Abstract
Strain-specific differences in susceptibility to mouse adenovirus type 1 (MAV-1) are linked to the quantitative trait locus Msq1 on mouse chromosome 15. This region contains 14 Ly6 or Ly6-related genes, many of which are known to be expressed on the surface of immune cells, suggesting a possible role in host defense. We analyzed these genes for polymorphisms between MAV-1-susceptible and MAV-1-resistant inbred mouse strains. Sequencing of cDNAs identified 12 coding region polymorphisms in 2010109I03Rik, Ly6e, Ly6a, Ly6c1, and Ly6c2, six of which were non-synonymous and five of which were previously unlisted in dbSNP Build 132. We also clarified sequence discrepancies in GenBank for the coding regions of I830127L07Rik and Ly6g. Additionally, Southern blotting revealed size polymorphisms within the DNA regions of Ly6e, Ly6a, and Ly6g. Collectively, these genetic variations have implications for the structure, function, and/or expression of Ly6 and Ly6-related genes that may contribute to the observed strain-specific differences in susceptibility to MAV-1.
INTRODUCTION
To more fully understand viral pathogenesis, it is pivotal to consider the underlying host genetic factors that influence disease progression. Mouse adenovirus type 1 (MAV-1) is an endotheliotropic virus that disrupts the blood-brain barrier and can cause fatal encephalitis in mice (Gralinski et al. 2009; Guida et al. 1995). Inbred strains of mice show varying degrees of susceptibility to MAV-1. Upon infection, resistant BALB/cJ mice show few pathological signs, low brain viral loads, and more restricted chemokine expression than susceptible strains (Charles et al. 1999; Guida et al. 1995; Spindler et al. 2010). In contrast, susceptible SJL/J, 129S6SvEv/Tac, and C57BL/6J mice exhibit high viral loads in the brain and spleen, with the infection ultimately leading to mortality (Guida et al. 1995; Spindler et al. 2001; Spindler et al. 2010). We recently identified the major quantitative trait locus for susceptibility to MAV-1 infection, Msq1, and it includes 14 Ly6 and Ly6-related genes on mouse Chr 15 (Spindler et al. 2010). The Ly6 genes have also been associated with host susceptibility to encephalitic West Nile virus, HIV-1, and avian herpesvirus Marek’s disease virus infections, suggesting a broader role for them in viral host defense (Brass et al. 2008; Krishnan et al. 2008; Liu et al. 2003; Loeuillet et al. 2008). A better understanding of these Ly6 and Ly6-related genes will provide further insight into the pathogenesis of MAV-1 infection and other viral diseases.
The LY6 gene products are glycosylphosphatidylinositol-anchored proteins that are expressed primarily on hematopoietic cells including monocytes, granulocytes, and lymphoid precursor cells (reviewed in Bamezai 2004). LY6 proteins have a LY6/urokinase-type plasminogen activator receptor domain (uPAR) consisting of 10 conserved cysteine residues with varying numbers of residues between them; the last cysteine is followed by an asparagine (Behrendt et al. 1991; Ploug and Ellis 1994). Limited data have been published on the biological significance of these genes and their gene products, particularly in relation to their potential immune function (Bamezai 2004). Notably, LY6E (also known as TSA-1 and Sca-2) is critical in T-cell differentiation and regulation of the T-cell receptor-signaling pathway (Saitoh et al. 1995). LY6C expressed on the surface of CD8+ T cell subsets is critical for endothelial adhesion and can help stimulate IL-2 production (Hänninen et al. 1997). In addition, LY6G has been used extensively as a marker of granulocytes (Fleming et al. 1993a). However, there is a lack of data about the entire spectrum of the Ly6 and Ly6-related genes and their varying immune-related function(s), particularly with respect to viral infection.
In this study, we sought to identify polymorphisms in the Ly6 and Ly6-related genes of the Msq1 interval that may contribute to susceptibility to MAV-1. We sequenced the coding region of transcripts from resistant (BALB/cJ) and susceptible (SJL/J) mice and identified protein sequence polymorphisms. Additionally, we analyzed the genomic DNA and identified three size polymorphisms in Ly6 genes between resistant and susceptible strains that may influence expression or splicing. These polymorphisms provide information about the genetic variation in the Ly6 and Ly6-related genes of the Msq1 interval that may relate to the differential host response during MAV-1 infection between resistant and susceptible strains.
METHODS
RNA Isolation
Inbred mice were obtained from Jackson Laboratory and Taconic. Strains used that are susceptible to MAV-1 were SJL/J, 129S6SvEv/Tac, C57BL/6J, and CSJL/J (BALB/cJ × SJL/J) F1 mice. The resistant strain used was BALB/cJ. Euthanized mice were perfused with cold 1X PBS, and brain, thymus, spleen, and kidney were removed and stored at −20°C. For each sample, 100 mg of tissue was placed in 1 ml of Tri-Reagent (Molecular Research Company). Samples were homogenized in a BioSpec bead-beater for 60 seconds twice, with an intervening 60 seconds on ice. RNA was isolated from cells using Tri-Reagent according to the manufacturer's instructions. The isolated RNA was resuspended in 100 µl of H2O and stored at −20 °C. RNA was quantified by NanoDrop spectrophotometry.
cDNA Preparation
5 µg of RNA was added to 3 µl of 1:10 diluted random hexamers (Roche) or 1:3 diluted oligo(dT) (Invitrogen), and brought to a final volume of 13 µl with H2O. This reaction was incubated at 65 °C for 10 minutes and cooled quickly on ice. 4 µl of 5× M-MLV buffer, 2 µl 100 mM DTT, 1 µl 10 mM dNTPs, 2.4 µl H2O, 0.3 µl SUPERase-In (Ambion), and 0.3 µl M-MLV reverse transcriptase (Invitrogen) were added to each mixture and incubated at 37 °C for 1 hour.
