Summary
The nuclear matrix associated hnRNP U/SAF-A protein has been implicated in diverse pathways from transcriptional regulation to telomere length control to X inactivation, but the precise mechanism underlying each of these processes has remained elusive. Here, we report hnRNP U as a regulator of SMN2 splicing from a custom RNAi screen. Genome-wide analysis by CLIP-seq reveals that hnRNP U binds virtually to all classes of regulatory non-coding RNAs, including all snRNAs required for splicing of both major and minor classes of introns, leading to the discovery that hnRNP U regulates U2 snRNP maturation and Cajal body morphology in the nucleus. Global analysis of hnRNP U-dependent splicing by RNA-seq coupled with bioinformatic analysis of associated splicing signals suggests a general rule for splice site selection through modulating the core splicing machinery. These findings exemplify hnRNP U/SAF-A as a potent regulator of nuclear ribonucleoprotein particles in diverse gene expression pathways.
Introduction
hnRNP U was initially characterized as a component of heterogeneous ribonucleoprotein (RNP) particles or as a nuclear scaffold attachment factor A (SAF-A) (Kiledjian and Dreyfuss, 1992; Romig et al., 1992). About 50% of the protein is tightly attached to operationally defined “nuclear matrix” and biochemical analysis suggests that hnRNP U/SAF-A preferentially binds to A/T-rich double-stranded DNA, known as scaffold attachment regions, and to G/U-rich heterogeneous RNA (Fackelmayer and Richter, 1994; Kiledjian and Dreyfuss, 1992). The N-terminal domain of hnRNP U/SAF-A mediates its binding to DNA, whereas its C-terminal RGG domain is responsible for its RNA binding activities (Kim and Nikodem, 1999). The ability of hnRNP U/SAF-A to bind to both DNA and RNA has been postulated to play a critical role in high order organization of the nucleus (Fackelmayer et al., 1994).
hnRNP U/SAF-A is required for cell viability and a hypomorphic mutation of the gene causes early embryonic lethality in mice, indicating an essential role of the gene in the cell (Roshon and Ruley, 2005). Indeed, hnRNP U/SAF-A has been linked to a plethora of regulated gene expression processes, including transcriptional initiation or elongation through its interaction with the glucocorticoid receptor (Eggert et al., 1997), nuclear actin and the C-terminal domain of Pol II (Kukalev et al., 2005; Obrdlik et al., 2008), the transcription co-activator p300 (Martens et al., 2002), and the heterochromatic protein HP1α (Ameyar-Zazoua et al., 2009). Most of these interactions, however, were based on yeast two-hybrid assays or through affinity purification. Thus, it has been unclear whether the interactions are direct or mediated by a third party, nor the precise mechanism for positive or negative regulation of various gene expression events (Kim and Nikodem, 1999; Kukalev et al., 2005).
hnRNP U/SAF-A has also been implicated in various aspects of RNA metabolism, including RNA transport on a viral system (Gupta et al., 1998; Valente and Goff, 2006), RNA stability control via its binding to the 3′UTR of TNFα (Yugami et al., 2007), and the regulation of telomere length (Fu and Collins, 2007; Jady et al., 2004). More recently, several reports documented a pivotal role of hnRNP U/SAF-A in X inactivation where hnRNP U/SAF-A is not only recruited to Xi (the X chromosome to be inactivated in female) via the non-coding RNA Xist, but is also required for Xist to bind to Xi to establish gene silencing (Hasegawa et al., 2010; Helbig and Fackelmayer, 2003; Pullirsch et al., 2010).
Interestingly, despite its original identification as an hnRNP protein, thus indicative of a potential role in regulated splicing, the evidence for this widely anticipated function has been lacking. Through mass spectrometric analysis, hnRNP U/SAF-A has been reported to associate with purified spliceosomes (Rappsilber et al., 2002). However, another group failed to detect such association in a similar analysis (Zhou et al., 2002), indicating that hnRNP U/SAF-A may not be a core component of the spliceosome. It is also interesting to note that the Dreyfuss lab initially used the C-terminal RGG domain of hnRNP U to isolate the Survival of Motor Neuron (SMN1) gene in a two-hybrid screen (Liu and Dreyfuss, 1996). They went on to study the function of SMN1, leading to the discovery of SMN1 as a key molecular chaperon for snRNP biogenesis (Pellizzoni et al., 2002), but the question remains open whether or not hnRNP U itself plays a role in snRNP biogenesis or RNA splicing.
Our present work began with an unbiased RNAi screen against a large panel of RNA binding proteins in an attempt to identify potential splicing regulators of SMN2, a paralog of the SMN1 gene in the human genome. SMN2 carries a point C-to-T transition on exon 7, causing ~80% skipping of the exon and the production of an unstable SMN protein, which is sufficient to support embryonic development, but insufficient to fulfill the functional requirement of SMN1 in motor neurons (Gavrilov et al., 1998; Hsieh-Li et al., 2000). The spared SMN2 gene in the human genome may thus serve as a target for developing therapeutic strategies against the motor neuron disease through boosting its splicing efficiency. Biochemical studies have indeed identified a number of RNA binding proteins in the regulation of SMN2 splicing, including SRSF1 (Cartegni and Krainer, 2002), hnRNP A1/A2 (Kashima and Manley, 2003), hTra2β (Hofmann and Wirth, 2002), Sam68 (Pedrotti et al., 2010), etc. We now show that hnRNP U/SAF-A is one of the most potent SMN2 splicing regulators. We found that hnRNP U/SAF-A interacts with many non-coding RNAs, including all snRNAs required for splicing of both major and minor classes of introns, which led to the elucidation of a key role of hnRNP U/SAF-A in regulating U2 snRNP maturation. These findings not only reveal an precedented regulatory paradigm for splicing control, but also illuminate a mechanism for this nuclear matrix protein to modulate diverse RNP-mediated activities in mammalian cells.
Results
Identification of hnRNP U as a SMN2 splicing regulator in an esiRNA screen
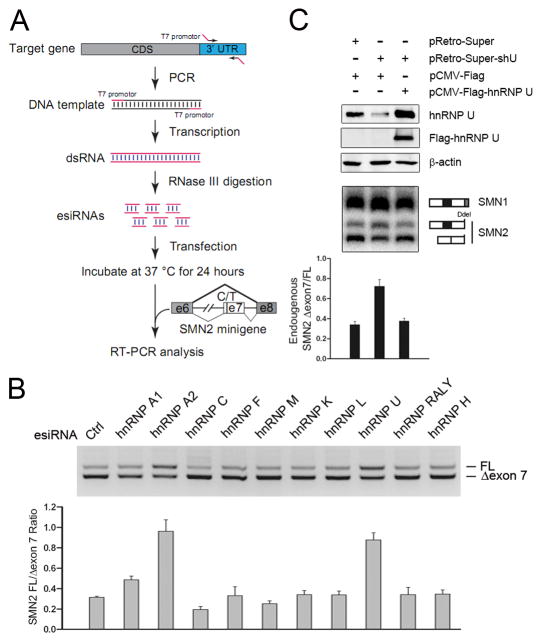
To systematically identify RNA binding proteins (RBPs) involved in SMN2 splicing control, we conducted a functional screen using a custom esiRNA library (Kittler et al., 2007) against 340 annotated RBPs encoded in the human genome (Fig. 1A, Table S1). We scored their effects on an SMN2-based splicing reporter in co-transfected HeLa cells (Fig. 1B). This analysis confirmed the reported role of hnRNP A1/A2 in suppressing SMN2 exon 7 inclusion (Kashima and Manley, 2003) where hnRNP A2 appears more potent than hnRNP A1 in the regulation (Fig. 1B, Fig. S1A–C). Several 3′ splice site recognition factors, including SF1, PUF60, and U2AF65, also scored positive in the screen (Fig. S1C–F), the latter two of which have been reported in previous studies (Hastings et al., 2007; Martins de Araujo et al., 2009). In this screen, we identified hnRNP U as one of the most potent inhibitors of SMN2 exon 7 inclusion (Fig. 1B). We confirmed the result using two independent synthetic siRNAs against hnRNP U, while a non-targeting control siRNA showed no effect (Fig. S1G–H).
