Summary
Pyruvate kinase isoform M2 (PKM2) is a glycolysis enzyme catalyzing conversion of phosphoenolpyruvate (PEP) to pyruvate with transferring a phosphate from PEP to ADP. We report here that PKM2 localizes to the cell nucleus. The levels of nuclear PKM2 correlate with cell proliferation. PKM2 activates transcription of MEK5 by phosphorylating stat3 at Y705. In vitro phosphorylation assays show that PKM2 is a protein kinase using PEP as phosphate donor. ADP competes with the protein substrate binding, indicating that the substrate may bind to the ADP site of PKM2. Our experiments suggest that PKM2 dimer is an active protein kinase, while the tetramer is an active pyruvate kinase. Expression a PKM2 mutant that exists as a dimer promotes cell proliferation, indicating that protein kinase activity of PKM2 plays a role in promoting cell proliferation. Our study reveals an important link between metabolism alteration and gene expression during tumor transformation and progression.
Keywords: Pyruvate kinase M2, Proliferation, stat3, protein kinase, glycolysis
Introduction
An important molecular feature of tumor development is the expression of glycolysis enzyme pyruvate kinase isoenzyme M2 (PKM2) (Elbers et al., 1991; Hacker et al., 1998). Catalytically active PKM2 exists as a tetramer and associates with a glycolytic enzyme complex (Altenberg and Greulich, 2004; Dombrauckas et al., 2005; Zwerschke et al., 1999). In tumor cells, PKM2 forms a dimer, and appears to be catalytically inactive for conversion of PEP to pyruvate (Ashizawa et al., 1991; Mazurek et al., 2005). It is believed that the inactive PKM2 actually provides growth advantage for tumor progression as it helps to channel the carbon source from glycolytic intermediates to biosynthesis, especially syntheses of nucleic acids, lipids, and proteins, to meet the demands for tumor cell proliferation. Recent studies have suggested that PKM2 changes its pyruvate kinase activity, consequently facilitating cell proliferation by binding to tyrosine phosphor-peptide, or becoming phosphorylated under growth stimulation (Christofk et al., 2008a; Christofk et al., 2008b; Hitosugi et al., 2009), indicating that growth signals regulate the activity of PKM2 in glucose metabolism.
It has long been noted that several enzymes that act in different metabolic pathways possess so-called moonlight activity (Jeffery, 1999). These moonlight activities may play important roles in a number of physiological and pathological processes (Sriram et al., 2005). Using the pyruvate kinase M2 purified from chromatin extracts of hepatoma tumor cells, Jan Ignacak and co-workers noticed that histone H1 is capable of activation of the enzymatic reaction of PKM2 of conversion of PEP to pyruvate in the absence of ADP (Ignacak and Stachurska, 2003). Although the study did not demonstrate the protein kinase activity of PKM2, it suggested that PKM2 may have an enzymatic activity that is not directly involved in transferring phosphate from PEP to ADP. Recently, several independent studies demonstrated that PKM2 localized in cell nucleus in response to different signals (Hoshino et al., 2007; Stetak et al., 2007). The nuclear PKM2 is shown to participate in regulation of gene transcriptions (Lee et al., 2008; Luo et al., 2011; Yang et al., 2011). Interestingly, PKM2 can function as a co-activator of HIF-1α (Luo et al., 2011). Thus, it is suggested that PKM2 may act in a feed-back loop in response to hypoxia condition to re-program cancer cell metabolism. Furthermore, growth stimulation by EGFR induces PKM2 nuclear translocation and activates the activity of PKM2 in the gene transcription regulation (Yang et al., 2011), suggesting that the role of PKM2 in gene transcription is regulated by growth stimulations.
We report here that PKM2 can act as a protein kinase. PKM2 dimer is an active protein kinase, while the tetramer is an active pyruvate kinase. The nuclear PKM2 is a dimeric form. The nuclear PKM2 directly phosphorylates stat3 independent of JaK2 and c-Src pathways. Phosphorylation of stat3 by nuclear PKM2 lead to activation of transcription of a number of stat3 targeted genes. Expression of a PKM2 mutant R399E that exist as dimer dramatically promotes cell proliferation and tumor growth, indicating the important role of the dimeric PKM2 in cancer progression.
Results
Nuclear PKM2 activates transcription of MEK5 by phosphorylating stat3
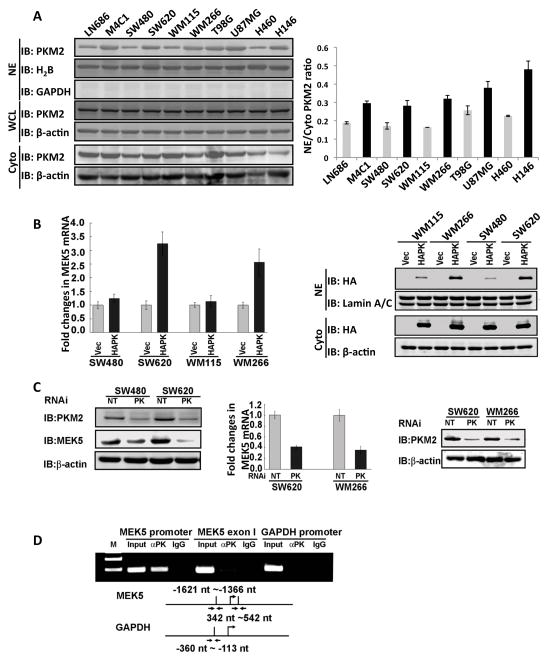
We and others have observed that the PKM2 localizes to the cell nucleus (Hoshino et al., 2007; Schneider et al., 2002; Stetak et al., 2007). To understand the functional significance of nuclear localization of PKM2, we examined nuclear PKM2 levels in ten cancer cell lines by immunoblot using an antibody raised against a peptide spanning aa 399 – 412 of PKM2 (PabPKM2). These ten cell lines represent different cancer progression stages and are derived from cancers of different organs/tissue types. Cell proliferation analyses demonstrated that M4C1, SW620, WM266, and H146 are more proliferated than their corresponding lines in the matched pairs (Fig. S1A). Immunoblot analyses indicated much higher nuclear levels of PKM2 in the more proliferated cancer cells than those in the corresponding less proliferated cells in the matched pair (Fig. 1A, and S1B), The higher nuclear PKM2 levels were not because of differences in PKM2 expression (Fig. 1A). The detection of PKM2 in the nuclear extracts was not due to contamination of cytoplasmic proteins as demonstrated by lack of GAPDH in the immunoblot analyses of the nuclear extracts (Fig. S1B).
Figure 1. PKM2 regulates MEK5 transcription.
(A) Nuclear localization of PKM2 in different cancer cells. (Left) Immunoblot analyses of PKM2 using the antibody PabPKM2 (IB:PKM2) in nuclear extracts (NE), cytoplasmic extracts (Cyto), and whole cell lysates (WCL) of ten cell lines (indicated). Blot of GAPDH (IB:GAPDH) in nuclear extracts is a control indicate no cytoplasmic protein contaminations. (Right) Quantization of the nuclear and cytoplasmic PKM2 in different cell lines based on the immunoblots. The results are presented as the ratio of nuclear vs cytoplasmic (NE/Cyto) levels. (B) (Left) RT-PCR analyses of cellular MEK5 mRNA in two pairs of cancer cells (indicated) in which PKM2 was expressed (HAPK) using the adeno-viral expression kit. The results are presented as fold increase in PCR products before and 48 hours after PKM2 expression in the same cells. The level of PCR product from each cell line before PKM2 expression was defined as 1. (Right) The levels of expressed HA-PKM2 (HAPK) in the nuclear (NE) and cytoplasm (Cyto) in four cell lines (indicated) were analyzed by immunoblot of nuclear or cytoplasmic extracts using anti-HA antibody (IB:HA). Vec means that the cells express vector alone. (C) (Left) Expressions of MEK5 (IB:MEK5) in SW480 and SW620 cells in which PKM2 was knocked down (PK) were analyzed by immunoblot using antibody against MEK5. The cellular levels of PKM2 were analyzed by immunoblot of PKM2 (IB:PKM2). NT means the cells were transfected with non-target siRNA. (Middle) RT-PCR analyses of cellular MEK5 mRNA levels and (Right) immunoblot analyses of the cellular PKM2 levels (IB:PKM2) in cancer cells (indicated) in which PKM2 was knocked down (PK). The results of mRNA levels are presented as fold increase in PCR products before and 48 hours after PKM2 expression in the same cells. The level of PCR product from each cell line before PKM2 expression was defined as 1. (D) ChIP of the MEK5 promoter (MEK5 promoter) using antibody against PKM2 in SW620 cells (αPK). ChIP using rabbit IgG was a negative control. ChIP using PCR primer pair targeting a region of exon 1 (MEK5 exon 1) of MEK5 gene (nt 342 – nt 542) and targeting GAPDH promoter (GAPDH promoter) using antibody against PKM2 were negative controls. The primer pair positions are indicated. Inputs were PCR products from DNA extracts without ChIP. Error bars in (B) and (C) are standard deviations of three measurements. In (A) (B), and (C), Immunoblot of β-actin (IB:β-actin), immunoblots of H2B (IB:H2B), and immunoblots of Lamin A/C (IB:Lamin A/C) are loading controls.