PCR amplification of cDNA
20–50 µl PCR reactions were performed using appropriate scaling of the following mixture: 4 µl of cDNA, 6 µl 5× Promega Green Buffer, 0.75 µl 10 mM dNTPs, 1.5 µl of each primer diluted to 10 µM, 0.15 µl of 5 units/µl Promega GoTaq, and 16.1 µl of H2O. The primers and conditions for these reactions are listed in Table 1. Primers were designed based on the NCBI 37/mm9 build (C57BL/6J sequence). Due to the potential for strain-specific polymorphisms within the primer regions altering annealing efficiency, additional primers at different loci were tested for those genes where product was not initially observed. PCR products were purified directly from the reaction or by gel purification from 1% agarose gels using the QIAGEN QIAquick system and quantified by NanoDrop spectrophotometry. Brain was the primary tissue used in these experiments due to the high MAV-1 viral load in this tissue during infection (Kring et al. 1995; Smith et al. 1998). If the expected product was not observed, thymus, spleen, and/or kidney cDNA preparations were substituted.
Table 1.
PCR primers and conditions for amplification from cDNA
| Genea | Primer 1 | Primer 2 | Anneal | Conditionsb |
|---|---|---|---|---|
| 2010109I03Rik | 5' CTGCTTGCTCTCATTGCCCA | 5' TTCAAGAAGTGCCCCACCCA | 62°C | 45/45/75 × 45 |
| Ly6e | 5' TCTTTGGCTTGCGAACCTTCA | 5' CTTTGCCCACCCCAGAATCC | 66°C | 30/30/60 × 40 |
| Ly6i | 5' TTGGATGGCTGAGCCGTGTG | 5' GGGTGGGGACCATCACATCA | 66°C | 30/30/60 × 40 |
| Ly6a | 5' GCAACCTGGTCAGAGAGGAA | 5' GGAGAACAAAGGGTTTATTGGA | 60°C | 30/30/60 × 40 |
| Ly6c1 | 5' TGGTGTCAGGAGGGAGCTGCTA | 5' GGCATTTACCAAGCAGGGGC | 66°C | 30/30/60 × 40 |
| Ly6c2 | 5' TGAGGATGGACAGTACTCACG | 5' GAGTGGTTGCACATGAGGAT | 60°C | 30/30/30 × 40 |
| I830127L07Rik | 5' AGGACTGGTGTCAGGAGGGA | 5' GCCAGGAATTGCAAGCCACA | 60°C | 30/30/60 × 45 |
| Ly6g | 5' GGTCAGAGAGGAACCCTTCTCC | 5' GGGTGCCTATACAGCAGGGGTT | 60°C | 30/30/60 × 45 |
| BC025446 | 5' GCCCAGGGATGTGATTGGTG | 5' CCAGCACCTTGGCCACATTC | 60°C | 30/30/60 × 40 |
| Ly6h | 5' AGCCAAAGCTCTTCCGAGAATCTG | 5' CAACAGGCTTAGGGGACAAGCTC | 67°C | 45/45/75 × 45 |
| Gpihbp1 | 5' AGCCTGAGCCAGGATGAAGG | 5' GCGTCCTGGTTGGAGGAGAA | 65°C | 30/30/60 × 40 |
Unique transcripts could not be amplified for Ly6f, Gm10238, or 9030619P08Rik with ≥ 2 primer sets (data not shown)
PCR reaction conditions listed as follows: denature(sec)/anneal(sec)/extension(sec) × number of cycles
Sequencing and analysis
PCR products were sequenced at the University of Michigan DNA Sequencing Core. Sequence identity was confirmed via in silico BLAST search (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Chromatograms were analyzed with FinchTV (Geospiza, Inc.). Sequence alignments were performed using the ClustalW method and analyzed with the DNASTAR Lasergene software suite.
Probe Preparation
Probes for Southern blotting were prepared by random primer labeling. Briefly, 400 ng of cDNA-derived PCR product was labeled with 50 µCi α-32P dATP (Perkin Elmer) by Klenow extension. Probes were purified with Sephadex G-25 Quick Spin Columns (Roche) and denatured by adding 0.1 volumes of 1 M NaOH at 37°C for 5 minutes. The hybridization mixture was prepared by adding 1–5 × 106 cpm/mL of radiolabeled probe to 10 mL of 7% SDS/0.5 M NaPO4/1 mM EDTA with 100–250 µg/mL of denatured herring sperm DNA.
