Abstract
Nucleocapsid protein (N) is an essential RNA binding protein in many RNA viruses. During replication, N protein encapsidates viral genomic and antigenomic RNA, but not viral mRNA or other cellular RNAs. To discriminate between different species of RNA in a host cell, it is likely that N interacts with specific sequences and/or secondary structures on its target RNA. In this study, we explore the RNA binding properties of N using both natural and artificially selected RNAs as ligands. We found that N binds to RNAs that resemble the terminal panhandle structures of RVFV genomic and antigenomic RNA. Furthermore, we used SELEX to isolate RNA aptamers that bound N with high affinity and determined that N specifically recognizes and binds to GAUU and pyrimidine/guanine motifs. Interestingly, BLAST analysis revealed the presence of these motifs within the coding region of the viral genome, suggesting that N may interact with non-terminal viral RNA sequences during replication. Finally, the aptamer RNAs were used to construct a sensitive fluorescence based sensor of N binding with potential applications for drug screening and imaging methodologies.
Keywords: Rift Valley fever virus, nucleocapsid, RNA aptamer, SELEX
1. Introduction
Rift Valley fever virus (RVFV) is a mosquito-borne bunyavirus (genus Phlebovirus) endemic to sub-Saharan Africa. It is responsible for widespread outbreaks of severe disease such as hepatitis, encephalitis and hemorrhagic fever in humans and abortion storms in livestock. In recent years the virus has spread as far north as Egypt and is now considered endemic in the Arabian Peninsula (Balkhy and Memish, 2003). The ability to cross extensive geographic barriers as well as the presence of competent mosquito vectors in Europe, Asia and the Americas signify the potential for global distribution. Currently there is no specific treatment for RVFV infection. Ribavirin, a nucleoside analog, is one of few drugs approved for treatment of viral hemorrhagic fevers. Although it has been administered to patients during past outbreaks, its use is limited due to adverse side effects (Kilgore et al., 1997; McCormick et al., 1986; Monath, 2008). T-705 is a pyrazinecarboxamide compound that inhibits viral replication post-infection in vitro and efficacy has been demonstrated for several RNA viruses in animal models (Gowen et al., 2008; Gowen et al., 2007). Despite these advances, T-705 is not licensed and therefore not a feasible treatment option for RVFV infection. RVF is an emerging infectious disease and a major zoonotic threat, and the characteristics described above emphasize the need for therapeutics to treat infected individuals.
RVFV has a tripartite single-stranded negative-sense RNA genome that encodes seven proteins. One of these gene products, the nucleocapsid protein, N, is an RNA binding protein required for the production of viable virus because of its involvement in several stages of viral replication. N protects the viral genome from degradation and prevents the formation of double stranded RNA intermediates during replication and transcription by encapsidating viral genomic and antigenomic RNA (Ruigrok et al., 2011). N also has RNA chaperone activity predicted to function during initiation of viral replication by unwinding an RNA helix that sequesters the self-complementary ends of the genome and allowing the polymerase to bind (Mir and Panganiban, 2006a, b). This activity could also remove higher order RNA structures that decrease the speed and efficiency of translation of viral mRNA.
RVFV is a member of the large bunyavirus family (Bunyaviridae), which is composed of five genera: Orthobunavirus, Hantavirus, Phlebovirus (including RVFV), Nairovirus and Tospovirus. A notable feature of all bunyavirus RNA segments is a conserved, self-complementary sequence present at the 5′ and 3′ ends of the genome and antigenome. This sequence undergoes base pairing to form panhandle-like structures and preferential binding of the termini of viral RNA segments by N has been demonstrated in Sin Nombre hantavirus (SNV) (Mir et al., 2006; Mir and Panganiban, 2004, 2005). Further analysis of the RNA binding properties of SNV N reveal that it binds with high affinity to UAG repeats located in the 5′ untranslated region of viral mRNA. It has been proposed that this sequence is a cis-acting signal that enhances the initiation step of translation and subsequently increases the production of viral proteins (Mir and Panganiban, 2010). The first 40 nucleotides present at the 5′ end of the viral genome and antigenome have also been shown to contain an RNA binding domain for N protein isolated from several members of the Bunyaviridae, such as Jamestown Canyon, Hantaan and Bunyamwera viruses (Ogg and Patterson, 2007; Osborne and Elliott, 2000; Severson et al., 2001). This region is thought to contain a stem-loop structure specifically recognized by N and binding to this site may initiate encapsidation of viral RNA (Osborne and Elliott, 2000; Raju and Kolakofsky, 1987). Thus, all of the known essential functions performed by N during an infection involve RNA binding. Furthermore, the multifunctional and viral specific nature of the N protein underscores its potential as an antiviral therapeutic target. Because nearly all RNA viruses, retroviruses, and hepadnaviruses encode RNA-binding nucleocapsid proteins, disrupting the N-RNA interaction represents a potentially powerful new antiviral therapeutic strategy with broad applicability. In this study, we explore the RNA binding properties of RVFV N using biochemical, biophysical, and combinatorial methods and describe construction of a fluorescent biosensor that reports on N-RNA interactions.
2. Materials and Methods
2.1. Overexpression and purification of Rift Valley fever virus N protein
Plasmid encoding (His6)-tagged RVFV N protein was transformed into E. coli strain BL21(DE3)pLysS. Transformed cells were grown at 37°C in LB media containing 100 μg/ml ampicillin and 50 μg/ml chloramphenicol until OD600=0.6-0.8. Protein expression was induced by addition of IPTG to a final concentration of 0.5mM and cultures were grown overnight at room temperature (24°C). Cells were harvested by centrifugation and stored at −80°C.
Cells were lysed by resuspension of thawed pellets in BugBuster (Novagen) in the presence of protease inhibitors. Benzonase (Novagen) was added to the lysis buffer at a concentration of 12 units/mL to degrade DNA and RNA. Insoluble cell debris was removed from the lysate by centrifugation at 12,000 RPM for 45 minutes at 4°C. The (His6)-tagged N was batch purified from the cell lysate by using Ni-charged IMAC resin (Bio-Rad). The resin was washed 5 times with 35mM imidazole/0.5M NaCl/1.5M urea in 50mM Tris-HCl buffer, pH 8.0 and the protein eluted with 300mM imidazole/0.5M NaCl in 50mM Tris-HCl buffer, pH 8.0. The eluate was concentrated and the buffer exchanged to 0.5M NaCl in 50mM Tris-HCl pH 8.0 using Amicon centrifugal filters (10K MWCO). After flash freezing with liquid nitrogen the N protein was stored at −80°C. The purity of the protein was checked by SDS-PAGE and its concentration was determined by the absorbance at 280 nm using extinction coefficient obtained from the ExPASy website (33,920 M−1cm−1).
2.2. Preparation of pool 0 DNA
In order to construct a pool of DNA template with a 30 nucleotide long randomized region flanked by constant regions, MMLV reverse transcriptase (Invitrogen) was used to perform an extension reaction on two chemically synthesized oligos that had complementary 3′ ends. The sense primer contained an EcoRI site and a T7 promoter near the 5′ end followed by a 13 nucleotide long constant region (Figure 1A, EcoT7SX5′). The antisense primer contained a XbaI site near the 5′ end and a 30 nucleotide randomized cassette flanked by constant regions (Figure 1A, XBbSx30). After extension the DNA was digested with EcoRI and XbaI to obtain templates of uniform size and purified by phenol/chloroform extraction and ethanol precipitation.
