Summary
Autophagy is a major catabolic pathway in eukaryotes associated with a broad spectrum of human diseases. In autophagy, autophagosomes carrying cellular cargoes fuse with lysosomes for degradation. However, the molecular mechanism underlying autophagosome maturation is largely unknown. Here we report that TECPR1 binds to the Atg12-Atg5 conjugate and phosphatidylinositol 3-phosphate (PtdIns(3)P) to promote autophagosome-lysosome fusion. TECPR1 and Atg16 form mutually exclusive complexes with the Atg12-Atg5 conjugate; and TECPR1 binds PtdIns(3)P upon association with the Atg12-Atg5 conjugate. Strikingly, TECPR1 localizes to and recruits Atg5 to autolysosome membrane. Consequently, elimination of TECPR1 leads to accumulation of autophagosomes and blocks autophagic degradation of LC3-II and p62. Finally, autophagosome maturation marked by GFP-mRFP-LC3 is defective in TECPR1 deficient cells. Thus, we propose that the concerted interactions among TECPR1, Atg12-Atg5 and PtdIns(3)P provide the fusion specificity between autophagosomes and lysosomes and that the assembly of this complex initiates the autophagosome maturation process.
Introduction
Autophagy is an important catabolic process for maintaining human health as it prevents various diseases including cancers, neurodegenerative disorders, and diabetes (Levine and Kroemer, 2008; Mizushima et al., 2008). During autophagy, a double membrane autophagosome engulfs cargo such as damaged organelles, cytosol and microorganisms. The autophagosome then fuses with the lysosome where its internal components are degraded by acid hydrolases (Levine and Klionsky, 2004). Current understanding of autophagy has been greatly expanded by S. cerevisiae genetic screening; 35 autophagy related (ATG) genes have been identified and most of them are conserved across higher eukaryotes (Inoue and Klionsky, 2010; Nazarko et al., 2011; Suzuki et al., 2010). Interestingly, very few ATG proteins are implicated in autophagosome maturation, albeit substantial evidence suggests that this process is highly regulated (Eskelinen, 2005; Kim and Klionsky, 2000). Recent studies in eukaryotes have implicated SNARE proteins (Abeliovich et al., 1999; Darsow et al., 1997; Ishihara et al., 2001; Moreau et al., 2011; Nair et al., 2011), endosomal COPs (Ishihara et al., 2001; Razi et al., 2009), the ESCRT III complex (Lee et al., 2007; Rusten et al., 2007), the HOPS complex (Nickerson et al., 2009), LAMP proteins (Eskelinen et al., 2002; Tanaka et al., 2000), small GTPase Rab proteins (Gutierrez et al., 2004; Jager et al., 2004; Kimura et al., 2007), the class III phosphatidylinositol 3-kinase (Liang et al., 2008; Matsunaga et al., 2009; Sun et al., 2010; Sun et al., 2011), and chaperon HSP70 family proteins (Leu et al., 2009) as critical players in autophagosome maturation. However, the detailed molecular mechanism dictating fusion specificity between autophagosomes and lysosomes is still unknown.
Among Atg proteins, Atg5 is an essential gene for autophagy that is constantly conjugated to an ubiquitin-like protein, Atg12, through an enzymatic cascade (Mizushima et al., 1998). The conjugated Atg12-Atg5 interacts with Atg16 and induces its oligomerization to form a ~350kd complex in yeast (Kuma et al., 2002) or a ~800kd complex in mammals (Mizushima et al., 2003). The Atg12-Atg5-Atg16 complex localizes to nascent autophagosomes (Mizushima et al., 2003; Mizushima et al., 2001), and functions upstream to the LC3 lipidation and its autophagosome targeting (Fujita et al., 2008; Hanada et al., 2007). The biological functions of Atg5 in mammalian development (Kuma et al., 2004) and a variety of physiological processes (Baerga et al., 2009; Cadwell et al., 2008; Hara et al., 2006; Jounai et al., 2007) have already been illustrated in mouse genetic studies.
TECPR1 is a component of the autophagy network that was identified through its interaction with Atg5 (Behrends et al., 2010). It has also been implicated in selective autophagy against bacterial pathogens (Ogawa et al., 2011). However, its function in canonical autophagy is not clear. We have identified TECPR1 as a binding partner of the Atg12-Atg5 conjugate in an independent study. We show that TECPR1 and Atg16 exist in two mutually exclusive complexes with the Atg12-Atg5 conjugate. TECPR1 resides on lysosomes and recruits Atg12-Atg5 to autolysosomes. Remarkably, we demonstrate that TECPR1 plays a crucial function in autophagosome maturation and promotes autophagosome-lysosome fusion by associating with both the Atg12-Atg5 conjugate and PtdIns(3)P. Through this study, we have uncovered a previously unknown critical function of the Atg12-Atg5-TECPR1 complex in autophagosome maturation and propose that the interaction between TECPR1 and Atg12-Atg5 provides fusion specificity between autophagosomes and lysosomes.
Results
TECPR1 interacts with the Atg12-Atg5 conjugate
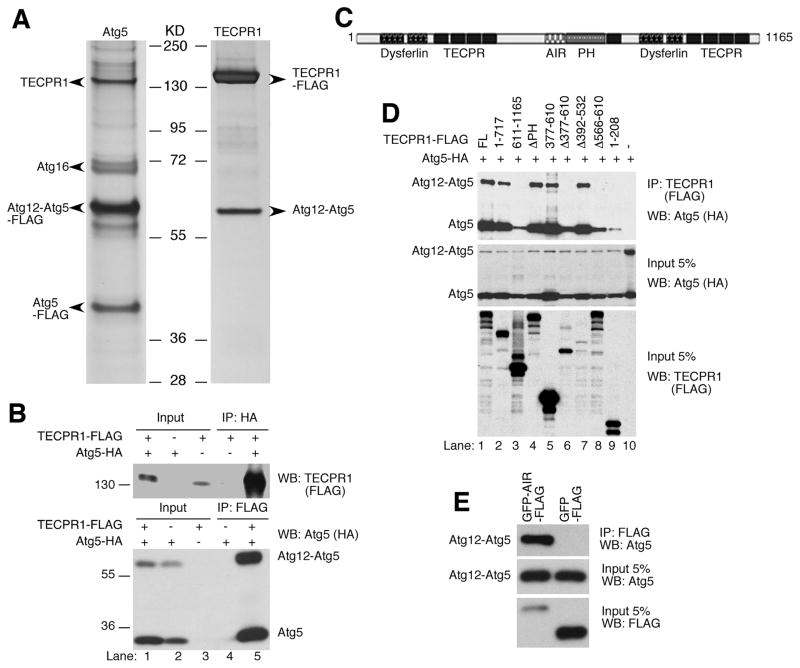
To dissect the regulation of Atg5 in autophagy, we performed a tandem affinity purification using tagged full-length Atg5 (ZZ-Atg5-FLAG) as the bait to purify the Atg5 complex. In the Atg5 complex, we identified free Atg5, the Atg12-Atg5 conjugate, Atg16 and TECPR1 (Tectonin beta-propeller repeat-containing protein 1, or KIAA1358) (Fig. 1A, left panel). TECPR1 has recently been identified in the autophagy interaction network through its associations with Atg5, Atg12, Atg3 and several other proteins (Behrends et al., 2010). Interestingly, a mass spectrometry analysis from our affinity purification did not detect these proteins. To consolidate the Atg5-TECPR1 complex formation, we performed reciprocal affinity purification for TECPR1 cellular complex; only the Atg12-Atg5 conjugate was present as a stoichiometric component of the TECPR1 complex (Fig. 1A, right panel). Surprisingly, Atg16 or other potential associated proteins failed to be detected in the TECPR1 complex, suggesting that they either do not exist in this complex or their interactions with TECPR1 are relatively weak.
