Abstract
Portable, low-cost and quantitative detection of a broad range of targets at home and in the field has the potential to revolutionize medical diagnostics and environmental monitoring. Despite many years of research, very few such devices are commercially available. Taking advantage of the wide availability and low cost of the pocket-sized personal glucose meter—used worldwide by diabetes sufferers—we demonstrate a method to use such meters to quantify non-glucose targets, ranging from a recreational drug (cocaine, 3.4 μM detection limit) to an important biological cofactor (adenosine, 18 μM detection limit), to a disease marker (interferon-gamma of tuberculosis, 2.6 nM detection limit) and a toxic metal ion (uranium, 9.1 nM detection limit). The method is based on the target-induced release of invertase from a functional-DNA–invertase conjugate. The released invertase converts sucrose into glucose, which is detectable using the meter. The approach should be easily applicable to the detection of many other targets through the use of suitable functional-DNA partners (aptamers DNAzymes or aptazymes).
The development of portable sensors for rapid, on-site and cost-effective detection of a broad range of targets has long been sought, because such sensors have the potential to revolutionize scientific research, environmental monitoring and personal healthcare in urban areas in developed countries as well as rural areas in developing countries1–10. Despite their great promise and many years of investigation, only a limited number of sensors are commercially available to the public at present. Perhaps the most successful example of such a sensor is the personal glucose meter (PGM)11,12, which is widely available in stores and has either saved or improved the quality of lives of millions of diabetic patients worldwide. However, it can only detect a single target, blood glucose. In this Article, we report a novel methodology that uses a commercially available PGM, but link it with functional DNA sensors to detect and quantify a broad range of non-glucose targets involved in diagnosis and environmental monitoring. These targets range from recreational drugs such as cocaine to important biological cofactors such as adenosine, as well as disease markers (for example, interferon-γ for tuberculosis) and toxic metal ions (for example, uranium).
The wide success of PGMs is largely due to their portable ‘pocket’ size, low cost, reliable quantitative results and simple operation. With their recent integration into mobile phones13 (for an example of a glucose meter integrated with the Apple iPhone, see http://ibgstar.com/web/ibgstar), PGMs are likely to find an even wider base of users. Although a number of other methods have been developed to quantitatively detect targets of interest using simple instruments, most such sensing devices have not been able to match the PGM in its wide commercial availability to the public. However, the PGM is still limited in that it can detect only a single target, glucose.
A major challenge in using a PGM as a portable meter for targets beyond glucose is to find a method that links the detection of glucose to that of other targets with a general method, while also avoiding changing the design and manufacturing process of state-of-the-art PGMs, because more than 30 years of scientific research and engineering have been devoted to making PGMs that are easy to use, with high performance and low cost. PGMs detect glucose by measuring the redox reaction of either glucose oxidase or dehydrogenase. The detection of other targets is possible if these enzymes can be replaced by those selective for other target analytes14. This is difficult, however, as enzymes selective for every possible target may not be available and it may also require modifications of the PGM. An additional challenge is that PGMs are designed to monitor glucose in blood and have a dynamic range of ~10–600 mg dl−1 or ~0.6–33 mM glucose12. As this is much higher than the nanomolar or micromolar concentration ranges of most targets relevant to medical diagnostics or environmental monitoring, a highly efficient signal amplification procedure with enhancement up to ×106 is thus required. In addition, for public use, the amplification must proceed without any laboratory-based devices or PGM add-ons.
To meet these challenges, we propose the use of functional-DNA-conjugated invertase to link glucose detection to the detection of other targets, and use the concentration of glucose to quantify the other targets of interest present in the samples. Functional DNAs include DNAzymes (also called deoxyribozymes, catalytic DNAs or DNA enzymes), which act as catalysts15,16, aptamers (which selectively bind targets17) and aptazymes (which are a combination of the two18). They are members of the functional nucleic acid family, which also includes RNA-based ribozymes, aptamers and aptazymes. The functional nucleic acids are selected from libraries of DNA or RNA with up to 1 × 1015 random sequences by means of a process known as in vitro selection15,16 or the systematic evolution of ligands by exponential enrichment (SELEX)17. The targets of functional DNA/RNAs can range from metal ions and small organic molecules to biomolecules, and even viruses or cells19, making functional DNA/RNA a general platform for recognizing a broad range of targets. Many functional DNA/RNA sensors have been developed for a broad range of targets20–22, and use laboratory-based devices to generate signal outputs including fluorescence23–27, colorimetry28–30, electrochemistry31–33 and magnetic resonance34. Colorimetric sensors28–30, including dipstick tests35, have been developed for qualitative or semi-quantitative detection that requires no instrumentation, similar to the ubiquitous enzyme-linked immunosorbent assay (ELISA) method widely used in diagnostics. However, numerous targets of interest require quantitative information, and laboratory-based devices are usually needed to provide this information. PGMs would be ideal alternatives to laboratory-based devices in detecting and quantifying these targets at home and in the field, but a link between glucose concentration and other target concentrations must be established before PGMs can be used.