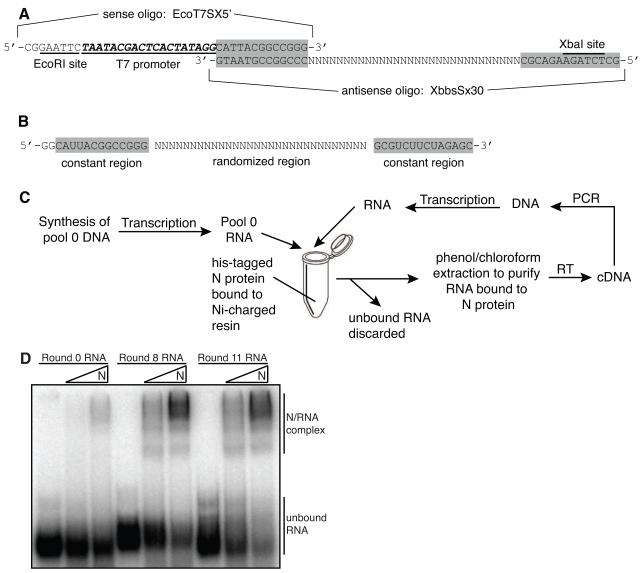
FIGURE 1. Construction of random sequence nucleic acid pools and in vitro selection methodology.
A. Oligos used to construct pool 0 DNA. The sense oligo (EcoT7SX5′) contained an EcoRI restriction site, a T7 promoter and a 3′ constant sequence that was complementary to the 3′ constant sequence on the antisense oligo (XBbsSx30). The antisense oligo contained the 30 nucleotide long randomized region flanked by constant regions and a 5′ Xba1 restriction site. Shaded boxes highlight constant regions present in both the oligo DNA and RNA. B. Randomized RNA sequence used for the in vitro selection. C. Schematic diagram of the technique used to select and amplify RNAs that bound with high affinity to RVFV N. Non-binding and binding RNAs were separated using N immobilized to Ni-charged resin as a target. Therefore, the resin also immobilized RNAs that bound to the N, allowing unbound RNAs to be washed away and selected RNAs were purified and amplified. RT-PCR was used to create more DNA template for transcription, generating more RNA for subsequent rounds of selection. This cycle was repeated 15 times. D. Evolution of RNAs with high affinity to N after repeated rounds of selection. The pools RNA binding affinity for N was analyzed by EMSA. The bands that migrate faster in the gel are unbound RNA species and the slower migrating bands represent N/RNA complexes. RNAs were internally labeled with 32P-UTP and the gels visualized by phosphorimaging.
2.3. Preparation of pool 0 RNA
Pool 0 RNA was generated using pool 0 DNA as a template and a MEGAshortscript Kit (Ambion) (Fig. 1B). The transcription reaction was DNase treated and purified by phenol/chloroform extraction and ethanol precipitation. The pool RNA was further purified by denaturing gel electrophoresis. The band corresponding to the RNA was visualized by UV shadowing, excised, eluted into 0.5M ammonium acetate, 1.0mM EDTA pH 8.0 and 0.2% SDS, and ethanol precipitated.
2.4. Selection of RNA aptamers to Rift Valley fever virus N protein
Pool RNAs were incubated at 30°C with Ni-charged IMAC resin (Bio-Rad) in binding buffer (10mM HEPES pH 7.5, 150mM NaCl and 5mM MgCl2) using a rotisserie in a hybridization oven. After incubation the reaction was placed on ice, the resin pelleted by gravity, and the unbound RNA in the supernatant collected and used for subsequent selections. This “pre-selection” step was necessary to prevent amplification of RNAs that bind with high affinity to Ni-charged resin. Next preselected RNAs were incubated with purified (His6)-tagged N protein that was pre-bound to Ni-charged resin in binding buffer with constant rotation at 30°C. The binding and nonbinding RNAs were partitioned by allowing the resin to settle to the bottom of the microfuge tube on ice. The unbound RNAs in the supernatant were removed and discarded. The Ni resin pellet containing the target RNA-N complexes was washed multiple times with binding buffer to remove weakly bound RNAs and then phenol/chloroform extracted and ethanol precipitated to purify RNAs that bound to N with high affinity.
The selected RNAs were then subjected to RT-PCR to obtain DNA templates for subsequent rounds of selection. The reverse transcription step was performed in a final volume of 20 μL using AMV reverse transcriptase (Promega) according to the manufacturer’s protocol. Approximately 200 ng XbaBbs primer (5′-GCT CTA GAA GAC GC-3′) and 400 ng RNA template (a 1:2 primer/template ratio was always used) were incubated at 70°C for 3 minutes and snap cooled on ice. Next, AMV RT buffer, dNTP mix (0.6mM of each NTP) and 0.3 μL AMV RT enzyme were added and the reaction incubated at 42°C for 1 hour. The PCR step was performed in a final volume of 50 μL containing 5 μL of the cDNA template, 0.2μM of EcoT7SX5′ and XbaBbs primers, dNTP mix (0.2mM of each dNTP), MgCl2 (1.5mM final), 1X PCR buffer, and 1.25 units of GoTaq Hot Start DNA polymerase. Twenty five cycles of amplification were used and the annealing temperature varied and was empirically determined for each round of selection by performing a small scale gradient PCR. The RT-PCR products were digested with EcoRI and XbaI and used as templates for T7 transcription to generate RNA for subsequent rounds of selection.
Fifteen rounds of selection for RNA aptamers to RVFV N were conducted using the protocol described above (Fig. 1C). We gradually increased the stringency of the selection each round by decreasing the amount of time allotted for RNA-protein complexes to form. Pool 0 RNAs and N were incubated together for 5 hours whereas pool 14 and 15 RNAs were incubated with N for less than 1 minute. The final (16th) round of selection was done by incubating N that had the His-tag removed by tobacco etch virus (TEV) protease digestion with a mixture of radioactive (200,000 cpm) and unlabeled (300 ng) pool 15 RNA for less than one minute at room temperature (24°C). The binding reactions were loaded with 2μL of glycerol loading dye onto a pre-chilled 6% acrylamide/TBE gel and samples migrated at 10V/cm on the gel in the cold room for 75 minutes. The gel was visualized using phosphorimagery (Fuji) and the bands corresponding to the N-RNA complex were excised from the gel. To obtain round 16 RNA, slices of the excised bands were subjected to RT-PCR followed by T7 transcription using essentially the same protocol described for previous selections. The only alteration involved performing the reverse transcription step in a final volume of 50 μL to synthesize cDNA from a 10 μL equivalent slice of gel. For comparative analysis the digested RT-PCR products of rounds 1, 8, 15 and 16 were ligated into the XbaI and EcoRI sites of the pUC18 plasmid and the DNA from individual clones isolated and sequenced.
2.5. Synthesis and purification of selected pool and clonal RNAs
RNA was synthesized using either clones digested with XbaI or pool DNA obtained from digested RT-PCR products as templates for T7 RNA polymerase. MEGAshortscript kits (Ambion) were used to synthesize non-radioactive RNA and MAXIscript kits (Ambion) were used to synthesize RNA in the presence of radioactive α-32P-UTP (800Ci/mmol; PerkinElmer). The transcription reactions were then DNase treated and purified by phenol/chloroform extraction and denaturing gel electrophoresis as described for the synthesis of pool 0 RNA.