To explore a possibility that nuclear PKM2 is involved in regulation of gene expressions, we carried out gene expression array analyses with SW620 cells expressing HA-PKM2. Expressions of 350 genes were upregulated, and expressions of 359 genes were downregulated, by at least two fold (Table S1). Among those affected genes, expression of MEK5 was affected by over six fold. Expression of MEK5 plays an important role in SW620 cell proliferation. Thus, we verified the role of PKM2 in expression of MEK5 by RT-PCR. Consistent with the gene array analyses, RT-PCR demonstrated a dramatic change in the MEK5 expression in SW620 cells, while only a marginal increase in SW480 cells, a colon cancer cell line that is derived from the same patient with substantially lower proliferation rate, following the expression of PKM2. Consistently, higher nuclear levels of exogenously expressed HA-PKM2 were observed in more proliferated cells (Fig. 1B). Similar patterns were also observed with another pair of melanoma cell lines, WM115 and WM266. To further verify the role of PKM2 in transcriptional regulation of MEK5, PKM2 was knocked down in SW620 and SW480 cells (Fig. 1C). Quantitative RT-PCR analyses demonstrated a strong reduction in cellular MEK5 mRNA (Fig. 1C). We further used chromatin immunoprecipitation (ChIP) to probe whether PKM2 interacted with the MEK5 promoter. The ChIP was carried out with SW620 cells using the PCR primer pair that spanned the region of nt −1,621 – −1,366 of MEK5 promoter. Our ChIP assays clearly demonstrated that PKM2 indeed interacted with the MEK5 promoter (Fig. 1D).
We next tested whether up-regulation of MEK5 by PKM2 contributed to cell proliferation. MEK5 was first knocked down in SW620 and WM266 cells. As a result, the cell proliferation rate was dramatically reduced (Fig. S1C). The increases in proliferation by expression of HA-PKM2 in the MEK5 knockdown SW620 and WM266 cells were also largely reduced (Fig. S1C), suggesting that up-regulation of MEK5, at least partially, mediated the effects of HA-PKM2 overexpression on cell proliferation. Moreover, we analyzed the MEK5 expression levels in these ten cancer cell lines. MEK5 was expressed in all ten cell lines, but there were substantially higher levels of MEK5 in the more proliferated cancer cells than in the less proliferated cancer cells within the matched pair (Fig. S1D). Clearly, there is a close correlation between cellular MEK5 levels and nuclear PKM2 levels, and both were correlated closely with the cell proliferation status of the cells (comparing Fig. S1D with Fig. 1A). To rule out a possibility that high cellular levels of MEK5 promote PKM2 nuclear localization, MEK5 was knocked down, and the nuclear/cytoplasmic PKM2 was examined. It was clear that knockdown of MEK5 did not affect the nuclear levels of PKM2 (Fig. S1E). We concluded from these studies that up-regulation of MEK5 by nuclear PKM2 contributed to cell proliferation.
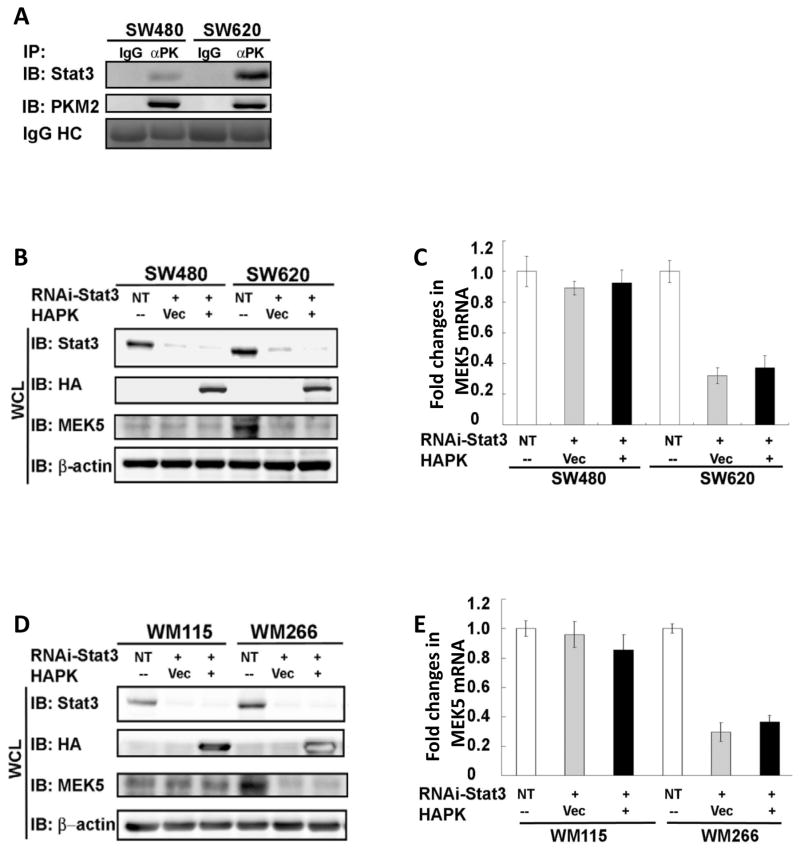
How would PKM2 function in regulation of gene transcription? Sequence analyses did not reveal any known DNA binding domain/motifs in PKM2. One possibility is that PKM2 may activate particular transcription factors. Thus, we attempted to probe the interaction of PKM2 with several known transcription factors in nuclear extracts of SW620 and SW480 cells by immunoprecipitation, such as Oct-1, Oct-4, Gadd45, SOX4-1, and stat3. Among these selected targets, stat3 and SOX4 were involved in transcription of MEK5 (Aaboe et al., 2006; Song et al., 2004), while Oct-4 was reported to interact with PKM2 in extracts made from ESC (Lee et al., 2008). Our experiments showed that stat3 was the only transcriptional factor among the selected targets that interacted with PKM2 (Data not shown, Fig. 2A, and Fig. S2 A&B). It might be due to the differences between ESC and cancer cells, we did not detect the PKM2 and Oct-4 interaction in the extracts prepared from cancer cells (data not shown). We noted that stat3 interacts with MEK5 promoter at −1776 nt – −1520 nt region by ChIP (Song et al., 2004). Interestingly, PKM2 also interacts with MEK5 promoter at the same region (Fig. 1D). To address whether the activation of stat3 mediates the effects of PKM2 on upregulation of MEK5 transcription, we first knocked down stat3 in SW620/SW480 and WM266/WM115 cells, and HA-PKM2 was subsequently expressed. It was clear that expression of HA-PKM2 could no longer upregulate MEK5 as measured by cellular levels of both mRNA and protein of MEK5 (Fig. 2B&C, D&E). To exclude a possibility that downregulation of MEK5 is solely due to stat3 knockdown, HA-PKM2 was expressed in these cells. Immunoblots indicated that MEK5 was up-regulated upon the HA-PKM2 expression (Fig. S2D). To further test the role of stat3 in mediating the effects of PKM2 on upregulation of MEK5 transcription, we used a dominant-negative mutant stat3 (Y705F, ref to as stat3-DN) (Xie et al., 2006). The mutant was co-expressed with HA-PKM2 in SW620/SW480 and WM266/WM115 cells. It was evident that expression of the stat3-DN largely diminished the effects of PKM2 on upregulating MEK5 transcription (Fig. S2 E&F, G&H). As a control, expression of the stat3-DN in PKM2 knockdown SW480/SW620 cells did not result in up-regulation of MEK5 expression (Fig. S2E). The activity of stat3 can be inhibited by specific inhibitor (Xu et al., 2008). We therefore tested the effects of a stat3 inhibitor on the upregulation of MEK5 by expression of HA-PKM2 in SW620 and WM266 cells. Treatment of the HA-PKM2 expressing cells with the inhibitor resulted in a decrease in MEK5 transcription (Fig. S2 I&J). Thus, we concluded that activation of stat3 mediated the regulatory effects of PKM2 on MEK5 transcription. Consistently, re-analyses of our expression array data revealed changes of a number of stat3 regulatory genes in the PKM2 over-expressing SW620 cells (Table S2).
