Abstract
Understanding the molecular mechanism of muscle contraction and its regulation has been greatly influenced and aided by studies of myofilament structure in invertebrate muscles. Invertebrates are easily obtained and cover a broad spectrum of species and functional specializations. The thick (myosin-containing) filaments from some invertebrates are especially stable and simple in structure and thus much more amenable to structural analysis than those of vertebrates. Comparative studies of invertebrate filaments by electron microscopy and image processing have provided important generalizations of muscle molecular structure and function. This article reviews methods for preparing thick and thin filaments from invertebrate muscle, for imaging filaments by electron microscopy, and for determining their three dimensional structure by image processing. It also highlights some of the key insights into filament function that have come from these studies.
Keywords: myosin filament, actin filament, three-dimensional reconstruction, structure, methodology, invertebrate
1. Introduction
All muscles contract by sliding of the thick (myosin-containing) filaments past the thin (actin-containing) filaments (for overviews see [1–3]). Essential to an understanding of contraction and its regulation is knowledge of the molecular structure of the myofilaments and the changes in their structure that occur in different states. Our anthropocentric view of biology and focus on the relevance of basic science to human disease means that most muscle studies have been carried out on vertebrates. However, some of our greatest insights into muscle structure and function have come from invertebrate studies. For example, the greater stability of invertebrate thick filaments compared with their vertebrate counterparts has made possible more detailed structural analysis in the invertebrates. The insights gained from these studies have provided an essential foundation for understanding vertebrate filament structure. In addition, invertebrates show a range of specializations in filament structure and organization related to their specific needs. This range has made it possible to derive useful generalizations concerning filament structure and function [2,3]. Several model systems, including spiders, horseshoe crabs, insects and mollusks, have played key roles in these advances. These systems have provided fundamental insights into both thick and thin filament structure, and the relation of this structure to the regulation of muscle contraction. The major techniques used in understanding filament structure include x-ray crystallography, X-ray fiber diffraction, spectroscopic methods such as electron paramagnetic resonance, and electron microscopy (EM1). In this article I focus on electron microscopy and image processing techniques that lead to an understanding of three-dimensional (3D) filament structure.
2. Filament preparation
The thick and thin filaments that make up striated muscle are arranged in overlapping arrays within contractile units known as myofibrils [1–3]. The state of muscle required to separate thick and thin filaments from each other, and to isolate them biochemically, is the relaxed state, in which filament interaction is minimal. The key step in isolating either thick or thin filaments is therefore homogenization of muscle in a relaxing medium, whose critical requirements are the presence of Mg.ATP and the absence of Ca2+. A typical relaxing solution contains 0.1 M salt, Mg.ATP (to dissociate actin from myosin), additional Mg2+ to stabilize the thin filaments, and ethylene glycol bis(β-aminoethyl ether) N,N,N′,N′-tetraacetic acid (EGTA) to chelate Ca2+; it is buffered to pH ~7.0 [4–7]. We also add NaN3 to inhibit bacterial growth. Our normal solution is: 0.1 M NaCI, 3 mM MgCl2, 5 mM Mg.ATP, 1 mM EGTA, 3 mM NaN3, 10 mM sodium phosphate, pH 7.0 [4].
2.1 Native thick filaments
In a typical preparation (Fig. 1; [4–7]), muscle fiber bundles 1–2 mm in diameter are removed from the freshly dissected animal and held at about rest length by securing to a frame with thread or by maintaining attachment to the rigid exoskeleton. The muscle is then placed in cold relaxing solution. 200 mg (wet weight) of muscle is finely chopped with a scalpel, the pieces placed in 3.0 ml ice-cold relaxing solution in a 15 ml tube, and the suspension homogenized briefly with cooling on ice. Typically we give 2 bursts each of 5 sec at setting 5 on a Polytron homogenizer fitted with a 12 mm diameter probe. The exact time and setting depends on the muscle and type of homogenizer, and is determined by trial and error; homogenization should be the minimum required to release sufficient filaments, as excess homogenization causes filament breakage and denaturation. The homogenate is centrifuged for 2 min at 15,000 g in a bench centrifuge at 4° C to remove large debris, leaving the filaments in the supernatant. At this stage the suspension is at fairly high concentration and appears milky. This crude suspension contains both thick and thin filaments as well as other cellular components such as fragmented organelles. For electron microscopy, the suspension is diluted 5–10 fold with relaxing solution to provide a good distribution of filaments on the EM grid without too much filament overlap; the dilution factor is determined by trial and error.
Figure 1.
Schematic of main steps in preparation of thick filaments.
A variation on this method that is often used [4–6] is to first chemically demembranate the muscle held at rest length by treating with detergent (e.g. 0.1% w/v saponin) in relaxing solution for ~ 3 hr at 4° C with agitation. This allows soluble protein to exit from the fibers, resulting in a cleaner filament preparation with less background protein. Use of relaxing solution maintains the filaments in a relaxing environment. The muscle is then chopped and homogenized as above.
If it is desired to purify the thick filaments by removing the thin filaments (for example for cleaner EM images, or to determine filament constituents by SDS-PAGE), further differential centrifugation would seem the obvious choice, due to the larger S-value of thick compared with thin filaments. In practice this works poorly, and sedimented thick filaments are invariably accompanied by many thin filaments. This appears most likely to be due to the trapping of thin filaments in the sedimenting mesh of thick filaments, or possibly to the presence of weak actin-myosin interaction even in relaxing conditions [8]. A simple solution to this problem has been devised, using the actin-severing protein gelsolin to cut the thin filaments into short segments, which are not readily enmeshed and which interact less readily with the thick filaments [8]. Gelsolin normally requires the presence of micromolar Ca2+ for activity, which is incompatible with the relaxing conditions required to keep filaments separated. Ca2+-insensitive gelsolin (the N-terminal half) can be prepared from plasma gelsolin by enzymatic digestion with thermolysin [8], or can be bacterially expressed [9]. Both methods yield a molecule that severs thin filaments into short segments under relaxing conditions. Following gelsolin treatment, the suspension is centrifuged (18,700 g for 15 min) to produce a pellet containing relatively pure thick filaments [8]. Re-suspension and re-centrifugation in relaxing solution can be carried out to further purify the filaments (Fig. 3 cf. Fig. 2a). In addition to treating filaments in suspension, gelsolin can also be used simply to rinse an EM grid with adherent filaments. The thin filaments rapidly disappear.
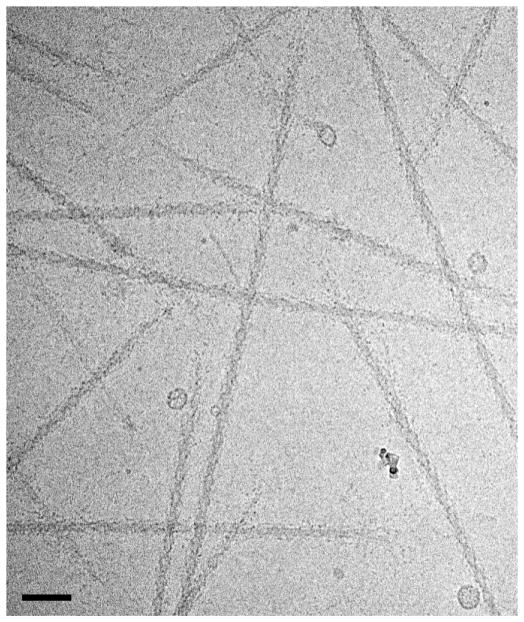
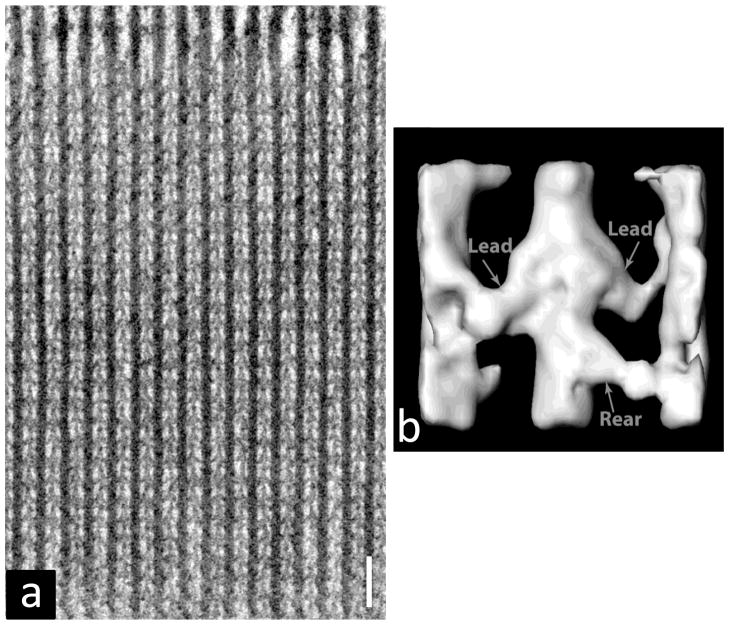
Figure 3.
Cryo-EM image of purified, frozen-hydrated relaxed thick filaments from tarantula muscle. Thin filaments have been removed by treatment with gelsolin, creating a cleaner image (cf. Fig. 2a). Bar = 100 nm. From [50].
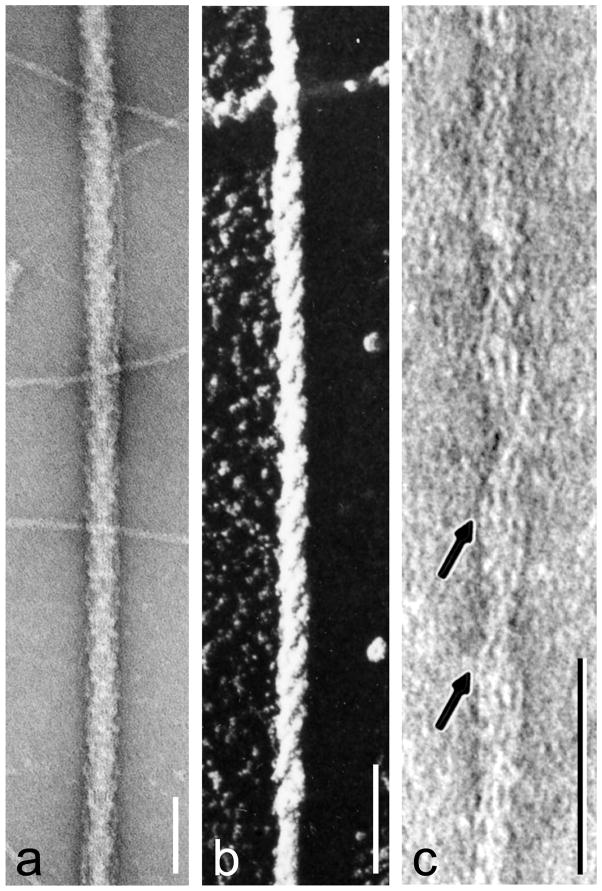
Figure 2.