2.6. Nitrocellulose filter binding assays
N was serial diluted to varying concentrations in a binding buffer (10mM HEPES pH 7.3, 150mM NaCl, 20mM KCl, and 5mM MgCl2 final concentration). Nine μL of various dilutions of N were added to each tube along with 1 μL of RNA that had been subjected to denaturation at 90°C for two minutes followed by snap cooling on ice. In each experiment 25,000 cpm/μL of reference RNA (wild type) was added. The cpm/μL of the other RNAs was adjusted based on their uridine content to ensure an equal number of molecules were used. After incubation at 30°C for one hour, reactions were diluted to 100 μL with ice-cold binding buffer and filtered through pre-soaked nitrocellulose filters (Millipore HAWP). The filters were washed twice with 500 μL ice-cold binding buffer, dried, and the radioactivity retained on filters was measured by scintillation counting.
2.7. Electrophoretic mobility shift assays
Samples for EMSAs were prepared as described for the filter binding assays. After incubation the reactions were placed on ice and 2 μL of glycerol loading dye was added to each tube. Samples were loaded onto pre-chilled 6% acrylamide/1X TBE gels that were run in the cold room at 10V/cm for 75 minutes. Gels were then dried, exposed to a phosphorimager screen, and visualized on a Fuji FLA3000G Image Analyzer.
2.8. Synthesis of mutant and truncated aptamer RNAs
DNA template that contained the desired mutations to the aptamer sequence was generated by performing a PCR extension reaction on two chemically synthesized oligos that had complementary 3′ ends. The PCR step was performed in a final volume of 50 μL containing 0.3μM of sense and antisense oligo, dNTP mix (0.2mM of each dNTP), MgCl2 (1.5mM final), 1XPCR buffer, and 1.25 units of GoTaq Hot Start DNA polymerase. Thirty cycles of amplification with an annealing temperature of 55°C were used. The PCR products were digested with EcoRI and XbaI and used as templates T7 transcription to generate mutant aptamer RNA. A MAXIscript kit (Ambion) was used to synthesize RNA in the presence of radioactive α-32P-UTP (800Ci/mmol; PerkinElmer) using the same protocol described for synthesis and purification of clonal RNAs.
2.9. RNA solution structure probing
RNAs were dephosphorylated using Antarctic phosphatase (New England BioLabs). The 5′ ends were labeled with γ-32P-ATP (6000Ci/mmol; PerkinElmer) using T4 polynucleotide kinase (Invitrogen) and the RNA purified by denaturing gel electrophoresis. Labeled RNAs were heated in water for two minutes at 90°C followed by snap cooling on ice. Binding buffer (10mM HEPES pH 8, 150mM NaCl, 20mM KCl and 5mM MgCl2) was added and samples incubated at 37°C for 10 minutes. The folded RNA (60,000 cpm/rxn) was incubated in binding buffer with 2 μg tRNA and either RNase T1 (0.25 units/rxn) or RNase U2 (10 units/rxn at 37°C for 10 minutes. A T1 ladder was prepared by incubating the RNA at 50°C in a denaturing buffer (6mM sodium citrate pH 5, 7M urea and 1 mM EDTA) prior to digestion with RNase T1. An alkaline ladder was prepared by incubating the RNA at 90°C for 4 minutes in 100mM sodium carbonate buffer pH 9.0. All samples were ethanol precipitated, resuspended in formamide loading dye and analyzed by denaturing gel electrophoresis.
2.10. Fluorescence polarization measurements
A 35 nucleotide long RNA was chemically synthesized, 3′ end labeled with FAM and HPLC purified by TriLink BioTechnologies. The fluorescent label was attached to the RNA using a six-carbon linker. N was serial diluted to varying concentrations in a binding buffer (10mM HEPES pH 8, 150mM NaCl, 20mM KCl and 5mM MgCl2). To determine the equilibrium dissociation constant of N/FAM-RNA complex, various dilutions of N were added to 10 nM FAM-labeled aptamer RNA that had been subjected to denaturation at 90°C for two minutes followed by snap cooling on ice. Reactions were incubated at 30°C for 1 hour. Samples for competitive binding experiments were prepared by incubating varying concentrations unlabeled competitor RNA (450 pM-5.0 μM) with 4.5 μM N for 40 minutes at 30°C. Next, 10 nM FAM-labeled RNA was added and the reaction incubated for 40 minutes at 30°C. Fluorescence polarization values were measured using a VICTOR X multilabel plate reader. Binding profiles were plotted and apparent Kd and Ki values were calculated using GraphPad Prism software.
3. Results
3.1. In vitro evolution of RNA aptamers to Rift Valley fever virus N protein
An 84 bp segment of DNA that contained a 30 bp random region flanked by constant sequences was used as a template for T7 RNA polymerase transcription. The template contained a 5′ EcoRI site, a T7 promoter, a 13 nucleotide-long constant region followed by a 30 nucleotide-long randomized region and then another constant region with a 3′ XbaI site (Fig. 1A). The presence of all 4 nucleotides in the randomized region was verified by batch sequencing of pool 0 DNA (data not shown) and by sequencing of individual clones from pool 1 DNA. The recombinant nucleocapsid protein (N) from the M12 strain, a clinical isolate of RVFV (Giorgi et al., 1991), was overexpressed in E. coli and purified using Ni-charged IMAC resin. Next, a selection/amplification technique (SELEX) was used to isolate several families of RNA aptamers that bind with high affinity and specificity to the N protein from a starting pool of approximately 1014 small RNAs (Fig. 1B). The basic methodology for the in vitro selection is outlined in Figure 1C and the technique described in detail in Materials and Methods.
The iterative selection step consisted of partitioning the unbound and bound RNA species by immobilizing the N protein onto Ni-charged IMAC resin. This allowed the bound RNAs to be pelleted with the resin beads and the unbound RNAs in the supernatant to be discarded. The selected RNAs were then purified from both N and Ni-resin by phenol/chloroform extraction and amplified using RT-PCR and primers specific to the constant sequences. Sixteen rounds of selection were conducted and negative selections were periodically performed to eliminate Ni-resin binding species and ensure isolation and amplification of N-specific aptamers. During the first rounds of selection N was allowed extended incubation with pool RNAs in order to capture and amplify a small number of functional molecules. In later rounds, the stringency of selection was increased using a greater RNA to protein ratio (100:1) and shorter incubation periods. The final (16th) round of selection was performed using non his-tagged N and the bound and unbound species of RNA were separated by electrophoretic mobility shift. The band on the gel corresponding to high molecular weight RNA/protein complexes was excised, and the RNA eluted from the gel slice and subjected to RT-PCR followed by T7 transcription.
3.2. Binding affinity of pool RNAs to N increases with repeated rounds of selection
Binding assays were performed at several intermediate rounds of the selection to assess the affinity of pool RNA sequences for N. Pool RNAs were transcribed from RT-PCR products and internally labeled with 32P-UTP. A constant amount of labeled RNAs (25,000 cpm) was incubated in binding buffer with either 1.2μM or 12μM RVFV N for 1 hour at 30°C and then subjected to non-denaturing gel electrophoresis (Fig. 1D). The range of N protein concentrations used in our study (0.6-35μM) ) was determined empirically. The bands that migrate faster on the gel correspond to the free RNA species and the higher migrating bands represent high molecular weight RNA/protein complexes. Round 0 RNAs exhibited low overall binding affinity to N whereas the binding affinity of round 8 RNAs had increased dramatically. When incubated with the highest concentration of N, most of round 8 and round 11 RNAs were present in the slowest migrating band, indicating that they were able to bind stably to N and that the affinity for N was increasing with subsequent rounds of selection. Although the binding affinity of round 11 RNAs for N was high, several further rounds of selection were carried out to decrease the population complexity.
