Abstract
Chemotherapy is the bedrock for the clinical management of cancer, and the tumor suppressor p53 has a central role in this therapeutic modality. This protein facilitates favorable antitumor drug response through a variety of key cellular functions, including cell cycle arrest, senescence, and apoptosis. These functions essentially cease once p53 becomes mutated, as occurs in ~50% of cancers, and some p53 mutants even exhibit gain-of-function effects, which lead to greater drug resistance. However, it is becoming increasingly evident that resistance is also seen in cancers harboring wild-type p53. In this review, we discuss how wild-type p53 is inactivated to render cells resistant to antitumor drugs. This may occur through various mechanisms, including an increase in proteasomal degradation, defects in post-translational modification, and downstream defects in p53 target genes. We also consider evidence that the resistance seen in wild-type p53 cancers can be substantially greater than that seen in mutant p53 cancers, and this poses a far greater challenge for efforts to design strategies that increase drug response in resistant cancers already primed with wild-type p53. Because the mechanisms contributing to this wild-type p53 “gain-of-resistance” phenotype are largely unknown, a concerted research effort is needed to identify the underlying basis for the occurrence of this phenotype and, in parallel, to explore the possibility that the phenotype may be a product of wild-type p53 gain-of-function effects. Such studies are essential to lay the foundation for a rational therapeutic approach in the treatment of resistant wild-type p53 cancers.
Keywords: Tumor suppressor p53, Antitumor drug response, Drug resistance, Gain-of-function mutants, Gain-of-resistance phenotype, Post-translational modifications
1. Introduction
Chemotherapy is a critical modality in the fight against cancer. Indeed, antitumor drugs are commonly used for cancer therapy, and their effectiveness is a clear indication that such agents have positive therapeutic indices; that is, successful drugs must have greater activity against tumor cells than against normal cells. Conversely, when activity against tumor cells falls below that against normal cells because of drug resistance, the clinical utility of the antitumor drug is essentially lost. Resistance of tumor cells can be intrinsic, when tumor cells are predisposed at the outset to minimal or negligible response to therapy, or acquired, when tumor cells may initially have a drug-sensitive phenotype that is lost during the course of therapy.
Genetic alterations are generally the reason for greater drug sensitivity of tumor cells relative to normal cells, but which specific genes or sets of genes are involved to induce this differential response will continue to be a subject of concerted laboratory investigations. However, such investigations are more difficult than previously thought since tumor cells demonstrate abnormal expression of over several hundred genes [1,2] and because no two cancers, even from the same tissue type, exhibit identical patterns of the abnormally expressed genes [3,4]. Therefore, the greater drug sensitivity of tumor cells may depend on a unique gene signature profile, which lowers the threshold for sensing cellular damage by antitumor agents and allows efficient transduction of signals for anti-proliferative and/or pro-apoptotic effects.
The lack of an explicit understanding of which dysfunctional genes promote greater drug sensitivity of tumor cells relative to normal cells has not hindered the progress toward identifying critical normal genes that work in concert with the tumor’s signaling network to enhance tumor sensitivity. One of these is the tumor suppressor p53, widely recognized as the “guardian of the genome,” which in its wild-type state has an essential role in facilitating antitumor drug response. Indeed, tumor cells harboring wild-type p53 are generally recognized as being sensitive to antitumor agents [5]. This premise is consistent with the understanding that activation of p53 is sufficient to induce cell death, even in the presence of strong survival signals from other deregulated genes that may be present [6]. Unfortunately, such tumor cells do eventually become resistant to therapy, and it is not surprising that inactivation of p53 function is an important resistance mechanism, both clinically and in tumor cell lines [7–11]. In this review, we discuss how wild-type p53 is inactivated to render cells resistant to antitumor drugs. We also consider evidence to support the notion of a gain-of-resistance phenotype, in which drug resistance in the presence of wild-type p53 can be greater than that anticipated from a simple loss of p53 function. Although this review uses reported examples of mechanisms in the context of resistance to specific agents, such as cisplatin, it should be noted that these mechanisms are also applicable to other antitumor drugs. This claim is supported by the observation that tumors resistant to cisplatin are generally cross-resistant to diverse, structurally unrelated cytotoxic agents [12].
2. Role of p53 in cancer and therapeutics
The p53 protein, identified as a tumor suppressor in 1989 [13], is encoded by the TP53 gene located on chromosome 17p13.1 and is composed of several domains: (i) a DNA-binding domain, (ii) a transactivation domain, (iii) an oligomerization domain, (iv) a proline-rich domain and v) a C-terminal regulatory domain [14]. The p53 protein functions mainly as a transcriptional activator by binding to specific DNA sequences of target genes involved in a broad range of biological functions, including cell cycle arrest, DNA repair, senescence, apoptosis, and inhibition of angiogenesis [15].
Cellular levels of p53 are usually kept low because of its short half-life of ~20 minutes. Degradation of p53 occurs mainly by binding to Mdm2, an E3 ligase that catalyzes mono-ubiquitination of p53 in the C-terminal domain and thereby targets it for proteasomal degradation [13,16,17]. A close homolog, Mdm4, can also regulate p53 function by binding to its transactivation domain, but unlike Mdm2 it cannot directly ubiquitinate p53 [18]. The E3 and E4 ubiquitin ligase p300 poly-ubiquitinates p53 in an Mdm2-dependent manner that also results in p53 degradation [16,19]. Several other molecules are involved in p53 regulation, including Pirh2, Parc, and p14ARF, which can also affect p53-dependent therapeutic response. Pirh2 binds to the DNA-binding domain of p53 and promotes its degradation, whereas Parc fixes p53 in the cytoplasm, preventing it from entering the nucleus and thus suppressing its transcriptional activity [16]. From the perspective of therapy, the tumor suppressor p14ARF is of significant interest given that it positively regulates p53 by directly inhibiting Mdm2 [17,20].
For tumor suppressive function or to facilitate a therapeutic response to an antitumor agent, p53 needs to be not only stabilized but also activated. This is accomplished by post-translational modifications, including phosphorylation and dephosphorylation at ~23 distinct sites by a network of kinases and phosphatases [21,22]. Thus, under conditions of stress, as may ensue after DNA damage, specific kinases, such as ATM, ATR, Chk1, and Chk2, become activated and phosphorylate p53 target sites, such as Ser15, Thr18, and Ser20, and this phosphorylation forces dissociation of p53 from the Mdm2–p53 complex. The resulting stabilization of p53 and its translocation to the nucleus enable p53 to bind as a tetramer to specific DNA sequences and transactivate target genes [22,23]. This transactivation by p53, driven by upstream activating pathways, is critical for its cellular functions, which include arresting the cell cycle to permit DNA repair and, if the damage is too severe to be repaired, activating cell death pathways [24]. Although the p53 tumor suppressive signaling cascade paradoxically fails to prevent cancer from developing in the first place, it can still be activated in many cancer cells with DNA damaging drugs to induce antitumor response [25,26]. However, the eventual failure of p53 to become activated in tumor cells during the course of chemotherapy is the most significant mechanism of drug resistance. This failure results from the loss of three of the most significant cellular pathways involved in antitumor response during chemotherapy: (i) induction of programmed cell death (apoptosis), (ii) induction of checkpoint response and cell cycle arrest, and (iii) induction of permanent cell cycle arrest (senescence); these pathways normally contribute independently or collectively to final therapeutic outcomes in patients. A brief discussion of these processes is warranted to better appreciate p53-dependent resistance mechanisms.
2.1. Induction of apoptosis
Although p53 can mediate apoptosis in a transcription-dependent manner, it is also involved in transcription-independent apoptosis. In the transcription-dependent process, two distinct signaling pathways are involved: extrinsic and intrinsic pathways. In the extrinsic pathway, p53 induces the transcription of death receptors of the tumor necrosis factor receptor (TNF-R) family: Fas, APO-1, CD95, DR5, and PERP [27]. After a ligand binds to its specific receptor, the formation of the death-inducing signaling complex (DISC) is accomplished by recruitment of the Fas-associated death domain (FADD) and caspase-8 and -10, leading to the activation of the effector caspases (e.g., caspase-3 and -7) and resultant DNA fragmentation as a hallmark of apoptosis. In contrast, the intrinsic pathway is activated through a DNA damage mechanism that involves mitochondrial apoptotic events, which are regulated largely by the Bcl-2 family of proteins [27]. Thus, upon stabilization and activation, p53 translocates to the nucleus, where it transactivates the pro-apoptotic genes Bax, Noxa, PUMA, and Bid [28]. Of these, Bax is the most important pro-apoptotic gene to be induced by p53, but its translocation and functional multimerization depend on other pro-apoptotic family members [29]. Bax can homo-multimerize or it can hetero-multimerize with Bak in the outer mitochondrial membrane and thereby induce the release of cytochrome c, Smac/DIABLO, and apoptosis-inducing factor (AIF) from the mitochondrial intermembrane space to the cytosol. The apoptosome complex, formed by cytochrome c, apoptotic protease-activating factor 1 (APAF-1), and pre-cleaved caspase-9, activates effector caspase-3, -6, and -7 to induce DNA fragmentation as the final stage of apoptosis [30]. Interestingly, the intrinsic and extrinsic pathways are connected via Bid, which upon cleavage to t-Bid by caspase-8 translocates to the mitochondria and activates the pro-apoptotic proteins Bax and Bak [28].
In the transcription-independent mechanism of apoptosis, some of the induced wild-type p53 translocates to the mitochondria and physically interacts with the anti-apoptotic proteins Bcl-xL and Bcl-2 to promote the pro-apoptotic homo- or hetero-multimerization between Bak and Bax [31,32]. This occurs rapidly and before p53 exhibits its transcriptional activity. In addition, p53 disrupts the inhibitory Bak–Mcl1 complex by binding to Bak directly [33]. Taken together, these actions of p53 permeabilize the outer mitochondrial membrane, allowing the release of pro-apoptotic factors into the cytoplasm. DNA damaging drugs may activate both membrane death receptors and the endogenous mitochondrial damage pathway, and because these apoptotic mechanisms are deregulated in cancers, proteins involved in these pathways are molecular targets of great interest in cancer therapy [34]. Indeed, most of the anticancer agents that act through DNA damage or stress-inducing mechanisms require wild-type p53 to exhibit the apoptotic phenotype [35].
2.2. Induction of checkpoint response and cell cycle arrest
The cell is endowed with elaborate checkpoint systems that, when activated by stressful stimuli such as DNA damage by antitumor agents, activate appropriate signal transduction pathways, which propagate information from the stress stimuli to the cell cycle machinery via sensor, transducer, and effector signaling proteins [36]. This integrated network of DNA damage signaling results in the inhibition of cyclin-dependent kinase (Cdk) activities, thereby impacting cell cycle kinetics. More specifically, inhibition of distinct Cdk/cyclin complexes that are present throughout the cell cycle prevents or slows G1/S transition, S-phase progression, G2/M transition, or all three [36,37]. Although many of the Cdks can be inhibited by members of the Cip/Kip family members of Cdk inhibitors (p21, p27, and p57), p21 as a critical target of p53 is the most significant for inhibiting G1-phase Cdk4/cyclin D and Cdk2/cyclin E. This significance is reflected in the fact that p21 deletion alone can prevent G1-phase arrest [37–39], whereas other mechanisms exist to inhibit S- and G2-phase Cdk activities [38]. Indeed, DNA damaging agents can induce S- and G2-phase arrest even if p53 function is not present [40].
Cell cycle arrest is considered a critical step to allow DNA repair and for cell survival, but if repair fails then apoptosis is induced. Whether persistent p21-dependent G1 arrest is actually a trigger for cell death is unclear. However, tumors retaining G1 checkpoint response appear to be more sensitive to therapeutic agents [5,41,42]. Indeed, a strong positive correlation has been demonstrated between the ability of tumor cells to arrest in G1 and their sensitivity to platinum-based drugs in the National Cancer Institute (NCI) panel of 60 tumor cell lines [43]. This correlation is also consistent with the finding that mutant p53 tumor cells transfected with a p21 expression vector are sensitized to antitumor drugs [44,45]. In addition, small-molecule inhibitors of G1-phase Cdks not only arrest cells but also induce apoptosis [46,47]. Moreover, cells from p21 knockout mice lack G1 checkpoint response [48], and because such mice develop tumors [49], the tumor suppressive function of p53 may be mediated in part through the downstream effects of p21 in checkpoint response.
2.3. Induction of senescence
Cellular senescence is defined as a permanent cell cycle arrest that prevents cell immortalization and transformation to a genetically unstable phenotype [50]. Therefore, senescence is identified as an additional tumor suppressive mechanism in benign or premalignant cancer lesions [51]. Senescent cells remain metabolically active but acquire distinct changes in morphology and physiology characterized by enlarged cell size, chromatin condensation, changes in gene expression, and high levels of senescence-associated β-galactosidase. The senescence phenotype occurs as a result of various forms of stress stimuli, such as telomerase shortening, oncogenic stimuli, ionizing radiation, and DNA damaging agents [52]. Not surprisingly, p53 is the pivotal player in regulating replicative as well as premature (stress-induced) senescence. In line with this evidence, tumor cells containing wild-type p53 are more likely to undergo senescence in response to chemotherapy [53].
Stress-induced senescence is driven via the ATM/ATR-Chk2/Chk1-p53 pathway, with the involvement of several p53-dependent downstream molecular markers, such as p21, PML, PAI-1, and DEC1 [53]. Of these, p21 is the seminal controller of the senescence program. In this respect, p21-dependent G1 arrest in senescence is similar to that after checkpoint response, but the significant difference is that activation of the senescence program requires prolonged p53-dependent expression of p21, whereas the checkpoint response pathway requires a relatively transient transactivation of p21 by p53 [15]. Despite this difference, premature senescence, like apoptosis and G1 checkpoint response, can significantly contribute to the antitumor effects mediated by p53 with a variety of anticancer agents [54–58]. Although apoptosis may have greater relevance in cancer chemotherapy, p53-dependent senescence may provide an important anti-proliferative option in cancer cells that have lost their ability to undergo p53-dependent apoptosis [54].