Negative staining and shadowing. (a) unpurified tarantula thick filaments, with thin filaments in background (from [4]); (b) metal-shadowed tarantula thick filament showing right-handed helical tracks (from [5] with permission); (c) negatively stained thin filament from frog cardiac muscle (from [103] with permission); this vertebrate image is chosen for the clarity with which it reveals tropomyosin (arrowed strands), although it was invertebrate filaments that first demonstrated the tropomyosin-shift mechanism unequivocally [12]. In (a,c), protein is white and stain dark; in (b) metal is white. Scale bars: a, b (100 nm), c (50 nm).
2.2 Native thin filaments
While thick filaments from different invertebrate species have varying helical parameters (and are thus useful for comparative studies; [3,10,11]), thin filaments from all muscles (both vertebrate and invertebrate) are remarkably similar in structure, and studies of invertebrate thin filaments have therefore played a less important role in our understanding of muscle molecular structure. An exception is a negative stain EM study of Limulus thin filaments, which was the first to directly visualize tropomyosin movement on actin that is the basis of thin filament regulation (see section 4.2.2) [12], strongly supporting earlier modeling studies of X-ray diffraction data [13–15]. Limulus filaments were chosen for this study because they have a larger (therefore more visible) troponin than vertebrates and exhibit excellent Ca2+-sensitivity of actin-activated myosin ATPase. They were therefore deemed most likely to reveal any changes in thin filament structure in response to Ca2+.
Thin filaments from Limulus and other invertebrates are prepared as described by [16] with modifications by [17] and [18]. Muscle fiber bundles are tied to sticks at close to rest length, treated overnight with 50% glycerol/water to permeabilize the cell membranes, teased into millimeter bundles while still attached to the sticks, and placed in 0.1 M KCl, 5 mM phosphate, 3 mM MgCl2, 3 mM EDTA (ethylenediamine-N,N,N′,N′-tetraacetic acid), pH 7.0 for 10 min. The fiber bundles are then homogenized in this solution. The rigor myofibrils thus produced are collected by centrifugation (5 min at 5,000 g), re-dispersed manually, and washed 3 times by centrifugation and resuspension. The myofibrils are then resuspended in a low salt relaxing solution (20 mM KCl, 0.1 mM EDTA, 1 mM MgCl2, 5 mM ATP, 5 mM phosphate, pH 6.0, homogenized to dissociate the thick and thin filaments, and centrifuged (5 min at 5,000 g) to remove coarse myofibrillar fragments. The supernatant is then centrifuged more strongly (1 hr at 25,000 g followed by 30 min at 50,000 g) to sediment the thick filaments. Although many thin filaments will also sediment, some are left in the supernatant, and these can be collected by sedimentation at 100,000 g for 3 hr. The pelleted filaments are resuspended and used for grid preparation.
3. EM observation
Filaments prepared as above can be studied by a variety of electron microscopic methods. These include negative staining, metal shadowing and cryo-electron microscopy.
3.1 Negative staining
3.1.1 Principle
Negative staining is a technique by which macromolecular assemblies such as filaments can be easily studied at moderately high resolution and with good contrast and structural preservation. Stain surrounds and supports the structure and penetrates its interstices, revealing detailed surface shape at resolutions approaching 2 nm. A drop of filament suspension is applied to a carbon-coated EM grid, followed by several drops of stain. The last drop is removed with filter paper and the remaining stain allowed to dry. The grid is then ready to image in the EM. The best negative stain for thick and thin filament observations is aqueous uranyl acetate (UA), generally 1–2% w/v. While UA also gives a degree of positive staining, its key advantage is its high contrast and the fact that it also acts as a rapid fixative [19]. Thus, when the stain dries, and its concentration increases, filament structure is not disrupted by the increased ionic strength. This is a key point, as thick filaments dissolve at high salt levels. The final thickness of the dried negative stain is an important factor in obtaining good images. Optimally the stain should be similar in thickness to the filament being studied, so that the whole depth of the filament is contrasted. All thin filaments are about 10 nm in diameter and require a relatively thin layer of stain. Thick filaments vary (depending on species) generally in the range 30–40 nm, and are best contrasted with a thicker layer of stain. This can be achieved by leaving a slightly thicker layer of the liquid stain on the grid before drying and/or by using a higher concentration of stain.
3.1.2 Uses
Negative staining of invertebrate thick and thin filaments has played a key role in our basic understanding of myofilament structure and function (Fig. 2). It was the first technique to reveal the bipolar organization of myosin molecules in thick filaments [20], and the 3D organization of actin in thin filaments [20,21]. Negatively stained Limulus thick filaments were the first in which the 3D helical organization of myosin heads was preserved, making possible the first 3D reconstruction of a thick filament [22,23]. Studies of other invertebrates, together with improved staining and 3D image processing techniques, yielded additional insights into thick filament structure (Fig. 2a) [5,6,24,25]. Negative staining has also provided key insights into the structure of thick filaments in invertebrate smooth muscles [26–28]. In addition, the technique has revealed the structural changes that occur in invertebrate thick filaments upon activation by Ca2+ binding or myosin phosphorylation [4,29,30]. Such routine negative staining techniques have been further developed to enable the capture of transient structural changes occurring on the millisecond timescale [19]. This time-resolved technique reveals the changes in scallop thick filament structure that occur in the first 10–30 ms after Ca2+ binding [31]. In the case of thin filaments, comparative studies of negatively stained invertebrate filaments revealed the common helical organization of actin present in diverse muscles [21], and it was negative staining of Limulus filaments in the presence and absence of Ca2+ [12,32] that first visualized the steric blocking model of thin filament regulation originally postulated on the basis of X-ray diffraction data [13–15] (see 2.2, 4.2.2; Figs. 2c, 5a-d).
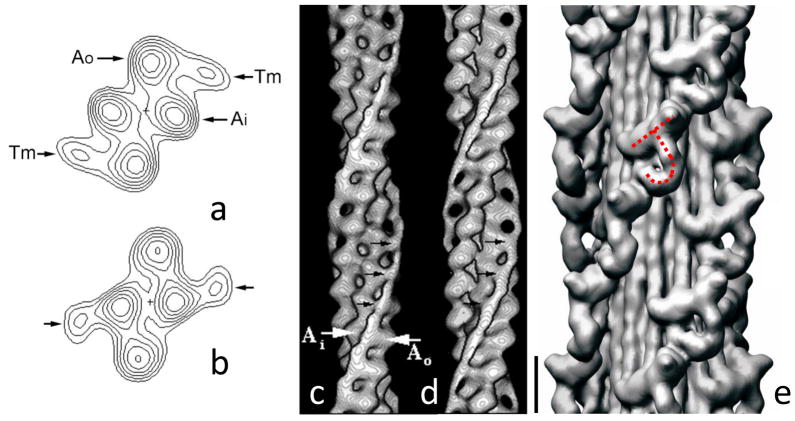
Figure 5.
Three-dimensional reconstruction of thin and thick filaments. (a–d) helical reconstruction of Limulus thin filament based on negative stain images. (a, b) Projections down long-pitch helices, showing outer and inner domains of actin subunits (Ao and Ai), and tropomyosin (Tm) associated with outer domain in low Ca2+ state (a) and inner domain in high Ca2+ state (b). (c, d) surface presentation of the same reconstruction at low (c) and high (d) Ca2+. (a–d) Modified from [32], with permission. (e) IHRSR reconstruction of tarantula thick filament based on cryo-EM images. The repeating “J”-shaped structures in 4 co-axial helices represent pairs of myosin heads (see Fig. 6). Scale bar for (c)–(e) = 10 nm. From [50].
3.1.3 Procedure
Technical details, and the many variations on negative staining technique applied to biological samples, can be found in [33,34]. Here the approaches used for studying muscle filaments in our laboratory are outlined. EM grids, generally 400 (or finer) mesh, are coated with carbon to act as a support for the filaments. This is a routine technique available in any biological EM lab. Carbon is first evaporated on to freshly cleaved mica in a vacuum evaporator, then floated from the mica on to a water surface. The floating carbon is picked up by lowering the water surface onto grids laid out on a wire mesh under the water, or by bringing individual grids held in forceps through the water surface to pick up the carbon from below. Image contrast is best when the carbon layer is thin, improving the signal over the background noise; however, thin carbon is fragile and easily breaks before or during observation under the electron beam, unless the support grid mesh is extremely fine. One way to achieve this is to first coat grids with a thick (therefore strong) “holey” carbon film, and use this as an extremely fine mesh (see Supplementary Information for holey film method). A very thin layer of carbon from mica can be picked up on such a grid and is stable over the narrow span of the holes (1–5 μm, compared with an ~ 40 μm opening in a 400 mesh grid). Filaments are imaged over the thin carbon on the holes. Structural detail is even better when there is no carbon underlying the filaments at all [20,35]. This can be achieved by applying filaments to a holey carbon film without any carbon over the holes. Filaments often remain in the film of stain stretching across the hole. However, this film is extremely fragile and usually breaks during observation in the electron beam (if not before), squashing or stretching the filaments. It can be stabilized by applying an extremely thin layer of carbon to the grid after staining [36]. Apart from improved contrast and lower background noise, an advantage of this method is that the top and bottom of the filament are more equally stained. A disadvantage is more work.
