Abstract
High-molecular-weight, branched N-(2-hydroxypropyl)methacrylamide (HPMA) copolymers were synthesized and conjugated with Fab’ fragments of the anti-CD20 antibody, 1F5. This produced multivalent conjugates with varying valency (amount of Fab’ per macromolecule) targeted to the B-cell antigen CD20. The apoptotic activity of the conjugates was screened against several B-cell lymphomas with varied expression levels of CD20 (Raji, Daudi, Ramos, Namalwa, and DG-75). The multivalent conjugates had the strongest activity against cells that had the highest expression of CD20, and failed to demonstrate any measurable activity against lymphomas that did not express the antigen. Furthermore, there was an apparent dose-dependent response to treatment with multivalent conjugates. At optimal valence and concentration, the apoptotic activity of HPMA copolymer-Fab’ conjugates superseded that of free anti-CD20 Ab that was hyper-crosslinked with a polyclonal, secondary Ab.
Keywords: HPMA copolymer, CD20, 1F5 antibody, apoptosis, B cells
Introduction
Increasingly the role of nanomedicine is not only to efficiently deliver a given drug to diseased tissue, but to also increase the efficiency of the drug through innate biological processes.1–3 In such approaches a biomimetic strategy, such as receptor clustering, can be used to sensitize diseased tissues to a therapeutic agent. Receptor clustering has also been shown to incite cellular events such as insulin uptake,4 cell adhesion,5 cell activation,6 and apoptosis7,8 among others.9 In particular, hypercrosslinking the CD20 antigen, which is achieved by first ligating the antigen with an anti-CD20 antibody (Ab) and then exposing the complex to a polyclonal secondary Ab results in clustered CD20 and efficiently induces apoptosis.10 Constructs of anti-CD20 monoclonal Ab (mAb) Rituximab with a hetero-bifunctional crosslinker or activated dextran have also demonstrated apoptotic activity.11–13 Incorporation of anti-CD20 mAb into a well- studied drug delivery platform to create a multivalent drug delivery construct would allow facile addition of a therapeutic drug that could synergistically treat diseased B-cell tissues.
HPMA polymers and copolymers have been extensively used in anticancer polymer-drug conjugate platforms that include Ab targeted polymer-drug conjugates.14 Their prolific use stems from the physicochemical and pharmacokinetic advantages the polymer renders to conjugated drugs. These advantages include increasing circulating half-life and accumulation in solid tumors,15,16 increasing solubility of hydrophobic drugs, and potentially evading cellular efflux pumps that mediate multi-drug resistance.17 Furthermore, chemical composition and architecture can be varied to meet robust design criteria.18–20 Star-like and branched HPMA-based polymers may be prepared able to accommodate multiple Fab’ antibody fragments.20 Another advantage of HPMA copolymers is that they assume dynamic random coiled conformations.21 This allows distortion within the polymeric backbone that is needed to maximize multimeric interactions with targeted receptors.8,20
In many ways the B-cell antigen CD20 is an ideal target for immunotherapies. It is an integral membrane protein22 that is expressed from pre-B cells to terminally differentiated plasma cells and is present on greater than 90% of B-cell malignancies.23,24 CD20 is not shed from the cell surface nor is it present in serum under standard physiological conditions. It is a cell cycle regulatory protein25 that either controls or functions as a store operated calcium channel. The protein forms dynamic dimers and tetramers26 constitutively associated in lipid rafts of the cell membrane.27 At present CD20 has no known ligand; however, there are several anti-CD20 mAb clones. Among them there is a remarkable diversity of epitopes28 with varied biological activities.29,30 Of the panel of FDA approved anti-CD20 mAb, two clones, Rituximab31,32 and B1,32 have been the most studied for the treatment of B-cell malignancies and B-cell mediated autoimmune diseases.33 Other well-studied anti-CD20 Ab clones include 1F534,35 and 2H7. Our study utilizes the clone 1F5, which is capable of activating resting tonsilar B-cells; however, after a secondary antibody has crosslinked CD20-bound 1F5, the mAb potently induced apoptosis.36 Similarly, Rituximab32 and B136 most efficiently induce apoptosis following crosslinking.37 This fact has clinical relevance since effector cells can crosslink ligated anti-CD20 Ab. Diminished responses to therapeutic anti-CD20 Ab have been attributed to the failure of effector cells to crosslink CD20.38,39
Noting the impracticality of hypercrosslinking receptor-bound IgG in vivo, two studies have undertaken efforts to demonstrate the utility of ex vivo crosslinked Rituximab. Ghetie et al. covalently crosslinked Rituximab with hetero-bifunctional crosslinkers making homodimers of either whole Rituximab or Fab’2 fragments.11 Treatment of cells with homodimers inhibited cell growth, induced apoptosis, and sensitized a drug resistant cell line to treatment with either doxorubicin or an immunotoxin. Zhang et al. followed up this study by constructing polymers of Rituximab that had been formed by binding Abs to oxidized dextran.13 Using this Rituximab-dextran conjugate, they demonstrated enhanced apoptotic activity in vivo and effectively inhibited xenogenic tumor growth in mice. In both approaches, covalent modification was non-specific with the sole function of crosslinking Rituximab for clustering CD20.
The evaluation of physicochemical properties of multivalent antiCD20 HPMA copolymer-Fab’ conjugates demonstrated their ability to interact with CD20 molecules in a multimeric manner.20 Since the multimeric interactions were in effect crosslinking CD20, we hypothesized that treatment of CD20-bearing lymphoma cells with such conjugates would efficiently induce apoptosis. To test this hypothesis the apoptotic activity of a panel of multivalent HPMA copolymer-Fab’ conjugates was tested on several CD20(+) lymphoma cell lines (Raji, Daudi, Ramos, and Namalwa) and a control CD20(-) cell line (DG-75). Additionally, caspase-3 activity and DNA fragmentation were also evaluated to confirm apoptotic activity
Experimental Part
Materials
Chemicals and solvents used were of reagent grade or better unless otherwise stated. N-(2-Hydroxypropyl)methacrylamide (HPMA) was prepared as previously described.40 N-(3-Aminopropyl)methacrylamide hydrochloride (AMA) was purchased from Polysciences (Warrington, PA). Succinimidyl-4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SMCC) was purchased from Soltec Ventures (Beverly, MA). Tris(2-carboxyethyl) phosphine hydrochloride (TCEP) and Iodogen tubes were purchased form Thermo Scientific (Waltham, MA). Phthalic dicerboxaldehyde (OPA), and 3-mercaptopropionic acid (MPA) were purchased from Sigma Aldrich (St. Louis, MO).
Cell Lines, Hybridomas, Fab' Fragment Preparation
Human Burkitt’s B-cell non-Hodgkin’s lymphoma Raji, Daudi, Ramos, Namalwa, and DG-75 cells lines (ATCC, Bethesda, MD) were cultured in RPMI-1640 medium (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). Cells were grown at 37 °C in a humidified atmosphere of 5% CO2 (v/v). All experiments were performed using cells in exponential growth. The anti-CD20 hybridoma clone 1F5 (ATCC, Bethesda, MD) was initially cultured and recloned through limiting dilution41 in RPMI 1640 supplemented with 10% FBS in a humidified environment with 5% CO2. The selected clone was adapted to chemically defined, serum-free media (Invitrogen, Carlsbad, CA). Adapted cells were used to seed a CellMax bioreactor (Spectrum Laboratories, Rancho Dominguez, CA) according to the manufacturer’s instructions. Anti-CD20 mAb was purified on a Protein G Sepharose 4 Fast Flow column (GE Healthcare, Piscataway, NJ) from bioreactor harvest supernatant. Whole IgG2a was digested into F(ab')2 with lysyl endopeptidase (Wako Chemicals USA, Richmond, VA) following the protocol of Fowers et al.42 Purity of harvested IgG2a and digested F(ab')2 were >95% as determined by SDS-PAGE and SEC (size exclusion chromatography). Immediately prior use, 5 mg/mL of F(ab')2 was reduced to Fab' with 4 mM TCEP in 0.1 M phosphate buffer (pH 6.5) at room temperature for 30 min. Some of the F(ab’)2 was labeled with tetramethylrhodamine (TAMRA) by dissolving a small amount of dye in DMSO (50µg/10µL) and adding it to a solution of F(ab’)2 (3mg/mL, 0.1 M borate buffer, pH 9.0) for 1 h while mixing. The labeling reaction produced 1.5 TAMRA dye per F(ab’)2, which was determined by comparing the concentration of Ab (UV absorbance of 280 nm) and the concentration of TAMRA (UV absorbance of 540 nm).