3. Mechanisms of resistance involving p53
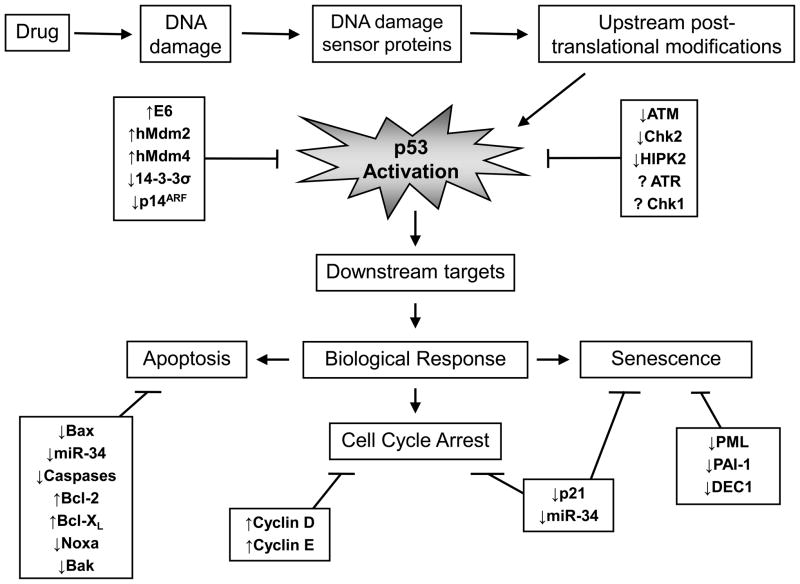
Induction of cell death by antitumor drugs requires (i) the recognition of DNA damage, (ii) activation of specific kinases, (iii) stabilization and activation of p53, (iv) activation of downstream p53-dependent pathways, and finally (v) execution of cytotoxic programs (Fig. 1). The p53 protein is central to this process, and its critical roles in activating anti-proliferative/pro-apoptotic pathways and facilitating the effects of antitumor agents also make it a vulnerable target; tumor cells will readily select for attenuated p53 pathways that lead to drug resistance and cell survival. Interestingly, resistance in tumor cells harboring wild-type p53 is observed with a large variety of agents, ranging from ionizing radiation to several classes of cytotoxic drugs, such as doxorubicin, paclitaxel, cisplatin, etoposide, and vinblastine [59]. This resistance can occur directly, via factors that reduce or negate the functional activity of p53, or indirectly, by deregulation of pathways downstream of p53 (Fig. 1). Although the purpose of this review is to focus on mechanisms that directly affect p53 function to induce drug resistance, either through mutation in the p53 protein or through the failure of upstream pathways that stabilize and post-translationally activate wild-type p53, it is appropriate to first briefly discuss resistance arising from deregulated pathways that affect p53-mediated signaling.
Fig. 1.
Deregulation of pathways attenuating p53-dependent drug response.
3.1. Deregulation of pathways downstream of p53
The indirect pathways that affect p53 signaling can involve downstream failure either in the activity of target gene products, such as p21, Bax, miRNAs, and caspases, or in the expression or mutation of proteins interacting with p53. A case in point is where p53 is functional and transactivates p21, but this Cdk inhibitor becomes inactivated through cytoplasmic sequestration after Akt-mediated phosphorylation, leading to resistance of testicular and ovarian cancers to antitumor agents such as cisplatin [60,61]. In this case, a possible mechanism for the resistance is the binding of phosphorylated form of p21 in the cytoplasm to procaspase-3, preventing its conversion to caspase-3 and thereby blocking apoptosis [62]. Although the p21 gene is rarely mutated, it can also influence therapeutic outcomes through other mechanisms. For instance, a recent report has demonstrated that a single nucleotide polymorphism at codon 31 of p21 in ~10% of chronic lymphocytic leukemia (CLL) patients is sufficient to cause greater aggressiveness and therapeutic resistance of the cancer [63]. Similarly, reduced p21 expression through p21 promoter hypermethylation in the corresponding acute lymphocytic leukemia (ALL) leads to a low survival rate of 6–8% in patients compared with a higher rate of ~60% when the promoter is hypomethylated and fully functional [64]. Bax expression is also known to be downregulated, but by mutation in the coding region, in ~50% of colorectal and gastric cancers, which then lose apoptotic activity and become drug-resistant, leading to poor patient survival [65–67]. Where Bax expression is normal, failure in its activation has also been reported as an alternative mechanism for Bax-related resistance in Hodgkin disease [68].
Recently, wild-type p53 has been reported to regulate the expression of microRNAs [69]. The most significant effect involves the induction by p53 of microRNAs miR-34a, miR-34b, and miR-34bc, which are involved in apoptosis, cell cycle arrest, and senescence, and defective expression of these targets has been associated with resistance to p53-dependent therapeutic agents [70]. The mechanism for this reduced expression has been attributed to gene silencing by CpG hypermethylation of miR-34a, miR-34b, and/or miR-34c promoters in 60–100% of renal cell carcinoma (RCC), pancreatic, breast, colorectal, ovarian, and urothelial cancers [71]. Conversely, ectopic expression of miR-34a has been demonstrated to sensitize medulloblastoma and prostate cells to chemotherapeutic agents [72,73].
In apoptosis, the ultimate goal of antitumor agents is to activate caspases, which degrade DNA and complete the process of cell death. Therefore, inhibition of caspase activity will also result in tumor resistance. Indeed, inactivation or downregulation of caspase-3, -8, and -9 has been reported and directly correlated with drug resistance in neuroblastoma and cervical, head and neck, ovarian, and breast cancers [74–79]. Moreover, >100 proteins can bind with p53 to potentially regulate cytotoxic outcome, and a defect in this interaction through alterations in expression or mutation in the binding partner can have serious therapeutic consequences. For instance, the p53-dependent nuclear export of BRCA1 is important in enhancing DNA damage response, but overexpression of BRCA1 in wild-type p53 MCF-7 tumor cells induced resistance to cisplatin as a result of increased BRCA1-dependent DNA repair, most likely due to a resultant increase in levels of BRCA1 retained in the nucleus [80,81]. Although Mdm2 and Mdm4 are the most prominent of the proteins known to interact with p53, their impact is more direct and will be discussed later.
3.2. Mutations in p53
The high risk of patients with Li–Fraumeni syndrome to develop fibrosarcomas and breast cancer is linked directly to the loss of tumor suppressive function as a result of heterozygous germline mutations in p53 [13,23,26,82]. Because p53 is mutated in ~50% of human cancers, this loss of function has a widespread impact on tumor incidence in many tissue types. Loss of p53 function from mutation also results in poor response of tumors to a wide range of clinical treatment regimens, in stark contrast to excellent prognosis and treatment outcome in tumors harboring wild-type p53 [18,83]. This contrast is clearly and perhaps uniquely exemplified in pediatric patients with choroid plexus carcinomas treated with chemoradiation; essentially, the treatment induced universal response and long-term survival (>5 years) in all patients with tumors expressing wild-type p53, whereas most patients (~80%) with tumors expressing mutant p53 died within 6 months [84]. Although not as dramatic, the response of patients treated for advanced breast cancer with paclitaxel or the 5-fluorouracil (5-FU)/epirubicin/cyclophosphamide combination was 45–64% when wild-type was expressed, but responses were totally absent when mutant p53 was expressed [85,86]. As another example, the 5-year survival rate in CLL patients with tumors expressing wild-type p53 was three-fold greater than in patients with tumors expressing mutant p53 (59% vs. 20%) [87]. Similarly, the exquisite curative sensitivity of testicular cancers to platinum-based therapy is widely attributed to the rarity of p53 mutations in this disease [88]. Consequently, the negative therapeutic association with mutant p53 has made the mutant form a popular molecular target, and thus drugs that may restore wild-type p53 conformation and function have been aggressively pursued [83,89].
Cytotoxic responses based on p53 genotype have also been studied in tumor cell lines. The most extensive study conducted to date was from NCI using a broad tumor panel comprising 39 mutant p53 and 18 wild-type p53 cell lines across 9 tissue types and exposed to 123 agents from a variety of antitumor drug classes [5]. That study demonstrated that the median resistance of cell lines in the mutant p53 group to cisplatin, 5-FU, and bleomycin was 3–10 times that in the group of wild-type p53 cell lines. This drug-dependent variation in resistance is not unexpected, and it is important to note that due to cell context p53 mutation may not necessarily render tumor cells resistant to every antitumor agent. Thus, the p53-R273H mutant expressed in H1299 lung adenocarcinoma cells induced a low level of resistance to etoposide but a substantially greater resistance to cisplatin (see Table 1), whereas ectopic expression of p53-V143A did not induce resistance to several antitumor agents, including cisplatin, paclitaxel, vincristine, and teniposide [90,91].
Table 1.
Cytotoxic drug response of tumor cell lines transfected with p53 gain-of-function mutant.
| p53-Null tumor model [Ref.] | p53 Mutation | Treatment | IC50 (μM)
|
|
|---|---|---|---|---|
| Control | Mutant p53 | |||
| H1299 [90] | R175H | Etoposide | <10 | >10 |
| R175H | Cisplatin | <8 | >8 | |
| R273H | Etoposide | >10 | <10 | |
| R273H | Cisplatin | <8 | >8 | |
| Saos-2 [107] | R273H | Methotrexate | 0.012 | 0.025 |
| R273H | Doxorubicin | 0.03 | >0.2 | |
Abbreviation: IC50, drug concentration that inhibits cell proliferation by 50%.
Somatic p53 mutations are mostly missense in nature and are located predominantly within the DNA-binding domain, where a single mutation is sufficient to cause loss of normal p53 function [82,92]. Four positions in p53 are frequently mutated; these “hot spots” are located at amino acid positions 175, 248, 249, and 273 and collectively account for >25% of all missense mutations in human cancers. Specific mutations in p53 are grouped into “contact” and “structural” classes, but both types impede the pro-apoptotic p53 transactivation functions by preventing binding of p53 with target promoter sites on DNA. Thus, mutations at amino acid positions in p53 that normally make direct contact with DNA (e.g., R248W and R273H) will disrupt promoter binding without necessarily altering p53 conformation. In contrast, mutations in noncontact positions (e.g., R175H and R249S) induce local and/or global structural distortions and thereby prevent p53 from binding to the specific DNA sites [93,94].
3.2.1. Mutations in p53 and the gain-of-function phenotype
It is noteworthy that specific mutations in p53 not only may result in loss of normal p53 functions but also can induce novel functions, leading to the so-called p53 gain-of-function phenotype, which still confers drug resistance [95]. More specifically, the novel biological effects of p53 mutations, such as R175H, R175P, R248W, R249S, R273C, and R273L, involve transcriptional activation or repression of genes whose expression is not normally regulated by wild-type p53, for example BFGF, EGFR, HSP70, and C-Myc [94,96]. These genes when deregulated have been implicated in the promotion of tumor cell growth and survival and the development of resistance to several chemotherapeutic agents by inhibiting apoptosis in several cell lines and in patients [97–100].
Evidence suggests that p53 gain-of-function mutants may also mediate their effects through inhibitory interactions with other transcriptional factors that normally bind to gene promoters [96,101]. An important example is the interaction of such p53 mutants with the family members p63 and p73, which are then prevented from transactivating their gene targets [94]. Moreover, protein–protein interactions with these p53 mutants can also have profound cellular consequences in a promoter-independent manner. For instance, the hot spot mutant R248W or R273H interacts with the nuclease Mre11, thus interfering with the formation and recruitment of the Mre11/Rad50/NBS1 (MRN) complex at the site of DNA double-strand breaks (DSBs) and thereby abolishing the co-recruitment and activation of ATM for DNA damage response [94,96,101–103].
From the perspective of therapeutic response, it is clear that mutations in p53 will inhibit both anti-proliferative and pro-apoptotic pathways, and induce resistance as a result. For example, p53 gene mutations may exist in only one allele, but if the mutant p53 molecule has a dominant-negative characteristic, its lone presence in the active tetrameric complex is sufficient to not only inhibit p53 function but also promote gain-of-function effects [104,105]. With p53 gain-of-function mutants, the resistance to DNA damaging agents arises both through downregulation of anti-proliferative and pro-apoptotic genes, such as FAS and MST-1, and through upregulation of pro-proliferative and anti-apoptotic genes, such as BAG-1 and NFKB2 [94,101]. This finding raises an important question of whether drug resistance from a combination of loss-of-function and gain-of-function effects will be greater than that from loss of p53 function alone. Investigations to address this question have used either transfection of p53-null cells (e.g., human lung adenocarcinoma H1299 and human osteosarcoma Saos-2 models) with p53 mutants or siRNA-dependent downregulation of p53 mutants. Interestingly, both approaches have demonstrated that tumor cells expressing p53 gain-of-function mutants, such as R175H, R179H, and R273H, exhibit significantly greater resistance to a number of antitumor drugs, including doxorubicin, cisplatin, etoposide, and 5-FU [90,94,95,103,106]. Examples of this effect are shown in Table 1. Note that increased resistance is not always observed but can be drug-dependent, as evidenced by the unexpectedly increased sensitivity to etoposide of H1299 tumor cells transfected with the p53-R273H mutant. With this mutant, the additional resistance to methotrexate or doxorubicin has been reported to arise through downregulation of procaspase-3 [107].
The potential of knocking down p53 gain-of-function mutants as a therapeutic option to sensitize tumor cells to antitumor agents is apparent from the above discussion and has been proposed independently by others [108]. However, it should be noted that this sensitization will be partial because p53-knockdown tumor cells will still retain resistance due to persistent loss of p53 functions. Therefore, approaches that convert the p53 gain-of-function mutant configuration to wild-type p53 are likely to be more successful. Indeed, substantial effort has been devoted by the research community toward the ultimate goal of identifying a viable therapeutic option for cancers expressing mutant p53. This concerted effort has led to the identification of several agents with clinical potential, such as the small molecules CP-31398, MIRA-1, STIMA-1, and PRIMA-1, and the PRIMA-1 analog APR-246 [109,110], which has recently entered phase I/II clinical trials [111].
3.3. Resistance in tumor cells harboring wild-type p53
The ~50% mutation rate of p53 in cancer and the associated failure in antitumor drug response has been pivotal in maintaining much interest in mutant p53 with the goal of developing more effective therapeutic solutions [112]. However, antitumor drug resistance is not limited to tumors expressing mutant or null p53. Indeed, in direct contradiction of the normal role of p53 in promoting apoptotic response with antitumor agents, as discussed here, it has become increasingly clear from the clinical literature that resistance is also observed in cancers harboring wild-type p53. The major evidence supporting this conclusion can be gleaned from the clinical reports summarized in Table 2. In the examples shown, the incidence of wild-type p53 is in the range from 40% to almost 100%. As mentioned earlier, cancers of the choroid plexus and germ cell tumors expressing wild-type p53 are exquisitely sensitive to therapeutic regimens, which are highly curative, as indicated by the 5-year survival rate of 90–100% [84,88,113]. In contrast, the presence of wild-type p53 in other advanced cancers, such as bladder, breast, CLL, mesothelioma, non-small cell lung cancer (NSCLC), ovarian, and RCC, results in significantly lower response and/or curative rates, ranging from 0% to ~60% [85,87,114–126]. A low p53 mutation rate is not predictive of greater therapeutic responses or long-term survival rates. This is best illustrated by comparing CLL, germ cell tumors, mesothelioma, and RCC. All of these cancers have a wild-type p53 incidence of ≥90%, but the response rate was best in germ cell tumors (~90%), intermediate in CLL (59–83%), and worst in mesothelioma and RCC (0–33%) [87,88,113,119–122,126,127].
