Abstract
Tumor location can profoundly affect morbidity and patient prognosis even for the same tumor type. Very little is known about whether tumor location is determined stochastically or whether genetic risk factors can affect where tumors arise within an organ system. We have taken advantage of the Nf1−/+;Trp53−/+cis mouse model of astrocytoma/glioblastoma to map genetic loci affecting whether astrocytomas are found in the spinal cord. We identify a locus on distal Chr 5, termed Scram1 for spinal cord resistance to astrocytoma modifier 1 with a LOD score of 5.0 and a genome-wide significance of P<0.004. Mice heterozygous for C57BL/6JX129S4/SvJae at this locus show less astrocytoma in the spinal cord compared to 129S4/SvJae homozygous mice, although we have shown previously that 129S4/SvJae mice are more resistant to astrocytoma than C57BL/6J. Furthermore, the astrocytomas that are found in the spinal cord of Scram1 heterozygous mice arise in older mice. Because spinal cord astrocytomas are very rare and difficult to treat, better understanding of the genetic factors that govern astrocytoma in the spine may suggest new targets of therapy or prevention.
Keywords: astrocytoma, glioma, spinal cord, Nf1, p53
Introduction
Malignant intramedullary spinal cord astrocytomas are very rare but devastating tumors, with an estimated age-adjusted incidence of 0.05 per 100,000 (Hsu et al. 2011) and a median survival time of less than 20 months (Milano et al. 2010). Because these tumors are so rare, very little is known about their molecular biology and genetics. It has been difficult to establish even basic prognostic factors and optimal treatment because samples must be collected over the course of decades, through periods of constantly evolving treatment options and from highly heterogeneous populations (Benes et al. 2009; Harrop et al. 2009). Because of the spinal cord anatomy, complete tumor resection must be balanced against the morbidity caused by loss of spinal cord function. Furthermore, radiation and chemotherapy have not shown clear benefit in these patients.
Mouse models of spinal cord astrocytoma can help to characterize the biology and genetics of these tumors, by allowing larger sample numbers to be collected over shorter time frames. In addition, mouse models can be used to reduce the heterogeneity of the population by controlling environment and diet, so that hypotheses can be tested on the risk factors for spinal cord astrocytoma and whether they differ from brain astrocytomas. A better understanding of spinal cord astrocytoma biology will identify directed hypotheses to be tested in the small number of human samples and may lead to new insights for targeted therapy of these tumors.
Very few animal models of spinal cord astrocytoma exist. A xenograft model of spinal gliosarcoma has been developed in the rat (Caplan et al. 2006) and a genetically engineered model of oligoastrocytoma has been developed in mouse (Hitoshi et al. 2008). We use a mouse model of astrocytoma/glioblastoma that develops tumors spontaneously in both the spine and brain. The model carries mutations in the Nf1 gene and Trp53 gene linked in cis on Chr 11 and initiates tumors through spontaneous loss of the wild-type copies of Nf1 and Trp53 on the opposite chromosome (Reilly et al. 2000). These Nf1−/+;Trp53−/+cis (NPcis) mice develop astrocytoma grades II–IV throughout the central nervous system depending on the strain background. NPcis mice on the C57BL/6J (B6) background are susceptible to astrocytoma, whereas NPcis mice on the 129S4/SvJae (129) background are resistant (Reilly et al. 2004). This model is well suited to screening for modifiers of astrocytoma because it has been inbred onto both the B6 and 129 strains and because the Nf1 and Trp53 mutations are tightly linked, allowing the model to be bred as a heterozygote to different strain backgrounds.
Anecdotal evidence suggests that the pathology of spinal cord astrocytoma is similar to brain astrocytoma in patients, although the spinal cord astrocytomas have not been rigorously studied using molecular techniques. Our observations of NPcis mouse astrocytomas suggest that tumors in the brain and spine are similar, as well. We therefore were interested in using this model to test the genetics of location-specific tumor growth. We used backcross mapping and linkage analysis to identify a modifier of astrocytoma specifically affecting tumors in the spine.
Materials and Methods
Mouse breeding
Mice for the mapping experiment were generated by crossing inbred 129-NPcis females to wild-type B6 males in the first generation to make F1(129XB6)-NPcis female progeny. Female F1s were backcrossed to wild-type 129 males in the second generation and NPcis male and female progeny were used for mapping modifier loci. Of 114 backcross progeny that were bred, 88 (37 females and 51 males) were successfully genotyped and phenotyped for binary linkage analysis. The C57BL/6J strain (Jackson Labs cat #000664) and the 129S4/SvJae strain maintained in our colony were used as the parental strains in the backcross. Mice were maintained at NCI-Frederick according to the guidelines and regulations of the Institutional Animal Care and Use Committee.
Phenotyping of astrocytoma
Mice were aged and euthanized according to predetermined criteria as described previously (Walrath et al. 2009), with the exception that only brains, spines, and visible masses were collected for histology. Half of the skull was fixed in Bouins and the contralateral half of the brain was fixed in neutral-buffered formalin. The Bouins-fixed brain was trimmed along the midline sagittal plane and cut parasagittally at the level of the eye. Three sagittal brain sections (1 midline Bouins, 1 contralateral formalin-fixed, and 1 parasagittal Bouins) were used for analysis of astrocytoma. The spine was cut at the cervical level below the brain stem. The entire spine was divided into ten pieces. Four cross-sections were taken at the cervical end, caudal end, and at the thoracic and lumbar levels to divide the remaining spine in thirds. The remaining thirds were cut longitudinally along the midline and then again parasagittally at the edge of the spinal cord through the nerve roots, giving rise to six longitudinal sections. All ten spinal cord sections and three brain sections were scored for the presence of astrocytoma grades II–IV by K.M.R. and independently scored by the Pathology/Histology Laboratory at Science Applications International Corporation, Inc (Frederick, MD). The presence or absence of astrocytoma (of any grade) in the ten spinal cord sections was taken as a binary trait (1 = present; 0 = absent) and used for linkage analysis.