Figure 2. PKM2 regulates MEK5 transcription via activation of stat3.
(A) Co-immunoprecipitation of PKM2 with stat3. PKM2 was immunoprecipitated from nuclear extracts of SW620 and SW480 cells using the antibody PabPKM2 (IP:αPK). The immunoprecipitates were analyzed by immunoblot using antibodies against stat3 (IB:Stat3) or PKM2 (IB:PKM2). Ponceau S staining of IgG heavy chain (IgG HC) in the immunoprecipitates is the loading control. Immunoprecipitation using rabbit IgG (IP:IgG) was the control immunoprecipitation. (B) & (D) Expressions of MEK5 (IB:MEK5) in SW620/SW480 (B) and WM115/WM266 cells (D) were analyzed by immunoblot of the cell lysate (WCL) of the cells in which stat3 was knocked down by RNAi (RNAi-Stat3, IB:Stat3). HA-PKM2 (HAPK) was expressed in the stat3 knockdown cells (IB:HA). (C) & (E) RT-PCR analyses of cellular MEK5 mRNA levels in SW620/SW480 (C) and WM266/WM115 cells (E) in which stat3 was knocked down (RNAi-Stat3, IB:Stat3). HA-PKM2 (HAPK) was expressed in the stat3 knockdown cells (IB:HA). The results are presented as fold changes in PCR products before and 48 hours after stat3 knockdown and HA-PKM2 expression in the same cells. The level of PCR products from each cell line that was treated with non-target RNAi was defined as 1. In (B), (C), (D), and (E), NT represents the cells treated with non-target RNAi. Vec were the cells infected with virus that carry the empty vector. Error bars in (C) and (E) are standard deviations of three measurements. In (B) and (D), Immunoblots of β-actin (IB:β-actin) are loading controls.
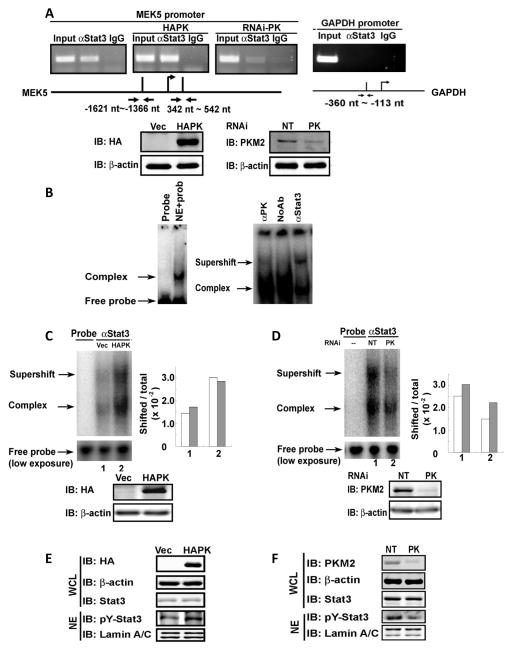
We analyzed whether expression of PKM2 affected the stat3 binding to its target DNA at the MEK5 promoter. To probe whether knockdown or expression of PKM2 affected the interaction of stat3 with the MEK5 promoter, we performed ChIP in SW620 cells in which the endogenous PKM2 was knocked down or HA-PKM2 was expressed. Clearly, the stat3 and MEK5 promoter interaction was strengthened by HA-PKM2 expression and was weakened by PKM2 knockdown (Fig. 3A). We then analyzed the effects of PKM2 on the interaction between stat3 and its target DNA by gel mobility shift assays using a 32P-labled oligonucleotide duplex containing a stat3 binding sequence (Xie et al., 2006). The experiments were carried out with nuclear extracts of SW620 cells with/without PKM2 knockdown or with/without HA-PKM2 expression. It was clear that a slow migration complex was assembled with the labeled probe and addition of the antibody against stat3 resulted in a supershift complex (Fig. 3B). Interestingly, knockdown of PKM2 substantially decreased the assembly of the oligo-protein complex and assembly of stat3 in the complex (the weak supershift) (Fig. 3D), while expression of HA-PKM2 increased assembled complex and assembly of stat3 in the complex (Fig. 3C).
Figure 3. PKM2 upregulates MEK5 transcription by promoting stat3 DNA interaction and phosphorylation of stat3.
(A) (Upper panels) ChIP of the MEK5 promoter (MEK5 promoter) using antibody against stat3 in SW620 cells (αStat3). The cells were treated non-target RNAi (left, NT) or with RNAi target PKM2 (right, RNAi-PK), or HA-PKM2 was expressed in the cells (middle, HAPK). Inputs were PCR products from DNA extracts without ChIP. The primer pair positions are indicated. ChIP using rabbit IgG (IgG) was a negative control. ChIP targeting GAPDH promoter (GAPDH promoter) using antibody against stat3 was another negative control. (Lower panels) the cellular PKM2 (right) and HA-PKM2 (left) levels in SW620 cells that were treated with RNAi target PKM2 (PK) or with non-target RNAi (NT), or infected with virus that carry HA-PKM2 expression vector (HAPK) or vector alone (Vec) were analyzed by immunoblots using anti-HA antibody (IB:HA) or anti-PKM2 antibody (IB:PKM2). (B) DNA-protein complex (Complex) assembled on a 32P-labeled oligo containing the stat3 targeting sequence in nuclear extracts of SW620 cells was detected by gel-shift. Free probe indicates the 32P-labeled oligo probe without addition of nuclear extracts. The antibodies against PKM2 (αPK), stat3 (αStat3), or no antibody (NoAb) was added to the complex to create supershift (Supershift). (C) & (D) Supershift complex assembled with the 32P-labeled oligo and anti-stat3 antibody (αStat3) in the nuclear extracts of SW620 cells in which (C) HA-PKM2 was expressed (HAPK) or (D) PKM2 was knocked down (PK) was detected by gel-shift. Probe only (Probe) is the free probe without addition of nuclear extracts. The free probe (low exposure) is the loading control with 1/10 of exposure time in autoradiography. The quantification of the assembled complex and super shift complex were presented as percentage (Shifted/total ×102) of probe in the shift complex calculated by intensities of complex [complex (grey bars) or supershift (open bars)] divided by Intensities of total [free probe + complex + supershift). Immunoblots at bottom of each panel indicate levels of HA-PKM2 (IB:HA) and endogenous PKM2 (IB:PKM2) in the cells from which the extracts were prepared for the above gel shift experiments. (E) & (F) The levels of Y705 phosphorylated stat3 (IB:pY-Stat3) in the cell nucleus were analyzed by immunoblot of nuclear extracts (NE) of SW620 cells in which HA-PKM2 was expressed (E. HAPK) or the PKM2 was knocked down (F, PK). The total cellular stat3 levels were analyzed by immunoblot analyses of stat3 (IB:Stat3) in whole cell lysate (WCL). In (E), immunoblot of HA-tag (IB:HA) indicates the HA-PKM2 expression levels in the cells. In (F), immunoblot of PKM2 (IB:PKM2) represents cellular PKM2 levels in the cells. Immunoblot of lamin A/C in (E) and (F), and β-actin in (A), (C), (D), (E), and (F) are the loading controls. NTs in (A), (C), (D), (E), and (F) mean the cells were treated with non-target RNAi. Vec in (A), (C), (D), (E), and (F) means the cells were infected with virus that carry the empty vector.
