Abstract
Plasma HIV-1 RNA was measured in 306 samples, collected from 273 highly active antiretroviral therapy (HAART)-experienced men, using both the Roche COBAS TaqMan (limit of detection [LD]=20 copies/mL) and Roche Amplicor (LD=50 copies/mL) assays. Mixtures of Gaussian distributions incorporating left-censored data were used in analyses. The more sensitive TaqMan assay estimated that 23% and 0.0003% of HIV-1 RNA values would be below 1 copy/mL and 1 copy/3L, respectively. This is in sharp contrast to the overestimation provided by the less sensitive Amplicor assay, whereby the corresponding predicted percentages were 51% and 1%. Both assays appropriately characterized sub-optimal virologic response as the rightmost peaks of both distributions provided an excellent fit to the observed data. Our results based on a widely available 20 copies/mL sensitive assay reproduce those obtained using customized assays that quantified HIV-1 RNA values as low as 1 copy/mL.
Keywords: limit of detection, HIV-1 RNA, bimodal distribution, HAART
INTRODUCTION
Plasma HIV-1 RNA levels among individuals receiving highly active antiretroviral therapy (HAART) have been shown to follow a bimodal distribution1. The rightmost peak of the distribution reflects individuals with sub-optimal virologic response (e.g., due to viral drug resistance or poor adherence to therapy), while the leftmost peak, which includes the great majority of individuals on HAART, reflects the maximal suppression that can be achieved. The latter peak has not been well characterized because it is comprised primarily of values that are below the limits of detection (LD) of assays that have been commercially available (e.g., 50 copies/mL). Recently, however, more sensitive assays have been developed, either research assays that can measure down to 1 copy/mL or commercial assays that can measure down to 20 copies/mL2–6. These assays have the potential to delineate more clearly the leftmost peak of the HIV-1 RNA distribution in treated populations. The characterization of this leftmost peak is important to understand not only the effectiveness of HAART, but also the biology of optimally treated HIV-1 infection.
Two assays from Roche Molecular Diagnostics used to measure HIV-1 RNA are the Ampliprep/Amplicor MONITOR® version 1.5 7, with LD of 50 copies/mL, and the recently FDA-approved COBAS® Ampliprep/COBAS® TaqMan version 2.0 8, whose LD is 20 copies/mL. The lower LD of the Taqman assay, allowing for the quantification of plasma HIV-1 RNA values between 20 and 49 copies/mL, reduces the number of left-censored observations and thus improves estimation of the distribution of HIV-1 RNA levels below 20 copies/mL. In 2010, the Multicenter AIDS Cohort Study (MACS), which had been measuring HIV-1 RNA using the Monitor assay, adopted the TaqMan assay for routine use. The goal of the current study was to characterize the distribution of suppressed HIV-1 RNA values among HAART-experienced individuals using an assay with an LD of 20 copies/mL and to show the misrepresentation provided by a less sensitive assay with an LD of 50 copies/mL.
METHODS
Study Population
The MACS was initiated in 1984 to study the natural history of HIV-1 infection among homosexual and bisexual men at four sites in the United States (Chicago; Baltimore/Washington, DC; Los Angeles; and Pittsburgh). The study design and characteristics of the study participants have been described previously9, 10. Briefly, a total of 3554 participants were either HIV-1-infected at enrollment (81%) or became HIV-1-infected during follow-up (19%). Of the 1854 HIV-1-infected men who were observed after January 1995, 1488 (80%) initiated HAART11 before October 2010. Appropriate Institutional Review Boards approved study protocols and consent forms, and each study participant gave written informed consent.