Figure 1. Identification of hnRNP U as a potent splicing regulator from an esiRNA screen.
(A) The scheme of the esiRNA screen strategy. Specific regions in the 3′UTR of individual RBPs (Table S1) were PCR-amplified and in vitro transcribed into dsRNAs. The long dsRNAs were digested with purified RNase III and small dsRNAs purified from native PAGE. Individual small dsRNAs were co-transfected with the SMN2 minigene reporter carrying the alternative exon 7 in HeLa cells and the products were analyzed by RT-PCR. (B) Results of a representative panel of hnRNP proteins from the esiRNA screen. The effects were quantified in the bottom panel. (C) Rescue of the splicing response to hnRNP U RNAi with a FLAG-tagged, RNAi-resistant version of hnRNP U. Cleavage by Dde I was used to distinguish the PCR products of the SMN2 pre-mRNA and spliced mRNA from that of spliced SMN1 mRNA. The results were quantified and shown at bottom. Data in B and C are shown as mean ± SD.
We next determined whether hnRNP U RNAi affected the splicing of the endogenous SMN2 gene by using an shRNA against hnRNP U (Fig. 1C). As the spliced products of SMN1 and SMN2 differ by only one nucleotide, we examined the spliced product of SMN2 by taking advantage of a unique restriction site (Dde I) present in exon 8 of SMN2. Cleavage of the PCR product generated two smaller fragments from the spliced SMN2 mRNA in addition to the uncleaved product from the spliced SMN1 mRNA (Fig. 1C). This analysis demonstrated that in vivo depletion of hnRNP U induced exon 7 inclusion from the endogenous SMN2 gene. The effect could be rescued by re-expressing an RNAi-resistant version of hnRNP U, thus ruling out potential off-target effect in the analysis (Fig. 1C). Together, these data established hnRNP U as a regulator of SMN2 splicing.
Cooperation of hnRNP U and hnRNP A in the regulation of SMN splicing
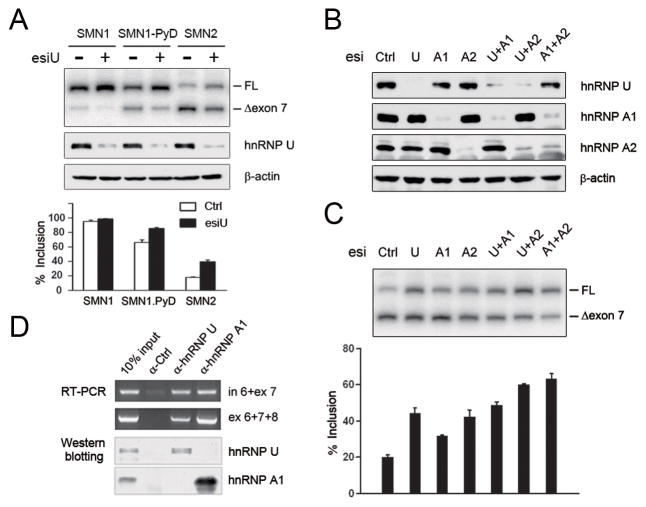
We next determined whether hnRNP U regulated SMN2 splicing was due to the point mutation in exon 7 by testing a mutant version of SMN1 containing a 3′ splice site mutation in its intron 6 (SMN1-PyD). Previous studies showed that the weakened 3′ splice site converted exon 7 of SMN1 from a constitutive to an alternative exon (Cartegni et al., 2006), thus permitting us to test the effect of hnRNP U knockdown. We found that hnRNP U depletion enhanced exon inclusion from both SMN1 and SMN2 minigenes, rather than selectively on SMN2 (Fig. 2A).
Figure 2. Involvement of hnRNP U in both SMN1 and SMN2 splicing independent of the effect of hnRNP A1/A2.
(A) Splicing response of SMN1, a mutant SMN1 containing a weakened polypyrimidine tract (SMN1-PyD), and SMN2 to hnRNP U knockdown. The results were quantified at bottom. (B) Single and double knockdown of hnRNP U and hnRNP A1/A2 analyzed by Western blotting. (C) Effects of single and double knockdown of hnRNP U and hnRNP A1/A2 on SMN2 splicing with quantified results shown at bottom. (D) Both hnRNP U and hnRNP A1 were associated with pre- and spliced SMN2 mRNA, but no interaction was detected between the two hnRNP proteins. Data in A and C are shown as mean ± SD.
This finding is reminiscent of the role of hnRNP A1/2 in regulating SMN splicing where these hnRNP proteins elicit a general influence on splicing of both SMN1 and SMN2, likely by competing with other general splicing regulators, such as SR proteins (Cartegni et al., 2006). To evaluate independent contribution of hnRNP U and hnRNP A to the regulation of SMN splicing, we knocked down these hnRNP proteins either individually or in combination in HeLa cells (Fig. 2B), finding that their effect on SMN2 splicing was not dependent on one another (Fig. 2C). Similarly, double knockdown of hnRNP A1 and A2 also exhibited independent effects in promoting SMN2 splicing (Fig. 2C). These results suggest that hnRNP U and hnRNP A1/A2 all act independently to regulate SMN2 splicing.
hnRNP A1 and A2 are known to regulate splicing by binding to pre-mRNA. To determine whether hnRNP U functions in a similar fashion, we performed a RiboIP experiment in which we separately immunoprecipitated hnRNP A1 and hnRNP U to determine whether they could each bind to endogenous SMN transcripts (Fig. 2D). We found that both interacted with the SMN1/2 pre-mRNA (indicated by the PCR product containing intron 6 and exon 7) as well as spliced mRNA (indicated by the PCR product containing exon 6 to exon 8). However, we did not detect any interaction by co-IP between hnRNP U and hnRNP A1, indicating that hnRNP U may not function in a stable complex with hnRNP A1 to modulate SMN splicing (Fig. 2D).
Genome-wide analysis of hnRNP U/RNA interactions by CLIP-seq
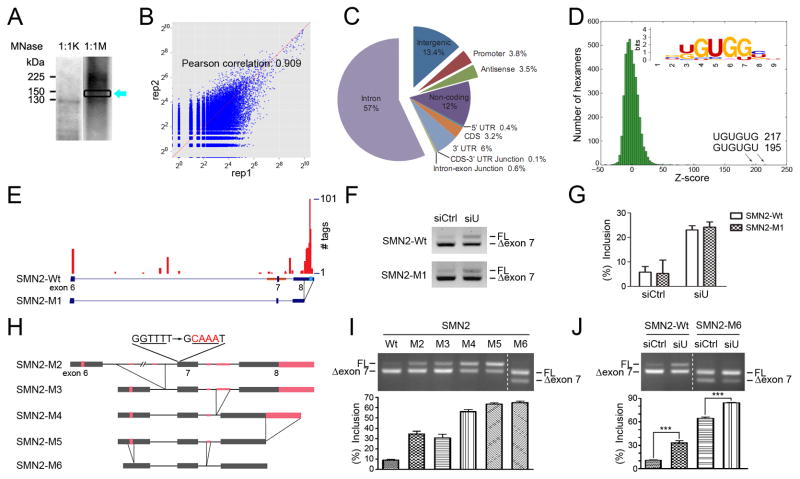
To address the mechanism underlying hnRNP U-regulated splicing, we set out to first pursue its in vivo RNA binding profile by performing CrossLinking ImmunoPrecipitation coupled with deep sequencing (CLIP-seq), which has been instrumental in identifying in vivo targets for RNA binding proteins and elucidating rules in regulated splicing (Licatalosi et al., 2008; Wang et al., 2010; Xue et al., 2009; Yeo et al., 2009). For this purpose, we UV-irradiated HeLa cells to induce crosslinking between proteins and nucleic acids followed by immunoprecipitation using a specific anti-hnRNP U antibody while a control IgG produced no signal (not shown). We isolated short smear above the position of hnRNP U (Fig. 3A). The resulting CLIP library was subjected to high throughput sequencing on an Illumina sequencer, yielding ~9 million tags that were uniquely mapped to the human genome. Two independent CLIP-seq experiments confirmed high reproducibility (R2=0.909) of the assay (Fig. 3B) and power analysis indicated that this level of tag density has reached ~80% saturation (Fig. S2A).