Table 2.
Clinical response of cancers harboring wild-type p53 to chemotherapy.
| Cancer type [Ref.] | Treatment | Incidence (%) | Response rate (%) | 5-Year survival rate (%) |
|---|---|---|---|---|
| Bladder [123] | Various | 49 | Not given | 45 |
| Breast [85] | Paclitaxel | 67 | 45 | Not given |
| Choroid plexus [84] | Various | 50 | Not given | 100 |
| CLL [87] | Chl/flu/cyc | 92 | 83 | 59 |
| Germ cell [88,113] | Platinum | ~100 | ~90 | ~90 |
| Mesothelioma [117–121] | Various | ~90 | 5–33 | 0–14 |
| NSCLC [114,115,125] | Various | 68 | 6–43 | 0–13 |
| Ovarian [116,124] | Paclitaxel/platinum | 40 | 47 | 43 |
| RCC [122,126,127] | Various | ~90 | Not given | ~30 |
Abbreviations: Chl/flu/cyc, chlorambucil/fludarabine/cyclophosphamide; CLL, chronic lymphocytic leukemia; NSCLC, non-small cell lung cancer; RCC, renal cell carcinoma.
The clinical data presented here clearly support the notion that the presence of wild-type p53 is a poor predictor of response and/or cure; many cancers harboring wild-type p53, as with mutant p53, are resistant to a variety of antitumor agents, including cisplatin, paclitaxel, and 5-FU. To explain this paradox requires a discussion of possible mechanisms that may downregulate wild-type p53 function. The mechanisms can involve deregulation of any component of the DNA damage pathway (see Fig. 1), but here we focus on mechanisms that directly impinge on p53: upregulation of pathways that promote p53 degradation and downregulation of pathways involved in post-translational modification.
3.3.1. Mechanisms promoting degradation of wild-type p53
Mechanisms that enhance degradation of wild-type p53 are the simplest way for tumor cells to select for attenuated p53 functions and demonstrate resistance to therapeutic agents. Indeed, accelerated proteasomal degradation of p53 and induction of resistance to anticancer therapy have been demonstrated through upregulation of specific p53-binding proteins, such as human papillomavirus type 16 (HPV-16) E6, Mdm2, and Mdm4 [22]. HPV-16 is found in several cancers, the most common of which is cervical cancer. HPV-16 expresses the E6 viral oncoprotein, which promotes ubiquitin-dependent proteasomal degradation of p53 and leads to low levels of this tumor suppressor in infected tumor cells [128]. As a result, HPV-positive cervical cancer cells are resistant to chemotherapeutic agents such as camptothecin and cisplatin [129].
As discussed here, p53 activation and stability are negatively regulated in a controlled manner via proteasomal degradation by Mdm2. However, Mdm2 gene expression can become deregulated, leading to overproduction of this oncoprotein in human and mouse tumors, as well as tumor cell lines, by one of three different mechanisms: gene amplification, increased transcription, or enhanced translation [130,131]. The overall frequency of Mdm2 amplification in human tumors is ~7–10%, but tissues such as osteosarcomas (16%), soft tissue tumors (31%), hepatocellular carcinoma (44%), and Hodgkin disease (67%) have some of the highest frequencies [22]. The overexpression of Mdm2 in cells substantially alters the stoichiometry with wild-type p53 and likely promotes cancer development [132]. Indeed, a 2-fold increase in Mdm2 has been reported as sufficient to attenuate p53 activation and thereby increase the risk of breast cancer [133]. In parallel, overexpression of Mdm2 leads to a reduction in therapeutic response. This is again well demonstrated with breast cancer, where in an extensive analysis of >2000 patients, those with tumors overexpressing Mdm2 had a significantly lower 10-year survival rate (58–61% vs. 73%) [134]. Such negative roles of Mdm2 in antitumor response have stimulated the search for small-molecule antagonists of p53–Mdm2 interactions to enhance the apoptotic activity of wild-type p53 [135]. Several molecules of interest have been identified, but probably the best known is Nutlin-3a, which displays p53-dependent activity against tumors overexpressing Mdm2, including those with upregulated Mdm2 as a result of ATM mutation [22,136–138]. Nutlin-3a can effectively stabilize wild-type p53 and induce antitumor activity not only as a single agent but also as a potent binary combination with other antitumor agents such as etoposide and cisplatin [139].
Alternative mechanisms involving protein–hprotein interactions can also modulate Mdm2 levels and contribute to the failure of antitumor agents to activate wild-type p53 and induce apoptosis. Mdm4 is one such protein that stabilizes Mdm2 by preventing its proteasomal degradation. Interestingly, 10–20% of cancers overexpress or amplify the Mdm4 gene, which results in a failure to disrupt the p53–Mdm2 complex and culminates in drug resistance [22,140]. This is clearly demonstrated by the ability of Mdm4 to attenuate p53-dependent transactivation of Bax and induction of apoptosis [141]. In contrast to Mdm4, 14-3-3σ normally negatively regulates Mdm2, but downregulation of 14-3-3σ in several cancers, such as ovarian, lung, breast, and prostate cancer, results in significant increases in Mdm2 levels and inhibition of antitumor drug response [142]. Similarly, deregulation of the tumor suppressor p14ARF by deletion, mutation, or epigenetic silencing in human tumors prevents its sequestration of Mdm2 in the nucleolus and thereby increases Mdm2 interactions with wild-type p53 to inhibit p53 functions and induce drug resistance [143–146]. In agreement with this finding, ectopic expression of p14ARF in tumor cells substantially increases p53-dependent G1-phase arrest, apoptosis, and sensitivity to antitumor drugs such as cisplatin [147].
3.3.2. Mechanisms downregulating post-translational activation of wild-type p53
To facilitate antitumor response to cytotoxic drugs, wild-type p53 must be transcriptionally active [148]. However, in drug resistance, this function is lost, and the two genes critically impeded are the checkpoint response gene p21 and the pro-apoptotic gene Bax. Defects in Bax expression have indeed been correlated with adverse drug responses in many human cancers, including pancreatic, colorectal, and breast tumors, irrespective of p53 status [65–67,149]. Similarly, loss of p21 transactivation after exposure of resistant wild-type p53 tumor cells to antitumor agents, such as those based on platinum, has also been reported [150–152]. This finding is consistent with the role of p21 as a tumor suppressor [49,153,154] and with correlations of p21 expression with superior therapeutic drug effects in patients with ovarian cancer and NSCLC [155,156] and in experimental tumor models [44,45,157–159].
Although loss of transactivation function of wild-type p53 can be explained by deregulated expression of Mdm2, Mdm4, 14-3-3σ, or p14ARF, this is not always the case. Thus, in breast cancer, the 45% incidence rate of resistant tumors harboring wild-type p53 (see Table 2) is not consistent with the lower (7–14%) rate of occurrence of Mdm2 overexpression or downregulation of p14ARF [160]. Similarly, the differential half-life of wild-type p53 in sensitive and resistant ovarian tumor cells after DNA damage with cisplatin does not correlate with Mdm2 levels [161]. Moreover, in RCC, in which p53 mutations are rare, wild-type p53 transactivation functions were still reported as defective when tumor cells were exposed to 5-FU, camptothecin, or doxorubicin, but interestingly no changes in Mdm2, Mdm4, or p14ARF were detected to explain these observations [162]. Therefore, it is clear that other mechanisms attenuating wild-type p53 function are also involved, and defects in post-translational modification are a primary mechanism to consider.
3.3.2.1. Significance of phosphorylation in post-translational modifications of p53
Stabilization and activation of p53 is governed by covalent post-translational modifications, such as phosphorylation, ubiquitination, acetylation, methylation, SUMOylation, neddylation, glycosylation, and ribosylation [163,164]. A total of ~50 sites on p53 are now known to be subject to modifications after a stress stimulus [165]. The N-terminus is modified primarily by phosphorylation, whereas the C-terminus can be the target of a variety of modifications, including phosphorylation [163]. Although an optimal combination of post-translational modifications is required for maximal p53 function, the antitumor effects of DNA damaging agents are dominantly governed by phosphorylation events [166,167]. There are ~23 possible sites on p53 that can be phosphorylated, and each site can be targeted by multiple kinases, suggesting possible redundancy [22]. However, it is likely that a specific antitumor agent only activates an individual kinase for each of the several sites targeted for phosphorylation. As a result, a DNA damaging agent produces a unique signature of p53-phosphorylated sites that induces p53 to adopt a specific structural conformation and thereby transactivate a select set of downstream target genes specific for that agent. It is highly likely that deregulation of any critical kinase will alter the unique p53 phosphorylation signature, impede gene transactivation, and attenuate antitumor response.
Of the multiple p53 phosphorylation sites that have been associated with its anti-proliferative and apoptotic functions, Ser15, Thr18, and Ser20 in the DNA-binding domain at the N-terminus are recognized as perhaps the most critical and are therefore the most extensively studied [22,168]. Indeed, these three sites are highly conserved between urochordates, where evolution of the p53–Mdm2 axis appeared, and humans [168]. Studies demonstrate that phosphorylations at these sites promote not only dissociation of p53 from Mdm2, as a prerequisite for p53 stabilization, but also its transactivation function, by inducing concomitant binding of the freed p53 with transcription co-activators, such as p300 and cAMP-response element-binding protein-binding protein (CBP) [165,169–171]. Moreover, several studies suggest that the effects of phosphorylation at these sites may be cooperative, particularly because Thr18 and Ser20 phosphorylations are strongly dependent on the priming phosphorylation at Ser15 [172–174]. Indeed, dual phosphorylation at Ser20 with either Ser15 or Thr18 is sufficient to induce apoptosis in glioma cells harboring wild-type p53 [175,176]. Further support is provided by studies in transgenic mice where mutations induced by replacing both Ser15 and Ser20 with alanine produced a more severe phenotype than a single mutation at either site, as evidenced by loss in apoptotic capacity, defects in replicative senescence, and latency in tumor appearance [163].
3.3.2.2. Downregulation of kinases involved in post-translational modifications of p53
Because of the seminal role of phosphorylation in regulating p53 stabilization and function, loss of this post-translational event at any one of the critical sites is likely to modulate cytotoxic response to antitumor agents. A case in point is with ATM, a transducer of DNA damage signals that upon exposure of tumor cells to antitumor agents phosphorylates p53 primarily at Ser15; this, in turn, stimulates phosphorylation at Thr18 and Ser20 by other kinases, such as Chk2. ATM also phosphorylates and activates homeodomain-interacting protein kinase 2 (HIPK2), a kinase that, in turn, modifies p53 at Ser46 [165]. Therefore, it is not surprising that mutation of ATM in ataxia telangiectasia leads to defects in p53 regulation and checkpoint response after DNA damage with ionizing radiation [177]. Interestingly, defective ATM function has been observed in ~20% of patients with B-cell CLL, which predominantly harbors wild-type p53, and ATM dysfunction correlates with resistance to fludarabine and with poor prognosis in patients [137,178]. In addition, deregulation of Ser376 phosphorylation of p53 has been observed in radioresistant melanoma, with loss of wild-type p53 function attributed to a loss in ATM-dependent signaling [179]. Because ATM can also affect the function of HIPK2 and, thereby, p53-dependent apoptosis, it would be expected that direct loss of HIPK2 would similarly induce drug resistance. Indeed, knockdown of HIPK2 by siRNA in wild-type p53 colorectal tumor cells resulted in resistance to doxorubicin and, more importantly, patients with colorectal cancers expressing low levels of HIPK2 had a significantly lower survival rate [180].
The DNA damage checkpoint kinase Chk2 is also a downstream target of ATM and has been identified as a tumor suppressor, the loss of which leads to multi-organ susceptibility to cancer [181]. Not surprisingly, suppression of either ATM or Chk2 in the presence of wild-type p53 decreased sensitivity of mouse embryonic fibroblasts (MEFs) or tumor cells to doxorubicin [182]. As a member of the calcium calmodulin kinase superfamily, Chk2 has the intrinsic ability to phosphorylate both the Thr18 and Ser20 sites, as well as several other sites in the C-terminus of p53 [183–185]. Interestingly, Chk2 can also be activated independently of ATM, and in the case of cisplatin, the homolog ATR appears to be involved in not only upregulating Chk2 to mediate the DNA damage signal to p53 but also phosphorylating p53 directly at Ser15 [186].
The importance of Chk2 in DNA damage response can be readily appreciated from reports that Chk2-null cells from genetically engineered mice are remarkably resistant to some DNA damaging agents [181,187,188]. The significance of these observations becomes clear when it is noted that Chk2 downregulation has been correlated with resistance to cisplatin in lung cancer cell lines [189]. Moreover, defects in Chk2 through epigenetic mechanisms have been observed clinically in 83% of tumor specimens from patients with NSCLC [189], a highly refractory cancer where cisplatin-based therapy is heavily used, but with a poor survival rate (see Table 2). In addition, 23% of clinical ovarian cancers are reported to be negative for Chk2 [190], a rate that is similar to that of cisplatin-resistant ovarian cancers harboring wild-type p53 (20–22%) [116,191]. It should also be noted as a caution that the results of independent studies with DNA damaging agents in knockout cell lines or in vivo murine models have been at odds with the roles of Chk2 and site-specific phosphorylation of the transactivation domain in p53 stabilization and stimulation and in cell death [22,192]. It has also been reported that defects in Chk2 are rare and, therefore, not relevant [193]. Nevertheless, other clinical assessments strongly indicate that a Chk2 defect is a significant occurrence [189]. In strong support of this is the recent finding that defects in p53 and Chk2 are mutually exclusive, as determined from an extensive analysis of epithelial malignancies, which revealed that 37% of tumors (124 of 335 cases) had either a p53 or a Chk2 defect but that a defect in both within the same tumor was rare (2/335) [182]. A similar mutual exclusivity between defects in either p53 or ATM was also demonstrated in that study. Thus, defects in ATM and Chk2 could account for drug resistance in tumor cells harboring wild-type p53.
