Abstract
The lipid droplets (LDs) are intracellular organelles mainly dedicated to the storage and provision of fatty acids. To accomplish these functions the LDs interact with other organelles and cytosolic proteins. In order to explore possible correlations between the physiological states of cells and the protein composition of LDs we have determined and compared the proteomic profiles of lipid droplets isolated from the fat bodies of 5th-instar larvae and adult Manduca sexta insects and from ovaries. These LD-rich tissues represent three clearly distinct metabolic states in regard to lipid metabolism: 1) Larval fat body synthesizes fatty acids (FA) and accumulates large amounts as triglyceride (TG); 2) Fat body from adult insects provides FA to support reproduction and flight; 3) Ovaries do not synthesize FA, but accumulate considerable amounts of TG in LDs. Major qualitative and semi-quantitative variations in the protein compositions of the LDs isolated from these three tissues were observed by MS/MS and partially validated by immuno-blotting. The differences observed included changes in the abundance of lipid droplet specific proteins, cytosolic proteins, mitochondrial proteins and also proteins associated with the machinery of protein synthesis. These results suggest that changes in the interaction of LDs with other organelles and cytosolic proteins are tightly related to the physiological state of cells. Herein, we summarize and compare the protein compositions of three subtypes of LDs and also describe for the first time the proteomic profile of LDs from an insect ovary. The compositions and compositional differences found among the LDs are discussed to provide a platform for future studies on the role of LDs, and their associated proteins, in cellular metabolism.
Keywords: Lipid Droplets, Proteomics, Fat Body, Ovaries, Manduca sexta, Metabolism
1. INTRODUCTION
The fat body in insects (Arrese and Soulages, 2010) and the adipose tissue in vertebrates (Frayn et al., 2003) play a major role in the homeostasis of energy metabolism. Adipocytes are the main cells found in these tissues and are characterized by the massive presence of lipid storage droplets, or lipid droplets (LDs) (Zweytick et al., 2000). LDs consist of a core of neutral lipids, predominately triglycerides (TG), surrounded by a layer of phospholipid and a number of embedded or peripherally associated proteins (Brasaemle, 2007a). LDs house organelle-specific proteins of the PAT family (Pfam 03036)(Lu et al., 2001; Miura et al., 2002), such as perilipin in vertebrates and the lipid droplet storage protein-1 and 2, Lsd1 and Lsd2, in insects. These proteins play a major role in the degradation of TG and its regulation as suggested by studies in vertebrates (Brasaemle, 2007b; Ducharme and Bickel, 2008) and insects (Beller et al., 2010b; Bickel et al., 2009). Although the proteins associated with the storage and/or hydrolysis of TG have been the main focus of studies centered on lipid droplets, several recent proteomic studies have shown that LDs are associated with a large number of cellular proteins, including proteins of typical cytosolic, mitochondrial, lyzosomal and endoplasmic reticulum location (Beller et al., 2010b; Murphy et al., 2009; Ohsaki et al., 2009; Welte, 2009). The protein composition of LDs suggests that these cannot be simply described as a fat globule surrounded by a monolayer of phospholipid and proteins. LDs are probably more accurately described if we think of them as a lipid enriched complex that involves interactions with other organelles and numerous cytosolic proteins (Murphy et al., 2009). The physical interactions between the lipid globule of the LDs and proteins from other compartments are likely to be at the center of the control of cellular metabolism (Goodman, 2008, 2009). Knowledge of the composition of these complexes is a first step toward the development of new hypotheses and tests to study and understand the role of LDs in the control of cellular lipid metabolism.
Two proteomic studies of LDs isolated from Drosophila, whole embryos (Cermelli et al., 2006) and larval fat body (Beller et al., 2006), have been previously reported. These are the only studies that have focused on the sub-proteome of insect LDs.
To explore possible correlations between the protein composition of LDs and the physiological state of fat body cells, in the present study we have determined and compared the proteomic profiles of lipid droplets isolated from the fat bodies of Manduca larvae and adult insects. The comparison of LDs from the fat bodies of larvae and adult insects is interesting because they represent two quite different metabolic states. During the fast growing larval stage, M. sexta accumulates massive amounts energy reserves as lipid droplets and this accumulation requires the synthesis of fat and proteins. Conversely, the adult stage of M. sexta is characterized by a massive depletion of energy reserves stored in the fat body. During its short life span, the adult insect uses most of its stored fat to support reproduction and flight. The metabolic differences between larvae and adult insects suggested that changes in the protein composition of LDs were possible and could be evidenced using a proteomic approach. The study was extended to LDs isolated from the ovaries of M. sexta because, as the fat body during the larval stage of M. sexta, the ovaries of adult females represent a stage of fat accumulation (Ziegler and Van Antwerpen, 2006). The main lipid form in the ovary is also TG, which is utilized to support the subsequent energy and membrane synthesis demands of the developing embryo. However, whereas the larval fat body can synthesize fat, the accumulation of fat by ovaries is dependent on the external supply of fatty acids stored in the fat body.
In this study we compare the protein compositions of three subtypes of LDs from Manduca sexta and also describe for the first time the proteomic profile of LDs from an insect ovary. The compositions and compositional differences found among LDs are discussed to provide a platform for future studies on the role of LDs, and their associated proteins, in cellular metabolism.
2. MATERIAL AND METHODS
2.1 Insects
Manduca sexta eggs were purchased from Carolina Biological supplies, and larvae were reared on artificial diet (Bell and Joachim, 1976). Adult insects were maintained at room temperature without food. Second day 5th instar larvae and adult females were used in the experiments.
2.2 Lipid droplet preparation
Fat body tissue from two insects was combined and homogenized with a Potter-Elvehjem glass homogenizer fitted with Teflon pestle, using 6ml of 0.25M sucrose buffer (20mM Tris, pH 7.4, 0.25M sucrose, 1mM EDTA, 1mM benzamidine, 1mM PMSF, 10mg/l leupeptine, 1mg/l aprotonin, 1 mM DTT). The homogenate was centrifuged at 1000xg for 10 min. The supernatant was adjusted to 1.17 M sucrose and transferred to a SW40 centrifuge tube to be subsequently overlaid with 1ml of each of 1.02M sucrose buffer, 0.87M sucrose buffer, 0.58M sucrose buffer, 0.29M sucrose buffer, 0.15M sucrose buffer, and 1.5ml of buffer without sucrose. Density gradients were centrifuged at 160,000g for 4 hr. The top fraction of the gradient, a distinctive thin white layer floating at the top that contains lipid droplets, was collected and mixed with 8ml of 0.25M sucrose buffer. The sample was transferred to a SW40 centrifuge tube and overlaid with 3 ml of buffer without sucrose and centrifuged at 111,000g for 1hr. The top layer (~1 ml), containing the lipid droplets, was collected. Samples were delipidated by acetone precipitation (85% acetone v/v, final concentration) at −20°C overnight. After centrifugation at 10,000g for 15 min, protein pellets were dissolved in Laemmli buffer containing 2.5% (w/v) SDS and 17 mM DTT and treated at 70°C for 10 min. Ovaries dissected from ten insects were extensively rinsed in phosphate buffer to completely remove fat body tissue. Ovaries were homogenized as described above and lipid droplets were isolated following the same procedure.
2.3 Sample processing
Samples were subjected to SDS-PAGE in 4–20% acrylamide gradient gels (Novex, Invitrogen). Each lane of the Coomassie Blue stained gel was divided in six regions. Each gel slice was finely minced. Proteins from each slice were reduced with tris(2-carboxyethyl)phosphine, alkylated with iodoacetamide, and digested for 6–16 hr with 8 μg/ml trypsin, using 50 mM ammonium bicarbonate as buffer. Digestion products were analyzed by LC-MS/MS.
2.4 LC-MS/MS
Samples were analyzed on a hybrid LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to a New Objectives PV-550 nanoelectrospray ion source and an Eksigent NanoLC-2D chromatography system. Peptides were analyzed by trapping on a 2.5 cm ProteoPrepII pre-column (New Objective) and separation on a 75 μm ID fused silica column, packed in house with 10-cm of Magic C18 AQ, terminated with an integral fused silica emitter pulled in house. Peptides were eluted using a 5–40% ACN/0.1% formic acid gradient performed over 40 min at a flow rate of 300 nl/min. During each one-second full-range FT-MS scan (nominal resolution of 60,000 FWHM, 300 to 2000 m/z), the three most intense ions were analyzed via MS/MS in the linear ion trap. MS/MS settings used a trigger threshold of 8000 counts, monoisotopic precursor selection, and rejection of parent ions that had unassigned charge states, were previously identified as contaminants on blank gradient runs, or were previously selected for MS/MS (data dependent acquisition using a dynamic exclusion for 150% of the observed chromatographic peak width). Column performance was monitored using trypsin autolysis fragments (m/z 421.76), and via blank injections between samples to assay for contamination. Data analysis: Centroided ion masses were extracted using the extract_msn.exe utility from Bioworks 3.3.1 and were used for database searching with Mascot v2.2.04 (Matrix Science) and X! Tandem v2007.01.01.1 (www.thegpm.org). A database containing the predicted proteins from all arthropods from NCBI (December 2009) was utilized for searching. MASCOT was set up to search NCBI. Searches used a fragment ion mass tolerance of 0.80Da and a parent ion tolerance of 10.0 ppm. Searches included the following potential peptide modifications: pyroglutamate modification of N-terminal glutamines, oxidation of methionine, acrylamide, and iodoacetamide adducts of cysteine, as well as potential formylation and acetylation of the N-terminus of the parent protein. Scaffold (v2.6.0; Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 99.9% probability as specified by the Peptide Prophet algorithm. Protein identifications were accepted if they could be established at greater than 99.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm.
2.5 Light microscopy
Samples of purified lipid droplets were imaged using a Leica TCS SP2 confocal microscope. Lipid droplets fluorescently labeled with Nile Red or without labeling were observed. Fat body tissue and ovaries from a female insect two days-old were embedded in paraffin and sections that were stained with Mallory’s Azure II Methylene Blue were imaged using a Leica TCS SP2 confocal microscope.
2.6 Electron microscopy
Portions of fat body dissected from a female insect two days-old were fixed for 12 hr in 2% glutaraldehyde in 200 mM sodium cacodylate at 4 °C. Tissue was washed three times in buffer A (60 mM sodium cacodylate, 180 mM sucrose) for 30 minutes each and post-fixed for 1.5 hr in 1% osmium tetroxide in 100 mM phosphate buffer (pH 7.2). Samples were washed three times in buffer A for 30 minutes each followed by dehydration with ethanol. Serial washes in 30%, 50%, 70%, 90%, 95% and 100% ethanol (v/v) for 30 minutes each step were followed by three washes in propylene oxide for 30 minutes each step. Tissue was infiltrated using EMBed 812 resin (Electron Microscopy Sciences, Hatfield, PA). Tissue was placed in 3:1 propylene oxide and EMBed for 4 days. Tissue was placed in 1:1 propylene oxide and EMBed for 2 days, then 1:3 propylene oxide and EMBed for 2 days. Tissue was placed in 100% embedding medium and placed in oven at 60°C for 48 hours. Embedded tissues were cut using an ultramicrotome into 80 nm thick sections, and placed on carbon film grids. Grids containing thin sections were stained using 2.5% uranyl acetate for 30 minutes then rinsed. Grids were then stained using Reynold’s lead citrate (Reynolds, 1963). Sections were imaged using a JEOL 2100 TEM operated at 120 kV.
2.7 Lipid analysis
Total lipids were extracted from 0.2ml of lipid droplets preparation by adding 5 volumes of chloroform-methanol (v/v) and 0.2 ml of phosphate buffer containing 150mM NaCl. Samples were vortexed and centrifuged. The organic phase was collected and subjected to lipid analysis by TLC using hexane-ethyl ether-formic acid 70/30/3 (v/v/v) as the developing solvent (Skipski and Barclay, 1969). Lipids were visualized by iodine and identified by comparing their retention time with that of the standards. A similar procedure was used to prepare total lipid extract from 100mg of whole tissue (fat body and ovaries).
2.8 Western Blot
Proteins were separated by SDS-PAGE (4–20%) and transferred to nitrocellulose membranes. Immunodetection was performed using the corresponding horseradish peroxidase-conjugated secondary antibody purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and ECL chemiluminescence reagents (Amersham Pharmacia, Piscataway, NJ) and exposed to X-ray films. Polyclonal antibodies against apoLp-I/apoLp-II and apoLp-III, were raised by Cocalico Biologicals Inc. (Reamstown, PA) in rabbits immunized with the corresponding proteins (HDLp and apoLpIII), which were purified from hemolymph. Polyclonal antibodies against Lsd1 and Lsd2 were obtained by Cocalico Biologicals Inc. (Reamstown, PA) from rabbits immunized with the recombinant proteins. M. sexta Lsd1 and Lsd2 recombinant proteins (GI: 238846407 and GI: 338858969, respectively) were expressed in E. coli and purified as previously described for Drosophila Lsd proteins (Arrese et al., 2008b; Arrese et al., 2008c). ACC was detected with commercial rabbit polyclonal antibodies from Cell Signaling (Billerica, MA). Lip-DH antibody was purchased from Abcam (Cambridge, MA). Mouse monoclonal anti-Hsp70 was a gift from Dr. Robert L. Matts.
