Abstract
Activation of innate immune signaling pathways through cytosolic RIG-I like receptors (RLR) is a critical response that is antagonized by many viruses. A variety of RNA related pathogen associated molecular patterns have been identified and their role in RLR activation has been examined. Recent studies suggest that several virally encoded components that antagonize RLR signaling interact with and inhibit the interferon (IFN)-α/β activation pathway using both RNA-dependent and RNA-independent mechanisms. The structural basis for these RLR inhibitory mechanisms, as well as the multifunctional nature of viral RLR antagonists, is reviewed in the context of recent biochemical and structural studies.
Keywords: innate immune inhibition, RIG-I, MDA-5, PRRs, PAMPs, viral inhibitors, IFNs
Recognition of viral ligands by the immune system
The host immune system is comprised of the innate and adaptive branches, with the innate immune response serving as the first line of defense against invading pathogens. Unlike adaptive immune responses, which require antigen specific receptors to be developed, innate immune responses are generated through a small number of germ-line encoded pattern recognition receptors (PRRs) that are constitutively expressed in most cells, including dendritic cells and macrophages. A key feature of PRRs that is distinct from antibody-antigen recognition by the adaptive immune system is that PRRs can recognize certain microbial molecular markers, known as pathogen-associated molecular patterns (PAMPs), without the help of additional cells or molecules. PAMP recognition by PRRs results in the activation of type I interferons (IFNs), which are cytokines that function in an autocrine and paracrine manner to activate the expression of a number of IFN stimulated response elements that combat viral infections. Lipids, nucleic acids, lipopolysaccharides, and CpG DNA are among the basic PAMPs that are recognized by PRRs. The Toll-like receptor (TLR) family of proteins (12 known members altogether; 10 in humans, 12 in mice) and the cytosolic PRRs known as the RIG-I like receptors (RLRs) [1, 2] are two key members of the PRR family.
Several recent reviews have highlighted studies that identify viral ligands recognized by PRRs, the nature of PRR activation, and downstream signaling, including TLR and NOD signaling; therefore, we will not discuss these [3, 4]. Here, we will discuss recently discovered molecular mechanisms of how virally encoded proteins from influenza, vaccinia, and Ebola viruses antagonize RLRs and focus on structural and biochemical mechanisms of RLR antagonism and the role of RNA ligands in this process.
RLRs play important roles in the detection of viral RNAs, similar to TLR3 and TLR7/8 that can detect viral double-stranded RNA (dsRNA) and single-stranded RNA (ssRNA), respectively. There are two main members of the RLR family, retinoic acid inducible gene-I (RIG-I) and melanoma associated differentiation factor-5 (MDA-5) [5]. Recent studies have shown that RLRs are critical for viral detection and for activation of IFN-α/β [1, 5]. Previous work has delineated various aspects of RLR regulation, including the identification of several ligands, such as 5′ppp, short poly I:C, and the panhandle structure formed by the 5′ and 3′ ends of negative-sense RNA genomes, that are capable of activating RLRs [1, 5–8]. Together, they demonstrate that many of the RLR activators are likely either part of viral genomes or products/byproducts of viral replication [9]. MDA-5 can be activated by long dsRNA, whereas much shorter dsRNA and those that contain 5′ppp can activate RIG-I more efficiently [10–13]. Together, these studies suggest that the type of RNA ligand can dictate the outcome in vivo [14, 15]. More recently, a third member of the family called laboratory of genetics and physiology 2 (LGP2) was identified [12, 16]. LGP2, similar to RIG-I and MDA-5, contains a helicase domain and RNA-binding domain (RBD), but lacks the N-terminal caspase activation and recruitment domains (CARDs). Subsequent studies indicate that LGP2 may function to regulate RIG-I and MDA-5 [16, 17].
Both RIG-I and MDA-5 are constitutively expressed, albeit at low levels, and their expression is enhanced by activation of IFN-α/β signaling. In the absence of activators, RIG-I and MDA-5 exist in an inactive conformation, which prevents effector access to the N-terminal CARDs and the helicase domain (Figure 1). Ligand binding to the C-terminal RBD serves to initiate activation, while subsequent RNA binding to the helicase domain is likely involved in RLR activation that result in conformational change(s) as indicated by recent structural studies of RIG-I proteins [18–21] (Figure 2a,b). In addition, RNA-bound RIG-I can also interact with polyubiquitin, a process mediated by tripartite motif-containing protein 25 (TRIM25), an ubiquitin E3 ligase, which promotes the N-terminal CARD interaction with IPS-1 (interferon-β promoter stimulator; also known as MAVS, VISA, and Cardif) [22–24]. This complex set of conformational changes, including RNA binding and ubiquitination, likely results in the formation of higher order RLRs, although the exact nature of these interactions requires additional studies. The transition from the inactive conformation to an active conformation facilitates interactions between the CARDs of RIG-I/MDA-5 and IPS-1 (Figure 3a) [25], which results in signaling to the IFN kinases TBK-1/IKKε, which phosphorylate IFN regulatory factors 3/7 (IRF3/7). IRF3/7 are transcription factors that dimerize and translocate to the nucleus upon phosphorylation in order to stimulate IFN-α/β production. A summary of these interactions are shown schematically in Figure 3. Subsequently, secreted IFN-α/β can activate the JAK/STAT pathway in self and neighboring cells, resulting in the upregulation and production of a large number of antiviral genes, including RIG-I/MDA-5, RNA dependent protein kinase (PKR), 2′,5′-oligoadenylate synthetase (OAS), and major histocompatibility complex (MHC) class I molecules (Figure 3b).