Fresh specimens from 200 consecutive HIV-1-infected participants (100 from the Baltimore/Washington DC site and 100 from the Los Angeles site) seen between June 2010 and September 2010 were tested with both TaqMan and Amplicor HIV-1 RNA assays. One hundred and twenty-seven samples of plasma stored at −70° C in local freezers and taken from participants seen between May 2009 and May 2010 were also tested with both HIV-1 RNA assays. Our analyses included the 306 (94% of 327) samples taken from 273 individuals who had initiated HAART. HAART was reported at 263 (86%) of the 306 study visits included in analyses; combination therapy at 15 (5%) study visits, monotherapy at 2 (<1%) study visits, and no therapy at 14 (5%) of study visits. At 12 (4%) of the 306 study visits, data on antiretroviral therapy used concomitantly with the HIV-1 RNA measurement was not available. At the 263 study visits in which HAART was reported, at least one protease inhibitor was reported in 133 (51%) instances and one non-nucleoside transcriptase inhibitor was 143 (54%) times. The median time since HAART had been initiated was 11.8 years (IQR = 7.6 – 13.4).
HIV-1 RNA Assays
Both the COBAS® TaqMan HIV-1 Test, Version 2.0, and the Roche Amplicor HIV-1 MONITOR® Test, Version 1.5, are tests for the quantification of HIV-1 RNA based on in vitro amplification of the highly conserved HIV-1 gag gene7, 8. The Amplicor is a semi-automated, conventional PCR assay that has a dynamic range of 50 – 750,000 copies/mL. The TaqMan is a completely automated, real-time PCR assay that has a broader dynamic range (20 – 10,000,000 copies/mL) and also targets the HIV-1 LTR region.
Statistical Analyses
Let Y denote the left-censored random variable log10(HIV-1 RNA) using the TaqMan assay with LD= log10(20) copies/mL, which hereafter we will refer to as L20. We allow Y to follow a bimodal mixture of two Gaussian distributions, one of which is the distribution of log10(HIV-1 RNA) measurements among individuals with lower levels (leftmost peak), and the second of which corresponds to the distribution of log10(HIV-1 RNA) measurements among individuals with higher levels (rightmost peak). Maximum likelihood methods were used as described by Chu et al.12 Similar methods were used for the measurements from the Amplicor assay with LD as L50= log10(50) copies/mL.
To determine the correlation between the TaqMan and Amplicor measurements in both the leftmost and rightmost peaks (i.e., two correlations), we allowed the random vector Y = (Y1, Y2) = (log10[TaqMan-measured HIV-1 RNA], log10[Amplicor-measured HIV-1 RNA]) to follow a bivariate mixture of two Gaussian distributions. Maximum likelihood methods were used as described by Chu et al.12
To determine the goodness of fit of each model, we compared the predicted percentage of HIV-1 RNA measurements falling below different levels with the observed percentages. More importantly, appropriately modeling the distribution of HIV-1 RNA below the limit of detection allows for the estimation of the percentage of individuals with lower than, for example, 1 copy per mL, as well as the percentage expected to have less than 1 copy in the whole plasma volume (~3 liters). Acknowledging the immunological finding that HIV cannot be completely eradicated from HIV-infected individuals with current therapies6, 13–15, the percentage of individuals with less than 1 copy per 3 liters of plasma is expected to be very low.
RESULTS
The median age of the 273 men was 52.4 years (inter-quartile range [IQR] = 46.4 – 58.6); 141 (52%) were White and 71 (26%) were African American. The median CD4 cell count was 568 cells/mm3 (IQR= 414 – 741). Forty-seven (17%) had a history of an AIDS-defining condition prior to the study visit at which their HIV-1 RNA was measured.
The HIV-1 RNA values and CD4 cell counts of the MACS men included in this convenience sample are internally consistent with the distributions of HIV-1 RNA and CD4 cell counts of the entire MACS HIV-infected population. Specifically, at their last study visit between October 2009 through September 2010, 79% of the entire cohort had undetectable HIV-RNA using the Amplicor assay (similar to the percentage of Amplicor measurements that were ≤50 copies/mL in our study population described below), and their median CD4 cell count was 557 cells/mm3 (IQR = 406, 728), similar to the median of 568 cells/mm3 [IQR = 414, 741] seen among individuals included in this study.