Invertase (β-fructofuranosidase) can provide this link. It is an enzyme that catalyses the hydrolysis of sucrose into fructose and glucose. Invertase can be used to link glucose to other target concentrations because sucrose is completely inert in a PGM, and the major hydrolysed product of sucrose—glucose—is detectable by the PGM. A direct relationship can therefore be established between the concentrations of invertase and glucose. More importantly, because of its highly efficient enzymatic turnover, even nano-molar levels of invertase are capable of converting a millimolar concentration of sucrose into glucose under ambient conditions, making it an ideal catalyst for the amplification of ‘turn-on’ signals in the PGM.
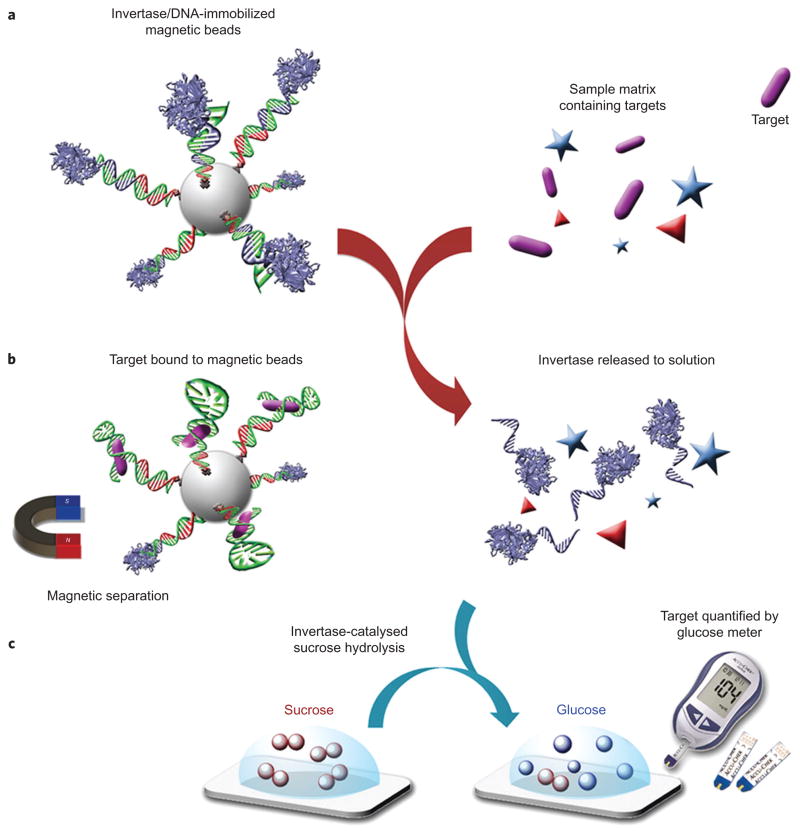
The next challenge is to establish the relationship between target concentration and invertase concentration. As illustrated in Fig. 1, this challenge can be met by designing a mechanism of target-induced release of DNA–invertase conjugate from functional DNA sequences that are immobilized on magnetic beads (see Supplementary Scheme S1 and Figs S1 and S2 for detailed synthesis and characterization of the DNA–invertase conjugate36, as well as experimental procedures for the preparations of sensors for target detection).
Figure 1.
Schematic of method using a PGM to detect a wide range of targets beyond glucose. a, DNA–invertase conjugates are immobilized onto magnetic beads (MBs) by DNA hybridization with a functional DNA that can specifically respond to the target of interest. b, Following addition of the target, interaction between the functional DNA and its target perturbs the DNA hybridization and causes the release of DNA–invertase conjugates from MBs into solution. c, After removal of MBs by a magnet, the DNA–invertase conjugates in solution can efficiently catalyse the hydrolysis of sucrose into glucose, which is quantified by a glucose meter. Because the concentration of DNA–invertase conjugate released into solution is proportional to the concentration of target present in the sample, the readout in the PGM can be used to quantify the target concentration. (More than one DNA strand is conjugated to each invertase, but the scheme here shows only one DNA strand on each invertase for reasons of clarity.).