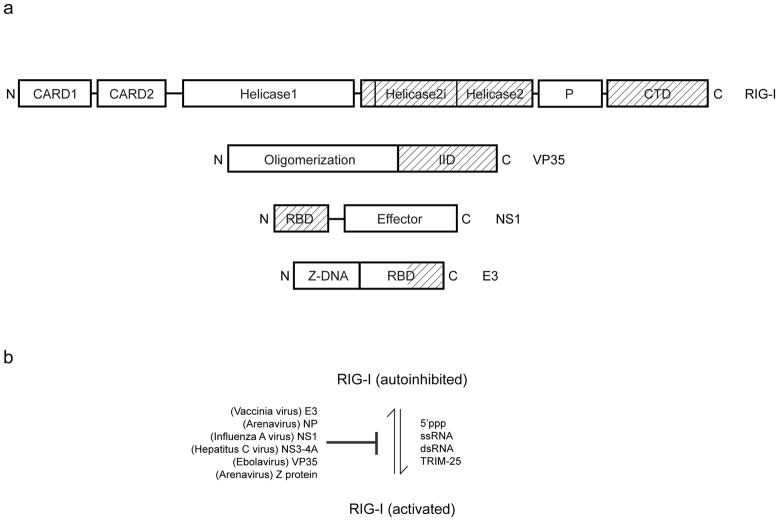
Figure 1.
Model for RLR activation and inhibition. A variety of viral and cellular factors regulate the activity of RLRs. Virally encoded proteins are largely responsible for inhibiting or inactivating RLRs, and viral RNA as well as host proteins such as TRIM25 are responsible for activating RLRs and downstream signaling events leading to IFN production. (a) Domain organization for RIG-I, Ebola virus VP35, influenza NS1 and vaccinia E3 proteins are shown. Regions important for dsRNA binding are highlighted (shaded). (b) Regulators of RIG-I activity.
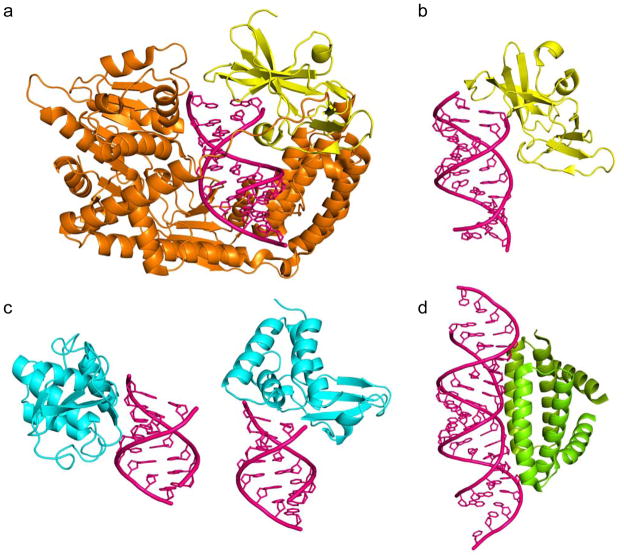
Figure 2.
RNA binding domains play an important role in IFN regulation. RNA binding regions are highlighted in the domain organization for RIG-I, VP35, NS1 and E3 proteins (see Figure 1). RNA binding by cellular and viral protein reveals similar recognition modes and reveal how structurally distinct proteins use similar RNA recognition modes. RNA is shown in magenta. (a) RIG-I protein (minus CARD domains) binding dsRNA (PDB: 2YKG). (b) RIG-I C-terminal domain bound to dsRNA (PDB: 3LRR). (c) Zaire Ebola virus VP35 interferon inhibitory domain (PDB: 3L25). (d) Influenza virus A NS1 RNA binding domain (PDB: 2ZKO).
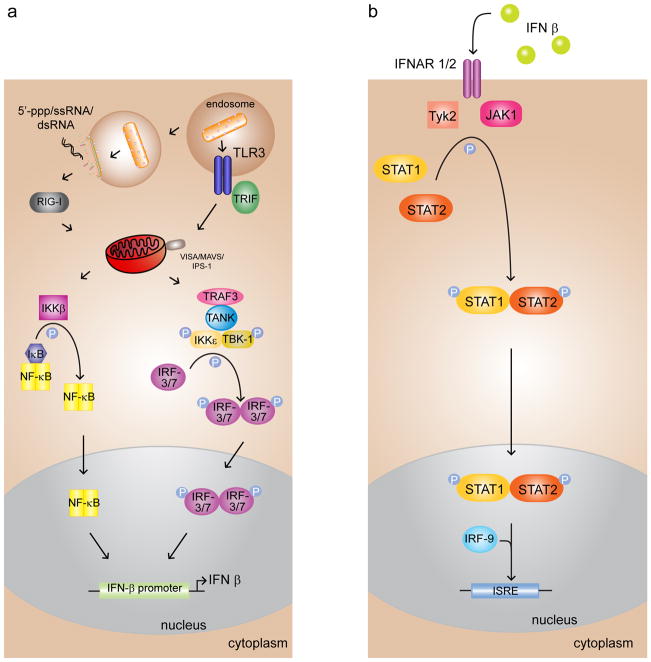
Figure 3.