A second critical member of the calcium calmodulin kinase superfamily involved in checkpoint response is Chk1, which is also activated by cisplatin in an ATR-dependent manner and targets p53 sites similar to those targeted by Chk2 [22,184]. However, the importance of the ATR/Chk1 pathway in p53 regulation and apoptosis has been difficult to demonstrate because ATR or Chk1 deficiency is embryonically lethal [194,195]. Other approaches using dominant-negative ATR mutants or the Chk1 kinase inhibitor UCN-01 have been possible, but the data have surprisingly demonstrated increased tumor cell sensitivity to DNA damaging agents [195–198]. It is notable that ATR and Chk1 mutations have been observed in 10–20% of gastric cancers [199], but studies to ascertain resultant effects on therapeutic outcomes and p53 functions are needed to determine whether defects in ATR and Chk1 can affect drug response in this disease.
4. Wild-type p53 gain-of-resistance phenotype
Given the seminal antitumor and apoptotic properties of p53, tumor cells resistant to antitumor agents in the presence of wild-type p53 represent somewhat of a contradiction; more importantly, this contradiction poses a major challenge in using p53 status as a prognostic factor in personalized medicine. However, the challenge may be far greater than envisaged so far, as it becomes apparent from sporadic reports in the literature that drug resistance in the context of a retained wild-type p53 genotype is greater than expected. Clinical evidence supports this notion, which is endorsed by data garnered from preclinical studies. For the purpose of this review, we term this observation a “gain-of-resistance” phenotype and consider whether it may be a direct result of a gain-of-function effect associated with wild-type p53.
4.1. Clinical data
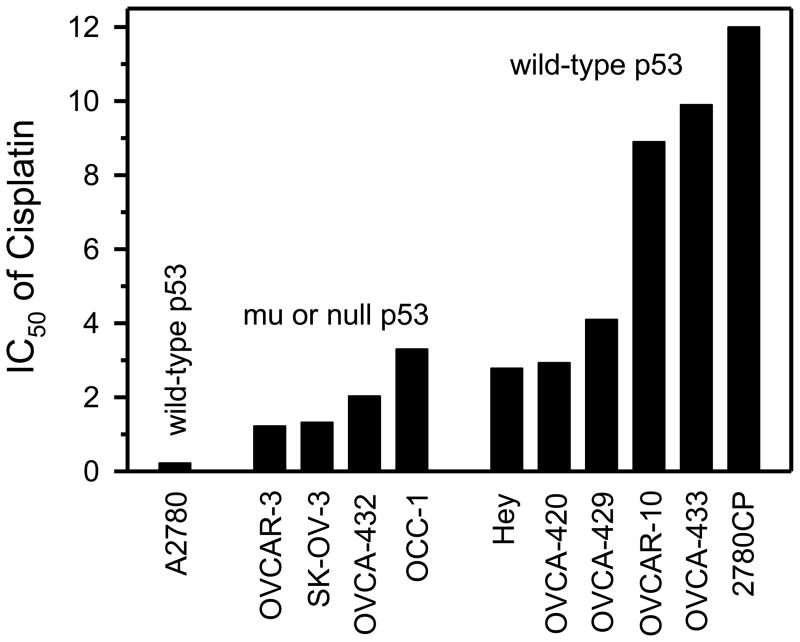
Several clinical studies have examined the role of p53 gene status in tumor response, and have concluded that the prognostic value of this tumor suppressor is low. This conclusion stems from clinical observation of therapeutic resistance in both mutant and wild-type p53 cancers, as discussed here. Similarly, tumor responses can also be observed independent of p53 status. For instance, in a clinical study of advanced ovarian cancer, cisplatin treatment induced a response in 46% of the patients in the wild-type p53 group and in a similar percentage (37%) of patients in the mutant p53 group [191]. In direct contrast, other studies have demonstrated that responses in the mutant p53 group are in fact significantly greater, as has been seen in advanced ovarian cancer with the platinum/paclitaxel combination (mutant, 86%; wild-type, 47%) [116], in NSCLC with paclitaxel (mutant, 75%; wild-type, 47%) [114], in metastatic bladder cancer with platinum-based combinations (mutant, 50%; wild-type, 27%) [200], and in advanced breast cancer with paclitaxel (mutant, 83%; wild-type, 38%) [86]. In non-inflammatory breast cancer, the contrast is even more striking. In this disease, >50% of the patients harboring mutant p53 responded to a dose-intense epirubicin–cyclophosphamide regimen, whereas none of the >100 patients with tumors harboring wild-type p53 responded to this regimen [14]. Interestingly, in a rare study involving serial sampling of ovarian cancer ascites and assessment in vitro of drug response, the intrinsic resistance of tumor cells to cisplatin increased 1.4-fold during therapy while cells retained wild-type p53 status; however, this resistance decreased below the intrinsic level after p53 became mutated as therapy progressed [201]. Moreover, because clinical responses are limited by the maximum tolerated dose, one may predict that responses would increase if higher doses were permissible. This appears to be the case with high-dose cisplatin or carboplatin therapy, where a 30% response rate has been observed in ovarian tumors already resistant to platinum therapy [202]. However, this response rate was concluded not to increase further with increasing dose on the basis of a critical analysis of carboplatin dose–response data from clinical investigations [203]. Thus, there appears to be a fraction of refractory ovarian tumors that are supra-resistant, and, on the basis of the discussion so far and from the preclinical data presented in Figure 2 (see below), these tumors are predicted to harbor wild-type p53.
Fig. 2.
Cisplatin resistance in an ovarian tumor panel according to p53 status. The A2780 cell line is the sensitive tumor model. The resistant models are grouped into mutant (mu)/null p53 (N = 4) and wild-type p53 (N = 6); the 2780CP cell line was developed in tissue culture from A2780 cells, whereas all others were established directly from patients’ biopsies. The IC50 value is the concentration of cisplatin that inhibits cell proliferation by 50% and is expressed as μM. Adapted from Hagopian et al. [40] and Siddik et al. [213].
Collectively, the clinical studies discussed here have been instrumental in justifying the use of appropriate therapeutic strategies involving selective antitumor agents (e.g., paclitaxel) against mutant p53 tumors in specific disease types. However, the underlying basis for the relatively poorer responses when wild-type p53 is present has been overlooked, even when it is clear that the activity of the agent (e.g., paclitaxel) is also dependent on the presence of wild-type p53 [204]. Nevertheless, these reports provide evidence for the existence of wild-type p53 cancers that are more resistant to therapy than mutant p53 cancers are; that is, the wild-type p53 cancers are displaying the gain-of-resistance phenotype.
4.2. Preclinical data
As in clinical studies, the effect of p53 status on response to antitumor agents has also been determined in tumor panels of various disease types. The cytotoxic data from head and neck squamous cell carcinoma (HNSCC), RCC, and ovarian cancer after exposure to cisplatin or paclitaxel have been reported in subgroups of mutant/null p53 or wild-type p53 models [40,205–207] and are listed in Table 3A. In these examples, the cytotoxicity is relatively lower (that is, the drug concentration that inhibits cell proliferation by 50% [IC50] is higher, or the resistance is higher) when p53 is wild-type. Interestingly, the degree of increased resistance with wild-type p53 was significant, and the ratio of IC50 between wild-type p53 and mutant/null p53 ranged from 2 to 77. More importantly, these data demonstrate the presence of additional drug resistance in wild-type p53 tumor models over and above that displayed in mutant p53 cancers.
Table 3.
Greater drug resistance in tumor panels and cell lines expressing wild-type p53.
| A. Studies in tumor panels
| |||||
|---|---|---|---|---|---|
| Tumor panel [Ref.] | Panel size (N) | Treatment (unit of IC50) | Mean IC50 (N)
|
Folda | |
| wt p53 | mu/null p53 | ||||
| HNSCC [205] | 23 | Cisplatin (μM) | 13.7 (10) | 6.8 (13) | 2 |
| Ovarian [40] | 9 | Cisplatin (μM) | 5.7 (5) | 2.0 (4) | 2.9 |
| Ovarian [206] | 15 | Paclitaxel (μM) | 0.077 (6) | 0.015 (9) | 5 |
| RCC [207] | 8 | Paclitaxel (μM) | 1.54 (6) | 0.02 (2) | 77 |
| B. Studies modulating wild-type p53 in cell lines by molecular approaches
| |||||
|---|---|---|---|---|---|
| Tumor type [Ref.] | Cell line | Treatment (unit of IC50) | Mean IC50 |
Folda | |
| wt p53 | dn/ko/kd p53 | ||||
| Glioma [208] | U87MG | Carmustine (μg/mL) | 7 | 3 | 2.3 |
| Colon [209] | HCT-116 | Trabectedin (nM) | 14.6 | 4.4 | 3.3 |
| Colon [210] | HCT-116 | Cisplatin (μM) | 3.9 | 0.9 | 4.3 |
| Breast [209] | MCF-7 | Trabectedin (nM) | >100 | 0.23 | >400 |
| Breast [211] | MCF-7 | Cisplatin (μM) | 3.38 | 1.69 | 2 |
Abbreviations: dn, dominant-negative; HNSCC, head and neck squamous cell carcinoma; IC50, drug concentration (in μg/ml, nM, or μM) that inhibits cell proliferation by 50%; kd, knockdown; ko, knockout; N, number of cell lines in group; RCC, renal cell carcinoma; wt, wild-type.
Fold, ratio of IC50 between wt p53 and mu/null p53 or dn/ko/kd p53 groups.
Additional data to support the wild-type p53 gain-of-resistance phenotype have been garnered from studies in isogenic systems after disruption of p53 with dominant-negative mutant p53, p53 knockout, or p53 knockdown by siRNA. Thus, if wild-type p53 indeed contributes to gain of resistance, then disruption of p53 should lower resistance. This is analogous to the studies with the p53 gain-of-function mutant, where the higher level of resistance to antitumor agents, such as cisplatin, was reduced by p53 siRNA [108]. Indeed, cell lines derived from gliomas, colon cancers, or breast cancers that were exposed to DNA damaging agents from three different structural classes displayed reduced IC50 (decreased resistance) by 2-fold to >400-fold after disruption of p53 by any of the three established procedures (Table 3B) [208–211]. The cell lines MCF-7 and HCT-116 have generally been considered to have a drug-sensitive phenotype, but reports indicate that they harbor a defect in apoptosis [210,211] and therefore may carry some intrinsic resistance that is reduced when wild-type p53 is disrupted. Sensitization of tumor cells to cisplatin by knockdown of wild-type p53 with siRNA has also been observed in Bcr/Abl-transformed murine hematopoietic cells [212]. In an alternative approach, these cells were transfected with temperature-sensitive p53, and resistance to cisplatin was found to be higher in cells expressing wild-type p53 at the permissive temperature of 32°C than in cells expressing mutant p53 at 37°C. A limitation of these data is the difficulty in ascertaining the level of resistance in mutant/null p53 versus wild-type p53 cell lines relative to a standard sensitive model. This limitation can be partially addressed using our previous findings in an ovarian tumor panel [40,213], which are shown in Figure 2. Compared with established cisplatin-sensitive A2780 cells, which harbor wild-type p53 and have intact checkpoint responses, all other models shown in the figure were resistant to cisplatin (had higher IC50 values), irrespective of their p53 status. However, it is notable that models harboring wild-type p53 had greater resistance than those harboring mutant p53.
The underlying basis of chemotherapy is that normal cells with wild-type p53 must have relatively greater resistance to DNA damaging drugs than tumor cells have. This is amply demonstrated by the report that the resistance of normal human foreskin fibroblasts to cisplatin, carboplatin, and paclitaxel is reduced 6- to 12-fold when wild-type p53 is disrupted by ectopic expression of the HPV-16 E6 oncogene [214]. Taken together, these findings are consistent with the premise that wild-type p53 tumor cells can indeed demonstrate supra-resistance, not unlike that in normal cells, and that wild-type p53 contributes to the greater resistance.
4.3. Underlying basis for wild-type p53 gain-of-resistance phenotype
Although the data presented here support the concept of a wild-type p53 gain-of-resistance phenotype in specific tumor systems, the underlying mechanisms are not known. However, the mechanism requires the presence of wild-type p53 because its knockdown reduces drug resistance in these systems. Thus, mechanisms that promote degradation may be eliminated from consideration. Indeed, such gain-of-resistance model systems do not necessarily demonstrate increased expression of Mdm2, Mdm4, or p14ARF. From an analysis of the available data, it is likely that defects in post-translational mechanisms play a role. This possibility is based in part on data from RCC, where wild-type p53 transactivation functions failed when tumor cells were exposed to antitumor agents even though levels of Mdm2, Mdm4, and p14ARF remained normal [162]. Further support comes from the demonstration that resistance to structurally distinct platinum drugs cisplatin and the trinuclear analog BBR3464 is associated with the inability of DNA damage to induce p21 expression in wild-type p53 models [151,215]. However, normal p21 expression could be restored if cells resistant to cisplatin were exposed to 1R,2R-diaminocyclohexane(trans-diacetato)(dichloro)platinum(IV) (DAP) or if cells resistant to BBR3464 were exposed to cisplatin; that is, the signal transduction pathways induced by BBR3464 and DAP are distinct from those induced by cisplatin. This is well demonstrated with DAP, which fails to phosphorylate p53 at Ser392; in contrast, cisplatin is very effective in modifying this site [215]. Moreover, in the ovarian panel shown in Figure 2, DAP was demonstrated to be highly effective against the wild-type p53 supra-resistant models but was not effective against the mutant/null models [40,213]. Therefore, it can be concluded that, at least for platinum-based drugs, there is a minimum of two parallel upstream pathways that independently converge on wild-type p53 to activate the tumor suppressor, and when one becomes defective and induces supra-resistance, the alternative intact pathway can be activated to bypass the defect and restore p53-dependent drug sensitivity. Furthermore, the transactivation of p21, the anti-proliferative activity of wild-type p53, and the cytotoxicity of antitumor drugs appear to be more dependent on phosphorylation of p53 at Ser15, Thr18, and Ser20 [175,216–218]. This finding supports post-translational defects as a likely cause of a failure to activate p53 and a likely mechanism of the greater resistance observed in tumor cells harboring the wild-type p53 genotype.