Using the Amplicor assay, 254 (83%) of the HIV-1 RNA measurements were less than or equal to 50 copies/mL. Two hundred and sixteen (71%) of the HIV-1 RNA measurements using the TaqMan assay were less than or equal to 20 copies/mL, and 30 (10%) were between 21 and 50 copies/mL. Of the 306 samples tested, 214 (70%) were ≤L20 for TaqMan and ≤L50 for Amplicor; 50 (16%) were >L20 and >L50; 40 (13%) were >L20 and ≤L50; and 2 (1%) were ≤L20 and >L50. The median duration of undetectable HIV-1 RNA in those who were ≤L20 for TaqMan and ≤L50 for Amplicor was approximately 4.5 years.
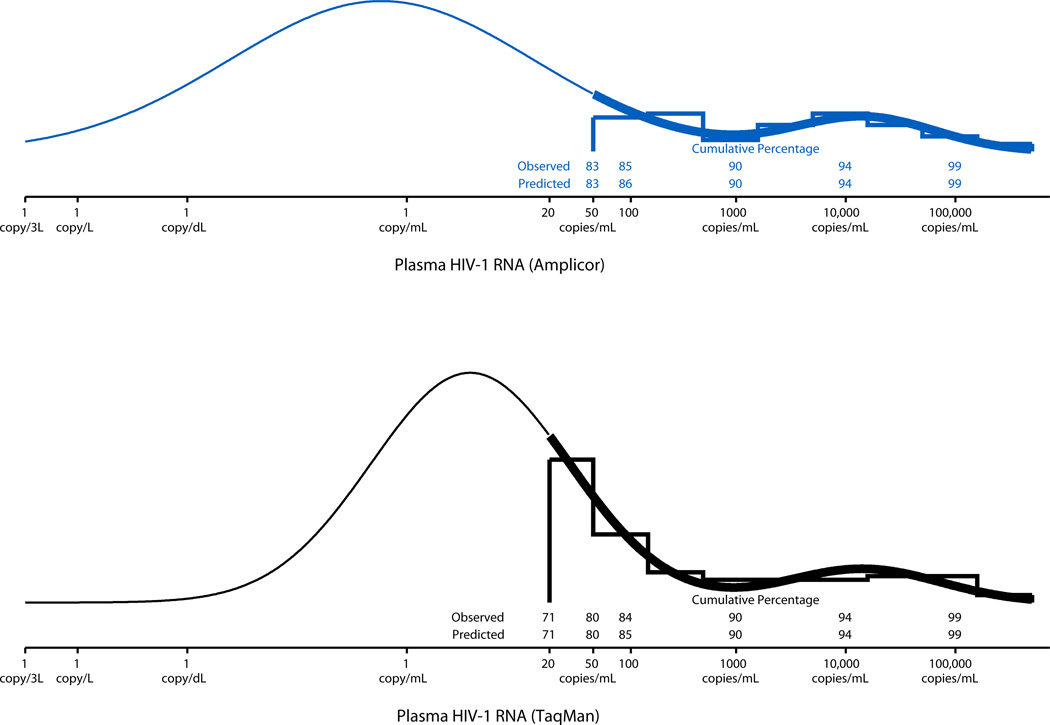
The bimodal mixture of two Gaussian distributions used to describe the distribution of log10(HIV-1 RNA) values for the Amplicor and TaqMan assays are shown in the top and bottom panels of Figure 1, respectively. The goodness of fit of the bimodal distributions to the observed data is readily apparent from the nearly perfect agreement between the observed and predicted cumulative percentages shown for both assays.
FIGURE 1.
