Abstract
Osteoarthritis (OA) is a degenerative disease of the joint and results in changes in the biochemical composition of cartilage. Studies have been undertaken in the past that have used high resolution NMR spectroscopy to study the biochemical composition of porcine, canine and bovine cartilage. In this study high resolution magical angle spinning (HRMAS) NMR spectroscopy at 11.7 T has been used to characterize metabolites and detect differences in the spectral signature of human knee articular cartilage from non-OA healthy cadaver knees and samples acquired from severe OA patients at the time of total knee replacement surgery. A statistically significant difference in the alanine (1.47 ppm), N-acetyl (2.04 ppm), choline (3.25 ppm) and glycine (3.55 ppm) metabolite levels is observed between healthy and OA specimens. The results of the study indicate that a decrease in the intensity of N-acetyl resonance occurs in later stages of OA. A positive correlation of the N-acetyl levels as measured by 1H HR-MAS NMR spectroscopy with the total proteoglycan content in the same cartilage specimens as measured by the GAG assay was observed. This indicates that N-acetyl can serve as an important bio-marker of OA disease progression. A decrease in the alanine concentration in OA may be attributed to the degradation of the collagen framework with disease progression and eventual loss of the degradation products that are transported from cartilage into the synovial cavity.
Keywords: 1H, HRMAS, osteoarthritis, cartilage, 1D, N-acetyl, collagen, proteoglycan
Introduction
Osteoarthritis (OA) is a degenerative disease of the joint and is primarily characterized by progressive loss of articular cartilage. It thereby impairs joint function and induces physical disability and is only second to cardiovascular disease in causing work disability [1]. OA affects approximately 12-16% of adults over the age of 60 in the United States [2]. It is a multi-factorial disease and the pathogenesis of OA is linked to many factors, including altered mechanical loading, injury, genetic dispositions, obesity, and age [3-4]. The current gold standard for radiological diagnosis of OA is a plain radiograph; it detects late stage OA pathologies such as joint space narrowing, joint deformity, and appearance of osteophytes [5]. However, a severe shortcoming of this method is that early biochemical or metabolic changes in articular cartilage cannot be detected or characterized.
Cartilage is an avascular tissue with low cellularity. Most of the tissue mass is derived from the extracellular matrix (ECM) consisting of a collagen network, proteoglycans (PG), and water. The proteoglycans are comprised of core proteins (aggrecan) that are covalently bonded to glycosaminoglycan (GAG) chains of chondroitin sulfate (CS) and keratan sulfate (KS). The GAG chains are negatively charged due to the presence of sulfate groups and as a result water is loosely bound to proteoglycans. The interactions between the different ECM components have an effect on the role of cartilage as a load bearing tissue. Healthy cartilage is in a state of metabolic stasis with components being degraded and synthesized constantly. Onset of OA perturbs this metabolic balance and leads to degradation and loss of these biochemical components, much earlier than these changes can be detected using conventional radiographs or MRIs. Magnetic Resonance Spectroscopy (MRS) can provide a biochemical profile of most tissues in vivo. However, the rigid nature of cartilage results in poor spectral resolution or broad line-width in case of in vivo NMR spectroscopy.
High-Resolution Magic Angle Spinning (HR-MAS) NMR spectroscopy is a promising technique for the characterization of intact tissue from ex vivo tissue samples. Sample preparation for HR-MAS is very straightforward and most NMR pulses can be easily used for HR-MAS acquisition. The tissue sample is rapidly spun (at about 2-5 kHz) at an angle of 54.7° relative to the B0 magnetic field reducing the line broadening effects caused by sample heterogeneity and anisotropic NMR parameters (such as chemical shift anisotropy and homonuclear dipolar couplings) that are normally averaged out in solution [6]. The MAS technique combined with high magnetic fields (typically 11.7 T, or 500 MHz for 1H) results in improved spectral resolution than in vivo MRS, comparable to that of NMR from tissue extracts [7]. A key advantage of HR-MAS over conventional NMR spectroscopy of tissue extracts in solution is that after spectral acquisition the tissue sample remains intact; the same sample can be used for direct comparison between the biochemical profile of the tissue and a downstream application such as histology or gene expression [8-9].
HR-MAS NMR has been previously used for the characterization of other tissues and pathologies in humans, e.g. the brain [10-11], breast [12], liver [13], prostate [14] and intervertebral disc [15-16]. HR-MAS of articular cartilage has been limited to animal studies, such as porcine or bovine cartilage digestion [17-19] or guinea pig model [20]. However, no studies have been performed to compare healthy and osteoarthritic articular cartilage from human subjects using HR-MAS NMR spectroscopy. The purpose of this study was to characterize the metabolites of normal human articular cartilage (from cadaver knees) using HR-MAS NMR spectroscopy, and to quantify any metabolite changes associated with OA. The secondary aim was to establish the reproducibility of this technique and its sensitivity in detecting differences in metabolite concentrations in cartilage samples taken from adjacent locations in the knee. The HR-MAS results were correlated with a biochemical GAG assay of cartilage and the results have been supported by histo-pathological analysis of both healthy and OA human articular cartilage.