Figure 3. Global analysis of hnRNP U binding to RNA by CLIP-seq and mutational analysis of hnRNP U binding sites on SMN2.
(A) SDS-PAGE analysis of immunoprecipitated hnRNP U linked to RNA. Samples were treated with two concentrations of micrococcal nuclease (1:1000 and 1:1 million dilutions). RNA UV-crosslinked to hnRNP U was 32P-labeled by T4 kinase and RNA-protein adducts above the position of hnRNP U (boxed) were recovered from the gel to construct CLIP libraries. (B) Comparison between two independent hnRNP U CLIP libraries. Mapped tags were compared on serial 500nt windows in the human genome. (C) The genomic distribution of hnRNP U CLIP tags with a significant fraction of hnRNP U binding events mapped to annotated non-coding RNAs. (D) Deduced hnRNP binding consensus based on Z-scoring. The two most enriched motifs are indicated by arrows and the top 20 motifs (Fig. S2B) were aligned by ClusterW to general the consensus shown in the insert. (E) The profile of hnRNP U binding on SMN2 around the alternative exon 7 (based on the first CLIP-seq data). The blue box indicates the G/U-rich sequence in exon 8, which was deleted to generate the SMN2-M1 mutant. The horizontal red line is the fragment containing exon 7 inserted into a split GFP gene in Fig S2C. (F and G) Both wt SMN2 and the SMN2-M1 mutant were analyzed in cells treated with a control or hnRNP U RNAi (F) and quantified (G). Errors are based on three independent experiments. (H) Constructs containing serial deletions of all mapped and potential hnRNP U binding sites in the SMN2 minigene. (I) The splicing efficiency of individual deletion mutants in transfected HeLa cells. (J) The response of the “all” deletion mutant (SMN2-M6) to hnRNP U RNAi. Data in E, I and J are shown as mean ± SD; *** in J indicates p<0.001.
The mapped tags are distributed among diverse primary RNA transcripts with the majority (57%) mapped to intronic regions of pre-mRNA (Fig. 3C), consistent with the association of hnRNP U with hnRNP particles. Interestingly, we also detected hnRNP U binding to various non-coding RNAs (12%), which appears more prevalent than several other RNA binding proteins we previously analyzed under the same conditions (Xue et al., 2009; Yeo et al., 2009). Motif analysis based on Z-scoring revealed highly enriched hexamers consisting of UG or UGG repeats (Fig. 3D), which agrees with reported high affinity binding of hnRNP U to G or U homopolymers (Kiledjian and Dreyfuss, 1992). The top 20 hexamers (Fig. S2B) were used to derive an U/G-rich hnRNP U binding consensus (Fig. 3D).
Evidence against hnRNP U-dependent splicing through direct binding to SMN2 pre-mRNA or via a transcription-coupled mechanism
The mapped hnRNP U binding profile provides initial clues to its role in regulated splicing. Interestingly, we detected significant binding of hnRNP U on SMN2 exon 8 at a G/U-rich region (Fig. 3E). As blocking the 3′ splice site of exon 8 or weakening the 3′ splice site recognition factors have been shown to enhance exon 7 inclusion (Hastings et al., 2007), such binding profile suggests that hnRNP U might suppress the recognition of the 3′ splice site of exon 8, thereby elevating the competitiveness of splicing signals around exon 7 of SMN2. To test this hypothesis, we removed the G/U-motif in exon 8 (SMN2-M1), and unexpectedly, we found that the mutant still responded to hnRNP U depletion, resulting in enhanced exon 7 inclusion (Fig. 3F–G).
Because of additional hnRNP U binding activities detected on other locations surrounding exon 7 of SMN2 (Fig. 3E), which may provide redundant regulatory signals, we constructed and tested a series of mutants (M2 to M6) that progressively removed all mapped and potential G/U-rich sites (Fig. 3H–I). We noted that M2 (which mutated a stretch of G/U sequence within exon 7) enhanced SMN2 splicing, but the mutated region corresponded precisely to a Sam68 binding site previously shown to inhibit SMN2 splicing (Pedrotti et al., 2010). We also found another mutant (M4) that further enhanced SMN2 splicing, which might be due, at least in part, to the removal of the two previously mapped hnRNP A1 binding sites (Kashima et al., 2007). Importantly, when all mapped and potential hnRNP U binding sites were eliminated, we found that the final mutant (M6) still responded to hnRNP U depletion (Fig. 3J). Furthermore, we transferred the regulatory exon (exon 7) along with its flanking intronic sequences from the SMN2 gene to a split GFP gene that carries consensus 5′ and 3′ splice sites (Wang et al., 2004) (Fig. S2C). In addition, we also introduced a series of mutations in the original SMN2 minigene (Fig. S2D), including three mutants that disrupt the previously identified intronic splicing silencers (Miyajima et al., 2002; Singh et al., 2006). We found that all of these mutants still responded to hnRNP U depletion (Fig. S2F–H).
Because hnRNP U has been implicated in transcription elongation (Kim and Nikodem, 1999; Obrdlik et al., 2008) and this gene expression step has been linked to splice site selection during co-transcriptional splicing (Kornblihtt, 2007), we explored a possibility that hnRNP U might influence SMN2 splicing in a transcription-dependent manner. For this purpose, we designed a series of PCR primer pairs that target multiple intronic locations along the SMN2 gene to detect potential elongation blockage in hnRNP U-depleted cells (Fig. S2I), a strategy we previously used to detect alterations in Pol II processivity (Lin et al., 2008). We did not detect any difference between wild-type and hnRNP U-depleted cells, indicating that the effect of hnRNP U on SMN2 splicing may not be related to its role in transcription elongation (Fig. S2I). Additionally, the transcription elongation inhibitor DRB has been reported to affect Fibronectin splicing (de la Mata et al., 2003), which we confirmed, but the drug showed little effect on SMN2 splicing (Fig. S2J–K). Collectively, these data indicate that hnRNP U may not regulate SMN2 splicing through direct binding to its pre-mRNA or via a transcription-coupled mechanism.
hnRNP U binding to diverse classes of non-coding RNAs, including all snRNAs
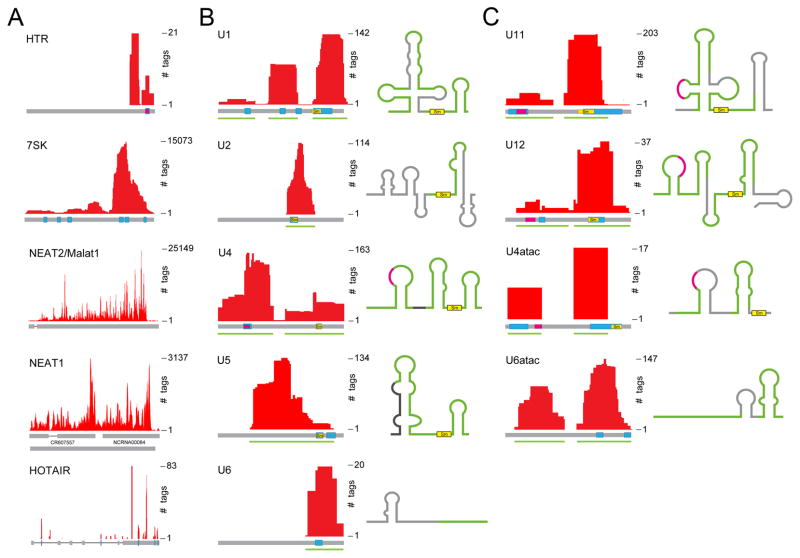
Having tentatively ruled out the splicing response through direct binding of hnRNP U to the SMN2 pre-mRNA or via a transcription-coupled mechanism, we turned to clue from other hnRNP U binding features in the human genome. Significantly, we observed prevalent hnRNP U interaction with literally all classes of regulatory non-coding RNAs in the nucleus (Fig. 4A), including hTR, which is in line with a critical role of hnRNP U in telomere length regulation (Fu and Collins, 2007). We also detected extensive interaction of hnRNP U with 7SK RNA, known to play a critical role as a molecular sink for transcription elongation regulators (Yik et al., 2003), with Malat-1/NEAT2, which colocalizes with nearly all components of the splicing machinery in nuclear speckles (Tripathi et al., 2010), with NEAT1 involved in organizing paraspeckles in the nucleus (Clemson et al., 2009), with HOTAIR, which serves as an integrator in transcriptional regulation (Tsai et al., 2010), and with numerous other non-coding RNAs (not shown). These observations raise an intriguing possibility that multiple functions previously recorded for hnRNP U/SAF-A in diverse regulatory pathways may be mediated through its interaction with various regulatory non-coding RNAs.