Figure 1. TECPR1 interacts with the conjugated Atg12-Atg5.
(A) Silver staining of the tandem affinity-purified Atg5 complexes or TECPR1 complexes in HEK293T cells. Proteins noted on the gel were identified by mass spectrometry. (B) Reciprocal co-immunoprecipitation of TECPR1 and Atg12-Atg5. HEK293T cells were transfected with TECPR1-FLAG and Atg5-HA. Whole cell lysate was immunoprecipitated with anti-FLAG or anti-HA agarose and analyzed by Western Blotting. (C) Domain structure of TECPR1. TECPR1 contains a pleckstrin homology (PH, aa611–717) domain, an Atg12-Atg5 interacting region (AIR, aa566–610), nine beta-propeller repeats (TECPR, aa209–240, 254–285, 301–332, 344–376, 729–756, 953–984, 1044–1075, 1087–1127), and two dysferlin domains (aa64–170, 816–922). (D) AA566–610 of TECPR1 is essential for Atg12-Atg5 binding. Atg5-HA was coexpressed with TECPR1-FLAG or different mutants as indicated. Whole cell lysate was immunoprecipitated with anti-FLAG agarose, followed by immunoblotting with anti-HA antibody. (E) The AIR domain of TECPR1 is sufficient for Atg12-Atg5 binding. Whole cell lysate was immunoprecipitated with anti-FLAG agarose. See also Figure S1.
The interaction of TECPR1 and Atg5 was further confirmed by co-immunoprecipiatation experiments. Full length TECPR1 precipitated with the Atg12-Atg5 conjugate, the free Atg5 and the free Atg12 (Fig. 1B and S1). However, Atg16 consistently failed to interact with TECPR1 (Fig. S1A), indicating that Atg16 and TECPR1 exist in distinct complexes with Atg12-Atg5.
Domain analysis indicated that TECPR1 contains a pleckstrin homology (PH) domain, nine beta-propeller repeats (TECPR), and two dysferlin domains (Fig. 1C). To investigate which region mediates the interaction between TECPR1 and Atg12-Atg5, deletion mutagenesis and co-immunoprecipitation assays were performed. Atg12-Atg5 interacts with the N-terminus of TECPR1 (Fig. 1D, lane 2 and 3). Neither the PH domain nor the TECPR repeats are required for Atg12-Atg5 binding (Fig. 1D, lane 4 and 5). However, a region spanning from aa566 to aa610 adjacent to the PH domain is necessary and sufficient to bind to Atg12-Atg5 (Fig. 1D, lane 8 and 1E). This region is therefore named as AIR (Atg12-Atg5 Interacting Region) (Fig. 1C). The TECPR1 mutant with AIR-deletion still binds free Atg5, but has no interaction with the Atg12-Atg5 conjugate (Fig. 1D, lane 8). In addition, a N-terminal region of TECPR1 (aa1–208) containing one of the dysferlin domains is able to interact with free Atg5 but not with the Atg12-Atg5 conjugate (Fig. 1D, lane 9). As a control, no Atg12-Atg5 is pulled down with beads alone (Fig. 1D, lane 10). In summary, TECPR1 binds to the Atg12-Atg5 conjugate via the AIR region, and interacts with free Atg5 likely through the dysferlin domain.
TECPR1 localizes to mature rather than early autophagic structures
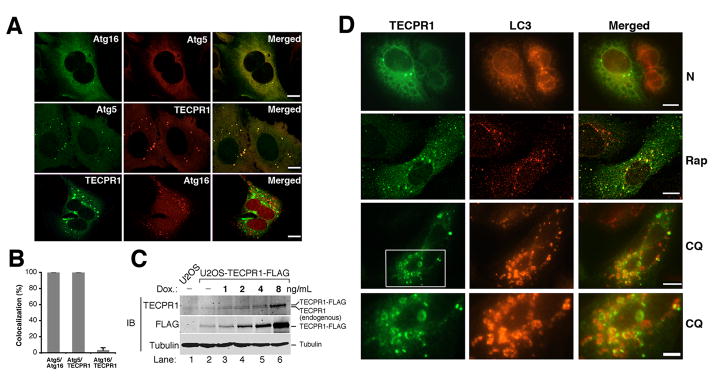
If TECPR1 and Atg16 exist in distinct Atg12-Atg5 complexes as indicated in Figure 1 and Fig. S1, we speculated their relative localization on autophagosomes might also be different. To investigate the subcellular distribution of TECPR1, we co-expressed Atg5-HA, Atg16-Myc and TECPR1-FLAG in different combinations in U2OS cells. As expected, Atg16 colocalized with Atg5 in cellular puncta (Fig. 2A, upper panel, Fig. 2B). Remarkably, TECPR1 also significantly colocalized with Atg5 in cellular puncta (Fig. 2A, middle panel, Fig. 2B). However, there was a limited overlap between TECPR1 and Atg16 (Fig. 2A, lower panel, Fig. 2B). To visualize the spatial relationship among Atg5, Atg16 and TECPR1, we co-expressed mCherry-Atg5, EGFP-Atg16 and TECPR1-FLAG in U2OS cells to examine their subcellular distributions. In cells expressing Atg16, Atg5 and TECPR1, the Atg5 signal either overlapped with Atg16 or TECPR1 (Fig. S2A).
Figure 2. TECPR1 localizes to late rather than early autophagic structures.
(A) Co-staining of TECPR1-FLAG, Atg16-Myc or Atg5-HA in transfected U2OS cells. Scale bar (10 μm). (B) Quantification analysis of (A). Results are representative of three independent experiments and shown as means ± SD. (C) TECPR1-FLAG expression in U2OS cells was inducibly expressed by titrating doxycycline (DOX) as indicated for one day. Endogenously and ectopically expressed TECPR1 were detected by an anti-TECPR1 antibody. (D) Co-staining of TECPR1 and LC3 in U2OS cells. TECPR1-FLAG expression was induced at 2 ng/ml in U2OS cells that were either mock treated (N) or treated with rapamycin (Rap) (2 μM for four hours) or CQ (200 μM for two hours). Endogenous LC3 was detected using an anti-LC3 antibody. Scale bar (10 μm). The scale bar for the bottom panel of (D) is 5 μm. See also Figure S2.
To exclude the possibility that the appearance of TECPR1 puncta is due to overexpression artifacts, we detected its subcellular localization at physiological levels. Since current commercially available or homemade TECPR1 antibodies are not suitable for immunostaining, we established a U2OS cell line that stably expresses TECPR1-FLAG in a doxycycline inducible manner. In this cell line, the expression of TECPR1-FLAG is tightly controlled by doxycycline supplementation. By using an extremely low concentration of doxycycline (2 ng/ml), TECPR1-FLAG was expressed at a level comparable to the endogenous TECPR1 level as detected by an anti-TECPR1 antibody (Fig. 2C, lane 4). Under this condition, TECPR1 was consistently stained as cytosolic puncta (Fig. 2D). After inducing autophagy by treating cells with the mTOR inhibitor rapamycin, the amount of TECPR1 cellular puncta increased and more than 50% of them colocalized with endogenous LC3 (Fig. 2D and S2B). Significant overlap between TECPR1 and LC3 was also observed when cells were treated with chloroquine (Fig. 2D and S2B). Chloroquine (CQ) is a lysosomotropic agent that raises the pH in the lumen of lysosomes and/or autolysosomes therefore compromising autophagic degradation and accumulating mature autophagic vacuoles(Poole and Ohkuma, 1981). Upon CQ treatment, TECPR1 and LC3 were colocalized on the ring-like autophagic structures (Fig. 2D). Since CQ did not promote the colocalization of TECPR1 with Atg16 (Fig. S2C), these ring-like autophagic structures must represent mature rather than early autophagic structures, indicating that TECPR1 might reside on mature autophagic structures.