Methods
Tissue Acquisition
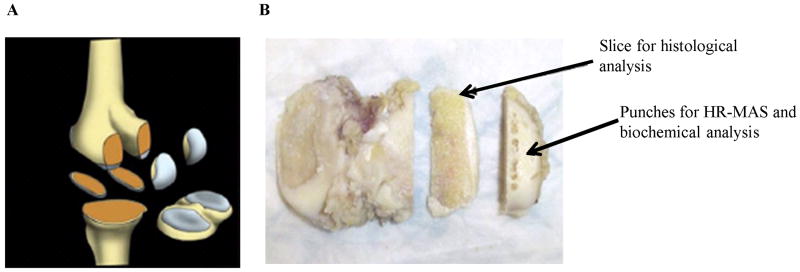
The study was approved by the university's Institutional Review Board and was performed in accordance with the regulations of the approved protocol. Osteoarthritic samples were excised from osteoarthritic patients (n= 12, range = 57- 81 years, 1 male, 11 females) undergoing total knee arthoplasties (TKA). In case of each patient five samples were collected: lateral and medial inferior femoral condyles, lateral and medial posterior medial condyles and the tibia plateau (Figure 1). Cadaveric knee specimens with no history of musculoskeletal diseases were obtained from the National Disease Research Interchange (NDRI) to serve as healthy controls (n= 4, range = 37- 48 years, 2 males, 2 females). Two adjacent 3 mm biopsy punches (mass = 14.55 ± 4.48 mg) were taken from each knee region during surgery or dissection for HR-MAS spectroscopy. The samples were flash frozen in dry ice at -80° C within 10 min after surgical removal until further experimental analysis.
FIG 1.
Cartilage specimen collection from advanced OA patients during TKA surgery (A). The diagram of the five specimens containing bone and cartilage sectioned during surgery: lateral/medial inferior femoral condyles (LIFC/MIFC), lateral/medial posterior femoral condyle (LPFC, MPFC) and tibial plateau containing lateral/medial tibia (LT/MT). (B) Location of the punches taken for HR-MAS and the slice for histology. Picture taken from [29].
HR-MAS data acquisition
HR-MAS spectra were acquired using a 11.7T (500 MHz for 1H) Varian INOVA spectrometer (Varian Inc., Palo Alto, CA, USA) equipped with a 4 mm gHX nanoprobe with HR-MAS capabilities. In addition to allowing the use of magic angle spinning, a nanoprobe was preferred to a “liquids” probe in order to reduce sample size and provide sufficient water suppression and consequently minimize baseline distortions that could in turn affect quantitation. Samples were analyzed in a custom-designed 20 μL leak-proof zirconium rotor containing oblate spheroid geometry to improve the magnetic field homogeneity across the sample. Approximately 3 μL of deuterium oxide containing 0.75 wt % 3-(trimethylsilyl) propionic-2,2,3,3-d4 acid (D2O + TSP; Sigma-Aldrich) was pipetted into the bottom of the rotor to achieve a field-frequency (D2O) lock signal and to establish a frequency reference (TSP). This was followed by introducing the tissue samples into the rotor. The weight of both D2O and the tissue samples were measured before acquiring data.
The data was acquired at a temperature of 1°C to minimize tissue degeneration during data acquisition. A spin rate of 2250 Hz was used as the spinning side bands generated from the water resonance at this spin rate did not overlap with metabolite resonances at 11.7 T. First order shimming was employed when necessary to ensure that the average line-width at half maximum of the water resonance was 7-8 Hz. Each spectrum was acquired with 40000 complex points over a 20000 Hz spectral width, a 90° pulse, a 2 s pre-saturation delay, TE = 144 ms and a TR = 4.2 s. A spin-echo rotor-synchronized Carr-Purcell-Meiboom-Gill (CPMG) sequence with an echo time of 144 ms was used to suppress the broad signals of large macromolecules. 512 transients were acquired per sample resulting in a total scan time of 35 minutes.
To quantify the metabolite signal, the Electronic Reference To access In-vivo Concentrations (ERETIC) method was used [21]. Using the ERETIC method, a synthesized RF pulse was added during the acquisition period to generate a signal in the NMR spectrum with an offset frequency to -0.5 ppm. As no reference compound was used to generate this signal, there were no concerns about chemical activity, binding, or visibility of the signal. Before starting the experiment, the ERETIC signal was phased to match the phase of the metabolites' signals. The ERETIC signal was calibrated against D2O+TSP on a weekly basis to maintain accuracy and reproducibility of the method.
The total time for data acquisition including sample preparation, tuning, shimming, pulse width calibration and data collection ranged from 1- 1.5 hours per sample. Overall, spectra of 22 healthy samples and 23 osteoarthritic samples were acquired. Reproducibility of the HR-MAS spectroscopy technique was examined by rescanning four tissue specimens on the same day without removing the sample from rotor but replacing both the rotor and the NMR probe in and out of the magnet. The spatial variability of the cartilage punches was measured by scanning four pairs of adjacent punches from the same knee region.