Preparation of Multivalent Conjugates
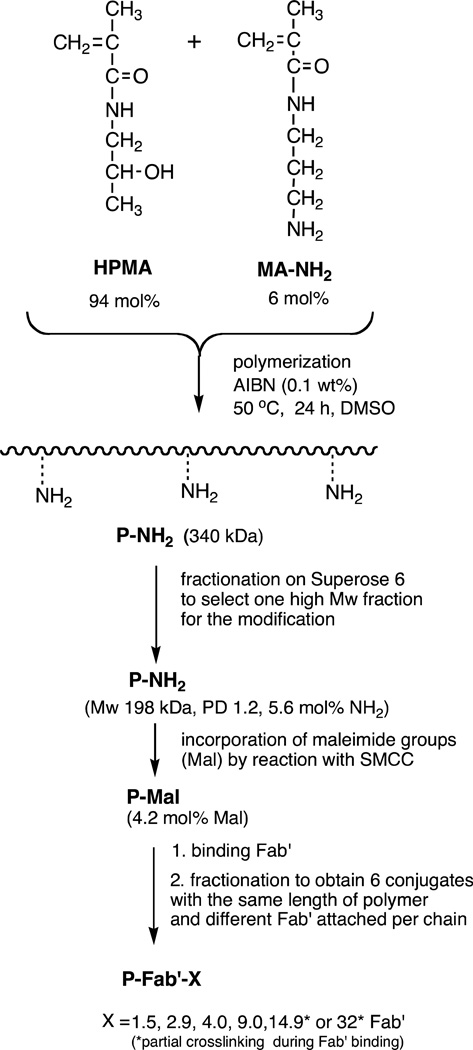
The synthesis of copolymers, their fractionation, polymer-analogous modification with SMCC, Fab' binding, and fractionation of the conjugates20 are depicted in Figure 1. The HPMA copolymer containing side chains terminated in NH2 groups (P-NH2) was prepared by radical polymerization using AIBN as the initiator. Briefly, HPMA (658.9 mg, 94 mol %), AMA (53.9 mg, 6 mol %), and AIBN (3.6 mg) were dissolved in 3.0 g of DMSO. After dissolution, the polymerization solution was bubbled with argon for 5 min, sealed in a glass ampoule, and then incubated at 50 °C for 24 h. After polymerization, P-NH2 was precipitated into acetone, filtered, and washed with acetone and diethyl ether. The precipitate was dried, dissolved in distilled water, and dialyzed against distilled water using dialysis tubing with a molecular weight cutoff of 12–14 kDa (Spectrum Laboratories, Rancho Dominguez, CA) to remove unreacted monomers. The P-NH2 was then lyophilized, dissolved in PBS, and fractionated on a Superose 6 HR16/60 preparative column (GE Healthcare, Piscataway, NJ) using the ÄKTA FPLC system (GE Healthcare, Piscataway, NJ) to produce a molecular weight “ladder” of ten fractions. One fraction (P-NH2: 198 kDa, PD 1.22, 5.6 mol % NH2, 27 wt %) was selected for the synthesis of conjugates. The P-NH2 fraction was dialyzed against water and then lyophilized.
Figure 1.
Scheme for the synthesis of multivalent HPMA copolymer-Fab’ conjugates targeted to the B-cell antigen CD20.
Polymeranalogous Conversion of Pendant Amine Groups into Maleimide Groups
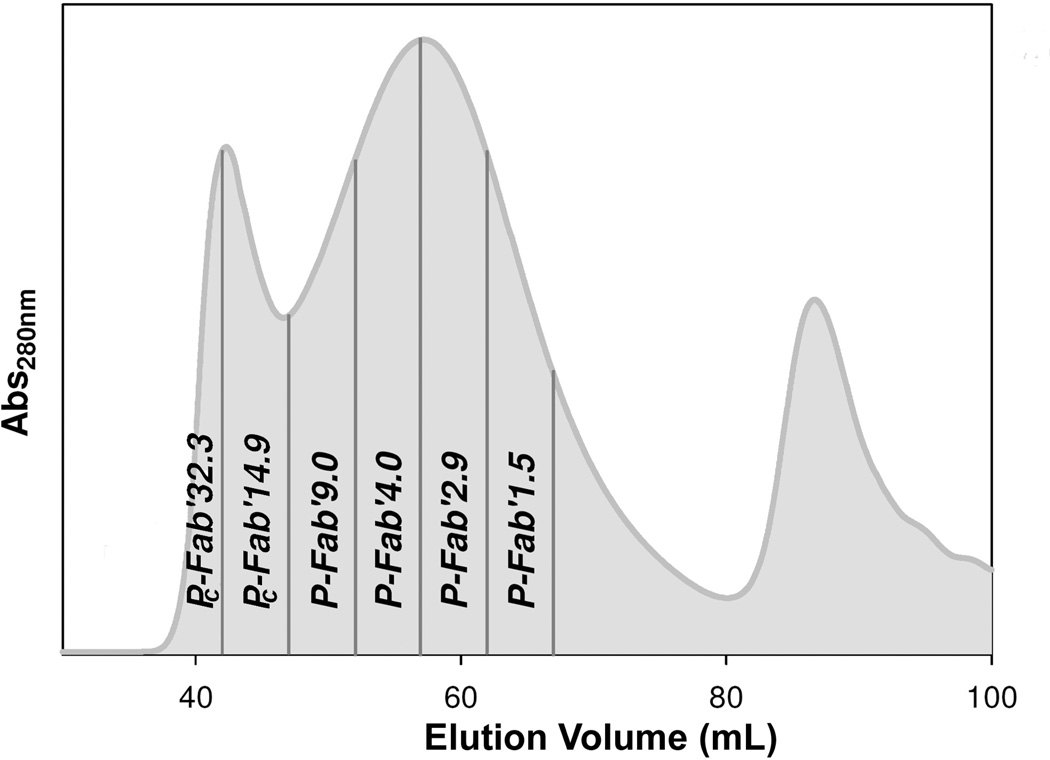
P-NH2 (86.9 mg, 65.2 µmol -NH2) was reacted with excess SMCC (40 mg, 127 µmol) in dry DMF in the presence of DIPEA (73 µL, 202 µmol) to generate pendant maleimide groups. The activated precursor was precipitated into an excess of acetone, filtered, and washed with ether to remove free SMCC. The conversion of amine groups into maleimide on the polymer proceeded with ~72% to 75% conversion, as determined by a modified Ellman’s assay. The product, P-Mal was then conjugated with reduced 1F5 Fab' fragments as previously described.20 Briefly, approximately 10 mg P-Mal (~4.6 µmol maleimide) was dissolved in 50 µL of DMF. Then, 20 mg of freshly reduced Fab' (0.4 µmol) in 4 mL of PBS (pH 6.5) was added to the P-Mal solution and allowed to react overnight at room temperature. The conjugate product was fractionated and unbound Fab' removed on a size-exclusion Superose 6 preparative column HR 16/60 to produce conjugates that varied in valence (Fab’ per polymer chain; fractionation was done to assure narrow polydispersities) (Figure 2). Conjugates made with TAMRA labeled Fab’ followed the same procedure.
Figure 2.
Fractionation of P-Fab’ conjugates by size exclusion chromatography. Elution profile of reaction mixture on a Superose S6 preparative column (HR 10/30, FPLC system); Buffer PBS pH 7.4; flow rate 1 mL/min; amount of sample applied 10 mg/2 mL; detection absorbance at 280 nm. The numerical suffix at the fraction designation is the average valence.
The conjugates are referred to by the fraction of polymeric precursor, presence of fluorogenic dye TAMRA, and valence determined by modified amino acid analysis. For example, P-TFab’9 indicates the TAMRA labeled polymer conjugate containing 9 Fab’ per polymer chain. Unlabeled equivalent would be referred to as P-Fab’9.