TECPR1 localizes to autolysosomes
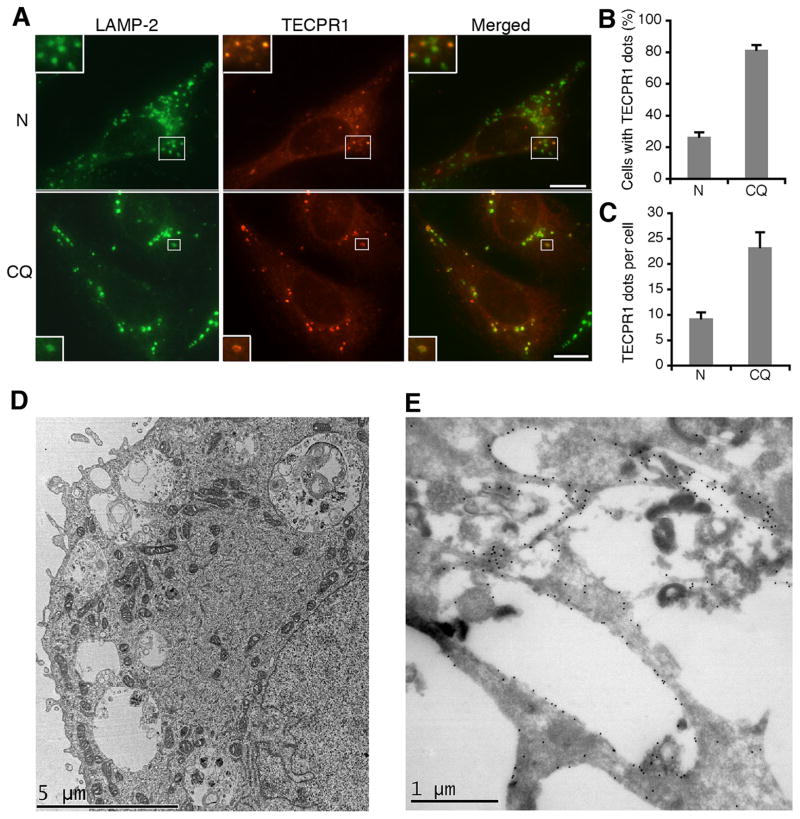
Upon formation, autophagosomes fuse with lysosomes to form autolysosomes that are marked by both autophagosome and lysosome markers. Because we suspected that TECPR1 might reside on late autophagic structures, we examined the colocalization of TECPR1 with lysosome markers. TECPR1 did not colocalize with the early endosome marker EEA1 (Fig. S3A), but significantly colocalized with the lysosome marker LAMP-2 in a subset of cellular puncta (Fig. 3A). The amount of cells exhibiting TECPR1 puncta and the ratio of TECPR1 puncta per cell significantly increased upon CQ treatment (Fig. 3B and 3C); essentially all TECPR1 puncta overlapped with LAMP-2 (Fig. S3B and S3C). These data consequently suggest that TECPR1 resides on lysosomes/autolysosomes.
Figure 3. TECPR1 localizes to the autolysosome membrane.
(A) Co-staining of TECPR1 and LAMP-2 (upper, untreated; bottom, CQ treated (200 μM for two hours)) in U2OS cells expressing TECPR1-FLAG by 2 ng/ml doxycycline addition. An anti-LAMP-2 antibody was used to detect endogenous LAMP-2. Scale bar (10 μm). (B) Quantification of the percentage of cells with TECPR1 puncta described in (A). Results are representative of three independent experiments and presented as means ± SD. (C) Quantification of TECPR1 puncta per cell described in (A). Results are representative of three independent experiments and presented as means ± SD. (D) CQ treated (20 μM overnight) U2OS cells expressing EGFP-TECPR1 were processed and observed under the electron microscope. (E) CQ treated (20 μM overnight) U2OS cells expressing EGFP-TECPR1 were processed for cryo-immunolabeling. The anti-GFP primary antibody and gold conjugated secondary antibody were used to detect EGFP signals. See also Figure S3.
The CQ-inducible TECPR1 lysosomal staining is not due to changes in protein levels, as TECPR1 expression was kept constant throughout treatments (Fig. S3D). TECPR1 membrane association was also confirmed using biochemical fractionation. In U2OS cell inducibly expressing TECPR1, LAMP-1 co-fractionated with TECPR1 in both light (P13) and heavy (P100) membrane fractions (Fig. S3E). TECPR1 also made the Atg12-Atg5 conjugate translocate to membrane fractions but had no effect on Atg16. As controls, GAPDH and LC3-I form were only present in the cytosolic fraction (S100), and LC3-II showed up in the light membrane fraction (Fig. S3E). Consequently, these data strongly support the notion that TECPR1 localizes to lysosome membranes.
TECPR1 puncta were significantly induced upon CQ treatment. We reasoned this inducibility was due to the accumulation of autolysosomes and tested it by imaging analysis. Colocalization between EGFP-LC3 and LAMP-2 was dramatically increased upon CQ treatment (Fig. S3F and S3G), suggesting that autolysosomes accumulate in the presence of CQ. EGFP is normally deprotonated in acidic lysosomes. However, the deprotonation and subsequent loss of EGFP fluorescence is attenuated in the CQ induced autolysosomes as CQ raises the pH in these organelles. To confirm autolysosome accumulation in CQ treated cells, we observed the ultrastructure of these cells using the electron microscope. Remarkably, in CQ treated U2OS cells, a massive accumulation of large (diameter 1–3 μm) autophagic vacuoles was observed (Fig. 3D). These vacuoles were surrounded by single membrane and contained undegraded materials, closely resembling autolysosomes accumulated in Gnptab−/− (UDP-N-acetylglucosamine-lysosomal enzyme) exocrine gland cells (Boonen et al., 2011). Hence, we conclude that CQ treatment leads to autolysosome accumulation.
We further examined the distribution of EGFP-TECPR1 in CQ treated cells by cryo-immunolabeling. In a procedure with a mild fixation condition and embedding without dehydration, we found that gold particles, representing EGFP-TECPR1, were surrounding the large autolysosomes in CQ treated U2OS cells (Fig. 3E). The pattern of these particles is similar to the LAMP-1 staining on the autolysosomes accumulated in Gnptab−/− exocrine gland cells (Boonen et al., 2011), therefore indicating that TECPR1 locates to the autolysosome membrane.