Genome-wide SNP genotyping
Tail DNA was prepared using the Promega Wizard SV Genomic DNA Purification System and concentrated by ethanol precipitation. Genomic tail DNA was resuspended in a range of 5 ng/µL to 155 ng/µL, with 75–150 ng/µL considered optimal. Thirty-five µL of DNA for each sample was sent to the Center for Inherited Disease Research (CIDR) and genotyped using the Illumina 1440K SNP Panel. This panel contains 880 SNPs polymorphic for B6 and 129.
Statistical analysis of linkage
Genotyping and phenotyping results were collated for each sample and binary trait interval mapping was used to identify the locations of quantitative trait loci (QTLs). Mapping was performed using the genetic mapping software R/qtl (Broman et al. 2003) and j/qtl (Smith et al. 2009), calculating LOD scores at genotyped markers and a grid of 1 cM pseudomarkers along each chromosome. Statistical significance was determined empirically by permutation testing with 1000 replicates, with a threshold of 0.05 considered significant. The locus was defined by a 1.5 LOD drop from the peak LOD score at position 77.3 cM. The nearest genotyped marker, rs4225536, at 75.7 cM was use to analyze samples based on their genotype at Scram1. Statistical analysis of contingency tables and survival was performed using Microsoft Excel 2008 for Mac (v12.2.9) and GraphPad Prism (v4.0a).
Bioinformatic analysis of Scram1
Polymorphisms between B6 and 129 in the Scram1 region were searched using the SNP Wizard of the Mouse Phenome Database (http://phenome.jax.org/SNP/), accessing data from mouse genome build 37.1/mm9, dbSNP128, and Ensembl 48. We considered SNPs polymorphic between B6 and either 129S1/SvImJ or 129X1/SvJ. Human synteny was analyzed using NCBI Homology Maps Viewer (http://www.ncbi.nlm.nih.gov/projects/homology/maps/) and NCBI Unigene (http://www.ncbi.nlm.nih.gov/unigene), accessing mouse genome build 37.1 and human genome build 37.2. Gene expression data from B6 and 129/SvImJ was downloaded from http://phenogen.ucdenver.edu/PhenoGen/index.jsp, and one probe from each gene was chosen based on the highest representation of the total exons in the gene. Expression in B6 and 129/SvImJ was compared and genes with ≥ 1.5-fold difference were graphed using GraphPad Prism.
Results
Spinal cord astrocytomas in Nf1−/+;Trp53−/+cis mice
We had noted from previous studies (Reilly et al. 2000; Reilly et al. 2004) that mice carrying the Nf1−/+;Trp53−/+cis (NPcis) mutations on the C57BL/6J (B6) background develop astrocytomas and glioblastomas (GBMs) throughout the central nervous system, including the spinal cord (Fig 1). The spinal cord astrocytomas show the same features as astrocytomas in the brain, including large, hyperchromatic, irregular nuclei; mitoses; hemorrhage; and occasional necrosis indicative of GBM. The spinal cord astrocytomas are diffusely infiltrative, sometimes spreading through the entire length of the spinal cord. We observed cases in which the spinal cord astrocytoma appeared to be the primary location of the tumor. In these cases either no astrocytoma cells were found in brain sections, or the spinal cord tumor appeared clearly focal and higher grade than tumor in the brain, suggesting the tumor infiltrated into the brain from a primary tumor in the spine. To examine the genetic factors controlling the spinal cord location of astrocytoma, we generated backcross progeny between the B6 strain, which is susceptible to astrocytoma, and the 129S4/SvJae (129) strain, which is resistant to astrocytoma (Reilly et al. 2000; Reilly et al. 2004).
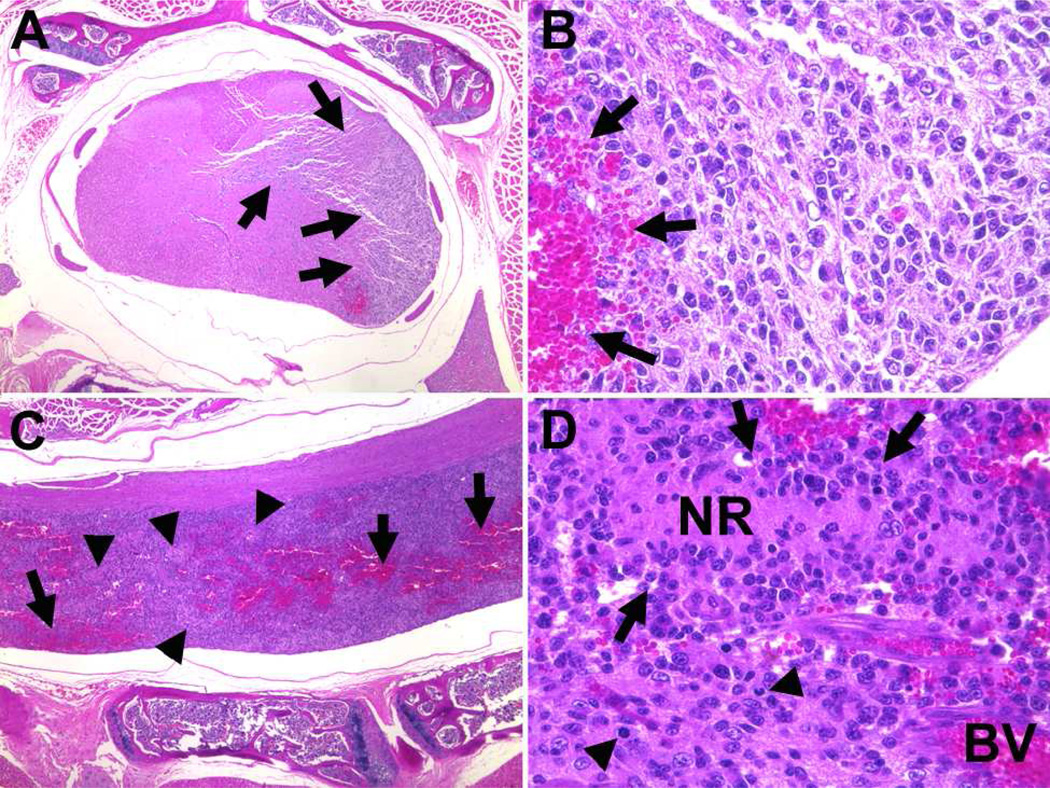
Figure 1.