The suspension of thick or thin filaments is applied to the coated grid in a 5 μl droplet, which is allowed to rest on the grid for about 30 s. The excess droplet is wicked off from the edge of the grid with filter paper (making sure that the thin layer left does not dry out!) and then several (6–10) drops of stain are run across the grid. The last one is also wicked off and the grid allowed to dry. To obtain optimal staining and structural detail, an appropriate filament concentration is required. Too many filaments and the stain tends to form dense patches and many filaments overlap. Too few and it is difficult to obtain the requisite good spreading of the stain. The optimal load is determined by trial and error, but a good starting concentration for both thick and thin filaments is about 1 μM.
3.1.4 Problems and fixes
The most common problem with negative staining of biological samples is ensuring that the stain spreads to a thin layer that surrounds the structure. Very frequently during drying, the stain pulls away from the grid surface to form a tiny droplet in one small area, due to the high surface tension of the aqueous stain and the hydrophobic nature of most carbon films. The result is poor filament structure and contrast (often the filament will appear positively stained). This problem can be avoided or minimized in several ways: one is glow discharge or UV treatment of the coated grids to make them hydrophilic [37]. We have also found that slow drying of the grid in a humidity chamber at about 80% relative humidity, or alternatively very rapid drying under a hair dryer can be very helpful. Because spreading of stain is aided by high humidity, labs in more humid environments have less trouble with negative staining than those in dry conditions (e.g. winter in the northern US).
A few special tricks have been developed that help with specific problems. To preserve thick filament structure optimally, it has been found that treatment with tannic acid before UA is useful [38,39]. Filaments are usually partially flattened during drying of the negative stain. This is lessened if (as mentioned above) the amount of stain used is such that the dried layer is roughly the same thickness as the filament (any thinner, and support is less and only part of the thickness may be contrasted; thicker, and contrast is reduced and noise added due to excess stain). Flattening is also reduced with tannic acid [39] or by including a trace of glycerol in the stain [40].
In the case of thick filaments, it is found that the helical ordering of the myosin heads, known to be present in the relaxed state, is easily disrupted. This precludes 3D image reconstruction (Section 4). There are several causes. One is the binding of the heads (which are flexibly attached to the filament) to the carbon substrate; this is worse with glow discharged or UV-treated grids which produce a charged carbon surface that attracts the heads [41]. Thus surface treatment to ensure spreading of stain must not be overdone, and the use of high humidity or rapid drying may be preferred over glow discharge or UV treatment of grids. Another, less obvious issue is the finding that UA staining of thick filaments that have been prepared in a chloride-based relaxing solution usually causes disordering of the heads. The reason is not known; Cl− is known to bind strongly to myosin filaments [42], but is present naturally inside cells only at low concentrations (~ 4 mM). This Cl− effect can be avoided by rinsing the filaments on the grid with a solution containing acetate in place of Cl− [5,7]. Thick filament helical order can sometimes be improved at lower pH (6.5 rather than 7 or 7.5) and at lower ionic strength, although at the expense of filament bundling. The most stable ordering of the heads is achieved in the presence of the myosin inhibitor blebbistatin [43]. In filaments regulated by myosin RLC phosphorylation, phosphorylation causes disordering of the heads [4]. Improved order should therefore also be obtained in the presence of kinase inhibitors or of phosphatase, although we have not yet determined whether this is actually the case. Making grids as soon as possible after filament preparation should also minimize unwanted phosphorylation.
Finally, the cleanest images of thick filaments (best suited for image reconstruction) are obtained if thin filaments are removed, so that filament superposition is minimized. While this is not essential (good images can be obtained by sufficiently diluting filaments), it is advantageous (see 2.1).
In the case of thin filaments, some of the best staining is obtained in the presence of phosphate ions (e.g. 5 mM) [35]. Again the reason is unclear. Phosphate (as the inorganic ion or as ATP) is often present in solutions either as a buffer (Pi) or in relaxing solutions (Mg.ATP). However, electron microscopists tend to substitute other buffers for Pi when negative staining with UA because the stain rapidly forms an insoluble precipitate with Pi, making grids dirty. This problem is easily overcome, so that one can take advantage of the UA/Pi combination, simply by ensuring that the two solutions do not mix for a significant length of time. If the grid, with filaments attached in a Pi-containing buffer, is rapidly flushed with 8–10 drops of UA, precipitation is not a problem [35].
3.1.5 Imaging
Following preparation, grids can be immediately observed in the microscope, or they can be stored for weeks to months without significant deterioration. Images can be obtained by routine methods if fine details of structure are not critical. The grid is usually searched at low magnification (2,000–3,000 X) to find areas of appropriate filament distribution and good negative staining (stain quality can vary a lot over the grid), then imaged at high magnification (30,000–60,000 X). If 3D reconstruction is desired and image detail is critical, then low dose imaging methods are preferable. Low dose microscopy minimizes damage to the specimen by electron irradiation. Typically electron micrographs are recorded after first focusing on the area of interest. This requires a relatively bright beam and several seconds of irradiation, during which time the filament will have been exposed to many thousands of electrons per square nm. Such irradiation causes the stain to migrate from its original fine distribution, to form larger pools that no longer delineate the finest details [44]. These details are best preserved if focusing is carried out to one side of the area to be imaged, so that the actual image recorded receives minimal electron doses <1000 electrons/nm2 [5]. Low dose software making this possible is available on many modern microscopes.
3.2 Shadowing
Filaments can also be imaged by metal shadowing techniques. While shadowing is usually less useful for understanding detailed structure, it has proved critical in determining one aspect of myofilament structure: the “handedness” (left or right) of the filament helix. Negative staining provides a projection image in which the top side and underside of the filament are superimposed. The hand of the helix is therefore unclear because the underside of a right-handed helix resembles the top side of a left-handed helix and vice versa. This issue is overcome by shadowing because only the top side of the filament is contrasted. In this way it has been shown that all myosin and actin filaments so far studied have long-pitch helices that are right-handed [5,6,45,46] (Fig. 2b). Shadowing has also been ingeniously employed to demonstrate myosin heads on the surface of invertebrate smooth muscle thick filaments (normally difficult to demonstrate by negative staining alone) [26,47].
Shadowing of thick and thin filaments can be carried out as follows. A drop of filament suspension is applied to a carbon-coated EM grid as for negative staining (mica, normally used for single molecule shadowing, is not desirable as it disrupts the helical organization of myosin heads). The filaments are fixed briefly by rinsing with 1% aqueous UA, rinsed with water then 30% (v/v) glycerol in water, and the excess wicked off to leave a thin film [6,46]. These steps help to strengthen the filaments against drying forces that would otherwise flatten them in the next step. The grid is then placed in a vacuum evaporator. Heavy metal (Pt) is evaporated onto the filaments at an angle of ~12–30°. Following shadowing, grids are observed in the EM. Low dose techniques are unnecessary here due to the robustness of the metal coating.
3.3 Cryo-EM
3.3.1 Principle
Cryo-EM is a method for observing macromolecular structures without fixation, staining or drying. “Frozen-hydrated” specimens are observed at low temperature in thin aqueous films suspended over holes in a carbon support film. Cryo-EM is thus free of the drying, staining and fixation artifacts present in other techniques. It provides the closest to in vivo filament images of biological structures available, although it comes with its own attendant difficulties.
3.3.2 Uses
Early studies showed that the helical order of invertebrate myosin filaments could be well preserved by cryo-EM [48], thus avoiding artifacts of negative staining. However, 3D reconstruction was not carried out. The first demonstration that thick filaments could be reconstructed from cryo-EM images was with scallop filaments [49], although head conformation could not be interpreted in detail in this work. This problem was overcome in studies of tarantula filaments, which have highly ordered head arrays: in this case cryo-EM played a crucial role in deciphering the organization of myosin heads, which had proved elusive for over twenty years using negative staining (Fig. 3; see 4.3.2, 4.4) [11,50]. Subsequent studies have shown that similar head arrangements are present in a variety of other invertebrates (Limulus [51], scallop [52,53]) and also vertebrates (mouse cardiac muscle [54]). While the earlier negative staining studies had preserved the helical organization of the heads, their relatively low resolution (5 nm) left the exact head arrangement ambiguous [55]. Most interpretations suggested that the heads of each myosin molecule were splayed apart from each other, extending up and down the filament to the next level in each direction [5,24,56]. Cryo-EM combined with single particle image processing showed unambiguously that this model was wrong: the two myosin heads from each molecule in fact interact with each other intramolecularly in a compact structure, in a way that immediately suggests how this might function to switch activity off in relaxed muscle (4.3.2) [50]. Cryo-EM has also played a key role in our current understanding of thin filament structure and regulation [57–60], although this work was not on invertebrate filaments. An invertebrate filament structure that has been elucidated by cryo-EM is that of arthrin, a mono-ubiquitinated form of actin found in some insect flight muscles [61].
3.3.3 Procedure
For cryo-EM, holey carbon films are generally used, as described earlier for negative staining (true holes are used, without thin carbon on top). The goal is to suspend the filaments in solution so that forces that might affect their structure (e.g. by binding to a carbon surface) are avoided or minimized. The holey films can be home-made (as described for negative staining) or purchased (Quantifoil (SPI Supplies, West Chester, PA) or C-flat (EMS, Hatfield, PA)). Quantifoil and C-flat grids have a regular array of holes of defined size rather than the random size and location in home-made grids – but the cost exceeds $5/grid. Specimens are prepared as follows (details can be found in [62]). A droplet of filament suspension (5 μl) is applied to a grid as for negative staining. The grid is held in forceps in a plunging device, ready to be dropped into liquid ethane (cooled by liquid nitrogen), to cause vitrification (freezing without ice crystal formation). After 30 s, the grid is blotted gently with filter paper (from one or both sides) to form a thin film. The grid is immediately plunged into the liquid ethane to freeze the thin film before it dries. The grid is then transferred to a pre-cooled grid box under liquid nitrogen and stored in a liquid nitrogen Dewar until ready for imaging. Liquid nitrogen and liquid ethane are both cryogens and must be handled with care. Liquid nitrogen can cause skin burns, and if used in a totally sealed room could cause suffocation. Liquid ethane also causes burns, and in the gaseous form is potentially explosive. It is therefore used in small amounts (~1–2 ml), and is disposed of by allowing it to evaporate in a fume hood.