Viral infection triggers the IFN-β signal transduction pathway of the host innate immune system, activating the antiviral state. (a) Viral RNAs are detected by cytosolic helicases RIG-I and MDA-5, leading to the phosphorylation and nuclear translocation of transcription factor IRF-3/7, which stimulates the production of the IFN-β cytokine. Activation of NF-κB, also resulting from PAMP recognition, can further enhance IFN-β production. (b) IFN-β activates the JAK/STAT pathway and IFN stimulated response elements (ISREs) or antiviral genes, such as PKR, MHC class I, and 2′5′ OAS.
Given the ability of RLRs to sense viral RNAs and activate IFN signaling cascades that eliminate viral infections, many viruses have developed various strategies to overcome detection by RLRs. A majority of these strategies can be considered as either immune evasion or immune inhibition mechanisms. The first category prevents host detection through modification of viral RNA genomes. This is carried out through modification of RNA. For example, some viruses engage in cap snatching (e.g. influenza virus), modification of 5′ppp to monophosphate through virally encoded phosphatases and nucleases (e.g. Borna disease virus, Lassa virus), 2′ O methylation, and use proteins to protect the 5′ ends (e.g. VPg protein from picornaviruses) or overhangs (e.g. arenavirus). The second category involves activity by virally encoded proteins that results in inhibition of RLR signaling. Some viruses encode dsRNA binding proteins that are likely to sequester dsRNA generated by viruses during transcription and replication. Generation of dsRNA likely occurs for many RNA viruses and often occurs in DNA viruses due to overlapping transcription of two open reading frames (ORFs) in the opposite direction. Below we summarize recent structural and functional characterizations of viral proteins from influenza virus, vaccinia virus, and Ebola viruses and their potential roles in mediating RLR antagonism. These examples were selected as recent structural studies highlight the role of RNA-dependent mechanisms, including a potential role for RNA sequestration, used by viral antagonists during RLR inhibition. However, it is important to note that many viral proteins also function via RNA-independent mechanisms. At present it is unclear what are the relative contributions of RNA-dependent and independent mechanisms that lead to RLR inhibition by these viral proteins.
Influenza A virus NS1
Influenza A viruses are enveloped viruses with genomes consisting of 8 negative-sense RNA molecules encoding at least 11 proteins [26]. Non-structural protein 1 (NS1) is a multifunctional dsRNA binding protein critical for evasion of innate immune responses and for virulence [27]. In addition to antiviral signaling, NS1 is also important for cellular gene expression and translation. Because NS1 can alter the responses of antigen presenting cells to infection, NS1 also influences adaptive immune responses [28]. Among the most critical mechanisms employed by NS1 proteins to counter host antiviral responses are the inhibition of host cell mRNA processing and transport, the binding of dsRNA molecules that may activate antiviral signaling, and the inhibition of RLR activation. These functions suppress IFN-α/β gene expression and induction of an IFN-induced antiviral state [29]. Lack of the NS1 protein IFN-antagonist function attenuates virus replication in systems where IFN is produced and diminishes virulence in vivo [30]
The NS1 effector domain and suppression of host cell gene expression
The NS1 protein possesses an N-terminal dsRNA binding domain (dsRBD) and a C-terminal effector domain (ED) (Figure 1a). NS1 interacts via the ED with host factors required for the processing, transport, and translation of cellular gene expression. NS1 can also inhibit 3′ end processing (resulting in a poly A tail) of cellular mRNAs, including IFN-α/β [31, 32].
NS1 inhibition of the RIG-I pathway
Some influenza A viruses, such as the 2009 pandemic H1N1 influenza virus, lack detectable cleavage and polyadenylation specificity factor 30 (CPSF30) binding activity, but NS1 can inhibit IFN-β production activated through RIG-I signaling [6, 33]. For example, the NS1 protein of influenza A/PR/8/34 (H1N1) virus directly interacts with RIG-I and prevent its activation [6, 34, 35]. These observations reveal the ability of NS1 to prevent IFN-α/β production induced by a constitutively active RIG-I mutant [34], and the ability of NS1 to inhibit virus-induced activation of the downstream targets of IRF-3, nuclear factor-kappa B (NF-κB) and activating transcription factor 2 (ATF-2)/c-Jun [36, 37].