Determination of Fab′ Content per Polymer Chain
The ratio of protein to polymer was determined by amino acid analysis.43 This method enabled measurement of both individual amino acids from Fab' as well as 1-amino-2-propanol (AP) released from HPMA monomer units. Calibration was done using unbound 1F5 Fab' and HPMA homopolymer. A small amount (>3 mg) of Fab', homopolymer, or conjugate was dissolved in 6 N HCl in a small ampoule, bubbled with argon for 5 min, and then the ampoule was sealed. The solution was incubated at 120 °C for 16 h, the ampoule was opened, and the hydrolyzed product was dried under vacuum at room temperature. Hydrolyzed products were dissolved in distilled H2O and then derivatized with OPA and MPA prior to being analyzed on a HPLC with an eclipse XDB-C8 column (Agilent, Santa Clara, CA). Fab' content was calibrated with a peak produced by hydrolyzed glutamate, which had an elution time of 9.4 min. Poly(HPMA) concentration was calibrated with a peak produced by AP, which had a retention time of 23.9 min.
The first two conjugates eluted in SEC prior to the theoretical retention limit (Figure 2) and were shown to be crosslinked products. In these molecules, residual amines from the polymeric precursor P-NH2 from the polymeric backbone reacted with inter-chain maleimide functional groups when the HPMA copolymers were conjugated with Fab’. There is also a possibility that minute amounts of amine groups of Fab’ molecule could be involved in crosslinking side reactions. SEC Fractions thought to contain a majority of crosslinked (branched) product have been referred to with the prefix PC. Amino acid analysis of the PC conjugates resulted in an incomplete description of the Fab’ loading of the conjugate. In order to determine the average amount of Fab’ per PC-Fab’14.9 and PC-Fab’32.3 conjugates, the absolute molecular weight of the aggregates was determined by laser light scattering and refractive index after SEC analysis. This was done by applying the crosslinked conjugates to a Shodex OHpak SB-804 HQ gel permeation column (Showa Denko K.K., Kawasaki, Japan) in line with a MiniDawn light scattering detector (Wyatt Technology, Santa Barbara, CA) and an Optilab rEX refractive index detector (Wyatt Technology, Santa Barbara, CA). In the analysis the dn/dc value, 0.179, was determined by mass fractions of Fab’ (dn/dc 0.185), and HPMA (dn/dc 0.169).
Analysis of Apoptotic Activity
Annexin V/Propidium Iodine Analysis
Apoptotic activity of the conjugates was studied by treating Raji cells with conjugates followed by Annexin V/Propidium iodide (PI) staining. For each assay, 5 × 105 Raji cells were suspended in 2 mL of fresh growth medium with an appropriate amount of conjugate (conjugates in Fab’ equivalent that ranged from 1 nM to 1000 nM). For treatments using hyper-crosslinked mAb, 5 × 105 cells were collected and incubated with anti-CD20 mAb (1F5) at concentrations that ranged from 1 nM to 1000 nM in growth medium for 1 h. The cells were then washed twice with PBS + 1% BSA and suspended in 2 mL of fresh growth medium with 10 µg per ml of goat anti-mouse Ab (GAM) (KPL, Gaithersburg, MD). The cells were treated for 18 h in a humidified environment at 37°C, and 5% CO2 prior to staining. Following treatment, Annexin V/PI staining was carried out using the RAPID protocol provided by the manufacturer (Oncogene Research Products, Boston, MA). Screening the conjugate’s activity against multiple cell lines followed the previously described method with the exception of each cell line (Raji, Daudi, Ramos, Namalwa, and DG-75) being treated with 200 nM of Fab’ equivalent of either conjugate or 200 nM of whole anti-CD20 mAb crosslinked with GAM. All experiments were carried out in triplicate.
Terminal Deoxynucleotide Mediated-dUTP Nick-End Labeling Assay (TUNEL)
Analysis of DNA fragmentation characteristic of apoptosis was conducted using the TUNEL assay. In these experiments 1 × 106 Raji cells were treated with 200 nM of Fab’ equivalent of conjugate in 2 mL of fresh growth medium and allowed to incubate for 20 h. When Raji cells were treated with 200 nM of Ab, they were allowed to incubate for 1 h. The cells were then washed twice with PBS + 1% BSA and suspended in 2 mL of fresh growth media with 10 µg/mL of GAM. Following treatment, the cells were fixed with 2% paraformaldehyde in PBS for 1 h at room temperature. Cells were then permeabilized in 70% ethanol overnight at 4° C. Nick-end labeling was done using an Apo Direct TUNEL kit (Phoenix Flow Systems, San Diego, CA) following the manufacturer’s protocol. Analysis was done by flow cytometry. All experiments were carried out in triplicate.
Treatment of Raji cells with TAMRA labeled conjugates followed by nick end labeling with the dUTP-BrdU allowed visualization of apoptotic cells by fluorescence microscopy following staining with anti-BrdU(FITC) mAb. Observation of treated cells was done under a confocal fluorescence microscope using a 40×oil immersion lens.
Caspase-3 Activation Assay
Caspase-3 activity was characterized by analysis of treated cells with the PhiPhixux kit (OncoImmunin, Gaithersburg, MD). Prior to the using the assay, 5 × 105 of exponentially growing Raji cells were collected and suspended in fresh growth medium with 200 nM of Fab’ equivalent of conjugates. The cells were treated for either 5 h or 24 h in a humidified atmosphere at 37°C and 5% CO2 and then analyzed for caspase-3 activation following the manufacturer’s protocol. All experiments were carried out in triplicate.
Results
Multivalent Conjugate Preparation
The conjugates were synthesized similarly as described in our previous study20 and shown on Figure 1. Briefly: In the first step a high molecular weight copolymer of HPMA with 6 mol % of N-(3-aminopropyl)methacrylamide (AMA) in feed composition was prepared by radical polymerization. The conditions of the polymerization were optimized to obtain a high Mw product: a high concentration of monomers (20 wt%) and a low concentration of initiator AIBN (0.1 wt%). The high-molecular weight polydisperse product (Mw 340 kDa; Mw/Mn = 1.8; amine content 5.6 mol %) was fractionated by size exclusion chromatography.
One fraction, P-NH2, with an Mw average of 198 kDa and a Mw/Mn of 1.22 was selected for conjugation with Fab’ fragments. To prepare a maleimide functionalized polymeric precursor, P-Mal, for conjugation with Fab’, primary amines of P-NH2 were reacted with SMCC. Maleimide content of P-Mal, was 4.2 mol %. Then, P-Mal was conjugated with freshly reduced Fab’ in an overnight reaction via a thioether bond. Both conjugation reactions with Fab’ and TAMRA labeled Fab’ produced similar elution profiles on SEC and were fractionated to produce conjugates used in multivalent studies. As was described in a previous study,20 loading of Fab’ onto a HPMA copolymer broadened the elution profile of the polymer on SEC, with a shift in the peak elution indicative of an increased size. The first two fractions collected eluted prior to the theoretical retention limit of the column (Figure 2). Dynamic light scattering indicated that Pc-Fab’32.3 was a heterogeneous mixture with an effective diameter of 103 nm in PBS. Pc-Fab’14.9 was also heterogeneous and had an effective diameter of 68.2 numb in PBS (Table 1). These two fractions resulted from of unintended interchain-crosslinking of residual free amines on the polymer (or amino groups on Fab’) with unreacted maleimides of P-Mal. Amino acid analysis indicated that each of these fractions had an average of 6.3 Fab’ fragments per original polymer chain of Mw 198 kDa. The degree of interchain crosslinking assessed by determining the absolute Mw of the crosslinked conjugates and comparing that value with a theoretical Mw of the original polymer chain (Mw 198 kDa) was as follows: 2 – 3 original polymer chains crosslinked in conjugate containing 14.9 Fab’ and 5 polymer chains in conjugate with 32 Fab’. This result is in good agreement with large increase in effective diameters determined by DLS. For all other conjugates the amount of Fab’ that had been loaded onto the polymer, or valence of the conjugates, was determined with modified amino acid analysis as shown in Table 1 and DLS appeared to confirm the degree of Fab’ loading onto the HPMA copolymer. In conjugates where Fab’ were labeled to TAMRA prior to conjugation, no apparent differences in elution time or profile was observed and the valence or each fraction was equal to corresponding, unlabeled conjugates.
Table 1.