2.9 Other Methods
SDS-PAGE was performed according to Laemmli (Laemmli, 1970) and proteins were visualized by Coomassie Brilliant Blue R staining. TG concentration from the gradient fractions was determined using the Infinity Triglyceride reagent kit as described by the manufacturer (Thermo Fisher Scientific Inc., Waltham, MA). Triolein was used as standard. Protein concentrations were determined by the Bradford dye-binding assay (Bradford, 1976) using bovine serum albumin as standard.
3. RESULTS AND DISCUSSION
3.1. Purification of Lipid Droplets
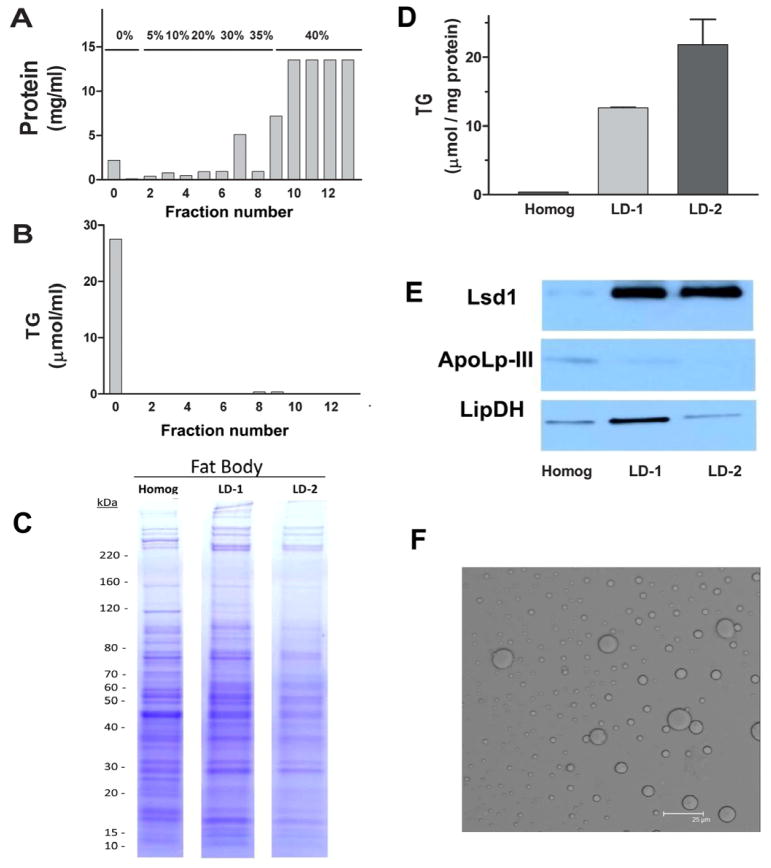
The first step for the isolation of lipid droplets (LDs) consisted in the centrifugation of homogenates in a sucrose density gradient. This method is commonly used to purify LDs from Drosophila fat body (Beller et al., 2006), Drosophila embryos (Cermelli et al., 2006) and from vertebrate adipose tissue (Brasaemle et al., 2004). The LDs accumulate in the top fraction of the gradient (0% sucrose) as reflected by the triglycerides (TG) levels (Fig 1B), whereas most of the cellular proteins accumulate at the bottom of the gradient (Fig 1A). LDs proteins accounted for less than 3% of total fat body homogenate proteins. The substantial removal of proteins, and the concomitantly high TG to protein ratio of the top fraction of the gradient, shows that this procedure leads to a significant purification of the LDs. The lipid droplet fraction (top) isolated from the sucrose gradient was re-purified by an additional high speed centrifugation step as indicated in section 2.2. Qualitative changes in the protein profiles from Coomassie Blue stained SDS-PAGE were observed between homogenates and LDs obtained after each centrifugation step. Fig 1C shows the profiles obtained for the larval LDs purification. The lipid to protein ratios of LDs after the first density gradient (12.6 ± 0.2 μmol TG/mg protein) and the second purification step (21.8 ± 3.7 μmol TG/mg protein) is shown in the Fig. 1D. Lipid to protein ratios of Manduca purified LDs are consistent with previous reports (Wolins et al., 2006). The total protein associated with purified LDs accounted for approximately 1.7% of total protein in the fat body homogenate. Lsd1, a protein that is found mostly associated with lipid droplets in Manduca sexta fat body (Arrese et al., 2008a), was used as fat body lipid droplet marker. As expected, the LDs fraction was enriched in this protein. Fig 1E shows that the top fraction of the gradient is highly enriched in Lsd1 after the first centrifugation and remains practically unchanged after the second centrifugation step. The progress of the purification was also assessed by monitoring the presence of apoLp-III and lipoamide dehydrogenase (LipDH) during the purification. ApoLp-III, a small soluble apolipoprotein, is undetectable by Western Blot after the second purification step (Fig 1E). On the other hand, the mitochondrial protein LipDH become less abundant after the second purification step, but it cannot be completely removed.
Figure 1. Isolation of Manduca Lipid Droplets from larval fat body.
Larval fat body homogenate was first fractionated in a sucrose density gradient as described in Methods. The concentration of protein and triglyceride in the fractions from top (0) to bottom (12) are shown in panels A) and B), respectively. The top fraction of the gradient (LD-1) was then subjected to a second centrifugation and the top fraction containing purified lipid droplets was obtained (LD-2). C) A Coomassie-stained 4–20% gel in which ~ 40 μg of protein from homogenate (homog), LD-1 and LD-2 were loaded. D) Panel shows TG to protein ratios ± SEM (n=3). E) Panel shows the abundance of Lsd1, apoLp-III and LipDH during purification: proteins were visualized by Western blot after separating 40μg of sample protein by SDS-PAGE. F) Panel shows a confocal microscopy image of purified lipid droplets (LD-2) (Bar: 25μm).
The morphology of purified LDs was assessed by confocal microscopy. Fig 1F shows an image of purified LDs from larval fat body. LDs showed a spherical shape with variable sizes and diameters ranging between 2μm and 12μm. This range of sizes is consistent with previously reported LDs from the fat body tissue of larval M. sexta (Willott et al., 1988).
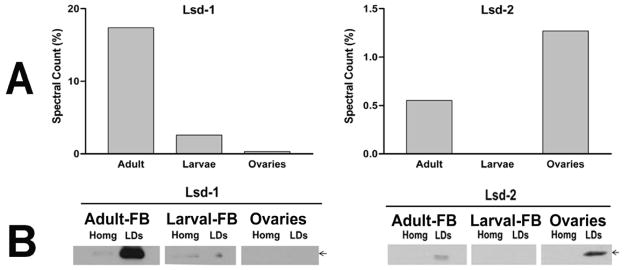
LDs purification from adult fat body and ovary homogenates proceeded in a very similar manner to that shown for the larval fat body. The lipid to protein ratio of LDs from the adult fat body after the first density gradient was 10.5 ±1.0 μmol TG/mg protein and 20.9 ± 1.9 μmol TG/mg protein after the second purification step. Likewise, the lipid to protein ratios for LDs from ovaries changed from 8.8 ±2.0 to 13.1 ±2.1 μmol TG/mg protein. The relative enrichment of Lsd1 and Lsd2 in purified LDs from adult fat body and ovaries are shown in Fig 5B.
Figure 5. Lsd proteins (Lsd1 and Lsd2).
A) Semi-quantitative estimates of Lsd1 and Lsd2 proteins calculated as spectral counts (%) for fat body and ovary LDs; B) The same LD-samples used for MS/MS were used to estimate Lsd1 and Lsd2 by western blotting. The figure also shows western blots of samples of the corresponding homogenates (Homg) used in the purification of LDs. All lanes were loaded with 30 μg of protein.
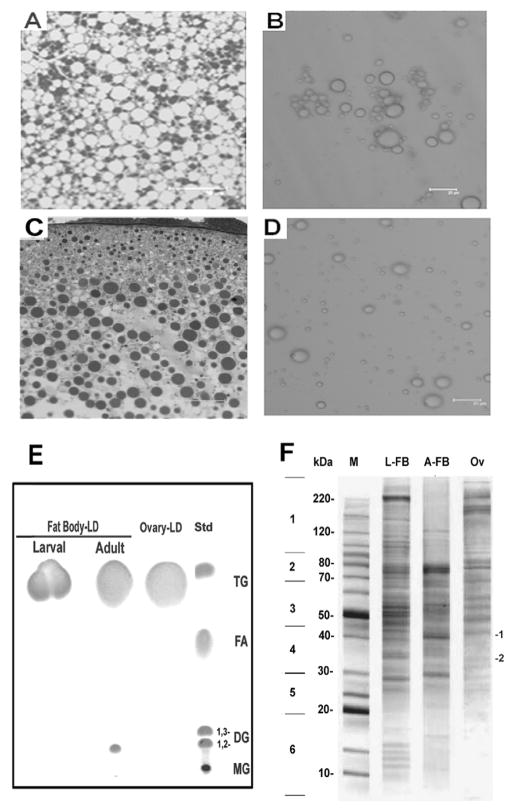
Images of adult fat body tissue and oocyte and their corresponding purified lipid droplets are shown in Fig 2A,B,C,D. Spherical LDs with diameters ranging between 2 μm and 22 μm and 1 and 13 μm were observed for LDs isolated from the adult fat body (Fig 2B) and ovaries (Fig 2D), respectively. The range of sizes of purified LDs was consistent with sizes observed in the tissues (Fig 2A and 2C).
Figure 2. Isolation of Manduca Lipid Droplets from adult fat body and ovaries.
Confocal microscopy image of adult fat body (panel A) and ovary sections (panel C), and corresponding purified lipid droplets (panel B, adult fat body; panel D, ovaries). E) Thin-layer chromatography of lipid extracts of purified LDs from fat bodies and ovaries; Std lane shows lipid standards (TG, triacylglycerols; FA, fatty acids; 1,2-DG and 1,3-DG, diacylglycerols; MG, monoacylglycerols). F) A Coomassie-stained 4–20% SDS-PAGE gel in which ~ 40 μg of delipidated lipid droplets (LD-2) obtained from fat body and ovaries of M. sexta were loaded: M, molecular weight marker; L-FB, Larval Fat body; A-FB, Adult female fat body; Ov, Ovaries. Each lane was excised in six regions as shown in the figure. Individual regions were subsequently analyzed by mass spectrometry. The approximate migration points where Lsd1 and Lsd2 are expected to appear were indicated with numbers 1 and 2, respectively.
The lipid analysis of isolated LDs confirmed that the integrity of lipids was preserved during the purification procedure. TG was the major lipid found in all preparations. A small amount of DG was also observed in LDs isolated from the adult fat body (Fig 2E).
To identify the proteins associated with LDs, the samples of purified LDs were delipidated, subjected to SDS-PAGE (Fig 2F) and subsequently analyzed by MS/MS, as indicated in section 2.4.
3.2. Proteomics of Lipid Droplets
A total of 229 different proteins were identified by MS/MS. The complete list of proteins in lipid droplets isolated from M. sexta fat body and ovaries is shown in Table 1. The larval fat body provided the largest number of identifications (172), followed by the LDs from female fat body (129), and ovaries (88). Some proteins were only found in one type of LDs (74 in larval fat body and 13 in LDs from ovaries). The number and variety of proteins identified in M. sexta LDs are consistent with previous proteomic studies of LDs from diverse origins. There are at least 13 papers specifically describing the proteomic composition of lipid droplets: two from insects (Beller et al., 2006; Cermelli et al., 2006), two from yeast (Binns et al., 2006; Fei et al., 2011) and the remaining of vertebrate origin (Brasaemle et al., 2004; Cavaletto et al., 2008; Cho et al., 2007; Fujimoto et al., 2004; Liu et al., 2004; Sato et al., 2006; Umlauf et al., 2004; Wan et al., 2007; Wu et al., 2000). All these studies have reported a number and diversity of proteins similar to those presented in our study. The proteins previously reported to be components of the LDs of Drosophila have been marked in the Table 1. Among the proteins found in M. sexta LDs approximately 80 proteins were also present in Dm LDs.
Table 1. Proteins present in the lipid droplets as identified by MudPIT ms/ms analysis. Delipidated samples of purified lipid droplets were subjected to SDS-PAGE and subsequently analyzed by mass spectrometry.