Spinal cord GBM in a BC(129XB6)X129-NPcis mouse. Panel A shows a cross-section view at low magnification with the GBM (arrows) diffusely infiltrating one side of the spinal cord. The high magnification view (B) shows densely packed irregular oblong nuclei and a region of hemorrhage (arrows). Panel C shows low-magnification, of a longitudinal section of the same tumor, highlighting the widespread extent of the tumor in the spinal cord with hemorrhage (arrows) and regions of necrosis (arrowheads). The high magnification view (D) shows a necrotic region (NR) with weakly pallisading tumor cells (arrows), blood vessels (BV) with evidence of microvascular proliferation, and atypical mitoses (arrowheads).
F1(129XB6)-NPcis females were crossed to wild-type 129 males to generate BC(129XB6)X129-NPcis progeny for mapping. In the cohort of 88 backcross mice 55% developed astrocytoma in the spinal cord. The majority of these also had astrocytoma in the brain, although in a few cases the astrocytoma was limited to the spinal cord. An additional 20% of the backcross mice had astrocytoma only in the brain.
Scram1 is located on distal Chr 5
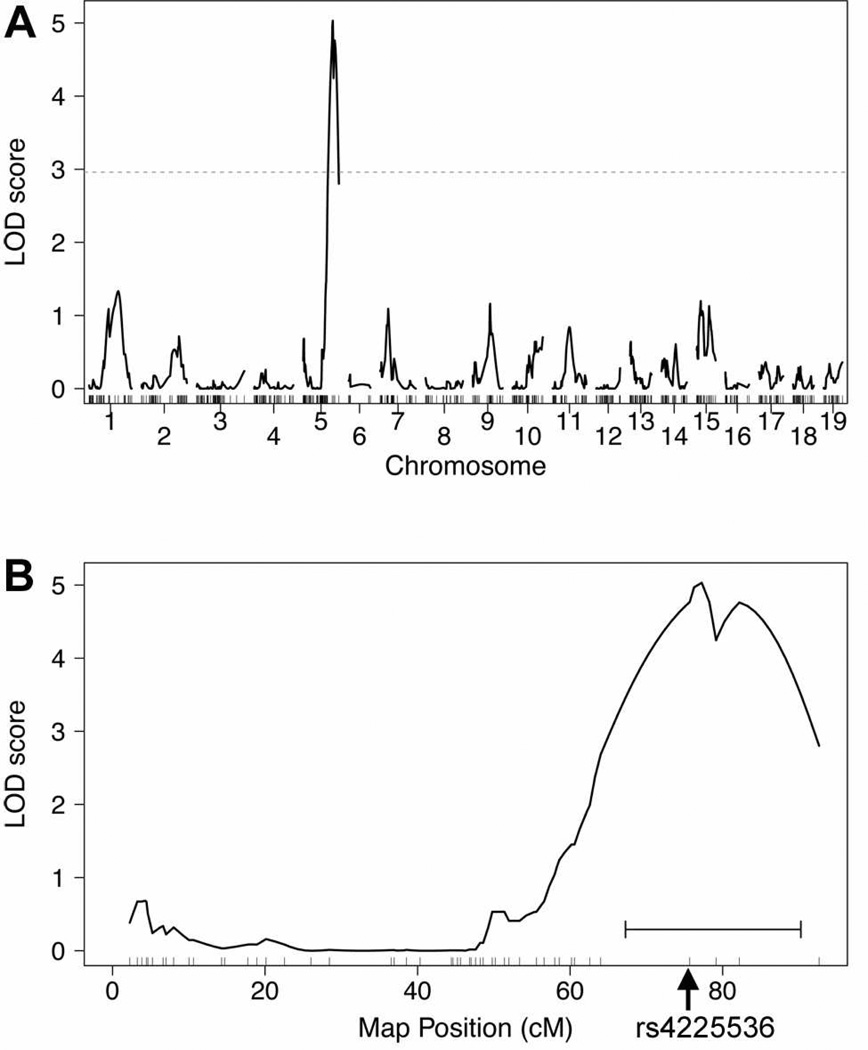
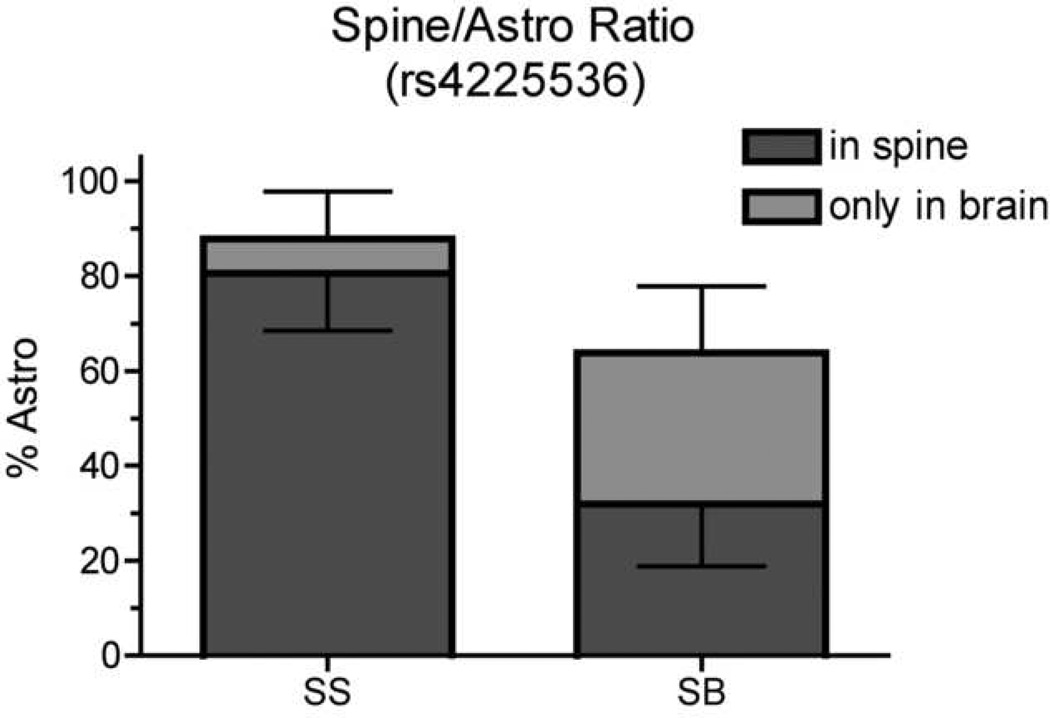
Binary trait interval mapping identified one significant linkage peak on the distal end of Chr 5 (Fig 2) with a LOD score of 5.03 and a genome-wide significance of P<0.004. We have named this locus Scram1 for spinal cord resistance to astrocytoma modifier 1. Although the inbred 129 strain is resistant to astrocytoma, in the BC(129XB6)X129 backcross the Scram1129/129 NPcis mice develop more spinal cord astrocytomas than Scram1129/B6 mice (Fig 3), suggesting the B6 Scram1 allele confers resistance to astrocytoma in the spinal cord. Scram1 also has an effect on total astrocytoma incidence (Fisher exact test P = 0.013), with 88% of Scram1129/129 NPcis mice developing astrocytoma anywhere in the central nervous system compared to 64% of Scram1129/B6 mice. However, the reduction in spinal cord astrocytomas in Scram1129/B6 mice is significantly greater than the drop in overall astrocytoma (Fisher exact test, P = 0.0002), with 92% of astrocytomas in Scram1129/129 mice being found in the spinal cord as opposed to 50% of astrocytomas in Scram1129/B6 mice (Fig 3).
Figure 2.