Stat3 is activated by phosphorylation at Y705. The phosphorylation increased its DNA binding affinity (Sehgal, 2008). We observed that PKM2 increased the stat3 binding to its target sequence both in vitro and in vivo. We suspected that PKM2 may play a role in stat3 phosphorylation at Y705. To test this conjecture, we analyzed the stat3 phosphorylation in the nuclear extracts prepared from SW620 cells in which the HA-PKM2 was expressed using an antibody against the Y705 phosphorylated stat3 (P-y705/stat3). Clearly, a significant increase in Y705 phosphorylation of stat3 was evident. Examination of the cellular levels of stat3 indicated that the expression levels of stat3 were not affected (Fig. 3E). We also observed a significant reduction in the stat3 phosphorylation in SW620 cells upon PKM2 knockdown (Fig. 3F). JAK2 and c-Src are the most common protein tyrosine kinases that phosphorylate stat3. We therefore probed whether JAK2 and c-Src became more activated by the HA-PKM2 expression. Using antibodies against JAK2, the phosphorylated JAK2, c-Src, and the phosphorylated c-Src, our experiments showed that there was no increase in JAK2 and c-Src activation (Fig. S3A). Furthermore, treatment of cells with JAK2 and c-Src inhibitors did not lead to any significant changes of the PKM2-depedent stat3 phosphorylations (Fig. S3 B&C&D), suggesting that stat3 phosphorylation was not due to activation of JAK2 and Src.
Since PKM2 acts in the glycolysis pathway, we analyzed cellular glucose, lactate, and pyruvate in two pairs of cells, SW620/SW480 and WM266/WM115, in which the HA-PKM2 was expressed. Upon expression of HA-PKM2, there were no significant changes in cellular glucose and production of lactate and pyruvate for either WM266 or SW620 cells (the more proliferated cell lines). Cellular pyruvate and lactate increased slightly in SW480 and WM115 cells (the less proliferated cell lines), indicating a slight increase in glycolytic pyruvate kinase activity in the less aggressive cancer cells, while expression of PKM2 did not lead to any changes in glycolytic pyruvate kinase activity in the more proliferated cancer cells. Overall, no substantial changes in cellular pyruvate and lactate were observed (Fig. S4 A-F), suggesting that the differences in stimulating cell proliferation by expression of HA-PKM2 were not simply due to the effects on the changes in carbohydrate metabolism.
Dimeric PKM2 is the active protein kinase
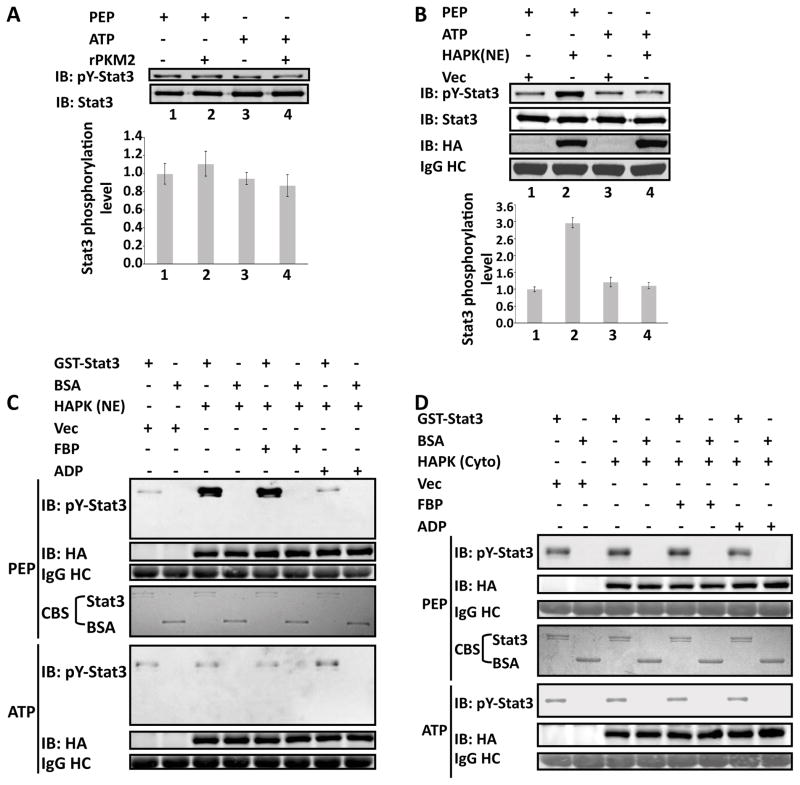
We next sought to test whether PKM2 could directly phosphorylates stat3. An in vitro phosphorylation assay using both the E. coli expressed recombinant PKM2 (rPKM2) and the HA-PKM2 immunopurified from nuclear extracts of SW620 cells in the presence of ATP did not yield phosphorylation of a commercially available GST-stat3. Since PKM2 uses PEP as phosphate donor to phosphorylate ADP in the glycolysis, we reasoned that the protein may use the same phosphate donor to phosphorylate a protein substrate. Thus, we replaced ATP by PEP in our in vitro reaction. Immunoblot using the antibody P-y705/stat3 demonstrated that the GST-stat3 was phosphorylated by the HA-PKM2 in the presence of PEP. Consistently, stat3 was not phosphorylated in the presence of ATP (Fig. 4A&B). These results indicated that PKM2 is a protein kinase using PEP as the phosphate donor.
Figure 4. Phosphorylation of GST-stat3 by the rPKM2.
Phosphorylation of GST-stat3 by the rPKM2 (A) and HA-PKM2 (HAPK(NE)) immunopurified from nuclear extracts of SW620 (B) in the presence of 5 mM ATP (ATP) or 5 mM PEP (PEP) was revealed by immunoblot assays using antibody against Y705 phosphorylated stat3 (IB:pY-stat3). Immunoblot analyses using antibody against stat3 (IB:Stat3) indicates the amounts of GST-stat3 used in each reaction. The bottom panels in (A) & (B) are the quantitative analyses of immunoblot signals. The error bars represent the standard deviations of four measurements. Phosphorylation of GST-stat3 by the HA-PKM2 immunopurified from the nuclear (HAPK (NE) in C) and the cytoplasmic (HAPK(Cyto) in D) extracts of SW620 cells in the presence of 5 mM ATP (ATP) or 5 mM PEP (PEP) was revealed by immunoblots using antibody against Y705 phosphorylated stat3 (IB:pY-stat3). The reactions were also carried out in the presence/absence of 5 mM FBP (FBP), or 5 mM ADP (ADP). The immunoblot of HA (IB:HA) indicates the amounts of HA-PKM2 used in each reaction. IgG HC is the ponceau S stain of antibody heavy chain, representing the amounts of antibody used in immunopurification of HA-PKM2. Coomassie blue staining (CBS) indicates the amounts of GST-stat3 and BSA used in each phosphorylation reaction. Vec were the cells infected with virus that carry an empty vector.
The kinase activity of the nuclear HA-PKM2 in phosphorylation of stat3 was substantially higher than that of the rPKM2 expressed in E. coli. The results led us to compare the protein kinase activity of the nuclear PKM2 and the cytoplasmic PKM2. The same in vitro phosphorylation reactions were performed with the HA-PKM2 immunopurified from nuclear or cytoplasmic extracts of SW620 cells in the presence of PEP or ATP. The HA-PKM2 from the nuclear extracts had much higher activity than that of protein from the cytoplasmic extracts (Fig. 4C&D). To test whether the Y705 of stat3 is the only phosphorylation site by PKM2 in cells, we expressed a stat3 mutant (Y705A) and GFP-PKM2 in SW620 cells. Phosphorylations of endogenous and exogenously expressed stat3 were examined by immunoprecipitation of HA-tagged stat3 mutant or endogenous stat3 followed by immunoblot using an antibody against phorpho-tyrosine. It was clear that the endogenous stat3 was phosphorylated, while the exogenously expressed mutant was not phosphorylated (Fig. S4G), indicating that Y705 is the only site.