As discussed here, part of the evidence supporting the gain-of-resistance phenotype comes from p53 knockdown studies. This phenotype is analogous to the increase in resistance that results from p53 gain-of-function mutants, where sensitivity to antitumor agents is increased after knockdown of the p53 mutant [108]. The supra-resistance associated with wild-type p53 is strongly suggestive of additional functions acquired by p53, although direct evidence for this conjecture is lacking at this stage. Hypothetically, if post-translational defects are indeed involved, as may be envisaged in the absence of ATM, Chk2, or HIPK2 activity, then wild-type p53 may assume a conformation not unlike that of p53 gain-of-function mutants. Whether the specific mechanism involved in the gain-of-resistance phenotype is indeed a result of gain-of-function effects is a question that must await rigorous studies, which are desperately needed not only for the rational design of novel drugs, but also for the clinical application of traditional antitumor agents.
5. Conclusions and future directions
From the limited available data presented in this review, it is evident that drug resistance can be significant, particularly when resistant tumor cells harbor wild-type p53. This indeed represents a paradox as it deviates from the established role of this genotype in facile antitumor drug response. Moreover, the resistance in tumor cells expressing wild-type p53 can be much greater than that in tumor cells expressing mutant p53, leading to the recognition of a wild-type p53 gain-of-resistance phenotype, and it is tempting to speculate that this resistance may be a result of gain-of-function effects associated with wild-type p53. It could be argued that the clinical and preclinical data discussed here represent an increase in the sensitivity of mutant/null p53 cancers to a specific drug through the phenomenon of synthetic lethality, rather than an increase in the resistance of wild-type p53 tumors. A concerted effort by the research community will be required to unravel this question and to define the underlying mechanism for the supra-resistance of wild-type p53 cancers. Such data will be of paramount importance to change the treatment paradigm and to increase responses and cure rates in wild-type p53 gain-of-resistance cancers. Effective prototypical small-molecule drugs (e.g., DAP) already exist and provide evidence that activity against such cancers is possible, but recognition of this gain-of-resistance phenotype in cancer is the first step toward defining the mechanisms involved and thus laying the foundation for rational-based therapies. Such a progression is vital to ultimately achieve success at the clinical level against resistant cancers harboring wild-type p53.
Acknowledgments
Grant support: This research is supported by U.S. Public Health Service grants CA127263 and CA160687 to ZHS and MD Anderson’s Cancer Center Support Grant (CA16672), awarded by the National Cancer Institute, and in part by the Megan McBride Franz Endowed Research Fund.
The authors are grateful to Karen R. Muller, Ph.D., of the Department of Scientific Publications, The University of Texas MD Anderson Cancer Center for professional assistance in the final editorial review of this manuscript.
Abbreviations
- ALL
acute lymphocytic leukemia
- APAF-1
apoptotic protease-activating factor 1
- AIF
apoptosis-inducing factor
- ATM
ataxia telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related
- CBP
cAMP-response element-binding protein-binding protein
- Cdk
cyclin-dependent kinase
- CLL
chronic lymphocytic leukemia
- DAP
1R,2R-diaminocyclohexane(trans-diacetato)(dichloro)platinum(IV)
- DISC
death-inducing signaling complex
- DSBs
DNA double-strand breaks
- FADD
Fas-associated death domain
- 5-FU
5-fluorouracil
- HIPK2
homeodomain-interacting protein kinase 2
- HNSCC
head and neck squamous cell carcinoma
- HPV-16
human papillomavirus type 16
- IC50
drug concentration that inhibits cell proliferation by 50%
- MEF
mouse embryonic fibroblasts
- MRN
Mre11/Rad50/NBS1 complex
- NSCLC
non-small cell lung cancer
- RCC
renal cell carcinoma
- TNF-R
tumor necrosis factor receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, et al. Gene expression profiles in normal and cancer cells. Science. 1997;276:1268–72. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 2.Ismail RS, Baldwin RL, Fang J, Browning D, Karlan BY, Gasson JC, et al. Differential gene expression between normal and tumor-derived ovarian epithelial cells. Cancer Res. 2000;60:6744–9. [PubMed] [Google Scholar]
- 3.Arango D, Wilson AJ, Shi Q, Corner GA, Aranes MJ, Nicholas C, et al. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br J Cancer. 2004;91:1931–46. doi: 10.1038/sj.bjc.6602215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roberts D, Schick J, Conway S, Biade S, Laub PB, Stevenson JP, et al. Identification of genes associated with platinum drug sensitivity and resistance in human ovarian cancer cells. Br J Cancer. 2005;92:1149–58. doi: 10.1038/sj.bjc.6602447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Connor PM, Jackman J, Bae I, Myers TG, Fan S, Mutoh M, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer Res. 1997;57:4285–300. [PubMed] [Google Scholar]
- 6.Kastan MB. Wild-type p53: tumors can’t stand it. Cell. 2007;128:837–40. doi: 10.1016/j.cell.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 7.van der Zee AG, Hollema H, Suurmeijer AJ, Krans M, Sluiter WJ, Willemse PH, et al. Value of P-glycoprotein, glutathione S-transferase pi, c-erbB-2, and p53 as prognostic factors in ovarian carcinomas. Journal of Clinical Oncology. 1995;13:70–8. doi: 10.1200/JCO.1995.13.1.70. [DOI] [PubMed] [Google Scholar]
- 8.Reles A, Wen WH, Schmider A, Gee C, Runnebaum IB, Kilian U, et al. Correlation of p53 mutations with resistance to platinum-based chemotherapy and shortened survival in ovarian cancer. Clin Cancer Res. 2001;7:2984–97. [PubMed] [Google Scholar]
- 9.Eliopoulos AG, Kerr DJ, Herod J, Hodgkins L, Krajewski S, Reed JC, et al. The control of apoptosis and drug resistance in ovarian cancer: influence of p53 and Bcl-2. Oncogene. 1995;11:1217–28. [PubMed] [Google Scholar]
- 10.Fan S, El Deiry WS, Bae I, Freeman J, Jondle D, Bhatia K, et al. p53 gene mutations are associated with decreased sensitivity of human lymphoma cells to DNA damaging agents. Cancer Res. 1994;54:5824–30. [PubMed] [Google Scholar]
- 11.Perego P, Giarola M, Righetti SC, Supino R, Caserini C, Delia D, et al. Association between cisplatin resistance and mutation of p53 gene and reduced bax expression in ovarian carcinoma cell systems. Cancer Res. 1996;56:556–62. [PubMed] [Google Scholar]
- 12.Ozols RF. Chemotherapy for advanced epithelial ovarian cancer. Hematol Oncol Clin North Am. 1992;6:879–94. [PubMed] [Google Scholar]
- 13.Smith ND, Rubenstein JN, Eggener SE, Kozlowski JM. The p53 tumor suppressor gene and nuclear protein: basic science review and relevance in the management of bladder cancer. J Urol. 2003;169:1219–28. doi: 10.1097/01.ju.0000056085.58221.80. [DOI] [PubMed] [Google Scholar]
- 14.Varna M, Bousquet G, Plassa LF, Bertheau P, Janin A. TP53 status and response to treatment in breast cancers. J Biomed Biotechnol. 2011;2011:284584. doi: 10.1155/2011/284584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang W, Rastinejad F, El Deiry WS. Restoring p53-dependent tumor suppression. Cancer Biol Ther. 2003;2:S55–S63. [PubMed] [Google Scholar]
- 17.Lain S. Protecting p53 from degradation. Biochem Soc Trans. 2003;31:482–5. doi: 10.1042/bst0310482. [DOI] [PubMed] [Google Scholar]
- 18.Mandinova A, Lee SW. The p53 pathway as a target in cancer therapeutics: obstacles and promise. Sci Transl Med. 2011;3:64rv1. doi: 10.1126/scitranslmed.3001366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu C, El Deiry WS. Targeting p53 for enhanced radio- and chemo-sensitivity. Apoptosis. 2009;14:597–606. doi: 10.1007/s10495-009-0330-1. [DOI] [PubMed] [Google Scholar]
- 20.Bell HS, Ryan KM. Targeting the p53 family for cancer therapy: ‘big brother’ joins the fight. Cell Cycle. 2007;6:1995–2000. doi: 10.4161/cc.6.16.4614. [DOI] [PubMed] [Google Scholar]
- 21.Meek DW. Mechanisms of switching on p53: a role for covalent modification? Oncogene. 1999;18:7666–75. doi: 10.1038/sj.onc.1202951. [DOI] [PubMed] [Google Scholar]
- 22.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 23.Lain S, Lane D. Improving cancer therapy by non-genotoxic activation of p53. Eur J Cancer. 2003;39:1053–60. doi: 10.1016/s0959-8049(03)00063-7. [DOI] [PubMed] [Google Scholar]
- 24.Al Ejeh F, Kumar R, Wiegmans A, Lakhani SR, Brown MP, Khanna KK. Harnessing the complexity of DNA-damage response pathways to improve cancer treatment outcomes. Oncogene. 2010;29:6085–98. doi: 10.1038/onc.2010.407. [DOI] [PubMed] [Google Scholar]
- 25.Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6:1006–10. doi: 10.4161/cc.6.9.4211. [DOI] [PubMed] [Google Scholar]
- 26.Lane DP, Lain S. Therapeutic exploitation of the p53 pathway. Trends Mol Med. 2002;8:S38–S42. doi: 10.1016/s1471-4914(02)02309-2. [DOI] [PubMed] [Google Scholar]
- 27.Schuler M, Green DR. Mechanisms of p53-dependent apoptosis. Biochem Soc Trans. 2001;29:684–8. doi: 10.1042/0300-5127:0290684. [DOI] [PubMed] [Google Scholar]
- 28.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis - the p53 network. J Cell Sci. 2003;116:4077–85. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Zhang L. No PUMA, no death: implications for p53-dependent apoptosis. Cancer Cell. 2003;4:248–9. doi: 10.1016/s1535-6108(03)00249-6. [DOI] [PubMed] [Google Scholar]
- 30.Henry H, Thomas A, Shen Y, White E. Regulation of the mitochondrial checkpoint in p53-mediated apoptosis confers resistance to cell death. Oncogene. 2002;21:748–60. doi: 10.1038/sj.onc.1205125. [DOI] [PubMed] [Google Scholar]
- 31.Erster S, Moll UM. Stress-induced p53 runs a direct mitochondrial death program: its role in physiologic and pathophysiologic stress responses in vivo. Cell Cycle. 2004;3:1492–5. doi: 10.4161/cc.3.12.1318. [DOI] [PubMed] [Google Scholar]
- 32.Galluzzi L, Morselli E, Kepp O, Tajeddine N, Kroemer G. Targeting p53 to mitochondria for cancer therapy. Cell Cycle. 2008;7:1949–55. doi: 10.4161/cc.7.13.6222. [DOI] [PubMed] [Google Scholar]
- 33.Erster S, Moll UM. Stress-induced p53 runs a transcription-independent death program. Biochem Biophys Res Commun. 2005;331:843–50. doi: 10.1016/j.bbrc.2005.03.187. [DOI] [PubMed] [Google Scholar]
- 34.Kaina B. DNA damage-triggered apoptosis: critical role of DNA repair, double-strand breaks, cell proliferation and signaling. Biochem Pharmacol. 2003;66:1547–54. doi: 10.1016/s0006-2952(03)00510-0. [DOI] [PubMed] [Google Scholar]
- 35.Brown JM, Attardi LD. The role of apoptosis in cancer development and treatment response. Nat Rev Cancer. 2005;5:231–7. doi: 10.1038/nrc1560. [DOI] [PubMed] [Google Scholar]
- 36.Iliakis G, Wang Y, Guan J, Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22:5834–47. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- 37.Samuel T, Weber HO, Funk JO. Linking DNA damage to cell cycle checkpoints. Cell Cycle. 2002;1:162–8. [PubMed] [Google Scholar]
- 38.Pietenpol JA, Stewart ZA. Cell cycle checkpoint signaling: cell cycle arrest versus apoptosis. Toxicology. 2002;181–182:475–81. doi: 10.1016/s0300-483x(02)00460-2. [DOI] [PubMed] [Google Scholar]
- 39.Gartel AL, Tyner AL. Transcriptional regulation of the p21((WAF1/CIP1)) gene. Exp Cell Res. 1999;246:280–9. doi: 10.1006/excr.1998.4319. [DOI] [PubMed] [Google Scholar]
- 40.Hagopian GS, Mills GB, Khokhar AR, Bast RC, Jr, Siddik ZH. Expression of p53 in cisplatin-resistant ovarian cancer cell lines: modulation with the novel platinum analogue (1R, 2R- diaminocyclohexane)(trans-diacetato)(dichloro)-platinum(IV) Clin Cancer Res. 1999;5:655–63. [PubMed] [Google Scholar]
- 41.O’Connor PM, Kohn KW. A fundamental role for cell cycle regulation in the chemosensitivity of cancer cells? Semin Cancer Biol. 1992;3:409–16. [PubMed] [Google Scholar]
- 42.O’Connor PM, Fan S. DNA damage checkpoints: implications for cancer therapy. Prog Cell Cycle Res. 1996;2:165–73. doi: 10.1007/978-1-4615-5873-6_16. [DOI] [PubMed] [Google Scholar]
- 43.Vekris A, Meynard D, Haaz MC, Bayssas M, Bonnet J, Robert J. Molecular determinants of the cytotoxicity of platinum compounds: the contribution of in silico research. Cancer Res. 2004;64:356–62. doi: 10.1158/0008-5472.can-03-2258. [DOI] [PubMed] [Google Scholar]
- 44.Lincet H, Poulain L, Remy JS, Deslandes E, Duigou F, Gauduchon P, et al. The p21(cip1/waf1) cyclin-dependent kinase inhibitor enhances the cytotoxic effect of cisplatin in human ovarian carcinoma cells. Cancer Lett. 2000;161:17–26. doi: 10.1016/s0304-3835(00)00586-3. [DOI] [PubMed] [Google Scholar]