TECPR1 recruits Atg12-Atg5 to autolysosomes
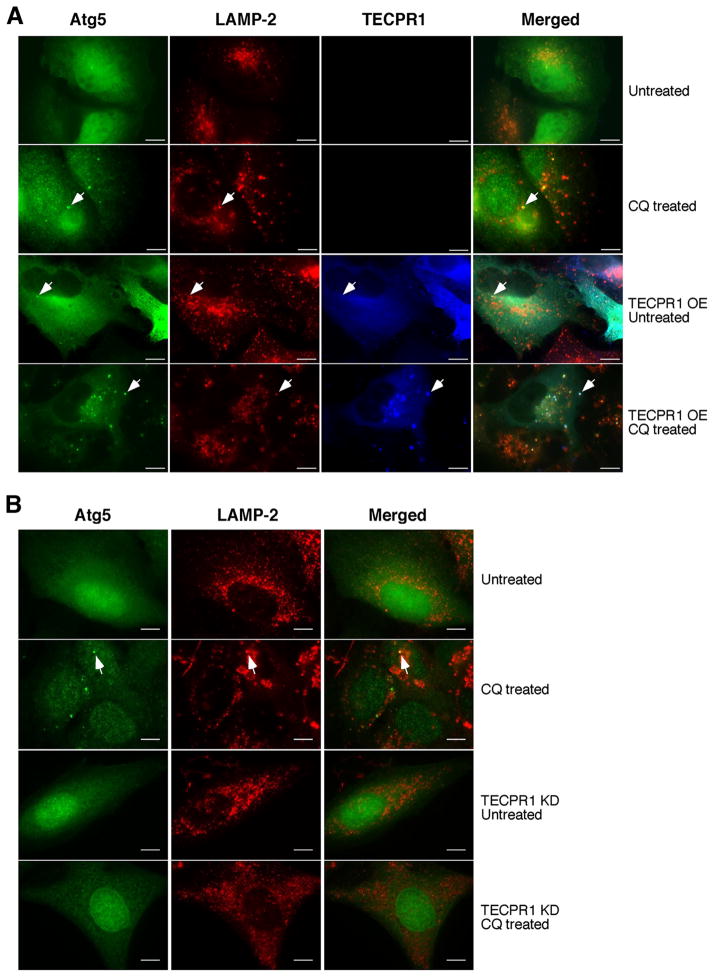
If TECPR1 localizes to autolysosome, its overlap with Atg5 implies that Atg5 may also be recruited to autolysosomes. Atg5 puncta were rarely detected in unstressed U2OS cells (Fig. 4A and S4A). However, in CQ treated cells, a moderate induction of Atg5 puncta was observed, and these puncta colocalized with the lysosome marker LAMP-2 (Fig. 4A and S4A). Remarkably, with TECPR1 overexpression and CQ treatment, the amount of Atg5 and TECPR1 puncta dramatically increased and these puncta colocalized with LAMP-2 (Fig. 4A and S4A). The inducibility of Atg5 autolysosomal localization is rather surprising, as it extends the classic idea that Atg5 localizes exclusively to phagophores or isolation membranes (Mizushima et al., 2001). Given the interaction between Atg5 and TECPR1, we speculated that Atg5 autolysosomal distribution is dependent on TECPR1. To investigate the requirement of TECPR1 in Atg5 recruitment, we established an inducible U2OS TECPR1 knockdown cell line. The autolysosomal localization of Atg5 upon CQ treatment was completely abolished in TECPR1 depleted cells (Fig. 4B and S4B), indicating that TECPR1 mediates the autolysosomal distribution of Atg5.
Figure 4. TECPR1 recruits Atg5 to autolysosomes.
(A) TECPR1-FLAG was inducibly expressed by doxycycline (1 μg/ml) in U2OS cells co-expressing Atg5-EGFP. These cells were left untreated or treated with CQ (200 μM for two hours). Endogenous LAMP-2 was stained with a LAMP-2 antibody. Scale bar (10 μm). Representative Atg5-positive autolysosomes are marked by arrows. (B) In TECPR1 wild type and RNAi knockdown cells with or without CQ (200 μM for two hours) treatment, Atg5-EGFP and LAMP-2 staining were examined. Scale bar (10 μm). Representative Atg5-positive autolysosomes are marked by arrows. See also Figure S4.
TECPR1 is required for autophagosome maturation
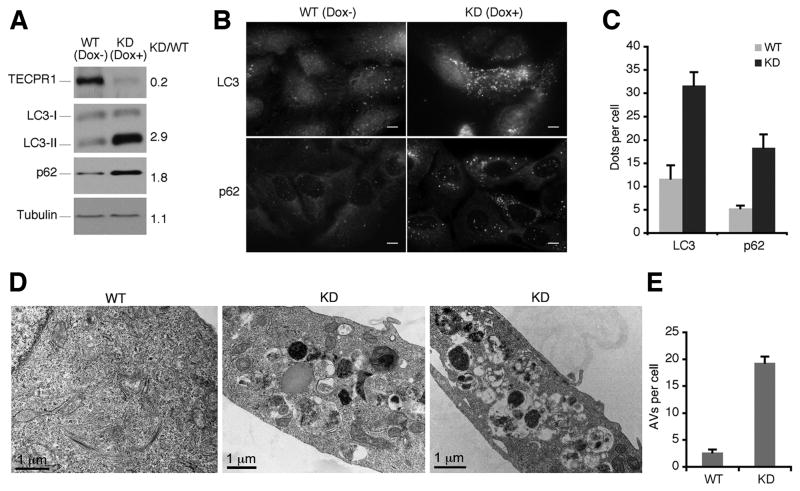
To further explore the role of TECPR1 in autophagy, we set up a doxycycline-inducible TECPR1 knockdown cell line in U2OS-TR cells. Upon depletion of TECPR1, we observed the accumulation of both the lipid conjugated form of LC3 (LC3-II), a faithful autophagosome marker, and p62, an autophagic substrate (Fig. 5A, 5B and 5C). This suggests that without TECPR1, autophagic degradation is impaired. To exclude the possibility of off-target effects, we also set up a TECPR1 depleted cell line in HeLa cells with TECPR1 shRNA lentiviral particles and a similar result was obtained (Fig. S5A).
Figure 5. Autophagosomes are accumulated in TECPR1 deficient cells.
(A) Doxycycline-inducible TECPR1 RNAi depletion in U2OS-TR cells. In U2OS cells inducibly expressing shRNA against TECPR1, endogenous TECPR1, LC3, p62 and Tubulin were detected in the presence or absence of doxycycline (1 μg/ml) for 5 days. (B) Endogenous LC3 or p62 was stained using anti-LC3 or anti-p62 antibody. Scale bar (10 μm). (C) Quantification result of at least 200 cells from three independent experiments described in (B). Data are presented as means ± SD. (D) Control or TECPR1 depleted U2OS cells were analyzed under transmission electron microscope. (E) Quantification result of at least 100 cells from three independent experiments described in (D). Data are presented as means ± SD. See also Figure S5.
To better understand the nature of accumulated vesicles upon TECPR1 knockdown, these cells were observed using transmission electron microscopy. Remarkably, we observed the accumulation of numerous autophagic vacuoles (Fig. 5D and 5E), further confirming that TECPR1 depletion impairs autophagic degradation.