3.3. Selected aptamer RNAs have a distinct primary sequence signature
The RT-PCR products from rounds 1, 8, 15 and 16 of the selection were cloned into pUC18 plasmid. Individual clones from rounds 1 and 8 and a total of 100 individual clones from rounds 15 and 16 were sequenced. Analysis of round 1 sequences showed a distribution of 27% A, 19% G, 32 % C, and 23 % T (Table 1). Interestingly, when these percentages were compared to the nucleotide distribution of sequences isolated from rounds 8, 15 and 16 of the selection, a progressive enrichment for both guanosine and thymidine residues was evident. While the enrichment observed for guanosine residues stabilized by round 8, the percentage of thymidine residues continued to increase from 31% at round 8 to 40% in rounds 15 and 16. Also, the percentage of adenosine and cytidine residues was reduced in sequences isolated from later rounds of selection compared to round 1 clones. Analysis of individual sequences from rounds 15 and 16 of the selection showed that 49 out of the 100 sequences occurred with multiple frequencies or were at least 90% similar (Table 2). These results suggest that, while identifiable sequence trends were evident throughout the population, several sequence families were beginning to dominate the pool.
TABLE 1.
Distribution of nucleotides within the RVFV genome and the randomized region of clonal RNAsa
| Sequence Analyzed | % A | % G | % C | % T |
|---|---|---|---|---|
| Round 1 RNA | 27 ± 9 | 19 ± 6 | 32 ± 11 | 23 ± 9 |
| Round 8 RNA | 17 ± 8 | 29 ± 9 | 19 ± 5 | 31 ± 7 |
| Rounds 15&16 RNA | 19 ± 6 | 28 ± 4 | 15 ± 5 | 40 ± 6 |
| RVFV S segment | 23.6 | 23.1 | 25.9 | 27.4 |
| RVFV M segment | 27.3 | 25.4 | 20.1 | 27.3 |
| RVFV L segment | 29.2 | 23.8 | 20.3 | 26.7 |
Sequences corresponding to the viral genome were obtained from the NCBI nucleotide database using the following accession numbers: NC_014395.1, NC_014396.1, and NC_014397.1 that correspond to the S, M, and L segments respectively. Eight sequences from round 1 and round 8 and a total of 100 sequences from rounds 15 and 16 were analyzed. ± deviation from the mean.
TABLE 2.
Alignment of the randomized region (5′-3′) of RNA aptamers isolated from the different rounds of selection
| RNA a | sequenceb | Frequency (100 clones) |
|---|---|---|
| MBE16.49 | -------AGUAAUGCUAUGAUGAUAAUGUGUCCCGGGG-- | 1 |
| MBE16.41 | -------AGUAACGCUAUGAUGAUAAUGUAUCCCGGGG-- | 1 |
| MBE15.12 | ----CUGUUUACUGAACUAUGAUACUUU---CUGGGG--- | 5 |
| MBE16.64 | -----AUGUUACUGACUUCUUCUGAUUU---CUAGUGG-- | 2 |
| MBE15.2 | ----UGUCGUACUGAU---UGAUGAUUUACUCUGGGG--- | 5 |
| MBE15.8 | ----UGUCGUACUGAU---UGAUGAUUUACUCGGGGG--- | 1 |
| MBE16.87 | AGCCAGUAUUACUGAU---UGAUGAUU----CUGUGG--- | 2 |
| MBE16.59 | -UGAUGCAGAACUGAU---UGAUUACUU---CUGGGG--- | 2 |
| MBE15.21 | --GCUACU--UCUGAU---UGAUUAUUU---CUCUGGUGG | 2 |
| MBE16.50 | ---AUGUCAUACUGAUUACUGAUGACUU----UGGGG--- | 2 |
| MBE16.74 | ---AUGUCAUACUGAUUACUGAUGAAUU----UAGGGG-- | 1 |
| MBE8.1 | --------CUUCUGAUUACUGAUUUCUUUCUCUAGUGG-- | NDc |
| MBE16.113 | -------AAUGUCGAUUACUGGUUGCUGACUCUGGGG--- | 1 |
| MBE16.114 | -------AAUGUCGAUUACUGAUUGCUGAUUCUGGGG--- | 2 |
| MBE15.7 | -----UGCAUGCUGAUUACUG---AUUUUAUCCUGGGG-- | 1 |
| MBE15.20 | -----UGCAUGUUGAUUACUG---AUUU-AUCCUGGGG-- | 1 |
| MBE15.14 | -----UGCAGACUGAUUACUG---ACUUUCUCUAGUGG-- | 3 |
| MBE16.40 | ---AAUGCGGACUGAUUACUG---AUACUCUGGGG----- | 5 |
| MBE8.6 | --GUGGACGUCCUGCGUUGUG--GGCUUG---UGUGG--- | ND |
| MBE15.3 | -------CGUCCCGUAGUGUCGGUACUGAUUGAUGUG--- | 9 |
| MBE16.57 | -------CGUCCCGUAGUGUCGGUACUGAUUAAUGUG--- | 1 |
| MBE8.3 | -------CGCCCCGUAGUGCAUAGUCUGAUUGAUGUG--- | ND |
| MBE16.112 | ----UGCGAUACUGAAU-AUGAAAGCUUUAGGUGG----- | 2 |
| MBE1.4 | --ACGAGUGCCAUGCACUACAUUGUCUGUUGA-------- | ND |
| MBE1.3 | ----CACCGUAACAAAGUUGUAAGACACCAACCA------ | ND |
| Others | Orphan sequence | 51 |
Aptamer RNAs are labeled using two numbers. The first number corresponds to the round the aptamer was first isolated from and the second number is a unique identifier.
The constant regions are not shown. Sequences from rounds 15 and 16 that occurred with multiple frequencies or were <90% similar are shown. The GAUU motif is in bold font and the pyrimidine/guanine motif is underlined.
Not determined
3.4. Characterization of binding affinity between selected aptamer RNAs and N protein
Several clones from rounds 1, 8, and 15 of the selection were chosen to assess the evolution of aptamer affinity (Table 2). The filter binding assay was used to compare the binding of individual clonal RNAs (Fig. 2A). As expected, the binding affinity of the round 1 RNA (MBE1.3) was very low while the binding affinity of round 15 RNA (MBE15.3) to N was high. Moreover, the persistence of RNAs with low binding affinity to N at round 8 underscored the need for further rounds of selection to cull lower-affinity binders from the pool. To confirm the high affinity of our aptamers cloned from the last selection rounds, we tested them in a competitive binding assay against a specific natural target, the terminal panhandle structure of RVFV genomic RNA.
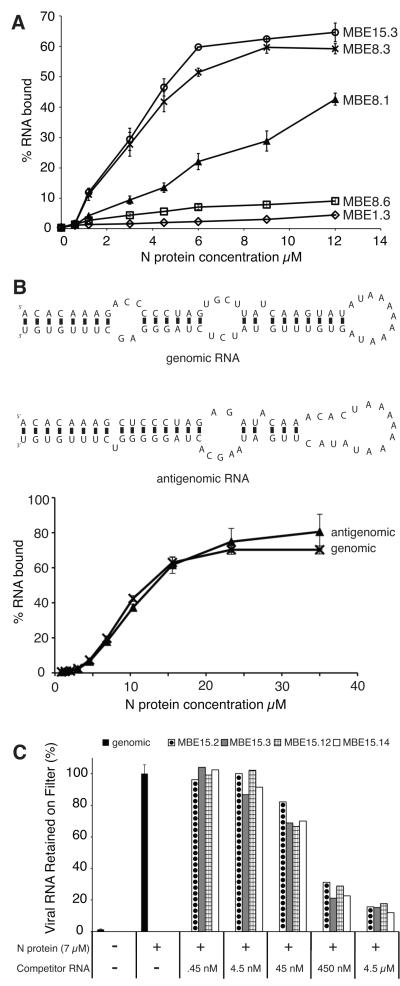
FIGURE 2. Characterization of binding interactions between selected RNAs and N.