- 45.Wu Q, Kirschmeier P, Hockenberry T, Yang TY, Brassard DL, Wang L, et al. Transcriptional regulation during p21WAF1/CIP1-induced apoptosis in human ovarian cancer cells. J Biol Chem. 2002;277:36329–37. doi: 10.1074/jbc.M204962200. [DOI] [PubMed] [Google Scholar]
- 46.Fry DW, Harvey PJ, Keller PR, Elliott WL, Meade M, Trachet E, et al. Specific inhibition of cyclin-dependent kinase 4/6 by PD 0332991 and associated antitumor activity in human tumor xenografts. Mol Cancer Ther. 2004;3:1427–38. [PubMed] [Google Scholar]
- 47.Soni R, O’Reilly T, Furet P, Muller L, Stephan C, Zumstein-Mecker S, et al. Selective in vivo and in vitro effects of a small molecule inhibitor of cyclin-dependent kinase 4. J Natl Cancer Inst. 2001;93:436–46. doi: 10.1093/jnci/93.6.436. [DOI] [PubMed] [Google Scholar]
- 48.Deng C, Zhang P, Harper JW, Elledge SJ, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–84. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 49.Martin-Caballero J, Flores JM, Garcia-Palencia P, Serrano M. Tumor susceptibility of p21(Waf1/Cip1)-deficient mice. Cancer Res. 2001;61:6234–8. [PubMed] [Google Scholar]
- 50.Itahana K, Dimri G, Campisi J. Regulation of cellular senescence by p53. Eur J Biochem. 2001;268:2784–91. doi: 10.1046/j.1432-1327.2001.02228.x. [DOI] [PubMed] [Google Scholar]
- 51.Stuhmer T, Bargou RC. Selective pharmacologic activation of the p53-dependent pathway as a therapeutic strategy for hematologic malignancies. Cell Cycle. 2006;5:39–42. doi: 10.4161/cc.5.1.2281. [DOI] [PubMed] [Google Scholar]
- 52.Schmitt CA. Cellular senescence and cancer treatment. Biochim Biophys Acta. 2007;1775:5–20. doi: 10.1016/j.bbcan.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. Tumour suppression by p53: the importance of apoptosis and cellular senescence. J Pathol. 2009;219:3–15. doi: 10.1002/path.2584. [DOI] [PubMed] [Google Scholar]
- 54.Gewirtz DA, Holt SE, Elmore LW. Accelerated senescence: an emerging role in tumor cell response to chemotherapy and radiation. Biochem Pharmacol. 2008;76:947–57. doi: 10.1016/j.bcp.2008.06.024. [DOI] [PubMed] [Google Scholar]
- 55.Wang X, Wong SC, Pan J, Tsao SW, Fung KH, Kwong DL, et al. Evidence of cisplatin-induced senescent-like growth arrest in nasopharyngeal carcinoma cells. Cancer Res. 1998;58:5019–22. [PubMed] [Google Scholar]
- 56.Gewirtz DA. Growth arrest and cell death in the breast tumor cell in response to ionizing radiation and chemotherapeutic agents which induce DNA damage. Breast Cancer Res Treat. 2000;62:223–35. doi: 10.1023/a:1006414422919. [DOI] [PubMed] [Google Scholar]
- 57.Chang BD, Broude EV, Dokmanovic M, Zhu H, Ruth A, Xuan Y, et al. A senescence-like phenotype distinguishes tumor cells that undergo terminal proliferation arrest after exposure to anticancer agents. Cancer Res. 1999;59:3761–7. [PubMed] [Google Scholar]
- 58.Chang BD, Swift ME, Shen M, Fang J, Broude EV, Roninson IB. Molecular determinants of terminal growth arrest induced in tumor cells by a chemotherapeutic agent. Proc Natl Acad Sci U S A. 2002;99:389–94. doi: 10.1073/pnas.012602599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wosikowski K, Regis JT, Robey RW, Alvarez M, Buters JT, Gudas JM, et al. Normal p53 status and function despite the development of drug resistance in human breast cancer cells. Cell Growth Differ. 1995;6:1395–403. [PubMed] [Google Scholar]
- 60.Koster R, di Pietro A, Timmer-Bosscha H, Gibcus JH, van den BA, Suurmeijer AJ, et al. Cytoplasmic p21 expression levels determine cisplatin resistance in human testicular cancer. J Clin Invest. 2010;120:3594–605. doi: 10.1172/JCI41939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xia X, Ma Q, Li X, Ji T, Chen P, Xu H, et al. Cytoplasmic p21 is a Potential Predictor for Cisplatin Sensitivity in Ovarian Cancer. BMC Cancer. 2011;11:399. doi: 10.1186/1471-2407-11-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suzuki A, Tsutomi Y, Miura M, Akahane K. Caspase 3 inactivation to suppress Fas-mediated apoptosis: identification of binding domain with p21 and ILP and inactivation machinery by p21. Oncogene. 1999;18:1239–44. doi: 10.1038/sj.onc.1202409. [DOI] [PubMed] [Google Scholar]
- 63.Johnson GG, Sherrington PD, Carter A, Lin K, Liloglou T, Field JK, et al. A novel type of p53 pathway dysfunction in chronic lymphocytic leukemia resulting from two interacting single nucleotide polymorphisms within the p21 gene. Cancer Res. 2009;69:5210–7. doi: 10.1158/0008-5472.CAN-09-0627. [DOI] [PubMed] [Google Scholar]
- 64.Roman-Gomez J, Castillejo JA, Jimenez A, Gonzalez MG, Moreno F, Rodriguez MC, et al. 5′ CpG island hypermethylation is associated with transcriptional silencing of the p21(CIP1/WAF1/SDI1) gene and confers poor prognosis in acute lymphoblastic leukemia. Blood. 2002;99:2291–6. doi: 10.1182/blood.v99.7.2291. [DOI] [PubMed] [Google Scholar]
- 65.Sturm I, Kohne CH, Wolff G, Petrowsky H, Hillebrand T, Hauptmann S, et al. Analysis of the p53/BAX pathway in colorectal cancer: low BAX is a negative prognostic factor in patients with resected liver metastases. J Clin Oncol. 1999;17:1364–74. doi: 10.1200/JCO.1999.17.5.1364. [DOI] [PubMed] [Google Scholar]
- 66.Krajewski S, Blomqvist C, Franssila K, Krajewska M, Wasenius VM, Niskanen E, et al. Reduced expression of proapoptotic gene BAX is associated with poor response rates to combination chemotherapy and shorter survival in women with metastatic breast adenocarcinoma. Cancer Res. 1995;55:4471–8. [PubMed] [Google Scholar]
- 67.Friess H, Lu Z, Graber HU, Zimmermann A, Adler G, Korc M, et al. bax, but not bcl-2, influences the prognosis of human pancreatic cancer. Gut. 1998;43:414–21. doi: 10.1136/gut.43.3.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kashkar H, Kronke M, Jurgensmeier JM. Defective Bax activation in Hodgkin B-cell lines confers resistance to staurosporine-induced apoptosis. Cell Death Differ. 2002;9:750–7. doi: 10.1038/sj.cdd.4401024. [DOI] [PubMed] [Google Scholar]
- 69.Wong MY, Yu Y, Walsh WR, Yang JL. microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review) Int J Oncol. 2011;38:1189–95. doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
- 70.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–9. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 71.Vogt M, Munding J, Gruner M, Liffers ST, Verdoodt B, Hauk J, et al. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011;458:313–22. doi: 10.1007/s00428-010-1030-5. [DOI] [PubMed] [Google Scholar]
- 72.Weeraratne SD, Amani V, Neiss A, Teider N, Scott DK, Pomeroy SL, et al. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011;13:165–75. doi: 10.1093/neuonc/noq179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fujita Y, Kojima K, Hamada N, Ohhashi R, Akao Y, Nozawa Y, et al. Effects of miR-34a on cell growth and chemoresistance in prostate cancer PC3 cells. Biochem Biophys Res Commun. 2008;377:114–9. doi: 10.1016/j.bbrc.2008.09.086. [DOI] [PubMed] [Google Scholar]
- 74.Kim PK, Mahidhara R, Seol DW. The role of caspase-8 in resistance to cancer chemotherapy. Drug Resist Updat. 2001;4:293–6. doi: 10.1054/drup.2001.0223. [DOI] [PubMed] [Google Scholar]
- 75.Duiker EW, Meijer A, van der Bilt AR, Meersma GJ, Kooi N, van der Zee AG, et al. Drug-induced caspase 8 upregulation sensitises cisplatin-resistant ovarian carcinoma cells to rhTRAIL-induced apoptosis. Br J Cancer. 2011;104:1278–87. doi: 10.1038/bjc.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding Z, Yang X, Pater A, Tang SC. Resistance to apoptosis is correlated with the reduced caspase-3 activation and enhanced expression of antiapoptotic proteins in human cervical multidrug-resistant cells. Biochem Biophys Res Commun. 2000;270:415–20. doi: 10.1006/bbrc.2000.2432. [DOI] [PubMed] [Google Scholar]
- 77.Mueller T, Voigt W, Simon H, Fruehauf A, Bulankin A, Grothey A, et al. Failure of activation of caspase-9 induces a higher threshold for apoptosis and cisplatin resistance in testicular cancer. Cancer Res. 2003;63:513–21. [PubMed] [Google Scholar]
- 78.Kojima H, Endo K, Moriyama H, Tanaka Y, Alnemri ES, Slapak CA, et al. Abrogation of mitochondrial cytochrome c release and caspase-3 activation in acquired multidrug resistance. J Biol Chem. 1998;273:16647–50. doi: 10.1074/jbc.273.27.16647. [DOI] [PubMed] [Google Scholar]
- 79.Kuwahara D, Tsutsumi K, Oyake D, Ohta T, Nishikawa H, Koizuka I. Inhibition of caspase-9 activity and Apaf-1 expression in cisplatin-resistant head and neck squamous cell carcinoma cells. Auris Nasus Larynx. 2003;30 (Suppl):S85–S88. doi: 10.1016/s0385-8146(02)00129-3. [DOI] [PubMed] [Google Scholar]
- 80.Wang H, Yang ES, Jiang J, Nowsheen S, Xia F. DNA damage-induced cytotoxicity is dissociated from BRCA1’s DNA repair function but is dependent on its cytosolic accumulation. Cancer Res. 2010;70:6258–67. doi: 10.1158/0008-5472.CAN-09-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Jiang J, Yang ES, Jiang G, Nowsheen S, Wang H, Wang T, et al. p53-dependent BRCA1 nuclear export controls cellular susceptibility to DNA damage. Cancer Res. 2011;71:5546–57. doi: 10.1158/0008-5472.CAN-10-3423. [DOI] [PubMed] [Google Scholar]
- 82.Petitjean A, Achatz MI, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–65. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 83.Goldstein I, Marcel V, Olivier M, Oren M, Rotter V, Hainaut P. Understanding wild-type and mutant p53 activities in human cancer: new landmarks on the way to targeted therapies. Cancer Gene Ther. 2011;18:2–11. doi: 10.1038/cgt.2010.63. [DOI] [PubMed] [Google Scholar]
- 84.Tabori U, Shlien A, Baskin B, Levitt S, Ray P, Alon N, et al. TP53 alterations determine clinical subgroups and survival of patients with choroid plexus tumors. J Clin Oncol. 2010;28:1995–2001. doi: 10.1200/JCO.2009.26.8169. [DOI] [PubMed] [Google Scholar]
- 85.Schmidt M, Bachhuber A, Victor A, Steiner E, Mahlke M, Lehr HA, et al. p53 expression and resistance against paclitaxel in patients with metastatic breast cancer. J Cancer Res Clin Oncol. 2003;129:295–302. doi: 10.1007/s00432-003-0430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kandioler-Eckersberger D, Ludwig C, Rudas M, Kappel S, Janschek E, Wenzel C, et al. TP53 mutation and p53 overexpression for prediction of response to neoadjuvant treatment in breast cancer patients. Clin Cancer Res. 2000;6:50–6. [PubMed] [Google Scholar]
- 87.Gonzalez D, Martinez P, Wade R, Hockley S, Oscier D, Matutes E, et al. Mutational status of the TP53 gene as a predictor of response and survival in patients with chronic lymphocytic leukemia: results from the LRF CLL4 trial. J Clin Oncol. 2011;29:2223–9. doi: 10.1200/JCO.2010.32.0838. [DOI] [PubMed] [Google Scholar]
- 88.Mayer F, Honecker F, Looijenga LH, Bokemeyer C. Towards an understanding of the biological basis of response to cisplatin-based chemotherapy in germ-cell tumors. Ann Oncol. 2003;14:825–32. doi: 10.1093/annonc/mdg242. [DOI] [PubMed] [Google Scholar]
- 89.Gallagher WM, Brown R. p53-oriented cancer therapies: current progress. Ann Oncol. 1999;10:139–50. doi: 10.1023/a:1008368500557. [DOI] [PubMed] [Google Scholar]
- 90.Blandino G, Levine AJ, Oren M. Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene. 1999;18:477–85. doi: 10.1038/sj.onc.1202314. [DOI] [PubMed] [Google Scholar]
- 91.Bartussek C, Naumann U, Weller M. Accumulation of mutant p53(V143A) modulates the growth, clonogenicity, and radiochemosensitivity of malignant glioma cells independent of endogenous p53 status. Exp Cell Res. 1999;253:432–9. doi: 10.1006/excr.1999.4654. [DOI] [PubMed] [Google Scholar]
- 92.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 93.Donzelli S, Biagioni F, Fausti F, Strano S, Fontemaggi G, Blandino G. Oncogenomic Approaches in Exploring Gain of Function of Mutant p53. Curr Genomics. 2008;9:200–7. doi: 10.2174/138920208784340713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701–13. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 95.Strano S, Dell’Orso S, Mongiovi AM, Monti O, Lapi E, Di Agostino S, et al. Mutant p53 proteins: between loss and gain of function. Head Neck. 2007;29:488–96. doi: 10.1002/hed.20531. [DOI] [PubMed] [Google Scholar]
- 96.Song H, Xu Y. Gain of function of p53 cancer mutants in disrupting critical DNA damage response pathways. Cell Cycle. 2007;6:1570–3. doi: 10.4161/cc.6.13.4456. [DOI] [PubMed] [Google Scholar]
- 97.Song S, Wientjes MG, Gan Y, Au JL. Fibroblast growth factors: an epigenetic mechanism of broad spectrum resistance to anticancer drugs. Proc Natl Acad Sci U S A. 2000;97:8658–63. doi: 10.1073/pnas.140210697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jackman D, Pao W, Riely GJ, Engelman JA, Kris MG, Janne PA, et al. Clinical definition of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. J Clin Oncol. 2010;28:357–60. doi: 10.1200/JCO.2009.24.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vargas-Roig LM, Gago FE, Tello O, Aznar JC, Ciocca DR. Heat shock protein expression and drug resistance in breast cancer patients treated with induction chemotherapy. Int J Cancer. 1998;79:468–75. doi: 10.1002/(sici)1097-0215(19981023)79:5<468::aid-ijc4>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 100.Leonetti C, Biroccio A, Candiloro A, Citro G, Fornari C, Mottolese M, et al. Increase of cisplatin sensitivity by c-myc antisense oligodeoxynucleotides in a human metastatic melanoma inherently resistant to cisplatin. Clin Cancer Res. 1999;5:2588–95. [PubMed] [Google Scholar]