Distributions of HIV-1 RNA measurements on 306 samples using the Amplicor assay (top panel) and Taqman (bottom panel) assay with fitted bimodal mixture distributions. The leftmost peaks comprised 91% and 90% of the Amplicor and TaqMan values, respectively. The leftmost peaks were centered at 0.59 copies/mL for Amplicor and 3.78 copies/mL for TaqMan; the rightmost peaks were centered at 14,673 copies/mL for Amplicor and 14,314 copies/mL for TaqMan. Thick lines are used over the interval of observed HIV-1 RNA levels above the LD; thin lines are used over the interval of estimated HIV-1 RNA levels below the LD. The predicted cumulative percentages of observations falling below a given HIV-1 RNA value were estimated using the curve for each assay.
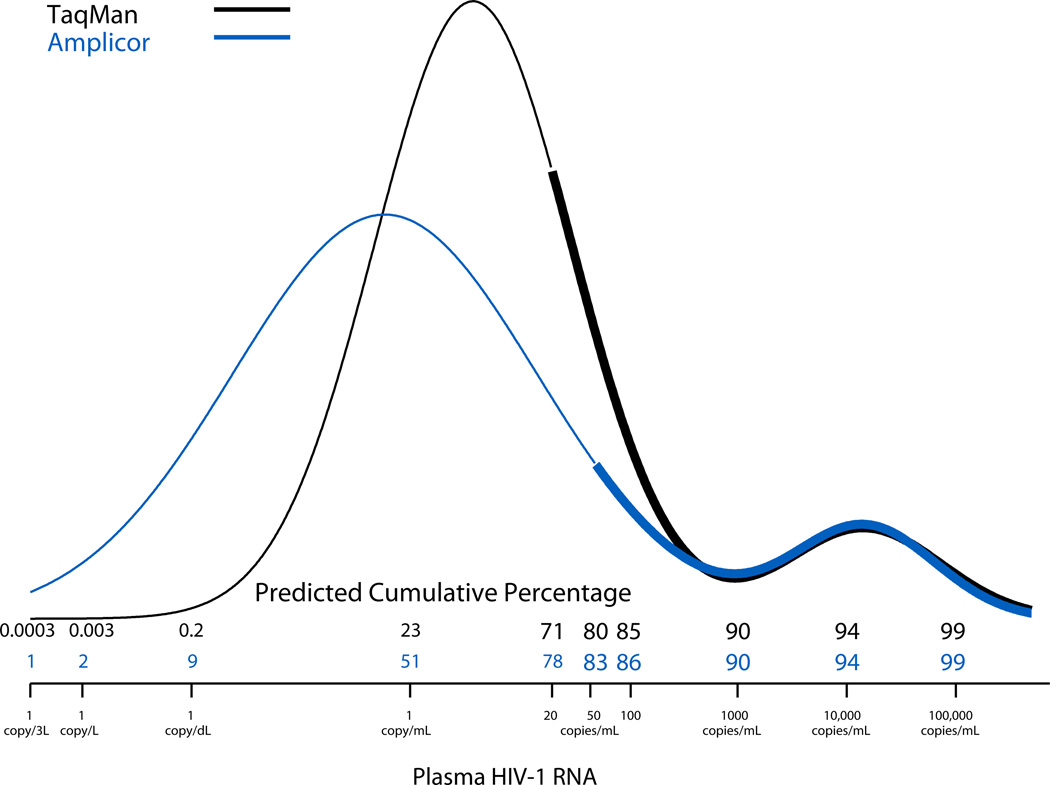
Figure 2 shows the fitted bimodal distributions for the Amplicor (blue line) and TaqMan (black line) assays. The 9% (=80% – 71%) of the HIV-1 RNA measurements that were between 21 and 50 copies/mL using the TaqMan assay represent the measurements that would have been undetectable if the LD had been 50 copies/mL instead of 20 copies/mL. The two distributions were very similar for values >1000 copies/mL (ρR = 0.99), but diverged substantially for HIV-1 RNA levels <50 copies/mL (ρL= 0.63). The leftmost peak of the TaqMan distribution was centered at a higher value (median = 3.78 copies/mL) than that of the Amplicor distribution (median = 0.59 copies/mL). Furthermore, the more sensitive TaqMan assay predicted that 23%, 0.2%, 0.003%, and 0.0003% of values would be <1 copy/mL, <1 copy/dL, <1 copy/L, and <1 copy/3L, respectively, consistent with clinical observations indicating that HIV-1 RNA cannot be completely eradicated by current HAART regimens. This is in sharp contrast to the higher estimates provided by the less sensitive Amplicor assay, whereby the corresponding predicted percentages were 51%, 9%, 2%, and 1%, respectively.