Identified proteins were separated in groups according to their function and/or location. The first three columns provide the protein names, gene identification numbers, and estimated masses, respectively. The last column indicates whether a protein was found in all (“ALL”) types of lipid droplets analyzed (Female fat body, F, Larval fat body, L, and ovaries, O).
| Lipid Metabolism | |||
| Lipid storage droplet protein 1* | gi|193876254 | 40 kDa | All |
| Lipid storage droplet protein 2 PLIN (Lsd2)* | gi|221579612 (+1) | 32 kDa | F,O |
| ATGL, Brummer lipase* | gi|338858968 | 62kDa | F,L |
| Apolipophorin I/II precursor* | gi|2498144 | 367 kDa | All |
| Apolipophorin-III; ApoLp-III | gi|114058 (+2) | 21 kDa | F |
| Lipophorin receptor isoform 1 | gi|162462017 (+2) | 101 kDa | F |
| High density lipoprotein binding protein/vigilin | gi|157118627 (+1) | 140 kDa | F,L |
| Acetyl-CoA carboxylase* | gi|157113072 (+1) | 285 kDa | F,L |
| ATP-citrate lyase GJ20089* | gi|195383876 | 121 kDa | F,L |
| Fatty acid synthase S-acetyltransferase* | gi|170038418 | 264 kDa | F |
| Long-chain-fatty-acid--CoA ligase, putative* | gi|212516281 (+1) | 79 kDa | All |
| NADPH--cytochrome P450 reductase* | gi|585549 | 76 kDa | F,L |
| Acyl-CoA-delta9-3a-desaturase* | gi|161407186 (+1) | 41 kDa | F,L |
| Tricarboxylate transport protein* | gi|157111903 (+5) | 35 kDa | All |
| 3-ketoacyl-(acyl-carrier-protein) reductase * | gi|195111777 | 27 kDa | L |
| Acyl-CoA dehydrogenase* | gi|157107359 (+8) | 69 kDa | F,O |
| Medium chain acyl-CoA dehydrogenase | gi|125978457 | 46 kDa | F |
| Short/branched chain acyl-CoA dehydrogenases* | gi|158298698 (+3) | 46 kDa | L,O |
| Trifunctional enzyme beta subunit (tp-beta) | gi|157127973 | 51 kDa | F |
| Enoyl-CoA hydratase, mitochondrial precursor | gi|225709666 (+2) | 30 kDa | L |
| FA oxidation complex Alpha subunit | gi|158297504 (+1) | 83 kDa | F,O |
| Acetoacetyl CoA Thiolase* | gi|146424692 | 40 kDa | F,L |
| Phorbol ester/diacylglycerol-binding protein unc-13 | gi|170040731 (+1) | 268 kDa | F,L |
| Storage and Cuticular Proteins | |||
| Arylphorin subunit alpha | gi|114240 | 84 kDa | All |
| Arylphorin subunit beta | gi|1168527 | 84 kDa | All |
| basic juvenile hormone sensitive hemolymph protein one | gi|156150 (+1) | 88 kDa | F,O |
| Basic juvenile hormone-suppressible protein 2 | gi|729864 | 90 kDa | F,O |
| methionine-rich storage protein | gi|228382 | 30 kDa | F |
| vitellogenin | gi|123299276 (+1) | 201 kDa | O |
| Microvitellogenin; | gi|138601 (+1) | 28 kDa | O |
| Insecticyanin-A | gi|124151 (+2) | 21 kDa | L,O |
| larval cuticle protein | gi|151301020 (+1) | 16 kDa | F,L |
| cuticle protein | gi|223671238 | 32 kDa | F,L |
| Cytoskeleton | |||
| Actin, clone 403* | gi|113255 (+33) | 42 kDa | All |
| Muscle myosin heavy chain * | gi|183979376 | 225 kDa | All |
| Spectrin alpha chain, putative* | gi|212515067 (+1) | 278 kDa | All |
| Spectrin beta chain * | gi|170038033 | 266 kDa | F,L |
| Myosin heavy chain | gi|197322814 (+2) | 66 kDa | L |
| Myosin heavy chain variant C | gi|234204034 | 105 kDa | F,L |
| Dynamin* | gi|157120582 | 81 kDa | L,O |
| Cytoplasmic dynein heavy chain, putative | gi|215504787 (+1) | 323 kDa | F |
| kl-2 1-beta dynein heavy chain | gi|190608814 (+2) | 517 kDa | F |
| Intermediate filament protein | gi|122920502 | 68 kDa | F,L |
| Tropomyosin isoform 4 | gi|153792609 (+1) | 30 kDa | L |
| Tropomyosin Isoform 2 | gi|114052272 | 32 kDa | F |
| paramyosin | gi|195963325 | 103 kDa | L |
| Tubulin beta-1 chain; | gi|12585365 (+4) | 50 kDa | F,O |
| Tubulin alpha chain * | gi|157113931 (+7) | 50 kDa | F |
| Protein Synthesis | |||
| Elongation factor-1 alpha* | gi|187234983 | 45 kDa | All |
| Elongation factor 1-beta* | gi|13124189 (+3) | 24 kDa | L |
| Translation elongation factor 2* | gi|122096234 | 95 kDa | L,O |
| Translation initiation factor 4 gamma | gi|157117248 (+3) | 109 kDa | O |
| Initiation factor 5C | gi|189031276 | 48 kDa | O |
| Translation initiation factor 4F, helicase subunit | gi|215492155 (+1) | 48 kDa | O |
| Translational activator GCN1 | gi|212505341 (+1) | 292 kDa | O |
| Aubergine | gi|166706856 | 101 kDa | O |
| Signal recognition particle receptor beta subunit | gi|111608123 | 32 kDa | L |
| Signal recognition particle 68 kDa protein | gi|148298713 | 69 kDa | L |
| Signal peptidase complex subunit 3; SPC22 | gi|17369766 (+3) | 20 kDa | L |
| t- RNA synth.ribosomal protein GA28351 | gi|125977176 (+16) | 23 kDa | L |
| Probable ribosome biogenesis protein RLP24 | gi|225709326 | 22 kDa | L |
| 40S ribosomal protein ATP synthase | gi|125773061 (+18) | 55 kDa | L |
| 40S ribosomal protein S2 AGAP003768-PA | gi|118780652 (+23) | 30 kDa | F |
| 40S ribosomal protein S4* | gi|74782218 | 30 kDa | All |
| 40S ribosomal protein S10 | gi|20140136 | 18 kDa | L |
| 40S ribosomal protein S11 * | gi|212518046 (+2) | 18 kDa | L |
| 40S ribosomal protein S13 * | gi|229487386 (+3) | 17 kDa | F,L |
| 40S ribosomal protein S14 | gi|112982701 (+4) | 16 kDa | All |
| 40S ribosomal protein S15 | gi|157129941 | 17 kDa | L |
| 40S ribosomal protein S15A | gi|212505805 (+3) | 15 kDa | L |
| 40S ribosomal protein S16* | gi|54039446 | 17 kDa | All |
| 40S ribosomal protein S17* | gi|20140135 (+2) | 15 kDa | All |
| 40S ribosomal protein S18* | gi|54039447 | 18 kDa | F,L |
| 40S ribosomal protein S19e | gi|125983482 (+19) | 17 kDa | L |
| 40S ribosomal protein S2 | gi|253981404 (+21) | 15 kDa | L |
| 40S ribosomal protein S20 | gi|212517174 (+3) | 14 kDa | L |
| 40S ribosomal protein S25 | gi|51316900 | 13 kDa | L |
| 40S s3a protein * | gi|251831260 (+1) | 30 kDa | L |
| 40S ribosomal protein S3 | gi|1350990 | 27 kDa | All |
| 40S ribosomal protein S5 | gi|172034641 (+237) | 20 kDa | L |
| 40S ribosomal protein S6 | gi|2500492 | 29 kDa | L |
| 40S ribosomal protein S7* | gi|1351005 | 22 kDa | F,L |
| 40S ribosomal protein S8 | gi|54039568 | 24 kDa | L |
| 60S ribosomal protein L4 * | gi|212514865 (+1) | 76 kDa | L,O |
| 60S ribosomal protein L8 | gi|170779023 (+2) | 28 kDa | All |
| 60S ribosomal protein L10* | gi|18202261 (+12) | 26 kDa | F,L |
| 60S ribosomal protein L10a* | gi|157135019 (+3) | 25 kDa | F,L |
| 60S ribosomal protein L11* | gi|31340369 | 22 kDa | L |
| 60S ribosomal protein L12 * | gi|195430854 | 18 kDa | L |
| 60S ribosomal protein L13 | gi|21759389 | 25 kDa | All |
| 60S ribosomal protein L13a | gi|31340317 (+1) | 23 kDa | F,L |
| 60S ribosomal protein L15 | gi|6831612 | 24 kDa | F,L |
| 60S ribosomal protein L19 | gi|23573623 | 9 kDa | All |
| 60S ribosomal protein L18* | gi|74910325 (+1) | 21 kDa | All |
| 60S ribosomal_L21e | gi|227262850 (+2) | 18 kDa | L |
| 60S ribosomal protein L22 * | gi|116833097 | 10 kDa | F,L |
| 60S ribosomal protein L23 | gi|212515267 (+1) | 15 kDa | F,L |
| 60S ribosomal protein L23A * | gi|183979309 | 31 kDa | F,L |
| 60S ribosomal protein L28 | gi|24418650 | 16 kDa | F,L |
| 60S ribosomal protein L3 | gi|84095076 | 47 kDa | All |
| 60S ribosomal protein L30 | gi|17368248 | 12 kDa | F,L |
| 60S ribosomal protein L31 | gi|51701794 (+1) | 14 kDa | F,L |
| 60S ribosomal protein L32 | gi|51701837 | 16 kDa | F,L |
| 60S ribosomal protein L39 | gi|209571492 (+4) | 6 kDa | L |
| 60S ribosomal protein L7 * | gi|209571477 (+1) | 29 kDa | L |
| 60S ribosomal protein L7A* | gi|229577209 | 30 kDa | L |
| 60S ribosomal protein L9 | gi|21759393 | 21 kDa | F,L |
| Ribosomal protein LP0 * | gi|187117172 (+14) | 34 kDa | L |
| Peroxisomes | |||
| Catalase | gi|112982683 | 57 kDa | L |
| Nuclear | |||
| Histone H2B * | gi|223670747 (+1) | 14 kDa | All |
| Histone H4 * | gi|10616 (+286) | 11 kDa | All |
| Histone H2A | gi|195356868 | 23 kDa | F,L |
| Histone H2A type 1-A * | gi|170059745 | 13 kDa | L |
| Fibrillarin | gi|157125583 | 33 kDa | L |
| Small nuclear ribonucleoprotein Sm D3 | gi|215504603 | 13 kDa | L |
| Small nuclear ribonucleoprotein E | gi|170065245 | 11 kDa | L |
| 116 kDa U5 small nuclear ribonucleoprotein component | gi|212516706 | 110 kDa | L |
| DNA binding (Histone) GM22498 | gi|195357506 | 9 kDa | L |
| Heterogeneous nuclear ribonucleoprotein k | gi|212518032 | 38 kDa | L |
| DNA repair protein xp-c/rad4 | gi|170060624 | 118 kDa | L |
| gag-pol polyprotein precursor | gi|21214752 | 206 kDa | L |
| RpII215 | gi|125983594 | 209 kDa | L |
| RNA_pol_Rpb8 GA10862 | gi|125978971 | 17 kDa | L |
| Chromosome segregation ATPases GI11335 | gi|195129665 | 103 kDa | L |
| Igf2 mRNA binding protein | gi|170036665 | 57kDa | L |
| H/ACA ribonucleoprotein complex subunit 4 | gi|170029441 (+1) | 58 kDa | L |
| Apoptosis-promoting RNA-binding protein TIA-1/TIAR | gi|215509072 (+1) | 75 kDa | L |
| pre-mRNA-processing-splicing factor | gi|212511480 (+1) | 277 kDa | L |
| pre-mRNA-splicing helicase BRR2 | gi|170055165 | 245 kDa | L |
| RNA-binding protein 1 | gi|112983196 (+3) | 19 kDa | L |
| ATP-dependent RNA helicase, putative* | gi|212509891 (+1) | 82 kDa | L |
| Vasa-like | gi|112983588 (+1) | 66 kDa | O |
| ATP-dependent RNA helicase DDX19B | gi|212515952 (+1) | 54 kDa | O |
| Membrane Proteins (Other) | |||
| Sarco/endoplasmic reticulum calcium ATPase * | gi|255661412 (+1) | 113 kDa | All |
| Calreticulin precursor* | gi|112983032 (+2) | 46 kDa | L |
| ER Transmembrane emp24 domain-containing protein 9 precursor (TMED) | gi|225713398 (+1) | 25 kDa | L |
| ER lumen protein retaining receptor GA18717 | gi|125986802 (+30) | 25 kDa | F,L |
| V-type proton ATPase subunit A | gi|401323 | 68 kDa | All |
| V-type proton ATPase subunit B | gi|401327 | 55 kDa | F,O |
| V-type proton ATPase subunit C* | gi|10886646 | 34 kDa | O |
| V-type proton ATPase subunit E | gi|401332 | 26 kDa | F |
| V-type proton ATPase subunit G | gi|253787068 | 14 kDa | All |
| ABC transporter* | gi|212518546 (+1) | 77 kDa | F |
| Folding and Postranslational modification | |||
| Glucose Regulated Protein (Bip) * | gi|170034715 | 72kDa | F,L |
| Protein disulfide isomerase* | gi|112984454 | 56 kDa | All |
| Methionine aminopeptidase | gi|157107323 (+3) | 56 kDa | L |
| HSP 90 GA18946* | gi|125773987 (+9) | 91 kDa | L,O |
| Heat shock protein 70 * | gi|219810308 (+2) | 73 kDa | All |
| Heat shock protein 60 * | gi|253993196 | 61 kDa | All |
| Lethal (2) 03709, isoform C * | gi|157400397 (+17) | 33 kDa | All |
| Dolichyl-diphosphooligosaccharide-glycosyltransferase subunit STT3A | gi|212506780 | 71 kDa | O |
| Ubiquitin carboxyl-terminal hydrolase 7 | gi|170051523 | 128 kDa | O |
| Ubiquitin | gi|118782538 (+81) | 26 kDa | All |
| Ubiquitin-conjugating enzyme E2 | gi|136643 | 17 kDa | O |
| Glutathione S-transferase 2 * | gi|1170107 | 24 kDa | F |
| Thioredoxin peroxidase | gi|114052210 | 25 kDa | F |
| Signal Transduction | |||
| 14-3-3zeta | gi|114050901 (+1) | 28 kDa | All |
| 14-3-3 epsilon protein | gi|148298752 | 30 kDa | F,O |
| Farnesoic acid O-methyl transferase | gi|148298754 | 25 kDa | F |
| Receptor for activated protein kinase C RACK 1 isoform 1 | gi|115345341 | 36 kDa | L |
| Juvenile hormone epoxide hydrolase | gi|223029806 | 52 kDa | L |
| Juvenile hormone-binding protein | gi|400079 | 25 kDa | O |
| Wingless | gi|256265057 (+16) | 15 kDa | L |