Grids for cryo-EM are first glow discharged in an atmosphere of amylamine within 2 h of use, to make the surface hydrophilic. This ensures that when the grid is blotted, the thin aqueous film remains spread. For muscle filaments and other structures that are salt-sensitive, blotting must be performed in a humid atmosphere in order to minimize evaporation between the times of blotting and freezing. Evaporation would otherwise lead to disruptive increases in ionic strength (high ionic strength for example, causes myosin heads to disorder [63] and filaments to dissolve). A Plexiglas chamber supplied with humid air from a domestic humidifier works well, or commercial, humidity-controlled plunge-freezing devices are available [64]. These devices, under computer control (Gatan (Pleasanton, CA) model CP3; or FEI (Hillsboro, OR) Vitrobot), are capable of giving reproducible ice thickness and humidity control, but are expensive (tens of thousands of dollars).
3.3.4 Imaging
Frozen grids are placed in a pre-cooled (to approximately liquid nitrogen temperature) cryo specimen holder (e.g. Gatan 626) in a cryo-transfer station. The holder is then inserted into the microscope while observing precautions to avoid ice contamination on the grid surface (by having the grid covered with a movable shutter during transfer, and carrying out the transfer operation quickly). The holder is left in the microscope for about 30 minutes to allow the vacuum and temperature to stabilize. The shutter is then opened and the grid searched at low magnification (~2,000 X) for areas of suitable ice thickness. Images are acquired under low dose conditions (< 1000 electrons/nm2), which are mandatory for cryo-EM, as aqueous specimens in the absence of stain are rapidly destroyed in the electron beam.
3.3.5 Issues
The key advantage of cryo-EM is its ability to capture and image close-to in vivo macromolecular structure. In addition, the entire structure (including the interior) is visualized, not just the surface (as with negative staining and shadowing). Cryo-EM has transformed our understanding of thick filament structure, and invertebrates have been essential to this success [50]; cryo-EM has also succeeded at imaging F-actin filaments at high resolution [59,60]. However, cryo-EM comes at a cost. It is slower, technically more difficult, and requires more advanced instrumentation. Image contrast is low due to the absence of stain. Images must therefore be recorded at higher defocus (~1.5–3.0 μm) than with stained specimens in order to generate some phase contrast – but this compromises resolution. Many grids are useless because the ice is too thick or too thin. Low dose imaging means that the operator is “shooting blind”, so that many images are later found to contain few or no filaments. When using film, the operator does not know whether success has been achieved until the film is processed: the advent of CCD cameras has diminished this difficulty greatly as images can be viewed immediately after acquisition, providing immediate feedback on specimen quality.
3.3.6 Choosing between negative stain and cryo-EM
Both techniques can yield high resolution insights into muscle filament structure. The key advantage of cryo-EM is the imaging of structure in its native form. However, negative stain is sufficient for answering most questions, and the added difficulty of cryo-EM means that in many cases, negative stain is to be preferred [65]. It is well established that in most instances, negative stain provides excellent fidelity of structural preservation, so that it can be used to answer moderate resolution questions. An example – negative stain has been used in many papers to study the effects of mutations in tropomyosin, actin or troponin on the regulatory movements of tropomyosin (e.g. [66,67]. Carrying out the same experiments using cryo-EM would have taken years longer. Any potential additional insights may not have justified the greater effort.
3.4 Sectioning techniques: conventional and cryo sectioning
3.4.1 Principle
If one is interested in understanding filament structure and interactions in the context of the filament lattice of the sarcomere, sectioning techniques are required. Muscle specimens (live or chemically demembranated) are fixed, embedded, and sectioned either transversely or longitudinally. Stained sections are observed in the microscope, and the structure, arrangement and interactions of filaments can be studied in this way.
3.4.2 Uses
Routine fixation (glutaraldehyde followed by osmium tetroxide) and routine embedding/sectioning (epoxy resin, room temperature sectioning) methods have been applied to both vertebrate and invertebrate muscles many times, leading to our knowledge of basic filament and sarcomere structure [2]. One invertebrate muscle stands out in going well beyond this basic information: asynchronous insect flight muscle. The quasi-crystalline organization of filaments in this muscle has led to critical insights into the interactions between actin and myosin in different physiological states – insights applicable to understanding the molecular basis of contraction in all muscles (Fig. 4a) [68,69]. Why is asynchronous insect flight muscle so useful? The answer lies primarily in its filament organization. In these muscles, used to generate wing movements, the sarcomere is organized with thin filaments in the same plane as nearest neighbor thick filaments, and all thick filaments in rotational register. Thus very thin longitudinal sections (~20 nm) can be cut, containing a single layer of actin and myosin filaments (myac layers), and the images are especially useful for image analysis and averaging. This contrasts with vertebrate and most other muscles (where thin filaments are not in the same plane as nearest neighbor thick filaments, and thick filaments can have varying rotations). The species most frequently used for studies of insect flight muscle is the giant water bug Lethocerus. Early studies of myac layers produced beautiful images of myosin heads in relaxed and rigor states, which led to some key concepts of the crossbridge cycle of contraction, including the idea that myosin heads went through a large change in angle while attached to the thin filament to generate the powerstroke [68]. While this idea has since been modified, it provided a simple framework for thinking about the contractile mechanism. More recent cryo studies of insect flight muscle have led to many other insights into the molecular basis of contraction (see 3.4.4).
Figure 4.
(a) Longitudinal section of rigor insect flight muscle, showing alternating thick and thin filaments with crossbridges joining them; M-line at top, protein black. (b) Class average of one chevron-like structure similar to those in (a) calculated from tomogram of rapidly frozen muscle, showing interpretation in terms of “lead” and “rear” heads (those closer to and further from the pointed end of the thin filament, respectively). The central filament is actin with partial myosin filaments on either side. After [72], with permission. Scale bar = 100 nm.
3.4.3 Conventional fixation and sectioning
Muscles are used fresh or chemically skinned (with detergent or glycerol), fixed with glutaraldehyde, postfixed with OsO4, stained with uranyl acetate, dehydrated with an ethanol or acetone series, embedded in epoxy (Araldite or Epon), then sectioned on a diamond knife [33,68]. The stunning contrast seen in many myac images of insect flight muscle results from the (unconventional) use of potassium permanganate as a section stain [69]. Several valuable variations on this procedure have been developed, including the use of tannic acid as an additional fixative, which improves both contrast and preservation [70].
3.4.4 Rapid freezing/freeze substitution
Although these routine methods are very powerful, they are limited in the level and fidelity of molecular detail they can provide. In general, crosslinking by glutaraldehyde appears to disrupt the helical organization of myosin heads in thick filaments, while molecular details of the thin filaments are also lost. Low temperature variants of these techniques applied to invertebrate muscles have proved very successful at overcoming some of these limitations. A powerful procedure for improving molecular preservation in sections is the use of fast physical fixation (rapid freezing) rather than slow chemical fixation (chemical crosslinking). The muscle fiber bundle is mounted on a specimen disc attached to a plunging device which is dropped on to a flat, polished copper block cooled with liquid helium [71,72]. The outermost 1–10 μm of the tissue are rapidly frozen (~ 1 ms) without damage due to ice crystal formation (immersion in liquid nitrogen, by comparison, is a slow method of freezing and results in unacceptable ice crystal damage). Following this rapid physical fixation, the specimen is freeze-substituted, a procedure carried out at ~ −80° C in which the ice in the specimen is slowly replaced by organic solvent (usually acetone) containing chemical fixative (OsO4 or tannic acid or UA, or some combination)[71,72]. In this way chemical fixation takes place at low temperature on tissue already stabilized by physical fixation. The result is superior preservation, even of the labile myosin head helical organization. Following freeze-substitution, specimens are embedded and sectioned conventionally. Using this approach, it has been possible to demonstrate the helical organization of myosin heads in thick filaments in situ in tarantula muscle and to determine directly the rotational symmetry of the head organization in both tarantula and scallop muscle by observing thick filaments in transverse section [73–75]. Similar detail has also been observed in vertebrate muscle [76–78] leading to improved insights into vertebrate sarcomeric structure. The most striking use of this technique has been in detailed studies of insect flight muscle [72], in which three-dimensional information has been obtained by electron tomography of sections and tomogram averaging. This is described in 4.1.2.
3.4.5 Cryosectioning
An alternative cryo approach involves low temperature sectioning (cryo-sectioning) followed by ambient temperature observation. This has proved extremely powerful in the analysis of thick filament structure in vertebrates [79]. In the case of invertebrates, detailed filament structure has also been observed. In one study, skinned muscles were put into ATP-free solution, creating strong rigor crosslinks between the myosin and actin filaments, then treated with cryo-protectant and plunge-frozen in liquid ethane [80]. Specimens were cryo-sectioned at −110° C [81], the sections collected on a grid, thawed, and negatively stained with uranyl acetate. Because the filaments were held together by rigor crosslinks, it was not necessary to fix before freezing. These sections made it possible to observe filament molecular detail (without fixation artifacts) at the high resolution possible with negative staining (see 3.1) but here in the context of the filament lattice.
The most faithful representation of filament structure in the filament lattice is obtained, as with isolated filaments, by cryo-EM. In this case, cryo-sectioning is carried out on unfixed, rapidly frozen tissue, and sections are observed in the frozen-hydrated state [82,83]. As with all unstained specimens, contrast is low and imaging must be carried out by low dose techniques. Excellent native filament detail has been obtained from sections of insect flight muscle [84], confirming inferences from more artifact-prone techniques involving fixation, staining or filament isolation. Cryo-sectioning of unfixed and un-cryoprotected tissue is difficult and comes with its own artifacts [82,83].
4. Three-dimensional reconstruction
Owing to the large depth of field in the electron microscope, images obtained are two-dimensional projections of 3D structures. Top sides and under sides of filaments are thus superimposed and cannot be distinguished in a single image. There are several ways to obtain 3D filament information, all requiring different angular views of the filament subunits. Three powerful 3D reconstruction techniques commonly used are electron tomography and two versions of helical reconstruction. These methods combined with the preparative techniques described earlier have provided key insights into myofilament structure in invertebrates.