A. A nitrocellulose filter binding assay was used to compare N binding to clonal RNAs from round 1(MBE1.3, open diamonds), round 8 (MBE8.1, filled triangles; MBE8.3, “X”; MBE8.6, open squares) and round 15 (MBE15.3, open circles). RNAs were internally labeled with 32P-UTP and radioactivity retained on filters was quantified by liquid scintillation counting. B. Top panel: mfold predicted secondary structure of viral genomic and antigenomic small RNA constructs. Bottom panel: a filter binding assay was used to test the binding ability of genomic (“X”) and antigenomic (filled triangles) RNA constructs to the N protein. C. A competitive filter binding assay was used to test the ability of RNAs from the final (15th and 16th) rounds of selection to inhibit binding of viral genomic RNA. The amount of radiolabeled (32P-UTP) viral RNA retained on the nitrocellulose filters in either the absence (black fill) or presence of unlabeled competitor was measured by liquid scintillation counting. The following aptamer RNAs were tested: MBE15.2, circle fill; MBE15.3, gray fill; MBE15.12, grid fill; MBE15.14; white fill.
3.5. Aptamer RNAs compete with viral RNA sequences to bind N
First, a binding experiment was performed to test the affinity of N for small RNA molecules that mimic the panhandle structures formed by the genomic or antigenomic RVFV RNAs (S segment). The stem-loop structures are formed by the base-pairing interaction of the 5′ and 3′ ends of one RNA molecule. For practical reasons, the long intervening sequence of the S segment was replaced with a stretch of adenine residues (Fig. 2B). The filter binding results showed that N binds well to both small RNA constructs (Fig. 2B).
Next, we performed competitive binding experiments using the internally radiolabeled genomic panhandle RNA construct and unlabeled aptamer RNAs. A constant amount of N (7μM) was incubated with varying amounts of unlabeled aptamer RNA (0.45 nM – 4.5 μM for 1 hour at 30°C. Labeled genomic RNA (100,000 cpm/rxn) was added and the binding reaction incubated for 5 more hours at 30°C. Binding of labeled RNA was then assessed by filter binding assay. The results from the competition filter binding assay showed that when increasing amounts of unlabeled aptamer RNA were present during the binding reaction, less genomic panhandle RNA was retained on the filter (Fig. 2C). Clearly, the presence of aptamer RNA inhibited the formation of genomic RNA-N complexes, suggesting the aptamer RNA binds to the same region on N as the viral panhandle.
3.6. Two conserved motifs are found in late-round aptamers
We then checked for the presence of conserved motifs among the high-affinity aptamers of rounds 15 and 16 using primary sequence alignment and visual inspection. The results are summarized in Table 2. A GAUU-containing motif was identified in 82% of the 49 sequences isolated with high frequency. This motif was variable in the number of GAUU repeats per RNA and the exact sequence observed. For example, MBE16.74 had a GAUUACUGAUGAAUUU motif while sequencing of another isolate (MBE16.87) revealed a GAUUGAUGAUU motif (Table 2). Despite these minor differences, the prevalence of variations of the GAUU motif in a high percentage of the RNAs isolated indicates that it may be an important recognition sequence for N. Also, a pyrimidine/guanine motif we call the U/G-rich motif, located near the 3′ end of the random region, was present in many of the sequences isolated from rounds 15 and 16 (Table 2). Interestingly, alignment of aptamer sequences with the RVFV genome and antigenome using BLAST revealed that the U/G-rich motif UUUCUGGGGGCG isolated 11 times in the selected RNAs was also present in the RVFV L antigenome (Table 3). The evolution of these two motifs could be responsible for the observed enrichment of guanosine and thymidine residues in sequences isolated from rounds 8, 15 and 16. There was a strong correlation between the emergence of guanosine and thymidine rich primary sequences during the selection and the evolution of RNAs that bind with high affinity to N. Together, these data suggest that N may preferentially bind to GAUU and/or U/G-rich sequences.
TABLE 3.
Regions of sequence similarity between RNA aptamers and the RVFV genome and antigenome identified using BLAST
|
3.7. Directed mutagenesis of GAUU and U/G-rich motifs in aptamer RNAs
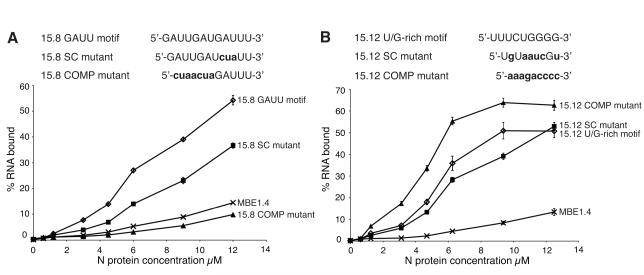
Mutant aptamer RNA constructs were generated to test the hypothesis that N specifically recognizes and binds to GAUU and U/G-rich motifs on selected RNAs. Two variants of the GAUUGAUGAUU motif, either obtained by scrambling the sequence or by introducing complementary nucleotides in place of the U/G-rich motif, were tested in the context of the MBE15.8 aptamer (Fig. 3A). The binding affinity of MBE15.12 RNA with similar disruptions to its U/G-rich motif (UUUCUGGGG) (Fig. 3B) was also tested.
FIGURE 3. Characterization of binding affinity of mutant aptamer RNAs to N.
A nitrocellulose filter binding assay was used to compare N binding to wild type and mutant aptamer RNAs where either the GAUU or U/G-rich primary sequence motifs were disrupted. RNAs were internally labeled with 32P-UTP and radioactivity retained on filters was quantified by liquid scintillation counting. A. Top panel: MBE15.8 aptamer RNA GAUU motif and mutant constructs. Changes to sequence motif are in bold and lowercase font. Bottom panel: Filter binding assay of wild type MBE15.8 aptamer, mutant constructs and round 1 MBE1.4 RNA. MBE15.8 wild type RNA; open diamonds, MBE15.8 SC mutant; filled squares, MBE15.8 COMP mutant; filled triangles, and round 1 MBE1.4 RNA; “X”. B. Top panel: MBE15.12 aptamer RNA U/G rich motif and mutant constructs. Changes to sequence motif are in bold and lowercase font. Bottom panel: Filter binding assay of wild type MBE15.12 aptamer, mutant constructs and round 1 MBE1.4 RNA. MBE15.12 wild type RNA; open diamonds, MBE15.12 SC mutant; filled squares, MBE15.12 COMP mutant; filled triangles, round 1 MBE1.4 RNA; “X”.
Filter binding assays were used to compare the affinity of mutant and wild type aptamer RNAs to N. Scrambling one GAUU repeat in the MBE15.8 sequence caused a one-third decrease in the level of RNA bound at 12 μM N. Complementary mutation of the first 7 nucleotides of the GAUU motif caused an even more dramatic decrease in binding. The binding affinity of the complementary MBE15.8 mutant (15.8 COMP mutant) to N was even lower than the affinity observed for a round 1 RNA (Fig. 3A). Changing the U/G-rich motif from MBE15.12 RNA from UUUCUGGGG to UGUAAUCGU caused a slight decrease in the binding affinity to N compared to the affinity of the original sequence. Unexpectedly, an RNA that contained a complementary mutation of the U/G-rich motif (15.12 COMP mutant) exhibited better binding than the original MBE15.12 aptamer (Fig. 3B). Interestingly, closer examination of this mutant revealed that it contained an 11 nucleotide sequence identical to the 5′ end of the S segment of viral genomic RNA (Table 3).