For the 2D total correlation spectroscopy (TOCSY) spectra, 4096 complex points were acquired with a 20 kHz spectral bandwidth in the direct dimension (F2) and 256 complex points were acquired over a 6500 Hz spectral width in the indirect dimension (F1). The TOCSY spectral acquisition was preceded by a 2 s HOD pre-saturation and relaxation delay followed by the adiabatic mixing period of 40 ms and then a 0.2 s acquisition period. A total of 64 transients were acquired for a total experiment time of 5 hours and 3 min. In order to minimize the effects of B0 and B1 inhomogeneities, rotor-synchronized constant-adiabaticity WURST-8 adiabatic pulses were used for isotropic mixing [16].
Post processing
The data was post processed with ACD Labs 1D NMR Processor Advanced Chemistry Development, Inc., Toronto, Canada). The FID's were zero filled to 128000 points, apodized using an exponential function with a line broadening factor of 0.5 Hz, and then Fourier transformed. The resulting spectra were then phase corrected, baseline corrected using a sixth degree polynomial function, and referenced to TSP at 0 ppm. Metabolite peaks with flat baselines were integrated manually, whereas other peaks were quantified using NMR Processors' peak-fitting function. Once the metabolite peaks were integrated, they were normalized to the ERETIC peak and tissue mass.
The TOCSY data was processed using ACD Labs 2D NMR processor using 3 × N forward linear predictions in F1 and apodized using the Gaussian weighting in both F1 and F2 dimensions. The assignment of the TOCSY cross peaks was partly based on previous work [17, 22-23] with some additions (glucuronic acid) based on TOCSY experiments on standards from Sigma-Aldrich. The presence of choline and glucuronic acid cross peaks were confirmed by running TOCSY experiments on choline and sodium salt of glucuronic acid (Sigma-Aldrich Co., St.Louis, MO, USA) respectively under experimental conditions identical to the TOCSY acquisition of the cartilage sample.
Biochemical Analysis
After HR-MAS acquisition, a subset of tissue samples (n = 17) including five healthy and twelve OA cartilage samples were analyzed for total proteoglycan content measurement. Samples were papain digested at 60°C for 20 hours. Then the samples were centrifuged and the digested supernatant was extracted. A DMMB assay (Blyscan Glycosaminoglycan Kit; Carrickfergus, UK) was performed as per manufacturer's instructions. The proteoglycan content in the sample was then measured by absorbance at 656 nm using a UV-VIS spectrometer and normalized to the wet weight of the sample.
Histology
Histology was performed on representative cartilage specimens from both control and OA groups to confirm the cartilage degeneration condition. Slices with 3 mm thickness adjacent to the HR-MAS biopsy punch locations were sectioned and stained using Hematoxylin and Eosin (H&E) and Safranin-O (AML Labs, Baltimore, MD). Histological slides were read by an experienced cartilage/bone pathologist.
Statistical Analysis
The mean and standard deviation (SD) for each metabolite of interest was calculated for both study groups; a Student's t-test (two-tailed, unequal variance) was performed to compare the metabolite concentration between two groups (the significance level was set to 0.05). The reproducibility and spatial variability of the data was measured using the coefficient of variation (CV). The correlation between the total proteoglycan content and quantified N-acetyl peak was measured using linear regression.
Results and Discussion
1H HR-MAS NMR spectroscopy of cartilage samples
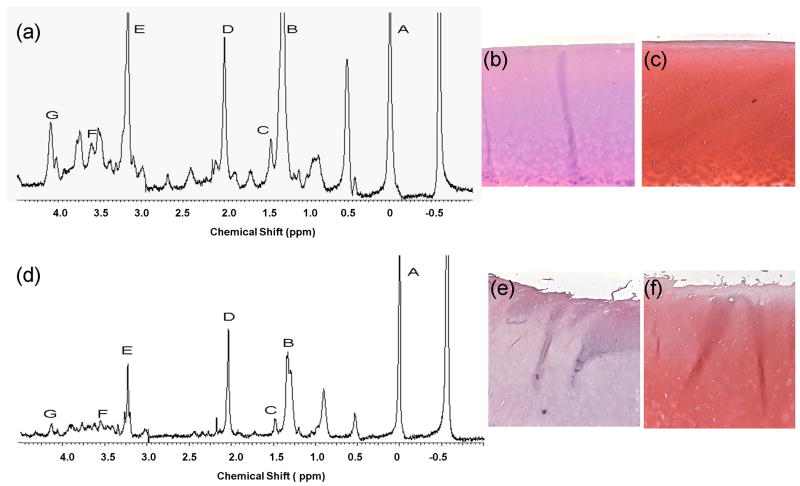
1H and 13C MAS studies have been used repeatedly to characterize nasal, articular and patellar cartilage in healthy and artificially induced degradation models in guinea pig [20] and bovine subjects [19]. A significant difference in collagen and polysaccharide content has been noted between pig nasal cartilage and bovine articular cartilage [24]. In order to better understand the degradation process in human cartilage, the work described here used 1H HR-MAS spectroscopy to characterize healthy and osteoarthritic cartilage from human subjects, with the aim to detect the presence of bio-markers of OA that may help stage disease progression and also understand the sequence of events involved in the cartilage degradation process. The focus was on N-acetyl resonance as a spectroscopic marker of proteoglycan content and alanine and glycine as markers for collagen content [15, 19]. Collagen moieties are the rigid and immobile components of cartilage, whereas the proteoglycan molecules have highly mobile side chains. The NMR signals from large and immobile components of chondroitin sulfate result in broad resonances with rapid transverse (T2) relaxation rates in the region of 3.5 – 4 ppm. The CPMG sequence with an echo time of 144 ms allows these relatively immobile spins to relax and provides high resolution spectra of the more mobile components like the proteoglycan side chain molecules. Figure 2 (a) and (d) shows the 1-dimensional (1D) 1H HR-MAS spectrum of healthy and OA cartilage specimens at 500 MHz. The assignment shown in Figure 2 is based in part on published work [16-17, 19, 22]. The HR-MAS spectra indicate a large N-acetyl resonance located at 2.04 ppm that arises from the CS side chains of proteoglycan. The lactate methyl resonance at 1.33 ppm (Lac-CH3) and the neighboring lipid alkyl methylene resonance at 1.31 ppm, several amino acids namely alanine (1.47 ppm), overlapping peaks of isoleucine, leucine and valine (0.9-1.04 ppm) are also observable. Earlier work also suggests that the resonances in the 3.21-3.25 ppm region characteristic of the choline trimethylammonium head group could arise from phosphatidylcholines, which are an integral component of cell membranes [17-18]. The resonances between 3.5 – 4 ppm can be attributed to the carbohydrate moieties in CS [22]. The glycine (3.55 ppm) originating from the collagen and the lactate methyne resonance (Lac-CH) is visible at 4.12 ppm, which may overlap with signal from CS groups in the same region [16, 19]. Elevated levels of lactate have been detected in synovial fluid rheumatoid arthritis (RA) patients [25]. This has been attributed to the high level of anaerobic metabolism that exists in the inflamed joint resulting in the accumulation of lactate as a by-product [26]. The high lactate levels are a characteristic feature of the hypoxic conditions that exist in the inflamed RA joint. However it has been observed in previous studies that a similar anaerobic and hypoxic physiological environment, although to a milder extent is present in healthy intra-articular space as well [25].
FIG 2.
Representative CPMG spectra of (a) healthy, (d) OA cartilage. Labeled peaks are as follows: A—TSP (0 ppm), B—Lactate/lipid (1.33 ppm), C—Alanine (1.47 ppm), D—N-acetyl (2.03 ppm), E—Choline (3.22 ppm), F—Glycine (3.55 ppm), G—Lactate methyne (4.12 ppm). The N-acetyl peak is a marker for PG [19], found in Keratan Sulfate (KS) and Chondroitin Sulfate (CS), constituents of PG. Alanine and Glycine are amino acids that constitute about 44% of collagen fibers [19]. (b), (c) and (e), (f) show the corresponding H & E and Safranin-O histology slides for healthy and OA cartilage respectively. The OA cartilage showed significant surface irregularities and loss of safranin-O staining ((e) and (f)) compared to the control ((b) and (c)).
Morphologically, the OA specimens appeared to be degenerated (less intact) to the naked eye and less rigid when compared to healthy control cartilage retrieved from cadaver knees. Histology from representative samples also confirmed the cartilage health status in control and OA samples, as shown in Figure 2. The OA cartilage showed significant surface irregularities and loss of safranin-O staining (Figure 2 (e) and (f)) compared to the control (Figure 2 (b) and (c)). Due to the degenerated nature of the OA cartilage the NMR spectral resolution of the OA specimens was higher than healthy cartilage specimens as can be observed visually from the representative spectra in figure 2 (a) and (d). This improved resolution enabled reliable quantification of smaller peaks like alanine (1.47 ppm) located in close proximity to larger resonances like lactate (1.33 ppm).
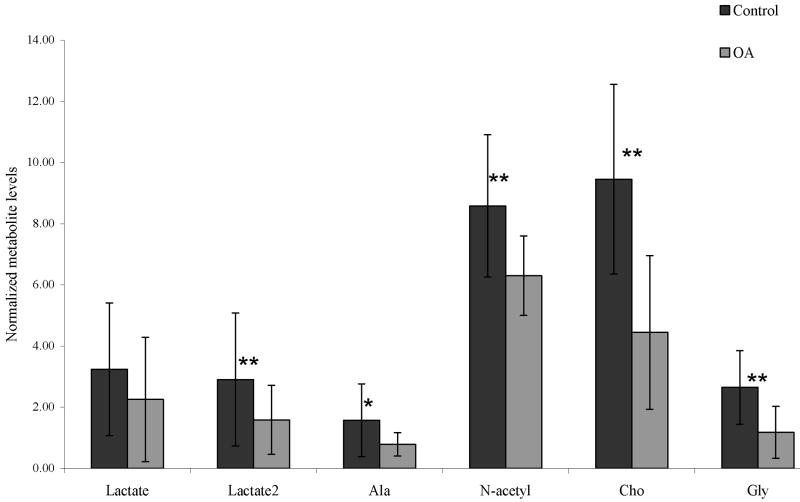
A statistically significant decrease in N-acetyl, alanine, choline, glycine and lactate methyne (4.12 ppm) was observed in OA cartilage compared to controls, as shown in Figure 3. Synovitis, which includes alterations of the joint capsule, inflammation of the synovial membrane, joint swelling and joint effusion are involved in OA pathophysiology [27]. We propose that joint effusion and increased joint fluid levels in OA patients may result in cartilage degradation products to be washed out into the synovial fluid thereby reducing the measured metabolite levels in OA cartilage. As a result a decrease in proteoglycan (PG) content in OA cartilage can be attributed to the decrease in N-acetyl levels which is considered a marker for PG in cartilage [19]. Depletion of the collagen framework in OA may result in decreased levels of alanine and glycine (amino acids of collagen). As mentioned earlier the choline signal may arise from the phosphatidylcholines which are an integral component of cell membranes. Washing out of the cartilage degradation by-products (disintegrated cell membrane components) into the synovial fluid might be the cause of decreased choline metabolite levels in OA.