Binary trait linkage analysis of spinal cord astrocytoma in BC(129XB6)X129)-NPcis mice. Panel A shows the linkage across the genome, with a peak on Chr 5. The dotted line shows the genome-wide significance level of P = 0.05 based on permutation testing. Panel B shows linkage on Chr 5 with the highest peak at 77.26 cM. The horizontal bracket below the peak indicates the 1.5 LOD support interval that defines the Scram1 locus. The rs4225536 marker that was used for subsequent analyses is indicated by the arrow at 75.7 cM.
Figure 3.
The B6 allele at Scram1 affects both the overall levels of astrocytoma and the ratio of astrocytomas found in the spinal cord in backcross mice. The incidence of spinal cord astrocytoma compared to all astrocytoma is shown at marker rs4225536, nearest to the Scram1 peak. Statistically significant differences were found between the incidence of total astrocytoma in the two groups (Fisher exact test P = 0.013), the incidence of spinal cord astrocytoma in the two groups (Fisher exact test P <0.0001), and the ratios of spinal cord astrocytoma to total astrocytoma in the two groups (Fisher exact test P = 0.0002). The upward error bar indicates the 95% CI for all astrocytomas and the downward error bar indicates the 95% CI for spinal cord astrocytomas (dark gray). SS indicates the 129/129 genotype at rs4225536 and SB indicates the 129/B6 genotype.
Scram1 affects timing of spinal cord astrocytomas
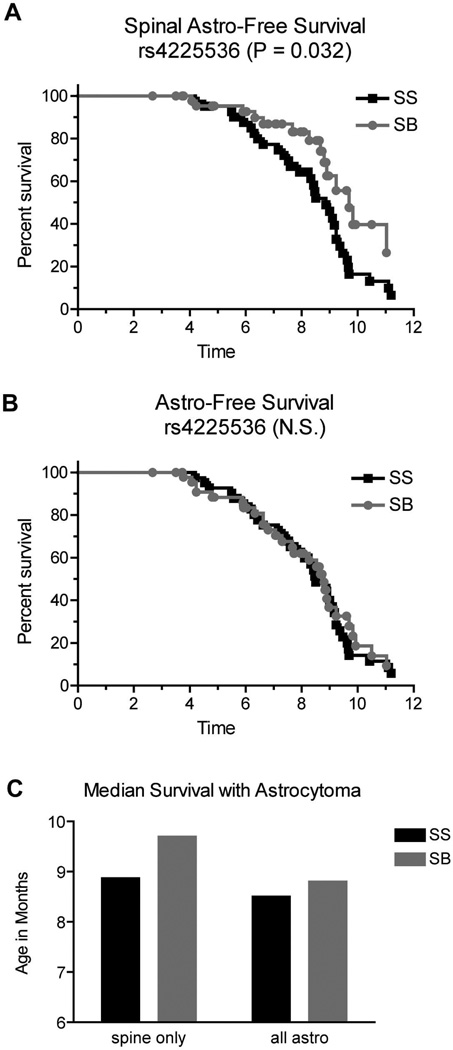
We examined the effect of the Scram1 genotype on the timing of spinal cord astrocytomas and on astrocytomas in general. Mice were divided based on the genotype at marker rs4225536 nearest Scram1 and plotted for spinal cord astrocytoma-free survival and total astrocytoma-free survival. The survival curves (truncated at 12 months) are shown in Fig 4. The time at which mice were euthanized due to any tumor type was taken as the date of death and mice were censored if they did not have astrocytoma in the spinal cord (Fig 4 A), or censored if they did not have astrocytoma in the central nervous system (brain and spine) (Fig 4 B). In the case of spinal cord astrocytoma, Scram1129/B6 mice are euthanized with spinal cord astrocytomas significantly later than Scram1129/129 mice (median age 129/B6 = 9.7 months; median age 129/129 = 8.9 months; Fig 4 C). There was no significant difference in the overall timing of astrocytoma at Scram1 (median age 129/B6 = 8.8 months; median age 129/129 = 8.5 months; Fig 4 C). This data could be consistent with either a delay in astrocytoma initiation in the spine, or alternatively a delay in spread of the astrocytoma from the brain to the spine. The data on incidence and latency by Scram1 genotype is summarized in Table 1.