These data show that the GAUU motif and, to a lesser extent, the U/G motif, are important for N binding to aptamer RNA and they likely facilitate interactions between N and other RNA molecules as well. The location of the motifs and the binding affinities of the RNA aptamers were then used to design truncated aptamers of diminished size.
3.8. Identification of truncated RNA aptamers that bind with high affinity to RVFV N
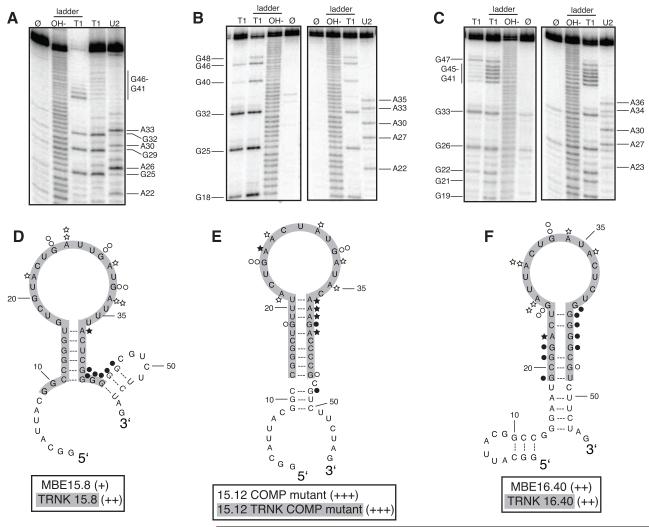
Starting with the sequences of several aptamers described above, a series of truncated RNA aptamers was generated in order to find one or more small RNAs that could eventually be chemically synthesized and fluorescently labeled. We chose to truncate RNAs that were either highly represented in the final round of selection, that contained GAUU or U/G-rich sequence motifs, or contained sequences that were also present in viral RNA (Fig. 4). During the truncation process we generally sought to preserve important sequence motifs and predicted secondary structure. A combination of RNase T1 and U2 structure probing and mfold were used to determine the conformation of several aptamer RNAs. Overall, results obtained from the structure probing experiments provide support for the mfold predicted structures. However, one common exception was that the mfold algorithm often predicted base pairing interactions at the apical stems of the aptamer RNAs. The RNase T1 and U2 structure probing data showed that guanosine and adenosine nucleotides at the apical stems were reactive (single-stranded) (Fig. 4A, B and C). All three RNAs presented here exhibit a strikingly similar stem loop structure that may serve as a binding site for N. Our results indicate that preservation of this structure is necessary for truncated RNAs to retain their ability to bind N (Fig. 4D, E and F).
Figure 4. Secondary structure model and binding affinity of full length and truncated RNA aptamers to RVFV N.
A. to C. Structure probing of full length RNA aptamers MBE15.8 (A), 15.12 COMP mutant (B) and MBE16.40 (C) was conducted to determine the reactivity of guanosine (lane T1) and adenosine (lane U2) residues to enzymes RNase T1 and RNase U2. Lane Ø represents undigested RNA. T1 and OH− ladders are used as RNA sequencing lanes. D. to F. Summary of nucleotide reactivity superimposed on secondary structure models of MBE 15.8 (D) 15.12COMP mutant (E) MBE16.40 (F). Circles represent results obtained from experiments conducted with the single-strand guanosine specific T1 nuclease and stars are representative of the single-strand adenosine specific U2 nuclease. The greater the number of symbols associated with a specific nucleotide, the stronger the reactivity. Open symbols indicate a lack of reactivity. Truncated aptamers are highlighted in gray. Aptamers were rated based on percent of input RNA bound at 12.5 μM N: 40-49% = (+), 50-59% = (++), and 60-69% = (+++).
MBE15.8 and MBE16.40 contained a GAUU motif that was shown above to be a requirement for N to bind MBE15.8 RNA. When these aptamers were truncated and the stem loop structure containing the GAUU motif was preserved, both retained their ability to bind N (Fig. 4D and F). Conversely, a truncation of MBE16.41 that disrupted the pyrimidine portion of U/G-rich motif caused the RNA to completely lose its affinity for N (Table 2; 7% input RNA bound at 12.5 μM). These results further support our hypothesis that the GAUU and U/G-rich motifs are important components of N-RNA recognition.
A truncated version of MBE15.14 that contained an 11 nucleotide-long sequence of RNA present in the L segment of viral genomic RNA bound N with high affinity (62% of input RNA bound at 12.5 μM). This U-rich sequence was isolated a total of 6 times from the final pool of RNA and was also found in two aptamers unrelated to MBE15.14 (Table 3). The 15.12 COMP mutant aptamer bound N better than the wild type aptamer MBE15.12 (Fig 3B). MBE15.12 RNA was of interest because it contained a U/G-rich motif also found in the L segment of viral antigenomic RNA (Table 3). Interestingly, the complement of the U/G-rich motif is a sequence found at the 5′ end of the S segment of viral genomic RNA. The truncated version of 15.12 COMP mutant (15.12 TRNK COMP mutant) retained this viral sequence, bound N with high affinity and was predicted by RNA structure probing experiments and mfold to be a stem loop structure (Fig 4E). Mutation of the large 15 nucleotide long loop in 15.12 TRNK COMP mutant to create RNAs with smaller loops (TRNK loop A and TRNK loop B) did not affect binding affinity to N (data not shown). TRNK loop A and TRNK loop B RNAs retained all and most of the viral RNA sequence, respectively, which suggests that this sequence may be important for N recognizing and binding to both viral and aptamer RNA.
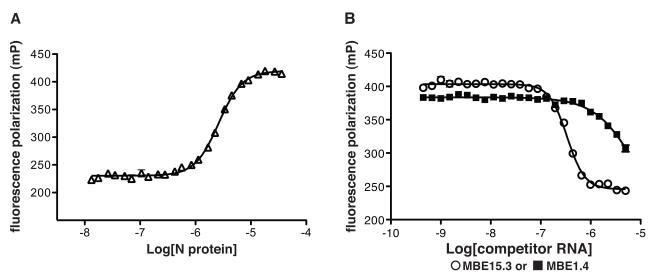
3.9. Fluorescence polarization detection of N-aptamer binding
A truncated RNA aptamer (15.12 TRNK COMP mutant, Fig. 4E) was chemically synthesized and 3′ end-labeled with fluorescein (TriLink BioTechnologies). Fluorescent RNA was incubated in binding buffer with varying concentrations of RVFV N at 30°C for 1 hour and fluorescence polarization (FP) measured. The results showed that the FP value was low (220 mP) when the label was attached to RNA alone and increased to 420 mP when the RNA was incubated with increasing concentrations of N (Fig. 5A). Titration of N with the 3′-FAM-labeled RNA aptamer gave an apparent Kd of 2.6 μM. Next we performed competitive binding experiments by incubating a constant amount of N (4.5 μM) with varying concentrations (450 pM-5.0 μM) unlabeled competitor RNA at 30°C for 40 minutes. Labeled aptamer RNA was added (10 nM) and the reaction incubated for 40 more minutes at 30°C. Fluorescence polarization measurements were taken and the results showed unlabeled aptamer RNA competed for N binding with labeled aptamer RNA. Competition experiments were conducted with four different aptamers and the apparent Ki values were all in the ~200 nM range. A nonbinding RNA (MBE1.4 RNA) was unable to effectively inhibit the labeled RNA from binding N (Fig 5B). These data demonstrate that fluorescence polarization can be used to report binding of fluorescently labeled aptamer RNA to N as a function of competitor binding and that this approach could find utility as a biosensor or drug-screening tool.