4.1 Electron tomography
4.1.1 Principle
With electron tomography, the specimen grid is tilted in the electron microscope and the information obtained from multiple tilt views is combined by back projection to create a 3D reconstruction [72,85]. The microscope must be equipped with an accurate (preferably motorized and automated) goniometer tilt stage (common on modern microscopes). Images are typically collected over a tilt range of +/− 60°, and typically at ≤2° intervals (dependent upon the thickness of the object being imaged and the desired resolution). The physical limitations of holders and stages prevent images being obtained at much greater than 70° tilt, resulting in loss of some information in the Z direction (corresponding to the thickness of the filament). Modern microscopes equipped with CCD cameras and software that automates specimen tilting, focusing and image acquisition enable a full set of images to be collected in an hour or so. Automation also makes possible a large decrease in the electron dose required to obtain a tilt series, resulting in improved resolution. Tomographic reconstruction programs are available at no cost (e.g. IMOD [86]) or are sold as a part of a tomography package with modern microscopes.
4.1.2 Uses
Some of the most impressive insights into actomyosin filament interaction have come from electron tomography of sectioned insect flight muscle (used because of its high degree of order; see 3.4.2). Many of these studies have used rapidly frozen, freeze substituted muscle, which not only preserves molecular structure optimally (3.4.4), but also makes possible the capture of intermediate structural states of myosin crossbridges during contraction [72]. A key innovation in this work has been the computational classification of different structural motifs (representing, for example, myosin heads attached to actin in different patterns and conformations) and the 3D averaging of motifs within classes, enabling detailed structural information to be extracted from heterogeneous populations (Fig. 4b). The resolution of these studies is often of the order of 5 nm, and individual myosin heads attached to actin, as well as troponin molecules on the thin filaments, are thought to have been visualized and their conformations elucidated by fitting of myosin head atomic structures (see 4.4). These studies have led to insights into the effect of adding different nucleotides to rigor muscle, the visualization of a wide range of myosin head structures during isometric contraction, and the ability to determine the effects of transient mechanical perturbations on myosin head conformation [72].
4.2 Helical reconstruction
4.2.1 Principle
Helical structures, such as actin and myosin filaments, display multiple views of their subunits in a single image, due to the intrinsic nature of a helix. If the symmetry of the helix (the axial rise and rotation per subunit) is known, then helical reconstruction techniques, taking advantage of these views, can be used to compute a 3D structure from a single image [87–89]. Such reconstructions reveal only the features in the structure that obey the helical symmetry. Individual reconstructions are typically noisy and vary one from another, for example due to variations in stain distribution in negatively stained specimens. Individual reconstructions are therefore averaged to create a reconstruction with a higher signal to noise ratio with improved resolution. To carry out a helical reconstruction, digitized electron micrograph images are computationally boxed, then straightened, and their Fourier transforms calculated. By combining the indexing information of the transform (a function of the helical symmetry) with the layer line data, a 3D reconstruction (density map) can be computed. Multiple reconstructions are averaged by aligning and averaging the filament data in Fourier space then computing the reconstruction from the averaged data [89].
4.2.2 Uses
Helical reconstruction techniques have played a key role in our understanding of both thin and thick filament structures. Early reconstructions of F-actin were based on vertebrate proteins, but, as described earlier, it was helical reconstruction of negatively stained invertebrate filaments (Limulus) that first directly revealed [12,32] the physical blocking mechanism by which tropomyosin switches off thin filaments, and the way in which Ca2+ acts to move tropomyosin, making previously blocked myosin binding sites accessible (Fig. 5a–d; Sections 2.2, 3.1.2) [13–15]. Reconstructions of thin filaments from Drosophila with engineered mutant troponin have provided insights into critical residues involved in the regulatory movement of tropomyosin [90].
Similarly, it was helical reconstruction (of negatively stained Limulus thick filaments) that provided the first visualization of the arrangement of myosin heads on helical tracks in a thick filament (3.1.2) [22]. In work on vertebrates, the helical order known (from X-ray diffraction) to be present in intact muscle [91] was lost during filament isolation and staining [20,41]. Success with Limulus was a result of the greater stability of the myosin head arrays compared with vertebrate filaments, and the development of gentle methods of filament isolation that preserved myosin head order [22]. Helical reconstruction of Limulus filaments directly revealed the 4-fold arrangement of myosin head helical tracks on the filament surface [22]. Subsequent studies of scallop, scorpion, tarantula and insect filaments yielded similarly striking reconstructions [5,6,24,25,49], and demonstrated some of the diversity of invertebrate filament structure. In the case of the closely related Limulus, scorpion and tarantula [5,22,24], it was shown that all filaments had a similar structure and 4-fold rotational symmetry. In contrast, scallop was shown to have a 7-fold arrangement of heads [6]. In the initial studies [6,22] the two heads of each myosin molecule were not resolved. Improvements to the reconstruction technique provided better resolution, but still left uncertain the organization of individual heads [5,24,55,56], a situation that was only corrected by the use of cryo-EM together with a new method of reconstruction (see 4.3). The demonstration that myosin head helical order could be preserved in invertebrates was followed by improvements in techniques for preserving helical order in vertebrate filaments as well, finally making possible 3D reconstructions of these more complex structures [39,54,92].
4.3 Iterative Helical Real Space Reconstruction (IHRSR)
4.3.1 Principle
Helical reconstructions are carried out in reciprocal space and require computation of Fourier transforms, and identification and indexing of layer lines. This process is tedious, and determination of helical symmetry can be problematic. An alternative, real-space method – Iterative Helical Real Space Reconstruction (IHRSR) – also takes advantage of the multiple subunit views present in a single image of a helix, but does so in a different way [93–95]. An initial reference model of the structure is generated based on known information, for example a preliminary helical reconstruction (4.2). Density projections of the model are computed for different rotations about its long axis (e.g. every 4°). Filament images are computationally boxed into short, overlapping segments, and each segment thus produced is compared with the different projections of the model by cross-correlation (typically the box length is chosen to contain a small number of subunits of the repeating structure, and the box is shifted longitudinally by one subunit between segments: the optimal parameters are determined by trial and error). The angle of the model that gives the best match to the filament segment is then assigned to that segment. A reconstruction is computed from the different segments by back projection using these assigned angles, and the reconstruction helically averaged. The resulting reconstruction becomes the model for a second round of matching, and the process is repeated until there are no further changes in the reconstruction. This iterative real space technique has some key advantages over traditional helical reconstruction. For example, filament straightening and layer line picking are not required, and filaments with disorder or with particular symmetries that are problematic for helical reconstruction (because information from different helical tracks overlaps in the Fourier transform) can often be readily reconstructed by IHRSR.
4.3.2 Uses
A major breakthrough in our understanding of thick filament structure was obtained using IHRSR to reconstruct cryo-EM images of relaxed tarantula filaments [50]. The reconstruction had a resolution of 2.5 nm (a 2-fold improvement over the negative stain helical reconstruction), revealing the myosin heads unambiguously and showing that the two heads of each myosin interacted with each other (Fig. 5e; see 4.4). Subsequent studies of other invertebrate thick filaments and isolated myosin molecules have shown that this interacting-head motif is likely to be widespread across the animal kingdom [50–53,96,97]. Comparable observations on vertebrate filaments (mouse cardiac) have been much more difficult and are at lower resolution due to the more complex structure and less stable myosin head arrays of vertebrates; however, they too appear to show a similar structure [54]. Interpretation of the complex vertebrate reconstruction depended strongly on the insights first obtained from the unambiguous results of the invertebrate (tarantula) filaments. In addition to revealing these head-head interactions, the tarantula reconstruction also showed the subfragment 2 (S2) region of the myosin tail, and the presence of subfilaments in the filament backbone. The assembly of myosin tails into subfilaments had been predicted 25 years earlier on the basis of model building using invertebrate muscle X-ray diffraction data [98]; the tarantula reconstruction provided compelling support for this model. Currently the best thick filament reconstruction has been limited to 2.5 nm resolution, due in part to the use of a conventional (non-field emission) microscope. Other filaments (e.g. F-actin) have been reconstructed to as high as 0.6 nm resolution (enabling the visualization of secondary structure such as α-helices) using field emission microscopy and more advanced image processing techniques [59,60]. Whether similar techniques applied to thick filaments are as successful will depend on whether inherent disorder in the heads caused by their flexible attachment to the tails limits resolution.
4.4 Atomic fitting
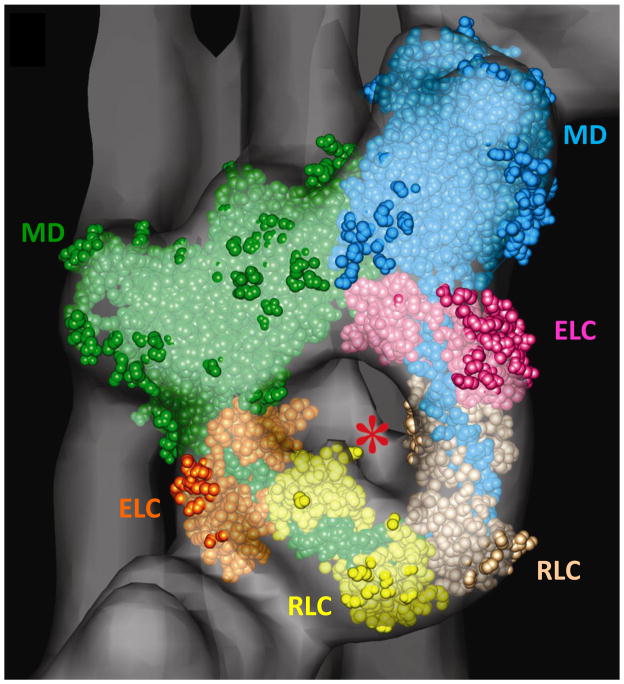
A final and key step in the interpretation of reconstructions of sufficient resolution is the computational fitting (docking) of atomic structures of filament subunits (e.g. myosin heads, actin monomers) into the reconstruction. If the fit is unambiguous, which is possible when subunits are asymmetric, this effectively extends the resolution of the reconstruction to near-atomic level. Fitting can be carried out using a number of (free) software packages, such as UCSF Chimera [99] and SITUS [100]. The excellent fit of the interacting-head structure present in crystals of smooth muscle heavy meromyosin [101] into the J-shaped motif (Fig. 5e) of the tarantula thick filament reconstruction directly demonstrated the presence of similar interactions in the intact filament (Fig. 6) [50]. In this structure, one head (the “free” head) physically blocks the actin-binding region of the other (the “blocked” head) – thus hindering binding of the blocked head to actin – while the blocked head was close to the converter domain and essential light chain of the free head, which could prevent the free head from functioning normally during ATP turnover [50,101,102]. Thus a 2.5 nm resolution reconstruction is interpreted in terms of specific polypeptide regions of the protein subunits, whose interaction suggests a simple structural model for the switching off of myosin head activity in relaxed muscle. Similarly, the position of tropomyosin on thin filaments in relation to specific actin residues was revealed by fitting of the atomic structure of actin into the 3D reconstruction of Limulus thin filaments [32]. The results showed that in the low Ca2+ state, tropomyosin physically blocked clusters of amino acid residues on actin known to be involved in myosin binding, while these residues became exposed when tropomyosin moved to its high Ca2+ position. These near-atomic level observations provided strong support to the steric blocking model for thin filament regulation.