| Inositol 1,4,5-trisphosphate receptor | gi|157106096 | 321 kDa | F |
| Putative death-receptor | gi|195163329 | 201 kDa | L |
| G(alpha)q | gi|156073243 | 41 kDa | All |
| G protein alpha subunit Go | gi|112982857 (+38) | 40 kDa | O |
| Guanine nucleotide-binding protein subunit beta-1 | gi|121007 (+30) | 37 kDa | F,L |
| SAR 1 | gi|125773595 (+23) | 23 kDa | All |
| Ras-related protein | gi|157117562 | 22 kDa | All |
| Rab 5-Like * | gi|125985719 (+30) | 24 kDa | O |
| Rab 11 * | gi|225710014 | 24 kDa | F, O |
| gtpase_rho | gi|157134619 (+21) | 22 kDa | F,O |
| Small GTPase * | gi|23573606 | 8 kDa | L |
| Predicted GTPase | gi|195120167 | 75 kDa | L |
| Vesicular Transport | |||
| Sec61 alpha subunit | gi|112983370 (+1) | 52 kDa | All |
| Sec61 gamma subunit | gi|157110041 (+1) | 8 kDa | L |
| Clathrin* | gi|218563475 (+1) | 192 kDa | F, O |
| Flotillin-1 | gi|157110506 (+15) | 46 kDa | All |
| Sec23-like | gi|194184156 (+12) | 87 kDa | L |
| Sec1 family er-golgi copII vesicle fusion/syntaxin binding | gi|195161348 (+2) | 72 kDa | L |
| Vesicle-fusing ATPase 1; NEM-sensitive fusion protein 1 | gi|1171772 (+23) | 83 kDa | L |
| Sec14p-like | gi|158288192 (+1) | 34 kDa | F |
| Vacuolar sorting protein 4A | gi|212516933 (+1) | 48 kDa | O |
| Carbohydrate/Amino acid Metabolism | |||
| Transketolase* | gi|114050833 | 67 kDa | F,O |
| Trehalose 6-phosphate synthase isoform I * | gi|198448627 | 93 kDa | F |
| PEPCK CG10924, isoform B | gi|157400394 (+14) | 62 kDa | L |
| Enolase* | gi|148298800 | 47 kDa | F |
| Fructose 1,6 –biphosphate aldolase | gi|148298685 | 40 kDa | F |
| UTP-glucose-1-phosphate uridylyltransferase 2 | gi|157110519 (+1) | 58 kDa | F,O |
| Glyceraldehyde-3-phosphate dehydrogenase * | gi|257133530 (+1) | 24 kDa | F |
| Serine hydroxymethyltransferase | gi|170052865 (+1) | 51 kDa | F,L |
| Aminoadipic semialdehyde synthase | gi|212505666 (+1) | 96 kDa | F,L |
| Arginine kinase | gi|256352409 | 40 kDa | F,O |
| Isovaleryl coenzyme A dehydrogenase | gi|208609173 (+2) | 46 kDa | L |
| Immunity | |||
| Leureptin | gi|110649252 | 46 kDa | L |
| Hemocyte aggregation inhibitor protein precursor | gi|259493819 | 48 kDa | F,L |
| Proteases and protease inhibitors | |||
| ATP-dependent metalloprotease FtsH | gi|195170272 (+9) | 81 kDa | F,L |
| 26S protease (S4) regulatory subunit | gi|157124490 | 67 kDa | L |
| C1A cysteine protease precursor | gi|254746340 | 38 kDa | F,L |
| Alaserpin; AltName: Serpin-1 | gi|134436 (+2) | 44 kDa | L |
| Serine proteinase-like protein 1 | gi|237861312 (+1) | 45 kDa | F |
| Mitochondria | |||
| ATP synthase beta subunit* | gi|157136033 | 54 kDa | All |
| ATP synthase alpha subunit mitochondrial* | gi|157131648 | 59 kDa | All |
| ATP synthase gamma* | gi|118789559 (+1) | 33 kDa | F,L |
| adenine nucleotide translocase * | gi|254946115 | 33 kDa | All |
| mitochondrial carrier protein * | gi|158287093 (+1) | 75 kDa | L |
| mitochondrial solute carrier | gi|157113249 (+1) | 75 kDa | F |
| Mitochondrial carrier protein (putative) | gi|125809463 (+10) | 41 kDa | F,L |
| Phosphate carrier protein, mitochondrial* | gi|6016596 | 38 kDa | F,L |
| Isocitrate dehydrogenase AGAP003168-PA* | gi|158291350 (+1) | 48 kDa | F,L |
| Citrate synthase 2 | gi|122067459 (+5) | 52 kDa | F,L |
| Succinate DH | gi|125807291 (+2) | 72 kDa | F |
| Glutamate dehydrogenase* | gi|114052462 | 61 kDa | All |
| Succinyl-CoA synthetase beta chain* | gi|157110382 (+14) | 46 kDa | L |
| ETF-b electron transfer* | gi|195113511 (+1) | 27 kDa | L |
| Pyruvate dehydrogenase E1* | gi|146738085 | 44 kDa | All |
| Dihydrolipoyl dehydrogenase;* | gi|6014978 | 53 kDa | All |
| Pyruvate carboxylase GF13141* | gi|194755401 | 133 kDa | All |
| P5CS: delta l-pyrroline-5-carboxylate synthetase | gi|118782666 (+13) | 80 kDa | All |
| NADH-ubiquinone reductase, putative * | gi|215494308 (+1) | 79 kDa | F,L |
| Cytochrome oxidase subunit 1 * | gi|188503652 | 19 kDa | L |
| Cytochrome c | gi|118005 | 12 kDa | L |
| Cytochrome c oxidase subunit 5A, mitochondrial | gi|6685345 | 7 kDa | F,L |
| NADH dehydrogenase ubiquinone Fe-S 8* | gi|114051372 (+22) | 25 kDa | F,L |
| Ubiquinol-cytochrome c reductase core protein II* | gi|163838684 | 46 kDa | F,L |
| (LETM-1) leucine zipper-EF-hand-containing transmemb protein 1 | gi|212510209 (+1) | 88 kDa | L |
| Prohibitin-2 | gi|212507492 (+1) | 33 kDa | F,L |
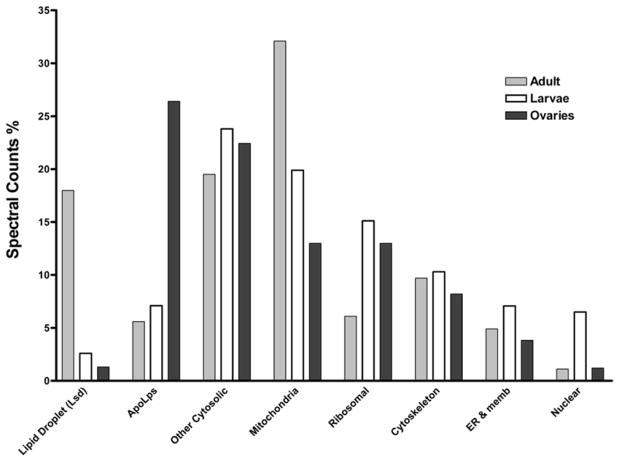
The list of proteins was classified in subgroups defined on the basis of the expected protein location. In order to have an estimate of the prevalence of each protein-subgroup in different lipid droplets, we performed a semi-quantitative analysis of the relative protein abundance based on the fraction of spectral counts obtained for each subgroup. Due to variations in the number of unique peptides theoretically derived from different proteins, and the peptide’s ionization efficiency and appropriate mass to charge ratio for detection, MS/MS does not provide a straightforward quantitative estimate of protein abundance. For instance, an over-estimation of the fraction of larger proteins present in a sample will be expected. In spite of these limitations, a comparison of the fractions of spectral counts for a given protein across different samples provides a good semi-quantitative estimate of possible changes in the abundance of the selected protein. For a given protein there is no bias due to the number of unique peptides, ionization efficiency, and other factors. Similarly, it is also possible to perform a semi-quantitative comparison of the abundance of groups of proteins by comparing the sum of their spectral counts across LDs samples. Fig 3 shows the comparison of the fraction of spectral counts of different groups of proteins for the three types of LDs analyzed. Mitochondrial and cytosolic proteins appear highly abundant in the three subtypes of LDs. Conversely, proteins of typical nuclear location and non-mitochondrial membrane proteins were weakly represented in all LDs. Moreover, clear differences among different LDs subtypes can be seen. Cytosolic and mitochondrial proteins dominate the distribution of spectral counts in the LDs of both larval and adult fat bodies. Lipid droplet specific proteins, Lsd1 and/or Lsd2, were only significantly abundant in LDs from the fat body of adult insects.
Figure 3. Relative Distribution of Spectral Counts among Protein Groups.
The total number of spectral counts of all identified proteins for each LDs sample was used as reference to calculate the corresponding percentage of spectral counts. Proteins were classified in eight subgroups defined on the basis of the expected protein location or function.
Most of the protein groups found in LDs from fat body adipocytes are also significantly represented in the LDs isolated from ovaries (Fig 3 and Table 1). The abundance of apolipophorin (apoLp), the protein component of the major lipoprotein in insects, is significant in all LDs. However, its high abundance in LDs from ovaries constitutes a unique feature of this subtype of LDs.
As observed in previous proteomic studies of lipid droplets of diverse origin (cited above), this proteomic study suggests that M. sexta LDs are also tightly associated with mitochondrial proteins, cytosolic proteins, and proteins of the machinery of protein synthesis. The interaction of LDs with other organelles is a topic that is receiving increasing attention by the scientific community (Goodman, 2008, 2009). These interactions are needed and must be at the center of the regulation of metabolism. Interactions between LDs and peroxisomes (Binns et al., 2006) have also been observed in Yeast, which carries out an active oxidation of fatty acids in these organelles. The ER synthesizes the TG that is stored in LDs and given the insolubility of TG the two apparently independent organelles must be physically connected. Studies in Yeast have shown that 96% of the LDs are permanently associated to ER (Szymanski et al., 2007). Ribosomal proteins were particularly abundant in LDs from larval fat body and from ovaries. The fast changes in the size of LDs for instance when the tissue accumulates TG are likely to require the coupling with protein synthesis and, thus, the presence of ribosomal proteins in LDs is not surprising.
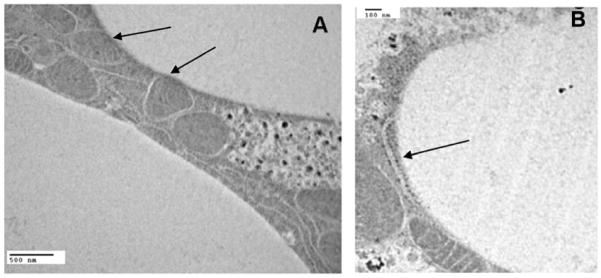
The physical interaction between mitochondria and LDs was observed in multiple studies (Cohen et al., 2004; Novikoff et al., 1980; Stemberger et al., 1984; Vock et al., 1996) and has been the subject of specific studies to determine nature and function of the proteins involved in the interaction (Pu et al., 2011). It has been recently shown that perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria (Wang et al., 2011). As shown in the electron micrographs the LDs of M. sexta fat body (Figures 4A–B) are also closely associated with both mitochondria and rough endoplasmic reticulum. Therefore, the presence of mitochondrial and ER proteins in the preparation of LDs is not surprising. Contrarily, the interaction and even fusion of ER and mitochondrial membranes with the LDs surface may be essential for the stability and function of LDs and also to allow a proper crosstalk between organelles.
Figure 4. Interaction between LDs, Mitochondria and RER.
Panel A The transmission electron micrograph (scale bars 500nm) shows the tight interactions, pointed by arrows, which are common between mitochondria and the LD surface in fat body of M. sexta. Panel B (scale bar 100nm) shows the presence of rough endoplasmic reticulum (pointed by arrow) surrounding the LD surface.
3.2.1. Proteins of Lipid Metabolism Associated with Lipid Droplets
3.2.1.1. Lipid droplet specific proteins
Only few proteins have shown a strong preference to associate with LDs in animal cells. This small set of proteins were grouped under the PAT family (Pfam 03036)(Lu et al., 2001; Miura et al., 2002), and comprises proteins such as perilipin, TIP47 and ADRP, in vertebrates, and lipid storage droplet protein-1 and -2, Lsd1 and Lsd2, in insects. These proteins share sequence similarity in the N-term region, a region called the PAT domain. The insect genomes encode two proteins of the PAT family (Bickel et al., 2009), Lsd1 and Lsd2. These proteins do not have a known enzymatic activity but, as suggested by studies in vertebrates (Brasaemle, 2007b; Ducharme and Bickel, 2008) and in insects (Beller et al., 2010a; Gronke et al., 2003; Teixeira et al., 2003), they play a major role in the degradation of TG and its regulation. PKA-dependent phosphorylation of Lsd1 is a major mechanism regulating the rate of lipolysis in M. sexta fat body (Arrese et al., 2008b; Patel et al., 2005). Moreover, Ca2+/calmodulin dependent phosphorylation of Lsd1 was also shown to be a key factor in the hydrolysis of TG associated with the production of the main Bombyx mori pheromone (Ohnishi et al., 2011). Lsd1 was also shown to control the accumulation of fat in Drosophila (Beller et al., 2010a) although the role of Lsd1 phosphorylation seems to be different to that observed in Lepidoptera.