NS1 dsRNA binding activity and inhibition of RIG-I
The capacity of NS1 to bind dsRNA may contribute to its inhibition of RIG-I signaling and IFN-β promoter activation. Studies of influenza A virus expressing a NS1 R38A/K41A mutant, which abrogates dsRNA binding, induced more IFN-α/β than wild-type virus and was attenuated in cell culture and mice [29, 38]. A phenotypic-revertant containing the NS1 R38A/K41A mutation acquired a S42G change, which lacked detectable dsRNA binding activity but restored virus replication and virulence [38]. Interestingly, the identity of residue 42 in H5N1 avian influenza viruses also influences pathogenesis in mice and IFN-α/β production in cell culture [39]. These data argue that NS1 dsRNA binding activity influences cellular IFN-α/β production and that residues 38 and 41 influence NS1 function by dsRNA-dependent and -independent mechanisms. One model to explain the dsRNA-dependent effects on IFN-β promoter activation is that NS1 sequesters dsRNAs from recognition by RIG-I. A recent study identified virus-derived RNAs, which potentially form panhandles, associated with RIG-I following infection with an influenza A virus lacking NS1 [40], but the functional relevance in virus infected cells remains to be determined.
The dsRNA binding activity of NS1 likely serves to counter additional antiviral mechanisms beyond inhibition of RIG-I activation. PKR and OAS are also antagonized by NS1 [41–46]. It has been argued that inhibition of OAS is the most critical role for NS1 dsRNA binding activity during virus replication [45], whereas sequestration of dsRNA likely contributes to inhibition of PKR [44]. However, NS1 also interacts with and inhibits PKR activation independently of dsRNA-binding [43].
NS1 interaction with TRIM25
NS1 interacts with the TRIM25 coiled-coil domain [47], disrupting the TRIM25 oligomerization required for RIG-I K63 ubiquitination. Mutations in NS1 that abrogate NS1–TRIM25 interaction lead to increased influenza A virus type I IFN production and attenuation in mice, demonstrating a role for TRIM25 inhibition in virus infection [47]. Interestingly, the use of TRIM25 knockout mouse embryo fibroblasts (MEFs) demonstrated that type I IFN production in MEFs is greatly reduced but not completely absent when TRIM25 is absent.
Structural basis for RIG-I inhibition and TRIM25 interaction
X-ray crystal structures for NS1 RBD, alone and in complex with dsRNA have been solved (Figure 2d); for the EDs of NS1s from multiple viruses, alone and in complex with portions of CPSF30 [48]. The RBD consists of two helices and forms a helical bundle in the dimer, where the top two helices interact with dsRNA in a sequence independent manner [49]. The RBD dimer in the absence of dsRNA is similar to the complex, although residue R38, which is required for dsRNA binding, reorients upon dsRNA binding. The ED also forms a dimer, but the structural elements that comprise the dimer interface vary among different NS1 proteins. For example, the dimer interface of the ED structure from a H5N1 influenza A virus is located between β-strands [48] whereas the dimer interface in the ED structure from the NS1 from influenza A/duck/Albany/76 (H12N5) occurs via helices [50]. Variations in the dimer interface likely reflect sequence differences in NS1, but the functional relevance is not known.
The X-ray crystal structure of a full-length NS1 [48] reveals that the RBD of one NS1 dimerizes with the RBD of an adjacent NS1 molecule, and the ED of one NS1 dimerizes with the ED of another adjacent NS1 molecule. The result is a chain of polymer-like structures that form a tubular structure with a ‘tunnel’ that could accommodate dsRNA [48]. Residues R38 and K41 are positioned within the tunnel to interact with dsRNA [48]. These data therefore suggest possible scenarios whereby NS1 might ‘hide’ dsRNA from RIG-I.
NS1 proteins from some influenza A viruses do not interact with TRIM25, but still inhibit IRF-3 activation. Thus, it is possible that TRIM25 inhibition is not a universal mechanism of IFN antagonism by these viruses. Nonetheless, the structural basis for interaction with and inhibition of TRIM25 remains important. Two mutant NS1s are defective for NS1–TRIM25 interaction. The first is the R38A/K41A mutant, previously demonstrated to abrogate binding to dsRNA. The fact that this mutant loses the ability to interact with TRIM25 suggests an alternate mechanism by which it loses ability to inhibit RIG-I. The second is the E96A/E97A mutant that adopts an acidic α-helix in the ED and also loses TRIM25 binding and inhibitory activity [47]. This acidic α-helix and residues 96 and 97 have previously been implicated in the activation of PI3 kinase (PI3K) holoenzyme. In the PI3K–NS1 structure, the NS1 acidic α-helix participates in a four-helix bundle and lies adjacent to a basic activation loop within the kinase. The acidic helix is proposed to modulate kinase activity via interaction with the basic activation loop [50]. These data demonstrate that NS1 residues E96 and E97 are available to engage in protein-protein interactions, and leave open the possibility that they mediate interaction with TRIM25.
Vaccinia virus E3
Vaccinia virus, a poxvirus, is a large enveloped DNA virus that replicates in the cytoplasm of infected cells [51]. It encodes many proteins that function to counter host innate immune responses, including the E3 protein (reviewed in [52]). E3 has long been known as a dsRNA binding protein that can inhibit PKR and OAS [53–56]. The ability of E3 to bind dsRNA has been proposed as a mechanism to explain these inhibitory activities [53, 54]. However, E3 has also been shown to directly interact with PKR to inhibit its function [57]. E3 also inhibits dsRNA activated adenosine deaminase activity [58]. Among its other anti-IFN related functions are the ability to bind to small ubiquitin-like modifier 1 (SUMO1) [59] and to impair the antiviral activity of the IFN-induced ubiquitin-like protein ISG15 [60].