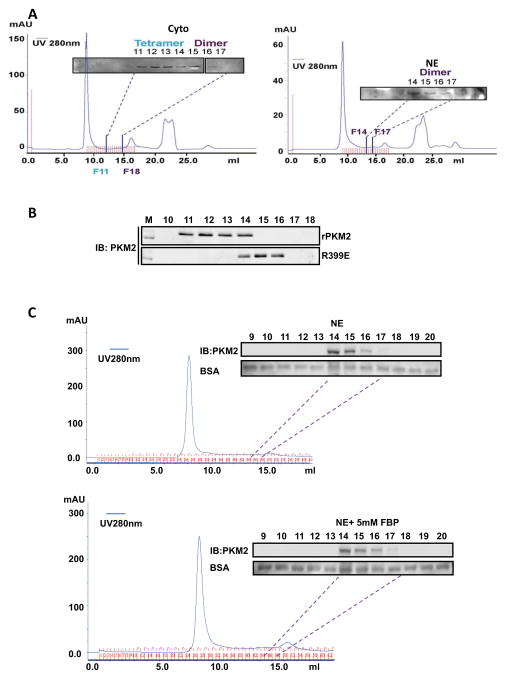
It was reported that the tetramer and dimer of PKM2 co-exist in proliferation cells (Mazurek et al., 2005). We therefore questioned whether the differences in the protein kinase activity of nuclear/cytoplasmic HA-PKM2 and the rPKM2 were due to dimer or tetramer of the protein. To investigate whether PKM2 is a dimer or a tetramer in the nucleus and in the cytoplasm, we first fractioned the nuclear and cytoplasmic extracts of SW620 cells by size exclusion chromatography. The levels of PKM2 in each fraction were examined by immunoblot using the antibody PabPKM2. Nuclear PKM2 was only detected in fractions 14 – 16, while cytoplasmic PKM2 was mainly detected in fractions 11 – 16 with the highest concentrations in fractions 11 – 13. According to the MW calibration standard (Fig. S5 A&B), fraction 11 co-elutes with a MW near 240 kDa, while fraction 14 co-elutes with a MW near 120 kDa (Fig. 5A). The gel-filtration chromatography suggested that nuclear PKM2 was completely dimer, while the cytoplasmic PKM2 existed in both dimer and tetramer. The same procedure was also employed to analyze whether the rPKM2 is a dimer or a tetramer. It was evident that the rPKM2 was mostly tetramer with very small amount of dimer (Fig. 5B). It is well documented that FBP functions as an allosteric regulatory factor that stabilizes the tetramer PKM2. We therefore asked whether FBP could convert the dimer nuclear PKM2 to a tetramer form. To this end, nuclear extracts of SW620 cells were incubated with 5 mM FBP at room temperature for 2 hours. The dimeric/tetrameric status of PKM2 in the nuclear extracts was analyzed by the same procedure. It was evident that FBP did not convert PKM2 from the dimeric to the tetrameric form (Fig. 5C).
Figure 5. Dimer and tetramer PKM2.
(A) Chromatography fractionation of nuclear (NE, right) and cytoplasmic (Cyto, left) extracts of SW620 cells. The fractions were collected at 300 μl per fraction. The fractions 6 to 17 from cytoplasmic extracts and 9 to 18 from nuclear extracts were immunoblotted using the antibody PabPKM2. Fraction 11 (F11) co-eluted with 240 kDa and fraction 14 (F14) co-eluted with 120 kDa on the same column under identical conditions determined by chromatography molecular weight calibration standard (see Fig. S3 C&D). (B) Chromatography fractionation of the wild-type rPKM2 and R399E mutant. The fractions F10 – F18 were collected. The fractions were analyzed by immunoblot using the antibody PabPKM2 (IB:PKM2). (C) Effects of FBP on the dimer/tetramer status of PKM2 in nuclear extracts. Chromatography fractionation of nuclear (NE) extracts of SW620 cells treated (bottom panel) or untreated (upper panel) with FBP (5mM). The fractions were collected at 300 μl per fraction. The fractions 6 to 20 from nuclear extracts were immunoblotted using the antibody PabPKM2 (IB:PKM2). Immunoblots of BSA are the loading controls. 2 μg of BSA was added to each fraction. After SDS-PAGE separation of 20 μl of each fraction, the BSA was analyzed by immunoblot (BSA).
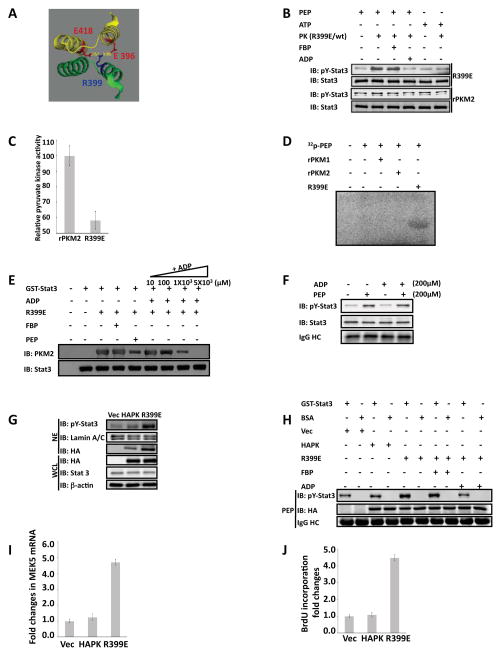
Close examination of the crystal structure of the tetramer human PKM2 (Dombrauckas et al., 2005) reveals that a positive charged residue R399 may plays a critical role in forming the tetramer of PKM2. It is notable that the R399 forms stable charge-charge interactions with residues E418 and E396 of PKM2 located on the other dimer of the tetramer PKM2 (Fig. 6A). We therefore created a mutant R399E to disrupt the interactions. Size exclusion chromatography analyses demonstrated that the R399E mutant was mostly dimer (Fig. 5B). We reasoned that the dimeric R399E would be more active in phosphorylating stat3. Thus, the in vitro phosphorylation reactions were carried out with the rPKM2 and the R399E. It was evident that the protein kinase activity of the R399E was substantially higher than that of the wild-type rPKM2 (Fig. 6B), while the pyruvate kinase activity of the mutant was dramatically lower than that of the rPKM2 (Fig. 6C). The results supported our speculation that dimeric PKM2 is an active protein kinase.
Figure 6. Dimeric PKM2 is active protein kinase and expression of the R399E mutant promotes cell proliferation.
(A) Part of x-ray crystal structure of human PKM2. The structure was obtained from PDB bank DOI: 10.1021/bi0474923. The residue R399 and its interactive residues E418, D357, and E396 are highlighted in color. (B) Phosphorylation of GST-stat3 by 10 μg/ml of rPKM2 (rPKM2) and R399E mutant (R399E) in the presence of 5 mM ATP (ATP), 5 mM PEP (PEP), 5 mM FBP (FBP), and/or 5 mM ADP (ADP) was revealed by immunoblot assays using antibody against Y705 phosphorylated stat3 (IB:pY-stat3). Immunoblot analyses using antibody against stat3 (IB:Stat3) indicates the amounts of GST-stat3 used in each reaction. (C) Pyruvate kinase activity of the rPKM2 or R399E (5 μg/ml) was analyzed by the method described by Christofk and coworkers. The pyruvate kinase activity was expressed as relative pyruvate kinase activity by define the activity in the rPKM2 as 100. (D) Phosphorylation of GST-stat3 by 10 μg/ml of rPKM2, the rPKM1, or the R399E in the presence of 32P-PEP (~0.002 μCi) and unlabeled PEP (5 mM). The reaction mixture were separated by SDS-PAGE and subjected to autoradiograph. (E) Interaction of GST-stat3 and R399E in the presence of FBP (5 mM), PEP (5 mM), or various concentrations of ADP (inducated) was analyzed by GST-pull-down. The co-precipitation of R399E with GST-stat3 was detected by immunoblot using the antibody against PKM2 (IB:PKM2). Immunoblots of precipitates using antibody against stat3 (IB:Stat3) indicate the amounts of GST-stat3 that was pulled-down by the glutathione beads. (F) Phosphorylation of GST-stat3 by the PKM2 (10 μg/ml) purified from nuclear extracts of SW620 cells in the presence of 200 μM PEP (PEP) and 200 μM ADP (ADP) was revealed by immunoblot assays using antibody against Y705 phosphorylated stat3 (IB:pY-stat3). Immunoblot analyses using antibody against stat3 (IB:Stat3) indicates the amounts of GST-stat3 used in each reaction. (G) Phosphorylation of stat3 in SW480 cells was examined by immunoblot analyses of the nuclear extracts (NE) using antibody against the Y705 phosphorylated stat3 (IB:pY-Stat3). PKM2 (HAPK) or the R399E (R399E) was expressed in the cells. Immunoblots using anti-HA antibody (IB:HA) and anti-stat3 antibody (IB:Stat3) in the whole cell lysate (WCL) indicate the levels of stat3 and HA-PKM2/HA-R399E in the cells. Immunoblot of lamin A/C (IB:Lamin A/C) and β-actin (IB:β-actin) are loading controls. (H) Phosphorylation of GST-stat3 by the HA-p68 and the HA-R399E that were immunopurified from cell lysate of SW480 cells in the presence of 5 mM PEP was revealed by immunoblot analyses using antibody against the Y705 phosphorylated stat3 (IB:pY-Stat3). The reactions were also carried out in the presence/absence of 5 mM FBP (FBP), or mM of ADP (ADP). The immunoblot of HA (IB:HA) indicates the amounts of HA-PKM2 or HA-R399E used in each reaction. (I) Expression of MEK5 mRNA in SW480 cells was analyzed by RT-PCR. HA-PKM2 (HAPK) or HA-R399E (R399E) was expressed in the cells. The results are presented as fold changes in PCR products before and 48 hours after HA-PKM2 or HA-R399E expression. (J) Cell proliferations of SW480 cells were measured using a proliferation kit. Proliferations were presented as fold changes in BrdU incorporation before and 48 hours after HA-PKM2 (HAPK) or HA-R399E (R399E) expression. The BrdU incorporation of the cells that were transfected with the empty vector (Vec) was defined as 1. In (F) and (H), IgG HC is the ponceau S stain of antibody heavy chain, representing the amounts of antibody used in immunopurification of HA-PKM2 from the extracts. Error bars in (C), (I), and (J) are standard deviations of three independent measurements. Vec in (G), (H), (I), and (J) are the cells transfected with the empty vector.