The MS/MS study showed that Lsd proteins are particularly abundant (~18% of total spectral counts) in LDs from the fat body of adult insects (Fig 5). Both proteins, Lsd1 and Lsd2, were found in LDs from adult fat body by MS/MS and western blotting. However, the MS/MS study suggests that Lsd1 is the predominant Lsd protein (97% of the spectral counts, Fig 5) in the fat body of adult insects.
Two independent estimates, MS/MS and western blotting, of the relative abundance of Lsd1 in different subtypes of LDs are shown in Fig 5. The higher abundance of Lsd1 in LDs from adult insects, as compared to LDs of larval fat body and ovaries, is consistent with the physiological state of adult insects, which are mobilizing FA from the fat body to the ovaries. Previous studies have linked the expression levels of Lsd1 (Arrese et al., 2008a) with the ability of M. sexta fat body to mobilize FA. Moreover, since adult insects are kept without food, they are also mobilizing and oxidizing FA to support basal metabolism. Similarly, the low levels of Lsd1 in LDs from larval fat body or from ovaries are also consistent with the notion that these tissues are accumulating rather than mobilizing fatty acids. We previously reported that Lsd1 was not detected in LDs isolated from young larval fat body by immunoblot analysis (Arrese et al., 2008a). In that study we used an antibody raised against two Lsd1 peptides. In the present study Lsd1 was detected with a different antibody, which was raised against the recombinant Lsd1 protein as described in Materials and Methods. This antibody detected Lsd1 in larval LDs. In agreement with this result, the MS/MS study also detected the presence of Lsd1 in the young larva.
Genetic studies in Drosophila have suggested that Lsd2 plays a role in the accumulation of fat. Flies overexpressing Lsd2 accumulated TG, whereas knockout flies were lean (Gronke et al., 2003; Teixeira et al., 2003). The level of spectral counts of Lsd2 (~1.5%) obtained in the MS/MS study suggests that Lsd2 represents a significant component of the LDs from ovaries. Since the developing ovaries of adult females accumulate lipids, Lsd2 could be involved in this process of TG accumulation. Ovaries accumulate lipid by endocytosis of lipophorin rather than by the novo fatty acid synthesis (Kawooya and Law, 1988). The larval stage of M. sexta is characterized as a frenzy feeding period during which there is a fast increase in body size and a concomitant accumulation of fat in the fat body. Thus, on the basis of the suggested role of Lsd2 in Drosophila’s energy metabolism, we expected to find high levels of expression of Lsd2 in larval LDs. However, and somewhat surprising, Lsd2 was undetectable in LDs from young larval fat body by MS/MS (Fig 5A) and western blotting (Fig 5B). These results could also indicate a difference between the roles of Lsd2 in flies and moths.
3.2.1.2. Lipogenic and lipolytic enzymes
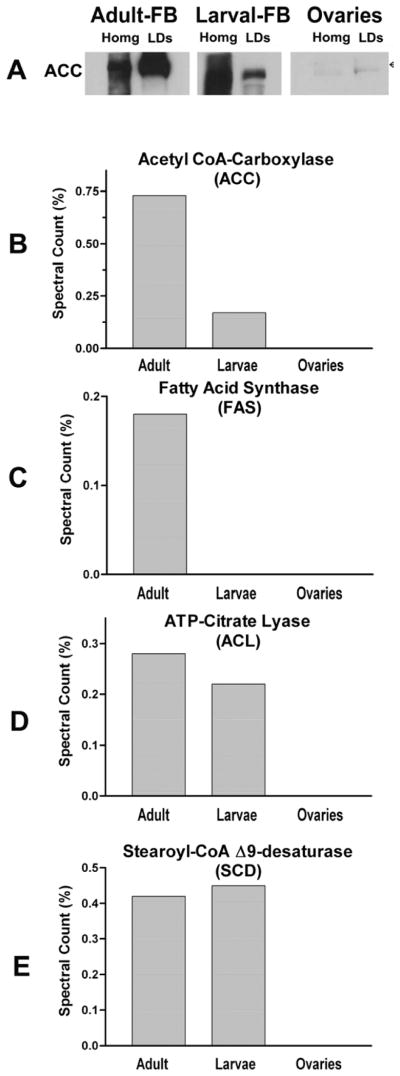
Several essential enzymes of the lipogenic pathway associate with the LDs (Table 1). Lipid droplets from the fat body of adult female insects showed the larger number of lipogenic enzymes, including acetyl-CoA carboxylase (ACC), FA-synthase (FAS), ATP-citrate lyase (ACL), pyruvate carboxylase (PC), Δ9 desaturase (SCD) and NADPH-cytochrome P450 reductase. Some proteins involved in lipogenesis were also found associated with LDs from larval fat body (Table I and Fig 6). ACL was detected in female and larva at similar levels. However, larval LDs lacked FAS (Fig 6D) and, as inferred from the spectral counts and western blotting, contained much lower levels of ACC than LDs from female FB. PC was also much higher in adult FB-LDs than in larval LDs (15–20 lower than in adult LDs).
Figure 6. Lipogenic Enzymes.
A) Semi-quantitative estimate of acetyl-CoA carboxylase, ACC. Samples (30 μg of protein) of tissue homogenates or purified LDs were separated by SDS-PAGE and analyzed by western blot. Panels B, C, D and E show the relative levels of ACC, ATP-CL, FAS and SCD in different types of LDs as inferred from the MS/MS study. The relative levels are expressed as spectral counts (%).
The rate of lipogenesis and fatty acid oxidation are controlled to a great extent by the expression levels and posttranslational modifications of ACC (Saggerson, 2008). The first step of the synthesis of FA consists in the synthesis of malonyl-CoA from acetyl-CoA. This reaction, which is catalyzed by ACC, is important not only for lipogenesis but also because malonyl-CoA is a major inhibitor of carnitine palmitoyltransferase (CPT) (Abu-Elheiga et al., 2001) and, therefore, ACC activity also controls FA oxidation.
The results of MS/MS suggested that ACC was present and highly abundant in fat body LDs. These results were confirmed by western blotting (Fig 6A). The immunoblot study also suggested that homogenates of larval fat bodies could have a higher content of ACC than fat bodies from adult insects (Fig 6A). This result would be consistent with the fact that the larval stage is the period of fat accumulation in M. sexta, whereas in the adult stage the moth uses most of the fat reserves to support flight and reproduction. Interestingly, the proportion of ACC associated with LDs of adult fat body was found to be much higher than that associated with LDs of larval fat body. The meaning of this change in distribution of ACC is not known at this time. It is known that human ACC is inhibited by binding of long chain fatty acyl-CoA (Faergeman and Knudsen, 1997). The association of ACC with LDs of adult insects could be a mechanism of posttranslational regulation of ACC activity. Binding to LDs perhaps inhibits ACC in adult insects. This could be needed to preserve the scarce reserves of glucose stored as glycogen. Alternatively, binding of ACC to LDs could constitute a mechanism of storage. Stored ACC could be released from the LDs and become active once the moth feeds on nectar. Clearly, further studies in this area could lead to interesting insights into the role of the association of ACC with the LDs in the regulation of lipogenesis in Lepidoptera. These findings suggest that LDs in adult insects could recruit lipogenic enzymes to prevent their degradation and preserve them for their use after an eventual nectar feeding period. The role of lipid droplets as protein storage depots has been recently reviewed (Hodges and Wu, 2010).
Although the potential role of ACC binding to LDs has not been previously discussed, it is important to note that association of ACC with LDs isolated from Drosophila (Beller et al., 2006) and from mammalian sources (Liu et al., 2004; Umlauf et al., 2004) has been previously reported. Moreover, it is well known that ACC2, one of the vertebrates’ isoforms of ACC, associates with mitochondria (Abu-Elheiga et al., 2000). ACC2 is assumed to interact with the mitochondrial membrane through a hydrophobic N-terminal region. However, we do not know if the N-terminal region of M. sexta ACC shares similarities with ACC2. On the other hand, we have not found homology between the N-terminal regions of insect ACCs and mouse or human ACC2. Further studies are required in both arthropods and mammalian systems to understand a possible physiological role of ACC binding to LDs.
ACC was not detected by MS/MS or by immunoblot in lipid droplets isolated from ovaries. In addition, western blot analysis of ovary homogenates also showed a very low level of ACC (Fig 6A). This result is in accord with previous studies indicating that de novo FA synthesis in ovaries is not relevant and only contributes 1% of the oocyte’s fat content (Kawooya and Law, 1988). The fact that the MS/MS study did not detect lipogenic enzymes in these LDs, Fig 6, is in agreement with the expected low lipogenic activity of ovaries.
Fatty acid mobilization is initiated by the action of lipases. Two lipases involved in the hydrolysis of TG are present in Manduca sexta fat body. These are TGL (Arrese et al., 2010; Arrese and Wells, 1994) and the adipose tissue TG lipase (ATGL), also known as Brummer lipase in Drosophila (Gronke et al., 2005; Gronke et al., 2007). ATGL, a highly conserved lipase present in all organisms from yeast to mammals, is expected to be associated with the LDs (Lass et al., 2011). Our MS/MS study detected very low levels of ATGL in LDs from fat bodies of larval and adult insects. Whether this is due to low expression levels of ATGL or to a particularly low sensitivity of MS/MS to detect tryptic peptides of ATGL remains to be seen. On the other hand, TGL was not detected by MS/MS in any of the LDs samples. This is in agreement with previous reports showing that TGL is a cytosolic enzyme (Arrese et al., 2006) and does not bind to the LDs with high affinity (Patel et al., 2005).
3.2.3. Lipid Transport Proteins Associated with Lipid Droplets
3.2.3.1. Apolipophorin
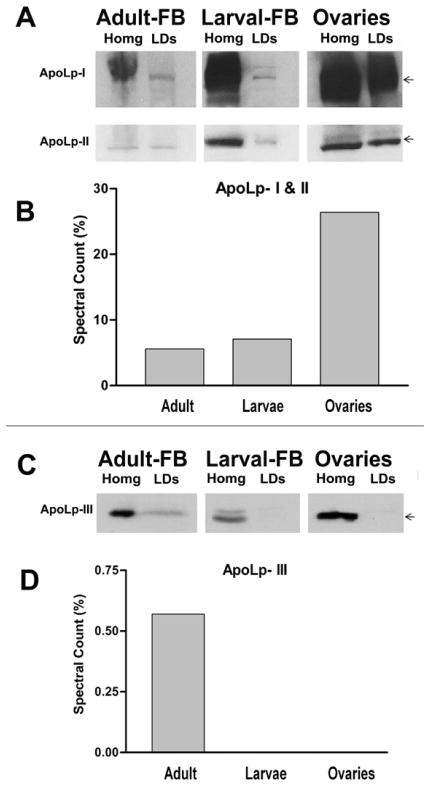
Lipophorin is the main lipoprotein found in insects (Soulages and Wells, 1994). Lipophorin transports lipids from the midgut to the fat body and from the fat body to other tissues, mainly muscles and ovaries (Arrese et al., 2001). The structural apolipoproteins of lipophorin (apoLp-I and apoLp-II), were found in all LDs studied. As shown in the Fig 7B, the MS/MS study suggested that apoLps were particularly abundant in LDs from ovaries, in which ~26% of all the spectral counts corresponded to apoLps. Confirming this result, Western blotting with anti-apoLp antibodies also showed that LDs isolated from ovaries contain the higher levels of apoLps (Fig. 7A).
Figure 7. Apolipophorins.
A) Relative levels of the structural lipoprotein subunits (apoLp-I and apoLp-II) as estimated by western blotting in tissue homogenates and corresponding LDs; B) The graph shows the relative levels (spectral counts %) of apolipophorins (apoLp-I + apoLp-II) in different types of LDs, as determined by MS/MS; C) Levels of the exchangeable apolipoprotein, apoLp-III, in tissue homogenates and LDs as determined by western blotting; D) Relative levels of apoLp-III in LDs as determined by MS/MS.
Lipid accounts for 40% of the dry weight of mature M. sexta eggs (Kawooya et al., 1988). Nearly all the oocyte’s lipid originates in the fat body and is transported to the ovary by lipophorin (95%) and to a lesser extent by vitellogenin (Kawooya and Law, 1988). Studies in M. sexta have shown that a significant fraction of circulating lipophorin is sequestered by the follicles and the lipophorin-DG converted to TG for storage (Kawooya and Law, 1988; Kawooya et al., 1988; Ziegler and Van Antwerpen, 2006). These processes take place without degradation of the apolipoproteins, which are also stored in the eggs (Kawooya et al., 1988; Sun et al., 2000). In agreement with these previous studies, a lipid analysis of ovaries showed a virtual absence of DG (Fig 1A). Given the large amounts of apolipophorin present in ovaries (homogenates and LDs), the fact that only TG is detected suggests that once the lipoprotein is taken up by the ovaries the lipoprotein bound DG is rapidly converted to TG. Since lipophorin represents the major source of lipids for the developing oocyte, the association of these apolipoproteins with the lipid droplets may be needed for the conversion of DG into TG and/or the subsequent transference of TG to the LDs core. The content of apolipophorins also suggests that they may constitute an important structural element of the LDs of ovaries.