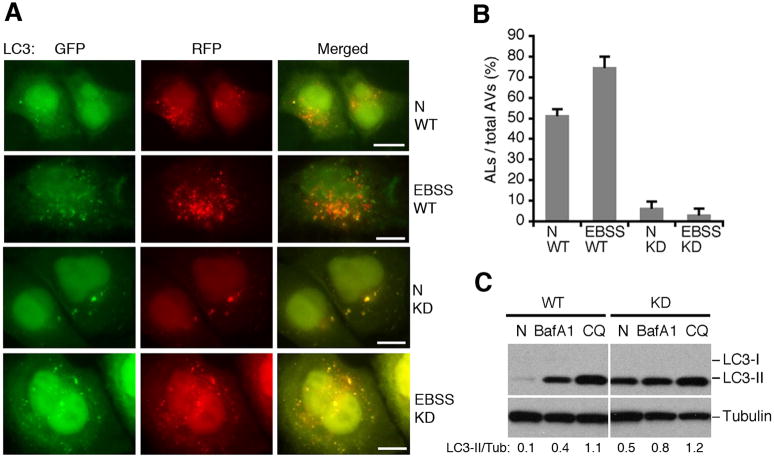
Lastly, we utilized a GFP-mRFP-LC3 marker to analyze autophagosome maturation in TECPR1 depleted cells (Kimura et al., 2009). Because GFP loses its fluorescence from deprotonation in acidic lysosomes, autophagosomes display both green and red fluorescence whereas autolysosomes are only red. Hence, autophagosome maturation is indicated by a dramatic increase in the number of red-only autolysosomes (Klionsky et al., 2008; Mizushima et al., 2010). In starved U2OS cells, about 70% of LC3 positive autophagic vacuoles are only red, suggesting that autolysosomes constitute the majority of autophagic vacuoles upon starvation (Fig 6A and 6B). However, upon starvation in TECPR1 depleted cells, approximately 90% of LC3 puncta are yellow (convergence of green and red signals). Therefore, the amount of red-only LC3 positive vacuoles representing autolysosomes is reduced to less than 10% (Fig. 6A and 6B). We also obtained a similar result in TECPR1 depleted HeLa cells (Fig. S5B and S5C). All these data indicate that autophagosome maturation is defective in the absence of TECPR1.
Figure 6. TECPR1 is required for autophagosome maturation.
(A) GFP-mRFP-LC3 was expressed and detected 48 hours after transfection in control cells or TECPR1 depleted U2OS cells treated with EBSS medium for one hour or mock treated. Scale bar (10 μm). (B) Quantification result of at least 200 cells from three independent experiments described in (A). Data are presented as means ± SD. AL, autolysosomes; AV, autophagic vacuoles. (C) The autophagic flux was examined in control and TECPR1 depleted U2OS cells mock treated (N), BafA1 or CQ treated. The intensity of LC3II/Tubulin was calculated as shown. See also Figure S6.
To consolidate the role of TECPR1 in autophagosome maturation, we looked for any defect in autophagic flux in both TECPR1 wild type and RNAi depleted cells. In wild type U2OS cells, LC3-II is dramatically accumulated upon CQ and Bafilomycin A1 (BafA1) (V-ATPase inhibitor) treatments due to the blockage in autophagic flux. In TECPR1 depleted U2OS cells, LC3-II accumulation also occurs in unstressed cells, and the fold induction of LC3-II upon CQ and BafA1 treatments is dramatically reduced (Fig. 6C), indicating that autophagic flux is impaired in TECPR1 depleted cells.
Although TECPR1 exclusively localizes to autolysosomes in our study, it has been implicated that TECPR1 might also have a function in phagophore biogenesis and maturation (Ogawa et al., 2011). To investigate a possible role for TECPR1 in phagophore biogenesis and maturation, we detected endogenous Atg16 in TECPR1 depleted cells. Compared to wild type cells, Atg16 puncta form but fail to accumulate in TECPR1 depleted cells (Fig. S6A and S6B), indicating that phagophore maturation is not affected when TECPR1 is depleted. Furthermore, the number of endogenous Atg16 puncta still increases upon Rapamycin treatment in TECPR1 depleted cells, suggesting that stress-induced phagophore biogenesis is not defective in the absence of TECPR1. In addition, the levels of Atg12-Atg5 remain unchanged in the absence of TECPR1 (Fig. S6C). Hence, phagophore biogenesis and maturation are not defective in the absence of TECPR1. To exclude another possibility that TECPR1 may be essential for the lysosomal activity therefore affects autophagosome consumption indirectly, we examined the lysosomal activity in TECPR1 depleted cells using cathepsin L processing as readout. The processing of cathepsin L from the precursor form to its mature form is only mildly reduced in the TECPR1 depleted cells compared to that in wild-type cells (Fig. S6D and S6E). In contrast, as a positive control, cathepsin L processing is dramatically reduced in the CQ-treated cells (Fig. S6F). These data suggest that TECPR1 is not likely to be essential for the lysosomal activity.
TECPR1 binds to PtdIns(3)P in an Atg12-Atg5 dependent manner
TECPR1 possesses a PH domain, which is known to bind phosphoinositides. In autophagy, the class III phosphatidylinositol 3-kinase plays a central role in autophagosome biogenesis and maturation (Backer, 2008; Funderburk et al., 2010; Simonsen and Tooze, 2009). The production of PtdIns(3)P on the autophagosome membrane is critical for developing a platform to recruit protein(s) for autophagosome formation, yet very few proteins that are involved in autophagy are known to bind PtdIns(3)P.
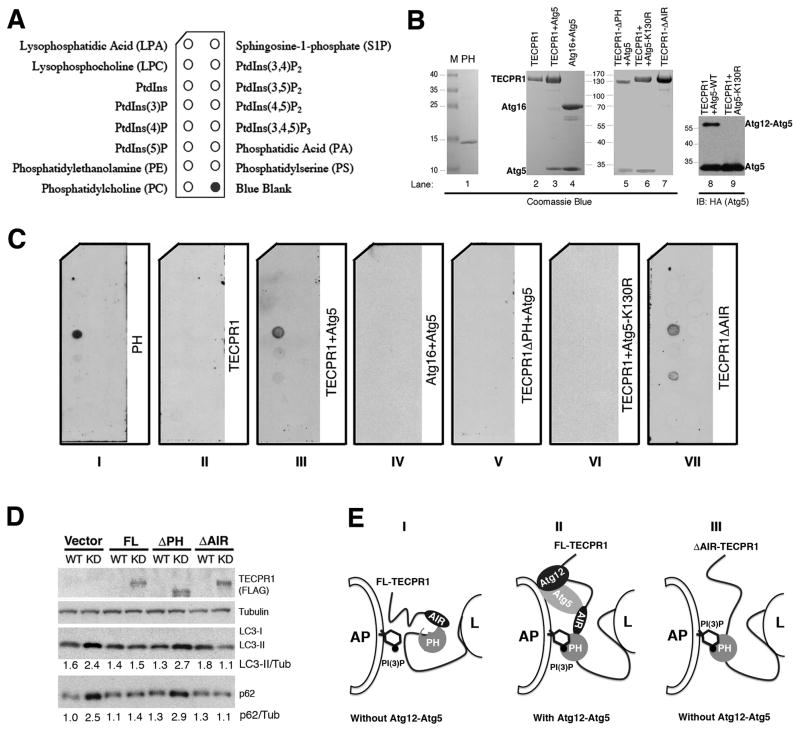
To investigate the type of lipid(s) that the PH domain of TECPR1 might bind, we performed a protein-lipid overlay assay (PIP Strips) containing eight phosphoinositides and other biologically active lipids (Fig. 7A). The recombinant PH domain of TECPR1 purified from HEK293T cells (Fig. 7B, lane 1) displays a strong and specific binding to PtdIns(3)P (Fig. 7C, panel I). Unexpectedly, the full-length TECPR1 alone purified from HEK293T cells (Fig. 7B, lane 2) loses its affinity for PtdIns(3)P (Fig. 7C, panel II). We reasoned that the association between PtdIns(3)P and TECPR1 might be regulated by Atg12-Atg5, given their physical interaction. To test this hypothesis, we co-expressed and purified the Atg12-Atg5-TECPR1 complex from HEK293T cells (Fig. 7B, lane 3) and tested its lipid binding activity. In the HEK293T overexpression system, only a portion of Atg5 is conjugated to Atg12 that could be readily detected by Western Blotting but barely detected by Coomassie Blue staining (Fig. 7B, lanes 3 and 8). Remarkably, this Atg12-Atg5-TECPR1 complex specifically binds PtdIns(3)P (Fig. 7C, panel III). As a control, the purified Atg12-Atg5-Atg16 complex (Fig. 7B, lane 4) has no detectable PtdIns(3)P binding activity (Fig. 7C, panel IV). The PtdIns(3)P binding of the Atg12-Atg5-TECPR1 complex is dependent on the PH domain of TECPR1 as deletion of this domain (Fig. 7B, lane 5) abolishes PtdIns(3)P association (Fig. 7C, panel V). A conjugation mutant form of Atg5 (Atg5 K130R) that lacks Atg12 conjugation (Fig. 7B, lanes 6 and 9), displays no detectable PtdIns(3)P binding activity (Fig. 7C, panel VI), indicating that Atg12 conjugation is required for the PtdIns(3)P binding of Atg5-TECPR1.
