, Merle P. Separation of mammalian cytochrome c oxidase into 13 polypeptides by a sodium dodecyl sulfate-gel electrophoresis procedure. Anal. Biochem. 1983;129(2):517–521. doi: 10.1016/0003-2697(83)90586-9. [DOI] [PubMed] [Google Scholar]
- Kadenbach B, Kuhn-Nentwig L, Buge U. Evolution of a regulatory enzyme: Cytochrome-c oxidase (complex IV). Current Topics in Bioenergetics. 1987;15:114–162. [Google Scholar]
- Kamisago M, Sharma SD, Depalma SR, Solomon S, Sharma P, Mcdonough B, Smoot L, Mullen MP, Woolf PK, Wigle ED, Seidman JG, Seidman CE. Mutations in sarcomere protein genes as a cause of dilated cardiomyopathy. New Eng. J. Med. 2000;343(23):1688–1696. doi: 10.1056/NEJM200012073432304. [DOI] [PubMed] [Google Scholar]
- Klement P, Nijtmans LG, Van Den Bogert C, Houstek J. Analysis of oxidative phosphorylation complexes in cultured human fibroblasts and amniocytes by blue-native-electrophoresis using mitoplasts isolated with the help of digitonin. Anal. Biochem. 1995;231(1):218–224. doi: 10.1006/abio.1995.1523. [DOI] [PubMed] [Google Scholar]
- Knoll R, Hoshijima M, Hoffman HM, Person V, Lorenzen-Schmidt I, Bang ML, Hayashi T, Shiga N, Yasukawa H, Schaper W, Mckenna W, Yokoyama M, Schork NJ, Omens JH, Mcculloch AD, Kimura A, Gregorio CC, Poller W, Schaper J, Schultheiss HP, Chien KR. The cardiac mechanical stretch sensor machinery involves a Z disc complex that is defective in a subset of human dilated cardiomyopathy. Cell. 2002;111(7):943–955. doi: 10.1016/s0092-8674(02)01226-6. [DOI] [PubMed] [Google Scholar]
- Kuhn-Nentwig L, Kadenbach B. Isolation and properties of cytochrome c oxidase from rat liver and quantification of immunological differences between isozymes from various rat tissues with subunit-specific antisera. Eur. J. Biochem. 1985a;149(1):147–158. doi: 10.1111/j.1432-1033.1985.tb08905.x. [DOI] [PubMed] [Google Scholar]
- Kuhn-Nentwig L, Kadenbach B. Orientation of rat liver cytochrome c oxidase subunits investigated with subunit-specific antisera. Eur. J. Biochem. 1985b;153(1):101–104. doi: 10.1111/j.1432-1033.1985.tb09273.x. [DOI] [PubMed] [Google Scholar]
- Lee I, Kadenbach B. Palmitate decreases proton pumping of liver-type cytochrome c oxidase. Eur. J. Biochem. 2001;268(24):6329–6334. doi: 10.1046/j.0014-2956.2001.02602.x. [DOI] [PubMed] [Google Scholar]
- Lee I, Pecinova A, Pecina P, Neel BG, Araki T, Kucherlapati R, Roberts AE, Huttemann M. A suggested role for mitochondria in Noonan syndrome. Biochim. Biophys. Acta. 2010;1802(2):275–283. doi: 10.1016/j.bbadis.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Salomon AR, Ficarro S, Mathes I, Lottspeich F, Grossman LI, Huttemann M. cAMP-dependent tyrosine phosphorylation of subunit I inhibits cytochrome c oxidase activity. J. Biol. Chem. 2005;280(7):6094–6100. doi: 10.1074/jbc.M411335200. [DOI] [PubMed] [Google Scholar]
- Lenka N, Vijayasarathy C, Mullick J, Avadhani NG. Structural organization and transcription regulation of nuclear genes encoding the mammalian cytochrome c oxidase complex. Prog. Nucleic Acid Res. Mol. Biol. 1998;61:309–344. doi: 10.1016/s0079-6603(08)60830-2. [DOI] [PubMed] [Google Scholar]
- Li Z, Colucci-Guyon E, Pincon-Raymond M, Mericskay M, Pournin S, Paulin D, Babinet C. Cardiovascular lesions and skeletal myopathy in mice lacking desmin. Dev. Biol. 1996;175(2):362–366. doi: 10.1006/dbio.1996.0122. [DOI] [PubMed] [Google Scholar]