Figure 4.
Scram1 modifies timing of astrocytoma in the spinal cord, but not astrocytoma overall. Samples were stratified by the rs4225536 marker nearest the Scram1 peak. Spinal cord astrocytoma-free survival is delayed in Scram1129/B6 (SB) mice compare to Scram1129/129 (SS) mice (A). Astrocytomas overall are observed at the same ages in Scram1129/B6 (SB) and Scram1129/129 (SS) mice (B). P values for Kaplan-Meier analysis are given in each panel. N.S. = not significant. Median survival age is graphed for mice specifically with spinal cord astrocytomas and for mice with astrocytomas anywhere in the central nervous system (C) demonstrating that Scram1129/B6 mice have a delay in the presentation of astrocytoma in the spinal cord.
Table 1.
Incidence and Latency of Astrocytomas in the Spine and Central Nervous System
| Spinal Cord Astrocytoma | All Astrocyotma (Spine + Brain) | ||||||
|---|---|---|---|---|---|---|---|
| Genotype** | N | Incidence (% ± CI) |
Median Age (mo) |
HR [95% CI] | Incidence (% ± CI) |
Median Age (mo) |
HR [95% CI] |
| Scram1129/129, all | 41 | 80 ± 12 | 8.9 | 1.9* [1.1–3.3] | 88 ± 10 | 8.5 | 1.1 [0.7–1.7] |
| Scram1B6/129, all | 47 | 32 ± 13 | 9.7 | 64 ± 14 | 8.8 | ||
| Scram1129/129, M | 22 | 86 ± 14 | 9.2 | 1.5 [0.7–3.1] | 91 ± 12 | 9.0 | 0.9 [0.5–1.7] |
| Scram1B6/129, M | 29 | 34 ± 17 | 9.8 | 62 ± 18 | 8.5 | ||
| Scram1129/129, F | 19 | 74 ± 20 | 8.5 | 2.8* [1.1–6.7] | 84 ± 16 | 8.5 | 1.4 [0.7–3.2] |
| Scram1B6/129, F | 18 | 28 ± 21 | 9.2 | 67 ± 22 | 8.8 | ||
P <0.05, Kaplan-Meier analysis
Genotype at rs4225536
The Scram1 locus is syntenic with human Chr 7p, 7q, and 12q
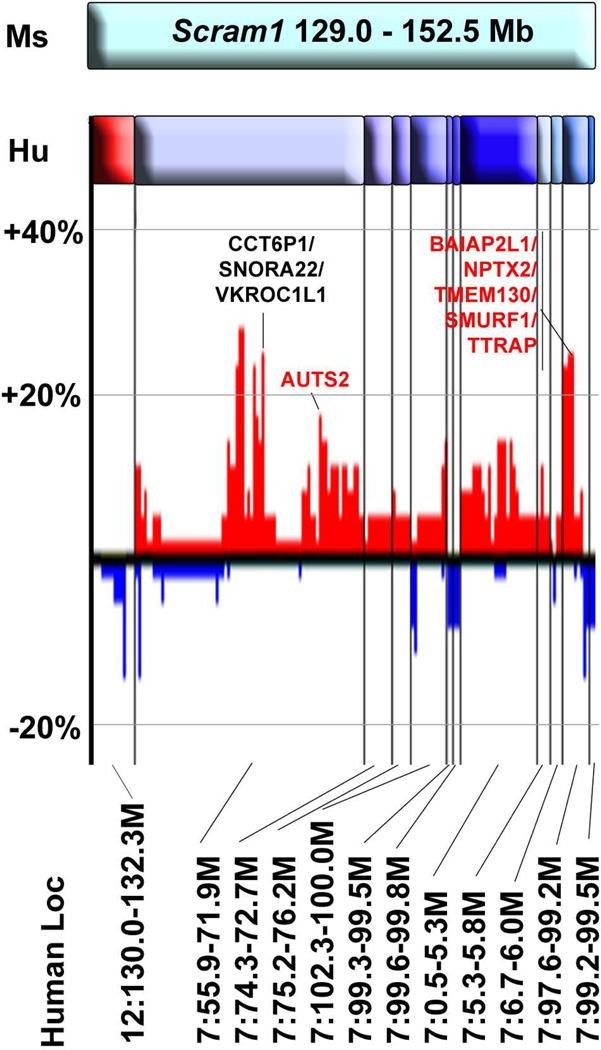
The Scram1 locus, defined by the 1.5 LOD support interval, covers from 67.26 cM to the distal end of mouse Chr 5 and corresponds to 129.0 – 152.5 Mb. The region contains 248 genes with SNPs between the B6 and 129S1/SvImJ and/or 129X1/SvJ, of which 46 genes have a SNP that is predicted to change the coding sequence of the protein (Mouse Phenome Database; http://phenome.jax.org/SNP/). The region is syntenic with human Chr 12q24, 7p11, 7q22-21, and 7p22 (Fig 5). Because spinal cord astrocytomas are so rare, there is very little available data on whether these regions are altered in spinal cord tumors. A small study of two brainstem glioblastomas and one spinal cord glioblastoma found amplification of Chr 7:69824947-qter that overlaps the region of the Scram1 locus (Sharma et al. 2010). We compared the Scram1 locus to amplifications and deletions found in human brain astrocytomas to determine if the region has been implicated in astrocytoma more generally. We used the Cancer Genome Workbench Heatmap Viewer (https://cgwb.nci.nih.gov/cgi-bin/heatmap) to examine Affymetrix SNP data from the REMBRANDT Astrocytoma Project for tumor DNA copy number changes. Human Chr 7 loci that are syntenic with Scram1 are amplified in up to 30% of astrocytoma cases, whereas Chr 12 loci are deleted in up to 16% of astrocytoma cases (Fig 5). Many of the genes within the amplified and deleted probes are polymorphic between B6 and 129 strains, suggesting they are candidates for the Scram1 modifier gene.