Southern Blotting
Tails and brains from BALB/cJ, SJL/J, C57BL/6J, and 129S6SvEv/Tac were digested overnight at 55°C with 15 µl of 10 mg/ml proteinase K in 350 µl tail buffer (0.5 M EDTA/1 M Tris pH 8.0/10% SDS). High quality genomic DNA was isolated by phenol/chloroform extraction (Schauwecker et al. 2004). 20 µµg of each genomic DNA sample was digested with AflII (for analysis of Ly6a), BsmI (Ly6c2, Ly6i, Ly6g, Ly6h), BspHI (Ly6e), PflFI (BC025446), SacI (Gpihbp1), or XcmI (2010109I03Rik, Ly6c1, I830127L07Rik). Digest products were ethanol precipitated and separated on 1% agarose gels at 40 V for 16 hrs. Gels were denatured for 30 minutes in 0.4 M NaOH/0.8 M NaCl, and alkaline transferred overnight in 0.4 M NaOH to Bio-Rad Zeta-Probe GT membranes. Blots were rinsed twice in 3X SSC and pre-hybridized with 10 mL of 7% SDS/0.5 M NaPO4/1 mM EDTA for 1 hr at 60°C. Pre-hybridization solution was replaced with the hybridization mixture and allowed to incubate overnight at 60°C. Blots were washed with varying concentrations of SSPE (6X–0.1X)/0.1% SDS at 60–65°C, exposed to X-ray film for 1–5 days with intensifying screens, and developed. Bacterial artificial chromosomes obtained from Children's Hospital Oakland Research Institute contained known portions of the Msq1 region and were used as positive and negative controls. These DNAs were originally generated from spleens of 129S6SvEv/Tac mice and cloned into the pBACe3.6 vector as described (http://bacpac.chori.org).
RESULTS
Coding region polymorphisms in Ly6 and Ly6-related genes
Using cDNA derived from brain, thymus, spleen, or kidney, we PCR amplified and sequenced the coding regions of 11 of the 14 Ly6 and Ly6-related genes in Msq1 (nt 74,680,379 to 75,432,404, mouse chromosome 15, NCBI genome build 37/mm9, July 2007) (see map, Fig. 1) and identified several polymorphisms between BALB/cJ and SJL/J mice (Table 2). These included both synonymous and non-synonymous single nucleotide polymorphisms, the latter causing amino acid substitutions as drastic as glycine to aspartate (LY6A) and glycine to glutamate (LY6C1). We were unable to identify unique transcripts in either BALB/cJ or SJL/J for three genes: Ly6f, Gm10238 (formerly Ensembl Accession AC116498.15-201), and 9030619P08Rik. For Ly6f, this corresponds with previous unsuccessful attempts to amplify a transcript from cDNA (Fleming et al. 1993b). Unique transcripts for Ly6i and I830127L07Rik were only identified in SJL/J cDNA pools. For these five genes from which we could amplify no transcript or only the SJL/J transcript, we attempted amplification with one or more additional primer sets, and with variations in PCR conditions, but were unsuccessful. These transcripts may have been below the limit of detection for this assay. The BALB/c sequence may be too degenerate relative to the C57BL/6 sequences (for the several primer sets we used) to be amplified, or the genes may be missing from the BALB/c genome. Alternatively, it is possible that these genes are not highly expressed in the tested tissues or that they are pseudogenes that do not produce transcripts. Data suggest this may be the case for Gm10238, which has been predicted as a pseudogene by computer analysis (Ensembl 63 Accession ENSMUSG00000068348). We sequenced Ly6a, Ly6e, Ly6h, and I830127L07Rik in another MAV-1-susceptible mouse strain, 129S6SvEv/Tac. The 129S6SvEv/Tac coding region sequences were identical to those of SJL/J for these genes (data not shown). Additionally, we compared the polymorphisms observed between BALB/cJ and SJL/J with the published C57BL/6J coding region sequences (NCBI Build 37/mm9). C57BL/6J, a MAV-1-susceptible strain, was identical to SJL/J for all polymorphic loci (data not shown). Thus, where examined, coding region differences among strains correlated with mouse strain susceptibility to MAV-1 infection.
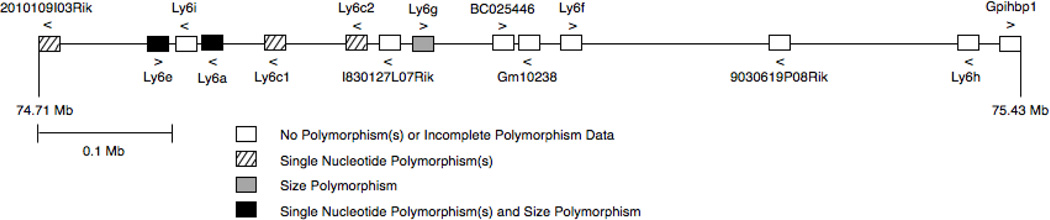
Figure 1. Msq1 map.
Polymorphism data from this study are displayed on the gene map of the Msq1 portion of the Ly6 region of Chr 15, based on NCBI Build 37/mm9. Boxes indicate Ly6 or Ly6-related genes. Arrowheads denote the direction of transcription of the gene along the chromosome (forward or reverse strand).
Table 2.