FIGURE 2.
Bimodal mixture distributions summarizing the HIV-1 RNA distributions derived from two assays used to measure HIV-1 RNA. Thick lines are used over the interval of observed HIV-1 RNA levels above the LD; thin lines are used over the interval of estimated HIV-1 RNA levels below the LD. The predicted cumulative percentage of observations falling below a given HIV-1 RNA value were estimated using the curve for each assay. The predicted cumulative percentages in smaller font correspond to predictions made at and below the LD.
DISCUSSION
The present study confirms our previous report that the distribution of post-HAART plasma log10(HIV-1 RNA) measurements is bimodal1. It also extends the previous report by incorporating data obtained using the TaqMan assay, which has an LD of 20 copies/mL rather than 50 copies/mL for the Amplicor assay used in the previous report, and thus allowed for a much better estimation of the distribution of HIV-1 RNA at very low values.
Use of the TaqMan assay provided data for 9% of the HIV-1 RNA distribution that were not measured using the Amplicor assay, i.e., the values between 21 and 50 copies/mL. With these added data, the leftmost peak of the distribution of TaqMan measurements was steeper and farther to the right (median= 3.78 copies/mL) than the distribution of HIV-1 RNA using the Amplicor assay (median = 0.59 copies/mL), and predicted many fewer individuals with levels of viremia <1 copy/mL than the Amplicor assay.
The improved estimates are consistent with the findings of studies indicating that HIV-1 RNA cannot be eradicated completely from circulating CD4 T cells by present HAART regimens13–15. In addition, using the TaqMan assay, our estimates of the leftmost peak and the percentage of observations predicted to be <1 copy/mL are strongly supported by the measurements obtained in two studies that used a specialized assay that quantified plasma HIV-1 RNA values as low as 1 copy/mL. Maldarelli et al. found a mean HIV-1 RNA of 3.1 copies/mL in people who had maintained HIV-1 RNA values <50 copies/mL for 1–2 years while receiving HAART, with >80% of samples having detectable viremia16. Similarly, Palmer et al. studied 293 plasma samples from 40 people who maintained HIV-1 RNA values <50 copies/mL for several years while receiving HAART, and found a median value of 3.34 copies/mL among the 77% of samples yielding values ≥1 copy/mL4. These estimates are in excellent agreement with the estimated median estimate of 3.78 copies/mL and 23% of values below 1 copy/mL in the present study. Taken together, these data suggest that the TaqMan assay with LD = 20 copies/mL is adequate for characterizing the distribution of suppressed HIV-1 RNA values. On the other hand, the Amplicor assay with LD= 50 copies/mL produces a stark misrepresentation of the distribution of suppressed HIV-1 RNA at very low levels.
The improved estimate of the distribution of HIV-1 RNA among individuals achieving viral suppression on HAART permits more precise quantitation of the effectiveness of HAART. Although plasma HIV-1 concentrations can now be directly measured down to 1 copy/mL using specialized assays, such assays require large amounts of plasma (e.g., 7 mL) and may not be feasible in large studies or at most clinical sites. Therefore, the methods and improved estimates provided in the present study may be useful in studies of the effect of residual HIV-1 viremia in people receiving HAART and in the assessment of medications or therapeutic approaches that may suppress HIV replication even more effectively than current HAART regimens, or that reduce the latent reservoirs of HIV-1.