Figure 5.
Synteny of human chromosomes to Scram1. The top cyan bar represents the mouse Scram1 locus. Human chromosome segments are shown below, with the Chr 12 segment in red and Chr 7 segments in shades of blue. The Mb coordinates of each segment are shown at the bottom of the diagram. The bar graph shows amplification (red) and deletion (blue) of these regions in human brain astrocytoma samples taken from the REMBRANDT Astrocytoma Project and visualized in the Cancer Workbench Heatmap Viewer (https://cgwb.nci.nih.gov/cgi-bin/heatmap). Vertical black lines show the division between the different chromosome segments aligned to mouse Scram1 and horizontal gray lines indicate amplification or deletion in 20% and 40% of samples. Genes within the regions of most frequent amplification are indicated, with the genes that are polymorphic between B6 and 129 in red type.
Genes within the Scram1 locus are differentially expressed in B6 and 129 mouse brains
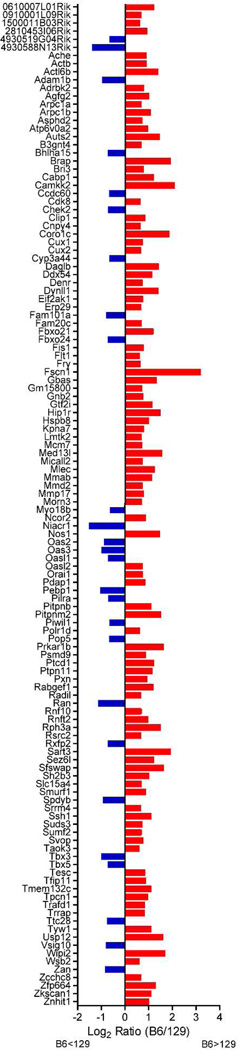
To gain further insight into candidate genes for the Scram1 locus, we examined publically available gene expression data. Expression of genes in spinal cords on different strain backgrounds is not currently available; however, a strain survey of 10–12 week old male mouse brains is publically available (http://phenogen.ucdenver.edu/PhenoGen/index.jsp). In the region of Scram1, 313 genes with probe data were examined and of those 124 genes showed a ≥1.5-fold difference in expression between B6 and the 129/SvImJ strain (Fig 6). The majority of the genes are expressed more highly in B6 than 129/SvImJ, with only 26 being more highly expressed in 129/SvImJ compared to B6. Further characterization of these differences in expression levels in the spinal cord will help prioritize the candidate genes. This data demonstrates that pre-existing gene expression differences in the nervous system between B6 and 129 may set up the conditions for tumorigenesis in the spinal cord vs the brain.
Figure 6.
Ratio of B6 expression levels to 129/SvImJ expression levels of Scram1 genes in normal male mouse brains. Publically available gene expression data (http://phenogen.ucdenver.edu/PhenoGen/index.jsp) was analyzed for genes in the Scram1 region that had ≥ 1.5-fold difference in expression level between B6 and 129/SvImJ. Only genes that met this threshold are shown. Red bars indicate genes that are more highly expressed in B6 compared to 129/SvImJ and blue bars indicate genes that are higher in 129/SvImJ. Gene symbols are listed in alphabetical order along the Y-axis.
Discussion
Our results demonstrate that region-specific tumorigenesis is under genetic control. The Scram1 locus on mouse Chr 5 affects the incidence and latency of astrocytoma in the spinal cord, but does not affect the latency of astrocytomas overall. Further studies are necessary to determine whether this is a direct effect on tumor initiation and progression within the spinal cord, or whether tumor-initiating cells travel down the spinal cord from the brain and Scram1 affects the ability of cells to migrate away from the brain. This could explain the longer tumor latency if spinal cord tumor initiating cells take longer to reach the spinal cord in Scram1B6/129 mice. Alternatively, a delay in initiation of resident tumor progenitor cells within the spinal cord could account for the longer latency.
Although we have found the 129 parental strain to be resistant to astrocytoma overall in comparison to the B6 strain, the locus identified here has the opposite effect on spinal cord astrocytoma, with the B6 allele providing resistance. This finding highlights the importance of mapping quantitative traits that are not predicted by the parental strains, because complex phenotypes can be governed by many, often conflicting, genetic contributions that can be uncovered in segregating crosses. This has been observed in other phenotypes as well, for example in a study of anxiety (Bailey et al. 2008) where strong effects were identified in segregating crosses of two strains with very little phenotypic difference, and in plasmocytomagenesis (Mock et al. 1993; Zhang et al. 2009) where a tumor resistance allele was found in a highly susceptible strain. Further studies are necessary to determine whether overall resistance in the 129 strain is due to the combinatorial effects of many different loci that did not reach the threshold of statistical significance in this study. Nonetheless, the identification of Scram1 will provide insight into the mechanisms of astrocytoma formation in the spinal cord as we begin to understand the mechanisms underlying its effect.