Coding Region Polymorphismsa
| Geneb | Locusc | dbSNP IDd | BALB/cJ | SJL/J | ||
|---|---|---|---|---|---|---|
| Nucleotide | Amino Acid | Nucleotide | Amino Acid | |||
| 2010109I03Rik | 74,711,196 | rs32400297 | A | Alanine | G | Alanine |
| 74,710,331 | Unlistede | A | Isoleucine | G | Valine | |
| 74,710,308 | rs32206043 | T | Leucine | C | Leucine | |
| Ly6e | 74,788,796 | rs13469390 | A | Threonine | G | Threonine |
| 74,789,008 | rs13469391 | A | Alanine | G | Alanine | |
| Ly6i | No Transcriptf | Confirmed Transcriptg | ||||
| Ly6a | 74,825,909 | rs32279213 | G | Glycine | A | Aspartate |
| 74,825,818 | Unlistede | T | Asparagine | C | Asparagine | |
| 74,825,780 | Unlistede | C | Alanine | T | Valine | |
| Ly6c1 | 74,878,906 | Unlistede | G | Leucine | C | Leucine |
| 74,878,877 | Unlistede | G | Glycine | A | Glutamate | |
| Ly6c2 | 74,939,115 | rs13464501 | G | Arginine | A | Lysine |
| 74,938,993 | rs13464500 | G | Valine | A | Isoleucine | |
| I830127L07Rik | No Transcriptf | Confirmed Transcriptg | ||||
| Ly6g | No Polymorphismsh | |||||
| BC025446 | No Polymorphismsh | |||||
| Ly6h | No Polymorphismsh | |||||
| Gpihbp1 | No Polymorphismsh | |||||
Polymorphisms were not analyzed for Ly6f, Gm10238, and 9030619P08Rik due to an inability to PCR amplify these genes from cDNA pools.
Genes are listed in the order they appear on the chromosome.
Nucleotide residue on mouse Chr 15, based on NCBI 37/mm9 build (July 2007)
NCBI dbSNP Build 132
Single nucleotide polymorphism previously unlisted in NCBI dbSNP (Build 132). For these SNPs the nucleotide pairs are listed for the coding strand.
Expected transcript could not be amplified in this mouse strain.
PCR amplification identified the expected transcript in this strain; BLAST search confirmed identity as the indicated Ly6 or Ly6-related gene by comparison with NCBI 37/mm9 build (C57BL/6J sequence).
Transcripts were amplified from both BALB/cJ and SJL/J mice; no coding region polymorphisms were observed.
Identification of N-terminal sequences of two Ly6 transcripts
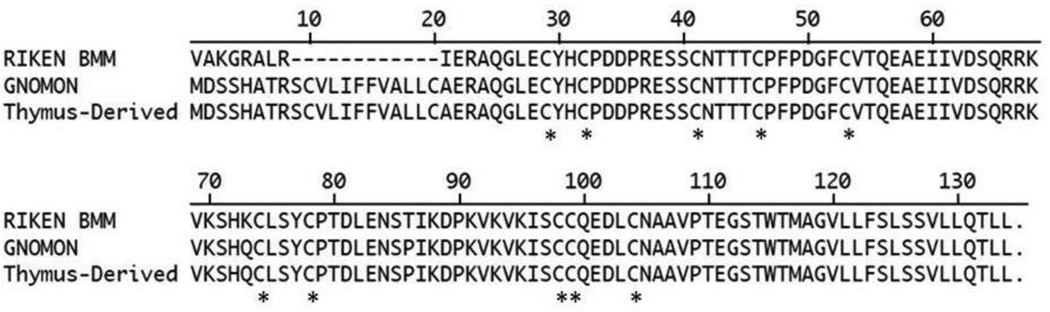
Discrepancies between predicted and cloned sequences for I830127L07Rik exist within NCBI’s GenBank. The RIKEN clone from bone marrow-derived macrophages (NCBI Accession AK153212.1) was reported to have a sequence with predicted homology to LY6A (previously termed Ly6A.2/Ly6E.1). The putative amino acid sequence from this clone lacked a start codon, suggesting that it is an incomplete or nonfunctional transcript. However, a separate computer-based GNOMON analysis found in GenBank predicted a full-length transcript for I830127L07Rik (NCBI Accession XM_001477102.1) that has an initial methionine and N-terminus homology to other Ly6 and Ly6-related genes. NCBI's GNOMON is a gene prediction system that identifies potential genes within a genome by comparison to intraspecies experimentally-derived cDNA clones or known proteins from other organisms (http://www.ncbi.nlm.nih.gov/projects/genome/guide/gnomon.shtml). While robust, computer-based genome analyses such as GNOMON can still yield errors at an appreciable rate (Nagy et al. 2008). We sought to resolve this discrepancy by determining whether the GNOMON-predicted full-length transcript existed in vivo. We isolated and sequenced an I830127L07Rik transcript from the thymus of SJL/J mice and aligned its predicted protein sequence with the bone marrow-derived macrophage and GNOMON-predicted sequences (Fig. 2). The alignment revealed a considerable divergence from the bone marrow-derived macrophage RIKEN clone sequence at the N-terminus of the protein. The thymus-derived clone corresponded to the GNOMON prediction at the N-terminus, confirming the existence of a transcript with the potential to produce a full-length LY6-like protein with a conserved LY6/urokinase-type plasminogen activator receptor domain and start codon.
Figure 2. I830127L07Rik protein alignments.
RIKEN BMM sequence is from a bone marrow macrophage-derived cDNA clone of I830127L07Rik (NCBI Accession AK153212.1). GNOMON is a predicted amino acid sequence for I830127L07Rik from a computer-based prediction using GNOMON analysis of the C57BL/6J genome (NCBI Accession XM_001477102.1). Thymus-Derived represents the putative amino acid sequence of the I830127L07Rik transcript that we amplified from the thymus of an SJL/J mouse. Asterisks (*) denote the ten conserved cysteine residues of the LY6/uPAR domain. DNASTAR Lasergene MegAlign was used to perform this ClustalW alignment. Dashes denote absent amino acids. Periods indicate a stop codon.