FIGURE 5. Analysis of aptamer RNA-N binding interactions using fluorescence polarization (FP).
A. Binding profile for the association of N with fluorescently labeled aptamer RNA (open triangles). B. A competition assay where a fixed concentration of N was incubated with varying concentrations of unlabeled competitor RNAs (round 1 MBE1.4 RNA; filled squares, MBE15.3 aptamer RNA; open circles). Fluorescently labeled aptamer RNA was added and the FP signal is plotted versus competitor RNA concentration.
4. Discussion
All known functions for N during viral replication involve binding RNA in either a non-specific or specific manner. On the one hand, N possesses sequence-independent binding properties that allow it to form N-RNA complexes along the entire length of viral RNA. On the other hand, specific recognition of cognate RNA by N has been observed in a number of viruses related to Rift Valley fever virus. In the present study, we characterize specific N-RNA interactions for Rift Valley fever virus.
RNA constructs that resemble the panhandle of RVFV viral RNA S segment bind with medium to high affinity to N. These results corroborate studies in Sin Nombre virus (SNV), a member of the Hantavirus genus, that show N specifically binds to a panhandle structure formed by the self-complementary 5′ and 3′ ends of the viral genome (Mir et al., 2006; Mir and Panganiban, 2004, 2005). The triplet repeat UAGUAGUAG is required for N to bind SNV mRNA and this sequence is also present at the 5′ end of all three segments of the SNV genome and antigenome (Mir and Panganiban, 2010). Together these data show that the binding specificity of N is influenced by both sequence and structural features of RNA. N from several other species of bunyavirus are known to preferentially bind to the 5′ end of the viral genome and antigenome (Ogg and Patterson, 2007; Osborne and Elliott, 2000; Severson et al., 2001). This binding event is thought to be an important signal that causes N to switch from a specific to a nonspecific mode of binding and initiates nucleocapsid assembly (Raju and Kolakofsky, 1987). Binding outside this region was also observed in Jamestown Canyon virus (Orthobunyavirus genus) but the sequences or secondary structures involved in this N-RNA interaction were not specifically identified (Ogg and Patterson, 2007).
To further characterize N-RNA binding motifs, we used an unbiased in vitro evolution approach to search for RNA ligands of N without any a priori assumptions of sequence or structural constraints. A library of randomized RNA molecules was incubated with N and the RNA species that bound N with high affinity were isolated, amplified and used as substrates for the next round of selection. Sequence analysis of 100 individual RNAs isolated from the final rounds of the in vitro selection showed enrichment of guanosine and thymidine residues compared to sequences analyzed from earlier rounds (Table 1). Identification of regions of similarity between aptamer and viral RNA sequences using BLAST revealed the presence of motifs located in different regions of all three viral RNA segments. The most frequent similarity is represented by an U/G-rich motif in the RVFV L antigenome, found in 11 out of the 100 sequenced aptamers (Table 3). Surprisingly, an RNA that contained a complementary mutation of the U/G-rich motif bound with even higher affinity to N than the wild type aptamer (Fig 3B). These data suggest that N is able to recognize both a specific sequence and its complement. The same phenomenon has been observed in Jamestown Canyon virus, where it was shown that N binds with high affinity a short segment of ssRNA that corresponds to the 5′ end of the viral genome, as well as its complement (Ogg and Patterson, 2007).
Several GAUU rich aptamer sequences aligned with sequences present on the M and L segments of the RVFV genome and the S antigenome (Table 3). Mutation of the GAUUGAUGAUU motif in the MBE15.8 aptamer showed that preservation of this sequence is a requirement for N binding (Fig. 3A). Although this sequence has not yet been shown to be important for N-RNA recognition in other bunyaviruses, the bacterial chaperone protein Hfq preferentially binds AU rich RNA (de Haseth and Uhlenbeck, 1980; Senear and Steitz, 1976). It is possible that like SNV, RVFV N has chaperone activity. If so, binding and unwinding GAUU rich stem loop structures in viral mRNA could increase the rate of production of viral proteins. Indeed, chaperone proteins have been shown to be essential for some RNA viruses, such as phage Qβ (Franze de Fernandez et al., 1968; Franze de Fernandez et al., 1972; Miranda et al., 1997).
Interestingly, the novel N-RNA binding motifs that we identified in aptamer sequences map to coding regions of the different viral RNA segments (Tables 2 and 3). Further analysis of these data could potentially shed light on N-RNA interactions that are important for N to function during other phases of the viral replication cycle. For example, interaction with host translational machinery, specifically ribosomal protein S19, suggests a role for N in translation of viral mRNA (Cheng et al., 2011; Haque and Mir, 2010). Also, accumulation of N is required to trigger the switch from transcription to full-length cRNA synthesis (Schmaljohn, 1996). It is possible that during these processes N interacts with non-terminal viral RNA sequences. Future work will involve determining if the N binding motifs identified in this study are important for regulation of viral replication.
Information obtained from sequence analysis of the RNA aptamers was also used to make a biosensor that reports on N-RNA binding. We chose the 15.12 TRNK COMP aptamer as the RNA sensor for two reasons. First, it was one of the RNA constructs that retained a high affinity for the N protein upon truncation (Fig 4E). Second, analysis of sequence similarities revealed that its 3′ end contained an 11-nucleotide motif present at the 5′ terminus of the S segment of RVFV RNA (Table 3). In fact, the first 6 nucleotides of this motif are highly conserved, as they are present at the 5′ end of all segments of the viral RNA genome and antigenome. Significant changes in fluorescence polarization (FP) of a labeled 15.12 TRNK COMP RNA were detected as a result of N binding and this change in FP could be inhibited by addition of an unlabeled RNA that also binds N with high affinity. Aptamer RNAs could thus be used as a sensitive fluorescence-based sensor of RVFV N binding with potential applications for drug screening and imaging methodologies.
Several lines of evidence suggest that targeting nucleocapsid protein is a viable therapeutic strategy. Experiments conducted in La Crosse virus (genus Orthobunyavirus) show that interferon-induced cellular MxA protein inhibits viral genome production by binding and depleting the infected cell of N. This binding interaction causes N to be sequestered in cytoplasmic inclusion bodies and inaccessible during viral replication (Kochs et al., 2002). Furthermore, RNA aptamers against HIV-1 Gag polyprotein have been shown to cause inhibition of viral replication in cell culture. Some of these aptamers function by targeting the nucleocapsid (NC) component of the polyprotein and competing with viral genomic RNA for binding (Ramalingam et al., 2011). Finally, the characterization of several novel antiviral agents that target HIV-1 and respiratory syncytial virus nucleocapsid proteins (Chapman et al., 2007; Rice et al., 1993) underscore the potential utility of understanding and exploiting N-RNA interactions for antiviral strategies.
Highlights-Ellenbecker et al.
-
>
In this study, we use SELEX to derive high affinity RNA ligands to Rift Valley fever virus nucleocapsid protein.
-
>
The sequences of the RNA aptamers inform about the RNA sequence and structural preferences of nucleocapsid protein.
-
>
We constructed a fluorescent biosensor that reports on nucleocapsid-RNA interactions.
-
>
This biosensor can be used as a molecular target assay for potential new antiviral drug screening.
Acknowledgments
We gratefully acknowledge Richard Elliott, Olve Peersen, and Su Chiang for helpful discussions and Brian Geiss, Doug Raiford, Sandy Ross, Labe Black, and Kie-Hoon Jung for help with several new protocols and fruitful discussions.