ApoLp-I and apoLp-II were also highly abundant in LDs of the fat bodies of insects in both larval and adult stages. The spectral counts in adult and larvae represented 5% and 6% of the total counts, respectively. Contrarily to the ovaries, which only take up lipophorin, the fat body is the place of synthesis of lipophorin and also the location for reloading of partially lipid depleted lipophorin particles. Biosynthesis of lipophorin is expected to involve the co-translational lipidation of apoLps (Dantuma et al., 1999). ApoLp-I and apoLp-II are coded by a single gene and synthesized as a long precursor that is subsequently cleaved. The interaction of fat body LDs with apoLps may be needed to allow an efficient loading of the nascent apoLp chain with neutral lipids and, thus, prevent misfolding, aggregation and degradation of the nascent apoLps. Apolipophorins share a number of similarities with apoB (Smolenaars et al., 2007), which has been previously shown to associate with LD in hepatome cell lines (Ohsaki et al., 2006; Ohsaki et al., 2008). The apoB-LD interaction could be needed in vertebrates for either/or both the degradation of apoB containing lipoproteins, LDL and VLDL, (Ohsaki et al., 2006) or for the biosynthesis of VLDL. In feeding and fast growing larvae the rate of de novo synthesis of lipophorin is high; thus, the association of apoLps with LDs is probably due to the coupling of apolipoprotein synthesis and lipid loading. In adult insects, lipophorin works to some extent as a lipid shuttle, or reusable lipoprotein, mobilizing DG from the fat body to other tissues. Therefore, the association of apoLps with fat body LDs is more likely due to the process of lipid loading of partially lipid-depleted lipophorin particles. The process of lipid loading of Lp particles in insects is not fully understood, yet. However, endocytosis followed by lipid-loading and resecretion of the lipophorin particles is possibly the most important mechanism (Van Hoof et al., 2005). Adult insects have circulating lipophorin particles of higher lipid content than larvae. Two Lps are found in adult insects, HDLp and LDLp. In addition to apoLp-I and apoLp-II, the adult Lps have, a third small (17kDa) apoLp, apoLp-III, which is needed to prevent the aggregation of the highly DG-loaded Lps (Weers and Ryan, 2006). LDLp contains more lipid and more molecules of apoLp-III than HDLp, which represents the lipid depleted Lp in adult insects (Soulages et al., 1996). Uptake of HDLp and its binding to the LDs could be needed to allow loading of HDLp with DG. The TG molecules stored in LDs are the precursors of DG. In accordance with this hypothesis, apoLp-III was found associated with LDs from the fat bodies of adult insects, but was absent in larval LDs. Moreover, contrasting with the absence of DG in LDs from ovaries and larval fat body, fat body-LDs from adult insects contain significant amounts of DG (Fig 2E). This difference is probably explained by the fact that lipophorin loads DG in fat body and unloads it in ovaries.
3.2.3.2. Lipophorin Receptor
A lipoprotein receptor with similarities to the mammalian LDL receptor family was initially purified from M. sexta (Tsuchida and Wells, 1990). This receptor is involved in endocytosis of HDLp (Dantuma et al., 1999) and its mRNA is expressed in several tissues (Ciudad et al., 2007; Gopalapillai et al., 2006; Seo et al., 2003). However, due to the fact that an extensive transference of lipids between lipophorin and tissues takes place in an endocytosis independent manner (Van der Horst and Rodenburg, 2010; Van der Horst et al., 2009), the physiological function of this receptor is not clear, yet. The presence of lipophorin receptor in LDs of adult insects (Table 1), which mobilize lipid mainly from the fat body to the ovaries and flight muscles, suggests that its main role could be the delivery of lipids from the fat body to other tissues.
Vitellogenin (Vg), the major egg yolk lipoprotein, was found associated with the LDs of ovaries. Vg belongs to the superfamily of lipid transfer proteins that includes mammalian apoB, insect apoLp-I/II and others (Smolenaars et al., 2007). Vg provides only ~5% of the total egg lipid (Kawooya and Law, 1988). However, it plays an essential role in embryo development. Vg and microvitellogenin (mVg) represented 1.6% of the total spectral counts of the LDs from ovaries. The role of Vg or mVg in the LDs is not known, but given their abundances and lipid binding capabilities they could play a structural role. It may be interesting to know whether these proteins can interact with Lsd2, the lipid droplet storage protein that is abundant in LDs from ovaries.
3.3. Lipid Droplets and Protein Synthesis
The subgroup of larval LD-proteins related to protein synthesis is particularly interesting because in addition to a large number of ribosomal proteins (Table 1 and Fig 1), larval LDs also contain several non-ribosomal proteins needed for protein synthesis, such as translation elongation factors, chaperons and subunits of the signal recognition particle and signal peptidase (Table I). As is the case for ribosomal proteins, chaperons were particularly abundant accounting for ~12% of all spectral counts associated to larval LDs. It must be noted that the association of HSP70 (Jiang et al., 2007), Bip (Prattes et al., 2000) and many other members of the chaperon family has been observed in lipid droplets isolated from Yeast (Binns et al., 2006), Drosophila (Beller et al., 2006; Cermelli et al., 2006) and mammalian tissues or cells (Liu et al., 2004; Turro et al., 2006; Umlauf et al., 2004; Wan et al., 2007; Wu et al., 2000). HSP70 was shown to co-localize with perilipin on the surface of LDs in adipocytes (Jiang et al., 2007). The association of HSP70 with the LDs of adipocytes has been proposed to play a role in the stabilization of LDs or in the refolding of LD-associated proteins (Jiang et al., 2007).
Among the chaperons associated with M. sexta LDs, the predominantly cytosolic HSP 70 was the most abundant protein (Fig 8). However, ER resident chaperons such as GRP78 (Bip), calreticulin and protein disulfide isomerase (PDI) also accounted for a significant proportion of the spectral counts in larval LDs (Fig 8).
Figure 8. Chaperons.
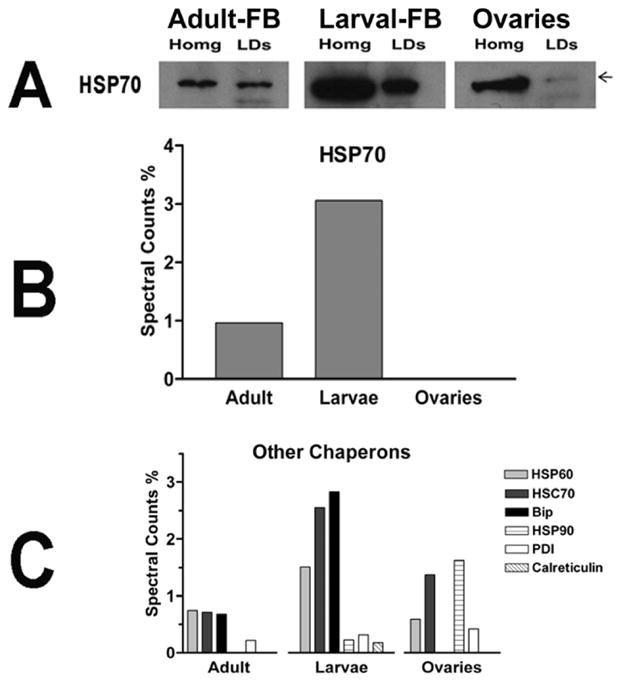
A) The content of HSP 70 in homogenates and LDs was estimated by western blotting; B) Levels of HSP70 in different types of LDs as determined by MS/MS (spectral counts %); C) Relative levels of six different chaperons associated with LDs as determined by MS/MS.
With the exception of HSP90 and PDI, which were more abundant in LDs from ovaries, all other chaperons were relatively more abundant in larval LDs (Fig 8B–C). The relative levels of HSP70 in all LDs studied were also evaluated by immunoblot. As shown in the Fig 8A, and in accordance with the spectral counts, LDs of larval fat body have a higher content of HSP70 than the LDs of the fat body and ovaries of adult insects.
The relative abundance of HSP70 in the homogenate of larval fat body, as compared to the homogenates of fat body and ovaries of adult insects (Fig 8A), also shows an overall higher abundance of HSP70 in larvae. The relative content of proteins involved in protein synthesis in larval fat body suggests that there is a tight association of the lipid droplets with rough endoplasmic reticulum (RER). This observation also suggests a possible involvement of LDs in the synthesis of membrane and secretory proteins in the fat body of larval stages.
A significant number of ribosomal proteins were also found in the lipid droplets of adult female M. sexta. However, the absence of translation elongation factors and other proteins characteristic of the RER suggests that lipid droplets of adult insects may not be tightly associated to RER or involved in protein synthesis. EF1α was the only translation elongation factor present in lipid droplets from adult insects. However, this is a ubiquitous abundant protein that plays roles not only in protein synthesis but also in proteolysis, organization of the cytoskeleton and apoptosis (Mateyak and Kinzy, 2010).
Ovary-LDs were also found to be associated with several ribosomal proteins and translation elongation factors. Two vasa-like, DEAD-box, proteins were exclusively found in LDs from ovaries. These putative RNA helicases have been involved in initiation of translation and in embryonic development in a number of animal species, including insects (Nakao et al., 2006). Aubergine, a protein involved in siRNA mediated control of translation in oocytes (Kennerdell et al., 2002) was also associated with LDs from ovaries. This set of proteins and the presence of ribosomal proteins would suggest a role of the LDs of ovaries in protein synthesis.
Overall, the analysis of this subgroup of proteins suggests that in the larval stage and, perhaps also in oocytes, the lipid droplets play a significant role in protein synthesis. These two stages of the insect development are characterized by the accumulation of lipid and a concomitant increase in the size and number of lipid droplets. This process involves protein synthesis to provide structural and regulatory proteins of the LDs. A tight association between RER and LDs may be needed to synchronize or allow the proper assembly of the growing number of lipid droplets. We have not investigated the presence of RNA and ribosomes in the LDs, yet. However, other studies have suggested the presence of RNA (Dvorak et al., 2003) and ribosomes (Wan et al., 2007) in lipid droplets and a possible role of these organelles in protein synthesis.
3.4. Histones and other Nuclear Proteins Associated with Lipid Droplets
LDs of Drosophila embryos were shown to contain large amounts of histones H2A and H2B (Cermelli et al., 2006). During the early hours of embryo development the LDs appear to supply the newly formed nuclei with histones. These observations suggested that the LDs play a role as maternal histone storage organelles in Drosophila (Cermelli et al., 2006).
Our study detected three histones (H2A, H2B and H4) in LDs from M. sexta. Histones H2B and H4 were observed in the LDs of all developmental stages. However, histone H2A was not detected in ovaries and it was the least abundant histone in the fat bodies of female and larval stages. Since we isolated ovaries from 2-days old adult and virgin insects, it would seem that, contrasting with Drosophila, the LDs from M. sexta ovaries do not accumulate H2A and, therefore, do not function as a storage site and source of H2A for the developing embryo.
On the other hand, the fact that histones H2A, H2B and H4 were found associated with the LDs from larval and adult fat bodies suggests that their association may be linked to cellular functions independent of embryo development. Based on the spectral counts, a semi-quantitative comparison of the levels of different histones in larvae and female LDs suggested that H2B is the most abundant LDs histone followed by H4. This relative abundance is reverted in the case of ovaries, which have a higher ratio of H4 to H2B.
In addition to histones, this study identified 22 proteins of typical nuclear localization. The vast majority of these proteins, twenty, were only found in LDs from larval fat body (Table 1). The large number of nuclear proteins exclusively found in larval LDs suggests differences in the structural organization and perhaps functions of larval LDs as compared to LDs from adult insects or from ovaries. The association of larval LDs with ER and nuclei may be related to the fast rate of fat body growth that takes places during the larval stage. These interactions would be needed to provide the proteins and lipids for both the LDs and the nuclear membrane envelope.
3.5. Mitochondrial proteins
Mitochondrial proteins were highly abundant components of the LDs in terms of both the number of proteins identified and the fraction of total spectral counts. Lipid droplets from the fat body of adult insects contained more mitochondrial proteins (24 proteins representing ~25% of the spectral counts) than LDs from larval fat body (19 proteins representing ~16% of the spectral counts) and ovaries (9 proteins, ~9% of spectral counts).
Early studies have described tight interactions between LDs and mitochondria in intact adipose cells (Novikoff et al., 1980; Stemberger et al., 1984) and muscle cells (Vock et al., 1996). The association of mitochondria with LDs was also observed to increase as the energy demand of muscle cells increased (Tarnopolsky et al., 2007). These and other studies have led to the notion that these inter-organelle interactions are needed for an efficient transport of FA to the mitochondria and subsequent oxidation. Thus, the extensive association of female fat body LDs with mitochondria could be related to the higher catabolic rate of the tissue in females, which are unfed and mobilizing, synthesizing and exporting, nutrients to the ovaries.