- 101.Xu Y. Induction of genetic instability by gain-of-function p53 cancer mutants. Oncogene. 2008;27:3501–7. doi: 10.1038/sj.onc.1211023. [DOI] [PubMed] [Google Scholar]
- 102.Liu DP, Song H, Xu Y. A common gain of function of p53 cancer mutants in inducing genetic instability. Oncogene. 2010;29:949–56. doi: 10.1038/onc.2009.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Roemer K. Mutant p53: gain-of-function oncoproteins and wild-type p53 inactivators. Biol Chem. 1999;380:879–87. doi: 10.1515/BC.1999.108. [DOI] [PubMed] [Google Scholar]
- 105.Willis A, Jung EJ, Wakefield T, Chen X. Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene. 2004;23:2330–8. doi: 10.1038/sj.onc.1207396. [DOI] [PubMed] [Google Scholar]
- 106.Pugacheva EN, Ivanov AV, Kravchenko JE, Kopnin BP, Levine AJ, Chumakov PM. Novel gain of function activity of p53 mutants: activation of the dUTPase gene expression leading to resistance to 5-fluorouracil. Oncogene. 2002;21:4595–600. doi: 10.1038/sj.onc.1205704. [DOI] [PubMed] [Google Scholar]
- 107.Wong RP, Tsang WP, Chau PY, Co NN, Tsang TY, Kwok TT. p53-R273H gains new function in induction of drug resistance through down-regulation of procaspase-3. Mol Cancer Ther. 2007;6:1054–61. doi: 10.1158/1535-7163.MCT-06-0336. [DOI] [PubMed] [Google Scholar]
- 108.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–9. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- 109.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282–8. doi: 10.1038/nm0302-282. [DOI] [PubMed] [Google Scholar]
- 110.Wiman KG. Pharmacological reactivation of mutant p53: from protein structure to the cancer patient. Oncogene. 2010;29:4245–52. doi: 10.1038/onc.2010.188. [DOI] [PubMed] [Google Scholar]
- 111.Cheok CF, Verma CS, Baselga J, Lane DP. Translating p53 into the clinic. Nat Rev Clin Oncol. 2011;8:25–37. doi: 10.1038/nrclinonc.2010.174. [DOI] [PubMed] [Google Scholar]
- 112.Maslon MM, Hupp TR. Drug discovery and mutant p53. Trends Cell Biol. 2010;20:542–55. doi: 10.1016/j.tcb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 113.Fossa SD, Cvancarova M, Chen L, Allan AL, Oldenburg J, Peterson DR, et al. Adverse prognostic factors for testicular cancer-specific survival: a population-based study of 27,948 patients. J Clin Oncol. 2011;29:963–70. doi: 10.1200/JCO.2010.32.3204. [DOI] [PubMed] [Google Scholar]
- 114.King TC, Akerley W, Fan AC, Moore T, Mangray S, Hsiu CM, et al. p53 mutations do not predict response to paclitaxel in metastatic nonsmall cell lung carcinoma. Cancer. 2000;89:769–73. doi: 10.1002/1097-0142(20000815)89:4<769::aid-cncr8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 115.Murakami I, Hiyama K, Ishioka S, Yamakido M, Kasagi F, Yokosaki Y. p53 gene mutations are associated with shortened survival in patients with advanced non-small cell lung cancer: an analysis of medically managed patients. Clin Cancer Res. 2000;6:526–30. [PubMed] [Google Scholar]
- 116.Lavarino C, Pilotti S, Oggionni M, Gatti L, Perego P, Bresciani G, et al. p53 gene status and response to platinum/paclitaxel-based chemotherapy in advanced ovarian carcinoma. J Clin Oncol. 2000;18:3936–45. doi: 10.1200/JCO.2000.18.23.3936. [DOI] [PubMed] [Google Scholar]
- 117.Mayall FG, Jacobson G, Wilkins R. Mutations of p53 gene and SV40 sequences in asbestos associated and non-asbestos-associated mesotheliomas. J Clin Pathol. 1999;52:291–3. doi: 10.1136/jcp.52.4.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Metcalf RA, Welsh JA, Bennett WP, Seddon MB, Lehman TA, Pelin K, et al. p53 and Kirsten-ras mutations in human mesothelioma cell lines. Cancer Res. 1992;52:2610–5. [PubMed] [Google Scholar]
- 119.Tsao AS, Wistuba I, Roth JA, Kindler HL. Malignant Pleural Mesothelioma. J Clin Oncol. 2009;27:2081–90. doi: 10.1200/JCO.2008.19.8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tomek S, Manegold C. Chemotherapy for malignant pleural mesothelioma: past results and recent developments. Lung Cancer. 2004;45 (Suppl 1):S103–S119. doi: 10.1016/j.lungcan.2004.04.020. [DOI] [PubMed] [Google Scholar]
- 121.Lee CW, Murray N, Anderson H, Rao SC, Bishop W. Outcomes with first-line platinum-based combination chemotherapy for malignant pleural mesothelioma: A review of practice in British Columbia. Lung Cancer. 2009:1–6. doi: 10.1016/j.lungcan.2008.09.008. (ePub) [DOI] [PubMed] [Google Scholar]
- 122.Noon AP, Vlatkovic N, Polanski R, Maguire M, Shawki H, Parsons K, et al. p53 and MDM2 in renal cell carcinoma: biomarkers for disease progression and future therapeutic targets? Cancer. 2010;116:780–90. doi: 10.1002/cncr.24841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lu ML, Wikman F, Orntoft TF, Charytonowicz E, Rabbani F, Zhang Z, et al. Impact of alterations affecting the p53 pathway in bladder cancer on clinical outcome, assessed by conventional and array-based methods. Clin Cancer Res. 2002;8:171–9. [PubMed] [Google Scholar]
- 124.Havrilesky L, Darcy M, Hamdan H, Priore RL, Leon J, Bell J, et al. Prognostic significance of p53 mutation and p53 overexpression in advanced epithelial ovarian cancer: a Gynecologic Oncology Group Study. J Clin Oncol. 2003;21:3814–25. doi: 10.1200/JCO.2003.11.052. [DOI] [PubMed] [Google Scholar]
- 125.Cosaert J, Quoix E. Platinum drugs in the treatment of non-small-cell lung cancer. Br J Cancer. 2002;87:825–33. doi: 10.1038/sj.bjc.6600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Haitel A, Wiener HG, Baethge U, Marberger M, Susani M. mdm2 expression as a prognostic indicator in clear cell renal cell carcinoma: comparison with p53 overexpression and clinicopathological parameters. Clin Cancer Res. 2000;6:1840–4. [PubMed] [Google Scholar]
- 127.Kawasaki T, Bilim V, Takahashi K, Tomita Y. Infrequent alteration of p53 pathway in metastatic renal cell carcinoma. Oncol Rep. 1999;6:329–33. doi: 10.3892/or.6.2.329. [DOI] [PubMed] [Google Scholar]
- 128.Talis AL, Huibregtse JM, Howley PM. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem. 1998;273:6439–45. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 129.Padilla LA, Leung BS, Carson LF. Evidence of an association between human papillomavirus and impaired chemotherapy-induced apoptosis in cervical cancer cells. Gynecol Oncol. 2002;85:59–66. doi: 10.1006/gyno.2002.6604. [DOI] [PubMed] [Google Scholar]
- 130.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453–9. doi: 10.1093/nar/26.15.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Freedman DA, Wu L, Levine AJ. Functions of the MDM2 oncoprotein. Cell Mol Life Sci. 1999;55:96–107. doi: 10.1007/s000180050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Onel K, Cordon-Cardo C. MDM2 and prognosis. Mol Cancer Res. 2004;2:1–8. [PubMed] [Google Scholar]
- 133.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299–309. doi: 10.1016/j.tcb.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Turbin DA, Cheang MC, Bajdik CD, Gelmon KA, Yorida E, De Luca A, et al. MDM2 protein expression is a negative prognostic marker in breast carcinoma. Mod Pathol. 2006;19:69–74. doi: 10.1038/modpathol.3800484. [DOI] [PubMed] [Google Scholar]
- 135.Klein C, Vassilev LT. Targeting the p53-MDM2 interaction to treat cancer. Br J Cancer. 2004;91:1415–9. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888–93. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kojima K, Konopleva M, McQueen T, O’Brien S, Plunkett W, Andreeff M. Mdm2 inhibitor Nutlin-3a induces p53-mediated apoptosis by transcription-dependent and transcription-independent mechanisms and may overcome Atm-mediated resistance to fludarabine in chronic lymphocytic leukemia. Blood. 2006;108:993–1000. doi: 10.1182/blood-2005-12-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shangary S, Wang S. Targeting the MDM2-p53 interaction for cancer therapy. Clin Cancer Res. 2008;14:5318–24. doi: 10.1158/1078-0432.CCR-07-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Barbieri E, Mehta P, Chen Z, Zhang L, Slack A, Berg S, et al. MDM2 inhibition sensitizes neuroblastoma to chemotherapy-induced apoptotic cell death. Mol Cancer Ther. 2006;5:2358–65. doi: 10.1158/1535-7163.MCT-06-0305. [DOI] [PubMed] [Google Scholar]
- 140.Lavin MF, Gueven N. The complexity of p53 stabilization and activation. Cell Death Differ. 2006;13:941–50. doi: 10.1038/sj.cdd.4401925. [DOI] [PubMed] [Google Scholar]
- 141.Wade M, Rodewald LW, Espinosa JM, Wahl GM. BH3 activation blocks Hdmx suppression of apoptosis and cooperates with Nutlin to induce cell death. Cell Cycle. 2008;7:1973–82. doi: 10.4161/cc.7.13.6072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Lee MH, Lozano G. Regulation of the p53-MDM2 pathway by 14-3-3 sigma and other proteins. Semin Cancer Biol. 2006;16:225–34. doi: 10.1016/j.semcancer.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 143.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 144.Humbey O, Pimkina J, Zilfou JT, Jarnik M, Dominguez-Brauer C, Burgess DJ, et al. The ARF tumor suppressor can promote the progression of some tumors. Cancer Res. 2008;68:9608–13. doi: 10.1158/0008-5472.CAN-08-2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tortora G, Caputo R, Damiano V, Bianco R, Chen J, Agrawal S, et al. A novel MDM2 anti-sense oligonucleotide has anti-tumor activity and potentiates cytotoxic drugs acting by different mechanisms in human colon cancer. Int J Cancer. 2000;88:804–9. doi: 10.1002/1097-0215(20001201)88:5<804::aid-ijc19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 146.Shen L, Kondo Y, Hamilton SR, Rashid A, Issa JP. P14 methylation in human colon cancer is associated with microsatellite instability and wild-type p53. Gastroenterology. 2003;124:626–33. doi: 10.1053/gast.2003.50102. [DOI] [PubMed] [Google Scholar]
- 147.Deng X, Kim M, Vandier D, Jung YJ, Rikiyama T, Sgagias MK, et al. Recombinant adenovirus-mediated p14(ARF) overexpression sensitizes human breast cancer cells to cisplatin. Biochem Biophys Res Commun. 2002;296:792–8. doi: 10.1016/s0006-291x(02)00948-8. [DOI] [PubMed] [Google Scholar]
- 148.Bargonetti J, Manfredi JJ. Multiple roles of the tumor suppressor p53. Curr Opin Oncol. 2002;14:86–91. doi: 10.1097/00001622-200201000-00015. [DOI] [PubMed] [Google Scholar]
- 149.Sturm I, Papadopoulos S, Hillebrand T, Benter T, Luck HJ, Wolff G, et al. Impaired BAX protein expression in breast cancer: mutational analysis of the BAX and the p53 gene. Int J Cancer. 2000;87:517–21. doi: 10.1002/1097-0215(20000815)87:4<517::aid-ijc9>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 150.Siddik ZH, Mims B, Lozano G, Thai G. Independent pathways of p53 induction by cisplatin and X-rays in a cisplatin-resistant ovarian tumor cell line. Cancer Res. 1998;58:698–703. [PubMed] [Google Scholar]
- 151.Perego P, Gatti L, Righetti SC, Beretta GL, Carenini N, Corna E, et al. Development of resistance to a trinuclear platinum complex in ovarian carcinoma cells. Int J Cancer. 2003;105:617–24. doi: 10.1002/ijc.11140. [DOI] [PubMed] [Google Scholar]
- 152.Delmastro DA, Li J, Vaisman A, Solle M, Chaney SG. DNA damage inducible-gene expression following platinum treatment in human ovarian carcinoma cell lines. Cancer Chemother Pharmacol. 1997;39:245–53. doi: 10.1007/s002800050568. [DOI] [PubMed] [Google Scholar]
- 153.Gartel AL, Radhakrishnan SK. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–5. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 154.Liu G, Lozano G. p21 stability: linking chaperones to a cell cycle checkpoint. Cancer Cell. 2005;7:113–4. doi: 10.1016/j.ccr.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 155.Shoji T, Tanaka F, Takata T, Yanagihara K, Otake Y, Hanaoka N, et al. Clinical significance of p21 expression in non-small-cell lung cancer. J Clin Oncol. 2002;20:3865–71. doi: 10.1200/JCO.2002.09.147. [DOI] [PubMed] [Google Scholar]
- 156.Rose SL, Goodheart MJ, DeYoung BR, Smith BJ, Buller RE. p21 expression predicts outcome in p53-null ovarian carcinoma. Clin Cancer Res. 2003;9:1028–32. [PubMed] [Google Scholar]
- 157.Gartel AL, Tyner AL. The role of the cyclin-dependent kinase inhibitor p21 in apoptosis. Mol Cancer Ther. 2002;1:639–49. [PubMed] [Google Scholar]
- 158.Liu S, Bishop WR, Liu M. Differential effects of cell cycle regulatory protein p21(WAF1/Cip1) on apoptosis and sensitivity to cancer chemotherapy. Drug Resist Updat. 2003;6:183–95. doi: 10.1016/s1368-7646(03)00044-x. [DOI] [PubMed] [Google Scholar]
- 159.Qin LF, Ng IO. Exogenous expression of p21(WAF1/CIP1) exerts cell growth inhibition and enhances sensitivity to cisplatin in hepatoma cells. Cancer Lett. 2001;172:7–15. doi: 10.1016/s0304-3835(01)00701-7. [DOI] [PubMed] [Google Scholar]