Chemical and Physical Analysis of Polymeric Precursors and Conjugates
| Conjugate | Valencea | Effective Diameterb | PDIc |
|---|---|---|---|
| P-Fab’1.5 | 1.5 | 17.8 | 0.203 |
| P-Fab’2.9 | 2.9 | 21.3 | 0.190 |
| P-Fab’4.0 | 4.0 | 30.2 | 0.168 |
| P-Fab’9.0 | 9.0 | 38.7 | 0.174 |
| Pc-Fab’14.9 | 14.9* | 68.2 | 0.209 |
| Pc-Fab’32.3 | 32.3* | 103.0 | 0.260 |
Fab’ per chain of conjugate or Fab’ per polymer chain as determined by modified amino acid analysis.
Effective diameter of conjugates as determined by dynamic light scattering (DLS).
Value of polydispersity index of the diameter of the conjugate determined by DLS.
Valence of the crosslinked fractions was determined by dividing the molecular weight determined by SEC and by the ratio of Fab’ to primary polymer chains that was determined by amino acid analysis.
Induction of Apoptosis by Treatment with Multivalent HPMA Copolymer-(anti-CD20) Fab’ Conjugates
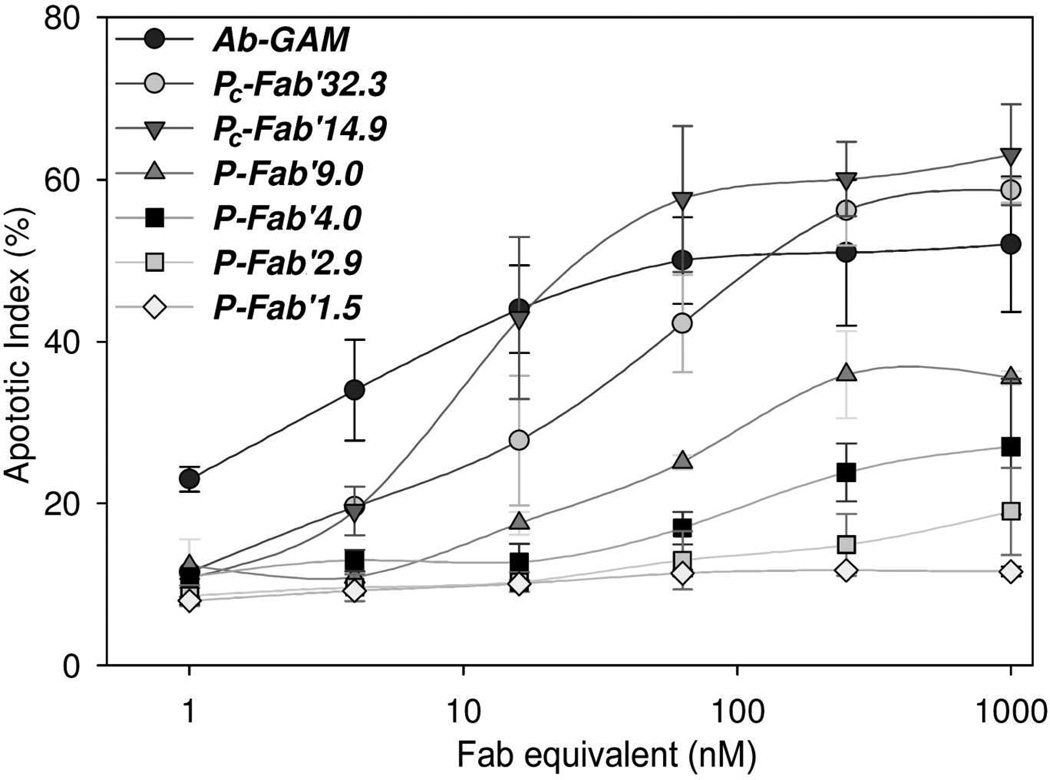
Apoptosis was examined using Annexin V/PI staining of Raji cells that had been treated at various concentrations of conjugates or hyper-crosslinked 1F5 antibodies (Figure 3; the term “hyper-crosslinked” infers treatment with a secondary Ab). In control studies, it was shown that neither the HPMA polymeric precursor, free anti-CD20 Ab (1F5), nor GAM secondary antibodies alone, induced significant amounts of apoptotic or necrotic cells compared with untreated cells. As a positive control various concentrations of 1F5 were hyper-crosslinked in situ with 10 µg/mL of GAM Ab after 1F5 was allowed to bind CD20 receptors on the cell surface. Apoptotic activity was observed at concentrations as low as 1 nM of anti-CD20 mAb. As the concentrations increased apoptotic activity was enhanced up to 50% apoptotic cells at 64 nM. Further increases of mAb concentration failed to produce significant increases in apoptosis. As the conjugates were screened, apoptosis was dependent on the degree of valence and the concentration of conjugate, in terms of Fab’ equivalent. The conjugate with the lowest valence, P-Fab’1.5, had no apparent apoptotic activity against CD20 expressing cells regardless of concentration and was, therefore, only screened against Raji cells. This result was expected since P-Fab’1.5 is analogous to free anti-CD20 Fab’ that has not been hyper-crosslinked. In contrast, Pc-Fab’14.9 and Pc-Fab’32.3 both potently induced apoptosis beyond concentrations of 5 nM of Fab equivalent conjugate. At 16 nM of Fab’ equivalent, Pc-Fab’14.9 activity superseded hyper-crosslinked 1F5 mAb and reached its peak activity at 64 nM with 58% of cells stained positive for apoptosis with no statistically significant changes in apoptotic activity at higher concentrations of conjugate (Figure 3). Potency of the other fractions diminished according to their valence and the concentration of the conjugate. P-Fab’9.0 reached peak activity at 250 nM of Fab’ equivalent with 36% the cells undergoing apoptosis. Conjugate P-Fab’2.9 induced apoptosis only at the highest concentrations tested (Figure 3).
Figure 3.
Dose dependent studies of hyper crosslinked Ab and multivalent HPMA copolymer-Fab’ conjugates targeting CD20. Apoptotic activity of the conjugates was determined by treating Raji cells for 18 h and then staining cells with annexing V and propidium iodide. Analysis was carried out using flow cytometry.