- Linder D, Freund R, Kadenbach B. Species-specific expression of cytochrome c oxidase isozymes. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 1995;112(3):461–469. doi: 10.1016/0305-0491(95)00093-3. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-ΔΔc(t)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lomax MI, Grossman LI. Tissue-specific genes for respiratory proteins. Trends Biochem. Sci. 1989;14(12):501–503. doi: 10.1016/0968-0004(89)90185-0. [DOI] [PubMed] [Google Scholar]
- Luo W, Grupp IL, Harrer J, Ponniah S, Grupp G, Duffy JJ, Doetschman T, Kranias EG. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ. Res. 1994;75(3):401–409. doi: 10.1161/01.res.75.3.401. [DOI] [PubMed] [Google Scholar]
- Marin-Garcia J, Goldenthal MJ. Mitochondrial centrality in heart failure. Heart Fail. Rev. 2008;13(2):137–150. doi: 10.1007/s10741-007-9079-1. [DOI] [PubMed] [Google Scholar]
- Mounkes LC, Kozlov SV, Rottman JN, Stewart CL. Expression of an LMNAN195K variant of A-type lamins results in cardiac conduction defects and death in mice. Hum. Mol. Genet. 2005;14(15):2167–2180. doi: 10.1093/hmg/ddi221. [DOI] [PubMed] [Google Scholar]
- Muthuchamy M, Pieples K, Rethinasamy P, Hoit B, Grupp IL, Boivin GP, Wolska B, Evans C, Solaro RJ, Wieczorek DF. Mouse model of a familial hypertrophic cardiomyopathy mutation in alpha-tropomyosin manifests cardiac dysfunction. Circ. Res. 1999;85(1):47–56. doi: 10.1161/01.res.85.1.47. [DOI] [PubMed] [Google Scholar]
- Nahrendorf M, Spindler M, Hu K, Bauer L, Ritter O, Nordbeck P, Quaschning T, Hiller KH, Wallis J, Ertl G, Bauer WR, Neubauer S. Creatine kinase knockout mice show left ventricular hypertrophy and dilatation, but unaltered remodeling post-myocardial infarction. Cardiovasc. Res. 2005;65(2):419–427. doi: 10.1016/j.cardiores.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Olson TM, Kishimoto NY, Whitby FG, Michels VV. Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J. Mol. Cell. Cardiol. 2001;33(4):723–732. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- Olson TM, Michels VV, Ballew JD, Reyna SP, Karst ML, Herron KJ, Horton SC, Rodeheffer RJ, Anderson JL. Sodium channel mutations and susceptibility to heart failure and atrial fibrillation. JAMA. 2005;293(4):447–454. doi: 10.1001/jama.293.4.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL, Vandenberg JI, Colledge WH, Grace AA. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene SCN5A. Proc. Natl. Acad. Sci. U. S. A. 2002;99(9):6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Chrzanowska-Lightowlers ZMA, Lightowlers RN. The tissue-specific rna-binding protein COLBP is differentially regulated during myogenesis. Biochim. Biophys. Acta. 1994;1221(3):286–289. doi: 10.1016/0167-4889(94)90252-6. [DOI] [PubMed] [Google Scholar]
- Preiss T, Lightowlers RN. Post-transcriptional regulation of tissue-specific isoforms - a bovine cytosolic RNA-binding protein, COLBP, associates with messenger RNA encoding the liver-form isopeptides of cytochrome-c oxidase. J. Biol. Chem. 1993;268(14):10659–10667. [PubMed] [Google Scholar]
- Preiss T, Sang AE, Chrzanowskalightowlers ZMA, Lightowlers RN. The mRNA-binding protein COLBP is glutamate dehydrogenase. FEBS Lett. 1995;367(3):291–296. doi: 10.1016/0014-5793(95)00569-u. [DOI] [PubMed] [Google Scholar]