While Scram1 contains many genes that could be involved in spinal cord astrocytoma, several are notable. Two of the genes cause familial cancer syndromes when mutated. Brca2 is associated with familial breast cancer (Stratton et al. 1994) and Pms2 is associated with a form of Turcot Syndrome that includes high-grade glioma (Lucci-Cordisco et al. 2003). Many genes in the Scram1 locus are important for invasion and migration of cancer cells, including Rac1 (Ridley 2006), Mmp17 (Huang et al. 2009), Pxn (Han et al. 2001), Trip6 (Chastre et al. 2009), and Serpine1 (Binder and Mihaly 2008). Finally, many genes in the Scram1 locus are important in signal transduction pathways, including Ephb4 (Noren and Pasquale 2007), Prkar1b (PKA) (Chen et al. 1998), Lmtk2 (Kawa et al. 2004), Epo (Cao et al. 2010; Mohyeldin et al. 2007; Peres et al. 2011; Wang et al. 2010), and Pdgfa (Calzolari and Malatesta 2010). Pdgfa encoding the Pdgfaa growth factor is of particular interest because its receptor, PDGFRa, has been found to be expressed in intramedullary astrocytoma and GBM (Ellis et al. 2011), and a closely related gene, Pdgfb, has been used to drive spinal gliomagenesis in mice (Hitoshi et al. 2008). Although Pdgfa is polymorphic between B6 and 129, it carries only one polymorphic SNP in intron 2 of unknown significance and is not differentially expressed in the brains of B6 and 129 mice. It is therefore not clear whether Pdgfa is the acting modifier gene in the Scram1 locus that differentially affects B6 and 129 mice. Future experiments will seek to narrow the Scram1 region to determine which of these many promising candidate genes modifies spinal cord tumorigenesis.
Astrocytomas and glioblastomas are complex tumors with many molecular and clinical subtypes. It is becoming increasingly clear that the genetics of astrocytoma/glioblastoma susceptibility is similarly complex. The data we present here in combination with our previous studies (Reilly et al. 2000; Reilly et al. 2004) show that combinatorial genetic effects can determine which individuals develop astrocytoma and where the tumors develop in the nervous system.
Acknowledgements
We thank M. Anvers, R. Tuskan, K. Smith, K. Fox, and E. Truffer for technical assistance and D. Louis and G. Jallo for helpful discussions. J.C.A.V.S. is a predoctoral student in the Graduate Partnership Program of the N.I.H. and the Institute for Biomedical Sciences at George Washington University. This work is from a dissertation to be prepared in partial fulfillment of the requirements for the PhD degree. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsements by the US Government.
Funding: Intramural Research Program of the National Institute of Health, National Cancer Institute [ZIA BC 010539 to K.M.R.]; Federal funds from the National Cancer Institute to SAIC Frederick [contract N01-CO-12400], Federal contract from the National Institutes of Health to The Johns Hopkins University [contract HHSN268200782096C]; Extramural funding from the National Institute of Health [R01 GM074244 to K.W.B.]
References
- Bailey JS, Grabowski-Boase L, Steffy BM, Wiltshire T, Churchill GA, Tarantino LM. Identification of quantitative trait loci for locomotor activation and anxiety using closely related inbred strains. Genes Brain Behav. 2008;7:761–769. doi: 10.1111/j.1601-183x.2008.00415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes V, 3rd, Barsa P, Benes V, Jr, Suchomel P. Prognostic factors in intramedullary astrocytomas: a literature review. Eur Spine J. 2009;18:1397–1422. doi: 10.1007/s00586-009-1076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binder BR, Mihaly J. The plasminogen activator inhibitor "paradox" in cancer. Immunol Lett. 2008;118:116–124. doi: 10.1016/j.imlet.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Broman KW, Wu H, Sen S, Churchill GA. R/qtl: QTL mapping in experimental crosses. Bioinformatics. 2003;19:889–890. doi: 10.1093/bioinformatics/btg112. [DOI] [PubMed] [Google Scholar]
- Calzolari F, Malatesta P. Recent insights into PDGF-induced gliomagenesis. Brain Pathol. 2010;20:527–538. doi: 10.1111/j.1750-3639.2009.00335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Lathia JD, Eyler CE, Wu Q, Li Z, Wang H, McLendon RE, Hjelmeland AB, Rich JN. Erythropoietin Receptor Signaling Through STAT3 Is Required For Glioma Stem Cell Maintenance. Genes Cancer. 2010;1:50–61. doi: 10.1177/1947601909356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caplan J, Pradilla G, Hdeib A, Tyler BM, Legnani FG, Bagley CA, Brem H, Jallo G. A novel model of intramedullary spinal cord tumors in rats: functional progression and histopathological characterization. Neurosurgery. 2006;59:193–200. doi: 10.1227/01.NEU.0000219276.44563.DA. discussion 193–200. [DOI] [PubMed] [Google Scholar]
- Chastre E, Abdessamad M, Kruglov A, Bruyneel E, Bracke M, Di Gioia Y, Beckerle MC, van Roy F, Kotelevets L. TRIP6, a novel molecular partner of the MAGI-1 scaffolding molecule, promotes invasiveness. Faseb J. 2009;23:916–928. doi: 10.1096/fj.08-106344. [DOI] [PubMed] [Google Scholar]
- Chen TC, Hinton DR, Zidovetzki R, Hofman FM. Up-regulation of the cAMP/PKA pathway inhibits proliferation, induces differentiation, and leads to apoptosis in malignant gliomas. Lab Invest. 1998;78:165–174. [PubMed] [Google Scholar]