FIG 3.
Comparison of mean normalized metabolite concentrations between control and OA cartilage. Results are expressed as means ± standard deviation. (Statistical significance between control and OA group, * p<0.05, **p<0.005)
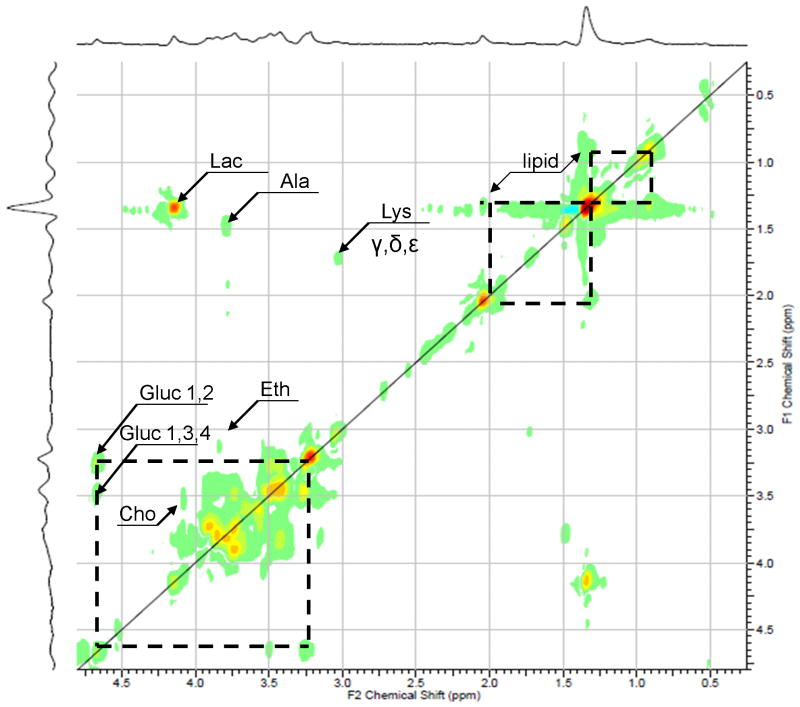
Figure 4 shows the HR-MAS TOCSY spectrum of healthy cartilage sample acquired at 500 MHz at 11.7 T. The choline cross peak (3.55 × 4.07 ppm) present in the TOCSY spectra was used to verify the assignment of the choline resonance in the 1D CPMG spectra. Prior work on prostrate and cartilage tissue and the respective chemical-shift values was used to assign this spectrum [16, 19, 23]. In addition to cross peaks corresponding to choline, ethanolamine, lactate, alanine, lysine and lipid the TOCSY reveals the presence of free glucuronic acid, one of the monomeric components of CS [28].
FIG 4.
A rotor-synchronized adiabatic 1H TOCSY spectrum of healthy cartilage specimen acquired with a mixing time of 40 ms. Cho, Choline; Lac, lactate; Ala, Alanine; Gluc, Glucuronic acid (constituent of chondroitin sulfate);
Reproducibility and spatial variation of HR-MAS measures in cartilage
Table 1 provides the root mean square (RMS) coefficients of variation (CV, %) between repeated measurements, indicating that most metabolites could be measured with reasonable precision, however the increase in CV in case of the lactate resonance at 1.3 ppm could be due to contamination from lipid resonances in the region and the high variability in case of the lactate methyne resonance at 4.12 ppm could be attributed to low-signal-to-noise ratio in the region. Also included in Table 1 is the difference in metabolite levels in cartilage samples taken from adjacent regions in a given condyle. The results indicate that cartilage tissue is extremely heterogeneous in nature. The high degree of spatial variation in alanine content in cartilage samples taken from adjacent locations might indicate the heterogeneous nature of collagen framework and its degradation in cartilage tissue. This heterogeneity in the collagen network may be a result of uneven loading of the joint and the condyles that may trigger a variation in the biochemical composition of cartilage and including the collagen framework.
Table 1.