Figure 4. Prevalent association of hnRNP U with non-coding RNAs, including all splicing snRNAs.
(A) Examples of hnRNP U-associated regulatory non-coding RNAs. (B and C) Interaction of hnRNP U with snRNAs required for splicing of both major (B) and minor (C) classes of introns. The secondary structure of each snRNA is diagrammed on the right. Blue boxes indicate the location of G/U-rich sequences and yellow box highlights the position of the Sm binding site in each snRNA. Pink line in folded RNA show the region for targeting individual snRNAs to Cajal bodies.
Particularly relevant to our current investigation of hnRNP U-regulated splicing, we noted its extensive association with all of the snRNAs required for splicing of both major and minor classes of introns in mammalian genomes (Fig. 4B–C). This is in contrast to the lack of such interaction with other splicing regulators we analyzed by CLIP-seq under the same conditions (e.g. Fox2 (Yeo et al., 2009) and PTB (Xue et al., 2009)). Notably, the tag density on individual major spliceosome snRNAs is relatively equal (with the exception of U6) compared to the total population of major snRNPs with U1 snRNP being the most abundant in the cell. In addition, hnRNP U seems to prefer U11 and U6atac over U12 and U4atac involved in splicing of the minor class of introns (note that snRNAs for minor introns are much lower in abundance than snRNAs for major introns, indicating that hnRNP U might play a critical role in regulating minor intron splicing, a subject for future studies). While the significance of these binding differences remains to be determined, the observed interaction of hnRNP U with all splicing snRNAs suggests that it might be involved in the regulation of snRNP biogenesis/maturation and/or their assembly into the spliceosome.
A role of hnRNP U in the regulation of U2 snRNP maturation
Following the clue on hnRNP U binding to splicing snRNAs, we next focused on the potential role of this nuclear matrix protein in the regulation of snRNP biogenesis in the cell. We first confirmed by Western blotting that hnRNP U depletion elevated the expression of the SMN protein, as expected from enhanced SMN2 splicing (Fig. 5A). Because SMN is known to function as a chaperon for the assembly of the Sm core onto individual snRNAs (Pellizzoni et al., 2002), we examined and found that levels of both total splicing snRNAs and those associated with Sm-containing snRNPs remained unaltered in hnRNP U-depleted cells compared to untreated cells (Fig. 5B). The lack of effect of enhanced SMN expression on the total population of Sm-containing snRNPs is consistent with the previous observations that SMN is not a rate-limiting factor for snRNP biogenesis because cells can tolerate a significant degree of SMN reduction (Zhang et al., 2008) and SMN overexpression had little effect in enhancing snRNP biogenesis (Jodelka et al., 2010).
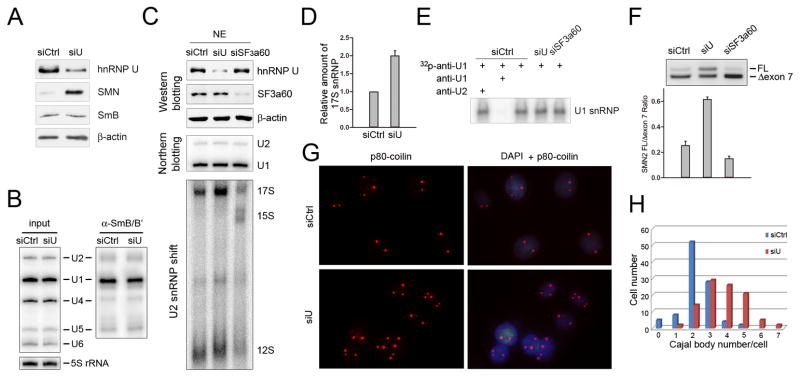
Figure 5. A critical role of hnRNP U in regulating U2 snRNP maturation.
(A) Induction of SMN in hnRNP U-depleted cells. No effect on SmB was detected. (B) Similar levels of both total and Sm-associated snRNAs before and after hnRNP U RNAi. (C) Induction and suppression of the 17S U2 snRNP complex in hnRNP U- and SF3a60-depeleted cells, respectively. The degree of knockdown down in each case was validated by Western blotting. β-actin detected by Western blotting and U1 and U2 snRNA detected by Northern blotting were used as loading controls. U2 snRNP complexes were detected in a gel shift assay using a radio-labeled anti-U2 2′-OMe probe. (D) Quantification of the induction of the 17S U2 snRNP complex in hnRNP U-depleted cells. (E) Analysis of U1 snRNP by gel shift as in C using an anti-U1 2′-OMe probe. (F) The opposite effects of hnRNP U and SF3a60 knockdown on SMN2 splicing. (G) Immunocytochemical analysis of Cajal bodies with anti-p80 coilin in control and hnRNP U RNAi-treated cells. (H) Quantification of Cajal Bodies in response to hnRNP U depletion based on counting of 100 nuclei. Data in D and F are shown as mean ± SD.
The lack of enhancement of Sm-containing snRNPs prompted us to explore the possibility that hnRNP U depletion might affect snRNP maturation in the nucleus. In particular, it has been previously documented that Sm-containing U2 snRNP is progressively converted from initial 12S to intermediate 15S complex with the addition of SF3b and then to the splicing competent 17S complex with the addition of SF3a, which can be monitored by using a radio-labeled anti-U2 2′-OMe oligo in a gel shift assay (Brosi et al., 1993a; Brosi et al., 1993b; Kramer et al., 1999; Will et al., 2002). It has also been reported that, while the 17S U2 snRNP is the dominant complex in the cell, the intermediates became detectable in SF3a60 RNAi-treated cells (Tanackovic and Kramer, 2005).
We therefore examined the impact of hnRNP U depletion on U2 snRNP biogenesis, finding that depletion of hnRNP U indeed significantly enhanced the formation of 17S U2 snRNP (Fig. 5C–D). In contrast, SF3a60 RNAi blocked the conversion of U2 snRNP from 15S to 17S (Fig. 5C). As a control to this experiment, we performed a similar gel shift assay to monitor U1 snRNP. We observed that a specific anti-U1 probe could detect U1 snRNP, whcih could be competed away with a cold anti-U1 probe, but not with a cold anti-U2 probe, and importantly, depletion of either hnRNP U or SF3a60 had little effect on levels of U1 snRNP (Fig. 5E). Finally, consistent with the reciprocal effects of hnRNP U and SF3a60 depletion on the levels of functional 17S U2 snRNP, we found that the reduction of hnRNP U enhanced SMN2 exon 7 inclusion whereas down-regulation of SF3a60 had the opposite effect (Fig. 5F). Together, these findings revealed a previously unrecognized regulatory step in U2 snRNP maturation and demonstrated a key role of hnRNP U in this process.