- Ellis JA, Canoll P, McCormick PC, 2nd, Feldstein NA, Anderson RC, Angevine PD, Kaiser MG, McCormick PC, Bruce JN, Ogden AT. Platelet-derived growth factor receptor (PDGFR) expression in primary spinal cord gliomas. J Neurooncol. 2011 doi: 10.1007/s11060-011-0666-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Stewart JE, Jr, Bellis SL, Benveniste EN, Ding Q, Tachibana K, Grammer JR, Gladson CL. TGF-beta1 up-regulates paxillin protein expression in malignant astrocytoma cells: requirement for a fibronectin substrate. Oncogene. 2001;20:7976–7986. doi: 10.1038/sj.onc.1204996. [DOI] [PubMed] [Google Scholar]
- Harrop JS, Ganju A, Groff M, Bilsky M. Primary intramedullary tumors of the spinal cord. Spine (Phila Pa 1976) 2009;34:S69–S77. doi: 10.1097/BRS.0b013e3181b95c6f. [DOI] [PubMed] [Google Scholar]
- Hitoshi Y, Harris BT, Liu H, Popko B, Israel MA. Spinal glioma: platelet-derived growth factor B-mediated oncogenesis in the spinal cord. Cancer Res. 2008;68:8507–8515. doi: 10.1158/0008-5472.CAN-08-1063. [DOI] [PubMed] [Google Scholar]
- Hsu S, Quattrone M, Ostrom Q, Ryken TC, Sloan AE, Barnholtz-Sloan JS. Incidence patterns for primary malignant spinal cord gliomas: a Surveillance, Epidemiology, and End Results study. J Neurosurg Spine. 2011;14:742–74. doi: 10.3171/2011.1.SPINE10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CH, Yang WH, Chang SY, Tai SK, Tzeng CH, Kao JY, Wu KJ, Yang MH. Regulation of membrane-type 4 matrix metalloproteinase by SLUG contributes to hypoxia-mediated metastasis. Neoplasia. 2009;11:1371–1382. doi: 10.1593/neo.91326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawa S, Fujimoto J, Tezuka T, Nakazawa T, Yamamoto T. Involvement of BREK, a serine/threonine kinase enriched in brain, in NGF signalling. Genes Cells. 2004;9:219–232. doi: 10.1111/j.1356-9597.2004.00714.x. [DOI] [PubMed] [Google Scholar]
- Lucci-Cordisco E, Zito I, Gensini F, Genuardi M. Hereditary nonpolyposis colorectal cancer and related conditions. Am J Med Genet A. 2003;122A:325–334. doi: 10.1002/ajmg.a.20475. [DOI] [PubMed] [Google Scholar]
- Milano MT, Johnson MD, Sul J, Mohile NA, Korones DN, Okunieff P, Walter KA. Primary spinal cord glioma: a Surveillance, Epidemiology, and End Results database study. J Neurooncol. 2010;98:83–92. doi: 10.1007/s11060-009-0054-7. [DOI] [PubMed] [Google Scholar]
- Mock BA, Krall MM, Dosik JK. Genetic mapping of tumor susceptibility genes involved in mouse plasmacytomagenesis. Proc Natl Acad Sci U S A. 1993;90:9499–9503. doi: 10.1073/pnas.90.20.9499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohyeldin A, Dalgard CL, Lu H, McFate T, Tait AS, Patel VC, Wong K, Rushing E, Roy S, Acs G, Verma A. Survival and invasiveness of astrocytomas promoted by erythropoietin. J Neurosurg. 2007;106:338–350. doi: 10.3171/jns.2007.106.2.338. [DOI] [PubMed] [Google Scholar]
- Noren NK, Pasquale EB. Paradoxes of the EphB4 receptor in cancer. Cancer Res. 2007;67:3994–3997. doi: 10.1158/0008-5472.CAN-07-0525. [DOI] [PubMed] [Google Scholar]
- Peres EA, Valable S, Guillamo JS, Marteau L, Bernaudin JF, Roussel S, Lechapt-Zalcman E, Bernaudin M, Petit E. Targeting the erythropoietin receptor on glioma cells reduces tumour growth. Exp Cell Res. 2011 doi: 10.1016/j.yexcr.2011.06.011. [DOI] [PubMed] [Google Scholar]
- Reilly KM, Loisel DA, Bronson RT, McLaughlin ME, Jacks T. Nf1;Trp53 mutant mice develop glioblastoma with evidence of strain-specific effects. Nat Genet. 2000;26:109–113. doi: 10.1038/79075. [DOI] [PubMed] [Google Scholar]
- Reilly KM, Tuskan RG, Christy E, Loisel DA, Ledger J, Bronson RT, Smith CD, Tsang S, Munroe DJ, Jacks T. Susceptibility to astrocytoma in mice mutant for Nf1 and Trp53 is linked to chromosome 11 and subject to epigenetic effects. Proc Natl Acad Sci U S A. 2004;101:13008–13013. doi: 10.1073/pnas.0401236101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 2006;16:522–529. doi: 10.1016/j.tcb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Sharma S, Free A, Mei Y, Peiper SC, Wang Z, Cowell JK. Distinct molecular signatures in pediatric infratentorial glioblastomas defined by aCGH. Exp Mol Pathol. 2010;89:169–174. doi: 10.1016/j.yexmp.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Smith R, Sheppard K, DiPetrillo K, Churchill G. Quantitative trait locus analysis using J/qtl. Methods Mol Biol. 2009;573:175–188. doi: 10.1007/978-1-60761-247-6_10. [DOI] [PubMed] [Google Scholar]
- Stratton MR, Ford D, Neuhasen S, Seal S, Wooster R, Friedman LS, King MC, Egilsson V, Devilee P, McManus R, et al. Familial male breast cancer is not linked to the BRCA1 locus on chromosome 17q. Nat Genet. 1994;7:103–107. doi: 10.1038/ng0594-103. [DOI] [PubMed] [Google Scholar]
- Walrath JC, Fox K, Truffer E, Gregory Alvord W, Quinones OA, Reilly KM. Chr 19(A/J) modifies tumor resistance in a sex- and parent-of-origin-specific manner. Mamm Genome. 2009;20:214–223. doi: 10.1007/s00335-009-9179-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yao M, Zhou C, Dong D, Jiang Y, Wei G, Cui X. Erythropoietin promotes spinal cord-derived neural progenitor cell proliferation by regulating cell cycle. Neuroscience. 2010;167:750–757. doi: 10.1016/j.neuroscience.2010.02.007. [DOI] [PubMed] [Google Scholar]
- Zhang K, Kagan D, DuBois W, Robinson R, Bliskovsky V, Vass WC, Zhang S, Mock BA. Mndal, a new interferon-inducible family member, is highly polymorphic, suppresses cell growth, and may modify plasmacytoma susceptibility. Blood. 2009;114:2952–2960. doi: 10.1182/blood-2009-01-198812. [DOI] [PMC free article] [PubMed] [Google Scholar]