Reproducibility measured by the root mean square coefficients of variation for quantification of metabolites using HR-MAS for n=4 in 2 trials, and the range of spatial variability for 4 pairs of cartilage punches taken from adjacent locations in the condyles expressed in terms of % coefficient of variation.
| Metabolite | Lactate (1.33 ppm) | Lactate methyne (4.12 ppm) | Alanine (1.47 ppm) | N-acetyl (2.04 ppm) | Choline (3.21 – 3.25 ppm) | Glycine (3.55 ppm) |
|---|---|---|---|---|---|---|
| Reproducibility (RMS CV %) | 9.8 | 17.3 | 2.0 | 4.5 | 9.2 | 6.0 |
| Range of Spatial variability (CV %) | 1-38 | 0-12 | 13-33 | 2-5 | 3-13 | 0-16 |
Biochemical Analysis of Cartilage Samples
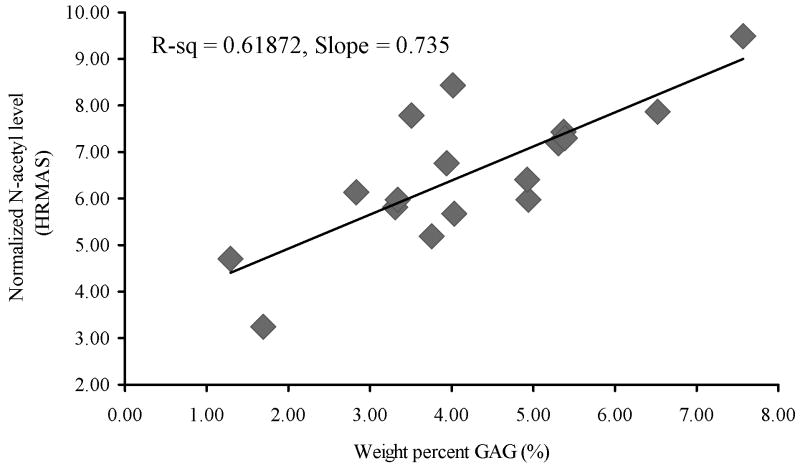
Figure 5 shows the correlation of N-acetyl levels with the proteoglycan content measured using DMMB assays in the same cartilage specimens following the spectroscopy measurements. A significant positive correlation with R-square value of 0.6187 was measured thereby confirming that N-acetyl can be considered as a marker for proteoglycan contents in cartilage.
FIG 5.
Correlation between N-acetyl as measured by HR-MAS and GAG levels measured by biochemical assay.
A previous HR-MAS study in vertebral disc also showed correlation between HR-MAS derived metabolic levels, including glycine and hydroxyproline and collagen contents measured by biochemical assays [15]. In addition, compared to conventional biochemical assays that provide only the total collagen content and cannot differentiate between the amounts of degraded collagen versus intact collagen, HR-MAS may provide more specific information related with different stages of collagen degradation by quantifying the resonances associated with the collagen content, their peak areas as well as line-width. The authors would like to acknowledge that one of the limitations of this study is that the metabolite resonances have not been corrected for their NMR relaxation times. However all healthy and OA cartilage samples were acquired with the same echo time (TE=144 ms) and as a result the comparison of metabolite concentrations between the two groups that have all been normalized to the ERETIC signal is still valid. The present study is also limited by the fact that no experiments were performed on quantifying collagen contents using biochemical assays or on evaluating collagen structures using techniques such as polarized light. Such measures are warranted in future studies to confirm the correlation between HR-MAS measures and collagen degradation in cartilage.
Conclusion
1H HR-MAS NMR is a non-destructive tool for analyzing the metabolite content in cartilage. This is particularly useful in studies wherein NMR analysis can be followed by bio-chemical, histological, immunohistochemical and genetic analyses of the same tissue specimens if necessary. The study described here presents findings of the application of 1H HR-MAS NMR spectroscopy at 11.7 T to characterize OA cartilage specimens obtained from advanced OA patients while they undergo knee replacement surgery and compare the spectral signature of OA cartilage with cartilage specimens harvested from non-osteoarthritic healthy cadaver knees (control). To our best knowledge, this is the first study documenting metabolic changes in human OA cartilage using HR-MAS NMR.
A statistically significant difference in the alanine (1.47 ppm), N-acetyl (2.04 ppm), choline (3.25 ppm) and glycine (3.55 ppm) metabolite levels is observed between healthy and OA specimens. The results of the study indicate that a decrease in the intensity of N-acetyl resonance occurs in later stages of OA. The positive correlation of the N-acetyl levels as measured by 1H HR-MAS NMR spectroscopy with the total proteoglycan content in the same cartilage specimens as measured by the GAG assay indicate that N-acetyl can serve as an important bio-marker of OA disease progression. A decrease in the alanine concentration in OA may be attributed to the degradation of the collagen framework with disease progression and thereby loss of the degradation products that are transported into the synovial cavity.
This work demonstrates that HR-MAS spectroscopic analysis is highly reproducible and is capable of detecting differences in levels of important metabolites between healthy and OA cartilage specimens from human subjects. High resolution NMR spectra allow identification of important metabolic markers involved in disease progression in OA. This enables a better understanding of the mechanism of cartilage degradation in OA. Information regarding detectable markers of disease progression will then make available novel techniques for evaluation of new treatment modalities for patients affected by the disease.