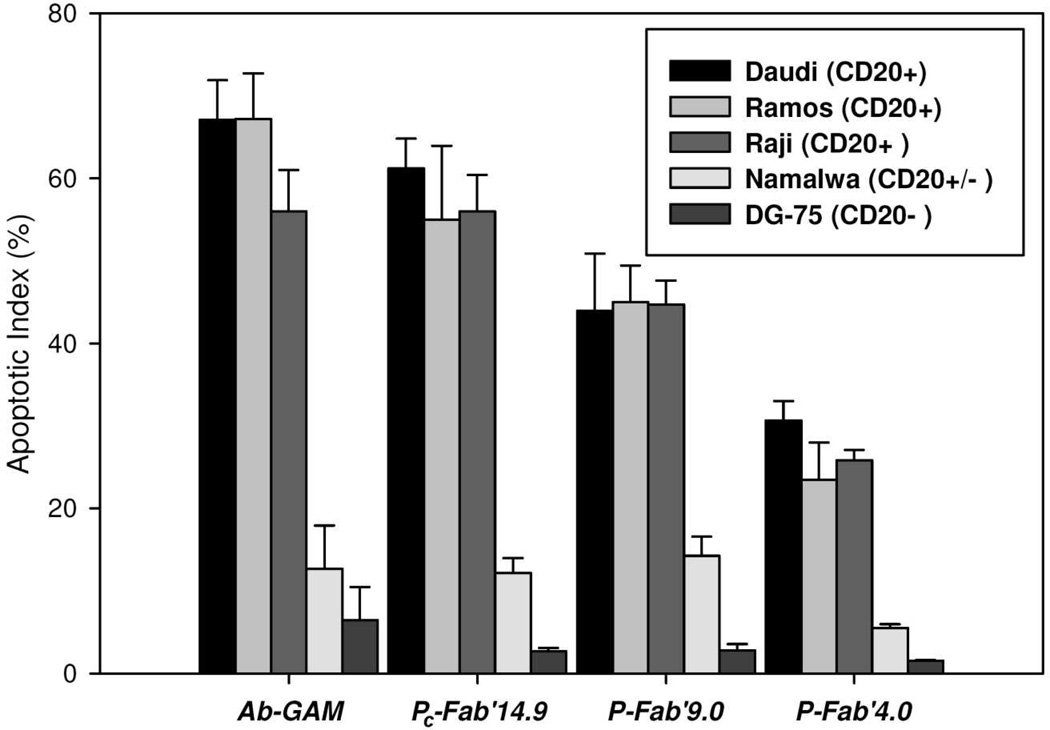
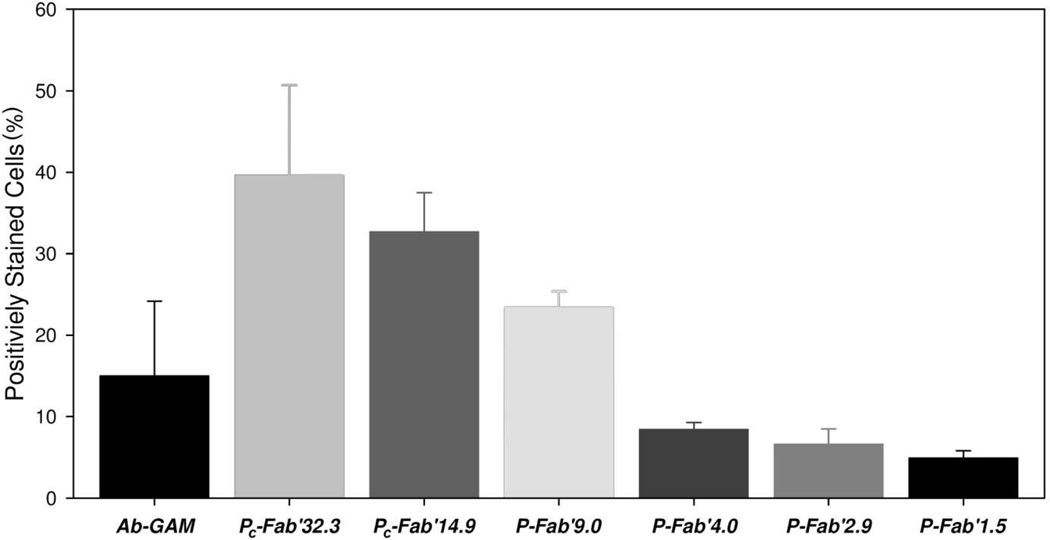
Apoptotic activities of the conjugates were also demonstrated in five different cell types after treating cells with 200 nM Fab’ equivalent of conjugates or with hyper-crosslinked anti-CD20 mAb. At this concentration, CD20 receptors are thought to be saturated on the surface of the cells.20 Of those cell types Raji, Ramos and, Daudi express CD20 strongly (CD20+). Namalwa had some expression of CD20 (CD20+/-), while DG-75 did not express CD20 (CD20-) (data not shown), which agreed with observations made in other studies.11,13 Responses in apoptotic activity were similar in all CD20 expressing cell lines. The strongest apoptotic signals correlated with the largest Fab’ per polymer chain ratios. None of the conjugates or hyper-crosslinked 1F5 produced a significant amount of apoptotic DG-75 cells. Treatment of Namalwa cells resulted in apoptosis between 13% and 14% of the cells, except went treated with P-Fab’4.0 where an insignificant amount of apoptotic activity was observed. The positive control, hyper-crosslinked anti-CD20 mAb, induced apoptosis in 50 % to 67 % of all cell lines that strongly expressed CD20. Activity of hyper-crosslinked anti-CD20 mAb treated Namalwa cells was reduced to 12%. PC-Fab’32.3 induced apoptosis in 55% to 62% of cells in Daudi, Ramos, and Raji cells. Apoptotic activity of P-Fab’9.0 fell to between 36% and 45% of cells, and apoptotic activity of P-Fab’4.0 fell to between 23% and 31% (Figure 4).
Figure 4.
Apoptotic activity of anti-CD20 multivalent HPMA copolymer-Fab’ conjugates. Five different cell lines were treated with 200 nM of Fab’ equivalent of conjugates. After 18 h of treatment cells were stained with annexin V and propidium iodide and then analyzed by flow cytometry to determine the amount of cells undergoing apoptosis. Raji, Ramos, and Daudi cell lines expressed CD20 well (CD20+). Namalwa cells had slight expression of CD20, and DG-75 did not express CD-20.
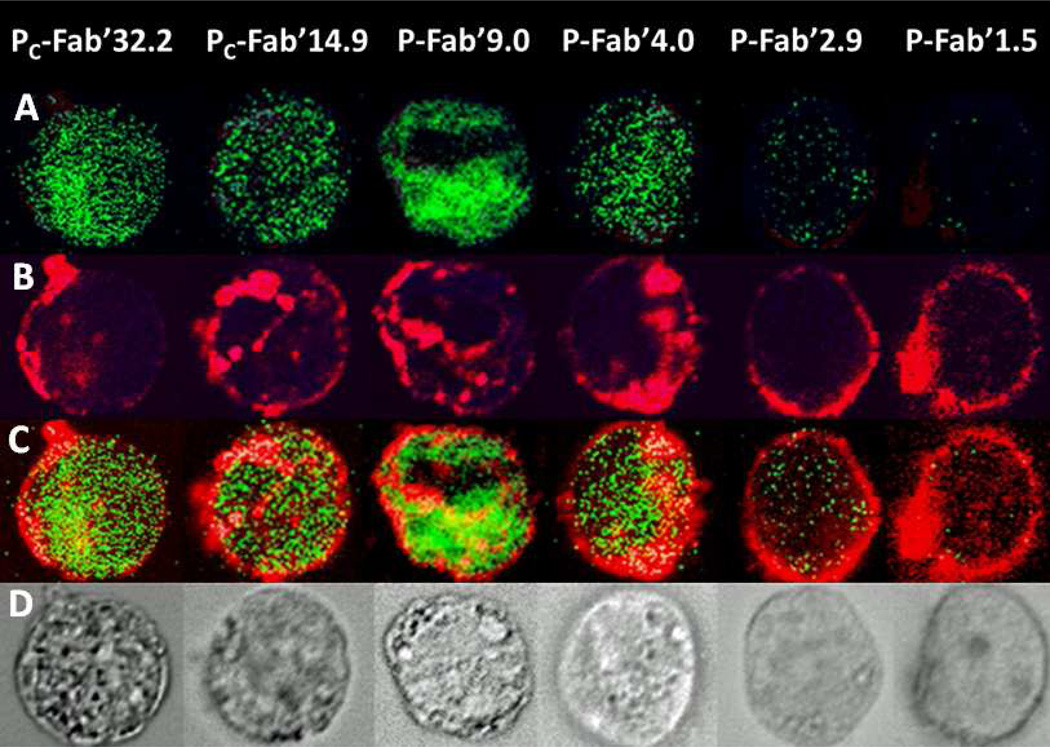
To confirm apoptotic activity observations, terminal deoxynucleotide transferase mediated dUTP nick end labeling (TUNEL) assay was performed and analyzed by flow cytometry and microscopy. Flow cytometry data was consistent with Annexin V/PI staining assays demonstrating a strong dependence of apoptotic activity on the valence of the conjugate as seen in Figure 5. Qualitative observation by fluorescence microscopy demonstrated that the majority of CD20(+) Raji cells treated with Pc-Fab’32.3 and Pc-Fab’14.9 were positively stained with anti-BrdU FITC. P-Fab’9.0 and P-Fab’4.0 had fewer positively stained cells corresponding to the valence of the conjugates. P-Fab2.9 and P-Fab’1.5 had virtually no TUNEL staining (Figure 5). Transmission light microscopy was used to confirm flow cytometry observations by correlating cells that stained positive with TUNEL stain and the appearance of apoptotic bodies. In addition, the TAMRA labeled conjugates allowed visualization of clustering patterns of conjugate bound receptors. Cluster patterns closely followed valence of conjugates. Conjugates with the highest degree of valence caused formation of the largest cluster patterns of receptor-conjugate complexes as seen in Figure 3. P-Fab’2.9 and P-Fab’1.5 had more homogenous staining of the cell membrane which indicated the absence of large receptor clusters (Figure 6).
Figure 5.
DNA fragmentation in cells treated with mutivalent HPMA copolymer-Fab’ conjugates. TUNEL assay of Raji cells treated with 200 nM Fab’ fractions for 18 h. Cells were stained with FITC labeled apo-BrdU antibodies. The number of positively stained cell indicates the presence of DNA fragmentation typical of apoptosis.