Cocaine aptamer-based sensor
Sensor performance
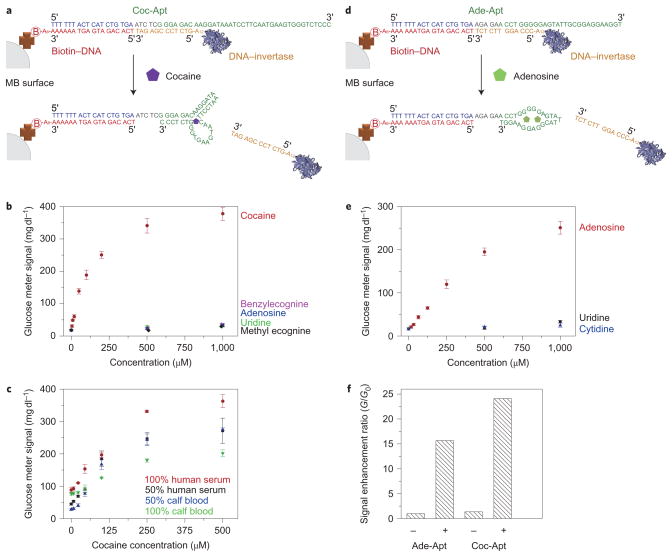
To test the ability of the PGM-based sensor to detect a broad range of targets, we first demonstrated the detection of cocaine using a cocaine aptamer33,37. Cocaine is an addictive drug, and its detection is important to prevent drug abuse in the workplace and to deter international drug trafficking. As shown in Fig. 2a, the biotin–DNA is immobilized onto magnetic beads (MBs) by means of streptavidin–biotin binding, and the DNA–invertase conjugate synthesized by the maleimidethiol reaction hybridizes with the cocaine DNA aptamer (Coc-Apt). The target-specific structure switching24,27 of the aptamer in the presence of cocaine can cause the DNA–invertase conjugate to release and amplify the signal in a PGM. Upon the addition of 1 mM cocaine and subsequent removal of the MBs, the PGM indicated that the resulting solution could yield nearly 20-fold more glucose from sucrose than a sample containing no cocaine or containing other compounds such as benzylecognine, methyl ecgonine, adenosine and uridine. This result suggests that the sensor has a high selectivity to cocaine (Fig. 2b). The concentration of glucose detected by the PGM was proportional to the concentration of cocaine in the sample until reaching a saturation at ~1 mM (Fig. 2b), because all the releasable DNA–invertase conjugates were released at this high concentration of cocaine. A detection limit of 3.4 μM (1.0 μg ml−1) in the detection range of ~0–500 μM cocaine was achieved based on a 3σb/slope, where σb is the standard deviation of five blank samples. This sensitivity is of the same order of magnitude as commercially available cocaine test kits such as the Emit kit (0.3 μg ml−1). More importantly, the present glucose-meter-based method can provide quantitative results, whereas most screen methods and commercial kits are semi-quantitative tests based on colour observation, the accuracy of which may be subject to variation for different people. To examine the role of the cocaine aptamer in the performance of the sensor, control experiments were carried out using a truncated cocaine aptamer as linker. The concentration of glucose detected by the PGM did not increase even in the presence of 1 mM cocaine (Fig. 2f).
Figure 2.
Design and performance of cocaine and adenosine detections using a PGM. a, Cocaine-induced release of immobilized DNA–invertase conjugates. b, Cocaine detection in buffer using the PGM. c, Cocaine detection in human serum and calf blood samples using the PGM. d, Adenosine-induced release of immobilized DNA–invertase conjugates. e, Adenosine detection using the PGM. f, Effects of active aptamers on the performance of the PGM in detecting adenosine and cocaine in the presence of 1 mM targets; −, with aptamer active site truncated; +, with no truncation. All error bars were obtained through the detection of three parallel samples.
Optimization and characterization of cocaine detection
Many factors can affect the performance and results of the above cocaine sensor system. The effects of incubation time with target (Supplementary Fig. S3), reaction time with sucrose (Supplementary Fig. S4), sucrose concentration (Supplementary Fig. S5), operation temperature (Supplementary Fig. S6), actual glucose concentration versus glucose signal from the PGM (Supplementary Fig. S7), stability of DNA–invertase-modified MBs upon storage (Supplementary Fig. S8) and the use of different brands of glucose meters (Supplementary Fig. S9) were all carefully investigated and optimized.
Detection in serum and blood
The sensor was also used to detect cocaine in 20% (Supplementary Fig. S10), 50% and undiluted human serum, and 50% and undiluted calf blood (Fig. 2c), and detection limits of 4.6 μM (1.4 μg ml−1), 6.9 μM (2.1 μg ml−1), 9.3 μM (2.8 μg ml−1), 16 μM (4.9 μg ml−1) and 24 μM (7.1 μg ml−1) were achieved, respectively. These detection limits are close to those obtained in buffer, suggesting that the other components of the serum or blood did not interfere significantly with sensor performance. The successful detection of cocaine in serum and blood as demonstrated here may not be straightforward when testing for cocaine clinically, because cocaine is metabolised into other species in the circulation. However, the excellent selectivity of our method for cocaine over its analogues in a complex sample matrix makes it very promising in applications where non-blood samples are used, such as in on-site and real-time screening of suspicious substances for the presence of cocaine by customs or law enforcement officials, avoiding false-positive results. In addition, our method (1.0 μg ml−1 sensitivity) is more sensitive than some currently used screen methods such as Scott’s test (up to mg ml−1 level sensitivity).