- Poetter K, Jiang H, Hassanzadeh S, Master SR, Chang A, Dalakas MC, Rayment I, Sellers, Fananapazir L, Epstein ND. Mutations in either the essential or regulatory light chains of myosin are associated with a rare myopathy in human heart and skeletal muscle. Nat Genet. 1996;13:63–9. doi: 10.1038/ng0596-63. [DOI] [PubMed] [Google Scholar]
- Radford NB, Wan B, Richman A, Szczepaniak LS, Li JL, Li K, Pfeiffer K, Schagger H, Garry DJ, Moreadith RW. Cardiac dysfunction in mice lacking cytochrome-c oxidase subunit VIaH. Am. J. Physiol. Heart Circ. Physiol. 2002;282(2):H726–H733. doi: 10.1152/ajpheart.00308.2001. [DOI] [PubMed] [Google Scholar]
- Radke MH, Peng J, Wu Y, Mcnabb M, Nelson OL, Granzier H, Gotthardt M. Targeted deletion of titin N2B region leads to diastolic dysfunction and cardiac atrophy. Proc. Natl. Acad. Sci. U. S. A. 2007;104(9):3444–3449. doi: 10.1073/pnas.0608543104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarnat HB, Marin-Garcia J. Pathology of mitochondrial encephalomyopathies. Can. J. Neurol. Sci. 2005;32(2):152–166. doi: 10.1017/s0317167100003929. [DOI] [PubMed] [Google Scholar]
- Satoh M, Takahashi M, Sakamoto T, Hiroe M, Marumo F, Kimura A. Structural analysis of the titin gene in hypertrophic cardiomyopathy: Identification of a novel disease gene. Biochem. Biophys. Res. Commun. 1999;262(2):411–417. doi: 10.1006/bbrc.1999.1221. [DOI] [PubMed] [Google Scholar]
- Sayen MR, Gustafsson AB, Sussman MA, Molkentin JD, Gottlieb RA. Calcineurin transgenic mice have mitochondrial dysfunction and elevated superoxide production. Am J Physiol Cell Physiol. 2003;284(2):C562–570. doi: 10.1152/ajpcell.00336.2002. [DOI] [PubMed] [Google Scholar]
- Schagger H. Blue-native gels to isolate protein complexes from mitochondria. Methods Cell Biol. 2001;65:231–244. doi: 10.1016/s0091-679x(01)65014-3. [DOI] [PubMed] [Google Scholar]
- Schagger H, Von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 1987;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Scheibye-Knudsen M, Quistorff B. Regulation of mitochondrial respiration by inorganic phosphate; comparing permeabilized muscle fibers and isolated mitochondria prepared from type-1 and type-2 rat skeletal muscle. Eur. J. Appl. Physiol. 2009;105(2):279–287. doi: 10.1007/s00421-008-0901-9. [DOI] [PubMed] [Google Scholar]
- Schillace R, Preiss T, Lightowlers RN, Capaldi RA. Developmental regulation of tissue-specific isoforms of subunit VIa of beef cytochrome c oxidase. Biochim. Biophys. Acta. 1994;1188(3):391–397. doi: 10.1016/0005-2728(94)90060-4. [DOI] [PubMed] [Google Scholar]
- Schmitt JP, Kamisago M, Asahi M, Li GH, Ahmad F, Mende U, Kranias EG, Maclennan DH, Seidman JG, Seidman CE. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299(5611):1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Silvestri G, Santorelli FM, Shanske S, Whitley CB, Schimmenti LA, Smith SA, DiMauro S. A new mtDNA mutation in the tRNA(leu(uur)) gene associated with maternally inherited cardiomyopathy. Hum. Mutat. 1994;3(1):37–43. doi: 10.1002/humu.1380030107. [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J. Biol. Chem. 2002;277(47):44784–44790. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- Szczesna-Cordary D, Guzman G, Zhao J, Hernandez O, Wei J, Diaz-Perez Z. The E22K mutation of myosin RLC that causes familial hypertrophic cardiomyopathy increases calcium sensitivity of force and ATPase in transgenic mice. J Cell Sci. 2005;118:3675–83. doi: 10.1242/jcs.02492. [DOI] [PubMed] [Google Scholar]