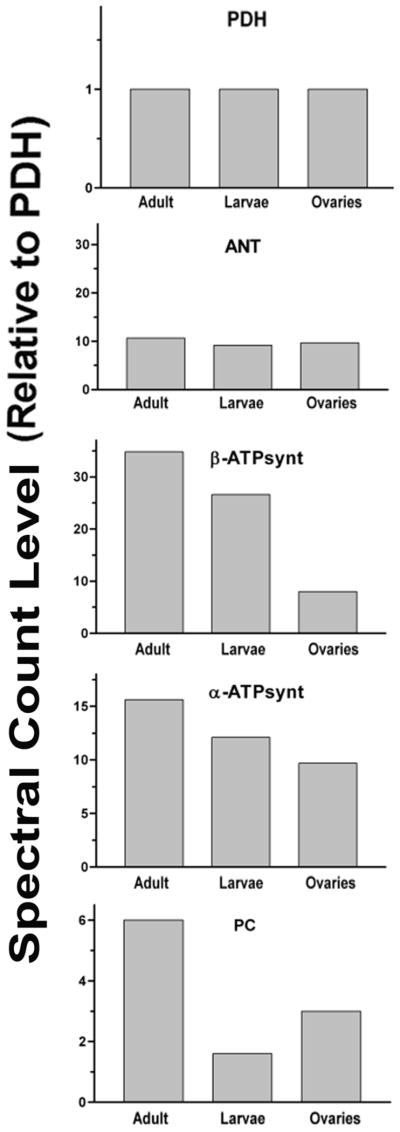
In addition to the apparently higher content of mitochondria in female fat body LDs, a comparative analysis of some mitochondrial proteins also suggests, that mitochondria associated with these LDs express higher levels of certain enzymes required for energy production. When the spectral counts are normalized using the α-subunit of PDH as a reference, the ratios of adenine nucleotide translocator (ANT) to α-PDH are similar for all three LDs subtypes (Fig 9). However, the abundance of the subunits of ATP synthase is greater in female fat body LDs, followed by larval fat body LDs and ovary-LDs. Also interesting is the fact that pyruvate carboxylase, PC, is significantly higher in female LDs. This could be related to the faster oxidation of FAs and the metabolic demand for a greater production of ATP. The increase in the production of oxalacetate would fuel the TCA cycle allowing a faster oxidation of the acetyl-CoA arising from FA-oxidation.
Figure 9. Spectral counts levels for some mitochondrial proteins associated with LDs.
The spectral counts were expressed relative to the level of spectral counts of pyruvate dehydrogenase (PDH). Adenine nucleotide translocase (ANT); β-subunit ATP synthase (β-ATPsynth); α-subunit ATP synthase (α-ATPsynth) and pyruvate carboxylase (PC).
4. CONCLUSIONS
This study compares the protein compositions of three subtypes of LDs that were purified by ultracentrifugation in density gradients. This purification procedure allows identifying proteins that constitute the natural interactome of the LDs. Marked differences among LDs isolated from tissues characterized by distinct metabolic states were observed. Significant variations in the abundance of lipid droplet specific proteins, mitochondrial proteins and proteins associated with the machinery of protein synthesis were observed. The differences in the protein composition of LDs strongly suggest that the interaction of LDs with other organelles and cytosolic proteins plays a major role defining the physiological state of cells. The protein composition of larval fat body LDs strongly indicates the physical association of LDs to the machinery of protein synthesis. Since the larval stage is characterized by an intensive accumulation of LDs and this process requires the synthesis of proteins, it would be worthwhile to investigate the role of LDs in protein synthesis. In particular, it would be interesting to determine the identity of the proteins whose synthesis is physically associated with the LDs. Similarly, the higher abundance of mitochondrial proteins in LDs from adult M. sexta fat body suggests a possible role of the LD-mitochondria association in the release and oxidation of fatty acids and the production of energy.
In accordance with previous LD subproteomes from other origins, the proteomic profiles of M. sexta LDs show that LDs are complex organelles that interact with a large number of cytosolic proteins and organelles. These numerous interactions suggest that we are just starting to envisage the complexity of the function of LDs in cellular metabolism.
Acknowledgments
This project was supported by Oklahoma Agricultural Experiment Station, Oklahoma State University and by Grant Number R01GM064677 from the National Institute Of General Medical Sciences. The content is solely the responsibility of the authors and does not necessarily represent the official use of the National Institute Of General Medical Sciences or the National Institutes of Health. The authors are grateful to Janet Rogers and Lisa Whitworth for their excellent technical assistance in the experiments of mass spectrometry and microscopy, respectively.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errorsmaybe discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abu-Elheiga L, Brinkley WR, Zhong L, Chirala SS, Woldegiorgis G, Wakil SJ. The subcellular localization of acetyl-CoA carboxylase 2. Proc Natl Acad Sci U S A. 2000;97:1444–1449. doi: 10.1073/pnas.97.4.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001;291:2613–2616. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Canavoso LE, Jouni ZE, Pennington JE, Tsuchida K, Wells MA. Lipid storage and mobilization in insects: current status and future directions. Insect Biochem Mol Biol. 2001;31:7–17. doi: 10.1016/s0965-1748(00)00102-8. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Howard AD, Patel RT, Rimoldi OJ, Soulages JL. Mobilization of lipid stores in Manduca sexta: cDNA cloning and developmental expression of fat body triglyceride lipase, TGL. Insect Biochem Mol Biol. 2010;40:91–99. doi: 10.1016/j.ibmb.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Mirza S, Rivera L, Howard AD, Chetty PS, Soulages JL. Expression of lipid storage droplet protein-1 may define the role of AKH as a lipid mobilizing hormone in Manduca sexta. Insect Biochem Mol Biol. 2008a;38:993–1000. doi: 10.1016/j.ibmb.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Patel RT, Soulages JL. The main triglyceride-lipase from the insect fat body is an active phospholipase A(1): identification and characterization. J Lipid Res. 2006;47:2656–2667. doi: 10.1194/jlr.M600161-JLR200. [DOI] [PubMed] [Google Scholar]
- Arrese EL, Rivera L, Hamada M, Mirza S, Hartson SD, Weintraub S, Soulages JL. Function and structure of lipid storage droplet protein 1 studied in lipoprotein complexes. Arch Biochem Biophys. 2008b;473:42–47. doi: 10.1016/j.abb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Rivera L, Hamada M, Soulages JL. Purification and characterization of recombinant lipid storage protein-2 from Drosophila melanogaster. Protein Pept Lett. 2008c;15:1027–1032. doi: 10.2174/092986608785849191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Soulages JL. Insect fat body: energy, metabolism, and regulation. Annu Rev Entomol. 2010;55:207–225. doi: 10.1146/annurev-ento-112408-085356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese EL, Wells MA. Purification and properties of a phosphorylatable triacylglycerol lipase from the fat body of an insect, Manduca sexta. J Lipid Res. 1994;35:1652–1660. [PubMed] [Google Scholar]
- Bell RA, Joachim FG. Techniques for rearing laboratory colonies of tobacco hornworms and pink bollworms. Annals of the Entomological Society of America. 1976;69:365–373. [Google Scholar]
- Beller M, Bulankina AV, Hsiao HH, Urlaub H, Jackle H, Kuhnlein RP. PERILIPIN-Dependent Control of Lipid Droplet Structure and Fat Storage in Drosophila. Cell Metab. 2010a;12:521–532. doi: 10.1016/j.cmet.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Beller M, Riedel D, Jansch L, Dieterich G, Wehland J, Jackle H, Kuhnlein RP. Characterization of the Drosophila lipid droplet subproteome. Mol Cell Proteomics. 2006;5:1082–1094. doi: 10.1074/mcp.M600011-MCP200. [DOI] [PubMed] [Google Scholar]
- Beller M, Thiel K, Thul PJ, Jackle H. Lipid droplets: a dynamic organelle moves into focus. FEBS Lett. 2010b;584:2176–2182. doi: 10.1016/j.febslet.2010.03.022. [DOI] [PubMed] [Google Scholar]
- Bickel PE, Tansey JT, Welte MA. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim Biophys Acta. 2009;1791:419–440. doi: 10.1016/j.bbalip.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D, Januszewski T, Chen Y, Hill J, Markin VS, Zhao Y, Gilpin C, Chapman KD, Anderson RG, Goodman JM. An intimate collaboration between peroxisomes and lipid bodies. J Cell Biol. 2006;173:719–731. doi: 10.1083/jcb.200511125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL. The perilipin family of structural lipid droplet proteins: Stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007a:R700014-JLR700200. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL. Thematic review series: adipocyte biology. The perilipin family of structural lipid droplet proteins: stabilization of lipid droplets and control of lipolysis. J Lipid Res. 2007b;48:2547–2559. doi: 10.1194/jlr.R700014-JLR200. [DOI] [PubMed] [Google Scholar]
- Brasaemle DL, Dolios G, Shapiro L, Wang R. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3-L1 adipocytes. J Biol Chem. 2004;279:46835–46842. doi: 10.1074/jbc.M409340200. [DOI] [PubMed] [Google Scholar]
- Cavaletto M, Giuffrida MG, Conti A. Milk fat globule membrane components--a proteomic approach. Adv Exp Med Biol. 2008;606:129–141. doi: 10.1007/978-0-387-74087-4_4. [DOI] [PubMed] [Google Scholar]
- Cermelli S, Guo Y, Gross SP, Welte MA. The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr Biol. 2006;16:1783–1795. doi: 10.1016/j.cub.2006.07.062. [DOI] [PubMed] [Google Scholar]
- Cho SY, Shin ES, Park PJ, Shin DW, Chang HK, Kim D, Lee HH, Lee JH, Kim SH, Song MJ, Chang IS, Lee OS, Lee TR. Identification of mouse Prp19p as a lipid droplet-associated protein and its possible involvement in the biogenesis of lipid droplets. J Biol Chem. 2007;282:2456–2465. doi: 10.1074/jbc.M608042200. [DOI] [PubMed] [Google Scholar]
- Ciudad L, Belles X, Piulachs MD. Structural and RNAi characterization of the German cockroach lipophorin receptor, and the evolutionary relationships of lipoprotein receptors. BMC Mol Biol. 2007;8:53. doi: 10.1186/1471-2199-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AW, Razani B, Schubert W, Williams TM, Wang XB, Iyengar P, Brasaemle DL, Scherer PE, Lisanti MP. Role of caveolin-1 in the modulation of lipolysis and lipid droplet formation. Diabetes. 2004;53:1261–1270. doi: 10.2337/diabetes.53.5.1261. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Potters M, De Winther MP, Tensen CP, Kooiman FP, Bogerd J, Van der Horst DJ. An insect homolog of the vertebrate very low density lipoprotein receptor mediates endocytosis of lipophorins. J Lipid Res. 1999;40:973–978. [PubMed] [Google Scholar]
- Ducharme NA, Bickel PE. Lipid droplets in lipogenesis and lipolysis. Endocrinology. 2008;149:942–949. doi: 10.1210/en.2007-1713. [DOI] [PubMed] [Google Scholar]
- Dvorak AM, Morgan ES, Weller PF. RNA is closely associated with human mast cell lipid bodies. Histol Histopathol. 2003;18:943–968. doi: 10.14670/HH-18.943. [DOI] [PubMed] [Google Scholar]
- Faergeman NJ, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323 (Pt 1):1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Zhong L, Ta MT, Shui G, Wenk MR, Yang H. The size and phospholipid composition of lipid droplets can influence their proteome. Biochem Biophys Res Commun. 2011 doi: 10.1016/j.bbrc.2011.10.091. [DOI] [PubMed] [Google Scholar]
- Frayn KN, Karpe F, Fielding BA, Macdonald IA, Coppack SW. Integrative physiology of human adipose tissue. Int J Obes Relat Metab Disord. 2003;27:875–888. doi: 10.1038/sj.ijo.0802326. [DOI] [PubMed] [Google Scholar]
- Fujimoto Y, Itabe H, Sakai J, Makita M, Noda J, Mori M, Higashi Y, Kojima S, Takano T. Identification of major proteins in the lipid droplet-enriched fraction isolated from the human hepatocyte cell line HuH7. Biochim Biophys Acta. 2004;1644:47–59. doi: 10.1016/j.bbamcr.2003.10.018. [DOI] [PubMed] [Google Scholar]
- Goodman JM. The gregarious lipid droplet. J Biol Chem. 2008;283:28005–28009. doi: 10.1074/jbc.R800042200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res. 2009;50:2148–2156. doi: 10.1194/jlr.R001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalapillai R, Kadono-Okuda K, Tsuchida K, Yamamoto K, Nohata J, Ajimura M, Mita K. Lipophorin receptor of Bombyx mori: cDNA cloning, genomic structure, alternative splicing, and isolation of a new isoform. J Lipid Res. 2006;47:1005–1013. doi: 10.1194/jlr.M500462-JLR200. [DOI] [PubMed] [Google Scholar]
- Gronke S, Beller M, Fellert S, Ramakrishnan H, Jackle H, Kuhnlein RP. Control of fat storage by a Drosophila PAT domain protein. Curr Biol. 2003;13:603–606. doi: 10.1016/s0960-9822(03)00175-1. [DOI] [PubMed] [Google Scholar]