Effect of original glucose in the sample matrix
If there already exists a detectable amount of glucose in the sample matrix, this original glucose concentration has to be taken into account in the accurate quantification of cocaine by the glucose meter signal from a PGM. Indeed, the detection of cocaine in serum and blood shown above includes the original glucose concentration in the glucose meter signal. For samples with known glucose concentrations, cocaine concentration may be calculated directly from the corresponding calibration curve. However, if the original glucose concentration in a sample is unknown or could vary (meaning that the calibration curve cannot be used directly), a simple calibration step is required in which the sample matrix is measured directly using the PGM before target detection. The user can obtain the glucose concentration in the original sample and store the number in the meter, and this value is then subtracted from the glucose meter signal obtained when performing the actual test. The result of cocaine detection in a buffer containing 130 mg dl−1 glucose after signal subtraction was found to be very similar to that in the glucose-free buffer (Supplementary Fig. S11). In addition, cocaine detection in buffers containing different concentrations of original glucose (~0–300 mg dl−1) gave similar trends and results after subtraction of the original glucose concentrations (Supplementary Fig. S11).
Reducing the time for cocaine detection
When using more concentrated MBs (8 mg ml−1), the 15 min incubation time with cocaine in the standard procedure could be reduced to as little as 5 min. For the reaction time with sucrose, because the DNA–invertase-modified MBs were more concentrated, there were more DNA–invertase conjugates released under the same conditions. The reaction time with sucrose was successfully reduced from 15 min to 8 min. Therefore, the time required for one typical cocaine test could be reduced to no more than 15 min (Supplementary Fig. S12).
Simplifying cocaine detection using a syringe and filter membrane approach
The above standard procedure for the detection of cocaine may not be very user-friendly, because the end user may find the use of a magnet for the separation of MBs and the transfer of a small volume of liquid to be complicated. To make the detection more user-friendly, we simplified the procedure into a syringe and filter membrane based protocol. In a typical test, the end user only needs to transfer the sample into the modified MBs using a syringe, push the syringe through a filter membrane, then carry out the test using a PGM (Supplementary Figs S12 and S13). All these steps can be performed within a period of 30 min, and the whole procedure is easy to accomplish by a layman at home or in the field.
Adenosine aptamer-based sensor
To demonstrate the generality of the design, we also used an adenosine aptamer to detect adenosine38, an important biological cofactor involved in many biological processes, such as those in the kidney and urine39. Similar to the cocaine aptamer sensor described above, the adenosine aptamer (Ade-Apt) was used as the linker between the immobilized biotin–DNA and the DNA–invertase conjugate to produce an adenosine sensor (Fig. 2d). Unlike the cocaine sensor, however, the DNA–invertase conjugate was obtained through the reaction of an amine-reactive homobifunctional linker, p-phenylene diisothiocyanate (PDITC), with the reactive amine groups on invertase and DNA. This successful conjugation demonstrates that our design method is compatible with different conjugation methods. In the presence of 1 mM adenosine, the solution left over after the removal of MBs exhibited a more than tenfold increase in glucose production from sucrose, as measured by a PGM (Fig. 2e). A titration curve using samples with increasing amounts of adenosine (~0–1 mM) showed a corresponding increase in the concentration of glucose detected by the PGM (Fig. 2e). The detection limit was estimated to be 18 μM in the detection range of ~0–1,000 μM adenosine by the definition of the 3σb/slope. This detection limit is higher than the typical adenosine levels in the kidney or urine39. The sensitivity may be improved by using an RNA aptamer40 that has a stronger binding affinity than the DNA aptamer38. Under the same conditions, other nucleotides such as uridine and cytidine did not produce enhanced readings of glucose in the PGM, suggesting that this sensor has a high selectivity for adenosine. The adenosine aptamer was essential for the performance of the sensor, because no PGM response was observed if a truncated adenosine aptamer was used (Fig. 2f).