Acknowledgments
This work was supported by the NIH/NIAMS R21AR056773 grant and seed grant funding from the Department of Radiology, University of California-San Francisco, San Francisco, California. The authors would like to thank Joseph Schooler and Fei Liang for helping with specimen collection.
Grant sponsor: NIH/NIAMS R21AR056773
Abbreviations used
- OA
osteoarthritis
- 1D
one-dimensional
- 2D
two-dimensional
- CS
chondroitin sulfate
- HRMAS
high resolution magic angle spinning
- PG
proteoglycan
- GAG
glycosaminoglycan
- TOCSY
total correlation spectroscopy
- ERETIC
Electronic Reference To access In-vivo Concentrations
- TKA
total knee arthoplasties
References
- 1.Jackson D, Simon T, Aberman H. Symptomatic articular cartilage degeneration: the impact in the new millennium. Clin Orthop Relat Res. 2001;391(suppl):14–25. [PubMed] [Google Scholar]
- 2.Holt HL, Katz JN, Reichmann WM, Gerlovin H, Wright EA, Hunter DJ, Jordan M, Kessler CL, Losina E. Forecasting the burden of advanced knee osteoarthritis over a 10-year period in a cohort of 60-64 year-old US adults. Osteoarthritis Cartilage. 2011;19:44–50. doi: 10.1016/j.joca.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aigner T, Rose J, Martin J, Buckwalter J. Aging theories of primary osteoarthritis: from epidemiology to molecular biology. Rejuvenation Res. 2004;7:134–145. doi: 10.1089/1549168041552964. [DOI] [PubMed] [Google Scholar]
- 4.Roberts J, Burch TA. Osteoarthritis prevalence in adults by age, sex, race, and geographic area. Vital Health Stat 11. 1966;15:1–27. [PubMed] [Google Scholar]
- 5.Milgram JW. Morphologic alterations of the subchondral bone in advanced degenerative arthritis. Clin Orthop Relat Res. 1983;173:293–312. [PubMed] [Google Scholar]
- 6.Andrew ER. The narrowing of NMR spectra of solids by high speed specimen rotation and the resolution of chemical shift and spin multiplet structures for solids. Progr Nucl Magn Reson Spectrosc. 1971;8:1–39. [Google Scholar]
- 7.Wilson M, Davies NP, Grundy RG, Peet AC. A quantitative comparison of metabolite signals as detected by in vivo MRS with ex vivo 1H HR-MAS for childhood brain tumours. NMR Biomed. 2009;22:213–219. doi: 10.1002/nbm.1306. [DOI] [PubMed] [Google Scholar]
- 8.Opstad KS, Bell BA, Griffiths JR, Howe FA. An investigation of human brain tumour lipids by high-resolution magic angle spinning 1H MRS and histological analysis. NMR Biomed. 2008;21:677–685. doi: 10.1002/nbm.1239. [DOI] [PubMed] [Google Scholar]
- 9.Tzika AA, Astrakas L, Cao H, Mintzopoulos D, Andronesi OC, Mindrinos M, Zhang J, Rahme LG, Blekas KD, Likas AC, Galatsanos NP, Carroll RS, Black PM. Combination of high-resolution magic angle spinning proton magnetic resonance spectroscopy and microscale genomics to type brain tumor biopsies. Int J Mol Med. 2007;20:199–208. [PubMed] [Google Scholar]
- 10.Opstad KS, Wright AJ, Bell BA, Griffiths JR, Howe FA. Correlations between in vivo (1)H MRS and ex vivo (1)H HRMAS metabolite measurements in adult human gliomas. J Magn Reson Imaging. 2010;31:289–297. doi: 10.1002/jmri.22039. [DOI] [PubMed] [Google Scholar]
- 11.Cheng LL, Ma MJ, Becerra L, Ptak T, Tracey I, Lackner A, Gonzalez RG. Quantitative neuropathology by high resolution magic angle spinning proton magnetic resonance spectroscopy. Proc Natl Acad Sci U S A. 1997;94(12):6408–13. doi: 10.1073/pnas.94.12.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sitter B, Sonnewald U, Spraul M, Fjosne HE, Gribbestad IS. High-resolution magic angle spinning MRS of breast cancer tissue. NMR Biomed. 2002;15:327–337. doi: 10.1002/nbm.775. [DOI] [PubMed] [Google Scholar]
- 13.Duarte IF, Stanley EG, Holmes E, Lindon JC, Gil AM, Tang H, Ferdinand R, McKee CG, Nicholson JK, Vilca-Melendez H, Heaton N, Murphy GM. Metabolic assessment of human liver transplants from biopsy samples at the donor and recipient stages using high-resolution magic angle spinning 1H NMR spectroscopy. Anal Chem. 2005;77:5570–5578. doi: 10.1021/ac050455c. [DOI] [PubMed] [Google Scholar]
- 14.Cheng LL, Wu C, Smith MR, Gonzalez RG. Non-destructive quantitation of spermine in human prostate tissue samples using HRMAS 1H NMR spectroscopy at 9.4 T. FEBS Lett. 2001;494:112–116. doi: 10.1016/s0014-5793(01)02329-8. [DOI] [PubMed] [Google Scholar]