Figure 7. Binding of PtdIns(3)P by TECPR1 requires Atg12-Atg5 association.
(A) Schematic of a PIP Strip spotted with eight phosphoinositides and other bioactive lipids. (B) Purification of recombinant PH domain (lane 1), full-length TECPR1 (lane 2), Atg12-Atg5-TECPR1 complex (lane 3), Atg12-Atg5-Atg16 complex (lane 4), Atg12-Atg5-TECPR1 (PH domain deleted) (lane 5), TECPR1-Atg5K130R (lane 6) and Atg12-Atg5-TECPR1 (AIR domain deleted) (lane 7) in transfected HEK293T cells. Purified recombinant Atg12-Atg5-TECPR1 complex (lane 8) and TECPR1-Atg5K130R complex (lane 9) were probed with anti-Atg5 (HA) antibody for Western Blotting. (C) PIP Strips were incubated with recombinant proteins (1 μg/ml) as indicated for overnight at 4°C and probed with anti-FLAG antibody. (D) Complementation of TECPR1 with the indicated shRNA-resistant TECPR1 expression plasmids in TECPR1 depleted U2OS-TR cells. The inducible U2OS cells were cultured in the presence or absence of doxycycline (1 μg/ml) for 5 days. Expression of TECPR1 wild-type and mutants was detected with the FLAG antibody. The intensities of LC3II/Tubulin and p62/Tubulin were calculated as shown. (E) Proposed models of the role of TECPR1 in autophagosome maturation through its association with Atg12-Atg5 and PtdIns(3)P. Panel I, full-length TECPR1 in the absence of the Atg12-Atg5 conjugate. Panel II, full-length TECPR1 in the presence of the Atg12-Atg5 conjugate. Panel III, AIR-deleted TECPR1 in the absence of the Atg12-Atg5 conjugate. AP, autophagosome; L, lysosome. See also Figure S7.
Since the Atg12-Atg5 associated TECPR1, but not TECPR1 alone, is able to bind PtdIns(3)P, it is likely that the PH domain of TECPR1 is normally embedded and the binding of the Atg12-Atg5 conjugate to the AIR region of TECPR1 may cause a conformational change to expose the PH domain for PtdIns(3)P binding. To test this possibility, we purified the AIR-deleted (ΔAIR) TECPR1 from the HEK293T cells (Fig. 7B, lane 7). As expected, the purified AIR-deleted (ΔAIR) TECPR1 exhibits strong PtdIns(3)P binding activity (Fig. 7C, panel VII) that is comparable to the Atg12-Atg5 associated TECPR1. These data suggest that the AIR domain of TECPR1 indeed blocks the access of the PH domain to PtdIns(3)P in vitro.
To evaluate the contribution of PtdIns(3)P binding to the function of TECPR1 in autophagy, we tested the requirement of different domains of TECPR1 in the complementation assay (Fig. 7D). The autophagosome maturation defects (shown as the accumulation of LC3-II and p62) in TECPR1 depleted cells could be fully rescued by the expression of wild-type TECPR1 but not the vector alone. The PH-deleted TECPR1 also failed to complement, suggesting that the PtdIns(3)P binding is required for the autophagic function of TECPR1. Interestingly, the AIR-deleted TECPR1 could fully complement the autophagic defects in TECPR1 depleted cells, which is consistent with its robust PtdIns(3)P binding activity. We further investigated if the subcellular localization of TECPR1 in unstressed or challenged cells is affected by PtdIns(3)P production. The lysosomal distribution of TECPR1 in the unstressed cells was not significantly affected by the treatment of a class III phosphatidylinositol 3-kinase (PI3K) inhibitor 3-Methyladenine (3-MA) (Fig. S7). Upon Rapamycin or CQ treatment, the autolysosomal appearance of TECPR1 was dramatically increased. Remarkably, autolysosomal localization of TECPR1 was severely impaired by 3-MA treatment, suggesting a requirement for the PI3K activity in TECPR1 localization (Fig. S7). As a positive control, stress-induced LC3 puncta were diminished in the 3-MA treated cells (data not shown). All these data support an essential role of PtdIns(3)P binding in the autophagic function of TECPR1.
Discussion
In this study, we demonstrate that TECPR1 controls autophagosome maturation by interacting with both Atg12-Atg5 and PtdIns(3)P. TECPR1 interacts with Atg12-Atg5 in a subcomplex that is mutually exclusive from the Atg12-Atg5-Atg16 complex. Additionally, TECPR1 specifically localizes to and recruits the Atg12-Atg5 conjugate to the autolysosome. TECPR1 depletion leads to accumulation of autophagic substrates including p62 and lipidated LC3 (LC3-II). TECPR1 deficient cells accumulate autophagic vacuoles. Moreover, autophagosome maturation and autophagic flux are blocked in TECPR1 depleted cells. Importantly, PtdIns(3)P binding of TECPR1 through the PH domain, which might be regulated by the Atg12-Atg5 conjugate in an auto-inhibitory manner (Fig. 7E), is essential for the autophagic function of TECPR1. Taken together, these data demonstrate that TECPR1 plays a critical role in autophagosome maturation. We anticipate that TECPR1 might function as a tethering factor to join autophagosome with lysosome through its association of Atg12-Atg5 and PtdIns(3)P; such action might initiate autophagosome-lysosome fusion.
TECPR1 was recently identified as a component of the autophagy interaction network which consists of hundreds of proteins interacting with the core autophagy machinery (Behrends et al., 2010). In this network, it was found that TECPR1 interacts with Atg5, which was coined by another recent report by Ogawa et al. (Ogawa et al., 2011). Interestingly, in both studies, TECPR1 has been reported to interact with proteins in addition to Atg5, including WIPI-2, Atg3, TTC15 (TPR coatomer ε) and the chaperonin complex. These proteins were not identified in our purified TECPR1 cellular complex probably due to relatively weak interactions. Behrends et al. reported that deletion of TECPR1 leads to LC3-II accumulation (Behrends et al., 2010). However, Ogawa et al. suggested that TECPR1 is more important in selective autophagy against pathogens rather than canonical autophagy, since LC3-II conjugation upon autophagic stresses remains efficient in TECPR1 knockout cells (Ogawa et al., 2011).