Figure 6.
Fluorescence images of cells treated with 200 nM Fab’ equivalent of HPMA copolymer-Fab’ conjugates labeled with TAMRA dye. In the first row (A) DNA fragmentation is indicated by TUNEL staining. In row (B), CD20 was visualized by TAMRA label if the HPMA-Fab’ conjugate. Clustering of CD20 is indicated by a change in the distribution of staining from peripheral stains on the surface of the cell to focal centers. Clustering increased with the valence of the conjugates. The next row (C) shows an overlay of both DNA fragmentation and CD20. The last row (D) is of transmission light images of the same cells with observable apoptotic bodies.
Caspase 3 Analysis
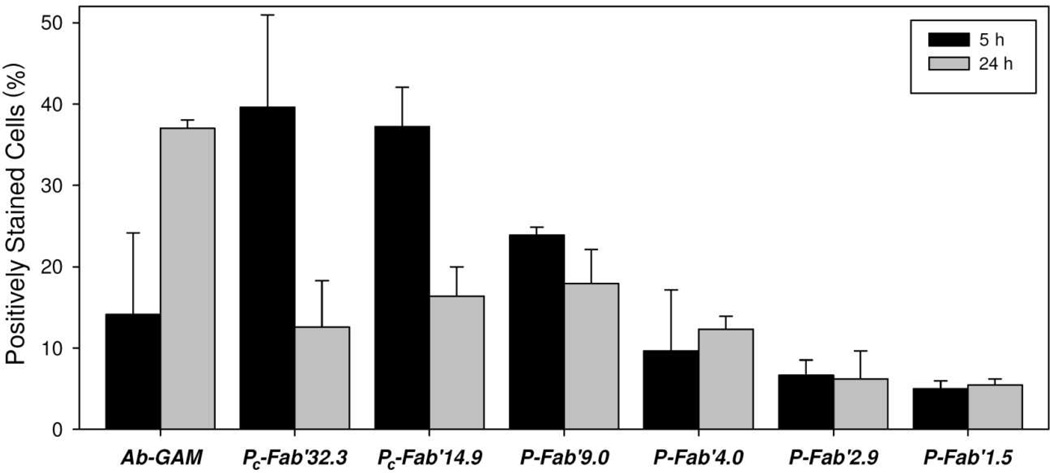
Induction of apoptosis was also monitored by caspase-3 activation in Raji cells treated with 200 nM of Fab’ equivalent of each conjugate or 1F5 anti-CD20 mAb crosslinked with GAM (Figure 7). Hyper-crosslinked 1F5 activated caspase-3 in 14% of treated cells after 5 h. The level of activated caspase-3 increased to 37% after 24 h. The most potent activators of caspase-3 at 5 h were Pc-Fab’14.9 and Pc-Fab’32.3, activating caspase-3 in nearly 40% of all cells. At 24 h, caspase-3 activity fell to 13% and 16%, respectively. The conjugate P-Fab’9.0 activated caspase-3 in 24% of treated cells, which fell to 18% after 24 h. The activity of P-Fab’4.0, P-Fab’2.9, and P-Fab’1.5 all had slight capase-3 activity. Hyper-crosslinked 1F5 mAb was around 15% at 5 h and increased to 37% after 24 h. Both P-Fab’2.9 and P-Fab’1.5 had trivial amounts of caspase-3 activity.
Figure 7.
Caspase activation in cells treated with multivalent HPMA copolymer-Fab’ conjugates. Raji cells treated with 200 nM of Fab’ equivalent and assayed (PhiPhiLux) after 5 and 24 h following treatment.
Discussion
Given that CD20 is a non-internalizing receptor23 but triggers apoptosis upon Ab ligation and crosslinking-induced clustering27,35,44,45, macromolecular therapeutics may offer novel means for design-based bioactivation and bioresponsiveness. Furthermore, studies that treated B-cell lymphoma’s with doxorubicin or immunotoxins after CD20 clustering have demonstrated enhanced therapeutic efficiencies and the ability to overcome P-glycoprotein efflux pump mediated multidrug resistance.46 These observations provide compelling motivation for macromolecular therapeutics targeting the B-cell antigen CD20 to include an intrinsic mechanism to cluster CD20. In the present study, we have proposed and demonstrated the utility of multivalent construction with HPMA copolymer conjugates targeting CD20. This report described their apoptotic activity and also formed a descriptive correlation between the valence of the conjugate and its ability to induce apoptosis in CD20 expressing B-cell lymphomas.
The physicochemical properties of a panel of similar, well-defined multivalent HPMA copolymer-(anti-CD20) Fab’ conjugates were described in an earlier report.20 These materials bound to CD20 following a negative cooperative model, with affinities that surpassed that of whole, unmodified Ab. One of the defining attributes of a negative cooperative model is at low concentrations of a multivalent ligand, the ligand interacts with targeted receptors multimerically,47–50 effectively clustering the receptors. Based on this observation, it was hypothesized that the constructs would efficiently induce apoptosis mediated through clustering CD20 in a concentration dependent manner. To test this hypothesis a panel of well-characterized, multivalent HPMA copolymer-(anti-CD20) Fab’ conjugates were screened against several B-cell lymphoma cell lines that had varied expression of CD20 to determine their apoptotic activities and relate them to valence of the conjugates. Typically, conjugates with the greatest amount of Fab’ per polymer chain were the most efficient at inducing apoptosis. However, a notable observation was that conjugate Pc-Fab’14.9 had stronger apoptotic activity then the Pc-Fab’32.3 counterpart (Figure 3). This suggests that Pc-Fab’14.9 may have reached an optimal valence and/or the size to facilitate multimeric interactions and, thereby, potently triggering apoptosis.
Another notable observation is the dose-dependent activity of the conjugates. In most conjugates apoptotic activity increased with the effective concentration of Fab’. This is striking since interactions involving the largest number of receptors occur at the lowest concentrations of multivalent ligands, according to the negative cooperative models.49 In theory, as concentration of the ligand increases, more receptors interact with the multivalent ligand but with lower avidity. This implies that the largest clusters of CD20 would form at the lowest concentrations of conjugates. However, apoptotic activity peaked at concentrations that would saturate CD20 present on the cell surface and result in smaller ligand-receptor complexes. In several examples in literature, kinetic studies of multivalent ligand off-rates (Koff) show that cell-bound receptors never completely release the multivalent ligand due to the avidity of the interactions.48,49 However there is a time dependent release of some of the multivalent ligands, while the targeted receptors remain saturated or nearly saturated. This suggests that monomerically bound ligands were displaced by multimerically interacting ligands,51–53 a phenomenon that provides a model of apoptosis triggered by CD20 clustering. While interactions would likely produce small clusters of CD20, the clusters would likely grow as monomerically interacting ligands were displaced by energetically more favorable multimeric interactions. Since this effect would occur at CD20-saturating concentrations of multivalent HPMA copolymer-Fab conjugates, elevated concentrations would neither enhance or diminish apoptosis of the cells as observed by a plateau in the dose activity relationship of some conjugates in Figure 3.
It is also worth noting the differences in apoptotic activity of the HPMA copolymer-Fab’ conjugates and hyper-crosslinked Ab as observed through Annexin V staining, caspase 3 activation, and fragmentation of DNA as determined by TUNEL. While Annexin V staining indicated comparable levels of apoptosis in cells treated with hyper-crosslinked Ab and conjugates with high valence (Pc-Fab’32.3, Pc-Fab’14.9, P-Fab’9.0), caspase 3 activation at 5 h and TUNEL staining was significantly higher in cells treated with multivalent conjugates. Interestingly, the caspase-3 activation at 24 h was much greater in cells treated by hyper-crosslinked Ab then the conjugates and may indicate that the multivalent conjugates were able to induce more potent apoptotic signals causing accelerated cell death. In this regard, the activity of the conjugates bodes well for further development of the conjugate. Combining an additional therapeutic element could further enhance the therapeutic potential of these materials. One interesting addition would include the use of radio-emitting elements such as 131I or 90Y as has been used in other CD20-targeting therapies such as Bexxar54 and Zevalin.55 These β-emitters are appealing since they are capable of potently killing targeted cells as well as neighboring malignant cells that may not express targeted antigens. Another strategy might be through the use of a prodrug,56 since CD20 constitutively remains on the surface of the cell. In each case the drug could also treat refractory cell populations through bystander effects lending the conjugate added functionality.