- 15.Keshari KR, Lotz JC, Kurhanewicz J, Majumdar S. Correlation of HR-MAS spectroscopy derived metabolite concentrations with collagen and proteoglycan levels and Thompson grade in the degenerative disc. Spine. 2005;30:2683–2688. doi: 10.1097/01.brs.0000188256.88859.9c. [DOI] [PubMed] [Google Scholar]
- 16.Keshari KR, Zektzer AS, Swanson MG, Majumdar S, Lotz JC, Kurhanewicz J. Characterization of intervertebral disc degeneration by high-resolution magic angle spinning (HR-MAS) spectroscopy. Magn Reson Med. 2005;53:519–527. doi: 10.1002/mrm.20392. [DOI] [PubMed] [Google Scholar]
- 17.Schiller J, Naji L, Huster D, Kaufmann J, Arnold K. 1H and 13C HR-MAS NMR investigations on native and enzymatically digested bovine nasal cartilage. MAGMA. 2001;13:19–27. doi: 10.1007/BF02668647. [DOI] [PubMed] [Google Scholar]
- 18.Schiller J, Huster D, Fuchs B, Naji L, Kaufmann J, Arnold K. Evaluation of cartilage composition and degradation by high-resolution magic-angle spinning nuclear magnetic resonance. Methods Mol Med. 2004;101:267–285. doi: 10.1385/1-59259-821-8:267. [DOI] [PubMed] [Google Scholar]
- 19.Ling W, Regatte RR, Schweitzer ME, Jerschow A. Characterization of bovine patellar cartilage by NMR. NMR Biomed. 2008;21:289–295. doi: 10.1002/nbm.1193. [DOI] [PubMed] [Google Scholar]
- 20.Borel M, Pastoureau P, Papon J, Madelmont JC, Moins N, Maublant J, Miot-Noirault E. Longitudinal profiling of articular cartilage degradation in osteoarthritis by high-resolution magic angle spinning 1H NMR spectroscopy: experimental study in the meniscectomized guinea pig model. J Proteome Res. 2009;8:2594–2600. doi: 10.1021/pr8009963. [DOI] [PubMed] [Google Scholar]
- 21.Swanson MG, Zektzer AS, Tabatabai ZL, Simko J, Jarso S, Keshari KR, Schmitt L, Carroll PR, Shinohara K, Vigneron DB, Kurhanewicz J. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn Reson Med. 2006;55:1257–1264. doi: 10.1002/mrm.20909. [DOI] [PubMed] [Google Scholar]
- 22.Mucci A, Schenetti L, Volpi N. 1H and 13C nuclear magnetic resonance identification and characterization of components of chondroitin sulfates of various origin. Carbohydrate Polym. 2000;41:37–45. [Google Scholar]
- 23.Swanson MG, Keshari KR, Tabatabai ZL, Simko JP, Shinohara K, Carroll PR, Zektzer AS, Kurhanewicz J. Quantification of choline- and ethanolamine-containing metabolites in human prostate tissues using 1H HR-MAS total correlation spectroscopy. Magn Reson Med. 2008;60:33–40. doi: 10.1002/mrm.21647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huster D, Schiller J, Naji L, Kaufmann J, Arnold K. NMR Studies of Cartilage - Dynamics, Diffusion, Degradation. In: Riehold H, Dieter M, Andreas P, editors. Molecules in interaction with surfaces and interfaces. Vol. 634. Springer; 2004. pp. 465–503. [Google Scholar]
- 25.Naughton DP, Haywood R, Blake DR, Edmonds S, Hawkes GE, Grootveld M. A comparative evaluation of the metabolic profiles of normal and inflammatory knee-joint synovial fluids by high resolution proton NMR spectroscopy. FEBS Lett. 1993;332:221–225. doi: 10.1016/0014-5793(93)80636-9. [DOI] [PubMed] [Google Scholar]
- 26.James MJ, Cleland LG, Rofe AM, Leslie AL. Intraarticular pressure and the relationship between synovial perfusion and metabolic demand. J Rheumatology. 1990;17:521–527. [PubMed] [Google Scholar]
- 27.Sellam J, Berenbaum F. The role of synovitis in pathophysilogy and clinical symptoms of osteoarthritis. Nat Rev Rheumatol. 2010;6:625–635. doi: 10.1038/nrrheum.2010.159. [DOI] [PubMed] [Google Scholar]
- 28.Richmond ME, Deluca S, Silbert JE. Biosynthesis of Chondroitin Sulfate. Microsomal Acceptors of Sulfate, Glucuronic Acid and N-acetylgalactosamine. Biochemistry. 1973;12:3898–3902. doi: 10.1021/bi00744a017. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Cheng J, Lin K, Saadat E, Bolbos RI, Ries MD, Hovai A, Link TM, Majumdar S. Quantitative MRI using T1rho and T2 in human osteoarthritic cartilage specimens: Correlation with biochemical measurements and histology. Magn Reson Imag. 2011;29(3):324–334. doi: 10.1016/j.mri.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]