- Tardiff JC, Factor SM, Tompkins BD, Hewett TE, Palmer BM, Moore RL, Schwartz S, Robbins J, Leinwand LA. A truncated cardiac troponin T molecule in transgenic mice suggests multiple cellular mechanisms for familial hypertrophic cardiomyopathy. J. Clin. Invest. 1998;101(12):2800–2811. doi: 10.1172/JCI2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierfelder L, Watkins H, Macrae C, Lamas R, Mckenna W, Vosberg HP, Seidman JG, Seidman CE. Alpha-tropomyosin and cardiac troponin T mutations cause familial hypertrophic cardiomyopathy: A disease of the sarcomere. Cell. 1994;77(5):701–712. doi: 10.1016/0092-8674(94)90054-x. [DOI] [PubMed] [Google Scholar]
- Tondera D, Grandemange S, Jourdain A, Karbowski M, Mattenberger Y, Herzig S, Da Cruz S, Clerc P, Raschke I, Merkwirth C, Ehses S, Krause F, Chan DC, Alexander C, Bauer C, Youle R, Langer T, Martinou JC. SLP-2 is required for stress-induced mitochondrial hyperfusion. EMBO J. 2009;28(11):1589–1600. doi: 10.1038/emboj.2009.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubata S, Bowles KR, Vatta M, Zintz C, Titus J, Muhonen L, Bowles NE, Towbin JA. Mutations in the human delta-sarcoglycan gene in familial and sporadic dilated cardiomyopathy. J. Clin. Invest. 2000;106(5):655–662. doi: 10.1172/JCI9224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science. 1995;269(5227):1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC. Neonatal lethality and lymphopenia in mice with a homozygous disruption of the c-abl protooncogene. Cell. 1991;65(7):1153–1163. doi: 10.1016/0092-8674(91)90011-m. [DOI] [PubMed] [Google Scholar]
- Van Kuilenburg ABP, Van Beeumen JJ, Van Der Meer NM, Muijsers AO. Subunits-VIIa,b,c of human cytochrome-c oxidase - identification of both heart-type and liver-type isoforms of subunit-VIIa in human heart. Eur. J. Biochem. 1992;203(1-2):193–199. doi: 10.1111/j.1432-1033.1992.tb19847.x. [DOI] [PubMed] [Google Scholar]
- Vatta M, Mohapatra B, Jimenez S, Sanchez X, Faulkner G, Perles Z, Sinagra G, Lin JH, Vu TM, Zhou Q, Bowles KR, Di Lenarda A, Schimmenti L, Fox M, Chrisco MA, Murphy RT, Mckenna W, Elliott P, Bowles NE, Chen J, Valle G, Towbin JA. Mutations in Cypher/ZASP in patients with dilated cardiomyopathy and left ventricular non-compaction. J. Am. Coll. Cardiol. 2003;42(11):2014–2027. doi: 10.1016/j.jacc.2003.10.021. [DOI] [PubMed] [Google Scholar]
- Villani G, Greco M, Papa S, Attardi G. Low reserve of cytochrome c oxidase capacity in vivo in the respiratory chain of a variety of human cell types. J. Biol. Chem. 1998;273(48):31829–31836. doi: 10.1074/jbc.273.48.31829. [DOI] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology. 2002;3(7):research0034.1–0034.11. doi: 10.1186/gb-2002-3-7-research0034. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Cheng H, Li X, Zhang J, Cui L, Ouyang K, Han L, Zhao T, Gu Y, Dalton ND, Bang ML, Peterson KL, Chen J. Cardiac-specific ablation of Cypher leads to a severe form of dilated cardiomyopathy with premature death. Hum. Mol. Genet. 2009;18(4):701–713. doi: 10.1093/hmg/ddn400. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.