Any future development will need to ensure biocompatibility of the conjugate. One limitation of multivalent HPMA copolymer-Fab’ conjugates is that the size of the conjugate is well above the renal threshold. Studies that examined large HPMA-drug conjugates have demonstrated enhanced blood circulation because the materials are above the renal threshold15,18. To some degree extended circulation would be desirable for a malignancy that does not form solid tumors, such as NHLs. However, prolonged circulation of large molecules inevitably limits the biocompatibility of the materials.15 To make the conjugate more biocompatible the polymeric precursor could be composed of primary HPMA copolymer chains connected via biodegradable linkers. This would, in effect, engineer degradation into the main chain of the polymeric precursor and facilitate the excretion of the material. In recent studies, enzymatic degradation was engineered into the main chain of linear HPMA copolymers using living radical (RAFT) polymerization and post-polymerization azide-alkyne or thiol-ene click reactions.16,59,60
The addition of multiple Fab’ fragments produced a remarkable increase in the effective diameters of the conjugates. Other studies of Ab-polymer and Ab-polymer-drug conjugates have demonstrated that conjugation can affect biodistribution of the Ab and drug.62,63 In the multivalent conjugates studied here, the effects of the increase in size on the biodistribution may be somewhat marginal given that the polymeric precursor is already above the renal threshold and B-cell malignancies generally do not form solid tumors. In a study that also evaluated constructs that crosslink-CD20 Wu et al. did not observe any toxicity in a multi-dose format61 suggesting that the constructs remained in circulation. Also, the size of the conjugates was in the range of nanoparticles that depend on the enhanced permeation/retention (EPR) effect within solid tumors, which may suggest applications of similar constructs to malignancies that produce solid tumors. Another issue which may arise is a net loss of conjugate activity in vivo,64 a phenomenon that urges further investigation of the multivalent HPMA copolymer-Fab’ conjugates with in vivo models.
Another concern regards the heterogeneity of the multivalent conjugates. In this study, producing multivalent conjugates with narrow polydispersity and chemical homogeneity required extensive post-polymerization fractionation steps and resulted in significant loss of starting materials. We have postulated that using physical interactions to coordinate multivalent loading would allow more homogenous conjugates to be produced. Biorecognition of heterodimeric coiled-coil domains have previously been used to produce hydrogels,59,60 and more recently has been used to create multivalent conjugates targeting CD20.20,61 Wu et el. utilized heterodimeric coiled-coils to self-assemble at the surface of B cells with concomitant crosslinking of CD20 receptors.20 They bound one peptide to the Fab’ fragment and the complementary peptide as multiple grafts to HPMA copolymer. These constructs efficiently induced apoptosis in vitro8 and were active on an animal model of non-Hodgkin lymphoma in vivo.61 Remarkably, the apoptotic response was comparable to that elicited by CD20-bound 1F5 that had been crosslinked by a secondary Ab, which acted as a positive control. This approach, named drug-free macromolecular therapeutics,8 may provide a method for producing multi-specific conjugates with well-controlled compositions.
Conclusions
In summary, apoptosis triggered by receptor clustering of CD20 offers the potential for novel designs of polymer antibody conjugates. Direct conjugation of an anti-CD20 to high molecular weight HPMA to produce a multivalent construct was studied. This included assessing the therapeutic potential of a panel of multivalent HPMA copolymer-Fab’ conjugates by investigating their ability to induce apoptosis in CD20-bearing lymphoma cells. Dose dependence of apoptotic activation was demonstrated for each conjugate and was compared with free Ab that had been hyper-crosslinked with a polyclonal secondary antibody. Apoptotic activity was confirmed by demonstrating caspase-3 activation and DNA fragmentation. Such materials may have potential in the development of new paradigms for anticancer treatment where the use of low molecular weight drugs may be avoided.61
Acknowledgments
This study was supported in part by NIH grants EB00588 and GM95606 (to JK).
References
- 1.Kopeček J. Polim. Med. 1977;7:191–221. [PubMed] [Google Scholar]
- 2.Couvreur P, Vauthier C. Pharmaceutical Res. 2006;23:1417–1450. doi: 10.1007/s11095-006-0284-8. [DOI] [PubMed] [Google Scholar]
- 3.Vicent MJ, Ringsdorf H, Duncan R. Adv. Drug Delivery Rev. 2009;61:1117–1120. doi: 10.1016/j.addr.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 4.Kahn CR, Baird KL, Jarrett DB, Flier JS. Proc. Natl. Acad. Sci. U. S. A. 1978;75:4209–4213. doi: 10.1073/pnas.75.9.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimizu Y, van Seventer GA, Ennis E, Newman W, Horgan KJ, Shaw S. J. Exp. Med. 1992;175:577–582. doi: 10.1084/jem.175.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fourcin M, Chevalier S, Guillet C, Robledo O, Froger J, Pouplard-Barthelaix A, Gascan H. J. Biol. Chem. 1996;271:11756–11760. doi: 10.1074/jbc.271.20.11756. [DOI] [PubMed] [Google Scholar]
- 7.Vallat LD, Park Y, Li C, Gribben JG. Blood. 2007;109:3989–3997. doi: 10.1182/blood-2006-09-045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu K, Liu J, Johnson RN, Yang J, Kopeček J. Angew. Chem. Int. Ed. 2010;49:1451–1455. doi: 10.1002/anie.200906232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiessling LL, Gestwicki JE, Strong LE. Curr. Opin. Chem. Biol. 2000;4:696–703. doi: 10.1016/s1367-5931(00)00153-8. [DOI] [PubMed] [Google Scholar]
- 10.Deans JP, Li H, Polyak MJ. Immunology. 2002;107:176–182. doi: 10.1046/j.1365-2567.2002.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghetie MA, Bright H, Vitetta ES. Blood. 2001;97:1392–1398. doi: 10.1182/blood.v97.5.1392. [DOI] [PubMed] [Google Scholar]
- 12.Ghetie MA, Podar EM, Ilgen A, Gordon BE, Uhr JW, Vitetta ES. Proc. Natl. Acad. Sci. U. S. A. 1997;94:7509–7514. doi: 10.1073/pnas.94.14.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang N, Khawli LA, Hu P, Epstein AL. Clin. Cancer Res. 2005;11:5971–5980. doi: 10.1158/1078-0432.CCR-05-0554. [DOI] [PubMed] [Google Scholar]
- 14.Kopeček J, Kopečková P. Adv. Drug Delivery Rev. 2010;62:122–149. doi: 10.1016/j.addr.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shiah JG, Dvořák M, Kopečková P, Sun Y, Peterson CM, Kopeček J. Eur. J. Cancer. 2001;37:131–139. doi: 10.1016/s0959-8049(00)00374-9. [DOI] [PubMed] [Google Scholar]