Enhanced appearance of Cajal bodies in hnRNP U-depleted cells
Enhanced U2 snRNP maturation detected in hnRNP U-depleted cells may result from two potential mechanisms, one of which may be mediated by a negative role of hnRNP U in the conversion of U2 snRNP to larger complexes. However, we did not detect any increase in the 17S U2 snRNP complex at the expense of the 12S complex, and this is in contrast to the accumulation of 15S complex in SF3a60 knockdown cells, which is known to participate in the conversion of U2 snRNP from 15S to 17S (Fig. 5C). Alternatively, as a nuclear matrix protein, hnRNP U may constraint trafficking of partially assembled U2 snRNP within the nucleus, therefore exerting a negative impact on U2 snRNP maturation. It has been proposed earlier that Cajal bodies might be key sites for U2 snRNP maturation in the nucleus, although neither SF3a nor SF3b could be localized in the structure unless under overexpression conditions (Nesic et al., 2004). In any case, Cajal bodies might reflect dynamic intranuclear trafficking of U2 snRNP to enhance its maturation at multiple stages (Cioce and Lamond, 2005).
To determine whether the number and/or structure of Cajal bodies might be altered in hnRNP U-depleted cells, we monitored Cajal bodies in response to hnRNP U RNAi using anti-p80 coilin, a marker for the nuclear structure (Andrade et al., 1991). Strikingly, we found that the number of Cajal bodies was significantly increased in hnRNP U-depleted cells compared to control siRNA-treated cells (Fig. 5G). Quantification of stained Cajal bodies revealed that they were nearly doubled in number with a significant fraction of nuclei even exhibiting 6 to 7 Cajal bodies in response to hnRNP U depletion (Fig. 5H). Such dramatic increase in Cajal bodies took place without any enhanced expression of the SmB antigen (see Fig. 5A), which was previously shown to boost Cajal body formation (Sleeman et al., 2001). These observations agree with the proposed role of Cajal bodies in U2 snRNP maturation (Nesic et al., 2004; Sleeman et al., 2001), suggesting that the reduced restriction of hnRNP U may permit enhanced flux of pre-matured U2 snRNP into the structure for further maturation.
hnRNP U as a global regulator of alternative splicing
The data presented above suggest that hnRNP U may function as a global splicing regulator by regulating the functional pool of U2 snRNPs in the nucleus. To test this possibility, we performed paired-end RNA-seq on RNA isolated from control and hnRNP U RNAi-treated HeLa cells to detect altered splicing events in an unbiased fashion. We generated ~28 million 75nt tags from both control and hnRNP U RNAi-treated cells, of which ~18 million tags were mapped to annotated genes under each condition. While “unmappable” tags may correspond to new isoforms, we first focused on the effect of hnRNP U RNAi on annotated mRNA isoforms, which are likely to provide sufficient events for us to deduce the regulatory mechanism. We therefore joined the tags at both ends of individual fragments if they are 50nt or less apart on KnownGenes because the average length of RNA fragments in our library is ~200nt. This generated a collection of ~14 million sequences uniquely mapped to known transcripts, ~60% of which (8.9 million tags from untreated cells and 8.1 million from hnRNP U-depleted cells) cover known splice junctions.
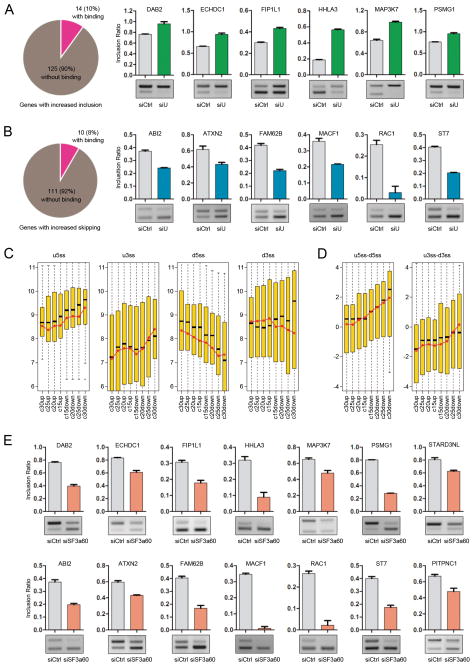
To calculate altered splicing events among known genes in response to hnRNP U knockdown, we determined the ratio of tags that correspond to included and skipped isoforms among annotated cassette exons. Using a stringent criterion, we obtained 139 induced exon inclusion events and 121 induced exon skipping events (Fig. 6A–B). We randomly selected 40 events (listed in Table S2) for validation by PCR, confirming the induced splicing changes in 34 events (the validation rate=85%, 6 events that could not be validated appear due to low tag counts from RNA-seq analysis). A representative set of these validated events are illustrated in Fig. 6A–B with the remaining events shown in Fig. S3A–B.
Figure 6. Global analysis of hnRNP U-regulated splicing.
(A and B) RNA-seq identification of exon inclusion (A) or skipping (B) in response to hnRNP U RNAi. The pie-chat in each case shows the altered splicing events and the hnRNP U binding evidence based on CLIP-seq analysis. Representative events validated by RT-PCR are shown on the right. The remaining events are shown in Fig. S3A–B. (C) The splicing signals associated with RNA-seq detected alternative splicing events were divided into 4 groups according to levels of induced exon inclusion or skipping. c30up, c25up, c20up and c15up indicate induced inclusion of cassette exon by =>30%, =>25%, =>20%, and =>15%, respectively. Similarly, c15down, c20down, c25down and c30down indicate induced skipping of cassette exon by =>15%, =>20%, =>25%, and =>30%, respectively. Black bars represent the medium and the red dots indicate the mean value in each group. (D) The differences between the upstream constitutive 5′ splice site and the downstream alternative 5′ splice sites (u5′ss-d5′ss) and the differences between the upstream alternative 3′ splice site and the downstream constitutive 3′ splice sites (u3′ss-d3′ss). (E) Representative splicing responses to SF30a60 RNAi. Data in A, B, and E are shown as mean ± SD
Comparison between hnRNP U binding and hnRNP U depletion-induced splicing reveals that the binding events are associated with detected splicing responses in only 8 to 10% cases (Fig. 6A–B), which is in agreement with the result of mutational analysis on the SMN2 minigene (Fig. 3), indicating that hnRNP U-dependent splicing may not be related to its binding to respective pre-mRNA. This observation therefore reinforces the possibility that hnRNP U-mediated snRNP maturation might be a major mechanism for the observed splicing changes in hnRNP U-depleted cells, which is also in line with published studies demonstrating the induction of alternative splicing by modeling the core splicing machinery (Saltzman et al., 2011).
Mechanistic insights into hnRNP U-regulated splicing
The puzzle is why a rise in the functional pool of U2 snRNP would cause exon inclusion on certain genes and exon skipping on others. To address this mechanistic issue, we analyzed the strength of splice signals associated with cassette splicing events and ranked them according to levels of induced exon inclusion or skipping based on the splice site strength scoring system (Yeo and Burge, 2004). This analysis revealed, while the mean (red dot) or medium (black bar) scores of the upstream 5′ splice site went up with diminishing levels of induced exon inclusion, the opposite was true with the downstream 5′ splice site associated with the alternative exon (Fig. 6C, compare panel 1 and 3). Less obvious trend was observed with the upstream and downstream 3′ splice sites (Fig. 6C, compare panel 2 and 4).
By linking the differences between the upstream and downstream alternative 5′ splice sites (u5′ss-d5′ss) and 3′ splice sites (u3′ss-d3′ss) to different degrees of induced exon inclusion and skipping (Fig. 6D), we observed that, among induced exon inclusion events, the differences between the 5′ and 3′ splice sites are relatively constant. In these cases, an increase in the functional U2 snRNP is expected to strengthen the recognition of both the internal and flanking 3′ splice sites, thereby leading to exon inclusion according to the long-established proximal rule for splice site selection (Reed and Maniatis, 1986). In contrast, the difference between the upstream and downstream 5′ splice sites is progressively enlarged with increasing levels of exon skipping (Fig. 6D). Because the downstream 5′ splice site is known to affect the recognition of the upstream 3′ splice site during exon definition (Kuo et al., 1991), the weak internal 5′ splice site may therefore render the upstream 3′ splice site insensitive to elevated U2 snRNP during the recognition of the alternative exon, and as a result, selective enhancement of the downstream 3′ splice site may result in skipping of the internal alternative exon.