Vaccinia virus inhibits IFN signaling
Vaccinia viruses potentially activate cellular antiviral responses via multiple PRRs. For example, vaccinia virus and the attenuated derivative, modified vaccinia virus Ankara (MVA) have been reported to induce signaling through TLRs, cytoplasmic sensors of DNA, PKR, and RLRs [61–64]. A number of studies point to the E3 protein as a critical suppressor of IFN-β gene expression, including IFN-β gene expression induced through RLRs. E3 inhibits the activation of IRF-3/7 and prevents type I IFN gene expression, when signaling was induced by Newcastle disease virus (NDV) infection, suggesting the capacity to antagonize RLRs. E3 inhibition of IRF-7 was abrogated by a single point mutation (K167A) that also abolished dsRNA binding. However, overexpression of either of two other dsRNA binding proteins, DRBP76 or Staufen, did not inhibit RLR activity, demonstrating that inhibition of RLR signaling is not a universal property of all dsRNA binding proteins. Finally, another vaccinia virus protein that inhibits PKR, K3L, did not inhibit IRF-7 activation, providing evidence that E3 mediated inhibition of IRF-7 activation does not necessarily occur through inhibition of PKR. Interestingly, E3 can also inhibit IFN-β activation by single-stranded 5′-triphosphate RNA or by dsDNA, although it is unclear if the single stranded RNA also contained some double stranded regions [65]. The E3 dsRNA binding domain is sufficient for this inhibition, and inhibition is lost if a dsRNA binding mutant E3 is used for any of the activators [65]. The relevance of this function for vaccinia virus infection is of interest given the multitude of potential PAMPs and the multiple reported functions of the E3 protein. In support of its relevance in vaccinia virus infected cells, an E3L deletion vaccinia virus was attenuated in cells deficient for RNase L, PKR, and Mx1, and this attenuation correlated with increased activation of IRF-3 and production of IFN-β by mutant virus, relative to the E3L wild-type virus.
Vaccinia E3 antagonizes RLR signaling
Several studies further suggest a role for the RLR pathways in E3-mediated suppression of IFN-β gene expression during vaccinia virus infection. Vaccinia virus signaling can proceed via an IPS-1-dependent (PKR-independent) pathway. IRF-3 activation requires IPS-1, and knockdown experiments implicated the MDA-5 and RIG-I pathway [66]. IFN-β gene expression can be induced by RLR-activating RNAs produced during vaccinia virus infection and is supported by the observation that RNAs purified from the vaccinia virus E3L deletion mutant can induce, upon introduction into HeLa or A549 cells, IFN-β gene expression in a manner that depends upon RLRs and IPS-1. Interestingly, the apparent contribution of RIG-I and MDA-5 seemed to depend upon the cell line tested [63].
However, IPS-1 was required for IFN-β and cytokine production and for IRF-3 activation, consistent with signaling through RLRs. Induction of IFN-β and cytokines was absent in a virus encoding an E3 lacking Z-DNA binding activity but retaining dsRNA binding activity, again consistent with its ability to block RLR signaling in a manner that requires dsRNA binding activity. Similarly, E3 was found to suppress NF-κB and IRF-3-dependent cytokine and IFN-β gene expression [63]. Given the presence of a Z-DNA binding domain in E3, the fact that dsDNA can also induce IFN-β gene expression is interesting. When IFN-β was induced by transfection of poly(dA-dT), E3 did not detectably bind the transfected DNA, but it inhibited poly(dA-dT) induced expression of IFN-β. One mechanism by which DNA can induce IFN-β is through RNA polymerase III-mediated transcription and detection of the transcripts by RIG-I. The inhibition was mediated by the C-terminal dsRNA binding domain, but not the N-terminal Z-DNA binding domain, suggesting that E3 might block RIG-I activation by sequestering transcripts from poly(dA-dT). These observations further demonstrate a role for E3-mediated inhibition of RLR signaling by preventing RLR activation by RNA. The precise mechanisms by which this inhibition occurs, including a potential role for dsRNA formed by transcripts that can activate RLRs remains to be elucidated.
Ebola virus VP35
Ebola virus is a negative stranded nonsegmented RNA virus that encodes seven structural proteins, including VP35. VP35 is a multifunctional virulence factor that serves as an innate immune antagonist and is required as a cofactor for the viral RNA polymerase complex, for viral assembly, and for RNAi silencing suppression. To date, the best characterized function of VP35 is its ability to inhibit host IFN-α/β responses. In a growth complementation assay, VP35 could functionally replace the influenza virus NS1 protein, a known IFN inhibitor [67]. VP35 also inhibits the activation of the IFN-β promoter as well as IFN stimulated response element (ISRE)-containing promoters induced by dsRNA or viral infection [67]. VP35 proteins contain an N-terminal oligomerization domain and a C-terminal RNA binding domain that is also important for IFN inhibition (Figure 1).