Funding
This work was funded by the NIH/RMRCE grant AI065357 (subaward G-7826 to JSL) and The University of Montana Small Grants program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Balkhy HH, Memish ZA. Rift Valley fever: an uninvited zoonosis in the Arabian peninsula. International Journal of Antimicrobial Agents. 2003;21:153–157. doi: 10.1016/s0924-8579(02)00295-9. [DOI] [PubMed] [Google Scholar]
- Chapman J, Abbott E, Alber DG, Baxter RC, Bithell SK, Henderson EA, Carter MC, Chambers P, Chubb A, Cockerill GS, Collins PL, Dowdell VC, Keegan SJ, Kelsey RD, Lockyer MJ, Luongo C, Najarro P, Pickles RJ, Simmonds M, Taylor D, Tyms S, Wilson LJ, Powell KL. RSV604, a novel inhibitor of respiratory syncytial virus replication. Antimicrobial Agents and Chemotherapy. 2007;51:3346–3353. doi: 10.1128/AAC.00211-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E, Haque A, Rimmer MA, Hussein IT, Sheema S, Little A, Mir MA. Characterization of the Interaction between hantavirus nucleocapsid protein (N) and ribosomal protein S19 (RPS19) J Biol Chem. 2011;286:11814–11824. doi: 10.1074/jbc.M110.210179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haseth PL, Uhlenbeck OC. Interaction of Escherichia coli host factor protein with Q beta ribonucleic acid. Biochemistry. 1980;19:6146–6151. doi: 10.1021/bi00567a030. [DOI] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Pruification and properties of host factor I, a ribonucleic acid-binding protein. The Journal of Biological Chemistry. 1972;247:824–831. [PubMed] [Google Scholar]
- Giorgi C, Accardi L, Nicoletti L, Gro MC, Takehara K, Hilditch C, Morikawa S, Bishop DH. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian Sandfly fever, and Uukuniemi viruses. Virology. 1991;180:738–753. doi: 10.1016/0042-6822(91)90087-r. [DOI] [PubMed] [Google Scholar]
- Gowen BB, Smee DF, Wong MH, Hall JO, Jung KH, Bailey KW, Stevens JR, Furuta Y, Morrey JD. Treatment of late stage disease in a model of arenaviral hemorrhagic fever: T-705 efficacy and reduced toxicity suggests an alternative to ribavirin. PloS One. 2008;3:e3725. doi: 10.1371/journal.pone.0003725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowen BB, Wong MH, Jung KH, Sanders AB, Mendenhall M, Bailey KW, Furuta Y, Sidwell RW. In vitro and in vivo activities of T-705 against arenavirus and bunyavirus infections. Antimicrobial Agents and Chemotherapy. 2007;51:3168–3176. doi: 10.1128/AAC.00356-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque A, Mir MA. Interaction of hantavirus nucleocapsid protein with ribosomal protein S19. Journal of Virology. 2010;84:12450–12453. doi: 10.1128/JVI.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilgore PE, Ksiazek TG, Rollin PE, Mills JN, Villagra MR, Montenegro MJ, Costales MA, Paredes LC, Peters CJ. Treatment of Bolivian hemorrhagic fever with intravenous ribavirin. Clinical Infectious Diseases. 1997;24:718–722. doi: 10.1093/clind/24.4.718. [DOI] [PubMed] [Google Scholar]
- Kochs G, Janzen C, Hohenberg H, Haller O. Antivirally active MxA protein sequesters La Crosse virus nucleocapsid protein into perinuclear complexes. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3153–3158. doi: 10.1073/pnas.052430399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM, Elliott LH, Belmont-Williams R. Lassa fever. Effective therapy with ribavirin. The New England Journal of Medicine. 1986;314:20–26. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- Mir MA, Brown B, Hjelle B, Duran WA, Panganiban AT. Hantavirus N protein exhibits genus-specific recognition of the viral RNA panhandle. Journal of Virology. 2006;80:11283–11292. doi: 10.1128/JVI.00820-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT. Trimeric hantavirus nucleocapsid protein binds specifically to the viral RNA panhandle. Journal of Virology. 2004;78:8281–8288. doi: 10.1128/JVI.78.15.8281-8288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT. The hantavirus nucleocapsid protein recognizes specific features of the viral RNA panhandle and is altered in conformation upon RNA binding. Journal of Virology. 2005;79:1824–1835. doi: 10.1128/JVI.79.3.1824-1835.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT. The bunyavirus nucleocapsid protein is an RNA chaperone: possible roles in viral RNA panhandle formation and genome replication. RNA. 2006a;12:272–282. doi: 10.1261/rna.2101906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT. Characterization of the RNA chaperone activity of hantavirus nucleocapsid protein. Journal of Virology. 2006b;80:6276–6285. doi: 10.1128/JVI.00147-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir MA, Panganiban AT. The triplet repeats of the Sin Nombre hantavirus 5′ untranslated region are sufficient in cis for nucleocapsid-mediated translation initiation. Journal of Virology. 2010;84:8937–8944. doi: 10.1128/JVI.02720-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda G, Schuppli D, Barrera I, Hausherr C, Sogo JM, Weber H. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: role of RNA interactions mediated by ribosomal proteins S1 and host factor. Journal of Molecular Biology. 1997;267:1089–1103. doi: 10.1006/jmbi.1997.0939. [DOI] [PubMed] [Google Scholar]
- Monath TP. Treatment of yellow fever. Antiviral Research. 2008;78:116–124. doi: 10.1016/j.antiviral.2007.10.009. [DOI] [PubMed] [Google Scholar]
- Ogg MM, Patterson JL. RNA binding domain of Jamestown Canyon virus S segment RNAs. Journal of Virology. 2007;81:13754–13760. doi: 10.1128/JVI.01492-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne JC, Elliott RM. RNA binding properties of bunyamwera virus nucleocapsid protein and selective binding to an element in the 5′ terminus of the negative-sense S segment. Journal of Virology. 2000;74:9946–9952. doi: 10.1128/jvi.74.21.9946-9952.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju R, Kolakofsky D. Unusual transcripts in La Crosse virus-infected cells and the site for nucleocapsid assembly. Journal of Virology. 1987;61:667–672. doi: 10.1128/jvi.61.3.667-672.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam D, Duclair S, Datta SA, Ellington A, Rein A, Prasad VR. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. Journal of Virology. 2011;85:305–314. doi: 10.1128/JVI.02626-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice WG, Schaeffer CA, Harten B, Villinger F, South TL, Summers MF, Henderson LE, Bess JW, Jr., Arthur LO, McDougal JS, et al. Inhibition of HIV-1 infectivity by zinc-ejecting aromatic C-nitroso compounds. Nature. 1993;361:473–475. doi: 10.1038/361473a0. [DOI] [PubMed] [Google Scholar]
- Ruigrok RW, Crepin T, Kolakofsky D. Nucleoproteins and nucleocapsids of negative-strand RNA viruses. Curr Opin Microbiol. 2011;14:504–510. doi: 10.1016/j.mib.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Schmaljohn CS. In: Virology, Raven. Fields BN, editor. New York: 1996. pp. 1447–1471. [Google Scholar]
- Senear AW, Steitz JA. Site-specific interaction of Qbeta host factor and ribosomal protein S1 with Qbeta and R17 bacteriophage RNAs. The Journal of Biological Chemistry. 1976;251:1902–1912. [PubMed] [Google Scholar]
- Severson WE, Xu X, Jonsson CB. cis-Acting signals in encapsidation of Hantaan virus S-segment viral genomic RNA by its N protein. Journal of Virology. 2001;75:2646–2652. doi: 10.1128/JVI.75.6.2646-2652.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]