Until this study, the most recently published sequence for Ly6g was only partial and lacked the region encoding the N-terminus of the protein product (Ensembl 63 Accession ENSMUST00000023246). GNOMON predictions, however, indicated that a complete transcript with sequence homology to other Ly6 genes was likely (NCBI Accessions XM_001475753.2 and XM_909927.3). We designed primers based on the GNOMON predictions, and amplified a transcript for Ly6g (Table 1). Sequencing revealed a Ly6g transcript that is consistent with the GNOMON predictions and published sequences and encodes a full-length LY6-like protein with a conserved LY6/urokinase-type plasminogen activator receptor domain and start codon.
Size polymorphisms in Ly6 and Ly6-related gene regions
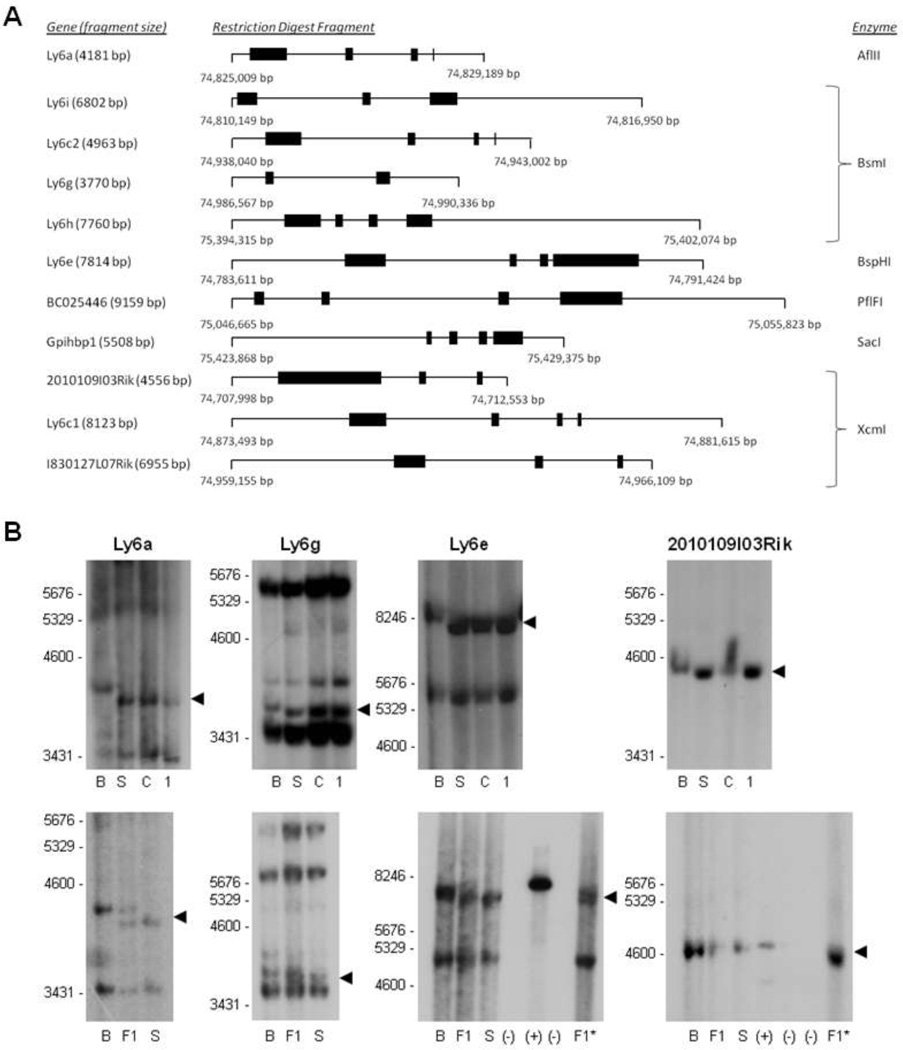
Sizable genetic polymorphisms in upstream, downstream, and intron regions can have significant implications for mRNA splicing and gene expression. We used Southern blotting to identify genomic size differences within and around the Ly6 and Ly6-related genes. Genomic DNA was isolated from BALB/cJ, SJL/J, C57BL/6J, and 129S6SvEv/Tac mice, and restriction digests were performed with enzymes that cleaved upstream and downstream of each gene (Fig. 3A). Probes were prepared from cDNA-derived PCR products, allowing us to analyze all genes except Ly6f, Gm10238, and 9030619P08Rik. Size polymorphisms were identified in the gene regions of Ly6a, Ly6e, and Ly6g in BALB/cJ relative to the other strains (Fig. 3B), which are susceptible to MAV-1 (Guida et al. 1995; Spindler et al. 2001; Spindler et al. 2010). The BALB/cJ Ly6 gene was larger than the others in each case. We confirmed these size polymorphisms with side-by-side analysis of DNAs from BALB/cJ, SJL/J and (BALBc/JxSJL/J) F1 mice (Fig. 3B, lower panel). As expected, the F1 DNA samples showed two distinct bands, corresponding to the sizes of the parental bands.
Figure 3. Genomic Size Polymorphisms.
Southern blotting was used to analyze size differences in genes in the Msq1 interval among mouse strains at the genomic level. (A) Annotations from UCSC Genome Browser, RefSeq, and Ensembl were assembled to create diagrams of the restriction fragments containing the appropriate Ly6 or Ly6-related gene. Black boxes indicate exons. Ly6e, Ly6i, and Ly6h have multiple spliced forms; the longest splice variant is depicted. Nucleotide positions of flanking restriction sites for the enzymes shown to the right are indicated below each line diagram (NCBI Build 37/mm9, Chr 15). (B) Radiolabeled probes were produced from cDNA-derived PCR products by Klenow extension for the Ly6 and Ly6-related genes indicated above each Southern blot. DNA size markers are indicated on the left of each blot. Arrowheads indicate bands of interest. B = BALB/cJ, S = SJL/J, C = C57BL/6J, and 1 = 129S6SvEv/Tac, F1 = tail-derived CSJL/J (BALB/cJ × SJL/J), F1* = brain-derived CSJL/J (BALB/cJ × SJL/J), (−) = negative control, (+) = positive control.