Interferon-gamma aptamer-based sensor
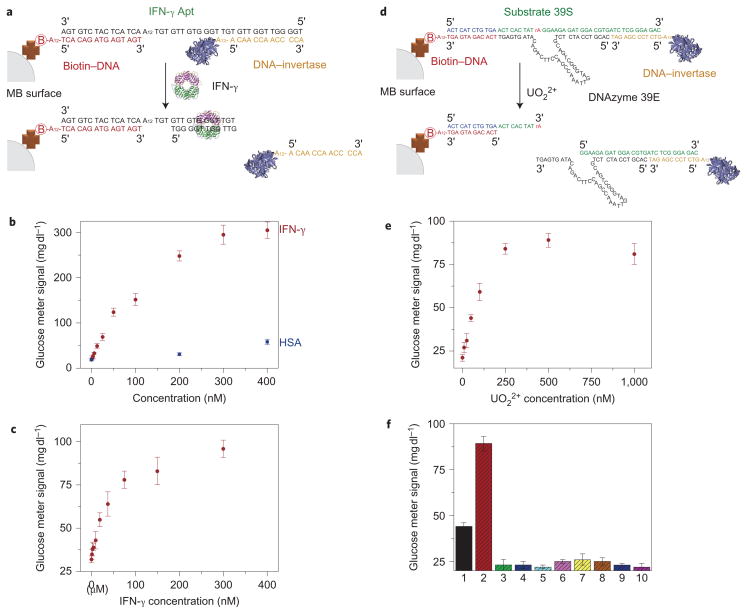
In addition to detecting the small molecules cocaine and adenosine, we also tested our method for its ability to detect a protein macromolecule, interferon-gamma (IFN-γ), using an IFN-γ aptamer41–43. IFN-γ is a cytokine released by the human immune system, and abnormal levels of IFN-γ are related to many infectious disease and pathogen-directed responses of the human body44. Currently, the IFN-γ release assay is used to diagnose tuberculosis45, which is an infectious disease estimated to be latent in one-third of the world’s population, with 10% of those who are latently infected possibly becoming actively infected during their lifetime. The design of the present IFN-γ sensor is similar to the cocaine and adenosine sensors shown above, but with an additional A12 linker between the biotin–DNA and the IFN-γ binding moiety of the aptamer (IFN-γ Apt) (Fig. 3a). This modification prevents the biotin–DNA from interfering with the binding between the large IFN-γ protein and the aptamer. In buffer solution, increasing amounts of glucose were detected by the PGM as the IFN-γ concentration in the solution increased in the range of ~0–400 nM, reaching a 15-fold signal enhancement in the presence of 200 nM IFN-γ. IFN-γ concentrations as low as 2.6 nM (44 ng ml−1) could be detected (Fig. 3b). This detection limit is similar to the binding affinity of the aptamer43, suggesting that the design has preserved the activity of the aptamer. In contrast, the addition of human serum albumin as a negative control to the sensor system resulted in a negligibly low reading from the PGM under the same conditions, indicating the high selectivity of the sensor. Furthermore, detection of IFN-γ in 20% human serum by the sensor was also investigated to show the sensor performance in a complex sample matrix containing numerous serum proteins (Fig. 3c). Interestingly, the PGM signal saturated at a lower IFN-γ concentration in 20% serum than in buffer. This phenomenon might be due to the fact that (i) the DNA–invertase conjugate is more easily released once the IFN-γ binds, because the DNA hybridization could be weaker in the diluted serum solution according to the Tm study (Supplementary Fig. S15), and (ii) the serum in the buffer could block the non-specific binding sites of the MBs, thus avoiding some IFN-γ non-specifically binding to the MBs with no DNA–invertase released (Supplementary Fig. S15). Nevertheless, the detection limit of 3.4 nM (58 ng ml−1) is similar to that obtained in buffer. Because T-cells from human blood can secret IFN-γ at levels as high as several hundreds of ng ml−1 after stimulation46,47, the sensitivity of the PGM-based method here should be sufficient for the IFN-γ release assay of T-cells. Because of the important role of IFN-γ in human immunity and the greater ease of diagnosing relevant diseases such as tuberculosis using PGMs, the sensor design in this work using a PGM without any other instrumentation can find potential applications in point-of-care diagnostics or household quantification of IFN-γ as a disease marker for tuberculosis.
Figure 3.
Design and performance of IFN-γ and UO22+ detections using a PGM. a, IFN-γ-mediated release of immobilized DNA–invertase conjugates. b, IFN-γ detection in buffer using the PGM. c, IFN-γ detection in 20% human serum samples using the PGM. d, UO22+-mediated release of immobilized DNA–invertase conjugates. e, UO22+ detection using the PGM. f, Selectivity of UO22+ detection using the PGM: (1) 50 nM UO22+; (2) 1 μM UO22+; (3) 1 μM Pb2+; (4) 1 μM Cd2+; (5) 100 μM Ca2+/Mg2+; (6) 1 μM Zn2+/Cu2+; (7) 1 μM Co2+/Ni2+; (8) 1 μM VO+; (9) 1 μM Th4+; (10) no metal ion. All error bars were obtained through the detection of three parallel samples.