To further verify the phosphorylation of stat3 by PKM2, we prepared [32P]-labeled PEP (Roossien et al., 1983). The same in vitro phosphorylation reaction was carried out with rPKM2, rPKM1, and R399E using the [32P]-PEP. Autoradiography indicated that the GST-stat3 was phosphorylated by the rR399E. The phosphorylation of the GST-stat3 by the rPKM2 was very weak (was only visualized by a substantial overexposure). The GST-stat3 was not phosphorylated by the rPKM1 in the presence of the [32P]-PEP (Fig. 6D). No phosphorylation can be detected even under very high overexposure. To ensure that the in vitro phosphorylation of stat3 by PKM2 and the R399E was at comparable physiological conditions, we compared the phosphorylation of the GST-stat3 by JAK2 and R399E. Clearly, very similar levels of stat3 phosphorylations were observed by both kinases (Fig. S6A). The R399E could not phosphorylate other proteins, such as stat5 and BSA under the same conditions (Fig. S6 B&C), indicating substrate specificity. Extensive analyses of enzyme kinetic of pyruvate kinase in various tissues (Km = 0.07 – 1.2 mM range) indicate that the physiological concentration of PEP is likely to be at μM - mM (Mazurek et al., 2007; van Veelen et al., 1978). To test whether PKM2 and the R399E would phosphorylate stat3 at the physiological PEP concentrations, we carried out the in vitro phosphorylation at various PEP concentrations. The levels of phosphorylated stat3 remained almost constant down to 100 μM of PEP. However, there was a clear decrease in stat3 phosphorylation when PEP concentration fell below 10 μM (Fig. S6D). These experiments indicated that phosphorylation of stat3 by PKM2 were at physiologically comparable conditions.
PKM2 protein kinase substrates may bind to the ADP binding site
One interesting question is how the protein substrate is bound to PKM2. In the catalysis of ADP phosphorylation in the glycolysis, PKM2 binds phosphate donor PEP at one site and substrate ADP at the other. Thus, a very logical assumption is that the stat3 may bind to the ADP site. To test this conjecture, we carried out competition binding analyses. The R399E and stat3 interaction was analyzed in the presence and absence of ADP, PEP, and FBP. The GST-stat3 was immobilized on the glutathione-agarose beads. Recombinant R399E in mixing with buffer, ADP, PEP, or FBP were incubated with the beads. Binding of the R399E to the immobilized GST-stat3 was then analyzed by immunoblot of R399E and stat3. It was clear that the interaction of R399E with GST-stat3 was not affected in the presence of 5 mM FBP, while the interaction was almost not detectable in the presence of 5 mM ADP. The R339E and stat3 interaction was also weakened in the presence of 5 mM PEP. This was most likely due to phosphorylation of stat3, as the reduction in stat3 and R399E interaction was not PEP concentration dependent, while the R399E and GST-stat3 interaction showed a clear ADP concentration dependent reduction (Fig. 6E), indicating the competition of ADP with stat3 in binding R399E. The notion that protein substrate is bound at the ADP binding site of PKM2 was also supported by our early observations that addition of ADP decreased the activity of PKM2 in phosphorylation of stat3, while addition of FBP did not (see Fig. 4C/D and 6B/C). Cellular ADP concentration is around 100 μM – 1 mM (Bradbury et al., 2000; Brosnan et al., 1990; Ronner et al., 2001). Thus, we asked whether PKM2 could phosphorylate stat3 under physiological PEP and ADP concentration. To this end, the same in vitro phosphorylation reaction was carried out with PKM2 purified from nuclear extracts of SW620 cells in the presence of 200 μM PEP and 200 μM ADP. Clearly, stat3 was phosphorylated by the PKM2 under the conditions (Fig. 6F).
Expression of R399E increased stat3 phosphorylation in cells and promoted cell proliferation
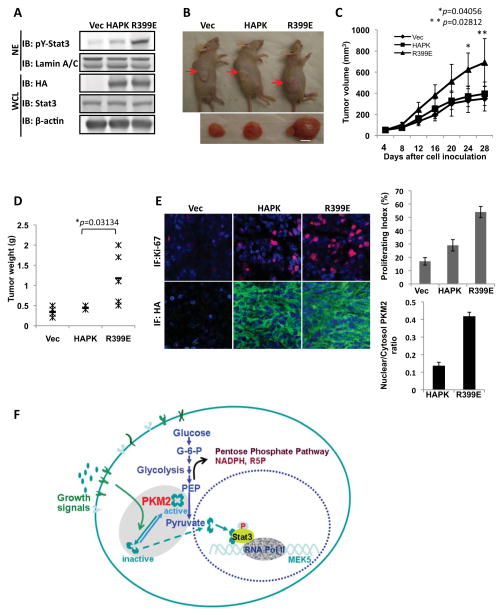
The recombinant R399E existed mostly as a dimer and exhibited strong activity in phosphorylation of GST-stat3. We therefore questioned whether the R399E would be active in phosphorylation of stat3 in cells. To this end, the HA-R399E was expressed in SW480 cells. The expressed R399E had substantially higher nuclear localization (Fig. 6G). Immunoblot of the nuclear extracts indicated a significant increase in the levels of phosphorylated stat3. This increase was not due to increase in cellular levels of stat3 (Fig. 6G). Size exclusion chromatography of cell lysate showed that, unlike the wild-type HA-PKM2, the HA-R399E was almost a complete dimer (Fig. S6E) In vitro phosphorylation assays indicated that the purified HA-R399E phosphorylated GST-stat3 (Fig. 6H). We also tested if expression of R399E in SW480 cells would lead to MEK5 up-regulation and cell proliferation. RT-PCR analyses clearly demonstrated that the mRNA of MEK5 increased significantly (Fig. 6I). Examination of cell proliferation demonstrated that expression of R399E led to a strong increase in cell proliferation (Fig. 6J). To test whether the stat3 phosphorylation contributes to cell proliferation promoted by expression of R399E, we measured cell proliferation rate in R399E expressing SW480 cells in which stat3 was knocked down or the stat3-DN was co-expressed. Clearly, expression of R399E did not lead to proliferation under condition of stat3 knockdown or expression of stat3-DN (Fig. S7 A&B), suggesting that stat3 phosphorylation, at least partially, mediated the effects of PKM2 in promoting cell proliferation. To further verify the role of PKM2 protein kinase activity in promoting cell proliferation, we created a cell line by stable expression of R399E in SW480 cells (Fig. 7A). The derived cells were implanted to nude mice by s.c. Tumors were grown for four weeks. Clearly, the R399E expressing tumors grew substantially bigger than the wild type PKM2 expression tumors did (Fig. 7B&C&D). Histology analyses using anti-Ki-67 antibody indicated that the R399E expressing tumors had much higher proliferation rates than the wild type PKM2 expressing tumors did (Fig. 7E). The results strongly support our hypothesis that protein kinase activity of PKM2 promotes tumor/cell proliferation.
Figure 7. The effects of PKM2 protein kinase activity on tumor growth.