Although 2010109I03Rik appeared to have a slight size polymorphism (Fig. 3B, upper), this was not confirmed in a Southern blot that included F1 DNA controls (Fig. 3B, lower). The apparent size difference (Fig. 3B upper) may be due to variances in the amount of DNA among the strains present on the blot or due to an artifact of the electrophoresis. Data were equivocal for I830127L07Rik (data not shown). High background around the expected size was observed for this gene, preventing us from identifying an I830127L07Rik-specific band for comparison in the four mouse strains by Southern blotting. For all the genes analyzed by Southern blots, due to the restriction enzyme digest array used to prepare the samples, cleavage sites were varying distances upstream and downstream of the targeted genes (Fig. 3A). Therefore, these data do not indicate whether the polymorphism was within an intron or flanking the gene. Additionally, the background present in several of the blots was not unexpected, with some of the Ly6 genes having >90% coding region homology. Care was taken in selecting restriction enzymes for these analyses to avoid the generation of unintended Ly6-containing restriction fragments at the same size as the targeted fragment.
DISCUSSION
Through molecular genetic analyses we were able to identify several polymorphisms in the Ly6 and Ly6-related genes between MAV-1-susceptible and resistant inbred mouse strains. Within the coding regions, there were numerous single nucleotide polymorphisms between susceptible SJL/J and resistant BALB/cJ mice. Six of these were silent mutations, while another six resulted in amino acid changes. The amino acid changes that are most likely to alter three-dimensional protein structure were glycine in BALB/cJ to aspartate in SJL/J at residue 63 in LY6A (G63D) and glycine in BALB/cJ to glutamate in SJL/J at residue 22 in LY6C1 (G22E). The difference in size and charge between these amino acids may have a critical impact on the secondary and tertiary structures of these proteins that affect protein function. Collectively, we identified five single nucleotide polymorphisms throughout 2010109I03Rik, Ly6a, and Ly6c1 that were not previously listed in Build 132 of the NCBI dbSNP database (Table 2).
Several of the amino acid polymorphisms we observed between BALB/cJ and SJL/J strains are consistent with previous analyses of the Ly6 genes. Specifically, the Ly6a transcripts for BALB/cJ and SJL/J showed amino acid polymorphisms (G63D and A106V) that are identical to the reported sequences for Ly6E.1 and Ly6A.2, respectively (LeClair et al. 1986; Reiser et al. 1988). Additionally, the Ly6c2 transcripts for BALB/cJ and SJL/J showed amino acid polymorphisms (R85K and V126I) that are consistent with the sequences for Ly6c.1 and Ly6c.2, respectively (Bothwell et al. 1988; Palfree et al. 1988).
We have used the standardized nomenclature of the International Committee on Standardized Genetic Nomenclature for Mice (www.informatics.jax.org, revised September 2010) for the Ly6 genes in this paper; however, previous studies have in many instances been less consistent in their terminology. Early immunological studies of LY6 tissue distribution revealed two unique haplotypes, Ly6.1 (Ly6a) and Ly6.2 (Ly6b), whose gene products show distinct cellular expression patterns (Potter et al. 1980). While early Southern blot data suggested the possibility of as many as 30 Ly6 genes existing within the mouse genome, early immunological studies were limited in their ability to clarify distinct gene products versus alleles of the same gene (Bothwell et al. 1988; LeClair et al. 1986; Reiser et al. 1988). For instance, Ly6E.1 and Ly6A.2, originally thought to be two separate genes, are actually both alleles of Ly6a (LeClair et al. 1986; Reiser et al. 1988). Additionally, comparisons of Ly6c.1 and Ly6c.2 transcript sequences indicate that they are both alleles of Ly6c2 (Bothwell et al. 1988; Palfree et al. 1988). Moreover, discrepancies in lymphocyte staining with LY6C and LY6C.2-specific antibodies suggest the possibility of highly homologous gene products (Schlueter et al. 1997). These data are consistent with the identification of two Ly6c loci, Ly6c1 and Ly6c2, which differ by only a few amino acids at the N-terminus.
We identified size polymorphisms between MAV-1-susceptible (SJL/J, 129S6SvEv/Tac, and C57BL/6J) and resistant (BALB/cJ) strains within the gene regions of Ly6a, Ly6e, and Ly6g. These variations may influence such things as mRNA expression and/or splicing. Specifically, Ly6e is known to have at least six splice variants in GenBank that vary in their 5' untranslated regions (NCBI Accessions NM_001164036.1, NM_001164037.1, NM_001164038.1, NM_001164039.1, NM_001164040.1, and NM_008529.3). The importance of these transcript variants is unknown, but they may be critical for the regulation of gene expression. Moreover, the human Ly6e homolog is also differentially spliced: two transcript variants have been reported in GenBank (NCBI Accessions NM_002346.2, NM_001127213.1). Both the mechanisms for generating these splice variants and their biological significance may be conserved between mice and humans. Furthermore, though no polymorphisms were observed, preliminary data suggest the existence of a BC025446 transcript variant with an additional 68 bp segment near the 3' end of the coding sequence in both BALB/cJ and SJL/J mice (M. Stier and K. Spindler, unpublished). The coding region and size polymorphisms observed in this study are summarized in Fig. 1.