- 16.Yang J, Luo K, Pan H, Kopečková P, Kopeček J. Reactive Functional Polym. 2011;71:294–302. doi: 10.1016/j.reactfunctpolym.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Minko T, Kopečková P, Kopeček J. Int. J. Cancer. 2000;86:108–117. doi: 10.1002/(sici)1097-0215(20000401)86:1<108::aid-ijc17>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 18.Dvořák M, Kopečková P, Kopeček J. J. Controlled Release. 1999;60:321–332. doi: 10.1016/s0168-3659(99)00087-5. [DOI] [PubMed] [Google Scholar]
- 19.Wang D, Kopečková P, Minko T, Nanayakkara V, Kopeček J. Biomacromolecules. 2000;1:313–319. doi: 10.1021/bm0000236. [DOI] [PubMed] [Google Scholar]
- 20.Johnson RN, Kopečková P, Kopeček J. Bioconjugate Chem. 2009;20:129–137. doi: 10.1021/bc800351m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bohdanecký M, Bažilová H, Kopeček J. Europ. Polym. J. 1974;10:405–410. [Google Scholar]
- 22.Einfeld DA, Brown JP, Valentine MA, Clark EA, Ledbetter JA. EMBO J. 1988;7:711–717. doi: 10.1002/j.1460-2075.1988.tb02867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Press OW, Farr AG, Borroz KI, Anderson SK, Martin PJ. Cancer Res. 1989;49:4906–4912. [PubMed] [Google Scholar]
- 24.Press OW, Howell-Clark J, Anderson S, Bernstein I. Blood. 1994;83:1390–1397. [PubMed] [Google Scholar]
- 25.Golay JT, Clark EA, Beverley PC. J. Immunol. 1985;135:3795–3801. [PubMed] [Google Scholar]
- 26.Bubien JK, Zhou LJ, Bell PD, Frizzell RA, Tedder TF. J. Cell Biol. 1993;121:1121–1132. doi: 10.1083/jcb.121.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deans JP, Li H, Polyak MJ. Immunology. 2002;107:176–182. doi: 10.1046/j.1365-2567.2002.01495.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polyak MJ, Deans JP. Blood. 2002;99:3256–3262. doi: 10.1182/blood.v99.9.3256. [DOI] [PubMed] [Google Scholar]
- 29.Holder M, Grafton G, MacDonald I, Finney M, Gordon J. Eur. J. Immunol. 1995;25:3160–3164. doi: 10.1002/eji.1830251126. [DOI] [PubMed] [Google Scholar]
- 30.Reff ME, Carner K, Chambers KS, Chinn PC, Leonard JE, Raab R, Newman RA, Hanna N, Anderson DR. Blood. 1994;83:435–445. [PubMed] [Google Scholar]
- 31.Leget GA, Czuczman MS. Curr. Opin. Oncol. 1998;10:548–551. doi: 10.1097/00001622-199811000-00012. [DOI] [PubMed] [Google Scholar]
- 32.Cardarelli PM, Quinn M, Buckman D, Fang Y, Colcher D, King DJ, Bebbington C, Yarranton G. Cancer Immunol. Immunother. 2002;51:15–24. doi: 10.1007/s00262-001-0247-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boye J, Elter T, Engert A. Ann. Oncol. 2003;14:520–535. doi: 10.1093/annonc/mdg175. [DOI] [PubMed] [Google Scholar]
- 34.Michel RB, Mattes MJ. Clin. Cancer Res. 2002;8:2701–2713. [PubMed] [Google Scholar]
- 35.Press OW, Appelbaum F, Ledbetter JA, Martin PJ, Zarling J, Kidd P, Thomas ED. Blood. 1987;69:584–591. [PubMed] [Google Scholar]
- 36.Hofmeister JK, Cooney D, Coggeshall KM. Blood Cells Mol. Dis. 2000;26:133–143. doi: 10.1006/bcmd.2000.0287. [DOI] [PubMed] [Google Scholar]
- 37.Cragg MS, Glennie MJ. Blood. 2004;103:2738–2743. doi: 10.1182/blood-2003-06-2031. [DOI] [PubMed] [Google Scholar]
- 38.Cartron G, Dacheux L, Salles G, Solal-Celigny P, Bardos P, Colombat P, Watier H. Blood. 2002;99:754–758. doi: 10.1182/blood.v99.3.754. [DOI] [PubMed] [Google Scholar]
- 39.de Haij S, Jansen JHM, Boross P, Beurskens FJ, Bakema JE, Bos DL, Martens A, Verbeek JS, Parren PWHI, van de Winkel JGJ, Leusen JHW. Cancer Res. 2010;70:3209–3217. doi: 10.1158/0008-5472.CAN-09-4109. [DOI] [PubMed] [Google Scholar]
- 40.Kopeček J, Bažilová H. Eur. Polym. J. 1973;9:7–14. [Google Scholar]
- 41.Ong GL, Elsamra SE, Goldenberg DM, Mattes MJ. Clin. Cancer Res. 2001;7:192–201. [PubMed] [Google Scholar]
- 42.Fowers KD, Callahan J, Byron P, Kopeček J. J. Drug Targeting. 2001;9:281–294. doi: 10.3109/10611860108997936. [DOI] [PubMed] [Google Scholar]
- 43.Irvine GB. Methods Mol. Biol. 1997;64:131–138. doi: 10.1385/0-89603-353-8:131. [DOI] [PubMed] [Google Scholar]
- 44.Taji H, Kagami Y, Okada Y, Andou M, Nishi Y, Saito H, Seto M, Morishima Y. Jpn. J. Cancer Res. 1998;89:748–756. doi: 10.1111/j.1349-7006.1998.tb03280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shan D, Ledbetter JA, Press OW. Blood. 1998;91:1644–1652. [PubMed] [Google Scholar]
- 46.Ghetie MA, Crank M, Kufert S, Pop I, Vitetta E. J. Immunother. 2006;29:536–544. doi: 10.1097/01.cji.0000211307.05869.6c. [DOI] [PubMed] [Google Scholar]
- 47.Hlavacek WS, Perelson AS, Sulzer B, Bold J, Paar J, Gorman W, Posner RG. Biophys. J. 1999;76:2421–2431. doi: 10.1016/s0006-3495(99)77397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crespo-Biel O, Lim CW, Ravoo BJ, Reinhoudt DN, Huskens J. J. Am. Chem. Soc. 2006;128:17024–17032. doi: 10.1021/ja0637705. [DOI] [PubMed] [Google Scholar]
- 49.Dower SK, Titus JA, Segal DM. Multivalent models. In: Perelson AS, Delisi C, Wiegel FW, editors. Cell Surface Dynamics: Concepts and Models (Receptors and Ligands in Intercellular Communication) Chapter 12. New York: Dekker; 1984. pp. 350–379. [Google Scholar]
- 50.Gestwicki JE, Cairo CW, Strong LE, Oetjen KA, Kiessling LL. J. Am. Chem. Soc. 2002;124:14922–14933. doi: 10.1021/ja027184x. [DOI] [PubMed] [Google Scholar]
- 51.Rossi EA, Goldenberg DM, Cardillo TM, Stein R, Chang C. Blood. 2009;113:6161–6171. doi: 10.1182/blood-2008-10-187138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Deyev SM, Lebedenko EN. BioEssays. 2008;30:904–918. doi: 10.1002/bies.20805. [DOI] [PubMed] [Google Scholar]
- 53.DeLisi C. Mol. Immunol. 1981;18:507–511. doi: 10.1016/0161-5890(81)90128-0. [DOI] [PubMed] [Google Scholar]
- 54.Horning SJ, Younes A, Jain V, Kroll S, Lucas J, Podoloff D, Goris M. J. Clin. Oncol. 2005;23:712–729. doi: 10.1200/JCO.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 55.Wiseman GA, White CA, Witzig TE, Gordon LI, Emmanouilides C, Raubitschek A, Janakiraman N, Gutheil J, Schilder RJ, Spies S, Silverman DH, Grillo-Lopez AJ. Clin. Cancer Res. 1999;5:3281s–3286s. [PubMed] [Google Scholar]
- 56.Devalapally H, Navath RS, Yenamandra V, Akkinepally RR, Devarakonda RK. Arch. Pharm. Res. 2007;30:723–732. doi: 10.1007/BF02977634. [DOI] [PubMed] [Google Scholar]
- 57.Pan H, Yang J, Kopečková P, Kopeček J. Biomacromolecules. 2011;12:247–252. doi: 10.1021/bm101254e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luo K, Yang J, Kopečková P, Kopeček J. Macromolecules. 2011;44:2481–2488. doi: 10.1021/ma102574e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang J, Xu C, Wang C, Kopeček J. Biomacromolecules. 2006;7:1187–1195. doi: 10.1021/bm051002k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kopeček J, Yang J. Acta Biomaterialia. 2009;5:805–816. doi: 10.1016/j.actbio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu K, Yang J, Liu J, Kopeček J. J. Controlled Release. 2012 doi: 10.1016/j.jconrel.2011.08.002. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pechar M, Ulbrich K, Jelínková M, Říhová B. Macromol. Biosci. 2011;3:364–372. [Google Scholar]
- 63.Flanagan PA, Duncan R, Šubr V, Ulbrich K, Kopečková P, Kopeček J. J. Controlled Release. 1992;18:25–38. [Google Scholar]
- 64.Říhová B, Kopečková P, Strohalm J, Rossmann P, Větvička V, Kopeček J. Clin. Immunol. Immunopathol. 1988;46:100–114. doi: 10.1016/0090-1229(88)90010-4. [DOI] [PubMed] [Google Scholar]