(A) PKM2 (HAPK) or R399E (R399E) was stably expressed in SW480 cells (IB:HA) and stat3 was phosphorylated in R399E expressing cells (IB: pY-stat3). (B) The representative examples of tumor-bearing mice and excised tumors from the derived (indicated) cell lines. (C) Tumor growth was monitored by measuring tumor volumes every two days. (D) At end of four weeks growth, tumors were sliced out and weighted. (E) (Left) Tissue sections were immune fluorescence stained with antibodies against Ki67 (Ab: Ki67, Red) and HA (Ab:HA, Green). The blue is DAPI stain of cell nucleus. (Right) Quantization of Ki-67 staining signals (Upper) and HA-staining signals (Lower) of the tissue sections. The proliferation index was percentage of the positive nucleus staining by counting the nucleus number with ki-67 positive staining in randomly selected three fields in each slide and randomly selected three slides from each tumor. The HA-stains are quantified by image-J software by manually define the areas of interest (e.g. nuclear or cytoplasmic based on the DAPI blue staining) with randomly selected five fields from randomly selected three slides of each tumor. In (A), (B), (C), (D), and (E), the Vec means the vector alone was expressed in the cells. The error bars in (C) and (E) are standard deviations from the measurements of six mice. The p values in (C) and (D) are calculated using pairwise student t-test. (F) A hypothetic model that illustrates the functional role of PKM2 in gene transcription.
Discussion
During tumor progression, growth signals stimulate the conversion of glycolytically active PKM2 to an inactive form, consequently regulating the glycolysis pathway to channel the carbon source from glucose for biosynthesis (Christofk et al., 2008a; Deberardinis et al., 2008; Garber, 2006; Mazurek and Eigenbrodt, 2003). It is conceivable that tumor cells need to coordinate the metabolic alterations with expression of genes that are related to cell proliferation. The functions of PKM2 in regulating expression of genes fulfill the role of feedback signaling from metabolic alterations to gene regulation during tumor malignancy transformation (see model in Fig. 7F).
It is intriguing that a glycolytic enzyme can function as a protein kinase and translocate to the nucleus acting on gene transcription. It was revealed from structural analyses of the tetramer PKM2 that the ADP binding site is formed by a large hydrophobic hole, indicating a great flexibility for nucleotide binding (Dombrauckas et al., 2005; Muirhead et al., 1986). The large hydrophobic hole at the nucleotide binding site is almost completely buried in a tetramer structure, while it would be completely accessible in the dimeric form. Thus, it is conceivable that the nucleotide binding site may be able to accommodate a protein substrate when PKM2 is converted to a dimer. It should be pointed out that our results could not exclude a possibility that ADP and stat3 do not bind at the same site. The competition could be due to allosteric effects of binding of ADP. Molecular mechanisms that regulate the conversion of the tetrameric PKM2 to dimer are interesting. Studies have revealed that multiple mechanisms co-exist to regulate this conversion, including interacting with tyrosine phosphor-peptide (Christofk et al., 2008b), phosphorylation modifications (Hitosugi et al., 2009), and direct interaction with oncogenic proteins (Mazurek et al., 2007). This phenomenon reflects that the conversion of dimer/tetramer PKM2 must be subjected to control of multiple growth signalings.
Stat3 is a transcription activator that is activated in response to inflammatory cytokines, such as IL-6 (Gao et al., 2007; Watson and Neoh, 2008). Activation of stat3 represents probably one of the most important molecular signatures involved in promoting cancer progression. It has been observed that activation of stat3 is detected in almost all cancer types (Frank, 2007; Groner et al., 2008; Huang, 2007; Kim et al., 2007). However, it is generally believed that activations of stat3 in response to growth factor and cytokines are usually transient. A long standing question is “How the malignant cancer cells maintain constitutive activation of stat3”. Currently, mutation(s) that lead to constitutive activation of stat3 have not been identified. Thus, activation of stat3 by PKM2 in malignant cancer cells potentially provides a highly plausible explanation for this long-standing question. Whether stat3 is the only PKM2 substrate for its protein kinase activity is an open question. PKM2 interacts with several other proteins (Garcia-Gonzalo et al., 2003; Lee et al., 2008; Spoden et al., 2009). It was also speculated that PKM2 purified from hepatoma tumor cells may be able to phosphorylate histone H1 (Ignacak and Stachurska, 2003). Thus, it will be interesting to identify other PKM2 protein kinase substrates and uncover the putative cellular function of the corresponding protein phosphorylations.
Materials and methods
In vitro protein kinase and pyruvate kinase assays
The bacterially expressed rPKM2/R399E or immunopurified HA-PKM2/HA-R399E (10 μg/ml) were incubated with GST-stat3 (10 μg/ml) under various conditions (indicated in the figure legends) with kinase buffer (50 mM Tris-HCl pH = 7.5, 100 mM KCl, 50 mM MgCl2, 1 mM Na3VO4, 1 mM PMSF, and 1 mM DTT) in 100 μl at RT for 1 hours. The reactions were terminated by addition of SDS-PAGE loading buffer and heated to 100°C. The reaction mixtures were then subjected to 10% SDS-PAGE analyses.
Pyruvate kinase activity was analyzed by following the experimental procedure similar to that was described by Christofk and co-workers (Christofk et al., 2008b).
Gel-mobility shift and super-shift assays
The mobility shift and supershift assays were carried out with oligonucleotide that harbors a stat3 binding sequence 5′-GATCCTTCTGGGAATTCCTAGATC-3′ (Xie et al., 2006) by standard procedure. The oligo was 5′-end 32P-labeled. For supershift analyses, a monoclonal antibody against stat3 or the antibody PabPKM2 was added to the oligo-nuclear extracts mixture and incubated for additional 45 minutes. The complexes in gel were transferred to a membrane and subjected to autoradiography.
Size-exclusion chromatography
Size exclusion chromatograph was performed with a Superdex 200 10/300GL column. The samples of cytoplasmic and nuclear extracts (5 – 7 mg/ml of total protein), the rPKM2 (~15 μM), the rR399E, and the immunopurified HA-PKM2/HA-R399E (~15 μM) were prepared in tris-HCl buffer. 6-AcA (600 mM) was added to the samples to disrupt the non-specific protein-protein interactions. 100 μl of the sample was loaded into the column and eluted with elution buffer (50 mM phosphate, 0.15M NaCl pH7.2). The fraction of 300 μl was collected, and 20 μl of each fraction was analyzed by immunoblot. The elution profiles were compared to that of a size exclusion chromatograph calibration kits (GE Healthcare) under identical conditions. The elution profile was plotted against LogMW according to vendor’s instructions.
Nude mice xenograft
Nude mice (nu/nu, Harlan Laboratory) were subcutaneously injected with 5 × 106 sub-lines of SW480 cells. Tumor formation and volumes were assessed every 2 days. Tumor volumes were measured by two perpendicular diameters of the tumors with the formula 4p/3 × (width/2)2 × (length/2). The tumors were collected and weighed at the end of the experiments. Tissue sections were prepared from harvested tumors, and stained using a commercially available antibody for Ki-67. Statistical analyses were done in comparison to the control group with a paired Student’s t test.
Supplementary Material
Highlights.
Pyruvate kinase M2 is also a protein kinase.