Figure 6.
Fitting of myosin head atomic structure into repeating J-motif in tarantula reconstruction (Fig. 5e). Motor domain (MD), essential light chain (ELC) and regulatory light chain (RLC) of blocked head are in green, orange and yellow; those of free head are blue, pink and beige. Asterisk indicates rod-like volume of density that represents the start of the S2 portion of the myosin tail. Modified from [50].
Acknowledgments
I thank Dr. Raúl Padrón for comments on the manuscript. The author’s work is supported by NIH grants AR034711 and HL059408.
Appendix
PREPARING HOLEY CARBON FILMS
-
1
Make up 100 ml of 0.5% (w/v) formvar in chloroform. Stir with magnetic stirrer. Takes a while to dissolve (15–20 min).
-
2
Thoroughly mix 10 ml glycerol with 10 ml distilled water.
-
3
Put 20 drops of glycerol/water mixture (from Pasteur pipette) into 50 ml of formvar solution. Use 50 ml stoppered graduated cylinder.
-
4
Immediately prior to sonication, shake vigorously for 30 sec. Shaking disperses the glycerol/water into fine droplets in the chloroform/formvar, but the droplets are not small enough and readily coalesce. The sonication step which follows makes the droplets much smaller and the right size for the holey film. This preliminary dispersion by shaking ensures that the glycerol/water is not floating as a “glob” at the surface when the sonication starts, in which case it is difficult to disperse.
-
4
Sonicate for 45–60 sec. at setting 6 using a microprobe sonicator (setting will depend on the sonicator and may be found by trial and error). After sonication, the mixture should feel warm and look uniformly milky (an emulsion of glycerol/water microdroplets in the formvar/chloroform).
-
5
Put solution into a staining (Coplin) jar. Clean several 3x1 inch glass microscope slides with Ross (lint-free) lens paper, dip them one at a time into formvar solution for a couple of seconds, remove, stand on their ends and leave until they appear dry by eye. Do all of the slides at one time because the dispersion is unstable and the droplets slowly coalesce. If it is a very dry day and the holes are few or small, it may help to dip and dry the slides in a humid chamber (about 60% RH).
-
6
As soon as one slide is dry, float the film off on water, pick up one test grid, dry it briefly (5 minutes), cut out the grid from the formvar with a razor blade, and check in the EM for hole size, number and distribution. For method, see (7) below. There should be plenty (minimum of 10–20 per 400-mesh grid square) of variable sized holes, often patchily distributed. To test whether the holes are large enough, check in the electron microscope at 50,000x: the larger holes should be large enough so that their edges are outside the field of view on the large fluorescent screen. The holes may have “ragged” edges and may be crossed by fine, very thin strands of formvar. These will be removed with the methanol etching step later. If the holes look good, proceed with the other slides, picking up 40–50 grids per slide (see (7) below). If they don’t look good, discard the slides and start again, varying sonication time, humidity etc until good holes are obtained on a test grid.
-
7
Fill crystallizing dish with distilled water. Wipe surface of water clean by dragging a sheet of Ross lens tissue across it. With a clean razor blade, score the film on one face of the slide near the edges so that the film will easily detach in the next step. Carefully float off the film on to the water surface by sliding the slide (scored side up) into the water at a 45° angle. “Huffing” on the slide first with moist breath helps to separate the film from the slide.
-
8
Place grids (no pre-cleaning necessary) face down on film on water surface. Pick up with Whatman #4 filter paper or Bibulous paper by touching the paper horizontally to floating formvar and then removing (these papers absorb water at the right rate to facilitate pick-up of grids and film). First make 90° bend at end of filter paper to act as handle.
-
9
After filter paper is dry, place it (with attached grids/formvar, grids uppermost) on a bed of filter paper saturated with methanol in a glass Petri dish for 10 min. (Use fume hood). The bottoms of the grids should be soaked from underneath, but the grids should not be immersed in the methanol. Cover dish so that methanol does not evaporate. This “etching” procedure removes glycerol/water droplets, dissolves any very thin plastic that might have been crossing “pseudo” holes, and gives a sharper edge to the real holes. After the grids are dry, check a grid in the EM to make sure that this has occurred. In particular, check holes at high magnification to make sure that they are real holes and not pseudo-holes (still having a thin coating of formvar stretching across the hole).
-
10
If the holes look good, coat the grids heavily with carbon (so that the supporting filter paper appears dark brown).
-
11
Set up a large glass Petri dish in hood with a bed of filter paper saturated with 1,2-dichloroethane (same procedure as for methanol) and place grids/filter paper/formvar/carbon on to the wet filter paper in the Petri dish for 2 hrs (grid side up, as in 9). This dissolves the formvar, leaving the holey carbon. Grids without the formvar are more stable in the electron beam (less drift, holes do not expand in beam, etc.).
-
12
Remove filter paper/grids and dry in a glass Petri dish. Store in a glass Petri dish in a refrigerator (glass dishes avoid electrostatic attraction of grids, which can be a problem with plastic dishes; in our experience, the grid surface remains cleaner and its surface properties remain more constant with storage in a refrigerator).
Footnotes
Abbreviations: EM, electron microscopy; 3D, three-dimensional; IHRSR, Iterative Helical Real Space Reconstruction; MLCK, myosin light chain kinase; EGTA, ethylene glycol bis(β-aminoethyl ether) N,N,N′,N′-tetraacetic acid; UA, uranyl acetate
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. Garland Science; New York: 2007. [Google Scholar]
- 2.Squire J. The Structural Basis of Muscular Contraction. Plenum Press; New York: 1981. [Google Scholar]
- 3.Squire JM. Muscle: Design, Diversity and Disease. Benjamin/Cummings Publishing Company, Inc; Menlo Park, CA: 1986. [Google Scholar]
- 4.Craig R, Padron R, Kendrick-Jones J. Structural changes accompanying phosphorylation of tarantula muscle myosin filaments. J Cell Biol. 1987;105:1319–1327. doi: 10.1083/jcb.105.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crowther RA, Padron R, Craig R. Arrangement of the heads of myosin in relaxed thick filaments from tarantula muscle. J Mol Biol. 1985;184:429–439. doi: 10.1016/0022-2836(85)90292-x. [DOI] [PubMed] [Google Scholar]
- 6.Vibert P, Craig R. Electron microscopy and image analysis of myosin filaments from scallop striated muscle. J Mol Biol. 1983;165:303–320. doi: 10.1016/s0022-2836(83)80259-9. [DOI] [PubMed] [Google Scholar]
- 7.Kensler RW, Levine RJ. An electron microscopic and optical diffraction analysis of the structure of Limulus telson muscle thick filaments. J Cell Biol. 1982;92:443–451. doi: 10.1083/jcb.92.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hidalgo C, Padron R, Horowitz R, Zhao FQ, Craig R. Purification of native myosin filaments from muscle. Biophys J. 2001;81:2817–2826. doi: 10.1016/S0006-3495(01)75923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaponnier C, Janmey PA, Yin HL. The actin filament-severing domain of plasma gelsolin. J Cell Biol. 1986;103:1473–1481. doi: 10.1083/jcb.103.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wray JS, Vibert PJ, Cohen C. Diversity of cross-bridge configurations in invertebrate muscles. Nature. 1975;257:561–564. doi: 10.1038/257561a0. [DOI] [PubMed] [Google Scholar]
- 11.Craig R, Woodhead JL. Structure and function of myosin filaments. Curr Opin Struct Biol. 2006;16:204–212. doi: 10.1016/j.sbi.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Lehman W, Craig R, Vibert P. Ca2+-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368:65–67. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 13.Huxley HE. Structural changes in the actin- and myosin-containing filaments during contraction. Cold Spr Harbor Symp Quant Biol. 1973;37:361–376. [Google Scholar]
- 14.Haselgrove JC. X-ray evidence for a conformational change in the actin-containing filaments of vertebrate striated muscle. Cold Spring Harbor Symp Quant Biol. 1973;37:341–352. [Google Scholar]
- 15.Parry DA, Squire JM. Structural role of tropomyosin in muscle regulation: analysis of the x-ray diffraction patterns from relaxed and contracting muscles. J Mol Biol. 1973;75:33–55. doi: 10.1016/0022-2836(73)90527-5. [DOI] [PubMed] [Google Scholar]
- 16.Kendrick-Jones J, Lehman W, Szent-Gyorgyi AG. Regulation in molluscan muscles. J Mol Biol. 1970;54:313–326. doi: 10.1016/0022-2836(70)90432-8. [DOI] [PubMed] [Google Scholar]