Uranium DNAzyme-based sensor
Finally, we extended our methodology from aptamers to DNAzymes for the on-site detection of toxic metal ions such as uranium (UO22+) (ref. 48), a radioactive heavy metal ion that is hazardous to both humans and the environment. To ensure the efficient release of the DNA–invertase conjugate from MBs for a DNAzyme-based sensor, the UO22+ sensor differed from the designs of aptamer sensors mentioned above. The biotin–DNA was immobilized on MBs, and the DNA–invertase conjugates were connected by the substrate (39S) of the UO22+-dependent DNAzyme (39E) via 12-base-pair hybridization (Fig. 3d). Further hybridization of 39E to 39S could not cause the cleavage of 39S and subsequent release of the DNA–invertase conjugate unless UO22+ was present. As expected, adding up to 1 μM UO22+ resulted in an approximately fourfold increase in glucose production over a blank without UO22+. As the concentration of UO22+ was increased from 0 to 1 μM in the samples, more glucose was detected by the PGM (Fig. 3e). A detection limit of 9.1 nM (2.4 μg l−1) in the detection range of ~0–200 nM UO22+ was achieved, which is approximately tenfold lower than the US Environmental Protection Agency (EPA) regulated level (30 μg l−1) for drinking water, suggesting that the method could be used for UO22+ detection in drinking water using a PGM at home or in the field. The sensor also exhibited excellent selectivity to UO22+ over other metal ions (Fig. 3f). This result demonstrates that the specificity of the original 39E DNAzyme is preserved in the sensor design48.
In summary, this study has demonstrated a novel method in which a commercially available PGM is linked with functional DNA sensors to achieve portable, low-cost and quantitative detection of many targets beyond glucose, including cocaine (3.4 μM detection limit (DL) and ~0–500 μM detection range (DR)), adenosine (18 μM DL and ~0–1,000 μM DR), interferon-γ (2.6 nM DL and ~0–400 nM DR) and uranium (9.1 nM DL and ~0–200 nM DR). The approach is based on the target-induced release of DNA–invertase conjugate from a functional DNA duplex immobilized on MBs, and subsequent conversion of PGM-inert sucrose into PGM-detectable glucose. Target-induced release was achieved either by structure switching of aptamers or substrate cleavage by DNAzymes. As more functional DNAs and RNAs are available or can be obtained through in vitro selection or SELEX to bind a broad range of targets, the method developed here can be used to quantify many other targets that functional DNAs and RNAs can recognize. In addition, it can be anticipated that nucleic acid detections and immunoassays of many targets should be achievable by means of a similar mechanism. The detection of non-glucose targets in this work was also simplified to a protocol that the general public could use without any professional knowledge. Although, in its current state, the method is not as simple as glucose detection using glucose meters, it can be further simplified and automated through more professional engineering, just as glucose meters have developed from complicated devices into pocket-sized meters over 30 years of progress. Given the wide availability of PGMs and the ability of functional DNAs to recognize many targets, the method demonstrated here has the potential to change environmental monitoring and medical diagnostics for the public at home and in the field.
Methods
The materials, methods for preparing DNA–invertase conjugate, additional characterization data, and detailed methods for sensor preparation and target detection are described in the Supplementary Information. Briefly, the DNA–invertase conjugates were prepared by using either sulfosuccinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (sulfo-SMCC) or p-phenylene diisothiocyanate (PDITC) as the linker between the DNA and invertase, and were further characterized by both native and sodium dodecyl sulfate polyacrylamide gel electrophoresis (~4–20% gradient). The MBs used for target detection were prepared by immobilization of the biotin-modified DNAs onto the streptavidin-coated MBs and subsequent addition of the functional DNAs and DNA–invertase conjugates. Target detection was conducted by adding the sample solution to the MBs prepared as above, separating the MBs using a magnet, mixing the remaining solution with sucrose, and then measuring the final solution using a commercially available PGM.
Supplementary Material
Acknowledgments
The authors thank the US Department of Energy (DE-FG02-08ER64568), National Institutes of Health (ES16865) and National Science Foundation (CTS-0120978) for financial support, and L.H. Tan and H.E. Ihms for the preparation of figures and proof reading of the manuscript.
Footnotes
Author contributions
Y.L. and Y.X. conceived and designed the experiments. Y.X. performed the experiments and analysed the data. Y.L. and Y.X. co-wrote the paper. Correspondence and requests for materials should be addressed to Y.L.
The authors declare no competing financial interests.
Additional information
Supplementary information accompanies this paper at www.nature.com/naturechemistry. Reprints and permission information is available online at http://www.nature.com/reprints.