- Gronke S, Mildner A, Fellert S, Tennagels N, Petry S, Muller G, Jackle H, Kuhnlein RP. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Gronke S, Muller G, Hirsch J, Fellert S, Andreou A, Haase T, Jackle H, Kuhnlein RP. Dual lipolytic control of body fat storage and mobilization in Drosophila. PLoS Biol. 2007;5:e137. doi: 10.1371/journal.pbio.0050137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges BD, Wu CC. Proteomic insights into an expanded cellular role for cytoplasmic lipid droplets. J Lipid Res. 2010;51:262–273. doi: 10.1194/jlr.R003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, He J, Pu S, Tang C, Xu G. Heat shock protein 70 is translocated to lipid droplets in rat adipocytes upon heat stimulation. Biochim Biophys Acta. 2007;1771:66–74. doi: 10.1016/j.bbalip.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kawooya JK, Law JH. Role of lipophorin in lipid transport to the insect egg. J Biol Chem. 1988;263:8748–8753. [PubMed] [Google Scholar]
- Kawooya JK, Osir EO, Law JH. Uptake of the major hemolymph lipoprotein and its transformation in the insect egg. J Biol Chem. 1988;263:8740–8747. [PubMed] [Google Scholar]
- Kennerdell JR, Yamaguchi S, Carthew RW. RNAi is activated during Drosophila oocyte maturation in a manner dependent on aubergine and spindle-E. Genes Dev. 2002;16:1884–1889. doi: 10.1101/gad.990802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50:14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, Anderson RG. Chinese hamster ovary K2 cell lipid droplets appear to be metabolic organelles involved in membrane traffic. J Biol Chem. 2004;279:3787–3792. doi: 10.1074/jbc.M311945200. [DOI] [PubMed] [Google Scholar]
- Lu X, Gruia-Gray J, Copeland NG, Gilbert DJ, Jenkins NA, Londos C, Kimmel AR. The murine perilipin gene: the lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm Genome. 2001;12:741–749. doi: 10.1007/s00335-01-2055-5. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Kinzy TG. eEF1A: thinking outside the ribosome. J Biol Chem. 2010;285:21209–21213. doi: 10.1074/jbc.R110.113795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Gan JW, Brzostowski J, Parisi MJ, Schultz CJ, Londos C, Oliver B, Kimmel AR. Functional conservation for lipid storage droplet association among Perilipin, ADRP, and TIP47 (PAT)-related proteins in mammals, Drosophila, and Dictyostelium. J Biol Chem. 2002;277:32253–32257. doi: 10.1074/jbc.M204410200. [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG. Lipid droplet-organelle interactions; sharing the fats. Biochim Biophys Acta. 2009;1791:441–447. doi: 10.1016/j.bbalip.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Nakao H, Hatakeyama M, Lee JM, Shimoda M, Kanda T. Expression pattern of Bombyx vasa-like (BmVLG) protein and its implications in germ cell development. Dev Genes Evol. 2006;216:94–99. doi: 10.1007/s00427-005-0033-8. [DOI] [PubMed] [Google Scholar]
- Novikoff AB, Novikoff PM, Rosen OM, Rubin CS. Organelle relationships in cultured 3T3-L1 preadipocytes. J Cell Biol. 1980;87:180–196. doi: 10.1083/jcb.87.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi A, Hull JJ, Kaji M, Hashimoto K, Lee JM, Tsuneizumi K, Suzuki T, Dohmae N, Matsumoto S. Hormone signaling linked to silkmoth sex pheromone biosynthesis involves Ca2+/calmodulin-dependent protein kinase II-mediated phosphorylation of the insect PAT family protein Bombyx mori lipid storage droplet protein-1 (BmLsd1) J Biol Chem. 2011;286:24101–24112. doi: 10.1074/jbc.M111.250555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Fujita A, Tokumoto T, Fujimoto T. Cytoplasmic lipid droplets are sites of convergence of proteasomal and autophagic degradation of apolipoprotein B. Mol Biol Cell. 2006;17:2674–2683. doi: 10.1091/mbc.E05-07-0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Suzuki M, Fujita A, Fujimoto T. Lipid droplets are arrested in the ER membrane by tight binding of lipidated apolipoprotein B-100. J Cell Sci. 2008;121:2415–2422. doi: 10.1242/jcs.025452. [DOI] [PubMed] [Google Scholar]
- Ohsaki Y, Cheng J, Suzuki M, Shinohara Y, Fujita A, Fujimoto T. Biogenesis of cytoplasmic lipid droplets: from the lipid ester globule in the membrane to the visible structure. Biochim Biophys Acta. 2009;1791:399–407. doi: 10.1016/j.bbalip.2008.10.002. [DOI] [PubMed] [Google Scholar]
- Patel RT, Soulages JL, Hariharasundaram B, Arrese EL. Activation of the lipid droplet controls the rate of lipolysis of triglycerides in the insect fat body. J Biol Chem. 2005;280:22624–22631. doi: 10.1074/jbc.M413128200. [DOI] [PubMed] [Google Scholar]
- Prattes S, Horl G, Hammer A, Blaschitz A, Graier WF, Sattler W, Zechner R, Steyrer E. Intracellular distribution and mobilization of unesterified cholesterol in adipocytes: triglyceride droplets are surrounded by cholesterol-rich ER-like surface layer structures. J Cell Sci. 2000;113 (Pt 17):2977–2989. doi: 10.1242/jcs.113.17.2977. [DOI] [PubMed] [Google Scholar]
- Pu J, Ha CW, Zhang S, Jung JP, Huh WK, Liu P. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2011;2:487–496. doi: 10.1007/s13238-011-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saggerson D. Malonyl-CoA, a key signaling molecule in mammalian cells. Annu Rev Nutr. 2008;28:253–272. doi: 10.1146/annurev.nutr.28.061807.155434. [DOI] [PubMed] [Google Scholar]
- Sato S, Fukasawa M, Yamakawa Y, Natsume T, Suzuki T, Shoji I, Aizaki H, Miyamura T, Nishijima M. Proteomic profiling of lipid droplet proteins in hepatoma cell lines expressing hepatitis C virus core protein. J Biochem. 2006;139:921–930. doi: 10.1093/jb/mvj104. [DOI] [PubMed] [Google Scholar]
- Seo SJ, Cheon HM, Sun J, Sappington TW, Raikhel AS. Tissue- and stage-specific expression of two lipophorin receptor variants with seven and eight ligand-binding repeats in the adult mosquito. J Biol Chem. 2003;278:41954–41962. doi: 10.1074/jbc.M308200200. [DOI] [PubMed] [Google Scholar]
- Skipski VP, Barclay M. Thin-layer chromatography of lipids. In: John ML, editor. Methods in Enzymology. Academic Press; 1969. pp. 530–598. [Google Scholar]
- Smolenaars MM, Madsen O, Rodenburg KW, Van der Horst DJ. Molecular diversity and evolution of the large lipid transfer protein superfamily. J Lipid Res. 2007;48:489–502. doi: 10.1194/jlr.R600028-JLR200. [DOI] [PubMed] [Google Scholar]
- Soulages JL, van Antwerpen R, Wells MA. Role of diacylglycerol and apolipophorin-III in regulation of physiochemical properties of the lipophorin surface: metabolic implications. Biochemistry. 1996;35:5191–5198. doi: 10.1021/bi952794d. [DOI] [PubMed] [Google Scholar]
- Soulages JL, Wells MA. Lipophorin: the structure of an insect lipoprotein and its role in lipid transport in insects. Adv Protein Chem. 1994;45:371–415. doi: 10.1016/s0065-3233(08)60644-0. [DOI] [PubMed] [Google Scholar]
- Stemberger BH, Walsh RM, Patton S. Morphometric evaluation of lipid droplet associations with secretory vesicles, mitochondria and other components in the lactating cell. Cell Tissue Res. 1984;236:471–475. doi: 10.1007/BF00214252. [DOI] [PubMed] [Google Scholar]
- Sun JX, Hiraoka T, Dittmer NT, Cho KH, Raikhel AS. Lipophorin as a yolk protein precursor in the mosquito, Aedes aegypti. Insect Biochemistry And Molecular Biology. 2000;30:1161–1171. doi: 10.1016/s0965-1748(00)00093-x. [DOI] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RG, Goodman JM. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci U S A. 2007;104:20890–20895. doi: 10.1073/pnas.0704154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarnopolsky MA, Rennie CD, Robertshaw HA, Fedak-Tarnopolsky SN, Devries MC, Hamadeh MJ. Influence of endurance exercise training and sex on intramyocellular lipid and mitochondrial ultrastructure, substrate use, and mitochondrial enzyme activity. Am J Physiol Regul Integr Comp Physiol. 2007;292:R1271–1278. doi: 10.1152/ajpregu.00472.2006. [DOI] [PubMed] [Google Scholar]
- Teixeira L, Rabouille C, Rorth P, Ephrussi A, Vanzo NF. Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech Dev. 2003;120:1071–1081. doi: 10.1016/s0925-4773(03)00158-8. [DOI] [PubMed] [Google Scholar]
- Tsuchida K, Wells MA. Isolation and characterization of a lipoprotein receptor from the fat body of an insect, Manduca sexta. J Biol Chem. 1990;265:5761–5767. [PubMed] [Google Scholar]
- Turro S, Ingelmo-Torres M, Estanyol JM, Tebar F, Fernandez MA, Albor CV, Gaus K, Grewal T, Enrich C, Pol A. Identification and characterization of associated with lipid droplet protein 1: A novel membrane-associated protein that resides on hepatic lipid droplets. Traffic. 2006;7:1254–1269. doi: 10.1111/j.1600-0854.2006.00465.x. [DOI] [PubMed] [Google Scholar]
- Umlauf E, Csaszar E, Moertelmaier M, Schuetz GJ, Parton RG, Prohaska R. Association of stomatin with lipid bodies. J Biol Chem. 2004;279:23699–23709. doi: 10.1074/jbc.M310546200. [DOI] [PubMed] [Google Scholar]
- Van der Horst DJ, Rodenburg KW. Lipoprotein assembly and function in an evolutionary perspective. BioMol Concepts. 2010;1:165–183. doi: 10.1515/bmc.2010.012. [DOI] [PubMed] [Google Scholar]
- Van der Horst DJ, Roosendaal SD, Rodenburg KW. Circulatory lipid transport: lipoprotein assembly and function from an evolutionary perspective. Mol Cell Biochem. 2009;326:105–119. doi: 10.1007/s11010-008-0011-3. [DOI] [PubMed] [Google Scholar]
- Van Hoof D, Rodenburg KW, Van der Horst DJ. Receptor-mediated endocytosis and intracellular trafficking of lipoproteins and transferrin in insect cells. Insect Biochem Mol Biol. 2005;35:117–128. doi: 10.1016/j.ibmb.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Vock R, Hoppeler H, Claassen H, Wu DX, Billeter R, Weber JM, Taylor CR, Weibel ER. Design of the oxygen and substrate pathways. VI. structural basis of intracellular substrate supply to mitochondria in muscle cells. J Exp Biol. 1996;199:1689–1697. doi: 10.1242/jeb.199.8.1689. [DOI] [PubMed] [Google Scholar]
- Wan HC, Melo RC, Jin Z, Dvorak AM, Weller PF. Roles and origins of leukocyte lipid bodies: proteomic and ultrastructural studies. FASEB J. 2007;21:167–178. doi: 10.1096/fj.06-6711com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Sreenevasan U, Hu H, Saladino A, Polster BM, Lund LM, Gong DW, Stanley WC, Sztalryd C. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J Lipid Res. 2011;52:2159–2168. doi: 10.1194/jlr.M017939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weers PM, Ryan RO. Apolipophorin III: role model apolipoprotein. Insect Biochem Mol Biol. 2006;36:231–240. doi: 10.1016/j.ibmb.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Welte MA. Fat on the move: intracellular motion of lipid droplets. Biochem Soc Trans. 2009;37:991–996. doi: 10.1042/BST0370991. [DOI] [PubMed] [Google Scholar]
- Willott E, Bew LK, Nagle RB, Wells MA. Sequential structural changes in the fat body of the tobacco hornworm, Manduca sexta, during the fifth larval stadium. Tissue and Cell. 1988;20:635–643. doi: 10.1016/0040-8166(88)90065-1. [DOI] [PubMed] [Google Scholar]
- Wolins NE, Quaynor BK, Skinner JR, Tzekov A, Croce MA, Gropler MC, Varma V, Yao-Borengasser A, Rasouli N, Kern PA, Finck BN, Bickel PE. OXPAT/PAT-1 is a PPAR-induced lipid droplet protein that promotes fatty acid utilization. Diabetes. 2006;55:3418–3428. doi: 10.2337/db06-0399. [DOI] [PubMed] [Google Scholar]
- Wu CC, Howell KE, Neville MC, Yates JR, 3rd, McManaman JL. Proteomics reveal a link between the endoplasmic reticulum and lipid secretory mechanisms in mammary epithelial cells. Electrophoresis. 2000;21:3470–3482. doi: 10.1002/1522-2683(20001001)21:16<3470::AID-ELPS3470>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Ziegler R, Van Antwerpen R. Lipid uptake by insect oocytes. Insect Biochem Mol Biol. 2006;36:264–272. doi: 10.1016/j.ibmb.2006.01.014. [DOI] [PubMed] [Google Scholar]
- Zweytick D, Athenstaedt K, Daum G. Intracellular lipid particles of eukaryotic cells. Biochim Biophys Acta. 2000;1469:101–120. doi: 10.1016/s0005-2736(00)00294-7. [DOI] [PubMed] [Google Scholar]