- 17.Lehman W, Szent-Gyorgyi AG. Regulation of muscular contraction. Distribution of actin control and myosin control in the animal kingdom. J Gen Physiol. 1975;66:1–30. doi: 10.1085/jgp.66.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lehman W, Regenstein JM, Ransom AL. The stoichiometry of the components of arthropod thin filaments. Biochim Biophys Acta. 1976;434:215–222. doi: 10.1016/0005-2795(76)90053-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhao FQ, Craig R. Capturing time-resolved changes in molecular structure by negative staining. J Struct Biol. 2003;141:43–52. doi: 10.1016/s1047-8477(02)00546-4. [DOI] [PubMed] [Google Scholar]
- 20.Huxley HE. Electron microscope studies on the structure of natural and synthetic protein filaments from striated muscle. J Mol Biol. 1963;7:281–308. doi: 10.1016/s0022-2836(63)80008-x. [DOI] [PubMed] [Google Scholar]
- 21.Hanson J, Lowy J. The structure of F-actin and of actin isolated from muscle. J Mol Biol. 1963;6:46–60. [Google Scholar]
- 22.Stewart M, Kensler RW, Levine RJ. Structure of Limulus telson muscle thick filaments. J Mol Biol. 1981;153:781–790. doi: 10.1016/0022-2836(81)90418-6. [DOI] [PubMed] [Google Scholar]
- 23.Kensler RW, Levine RJ. An electron microscopic and optical diffraction analysis of the structure of Limulus telson muscle thick filaments. J Cell Biol. 1982;92:443–451. doi: 10.1083/jcb.92.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stewart M, Kensler RW, Levine RJ. Three-dimensional reconstruction of thick filaments from Limulus and scorpion muscle. J Cell Biol. 1985;101:402–411. doi: 10.1083/jcb.101.2.402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morris EP, Squire JM, Fuller GW. The 4-stranded helical arrangement of myosin heads on insect (Lethocerus) flight muscle thick filaments. J Struct Biol. 1991;107:237–249. [Google Scholar]
- 26.Elliott A. The arrangement of myosin on the surface of paramyosin filaments in the white adductor muscle of Crassostrea angulata. Proc R Soc Lond B Biol Sci. 1974;186:53–66. doi: 10.1098/rspb.1974.0034. [DOI] [PubMed] [Google Scholar]
- 27.Elliott A. Structure of molluscan thick filaments: a common origin for diverse appearances. J Mol Biol. 1979;132:323–341. doi: 10.1016/0022-2836(79)90263-8. [DOI] [PubMed] [Google Scholar]
- 28.Castellani L, Vibert P, Cohen C. Structure of myosin/paramyosin filaments from a molluscan smooth muscle. J Mol Biol. 1983;167:853–872. doi: 10.1016/s0022-2836(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 29.Vibert P, Craig R. Structural changes that occur in scallop myosin filaments upon activation. J Cell Biol. 1985;101:830–837. doi: 10.1083/jcb.101.3.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levine RJ, Chantler PD, Kensler RW, Woodhead JL. Effects of phosphorylation by myosin light chain kinase on the structure of Limulus thick filaments. J Cell Biol. 1991;113:563–572. doi: 10.1083/jcb.113.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao FQ, Craig R. Ca2+ causes release of myosin heads from the thick filament surface on the milliseconds time scale. J Mol Biol. 2003;327:145–158. doi: 10.1016/s0022-2836(03)00098-6. [DOI] [PubMed] [Google Scholar]
- 32.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 33.Hayat MA. Principles and Techniques of Electron Microscopy, Biological Applications. Cambridge University Press; New York: 2000. [Google Scholar]
- 34.Harris JR. Negative staining of thinly spread biological samples. Methods Mol Biol. 2007;369:107–142. doi: 10.1007/978-1-59745-294-6_7. [DOI] [PubMed] [Google Scholar]
- 35.Craig R, Szent-Gyorgyi AG, Beese L, Flicker P, Vibert P, Cohen C. Electron microscopy of thin filaments decorated with a Ca2+-regulated myosin. J Mol Biol. 1980;140:35–55. doi: 10.1016/0022-2836(80)90355-1. [DOI] [PubMed] [Google Scholar]
- 36.Vibert P, Craig R. Three-dimensional reconstruction of thin filaments decorated with a Ca2+-regulated myosin. J Mol Biol. 1982;157:299–319. doi: 10.1016/0022-2836(82)90236-4. [DOI] [PubMed] [Google Scholar]
- 37.Burgess SA, Walker ML, Thirumurugan K, Trinick J, Knight PJ. Use of negative stain and single-particle image processing to explore dynamic properties of flexible macromolecules. J Struct Biol. 2004;147:247–258. doi: 10.1016/j.jsb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 38.Kensler RW, Levine RJ, Stewart M. Electron microscopic and optical diffraction analysis of the structure of scorpion muscle thick filaments. J Cell Biol. 1985;101:395–401. doi: 10.1083/jcb.101.2.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart M, Kensler RW. Arrangement of myosin heads in relaxed thick filaments from frog skeletal muscle. J Mol Biol. 1986;192:831–851. doi: 10.1016/0022-2836(86)90032-x. [DOI] [PubMed] [Google Scholar]
- 40.Kensler RW. The mammalian cardiac muscle thick filament: crossbridge arrangement. J Struct Biol. 2005;149:303–312. doi: 10.1016/j.jsb.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 41.Knight P, Trinick J. Structure of the myosin projections on native thick filaments from vertebrate skeletal muscle. J Mol Biol. 1984;177:461–482. doi: 10.1016/0022-2836(84)90295-x. [DOI] [PubMed] [Google Scholar]
- 42.Elliott GF. Measurements of the electric charge and ion-binding of the protein filaments in intact muscle and cornea, with implications for filament assembly. Biophys J. 1980;32:95–97. doi: 10.1016/S0006-3495(80)84927-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao FQ, Padron R, Craig R. Blebbistatin stabilizes the helical order of myosin filaments by promoting the switch 2 closed state. Biophys J. 2008;95:3322–3329. doi: 10.1529/biophysj.108.137067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unwin PN. Electron microscopy of the stacked disk aggregate of tobacco mosaic virus protein. II. The influence of electron irradiation of the stain distribution. J Mol Biol. 1974;87:657–670. doi: 10.1016/0022-2836(74)90076-x. [DOI] [PubMed] [Google Scholar]
- 45.DePue RH, Rice RV. F-actin is a right-handed helix. J Mol Biol. 1965;12:302–303. doi: 10.1016/s0022-2836(65)80306-0. [DOI] [PubMed] [Google Scholar]
- 46.Kensler RW, Levine RJ. Determination of the handedness of the crossbridge helix of Limulus thick filaments. J Muscle Res Cell Motil. 1982;3:349–361. doi: 10.1007/BF00713042. [DOI] [PubMed] [Google Scholar]
- 47.Elliott A. Direct demonstration of the helical nature of paramyosin filaments. Phil Trans Roy Soc Lond B. 1971;261:197–199. [Google Scholar]
- 48.Menetret JF, Schroder RR, Hofmann W. Cryo-electron microscopic studies of relaxed striated muscle thick filaments. J Muscle Res Cell Motil. 1990;11:1–11. doi: 10.1007/BF01833321. [DOI] [PubMed] [Google Scholar]
- 49.Vibert P. Helical reconstruction of frozen-hydrated scallop myosin filaments. J Mol Biol. 1992;223:661–671. doi: 10.1016/0022-2836(92)90982-p. [DOI] [PubMed] [Google Scholar]
- 50.Woodhead JL, Zhao FQ, Craig R, Egelman EH, Alamo L, Padron R. Atomic model of a myosin filament in the relaxed state. Nature. 2005;436:1195–1199. doi: 10.1038/nature03920. [DOI] [PubMed] [Google Scholar]
- 51.Zhao FQ, Craig R, Woodhead JL. Head-head interaction characterizes the relaxed state of Limulus muscle myosin filaments. J Mol Biol. 2009;385:423–431. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao FQ, Woodhead JL, Craig R. Head-head interaction characterizes the relaxed state of scallop and Limulus muscle myosin filaments. Biophys J. 2008;94:630-Pos. doi: 10.1016/j.jmb.2008.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Khayat HA, Morris EP, Squire JM. The 7-stranded structure of relaxed scallop muscle myosin filaments: support for a common head configuration in myosin-regulated muscles. J Struct Biol. 2009;166:183–194. doi: 10.1016/j.jsb.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 54.Zoghbi ME, Woodhead JL, Moss RL, Craig R. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc Natl Acad Sci U S A. 2008;105:2386–2390. doi: 10.1073/pnas.0708912105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Squire JM, Luther PK, Morris EP. Organisation and properties of the striated muscle sarcomere. In: Squire JM, editor. Molecular Mechanisms in Muscular Contraction. CRC Press, Inc; Boca Raton: 1990. pp. 1–48. [Google Scholar]
- 56.Offer G, Knight PJ, Burgess SA, Alamo L, Padron R. A new model for the surface arrangement of myosin molecules in tarantula thick filaments. J Mol Biol. 2000;298:239–260. doi: 10.1006/jmbi.2000.3664. [DOI] [PubMed] [Google Scholar]
- 57.Milligan RA, Whittaker M, Safer D. Molecular structure of F-actin and location of surface binding sites. Nature. 1990;348:217–221. doi: 10.1038/348217a0. [DOI] [PubMed] [Google Scholar]
- 58.Xu C, Craig R, Tobacman L, Horowitz R, Lehman W. Tropomyosin positions in regulated thin filaments revealed by cryoelectron microscopy. Biophys J. 1999;77:985–992. doi: 10.1016/S0006-3495(99)76949-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oda T, Iwasa M, Aihara T, Maeda Y, Narita A. The nature of the globular- to fibrous-actin transition. Nature. 2009;457:441–445. doi: 10.1038/nature07685. [DOI] [PubMed] [Google Scholar]
- 60.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 61.Burgess S, Walker M, Knight PJ, Sparrow J, Schmitz S, Offer G, Bullard B, Leonard K, Holt J, Trinick J. Structural studies of arthrin: monoubiquitinated actin. J Mol Biol. 2004;341:1161–1173. doi: 10.1016/j.jmb.2004.06.077. [DOI] [PubMed] [Google Scholar]
- 62.De Carlo S. Plunge-freezing (holey carbon method) In: Cavalier A, Spehner D, Humbel BM, editors. Hanbook of Cryo-preparation Methods for Electron Microscopy. CRC Press; Boca Raton: 2009. pp. 49–68. [Google Scholar]