To provide independent evidence for this working model, we examined the splicing response on a set of hnRNP U-regulated splicing events by reducing the functional pool of U2 snRNP through SF3a60 RNAi (Fig. 5C). A reduction of U2 snRNP is expected to weaken both the alternative exon and the flanking constitutive 3′ splice sites. Because U2 snRNP binding to the internal 3′ splice site is generally weaker relative to the flanking constitutive 3′ splice site (Fig. 6C), further reduction of functional U2 snRNP would exacerbate the competitiveness of the internal 3′ splice site with the downstream 3′ splice site, thus causing skipping of the alternative exon regardless of whether the alternative exon is induced to include or skip by elevated U2 snRNP in hnRNP U-depleted cells. This was precisely observed in SF3a60-depleted cells (Fig. 6E).
As specificity controls for these experiments, knocking down U1-70K or the SR protein SRSF1 (Fig. S3C) produced responses in both directions among hnRNP U-regulated splicing events (Fig. S3D–E). This is also expected because U1-70K may differentially affect exon definition for the internal alternative exon or flanking competing exons that depends on other regulatory events on the competing exons. Similarly, SRSF1 has also been shown to differentially module the splice site associated with either the internal or flanking exons to cause exon inclusion or skipping (Han et al., 2011). Together, these data provide compelling evidence that elevated U2 snRNP in conjunction with exon definition is a plausible mechanism to explain the observed splicing changes in hnRNP U-depleted cells.
Discussion
hnRNP U has been biochemically characterized as an RNA binding protein, but its putative role in regulated splicing has not been established (Kiledjian and Dreyfuss, 1992). Here, we demonstrated that hnRNP U functions as a global splicing regulator. Using CLIP-seq, we identified thousands of direct hnRNP U binding targets in the human genome and deduced its GU-rich binding consensus. While the analysis revealed the global landscape of hnRNP U/RNA interactions, including its direct binding on SMN 1/2 exon 8, we found no evidence that hnRNP U-dependent splicing response is mediated through its direct binding to pre-mRNA on the SMN2 minigene model. Instead, we uncovered a previously unknown regulatory pathway in U2 snRNP maturation and a key role of hnRNP U in this process. While we leave open the possibility that hnRNP U may contribute to regulated splicing through multiple other mechanisms, including its binding to other pre-mRNAs or potential indirect effects through altered expression of other splicing regulators, we provide a series of evidence suggesting that hnRNP U may regulate alternative splicing mainly through modulating U2 snRNP maturation.
snRNP biogenesis has been well characterized. Newly synthesized snRNAs are first exported to the cytoplasm where the SMN protein serves as a key chaperon for specific assembly of the Sm core onto snRNAs and the assembled RNP particles are reimported into the nucleus to participate in pre-mRNA splicing (Will and Luhrmann, 2001). Within the nucleus, the splicing snRNPs appear to recycle between Cajal bodies, nuclear speckles, and nascent RNAs (Cioce and Lamond, 2005). Among all splicing snRNPs, the maturation pathway for U2 snRNP has been best characterized. The core U2 snRNP is of 12S in size, which is joined by the multi-subunit SF3b complex to give rise to the 15S complex and then with another multi-subunit SF3a complex to produce the final, splicing competent 17S U2 snRNP (Brosi et al., 1993a; Brosi et al., 1993b; Kramer et al., 1999; Will et al., 2002). However, it has been unclear whether this U2 snRNP maturation pathway is subject to regulation.
Our current study reveals that hnRNP U plays a critical role in the regulation of the U2 snRNP maturation pathway. Previous biochemical studies did not detect hnRNP U as a component of U2 snRNP at any maturation stage (Will et al., 2002) and we did not detect increased 17S U2 snRNP at the expense of the 12S complex. These data indicate that hnRNP U may not interfere with U2 snRNP assembly at a specific stage; rather, given hnRNP U as a nuclear matrix factor, it may constraint dynamic intracellular trafficking of U2 snRNP during its maturation in the nucleus. Consistent with this possibility, hnRNP U depletion led to significantly enhanced Cajal bodies, indicative of dynamic flux of U2 snRNP through the nuclear structure for further maturation or recycling, as proposed earlier (Nesic et al., 2004; Sleeman et al., 2001).
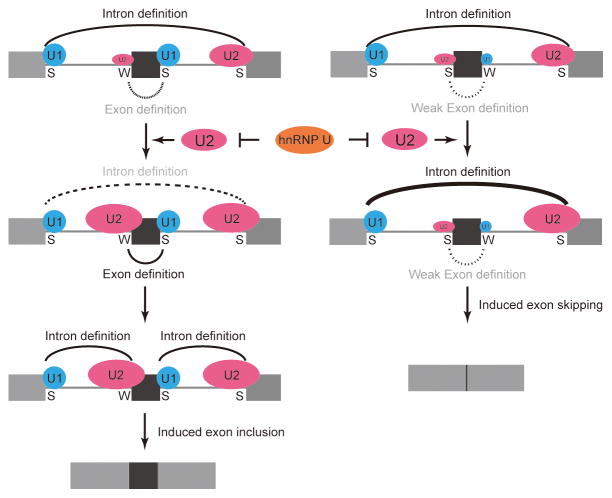
It has long been recognized that pre-mRNA splicing can be regulated by both sequence-specific RNA binding proteins as well as by specific components of the core splicing machinery. Are these regulatory strategies each follow distinct principles or could they operate under a uniform rule in the regulation of alternative splicing? Taking advantage of the elucidated function of hnRNP U in modulating U2 snRNP maturation, we determined its global impact on alternative splicing by using the unbiased RNA-seq approach. Analysis of the splice sites associated with both regulated exons and flanking constitutive exons suggests a working model (Fig. 7): If the competitiveness of the splice sites involved is relatively equal in strength, elevated U2 snRNP would enhance the recognition of both competing 3′ splice sites, thus inducing the inclusion of the alternative exon based on the proximal rule in splice site selection (Reed and Maniatis, 1986). On the other hand, if the internal 3′ splice site is insensitive to elevated U2 snRNP due to a weak downstream 5′ splice site that spoils exon definition, selective strengthening of the competing flanking 3′ splice site would then induce skipping of the internal alternative exon.
Figure 7. Model for splice site competition modulated by differential recruitment of U2 snRNP to competing 3′ splice sites.
On the left, the internal 3′ splice site is relatively weak (W) compared to the relatively strong (S) 3′ splice site associated with the flanking exon. Elevated U2 snRNP would enhance the recognition of the internal 3′ splice site, leading to the inclusion of the alternative exon. On the right, the internal 5′ splice site is relatively weak compared to the upstream 5′ splice site associated with the flanking constitutive exon. As a result, U2 recognition of the internal 3′ splice site is inefficient due to ineffective exon definition despite that the internal 3′ splice site may have sufficient strength. Elevated U2 snRNP in these cases would thus only enhance the recognition of the downstream 3′ splice site, leading to induced exon skipping.
Our present genomic and biochemical analyses of hnRNP U have also shed critical lights on its regulatory activities in other gene expression pathways in the nucleus. hnRNP U binds to many different classes of non-coding RNAs, consistent with its tight association with operationally defined “nuclear matrix”. Importantly, our study revealed a key role of hnRNP U in regulating a specific RNP function (i.e. U2 snRNP maturation), suggesting that this matrix-associated RNA binding protein may act at the level of RNPs by directing or sequestrating specific RNPs to or from some defined cellular locations, which may be mediated by its RNA and DNA binding activities through distinct domains. This is consistent with the reported role of hnRNP U in targeting Xist-containing RNPs to Xi (Hasegawa et al., 2010; Helbig and Fackelmayer, 2003; Pullirsch et al., 2010). Our work has therefore provided a conceptual framework to understand the diverse function of hnRNP U/SAF-A in regulated gene expression in mammalian cells.