Filoviruses, similar to other negative strand RNA viruses, such as influenza A virus, rabies virus, and vesicular stomatitis virus, trigger IFN-α/β production through RNA PAMP recognition by RLRs. VP35 inhibits activation of the IFN-β promoter that is stimulated by overexpression of several components of the RIG-I signaling pathway, including RIG-I, RIG-I CARD domain, IPS-1, IKKε, and TBK-1 [68, 69]. VP35 also blocks IRF-3 phosphorylation/dimerization but does not inhibit IFN-β promoter activation by a constitutively active IRF-3 mutant [68, 70]. Altogether, these studies strongly suggest that VP35 inhibits signaling through the RIG-I pathway that triggers IRF-3 phosphorylation by the IKKε/TBK-1 kinases [68, 70].
Ebola virus VP35 inhibits IFN-β activation
VP35 can antagonize IFN-β signaling by functioning as an alternative substrate for the IKKε/TBK-1 kinases, thereby interfering with the ability of these kinases to interact and activate IRF-3/7 [71]. The amino-terminal kinase domains of IKKε or TBK-1 are sufficient to coprecipitate with VP35 and phosphorylate several serine residues in VP35 in vitro. Overexpression of VP35 prevents coprecipitation of the IKKε kinase domain with IRF-3 and of the IKKε kinase domain with IPS-1. In vitro kinase assays show that VP35 prohibits IKKε phosphorylation of IRF-3 and that it prevents dimerization and nuclear accumulation of IRF-3 induced by viral infection [68, 70, 71]. Thus, one mechanism of Ebola virus suppression of IFN-β production is through VP35 inhibition of IRF-3/7 phosphorylation by the IKKε and TBK-1 kinases.
Unique structure of Ebola virus VP35 IID reveals the basis for dsRNA binding
Recent biochemical and structural data have begun to elucidate how VP35 binds dsRNA and contributes to host innate immune antagonism. VP35 contains an N-terminal oligomerization domain and a C-terminal dsRNA binding domain, also known as the IFN inhibitory domain (IID) [68, 69, 72]. The structure of Zaire Ebola virus VP35 IID consists of a unique fold, composed of an α-helical subdomain and a β-sheet subdomain, that binds dsRNA [73]. The α-helical subdomain contains an anti-parallel four helical bundle that spans approximately 70 residues. The β-sheet subdomain contains a four-stranded mixed β-sheet, an α-helix, and a left-handed type II polyproline helix. The inter-subdomain interface is formed by residues from helices 2 and 4 and βsheets 3 and 4, and the polyproline region [73]. Both subdomains are required to maintain the overall fold and function [69, 73].
Alignments of filoviral VP35 protein sequences identified highly conserved basic residues within VP35 IID that are arranged primarily in two patches, the first basic patch (FBP) and the central basic patch (CBP) [73]. The FBP is located in the α-helical subdomain and is critical for interaction with nucleoprotein (NP) and the replication complex [74]. Mutation of basic residues R225, K248, and K251 leads to loss of VP35 polymerase cofactor function, likely through impaired interactions with NP as VP35 mutants fail to coprecipitate NP. Furthermore, the VP35 IID alone is sufficient to interact with NP. The CBP is located within the β-sheet subdomain and basic residues R305, K309, and R312 are important for dsRNA binding and IFN inhibition [68, 72]. Highly conserved residues K319, R322, and K339 are also part of the CBP. Consistent with these studies, the structures of Zaire and Reston Ebola virus VP35 IIDs bound to dsRNA reveal that these basic residues play important roles in dsRNA recognition and protein-protein interactions, potentially with IKKε/TBK-1 [69, 75]. In the Zaire Ebola virus VP35–dsRNA complex, VP35 IID interacts with the dsRNA backbone and end-caps the blunt end of the dsRNA. These interactions are carried out by two different VP35 IID molecules. While both types of interactions include protein–backbone interactions, only one type of interaction is observed between the protein and the blunt end of the dsRNA. Interestingly, all residues that are important for dsRNA binding are also important for IFN inhibition. Comparison of the VP35 IID dsRNA complex structure with the RIG-I like helicase RNA binding domains suggests that VP35 IID mimics interactions between viral RNA and RIG-I, suggesting a potential mechanism for RIG-I antagonism [69, 75]. In vitro experiments suggest that VP35 IID can compete with RIG-I dsRNA binding domains. Furthermore, transfection of dsRNA and RIG-I with and without VP35 also show that VP35 can inhibit dsRNA-dependent RIG-I activation [69]. Taken together, these studies show that the Ebola virus VP35 protein can antagonize RLR signaling in a both RNA-dependent and RNA-independent manner. Such versatility likely makes significant contributions to the essential role that VP35 protein plays in Ebola virus pathogenesis.