The Ly6 and Ly6-related genes have roles in host responses to other viral infections. The Ly6 locus on human Chr 8q24 was identified as a region conferring cellular susceptibility to HIV-1 in vitro (Brass et al. 2008; Loeuillet et al. 2008). Ly6e appears to be vital for resistance to avian herpesvirus Marek’s disease virus in chickens (Liu et al. 2003). In addition, human LY6E was identified in a screen for proteins associated with West Nile virus infection (Krishnan et al. 2008). Therefore, the identification of a size polymorphism in the gene region of Ly6e is significant and warrants further investigation. It is interesting to note that some of the Ly6 genes found in the mouse are not found in syntenic regions of the human or rat chromosomes (Holmes and Stanford 2007). For example, genes in the mouse between Ly6e and Ly6h (see Table 2) are not found in the human genome. This may have implications for identification of specific gene(s) involved in susceptibility to MAV-1. Specifically, if the mechanism for Ly6-associated viral susceptibility is conserved between species, then the number of candidate genes for MAV-1 susceptibility would be significantly reduced.
Ly6a has been studied more extensively. In models of skeletal muscle regeneration, LY6A is an important mediator of extracellular matrix breakdown and remodeling (Kafadar et al. 2009). Ly6a is expressed in endothelial cells of the brain vasculature (van de Rijn et al. 1989; T.-H. Hsu and K. Spindler, unpublished). During MAV-1 infection, loss of integrity of the blood-brain barrier correlates with morbidity and mortality in susceptible mouse strains (Gralinski et al. 2009). Accordingly, LY6A may also play a role in viral-induced breakdown of the extracellular matrix and blood-brain barrier disruption. If this hypothesis is true, the identification of Ly6a coding region polymorphisms and genomic DNA size differences between MAV-1-susceptible and resistant mice may explain the strain-specific differences in blood-brain barrier disruption and MAV-1 susceptibility.
Through the identification of coding region and size polymorphisms in the Ly6 and Ly6-related genes, we have characterized several variations that may explain the mouse strain-specific differences in susceptibility to MAV-1. Moreover, these polymorphisms may also be critical for the host response in other viral infections as evidenced by the involvement of the Ly6 genes in susceptibility to other viruses (Brass et al. 2008; Krishnan et al. 2008; Liu et al. 2003; Loeuillet et al. 2008). Collectively, these data provide insights into the Ly6 and Ly6-related genes and provide a foundation for further characterization of the basis for MAV-1 susceptibility.
ACKNOWLEDGMENTS
We thank Dave Burke, Tien-Huei Hsu, and Mike Imperiale for critical comments on the manuscript. We thank Jason Weinberg for helpful discussions. We are grateful to Shanna Ashley for technical assistance. This work was supported by National Institutes of Health Grant AI068645 to K.R.S.
REFERENCES
- Bamezai A. Mouse Ly-6 proteins and their extended family: Markers of cell differentiation and regulators of cell signaling. Arch Immunol Ther Exp (Warsz) 2004;52:255–266. [PubMed] [Google Scholar]
- Behrendt N, Ploug M, Patthy L, Houen G, Blasi F, Dano K. The ligand-binding domain of the cell surface receptor for urokinase-type plasminogen activator. J Biol Chem. 1991;266:7842–7847. [PubMed] [Google Scholar]
- Bothwell A, Pace PE, LeClair KP. Isolation and expression of an IFN-responsive Ly-6C chromosomal gene. J Immunol. 1988;140:2815–2820. [PubMed] [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Charles PC, Chen X, Horwitz MS, Brosnan CF. Differential chemokine induction by the mouse adenovirus type-1 in the central nervous system of susceptible and resistant strains of mice. J Neurovirol. 1999;5:55–64. doi: 10.3109/13550289909029746. [DOI] [PubMed] [Google Scholar]
- Fleming TJ, Fleming ML, Malek TR. Selective expression of Ly-6G on myeloid lineage cells in mouse bone marrow. RB6-8C5 mAb to granulocyte-differentiation antigen (Gr1) detects members of the Ly-6 family. J Immunol. 1993a;151:2399–2408. [PubMed] [Google Scholar]
- Fleming TJ, O'HUigin C, Malek TR. Characterization of two novel Ly-6 genes. Protein sequence and potential structural similarity to alpha-bungarotoxin and other neurotoxins. J Immunol. 1993b;150:5379–5390. [PubMed] [Google Scholar]
- Gralinski LE, Ashley SL, Dixon SD, Spindler KR. Mouse adenovirus type 1-induced breakdown of the blood-brain barrier. J Virol. 2009;83:9398–9410. doi: 10.1128/JVI.00954-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guida JD, Fejer G, Pirofski LA, Brosnan CF, Horwitz MS. Mouse adenovirus type 1 causes a fatal hemorrhagic encephalomyelitis in adult C57BL/6 but not BALB/c mice. J Virol. 1995;69:7674–7681. doi: 10.1128/jvi.69.12.7674-7681.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hänninen A, Jaakkola I, Salmi M, Simell O, Jalkanen S. Ly-6C regulates endothelial adhesion and homing of CD8(+) T cells by activating integrin-dependent adhesion pathways. Proc Natl Acad Sci U S A. 1997;94:6898–6903. doi: 10.1073/pnas.94.13.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes C, Stanford WL. Concise review: Stem cell antigen-1: Expression, function, and enigma. Stem Cells. 2007;25:1339–1347. doi: 10.1634/stemcells.2006-0644. [DOI] [PubMed] [Google Scholar]