References
- 1.Daar AS, et al. Top ten biotechnologies for improving health in developing countries. Nat Genet. 2002;32:229–232. doi: 10.1038/ng1002-229. [DOI] [PubMed] [Google Scholar]
- 2.Martinez AW, Phillips ST, Butte MJ, Whitesides GM. Patterned paper as a platform for inexpensive, low-volume, portable bioassays. Angew Chem Int Ed. 2007;46:1318–1320. doi: 10.1002/anie.200603817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fan C, Plaxco KW, Heeger AJ. Biosensors based on binding-modulated donor–acceptor distances. Trends Biotechnol. 2005;23:186–192. doi: 10.1016/j.tibtech.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Xia F, et al. Colorimetric detection of DNA, small molecules, proteins, and ions using unmodified gold nanoparticles and conjugated polyelectrolytes. Proc Natl Acad Sci USA. 2010;107:10837–10841. doi: 10.1073/pnas.1005632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 6.Giepmans BNG, Adams SR, Ellisman MH, Tsien RY. Review–the fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 7.Wegner SV, Okesli A, Chen P, He C. Design of an emission ratiometric biosensor from MerR family proteins: a sensitive and selective sensor for Hg2+ J Am Chem Soc. 2007;129:3474–3475. doi: 10.1021/ja068342d. [DOI] [PubMed] [Google Scholar]
- 8.Murray RW. Nanoelectrochemistry: metal nanoparticles, nanoelectrodes, and nanopores. Chem Rev. 2008;108:2688–2720. doi: 10.1021/cr068077e. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, et al. Simultaneous phase and size control of upconversion nanocrystals through lanthanide doping. Nature. 2010;463:1061–1065. doi: 10.1038/nature08777. [DOI] [PubMed] [Google Scholar]
- 10.Favier F, Walter EC, Zach MP, Benter T, Penner RM. Hydrogen sensors and switches from electrodeposited palladium mesowire arrays. Science. 2001;293:2227–2231. doi: 10.1126/science.1063189. [DOI] [PubMed] [Google Scholar]
- 11.Clark LC, Lyons C. Electrode systems for continuous monitoring in cardiovascular surgery. Ann NY Acad Sci. 1962;102:29–45. doi: 10.1111/j.1749-6632.1962.tb13623.x. [DOI] [PubMed] [Google Scholar]
- 12.Montagnana M, Caputo M, Giavarina D, Lippi G. Overview on self-monitoring of blood glucose. Clin Chim Acta. 2009;402:7–13. doi: 10.1016/j.cca.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Carroll AE, Marrero DG, Downs SM. The HealthPia GlucoPack (TM) diabetes phone: a usability study. Diabetes Technol Ther. 2007;9:158–164. doi: 10.1089/dia.2006.0002. [DOI] [PubMed] [Google Scholar]
- 14.Nie Z, Deiss F, Liu X, Akbulut O, Whitesides GM. Integration of paper-based microfluidic devices with commercial electrochemical readers. Lab Chip. 2010;10:3163–3169. doi: 10.1039/c0lc00237b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robertson DL, Joyce GF. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded-DNA. Nature. 1990;344:467–468. doi: 10.1038/344467a0. [DOI] [PubMed] [Google Scholar]
- 16.Breaker RR, Joyce GF. A DNA enzyme that cleaves RNA. Chem Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- 17.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 18.Breaker RR. Engineered allosteric ribozymes as biosensor components. Curr Opin Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- 19.Lee JF, Hesselberth JR, Meyers LA, Ellington AD. Aptamer database. Nucleic Acids Res. 2004;32:D95–D100. doi: 10.1093/nar/gkh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Navani NK, Li Y. Nucleic acid aptamers and enzymes as sensors. Curr Opin Chem Biol. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 21.Song S, Wang L, Li J, Fan C, Zhao J. Aptamer-based biosensors. Trends Anal Chem. 2008;27:108–117. [Google Scholar]
- 22.Liu J, Cao Z, Lu Y. Functional nucleic acid sensors. Chem Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Lu Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J Am Chem Soc. 2000;122:10466–10467. [Google Scholar]
- 24.Nutiu R, Li Y. Structure-switching signaling aptamers. J Am Chem Soc. 2003;125:4771–4778. doi: 10.1021/ja028962o. [DOI] [PubMed] [Google Scholar]
- 25.Yang CJ, Jockusch S, Vicens M, Turro NJ, Tan W. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc Natl Acad Sci USA. 2005;102:17278–17283. doi: 10.1073/pnas.0508821102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Csordas A, et al. Detection of proteins in serum by micromagnetic aptamer PCR (MAP) technology. Angew Chem Int Ed. 2010;49:355–358. doi: 10.1002/anie.200904846. [DOI] [PubMed] [Google Scholar]