In contrast, through a comprehensive approach, we have established a rather surprising role for TECPR1 in autophagosome maturation. We observed that phagophore and autophagosome formation are not defective upon TECPR1 depletion, suggesting that TECPR1 does not play a critical role in autophagosome biogenesis. Strikingly, autophagosomes indicated by LC3-II and p62 staining are significantly accumulated in TECPR1 deficient cells, indicating a blockage in autophagosome maturation instead. It is currently unclear what causes these discrepancies, autophagosome maturation defects might be overlooked by the other groups.
Phosphoinositides are critical for signal transduction of membrane trafficking. Hence, PtdIns(3)P production is essential for autophagosome biogenesis (Backer, 2008; Funderburk et al., 2010). Several pieces of evidence have also suggested a role for PtdIns(3)P in autophagosome maturation. PtdIns(3)P is important for membrane curvature sensing and maintenance by the PI3KC3 (Fan et al., 2011; Sun et al., 2008). Additionally, depletion of PtdIns(3)P decreases membrane curvature and impedes membrane expansion (Hubner et al., 1998; Kovacs et al., 2000). Conversely, accumulation of PtdIns(3)P either by mutation of phosphatases (Taguchi-Atarashi et al., 2010) or blockage of vacuolar degradation (Wurmser and Emr, 1998) affects autophagosome maturation. It has been reported that phagophore localization of the Atg12-Atg5-Atg16 complex is dependent on the presence of PtdIns(3)P (Fujita et al., 2008). Here we show that PtdIns(3)P contributes to autophagosome maturation through the Atg12-Atg5-TECPR1 complex and is critical for the autolysosome localization of TECPR1. TECPR1 contains a PH domain that specifically interacts with PtdIns(3)P in vitro. Although most characterized PH domains bind PtdIns(3,4,5)P3, PtdIns(4,5)P2 and PtdIns(3,4)P2 with high affinity, some PH domains indeed display PtdIns(3)P binding specificity (Dowler et al., 2000). Interestingly, the TECPR1 PtdIns(3)P binding requires not only its PH domain, but also its interaction with the conjugated Atg12-Atg5 (Fig. 7). Both Ogawa et al. and our group showed that full-length TECPR1 fails to interact with PtdIns(3)P by itself. Ogawa et al. implied that TECPR1 might bind PtdIns(3)P indirectly by interacting with WIPI2, a PtdIns(3)P binding protein (Ogawa et al., 2011). However, using purified recombinant proteins, we showed that the PH domain alone, Atg12-Atg5 associated TECPR1 and AIR-deleted TECPR1 are capable of binding PtdIns(3)P, suggesting that the interaction of Atg12-Atg5 might expose the PH domain of TECPR1 and allow TECPR1 to bind PtdIns(3)P directly (Fig. 7E).
The AIR domain of TECPR1 mediates the interaction with the Atg12-Atg5 conjugate, but this domain might also serves as an auto-inhibitory domain to block the PH domain from PtdIns(3)P binding. The pleiotropic functions of the AIR domain make it difficult to further dissect the requirement of the Atg12-Atg5-TECPR1 complex in autophagy. It is crucial in the future study to identify a mutation of TECPR1 in the AIR domain that only disrupts the Atg12-Atg5 interaction without affecting the protein conformation, which probably requires more detailed information about the binding surface through crystallographic analysis.
Autophagic vacuole maturation is defective in certain human diseases including lysosomal storage disease (Cao et al., 2006; Chevrier et al., 2010; Raben et al., 2008) and Danon disease (Nishino et al., 2000). Autophagosome maturation appears not to be essential for animal development, which is best illustrated in LAMP-2 knockout mice. In these cells, there is an extensive accumulation of autophagic vacuoles yet LAMP-2 knockout mice could have an almost normal life span (Tanaka et al., 2000). However, autophagic vacuole accumulation is still problematic as the deficiency of LAMP-2 leads to multiple human diseases including cardiomyopathy and Danon disease (Nishino et al., 2000; Tanaka et al., 2000). Also, LAMP-2 deficient cells showed a reduced bacterial killing capacity caused by an impaired fusion of endo-lysosome with phagosomes (Beertsen et al., 2008; Binker et al., 2007). It is possible that TECPR1 and LAMP-2 share common mechanisms in autophagosome maturation. Understanding the molecular mechanism of this process should provide insights into the pathogenesis of these diseases, and pave the way for therapeutic drug design.
Experimental Procedures
Reagents
The anti-TECPR1 antibody was generated by Cell Signaling Technology. Other antibodies used in this study include anti-FLAG (Sigma), anti-HA (Roche), anti-Myc (Santa Cruz), anti-EEA1 (Abcam), anti-LAMP-1 (Abcam), anti-LAMP-2 (Santa Cruz), anti-LC3 (Sigma), anti-Atg5 (MBL International), anti-Atg16 (MBL International) and anti-p62 (MBL International).
Tandem Affinity Purification (TAP)
The Atg5 coding sequence was inserted between Bam HI and Not I cleavage sites to generate the reading frame of ZZ-Atg5-FLAG. The tandem affinity purification was performed as described before (Sun et al., 2008).
Immunofluorescence Staining
Cells seeded into 6-well plates with coverslips were fixed with cold methanol (−20 °C) and blocked with blocking buffer (2.5% BSA+0.1% Triton X-100 in PBS). After incubation with primary and secondary antibodies, samples were examined under a laser scanning confocal microscope (Zeiss LSM 510 META UV/Vis).
Electron Microscope Analysis
For transmission electron microscopy, cells were fixed in 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.4). For immuno-electron microscopy (Tokuyasu method), cells were fixed in freshly made 2% paraformaldehyde in 0.1 M PBS containing 0.2% glutaraldehyde, pH 7.2–7.4.
Protein–lipid overlay assay
The FLAG tagged recombinant proteins were purified from mammalian cells as described (Sun et al., 2010; Sun et al., 2011). PIP Strips (Echelon Biosciences Inc.) were blocked in blocking buffer (PBS with 1% nonfat dry milk) for 1 hour at room temperature and then incubated with 1.0 μg/ml indicated FLAG tagged protein in blocking buffer at 4 °C overnight. After 3 washes with PBS-T, strips were immunoblotted with anti-FLAG antibody.
Please see supplemental information for more experimental details.
Supplementary Material
Acknowledgments
We thank Alice Liang at OCS Microscopy Core of New York University Langone Medical Center for EM analysis and Cell Signaling Technology for TECPR1 antibody production. We also thank all Zhong lab members for helpful discussion, Qiming Sun and Gary Ma for technical assistance, Livy Wilz for the critical reading, and Jeremy Thorner for suggestions. The work is supported by University of California Cancer Research Coordinating Committee funds, a New Investigator Award for Aging from the Ellison Medical Foundation, Hellman Family Fund, American Cancer Society Research Scholar Grant (RSG-11-274-01-CCG) and NIH R01 (CA133228) to Q. Z..
Footnotes
D. C. performed most of the experiments. W. F. and Y. L. helped. X. D. and S. C. performed the mass spectrometry analysis. D. C. and Q. Z. wrote the paper. Q. Z. directed the work. All authors discussed the results and commented on the manuscript.