- Kafadar KA, Yi L, Ahmad Y, So L, Rossi F, Pavlath GK. Sca-1 expression is required for efficient remodeling of the extracellular matrix during skeletal muscle regeneration. Dev Biol. 2009;326:47–59. doi: 10.1016/j.ydbio.2008.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kring SC, King CS, Spindler KR. Susceptibility and signs associated with mouse adenovirus type 1 infection of adult outbred Swiss mice. J Virol. 1995;69:8084–8088. doi: 10.1128/jvi.69.12.8084-8088.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F, Montgomery RR, Lev S, Mason PW, Koski RA, Elledge SJ, Xavier RJ, Agaisse H, Fikrig E. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242–245. doi: 10.1038/nature07207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeClair KP, Palfree RG, Flood PM, Hammerling U, Bothwell A. Isolation of a murine Ly-6 cDNA reveals a new multigene family. EMBO J. 1986;5:3227–3234. doi: 10.1002/j.1460-2075.1986.tb04633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HC, Niikura M, Fulton JE, Cheng HH. Identification of chicken lymphocyte antigen 6 complex, locus E (LY6E, alias SCA2) as a putative Marek's disease resistance gene via a virus-host protein interaction screen. Cytogenet Genome Res. 2003;102:304–308. doi: 10.1159/000075767. [DOI] [PubMed] [Google Scholar]
- Loeuillet C, Deutsch S, Ciuffi A, Robyr D, Taffe P, Munoz M, Beckmann JS, Antonarakis SE, Telenti A. In vitro whole-genome analysis identifies a susceptibility locus for HIV-1. PLoS Biol. 2008;6:e32. doi: 10.1371/journal.pbio.0060032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy A, Hegyi H, Farkas K, Tordai H, Kozma E, Banyai L, Patthy L. Identification and correction of abnormal, incomplete and mispredicted proteins in public databases. BMC Bioinformatics. 2008;9:353. doi: 10.1186/1471-2105-9-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palfree RG, Sirlin S, Dumont FJ, Hammerling U. N-terminal and cDNA characterization of murine lymphocyte antigen Ly-6C.2. J Immunol. 1988;140:305–310. [PubMed] [Google Scholar]
- Ploug M, Ellis V. Structure-function relationships in the receptor for urokinase-type plasminogen activator. Comparison to other members of the Ly-6 family and snake venom alpha-neurotoxins. FEBS Lett. 1994;349:163–168. doi: 10.1016/0014-5793(94)00674-1. [DOI] [PubMed] [Google Scholar]
- Potter TA, McKenzie IF, Morgan GM, Cherry M. Murine lymphocyte alloantigens. I. The Ly-6 locus. J Immunol. 1980;125:541–545. [PubMed] [Google Scholar]
- Reiser H, Coligan J, Palmer E, Benacerraf B, Rock KL. Cloning and expression of a cDNA for the T-cell-activating protein TAP. Proc Natl Acad Sci U S A. 1988;85:2255–2259. doi: 10.1073/pnas.85.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh S, Kosugi A, Noda S, Yamamoto N, Ogata M, Minami Y, Miyake K, Hamaoka T. Modulation of TCR-mediated signaling pathway by thymic shared antigen-1 (TSA-1)/stem cell antigen-2 (Sca-2) J Immunol. 1995;155:5574–5581. [PubMed] [Google Scholar]
- Schauwecker PE, Williams RW, Santos JB. Genetic control of sensitivity to hippocampal cell death induced by kainic acid: a quantitative trait loci analysis. J Comp Neurol. 2004;477:96–107. doi: 10.1002/cne.20245. [DOI] [PubMed] [Google Scholar]
- Schlueter AJ, Malek TR, Hostetler CN, Smith PA, deVries P, Waldschmidt TJ. Distribution of Ly-6C on lymphocyte subsets: I. Influence of allotype on T lymphocyte expression. J Immunol. 1997;158:4211–4222. [PubMed] [Google Scholar]
- Smith K, Brown CC, Spindler KR. The role of mouse adenovirus type 1 early region 1A in acute and persistent infections in mice. J. Virol. 1998;72:5699–5706. doi: 10.1128/jvi.72.7.5699-5706.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler KR, Fang L, Moore ML, Hirsch GN, Brown CC, Kajon A. SJL/J mice are highly susceptible to infection by mouse adenovirus type 1. J Virol. 2001;75:12039–12046. doi: 10.1128/JVI.75.24.12039-12046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler KR, Welton AR, Lim ES, Duvvuru S, Althaus IW, Imperiale JE, Daoud AI, Chesler EJ. The major locus for mouse adenovirus susceptibility maps to genes of the hematopoietic cell surface-expressed LY6 family. J Immunol. 2010;184:3055–3062. doi: 10.4049/jimmunol.0903363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Rijn M, Heimfeld S, Spangrude GJ, Weissman IL. Mouse hematopoietic stem-cell antigen Sca-1 is a member of the Ly-6 antigen family. Proc Natl Acad Sci U S A. 1989;86:4634–4638. doi: 10.1073/pnas.86.12.4634. [DOI] [PMC free article] [PubMed] [Google Scholar]