- 27.Oh SS, Plakos K, Lou XH, Xiao Y, Soh HT. In vitro selection of structure-switching, self-reporting aptamers. Proc Natl Acad Sci USA. 2010;107:14053–14058. doi: 10.1073/pnas.1009172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, Lu Y. A colorimetric lead biosensor using DNAzyme-directed assembly of gold nanoparticles. J Am Chem Soc. 2003;125:6642–6643. doi: 10.1021/ja034775u. [DOI] [PubMed] [Google Scholar]
- 29.Willner I, Shlyahovsky B, Zayats M, Willner B. DNAzymes for sensing, nanobiotechnology and logic gate applications. Chem Soc Rev. 2008;37:1153–1165. doi: 10.1039/b718428j. [DOI] [PubMed] [Google Scholar]
- 30.Xue X, Wang F, Liu X. One-step, room temperature, colorimetric detection of mercury (Hg2+) using DNA/nanoparticle conjugates. J Am Chem Soc. 2008;130:3244–3245. doi: 10.1021/ja076716c. [DOI] [PubMed] [Google Scholar]
- 31.Willner I, Zayats M. Electronic aptamer-based sensors. Angew Chem Int Ed. 2007;46:6408–6418. doi: 10.1002/anie.200604524. [DOI] [PubMed] [Google Scholar]
- 32.Lubin AA, Plaxco KW. Folding-based electrochemical biosensors: the case for responsive nucleic acid architectures. Acc Chem Res. 2010;43:496–505. doi: 10.1021/ar900165x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swensen JS, et al. Continuous, real-time monitoring of cocaine in undiluted blood serum via a microfluidic, electrochemical aptamer-based sensor. J Am Chem Soc. 2009;131:4262–4266. doi: 10.1021/ja806531z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yigit MV, Mazumdar D, Lu Y. MRI detection of thrombin with aptamer functionalized superparamagnetic iron oxide nanoparticles. Bioconj Chem. 2008;19:412–417. doi: 10.1021/bc7003928. [DOI] [PubMed] [Google Scholar]
- 35.Liu J, Mazumdar D, Lu Y. A simple and sensitive ’dipstick’ test in serum based on lateral flow separation of aptamer-linked nanostructures. Angew Chem Int Ed. 2006;45:7955–7959. doi: 10.1002/anie.200603106. [DOI] [PubMed] [Google Scholar]
- 36.Niemeyer CM. Semisynthetic DNA–protein conjugates for biosensing and nanofabrication. Angew Chem Int Ed. 2010;49:1200–1216. doi: 10.1002/anie.200904930. [DOI] [PubMed] [Google Scholar]
- 37.Stojanovic MN, de Prada P, Landry DW. Aptamer-based folding fluorescent sensor for cocaine. J Am Chem Soc. 2001;123:4928–4931. doi: 10.1021/ja0038171. [DOI] [PubMed] [Google Scholar]
- 38.Huizenga DE, Szostak JW. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995;34:656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- 39.Vallon V, Muhlbauer B, Osswald H. Adenosine and kidney function. Physiol Rev. 2006;86:901–940. doi: 10.1152/physrev.00031.2005. [DOI] [PubMed] [Google Scholar]
- 40.Sassanfar M, Szostak JW. An RNA motif that binds ATP. Nature. 1993;364:550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- 41.Lee PP, Ramanathan M, Hunt CA, Garovoy MR. An oligonucleotide blocks interferon-γ signal transduction. Transplantation. 1996;62:1297–1301. doi: 10.1097/00007890-199611150-00021. [DOI] [PubMed] [Google Scholar]
- 42.Balasubrananian V, Nguyen LT, Balasubramanian SV, Ramanathan M. Interferon-γ-inhibitory oligodeoxynucleotides alter the conformation of interferon-γ. Mol Pharmacol. 1998;53:926–932. [PubMed] [Google Scholar]
- 43.Tuleuova N, et al. Development of an aptamer beacon for detection of interferon-γ. Anal Chem. 2010;82:1851–1857. doi: 10.1021/ac9025237. [DOI] [PubMed] [Google Scholar]
- 44.Boehm U, Klamp T, Groot M, Howard JC. Cellular responses to interferon-γ. Annu Rev Immunol. 1997;15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 45.Pai M, Riley LW, Colford JM. Interferon-γ assays in the immunodiagnosis of tuberculosis: a systematic review. Lancet Infect Dis. 2004;4:761–776. doi: 10.1016/S1473-3099(04)01206-X. [DOI] [PubMed] [Google Scholar]
- 46.Zhu H, et al. A microdevice for multiplexed detection of T-cell-secreted cytokines. Lab Chip. 2008;8:2197–2205. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- 47.Zhu H, et al. Detecting cytokine release from single T-cells. Anal Chem. 2009;81:8150–8156. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, et al. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc Natl Acad Sci USA. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.