These authors have no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeliovich H, Darsow T, Emr SD. Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. Embo J. 1999;18:6005–6016. doi: 10.1093/emboj/18.21.6005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J. 2008;410:1–17. doi: 10.1042/BJ20071427. [DOI] [PubMed] [Google Scholar]
- Baerga R, Zhang Y, Chen PH, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009;5:1118–1130. doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boonen M, van Meel E, Oorschot V, Klumperman J, Kornfeld S. Vacuolization of mucolipidosis type II mouse exocrine gland cells represents accumulation of autolysosomes. Mol Biol Cell. 2011;22:1135–1147. doi: 10.1091/mbc.E10-07-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Espinola JA, Fossale E, Massey AC, Cuervo AM, MacDonald ME, Cotman SL. Autophagy is disrupted in a knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. The Journal of biological chemistry. 2006;281:20483–20493. doi: 10.1074/jbc.M602180200. [DOI] [PubMed] [Google Scholar]
- Chevrier M, Brakch N, Lesueur C, Genty D, Ramdani Y, Moll S, Djavaheri-Mergny M, Brasse-Lagnel C, Laquerriere A, Barbey F, Bekri S. Autophagosome maturation is impaired in Fabry disease. Autophagy. 2010;6:589–599. doi: 10.4161/auto.6.5.11943. [DOI] [PubMed] [Google Scholar]
- Darsow T, Rieder SE, Emr SD. A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. The Journal of cell biology. 1997;138:517–529. doi: 10.1083/jcb.138.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler S, Currie RA, Campbell DG, Deak M, Kular G, Downes CP, Alessi DR. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem J. 2000;351:19–31. doi: 10.1042/0264-6021:3510019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL. Maturation of autophagic vacuoles in Mammalian cells. Autophagy. 2005;1:1–10. doi: 10.4161/auto.1.1.1270. [DOI] [PubMed] [Google Scholar]
- Eskelinen EL, Illert AL, Tanaka Y, Schwarzmann G, Blanz J, Von Figura K, Saftig P. Role of LAMP-2 in lysosome biogenesis and autophagy. Mol Biol Cell. 2002;13:3355–3368. doi: 10.1091/mbc.E02-02-0114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proc Natl Acad Sci U S A. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex - at the crossroads of autophagy and beyond. Trends Cell Biol. 2010;20:355–362. doi: 10.1016/j.tcb.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Hanada T, Noda NN, Satomi Y, Ichimura Y, Fujioka Y, Takao T, Inagaki F, Ohsumi Y. The Atg12-Atg5 conjugate has a novel E3-like activity for protein lipidation in autophagy. The Journal of biological chemistry. 2007;282:37298–37302. doi: 10.1074/jbc.C700195200. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Hubner S, Couvillon AD, Kas JA, Bankaitis VA, Vegners R, Carpenter CL, Janmey PA. Enhancement of phosphoinositide 3-kinase (PI 3-kinase) activity by membrane curvature and inositol-phospholipid-binding peptides. European journal of biochemistry/FEBS. 1998;258:846–853. doi: 10.1046/j.1432-1327.1998.2580846.x. [DOI] [PubMed] [Google Scholar]
- Inoue Y, Klionsky DJ. Regulation of macroautophagy in Saccharomyces cerevisiae. Semin Cell Dev Biol. 2010;21:664–670. doi: 10.1016/j.semcdb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, Ishii KJ, Kawai T, Akira S, Suzuki K, Okuda K. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Klionsky DJ. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annual review of biochemistry. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- Kimura S, Fujita N, Noda T, Yoshimori T. Monitoring autophagy in mammalian cultured cells through the dynamics of LC3. Methods Enzymol. 2009;452:1–12. doi: 10.1016/S0076-6879(08)03601-X. [DOI] [PubMed] [Google Scholar]
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs AL, Rez G, Palfia Z, Kovacs J. Autophagy in the epithelial cells of murine seminal vesicle in vitro. Formation of large sheets of nascent isolation membranes, sequestration of the nucleus and inhibition by wortmannin and 3-ethyladenine. Cell Tissue Res. 2000;302:253–261. doi: 10.1007/s004410000275. [DOI] [PubMed] [Google Scholar]
- Kuma A, Hatano M, Matsui M, Yamamoto A, Nakaya H, Yoshimori T, Ohsumi Y, Tokuhisa T, Mizushima N. The role of autophagy during the early neonatal starvation period. Nature. 2004;432:1032–1036. doi: 10.1038/nature03029. [DOI] [PubMed] [Google Scholar]
- Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. The Journal of biological chemistry. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Leu JI, Pimkina J, Frank A, Murphy ME, George DL. A small molecule inhibitor of inducible heat shock protein 70. Mol Cell. 2009;36:15–27. doi: 10.1016/j.molcel.2009.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nature cell biology. 2008;10:776–787. doi: 10.1038/ncb1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nature cell biology. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. J Cell Sci. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, Klionsky DJ, Ohsumi M, Ohsumi Y. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. The Journal of cell biology. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, et al. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarko VY, Nazarko TY, Farre JC, Stasyk OV, Warnecke D, Ulaszewski S, Cregg JM, Sibirny AA, Subramani S. Atg35, a micropexophagy-specific protein that regulates micropexophagic apparatus formation in Pichia pastoris. Autophagy. 2011;7:375–385. doi: 10.4161/auto.7.4.14369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson DP, Brett CL, Merz AJ. Vps-C complexes: gatekeepers of endolysosomal traffic. Curr Opin Cell Biol. 2009;21:543–551. doi: 10.1016/j.ceb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, Mora M, Riggs JE, Oh SJ, Koga Y, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease) Nature. 2000;406:906–910. doi: 10.1038/35022604. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, Piao Z, Ashida H, Yoshida M, Kakuta S, et al. A tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell host & microbe. 2011;9:376–389. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- Poole B, Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J Cell Biol. 1981;90:665–669. doi: 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raben N, Hill V, Shea L, Takikita S, Baum R, Mizushima N, Ralston E, Plotz P. Suppression of autophagy in skeletal muscle uncovers the accumulation of ubiquitinated proteins and their potential role in muscle damage in Pompe disease. Hum Mol Genet. 2008;17:3897–3908. doi: 10.1093/hmg/ddn292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M, Chan EY, Tooze SA. Early endosomes and endosomal coatomer are required for autophagy. The Journal of cell biology. 2009;185:305–321. doi: 10.1083/jcb.200810098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. The Journal of cell biology. 2009;186:773–782. doi: 10.1083/jcb.200907014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Fan W, Chen K, Ding X, Chen S, Zhong Q. Identification of Barkor as a mammalian autophagy-specific factor for Beclin 1 and class III phosphatidylinositol 3-kinase. Proc Natl Acad Sci U S A. 2008;105:19211–19216. doi: 10.1073/pnas.0810452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Westphal W, Wong KN, Tan I, Zhong Q. Rubicon controls endosome maturation as a Rab7 effector. Proc Natl Acad Sci U S A. 2010;107:19338–19343. doi: 10.1073/pnas.1010554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhang J, Fan W, Wong KN, Ding X, Chen S, Zhong Q. The RUN domain of rubicon is important for hVps34 binding, lipid kinase inhibition, and autophagy suppression. The Journal of biological chemistry. 2011;286:185–191. doi: 10.1074/jbc.M110.126425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. The Journal of biological chemistry. 2010;285:30019–30025. doi: 10.1074/jbc.M110.143511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic (Copenhagen, Denmark) 2010;11:468–478. doi: 10.1111/j.1600-0854.2010.01034.x. [DOI] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. Embo J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.