Concluding remarks and future directions
Identification of PRRs and the discovery of how IFNs are triggered by PAMP recognition paved the way for recent discoveries of viral antagonism of RLRs. Recent structural studies of influenza virus NS1, Ebola virus VP35 as well as biochemical studies of vaccinia virus E3 suggest that these proteins play key roles in IFN antagonism that is directly linked to their ability to bind RNA, despite major differences in their RNA binding modes as well as overall fold (Figure 2). Moreover, recent structural characterization of RLRs by several groups has shown that RNA recognition and activation is directly linked to the ability of RLRs to recognize and bind dsRNA. Therefore, one potential mechanism by which these proteins antagonize RLRs is to bind and sequester RNA. Although this model is supported by recent studies, which show that mutation of residues that are important for RNA binding results in loss of IFN antagonism by viral proteins, it is likely that additional mechanisms that involve protein mediated recognition elements (RNA-independent) are at play. In addition to the viral antagonists described above, there are many viruses that encode components that antagonize type I IFN activation by RLRs. For example, the reovirus s3 protein, Borna disease virus P protein, HIV-1 Tat protein, and rotavirus NSP1 and NSP3 proteins are also known to sequester viral RNA. While the pathogenesis of these viruses is well described, detailed structural mechanisms are currently lacking for many of these systems. Future studies directed at developing a better understanding of these regulatory mechanisms will not only provide further validation of biochemical and cell biological observations regarding the importance of RNA recognition by RLR antagonist, but will also serve as potential starting points for the development of structure-based therapeutics.
Acknowledgments
Work in the authors’ laboratories are supported by NIH grants (grant 1R56AI089547 to C.F.B. and G.K.A., grant 1F32AI084324 to D.W.L., grants R01AI059536 and AI057158 [Northeast Biodefense Center-Lipkin] to C.F.B., grant 1F32AI084453 to R.S.S., and grant R01AI081914 to G.K.A.), an MRCE developmental grant (grant U54AI057160-Virgin to G.K.A.), and the Roy J. Carver Charitable Trust (grant 09-3271 to G.K.A.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kumar H, et al. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30:16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 2.Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010;32:305–315. doi: 10.1016/j.immuni.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins C, Gale M., Jr Recognition of viruses by cytoplasmic sensors. Curr Opin Immunol. 2010;22:41–47. doi: 10.1016/j.coi.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 5.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 6.Pichlmair A, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 7.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saito T, Gale M., Jr Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J Exp Med. 2008;205:1523–1527. doi: 10.1084/jem.20081210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pichlmair A, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83:10761–10769. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 12.Yoneyama M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 13.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlee M, et al. Approaching the RNA ligand for RIG-I? Immunol Rev. 2009;227:66–74. doi: 10.1111/j.1600-065X.2008.00724.x. [DOI] [PubMed] [Google Scholar]
- 15.Schlee M, Hartmann G. The chase for the RIG-I ligand--recent advances. Mol Ther. 2010;18:1254–1262. doi: 10.1038/mt.2010.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rothenfusser S, et al. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol. 2005;175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 17.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Civril F, et al. The RIG-I ATPase domain structure reveals insights into ATP-dependent antiviral signalling. EMBO Rep. 2011;12:1127–1134. doi: 10.1038/embor.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 20.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gack MU, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 23.Inn KS, et al. Linear ubiquitin assembly complex negatively regulates RIG-I- and TRIM25-mediated type I interferon induction. Mol Cell. 2011;41:354–365. doi: 10.1016/j.molcel.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng W, et al. Reconstitution of the RIG-I pathway reveals a signaling role of unanchored polyubiquitin chains in innate immunity. Cell. 2010;141:315–330. doi: 10.1016/j.cell.2010.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, et al. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 26.Shaw ML, Palese P. Orthomyxoviridae: The viruses and their replication. In: Knipe DM, et al., editors. Fields Virology. 5. Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 27.Ehrhardt C, et al. Interplay between influenza A virus and the innate immune signaling. Microbes Infect. 2010;12:81–87. doi: 10.1016/j.micinf.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez-Sesma A, et al. Influenza virus evades innate and adaptive immunity via the NS1 protein. J Virol. 2006;80:6295–6304. doi: 10.1128/JVI.02381-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Sastre A, et al. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 30.Fernandez-Sesma A. The influenza virus NS1 protein: inhibitor of innate and adaptive immunity. Infect Disord Drug Targets. 2007;7:336–343. doi: 10.2174/187152607783018754. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z, et al. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nemeroff ME, et al. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 33.Hornung V, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–997. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 34.Mibayashi M, et al. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J Virol. 2007;81:514–524. doi: 10.1128/JVI.01265-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opitz B, et al. IFNbeta induction by influenza A virus is mediated by RIG-I which is regulated by the viral NS1 protein. Cell Microbiol. 2007;9:930–938. doi: 10.1111/j.1462-5822.2006.00841.x. [DOI] [PubMed] [Google Scholar]