The tetramer PKM2 is a pyruvate kinase, while the dimer is a protein kinase
PKM2 regulates gene transcriptions by directly phosphorylation of transcription activators
The protein kinase activity of PKM2 is important for cell proliferation
Acknowledgments
We thank comments from Michael Kirberger, Natalie White, Christie Carter, Phang C. Tai and Yun Huang. This work is supported in part by research grants from NCI (CA118113) and Georgia Cancer Coalition to ZR Liu. X. Gao and H. Wang are supported by a MBD fellowship, GSU.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sorensen FB, Thykjaer T, Sauter G, Jensen KM, Dyrskjot L, Orntoft T. SOX4 expression in bladder carcinoma: clinical aspects and in vitro functional characterization. Cancer Res. 2006;66:3434–3442. doi: 10.1158/0008-5472.CAN-05-3456. [DOI] [PubMed] [Google Scholar]
- Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Ashizawa K, Willingham MC, Liang CM, Cheng SY. In vivo regulation of monomer-tetramer conversion of pyruvate kinase subtype M2 by glucose is mediated via fructose 1,6-bisphosphate. J Biol Chem. 1991;266:16842–16846. [PubMed] [Google Scholar]
- Bradbury DA, Simmons TD, Slater KJ, Crouch SP. Measurement of the ADP:ATP ratio in human leukaemic cell lines can be used as an indicator of cell viability, necrosis and apoptosis. J Immunol Methods. 2000;240:79–92. doi: 10.1016/s0022-1759(00)00178-2. [DOI] [PubMed] [Google Scholar]
- Brosnan MJ, Chen L, Van Dyke TA, Koretsky AP. Free ADP levels in transgenic mouse liver expressing creatine kinase. Effects of enzyme activity, phosphagen type, and substrate concentration. J Biol Chem. 1990;265:20849–20855. [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008a;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. 2008b;452:181–186. doi: 10.1038/nature06667. [DOI] [PubMed] [Google Scholar]
- Deberardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44:9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- Elbers JR, van Unnik JA, Rijksen G, van Oirschot BA, Roholl PJ, Oosting J, Staal GE. Pyruvate kinase activity and isozyme composition in normal fibrous tissue and fibroblastic proliferations. Cancer. 1991;67:2552–2559. doi: 10.1002/1097-0142(19910515)67:10<2552::aid-cncr2820671027>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Frank DA. STAT3 as a central mediator of neoplastic cellular transformation. Cancer Lett. 2007;251:199–210. doi: 10.1016/j.canlet.2006.10.017. [DOI] [PubMed] [Google Scholar]
- Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garber K. Energy deregulation: licensing tumors to grow. Science. 2006;312:1158–1159. doi: 10.1126/science.312.5777.1158. [DOI] [PubMed] [Google Scholar]
- Garcia-Gonzalo FR, Cruz C, Munoz P, Mazurek S, Eigenbrodt E, Ventura F, Bartrons R, Rosa JL. Interaction between HERC1 and M2-type pyruvate kinase. FEBS Lett. 2003;539:78–84. doi: 10.1016/s0014-5793(03)00205-9. [DOI] [PubMed] [Google Scholar]
- Groner B, Lucks P, Borghouts C. The function of Stat3 in tumor cells and their microenvironment. Semin Cell Dev Biol. 2008;19:341–350. doi: 10.1016/j.semcdb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Hacker HJ, Steinberg P, Bannasch P. Pyruvate kinase isoenzyme shift from L-type to M2-type is a late event in hepatocarcinogenesis induced in rats by a choline-deficient/DL-ethionine-supplemented diet. Carcinogenesis. 1998;19:99–107. doi: 10.1093/carcin/19.1.99. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Hirst JA, Fujii H. Regulation of cell proliferation by interleukin-3-induced nuclear translocation of pyruvate kinase. J Biol Chem. 2007;282:17706–17711. doi: 10.1074/jbc.M700094200. [DOI] [PubMed] [Google Scholar]
- Huang S. Regulation of metastases by signal transducer and activator of transcription 3 signaling pathway: clinical implications. Clin Cancer Res. 2007;13:1362–1366. doi: 10.1158/1078-0432.CCR-06-2313. [DOI] [PubMed] [Google Scholar]
- Ignacak J, Stachurska MB. The dual activity of pyruvate kinase type M2 from chromatin extracts of neoplastic cells. Comp Biochem Physiol B Biochem Mol Biol. 2003;134:425–433. doi: 10.1016/s1096-4959(02)00283-x. [DOI] [PubMed] [Google Scholar]
- Jeffery CJ. Moonlighting proteins. Trends Biochem Sci. 1999;24:8–11. doi: 10.1016/s0968-0004(98)01335-8. [DOI] [PubMed] [Google Scholar]
- Kim DJ, Chan KS, Sano S, Digiovanni J. Signal transducer and activator of transcription 3 (Stat3) in epithelial carcinogenesis. Mol Carcinog. 2007;46:725–731. doi: 10.1002/mc.20342. [DOI] [PubMed] [Google Scholar]
- Lee J, Kim HK, Han YM, Kim J. Pyruvate kinase isozyme type M2 (PKM2) interacts and cooperates with Oct-4 in regulating transcription. Int J Biochem Cell Biol. 2008;40:1043–1054. doi: 10.1016/j.biocel.2007.11.009. [DOI] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O’Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Mazurek S, Drexler HC, Troppmair J, Eigenbrodt E, Rapp UR. Regulation of pyruvate kinase type M2 by A-Raf: a possible glycolytic stop or go mechanism. Anticancer Res. 2007;27:3963–3971. [PubMed] [Google Scholar]
- Mazurek S, Eigenbrodt E. The tumor metabolome. Anticancer Res. 2003;23:1149–1154. [PubMed] [Google Scholar]
- Muirhead H, Clayden DA, Barford D, Lorimer CG, Fothergill-Gilmore LA, Schiltz E, Schmitt W. The structure of cat muscle pyruvate kinase. The EMBO journal. 1986;5:475–481. doi: 10.1002/j.1460-2075.1986.tb04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronner P, Naumann CM, Friel E. Effects of glucose and amino acids on free ADP in betaHC9 insulin-secreting cells. Diabetes. 2001;50:291–300. doi: 10.2337/diabetes.50.2.291. [DOI] [PubMed] [Google Scholar]
- Roossien FF, Brink J, Robillard GT. A simple procedure for the synthesis of [32P]phosphoenolpyruvate via the pyruvate kinase exchange reaction at equilibrium. Biochim Biophys Acta. 1983;760:185–187. doi: 10.1016/0304-4165(83)90141-1. [DOI] [PubMed] [Google Scholar]
- Schneider J, Neu K, Grimm H, Velcovsky HG, Weisse G, Eigenbrodt E. Tumor M2-pyruvate kinase in lung cancer patients: immunohistochemical detection and disease monitoring. Anticancer Res. 2002;22:311–318. [PubMed] [Google Scholar]
- Sehgal PB. Paradigm shifts in the cell biology of STAT signaling. Semin Cell Dev Biol. 2008;19:329–340. doi: 10.1016/j.semcdb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Jin X, Lin J. Stat3 upregulates MEK5 expression in human breast cancer cells. Oncogene. 2004;23:8301–8309. doi: 10.1038/sj.onc.1208026. [DOI] [PubMed] [Google Scholar]
- Spoden GA, Morandell D, Ehehalt D, Fiedler M, Jansen-Durr P, Hermann M, Zwerschke W. The SUMO-E3 ligase PIAS3 targets pyruvate kinase M2. Journal of cellular biochemistry. 2009;107:293–302. doi: 10.1002/jcb.22125. [DOI] [PubMed] [Google Scholar]
- Sriram G, Martinez JA, McCabe ER, Liao JC, Dipple KM. Single-gene disorders: what role could moonlighting enzymes play? Am J Hum Genet. 2005;76:911–924. doi: 10.1086/430799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetak A, Veress R, Ovadi J, Csermely P, Keri G, Ullrich A. Nuclear translocation of the tumor marker pyruvate kinase M2 induces programmed cell death. Cancer Res. 2007;67:1602–1608. doi: 10.1158/0008-5472.CAN-06-2870. [DOI] [PubMed] [Google Scholar]
- van Veelen CW, Verbiest H, Vlug AM, Rijksen G, Staal GE. Isozymes of pyruvate kinase from human brain, meningiomas, and malignant gliomas. Cancer Res. 1978;38:4681–4687. [PubMed] [Google Scholar]
- Watson CJ, Neoh K. The Stat family of transcription factors have diverse roles in mammary gland development. Semin Cell Dev Biol. 2008;19:401–406. doi: 10.1016/j.semcdb.2008.07.021. [DOI] [PubMed] [Google Scholar]
- Xie TX, Huang FJ, Aldape KD, Kang SH, Liu M, Gershenwald JE, Xie K, Sawaya R, Huang S. Activation of stat3 in human melanoma promotes brain metastasis. Cancer Res. 2006;66:3188–3196. doi: 10.1158/0008-5472.CAN-05-2674. [DOI] [PubMed] [Google Scholar]
- Xu J, Cole DC, Chang CP, Ayyad R, Asselin M, Hao W, Gibbons J, Jelinsky SA, Saraf KA, Park K. Inhibition of the signal transducer and activator of transcription-3 (STAT3) signaling pathway by 4-oxo-1-phenyl-1,4-dihydroquinoline-3-carboxylic acid esters. Journal of medicinal chemistry. 2008;51:4115–4121. doi: 10.1021/jm701271y. [DOI] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerschke W, Mazurek S, Massimi P, Banks L, Eigenbrodt E, Jansen-Durr P. Modulation of type M2 pyruvate kinase activity by the human papillomavirus type 16 E7 oncoprotein. Proc Natl Acad Sci U S A. 1999;96:1291–1296. doi: 10.1073/pnas.96.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.