ACKNOWLEDGMENTS
Data in this manuscript were collected by the Multicenter AIDS Cohort Study (MACS) with centers (Principal Investigators) located at: The Johns Hopkins Bloomberg School of Public Health (Joseph B. Margolick); Howard Brown Health Center and Northwestern University Medical School (John P. Phair); University of California, Los Angeles (Roger Detels); University of Pittsburgh (Charles R. Rinaldo); and Data Analysis Center (Lisa Jacobson).
The study was supported by funding from the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute. UO1-AI-35042, UL1-RR025005, UO1-AI-35043, UO1-AI-35039, UO1-AI-35040, and UO1-AI-35041.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Li X, Chu H, Gallant JE, et al. Bimodal virological response to antiretroviral therapy for HIV infection: an application using a mixture model with left censoring. J Epidemiol Community Health. 2006;60:811–818. doi: 10.1136/jech.2005.044644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sizmann D, Glaubitz J, Simon CO, et al. HIV-1 RNA quantitation by COBAS AmpliPrep/COBAS TaqMan HIV-1 Test, v2.0 using a novel dual-target approach. J Clin Virol. 2010;49:41–46. doi: 10.1016/j.jcv.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Glaubitz J, Sizmann D, Simon CO, et al. Accuracy to 2ndInternational HIV-1 RNA WHO Standard: assessment of three generations of quantitative HIV-1 RNA nucleic acid amplification tests. J Clin Virol. 2011;50:119–124. doi: 10.1016/j.jcv.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Palmer S, Maldarelli F, Wiegand A, et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci. USA. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinoso JB, Kim SY, Siliciano RF, et al. A comparison of viral loads between HIV-1-infected elite suppressors and individuals who receive suppressive highly active antiretroviral therapy. Clin Infect Dis. 2008;47:102–104. doi: 10.1086/588791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dinoso JB, Kim SY, Wiegand AM, et al. Treatment intensification does not reduce residual HIV-1 viremia in patients on highly active antiretroviral therapy. Proc Natl Acad Sci USA. 2009;106:9403–9408. doi: 10.1073/pnas.0903107106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amplicor HIV-1 MONITOR®Test, version 1.5 [package insert] Branchburg, NJ: Roche Molecular systems, Inc.; 2002. [Google Scholar]
- 8.COBAS® AmpliPrep/COBAS® TaqMan® HIV-1 Test, version 2.0 [package insert] Branchburg, NJ: Roche Molecular systems, Inc.; 2010. [Google Scholar]
- 9.Kaslow RA, Ostrow DG, Detels R, et al. The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am J Epidemiol. 1987;126:310–318. doi: 10.1093/aje/126.2.310. [DOI] [PubMed] [Google Scholar]
- 10.Dudley J, Jin S, Hoover D, et al. The Multicenter AIDS Cohort Study: retention after 9 ½ years. Am J Epidemiol. 1995;142:323–330. doi: 10.1093/oxfordjournals.aje.a117638. [DOI] [PubMed] [Google Scholar]
- 11.Guidelines for the use of antiretroviral agents in HIV-1 infected adults and adolescents. Department of Health and Human Services; 2011. Jan 10, pp. 1–166. Available at: http://aidsinfonihgov/contentfiles/AdultandAdolescentGL.pdf. [Google Scholar]
- 12.Chu H, Moulton LH, Mack WJ, et al. Correlating two continuous variables subject to detection limits in the context of mixture distributions. Appl Statist. 2005;54:831–845. [Google Scholar]
- 13.Finzi D, Hermankova M, Pierson T, et al. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 14.Chun TW, Stuyver L, Mizell SB, et al. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci USA. 1997;94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey JR, Sedaghat AR, et al. Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J Virol. 2006;80:6441–6457. doi: 10.1128/JVI.00591-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maldarelli F, Palmer S, King MS, et al. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]