- 36.Ludwig S, et al. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J Virol. 2002;76:11166–11171. doi: 10.1128/JVI.76.21.11166-11171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Talon J, et al. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donelan NR, et al. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiao P, et al. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J Virol. 2008;82:1146–1154. doi: 10.1128/JVI.01698-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baum A, et al. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A. 107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergmann M, et al. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hatada E, et al. Mutant influenza viruses with a defective NS1 protein cannot block the activation of PKR in infected cells. J Virol. 1999;73:2425–2433. doi: 10.1128/jvi.73.3.2425-2433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, et al. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology. 2006;349:13–21. doi: 10.1016/j.virol.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Lu Y, et al. Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology. 1995;214:222–228. doi: 10.1006/viro.1995.9937. [DOI] [PubMed] [Google Scholar]
- 45.Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006;103:7100–7105. doi: 10.1073/pnas.0602184103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan SL, Katze MG. Biochemical and genetic evidence for complex formation between the influenza A virus NS1 protein and the interferon-induced PKR protein kinase. J Interferon Cytokine Res. 1998;18:757–766. doi: 10.1089/jir.1998.18.757. [DOI] [PubMed] [Google Scholar]
- 47.Gack MU, et al. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe. 2009;5:439–449. doi: 10.1016/j.chom.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bornholdt ZA, Prasad BV. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature. 2008;456:985–988. doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng A, et al. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19:187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- 50.Hale BG, et al. Structure of an avian influenza A virus NS1 protein effector domain. Virology. 2008;378:1–5. doi: 10.1016/j.virol.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 51.Moss B. Poxviridae: The Viruses and Their Replication. In: Knipe DM, et al., editors. Fields Virology. 5. Lippincott Williams & Wilkins; 2007. [Google Scholar]
- 52.Perdiguero B, Esteban M. The interferon system and vaccinia virus evasion mechanisms. J Interferon Cytokine Res. 2009;29:581–598. doi: 10.1089/jir.2009.0073. [DOI] [PubMed] [Google Scholar]
- 53.Chang HW, Jacobs BL. Identification of a conserved motif that is necessary for binding of the vaccinia virus E3L gene products to double-stranded RNA. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- 54.Romano PR, et al. Inhibition of double-stranded RNA-dependent protein kinase PKR by vaccinia virus E3: role of complex formation and the E3 N-terminal domain. Mol Cell Biol. 1998;18:7304–7316. doi: 10.1128/mcb.18.12.7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Davies MV, et al. The E3L and K3L vaccinia virus gene products stimulate translation through inhibition of the double-stranded RNA-dependent protein kinase by different mechanisms. J Virol. 1993;67:1688–1692. doi: 10.1128/jvi.67.3.1688-1692.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rivas C, et al. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- 57.Sharp TV, et al. The vaccinia virus E3L gene product interacts with both the regulatory and the substrate binding regions of PKR: implications for PKR autoregulation. Virology. 1998;250:302–315. doi: 10.1006/viro.1998.9365. [DOI] [PubMed] [Google Scholar]
- 58.Liu Y, et al. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology. 2001;289:378–387. doi: 10.1006/viro.2001.1154. [DOI] [PubMed] [Google Scholar]
- 59.Rogan S, Heaphy S. The vaccinia virus E3L protein interacts with SUMO-1 and ribosomal protein L23a in a yeast two hybrid assay. Virus Genes. 2000;21:193–195. doi: 10.1023/a:1008139514123. [DOI] [PubMed] [Google Scholar]
- 60.Guerra S, et al. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 2008;4:e1000096. doi: 10.1371/journal.ppat.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Delaloye J, et al. Innate immune sensing of modified vaccinia virus Ankara (MVA) is mediated by TLR2-TLR6, MDA-5 and the NALP3 inflammasome. PLoS Pathog. 2009;5:e1000480. doi: 10.1371/journal.ppat.1000480. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Valentine R, Smith GL. Inhibition of the RNA polymerase III-mediated dsDNA-sensing pathway of innate immunity by vaccinia virus protein E3. J Gen Virol. 2010;91:2221–2229. doi: 10.1099/vir.0.021998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Myskiw C, et al. Vaccinia virus E3 suppresses expression of diverse cytokines through inhibition of the PKR, NF-kappaB, and IRF3 pathways. J Virol. 2009;83:6757–6768. doi: 10.1128/JVI.02570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang P, et al. Protein kinase PKR-dependent activation of mitogen-activated protein kinases occurs through mitochondrial adapter IPS-1 and is antagonized by vaccinia virus E3L. J Virol. 2009;83:5718–5725. doi: 10.1128/JVI.00224-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Marq JB, et al. The double-stranded RNA binding domain of the vaccinia virus E3L protein inhibits both RNA- and DNA-induced activation of interferon beta. J Biol Chem. 2009;284:25471–25478. doi: 10.1074/jbc.M109.018895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang P, Samuel CE. Induction of protein kinase PKR-dependent activation of interferon regulatory factor 3 by vaccinia virus occurs through adapter IPS-1 signaling. J Biol Chem. 2008;283:34580–34587. doi: 10.1074/jbc.M807029200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Basler CF, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci U S A. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cardenas WB, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leung DW, et al. Structural basis for dsRNA recognition and interferon antagonism by Ebola VP35. Nat Struct Mol Biol. 2010;17:165–172. doi: 10.1038/nsmb.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Basler CF, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prins KC, et al. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hartman AL, et al. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 73.Leung DW, et al. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci U S A. 2009;106:411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Prins KC, et al. Mutations abrogating VP35 interaction with double-stranded RNA render ebola virus avirulent in guinea pigs. J Virol. 2010;84:3004–3015. doi: 10.1128/JVI.02459-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kimberlin CR, et al. Ebolavirus VP35 uses a bimodal strategy to bind dsRNA for innate immune suppression. Proc Natl Acad Sci U S A. 2010;107:314–319. doi: 10.1073/pnas.0910547107. [DOI] [PMC free article] [PubMed] [Google Scholar]