- 63.Wray JS, Vibert PJ, Cohen C. Cross-bridge arrangements in Limulus muscle. J Mol Biol. 1974;88:343–348. doi: 10.1016/0022-2836(74)90486-0. [DOI] [PubMed] [Google Scholar]
- 64.Frederik PM, de Haas F, Storms MMH. Controlled vitrification. In: Cavalier A, Spehner D, Humbel BM, editors. Handbook of Cryo-preparation Methods for Electron Microscopy. CRC Press; Boca Raton: 2009. pp. 69–99. [Google Scholar]
- 65.Ohi M, Li Y, Cheng Y, Walz T. Negative staining and image classification - powerful tools in modern electron microscopy. Biol Proced Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korman VL, Hatch V, Dixon KY, Craig R, Lehman W, Tobacman LS. An actin subdomain 2 mutation that impairs thin filament regulation by troponin and tropomyosin. J Biol Chem. 2000;275:22470–22478. doi: 10.1074/jbc.M002939200. [DOI] [PubMed] [Google Scholar]
- 67.Burhop J, Rosol M, Craig R, Tobacman LS, Lehman W. Effects of a cardiomyopathy-causing troponin t mutation on thin filament function and structure. J Biol Chem. 2001;276:20788–20794. doi: 10.1074/jbc.M101110200. [DOI] [PubMed] [Google Scholar]
- 68.Reedy MK, Holmes KC, Tregear RT. Induced changes in orientation of the cross-bridges of glycerinated insect flight muscle. Nature. 1965;207:1276–1280. doi: 10.1038/2071276a0. [DOI] [PubMed] [Google Scholar]
- 69.Reedy MK. Ultrastructure of insect flight muscle. I. Screw sense and structural grouping in the rigor cross-bridge lattice. J Mol Biol. 1968;31:155–176. doi: 10.1016/0022-2836(68)90437-3. [DOI] [PubMed] [Google Scholar]
- 70.Schmitz H, Lucaveche C, Reedy MK, Taylor KA. Oblique section 3-D reconstruction of relaxed insect flight muscle reveals the cross-bridge lattice in helical registration. Biophys J. 1994;67:1620–1633. doi: 10.1016/S0006-3495(94)80635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Padron R, Alamo L, Craig R, Caputo C. A method for quick-freezing live muscles at known instants during contraction with simultaneous recording of mechanical tension. J Microsc. 1988;151( Pt 2):81–102. doi: 10.1111/j.1365-2818.1988.tb04616.x. [DOI] [PubMed] [Google Scholar]
- 72.Taylor KA, Wu S, Reedy MC, Reedy MK. Imaging actomyosin in situ. Methods Cell Biol. 2007;79:321–368. doi: 10.1016/S0091-679X(06)79013-6. [DOI] [PubMed] [Google Scholar]
- 73.Padron R, Guerrero JR, Alamo L, Granados M, Gherbesi N, Craig R. Direct visualization of myosin filament symmetry in tarantula striated muscle by electron microscopy. J Struct Biol. 1993;111:17–21. doi: 10.1006/jsbi.1993.1031. [DOI] [PubMed] [Google Scholar]
- 74.Craig R, Padron R, Alamo L. Direct determination of myosin filament symmetry in scallop striated adductor muscle by rapid freezing and freeze substitution. J Mol Biol. 1991;220:125–132. doi: 10.1016/0022-2836(91)90386-k. [DOI] [PubMed] [Google Scholar]
- 75.Padron R, Alamo L, Guerrero JR, Granados M, Uman P, Craig R. Three-dimensional reconstruction of thick filaments from rapidly frozen, freeze-substituted tarantula muscle. J Struct Biol. 1995;115:250–257. doi: 10.1006/jsbi.1995.1049. [DOI] [PubMed] [Google Scholar]
- 76.Craig R, Alamo L, Padron R. Structure of the myosin filaments of relaxed and rigor vertebrate striated muscle studied by rapid freezing electron microscopy. J Mol Biol. 1992;228:474–487. doi: 10.1016/0022-2836(92)90836-9. [DOI] [PubMed] [Google Scholar]
- 77.Hirose K, Lenart TD, Murray JM, Franzini-Armstrong C, Goldman YE. Flash and smash: rapid freezing of muscle fibers activated by photolysis of caged ATP. Biophys J. 1993;65:397–408. doi: 10.1016/S0006-3495(93)81061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sosa H, Popp D, Ouyang G, Huxley HE. Ultrastructure of skeletal muscle fibers studied by a plunge quick freezing method: myofilament lengths. Biophys J. 1994;67:283–292. doi: 10.1016/S0006-3495(94)80479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sjostrom M, Squire JM. Fine structure of the A-band in cryo-sections. The structure of the A-band of human skeletal muscle fibres from ultra-thin cryo-sections negatively stained. J Mol Biol. 1977;109:49–68. doi: 10.1016/s0022-2836(77)80045-4. [DOI] [PubMed] [Google Scholar]
- 80.Menetret JF, Craig R. Unfixed cryosections of striated muscle to study dynamic molecular events. Biophys J. 1994;67:1612–1619. doi: 10.1016/S0006-3495(94)80634-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Humbel BM, Stierhof YD. Cryo-sectioning according to Tokuyasu. In: Cavalier A, Spehner D, Humbel BM, editors. Handbook of Cryo-preparation Methods for Electron Microscopy. CRC Press; Boca Raton: 2009. pp. 467–497. [Google Scholar]
- 82.Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlen LP, Richter K, Blanc NS, Studer D, Dubochet J. Cryo-electron microscopy of vitreous sections. EMBO J. 2004;23:3583–3588. doi: 10.1038/sj.emboj.7600366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dubochet J, Al-Amoudi A, Bouchet-Marquis C, Eltsov M, Zuber B. CEMOVIS: Cryo-electron microscopy of vitreous sections. In: Cavalier A, Spehner D, Humbel BM, editors. Handbook of Cryo-preparation Methods for Electron Microscopy. CRC Press; Boca Raton: 2009. pp. 259–289. [Google Scholar]
- 84.McDowall AW, Hofmann W, Lepault J, Adrian M, Dubochet J. Cryo-electron microscopy of vitrified insect flight muscle. J Mol Biol. 1984;178:105–111. doi: 10.1016/0022-2836(84)90233-x. [DOI] [PubMed] [Google Scholar]
- 85.Lucic V, Forster F, Baumeister W. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem. 2005;74:833–865. doi: 10.1146/annurev.biochem.73.011303.074112. [DOI] [PubMed] [Google Scholar]
- 86.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 87.DeRosier DJ, Moore PB. Reconstruction of three-dimensional images from electron micrographs of structures with helical symmetry. J Mol Biol. 1970;52:355–369. doi: 10.1016/0022-2836(70)90036-7. [DOI] [PubMed] [Google Scholar]
- 88.Owen CH, Morgan DG, DeRosier DJ. Image analysis of helical objects: the Brandeis Helical Package. J Struct Biol. 1996;116:167–175. doi: 10.1006/jsbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 89.Stewart M. Computer image processing of electron micrographs of biological structures with helical symmetry. J Electron Microsc Tech. 1988;9:325–358. doi: 10.1002/jemt.1060090404. [DOI] [PubMed] [Google Scholar]
- 90.Cammarato A, Hatch V, Saide J, Craig R, Sparrow JC, Tobacman LS, Lehman W. Drosophila muscle regulation characterized by electron microscopy and three-dimensional reconstruction of thin filament mutants. Biophys J. 2004;86:1618–1624. doi: 10.1016/S0006-3495(04)74229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Huxley HE, Brown W. The low-angle x-ray diagram of vertebrate striated muscle and its behaviour during contraction and rigor. J Mol Biol. 1967;30:383–434. doi: 10.1016/s0022-2836(67)80046-9. [DOI] [PubMed] [Google Scholar]
- 92.Al-Khayat HA, Morris EP, Kensler RW, Squire JM. 3D structure of relaxed fish muscle myosin filaments by single particle analysis. J Struct Biol. 2006;155:202–217. doi: 10.1016/j.jsb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 93.Egelman EH. A robust algorithm for the reconstruction of helical filaments using single-particle methods. Ultramicroscopy. 2000;85:225–234. doi: 10.1016/s0304-3991(00)00062-0. [DOI] [PubMed] [Google Scholar]
- 94.Egelman EH. Single-particle reconstruction from EM images of helical filaments. Curr Opin Struct Biol. 2007;17:556–561. doi: 10.1016/j.sbi.2007.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Egelman EH. The iterative helical real space reconstruction method: surmounting the problems posed by real polymers. J Struct Biol. 2007;157:83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 96.Jung HS, Komatsu S, Ikebe M, Craig R. Head-head and head-tail interaction: a general mechanism for switching off myosin II activity in cells. Mol Biol Cell. 2008;19:3234–3242. doi: 10.1091/mbc.E08-02-0206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jung HS, Burgess SA, Billington N, Colegrave M, Patel H, Chalovich JM, Chantler PD, Knight PJ. Conservation of the regulated structure of folded myosin 2 in species separated by at least 600 million years of independent evolution. Proc Natl Acad Sci U S A. 2008;105:6022–6026. doi: 10.1073/pnas.0707846105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wray JS. Structure of the backbone in myosin filaments of muscle. Nature. 1979;277:37–40. doi: 10.1038/277037a0. [DOI] [PubMed] [Google Scholar]
- 99.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 100.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wendt T, Taylor D, Trybus KM, Taylor K. Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proc Natl Acad Sci U S A. 2001;98:4361–4366. doi: 10.1073/pnas.071051098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Alamo L, Wriggers W, Pinto A, Bartoli F, Salazar L, Zhao FQ, Craig R, Padron R. Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. J Mol Biol. 2008;384:780–797. doi: 10.1016/j.jmb.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lehman W, Vibert P, Uman P, Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J Mol Biol. 1995;251:191–196. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]