- 160.Ho GH, Calvano JE, Bisogna M, Abouezzi Z, Borgen PI, Cordon-Cardo C, et al. Genetic alterations of the p14ARF -hdm2-p53 regulatory pathway in breast carcinoma. Breast Cancer Res Treat. 2001;65:225–32. doi: 10.1023/a:1010686518990. [DOI] [PubMed] [Google Scholar]
- 161.Yazlovitskaya EM, DeHaan RD, Persons DL. Prolonged wild-type p53 protein accumulation and cisplatin resistance. Biochem Biophys Res Commun. 2001;283:732–7. doi: 10.1006/bbrc.2001.4849. [DOI] [PubMed] [Google Scholar]
- 162.Gurova KV, Hill JE, Razorenova OV, Chumakov PM, Gudkov AV. p53 pathway in renal cell carcinoma is repressed by a dominant mechanism. Cancer Res. 2004;64:1951–8. doi: 10.1158/0008-5472.can-03-1541. [DOI] [PubMed] [Google Scholar]
- 163.Dai C, Gu W. p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med. 2010;16:528–36. doi: 10.1016/j.molmed.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–16. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 165.Meek DW, Anderson CW. Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb Perspect Biol. 2009;1:a000950. doi: 10.1101/cshperspect.a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Colman MS, Afshari CA, Barrett JC. Regulation of p53 stability and activity in response to genotoxic stress. Mutat Res. 2000;462:179–88. doi: 10.1016/s1383-5742(00)00035-1. [DOI] [PubMed] [Google Scholar]
- 167.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–72. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 168.MacLaine NJ, Hupp TR. The regulation of p53 by phosphorylation: a model for how distinct signals integrate into the p53 pathway. Aging (Albany NY) 2009;1:490–502. doi: 10.18632/aging.100047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.MacLaine NJ, Hupp TR. How phosphorylation controls p53. Cell Cycle. 2011;10:916–21. doi: 10.4161/cc.10.6.15076. [DOI] [PubMed] [Google Scholar]
- 170.Dornan D, Hupp TR. Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300. EMBO Rep. 2001;2:139–44. doi: 10.1093/embo-reports/kve025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Craig AL, Burch L, Vojtesek B, Mikutowska J, Thompson A, Hupp TR. Novel phosphorylation sites of human tumour suppressor protein p53 at Ser20 and Thr18 that disrupt the binding of mdm2 (mouse double minute 2) protein are modified in human cancers. Biochem J. 1999;342 (Pt 1):133–41. [PMC free article] [PubMed] [Google Scholar]
- 172.Meek DW. The p53 response to DNA damage. DNA Repair (Amst) 2004;3:1049–56. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 173.Teufel DP, Bycroft M, Fersht AR. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene. 2009;28:2112–8. doi: 10.1038/onc.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ferreon JC, Lee CW, Arai M, Martinez-Yamout MA, Dyson HJ, Wright PE. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci U S A. 2009;106:6591–6. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nakamizo A, Amano T, Zhang W, Zhang XQ, Ramdas L, Liu TJ, et al. Phosphorylation of Thr18 and Ser20 of p53 in Ad-p53-induced apoptosis. Neuro Oncol. 2008;10:275–91. doi: 10.1215/15228517-2008-015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Amano T, Nakamizo A, Mishra SK, Gumin J, Shinojima N, Sawaya R, et al. Simultaneous phosphorylation of p53 at serine 15 and 20 induces apoptosis in human glioma cells by increasing expression of pro-apoptotic genes. J Neurooncol. 2009;92:357–71. doi: 10.1007/s11060-009-9844-1. [DOI] [PubMed] [Google Scholar]
- 177.Lavin MF, Kozlov S. DNA damage-induced signalling in ataxia-telangiectasia and related syndromes. Radiother Oncol. 2007;83:231–7. doi: 10.1016/j.radonc.2007.04.032. [DOI] [PubMed] [Google Scholar]
- 178.Ripolles L, Ortega M, Ortuno F, Gonzalez A, Losada J, Ojanguren J, et al. Genetic abnormalities and clinical outcome in chronic lymphocytic leukemia. Cancer Genet Cytogenet. 2006;171:57–64. doi: 10.1016/j.cancergencyto.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 179.Satyamoorthy K, Chehab NH, Waterman MJ, Lien MC, El Deiry WS, Herlyn M, et al. Aberrant regulation and function of wild-type p53 in radioresistant melanoma cells. Cell Growth Differ. 2000;11:467–74. [PubMed] [Google Scholar]
- 180.Puca R, Nardinocchi L, Gal H, Rechavi G, Amariglio N, Domany E, et al. Reversible dysfunction of wild-type p53 following homeodomain-interacting protein kinase-2 knockdown. Cancer Res. 2008;68:3707–14. doi: 10.1158/0008-5472.CAN-07-6776. [DOI] [PubMed] [Google Scholar]
- 181.Antoni L, Sodha N, Collins I, Garrett MD. CHK2 kinase: cancer susceptibility and cancer therapy - two sides of the same coin? Nat Rev Cancer. 2007;7:925–36. doi: 10.1038/nrc2251. [DOI] [PubMed] [Google Scholar]
- 182.Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, et al. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895–909. doi: 10.1101/gad.1815309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Craig AL, Chrystal JA, Fraser JA, Sphyris N, Lin Y, Harrison BJ, et al. The MDM2 ubiquitination signal in the DNA-binding domain of p53 forms a docking site for calcium calmodulin kinase superfamily members. Mol Cell Biol. 2007;27:3542–55. doi: 10.1128/MCB.01595-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shieh SY, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- 185.Craig AL, Bray SE, Finlan LE, Kernohan NM, Hupp TR. Signaling to p53: the use of phospho-specific antibodies to probe for in vivo kinase activation. Methods Mol Biol. 2003;234:171–202. doi: 10.1385/1-59259-408-5:171. [DOI] [PubMed] [Google Scholar]
- 186.Pabla N, Huang S, Mi QS, Daniel R, Dong Z. ATR-Chk2 signaling in p53 activation and DNA damage response during cisplatin-induced apoptosis. J Biol Chem. 2008;283:6572–83. doi: 10.1074/jbc.M707568200. [DOI] [PubMed] [Google Scholar]
- 187.Takai H, Naka K, Okada Y, Watanabe M, Harada N, Saito S, et al. Chk2-deficient mice exhibit radioresistance and defective p53-mediated transcription. EMBO J. 2002;21:5195–205. doi: 10.1093/emboj/cdf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.MacLaren A, Slavin D, McGowan CH. Chk2 protects against radiation-induced genomic instability. Radiat Res. 2009;172:463–72. doi: 10.1667/RR1603.1. [DOI] [PubMed] [Google Scholar]
- 189.Zhang P, Wang J, Gao W, Yuan BZ, Rogers J, Reed E. CHK2 kinase expression is down-regulated due to promoter methylation in non-small cell lung cancer. Mol Cancer. 2004;3:14. doi: 10.1186/1476-4598-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Williams LH, Choong D, Johnson SA, Campbell IG. Genetic and epigenetic analysis of CHEK2 in sporadic breast, colon, and ovarian cancers. Clin Cancer Res. 2006;12:6967–72. doi: 10.1158/1078-0432.CCR-06-1770. [DOI] [PubMed] [Google Scholar]
- 191.Righetti SC, Della TG, Pilotti S, Menard S, Ottone F, Colnaghi MI, et al. A comparative study of p53 gene mutations, protein accumulation, and response to cisplatin-based chemotherapy in advanced ovarian carcinoma. Cancer Res. 1996;56:689–93. [PubMed] [Google Scholar]
- 192.Jallepalli PV, Lengauer C, Vogelstein B, Bunz F. The Chk2 tumor suppressor is not required for p53 responses in human cancer cells. J Biol Chem. 2003;278:20475–9. doi: 10.1074/jbc.M213159200. [DOI] [PubMed] [Google Scholar]
- 193.Miller CW, Ikezoe T, Krug U, Hofmann WK, Tavor S, Vegesna V, et al. Mutations of the CHK2 gene are found in some osteosarcomas, but are rare in breast, lung, and ovarian tumors. Genes Chromosomes Cancer. 2002;33:17–21. doi: 10.1002/gcc.1207. [DOI] [PubMed] [Google Scholar]
- 194.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 195.Zhou BB, Sausville EA. Drug discovery targeting Chk1 and Chk2 kinases. Prog Cell Cycle Res. 2003;5:413–21. [PubMed] [Google Scholar]
- 196.Burdak-Rothkamm S, Rothkamm K, Prise KM. ATM acts downstream of ATR in the DNA damage response signaling of bystander cells. Cancer Res. 2008;68:7059–65. doi: 10.1158/0008-5472.CAN-08-0545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, et al. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci U S A. 1998;95:7445–50. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, et al. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–69. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61:7727–30. [PubMed] [Google Scholar]
- 200.Sengelov L, Horn T, Steven K. p53 nuclear immunoreactivity as a predictor of response and outcome following chemotherapy for metastatic bladder cancer. J Cancer Res Clin Oncol. 1997;123:565–70. doi: 10.1007/s004320050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gibb R, O’Connor D, Katsanis W, Taylor D, Garcel-Taylor C. p53 mutational change in sequentially isolated cells in ovarian cancer. Proc Am Assoc Cancer Res. 1997;38:271. [Google Scholar]
- 202.Ozols RF, Behrens BC, Ostchega Y, Young RC. High dose cisplatin and high dose carboplatin in refractory ovarian cancer. Cancer Treat Rev. 1985;12 (Suppl A):59–65. doi: 10.1016/0305-7372(85)90019-2. [DOI] [PubMed] [Google Scholar]
- 203.Jodrell DI, Egorin MJ, Canetta RM, Langenberg P, Goldbloom EP, Burroughs JN, et al. Relationships between carboplatin exposure and tumor response and toxicity in patients with ovarian cancer [see comments] J Clin Oncol. 1992;10:520–8. doi: 10.1200/JCO.1992.10.4.520. [DOI] [PubMed] [Google Scholar]
- 204.Lanni JS, Lowe SW, Licitra EJ, Liu JO, Jacks T. p53-independent apoptosis induced by paclitaxel through an indirect mechanism. Proc Natl Acad Sci U S A. 1997;94:9679–83. doi: 10.1073/pnas.94.18.9679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Bradford CR, Zhu S, Ogawa H, Ogawa T, Ubell M, Narayan A, et al. P53 mutation correlates with cisplatin sensitivity in head and neck squamous cell carcinoma lines. Head Neck. 2003;25:654–61. doi: 10.1002/hed.10274. [DOI] [PubMed] [Google Scholar]
- 206.Debernardis D, Sire EG, De Feudis P, Vikhanskaya F, Valenti M, Russo P, et al. p53 status does not affect sensitivity of human ovarian cancer cell lines to paclitaxel. Cancer Res. 1997;57:870–4. [PubMed] [Google Scholar]
- 207.Reinecke P, Kalinski T, Mahotka C, Schmitz M, Dejosez M, Gabbert HE, et al. Paclitaxel/Taxol sensitivity in human renal cell carcinoma is not determined by the p53 status. Cancer Lett. 2005;222:165–71. doi: 10.1016/j.canlet.2004.09.045. [DOI] [PubMed] [Google Scholar]
- 208.Xu GW, Nutt CL, Zlatescu MC, Keeney M, Chin-Yee I, Cairncross JG. Inactivation of p53 sensitizes U87MG glioma cells to 1,3-bis(2-chloroethyl)-1-nitrosourea. Cancer Res. 2001;61:4155–9. [PubMed] [Google Scholar]
- 209.Moneo V, Serelde BG, Fominaya J, Leal JF, Blanco-Aparicio C, Romero L, et al. Extreme sensitivity to Yondelis (Trabectedin, ET-743) in low passaged sarcoma cell lines correlates with mutated p53. J Cell Biochem. 2007;100:339–48. doi: 10.1002/jcb.21073. [DOI] [PubMed] [Google Scholar]
- 210.Fan S, Chang JK, Smith ML, Duba D, Fornace AJ, Jr, O’Connor PM. Cells lacking CIP1/WAF1 genes exhibit preferential sensitivity to cisplatin and nitrogen mustard. Oncogene. 1997;14:2127–36. doi: 10.1038/sj.onc.1201052. [DOI] [PubMed] [Google Scholar]
- 211.Fan S, Smith ML, Rivet DJ, Duba D, Zhan Q, Kohn KW, et al. Disruption of p53 function sensitizes breast cancer MCF-7 cells to cisplatin and pentoxifylline. Cancer Res. 1995;55:1649–54. [PubMed] [Google Scholar]
- 212.Stoklosa T, Slupianek A, Datta M, Nieborowska-Skorska M, Nowicki MO, Koptyra M, et al. BCR/ABL recruits p53 tumor suppressor protein to induce drug resistance. Cell Cycle. 2004;3:1463–72. doi: 10.4161/cc.3.11.1229. [DOI] [PubMed] [Google Scholar]
- 213.Siddik ZH, Hagopian GS, Thai G, Tomisaki S, Toyomasu T, Khokhar AR. Role of p53 in the ability of 1,2-diaminocyclohexane-diacetato-dichloro- Pt(IV) to circumvent cisplatin resistance. J Inorg Biochem. 1999;77:65–70. doi: 10.1016/s0162-0134(99)00144-0. [DOI] [PubMed] [Google Scholar]
- 214.Hawkins DS, Demers GW, Galloway DA. Inactivation of p53 enhances sensitivity to multiple chemotherapeutic agents. Cancer Res. 1996;56:892–8. [PubMed] [Google Scholar]
- 215.Mujoo K, Watanabe M, Nakamura J, Khokhar AR, Siddik ZH. Status of p53 phosphorylation and function in sensitive and resistant human cancer models exposed to platinum-based DNA damaging agents. J Cancer Res Clin Oncol. 2003;129:709–18. doi: 10.1007/s00432-003-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jabbur JR, Huang P, Zhang W. DNA damage-induced phosphorylation of p53 at serine 20 correlates with p21 and Mdm-2 induction in vivo. Oncogene. 2000;19:6203–8. doi: 10.1038/sj.onc.1204017. [DOI] [PubMed] [Google Scholar]
- 217.Jabbur JR, Zhang W. p53 Antiproliferative function is enhanced by aspartate substitution at threonine 18 and serine 20. Cancer Biol Ther. 2002;1:277–83. doi: 10.4161/cbt.81. [DOI] [PubMed] [Google Scholar]
- 218.Fraser M, Bai T, Tsang BK. Akt promotes cisplatin resistance in human ovarian cancer cells through inhibition of p53 phosphorylation and nuclear function. Int J Cancer. 2008;122:534–46. doi: 10.1002/ijc.23086. [DOI] [PubMed] [Google Scholar]