Materials and Methods
Plasmids
All SMN constructs were derived from pCI-SMN1/2 (Lorson et al., 1999). pCI-SMN1-PyD mutant was constructed as described (Cartegni et al., 2006). To construct pGFP-SMN2, we inserted SMN2 exon 7 with flank intronic regions into the Xho I and Sac II sites in pZW4 (Wang et al., 2004). pRetro-Super-shU was generated by cloning an annealed double-stranded DNA oligo (5′-GAUGAACACUUCGAUGACAuucaagagaUGUCAUCGAAGUGUUCAUCuu-3′ downstream of the H1 promoter of pRetro-Super (Yugami et al., 2007). To generate pCMV-Flag-hnRNP U, the hnRNP U coding region was cloned from pGEM4-hnRNP U (Kiledjian and Dreyfuss, 1992) to the EcoR I site in pCMV-Flag2B (Clontech) and the shRNA targeting site was mutated without changing encoded amino acids. All primers used for plasmid construction are listed in Table S3.
esiRNA screen
We extracted RNA binding proteins containing known RNA binding motifs from the EBI database. The primers for preparing eiRNAs against these RBPs are listed in Table S1. esiRNA preparation was carried out largely as previously described (Kittler et al., 2007) with the following modifications. Most of the esiRNAs were designed to target the 3′ UTR of individual genes. After RNase III digestion, we recovered 20–30 bp esiRNA from native PAGE to minimize potential interferon responses. Specific siRNAs used include siU-1 (5′-GAUGAACACUUCGAUGACAdTdT-3′), siU-2, 5′-AGACCACGAGAAGAUCAUdTdT-3′), and a negative control siRNA (5′-UUCUCCGAACGUGUCACGUdTdT-3′), which were purchased from Genepharm. Oligofectamine/RNAimax and lipofectamine 2000 (Invitrogen) were used for esiRNA/siRNA and plasmid transfection, respectively.
Antibodies for immunoblotting
Immunoblotting was performed as described (Xue et al., 2009). The following antibodies were used to detect prospective proteins: mouse monoclonal anti-hnRNP U (3G6, Santa Cruz Biotech), anti-hnRNP A1 (4B10, Santa Cruz Biotech), anti-hnRNP A2 (DP3B3, Santa Cruz Biotech), anti-β-actin (AC-15, Sigma), anti-FLAG (M2, Sigma), anti-SmB/B′ (12F5, Santa Cruz Biotech), anti-SMN (2B1, Santa Cruz Biotech), Anti-U2AF65 (MC3, sigma), rabbit polyclone anti-U2AF35 (Aviva), rabbit polyclone anti-SF1(Aviva), and anti-p80 coilin (F-7, Santa Cruz Biotech). Secondary HRP-conjugated goat anti-mouse IgG (Sigma) was detected with the SuperSignal kit (Thermo).
CLIP-seq and RNA-seq
CLIP-seq and bioinformatics analysis were performed as described (Xue et al., 2009). We carried out immunoprecipitation in 1% NP-40 wash buffer to improve the IP efficiency. Paired-end RNA-seq (75 nt from each end) was performed on RNA isolated from control siRNA and hnRNP U siRNA-treated cells. Detailed information on tag mapping, calculation of tag distribution, peak identification, and calculation of splicing ratio is described in the supplementary experimental procedure.
Assay for snRNP maturation
Splicing snRNPs were immunoprecipitated with anti-Sm and anti-hnRNP U and extracted RNA was detected by 3′ pCp-labeling or by Northern blotting. Labeled snRNAs were resolved on 8% acrylamide/7 M urea gels and quantified by phosphoimaging. Analysis of U2 snRNP at different maturation stages was performed as described (Brosi et al., 1993a). A similar strategy was used to detect U1 snRNP.
Supplementary Material
Acknowledgments
The authors are grateful to Drs. G. Dreyfuss, A. Kramer, E. Androphy, and C. Burge for providing various plasmids and protocols. This work was supported by the China 863 program (2007AA02Z112) and NSFC (30770422) to Y.Z., the China 973 program (2005CB724604) to Y.Z. and X-D.F., the Chinese 111 project (B06018) and NIH grants (HG004659, GM049369 and GM052872) to X-D.F.
Footnotes
Accession numbers
The CLIP-seq and RNA-seq data for hnRNP U binding and regulated splicing are available at the Gene Expression Omnibus under the accession number GSE34491.
Supplemental Data include 3 figures, 3 tables, and extended data treatment procedures can be found with this article online at http://www.cell.com/molecular-cell/supplemental/...
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ameyar-Zazoua M, Souidi M, Fritsch L, Robin P, Thomas A, Hamiche A, Percipalle P, Ait-Si-Ali S, Harel-Bellan A. Physical and functional interaction between heterochromatin protein 1alpha and the RNA-binding protein heterogeneous nuclear ribonucleoprotein U. J Biol Chem. 2009;284:27974–27979. doi: 10.1074/jbc.M109.037929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade LE, Chan EK, Raska I, Peebles CL, Roos G, Tan EM. Human autoantibody to a novel protein of the nuclear coiled body: immunological characterization and cDNA cloning of p80-coilin. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosi R, Groning K, Behrens SE, Luhrmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993a;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- Brosi R, Hauri HP, Kramer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993b;268:17640–17646. [PubMed] [Google Scholar]
- Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR. Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet. 2006;78:63–77. doi: 10.1086/498853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- Cioce M, Lamond AI. Cajal bodies: a long history of discovery. Annu Rev Cell Dev Biol. 2005;21:105–131. doi: 10.1146/annurev.cellbio.20.010403.103738. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Mata M, Alonso CR, Kadener S, Fededa JP, Blaustein M, Pelisch F, Cramer P, Bentley D, Kornblihtt AR. A slow RNA polymerase II affects alternative splicing in vivo. Mol Cell. 2003;12:525–532. doi: 10.1016/j.molcel.2003.08.001. [DOI] [PubMed] [Google Scholar]
- Eggert M, Michel J, Schneider S, Bornfleth H, Baniahmad A, Fackelmayer FO, Schmidt S, Renkawitz R. The glucocorticoid receptor is associated with the RNA-binding nuclear matrix protein hnRNP U. J Biol Chem. 1997;272:28471–28478. doi: 10.1074/jbc.272.45.28471. [DOI] [PubMed] [Google Scholar]
- Fackelmayer FO, Dahm K, Renz A, Ramsperger U, Richter A. Nucleic-acid-binding properties of hnRNP-U/SAF-A, a nuclear-matrix protein which binds DNA and RNA in vivo and in vitro. Eur J Biochem. 1994;221:749–757. doi: 10.1111/j.1432-1033.1994.tb18788.x. [DOI] [PubMed] [Google Scholar]
- Fackelmayer FO, Richter A. Purification of two isoforms of hnRNP-U and characterization of their nucleic acid binding activity. Biochemistry. 1994;33:10416–10422. doi: 10.1021/bi00200a024. [DOI] [PubMed] [Google Scholar]
- Fu D, Collins K. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28:773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov DK, Shi X, Das K, Gilliam TC, Wang CH. Differential SMN2 expression associated with SMA severity. Nat Genet. 1998;20:230–231. doi: 10.1038/3030. [DOI] [PubMed] [Google Scholar]
- Gupta AK, Drazba JA, Banerjee AK. Specific interaction of heterogeneous nuclear ribonucleoprotein particle U with the leader RNA sequence of vesicular stomatitis virus. J Virol. 1998;72:8532–8540. doi: 10.1128/jvi.72.11.8532-8540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Ding JH, Byeon CW, Kim JH, Hertel KJ, Jeong S, Fu XD. SR proteins induce alternative exon skipping through their activities on the flanking constitutive exons. Mol Cell Biol. 2011;31:793–802. doi: 10.1128/MCB.01117-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Brockdorff N, Kawano S, Tsutui K, Tsutui K, Nakagawa S. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]
- Hastings ML, Allemand E, Duelli DM, Myers MP, Krainer AR. Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF(65) PLoS One. 2007;2:e538. doi: 10.1371/journal.pone.0000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig R, Fackelmayer FO. Scaffold attachment factor A (SAF-A) is concentrated in inactive X chromosome territories through its RGG domain. Chromosoma. 2003;112:173–182. doi: 10.1007/s00412-003-0258-0. [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Wirth B. hnRNP-G promotes exon 7 inclusion of survival motor neuron (SMN) via direct interaction with Htra2-beta1. Hum Mol Genet. 2002;11:2037–2049. doi: 10.1093/hmg/11.17.2037. [DOI] [PubMed] [Google Scholar]
- Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H. A mouse model for spinal muscular atrophy. Nat Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- Jady BE, Bertrand E, Kiss T. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodelka FM, Ebert AD, Duelli DM, Hastings ML. A feedback loop regulates splicing of the spinal muscular atrophy-modifying gene, SMN2. Hum Mol Genet. 2010;19:4906–4917. doi: 10.1093/hmg/ddq425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashima T, Manley JL. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, Manley JL. An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci U S A. 2007;104:3426–3431. doi: 10.1073/pnas.0700343104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. Embo J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Nikodem VM. hnRNP U inhibits carboxy-terminal domain phosphorylation by TFIIH and represses RNA polymerase II elongation. Mol Cell Biol. 1999;19:6833–6844. doi: 10.1128/mcb.19.10.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Surendranath V, Heninger AK, Slabicki M, Theis M, Putz G, Franke K, Caldarelli A, Grabner H, Kozak K, et al. Genome-wide resources of endoribonuclease-prepared short interfering RNAs for specific loss-of-function studies. Nat Methods. 2007;4:337–344. doi: 10.1038/nmeth1025. [DOI] [PubMed] [Google Scholar]
- Kornblihtt AR. Coupling transcription and alternative splicing. Adv Exp Med Biol. 2007;623:175–189. doi: 10.1007/978-0-387-77374-2_11. [DOI] [PubMed] [Google Scholar]
- Kramer A, Gruter P, Groning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol. 1999;145:1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukalev A, Nord Y, Palmberg C, Bergman T, Percipalle P. Actin and hnRNP U cooperate for productive transcription by RNA polymerase II. Nat Struct Mol Biol. 2005;12:238–244. doi: 10.1038/nsmb904. [DOI] [PubMed] [Google Scholar]
- Kuo HC, Nasim FH, Grabowski PJ. Control of alternative splicing by the differential binding of U1 small nuclear ribonucleoprotein particle. Science. 1991;251:1045–1050. doi: 10.1126/science.1825520. [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–826. doi: 10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. Embo J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci U S A. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens JH, Verlaan M, Kalkhoven E, Dorsman JC, Zantema A. Scaffold/matrix attachment region elements interact with a p300-scaffold attachment factor A complex and are bound by acetylated nucleosomes. Mol Cell Biol. 2002;22:2598–2606. doi: 10.1128/MCB.22.8.2598-2606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins de Araujo M, Bonnal S, Hastings ML, Krainer AR, Valcarcel J. Differential 3′ splice site recognition of SMN1 and SMN2 transcripts by U2AF and U2 snRNP. Rna. 2009;15:515–523. doi: 10.1261/rna.1273209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyajima H, Miyaso H, Okumura M, Kurisu J, Imaizumi K. Identification of a cis-acting element for the regulation of SMN exon 7 splicing. J Biol Chem. 2002;277:23271–23277. doi: 10.1074/jbc.M200851200. [DOI] [PubMed] [Google Scholar]
- Nesic D, Tanackovic G, Kramer A. A role for Cajal bodies in the final steps of U2 snRNP biogenesis. J Cell Sci. 2004;117:4423–4433. doi: 10.1242/jcs.01308. [DOI] [PubMed] [Google Scholar]
- Obrdlik A, Kukalev A, Louvet E, Farrants AK, Caputo L, Percipalle P. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol. 2008;28:6342–6357. doi: 10.1128/MCB.00766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedrotti S, Bielli P, Paronetto MP, Ciccosanti F, Fimia GM, Stamm S, Manley JL, Sette C. The splicing regulator Sam68 binds to a novel exonic splicing silencer and functions in SMN2 alternative splicing in spinal muscular atrophy. Embo J. 2010;29:1235–1247. doi: 10.1038/emboj.2010.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellizzoni L, Yong J, Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- Pullirsch D, Hartel R, Kishimoto H, Leeb M, Steiner G, Wutz A. The Trithorax group protein Ash2l and Saf-A are recruited to the inactive X chromosome at the onset of stable X inactivation. Development. 2010;137:935–943. doi: 10.1242/dev.035956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed R, Maniatis T. A role for exon sequences and splice-site proximity in splice-site selection. Cell. 1986;46:681–690. doi: 10.1016/0092-8674(86)90343-0. [DOI] [PubMed] [Google Scholar]
- Romig H, Fackelmayer FO, Renz A, Ramsperger U, Richter A. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. Embo J. 1992;11:3431–3440. doi: 10.1002/j.1460-2075.1992.tb05422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshon MJ, Ruley HE. Hypomorphic mutation in hnRNP U results in post-implantation lethality. Transgenic Res. 2005;14:179–192. doi: 10.1007/s11248-004-8147-8. [DOI] [PubMed] [Google Scholar]
- Saltzman AL, Pan Q, Blencowe BJ. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011;25:373–384. doi: 10.1101/gad.2004811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NK, Singh NN, Androphy EJ, Singh RN. Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol Cell Biol. 2006;26:1333–1346. doi: 10.1128/MCB.26.4.1333-1346.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman JE, Ajuh P, Lamond AI. snRNP protein expression enhances the formation of Cajal bodies containing p80-coilin and SMN. J Cell Sci. 2001;114:4407–4419. doi: 10.1242/jcs.114.24.4407. [DOI] [PubMed] [Google Scholar]
- Tanackovic G, Kramer A. Human splicing factor SF3a, but not SF1, is essential for pre-mRNA splicing in vivo. Mol Biol Cell. 2005;16:1366–1377. doi: 10.1091/mbc.E04-11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, Shi Y, Segal E, Chang HY. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente ST, Goff SP. Inhibition of HIV-1 gene expression by a fragment of hnRNP U. Mol Cell. 2006;23:597–605. doi: 10.1016/j.molcel.2006.07.021. [DOI] [PubMed] [Google Scholar]
- Wang Z, Kayikci M, Briese M, Zarnack K, Luscombe NM, Rot G, Zupan B, Curk T, Ule J. iCLIP predicts the dual splicing effects of TIA-RNA interactions. PLoS Biol. 2010;8:e1000530. doi: 10.1371/journal.pbio.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Spliceosomal UsnRNP biogenesis, structure and function. Curr Opin Cell Biol. 2001;13:290–301. doi: 10.1016/s0955-0674(00)00211-8. [DOI] [PubMed] [Google Scholar]
- Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R. Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD-box protein. EMBO J. 2002;21:4978–4988. doi: 10.1093/emboj/cdf480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhou Y, Wu T, Zhu T, Ji X, Kwon YS, Zhang C, Yeo G, Black DL, Sun H, et al. Genome-wide analysis of PTB-RNA interactions reveals a strategy used by the general splicing repressor to modulate exon inclusion or skipping. Mol Cell. 2009;36:996–1006. doi: 10.1016/j.molcel.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J Comput Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- Yeo GW, Coufal NG, Liang TY, Peng GE, Fu XD, Gage FH. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat Struct Mol Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]
- Yugami M, Kabe Y, Yamaguchi Y, Wada T, Handa H. hnRNP-U enhances the expression of specific genes by stabilizing mRNA. FEBS Lett. 2007;581:1–7. doi: 10.1016/S0014-5793(07)01283-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Licklider LJ, Gygi SP, Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.