Abstract
Translation of the basolateral zinc transporter ZIP5 is repressed during zinc deficiency but Zip5 mRNA remains associated with polysomes and can be rapidly translated when zinc is repleted. Herein, we examined the mechanisms regulating translation of Zip5. The 3′-untranslated region (UTR) of Zip5 mRNA is well conserved among mammals and is predicted by mFOLD to form a very stable stem-loop structure. Three algorithms predict this structure to be flanked by repeated seed sites for miR-328 and miR-193a. RNAse footprinting supports the notion that a stable stem-loop structure exists in this 3′-UTR and electrophoretic mobility shift assays detect polysomal protein(s) binding specifically to the stem-loop structure in the Zip5 3′-UTR. miR-328 and miR-193a are expressed in tissues known to regulate Zip5 mRNA translation in response to zinc availability and both are polysome-associated consistent with Zip5 mRNA localization. Transient transfection assays using native and mutant Zip5 3′-UTRs cloned 3′ to luciferase cDNA revealed that the miRNA seed sites and the stem-loop function together to augment translation of Zip5 mRNA when zinc is replete.
Electronic supplementary material
The online version of this article (doi:10.1007/s10534-011-9508-4) contains supplementary material, which is available to authorized users.
Keywords: miRNA, mRNA, RNA stem-loop, Translation, Zinc, Zinc transporter
Introduction
Zinc homeostasis in mammals involves controlled uptake, sequestration and efflux of this essential metal. We have hypothesized that the zinc transporter Slc39a5 (Zip5) plays a role in the efflux of zinc because it localizes to the basolateral membranes of enterocytes, pancreatic acinar cells and embryonic visceral endoderm cells when zinc levels are adequate (Dufner-Beattie et al. 2004). When dietary zinc is adequate in the adult, enterocytes and pancreatic acinar cells play a key role in zinc efflux while enterocytic zinc uptake is reduced. In mice, during pregnancy, the visceral yolk sac is also important for zinc homeostasis of the developing conceptus. Zinc efflux is attenuated and ZIP5 is internalized and degraded in each of these cell-types during zinc deficiency (Weaver et al. 2007). Interestingly, we found in wild-type animals that zinc availability does not affect Zip5 mRNA abundance or its association with polysomes, and ZIP5 protein is rapidly translated following zinc repletion in vivo and in vitro (Weaver et al. 2007). Cocktails of proteasomal or lysosomal inhibitors in visceral yolk sac explant cultures did not appear to enhance the accumulation of ZIP5 during zinc deficiency suggesting that futile degradation of ZIP5 is not a primary mechanism controlling ZIP5 abundance under these conditions (Weaver et al. 2007). Our previous results imply that a zinc-responsive translational stall mechanism may control the abundance of ZIP5 during zinc deficiency and allow for its rapid resynthesis when zinc is repleted.
Several mechanisms regulating translational activity have been described (Afonyushkin et al. 2005; Allard et al. 2005; Altuvia et al. 1998; Arrick et al. 1991; Ashizuka et al. 2002; Brengues et al. 2005; Ceman et al. 2000; Gray et al. 1996; Hess and Duncan 1996; Laggerbauer et al. 2001; Muralidharan et al. 2007; Parker and Sheth 2007), many of which function at the level of translation initiation (Kapp and Lorsch 2004; Kong et al. 2008). Iron-responsive mRNAs possess iron-regulatory elements (IREs) in their 5′ or 3′-untranslated regions (UTRs). IREs are stem-loop structures bound by either iron-regulatory protein 1 or 2 (IRPs1 or 2) during iron deficiency, when iron is lost from the Fe–S cluster (Leibold et al. 1990; Walden et al. 2006). IRPs either block translation by binding to the 5′-UTR, such as with ferritin heavy chain and light chain mRNAs (Leibold et al. 1990; Leibold and Guo 1992; Leibold and Munro 1988; Munro et al. 1988) or stabilize mRNAs by binding to the 3′-UTR, such as with transferrin receptor mRNA (Mullner et al. 1989). In this way, diminished iron storage and enhanced iron acquisition, respectively, are coordinated during iron deficiency. Such a mechanism has not been described for regulation of gene expression by other essential metals.
MicroRNA (miRNA)-mediated translational regulation has recently emerged as a widely distributed control mechanism (Reviewed by Dignam et al. 1983). miRNAs are thought to imperfectly base-pair to the target mRNA 3′-UTR resulting in altered protein synthesis by as yet poorly understood mechanisms. miRNA ribonucleoprotein (miRNP) complexes can inhibit translation initiation, cause translational stall (Valencia-Sanchez et al. 2006; Wang et al. 2006), initiate target mRNA degradation (Friedman et al. 2009; Grimson et al. 2007), or even stimulate translation (Vasudevan et al. 2007, 2008). miRNAs are predicted to control the activity of over 60% of protein coding mRNAs (Dignam et al. 1983). To date, a role for miRNAs in the homeostasis of essential metal ions has not been described in mammals but a recent report implicates miRNAs in the regulation of copper homeostasis in Arabadposis (Yamasaki et al. 2007).
In this report, we set out to evaluate the mechanisms of post-transcriptional regulation of ZIP5 in response to zinc availability. We hypothesized that the 3′-UTR of Zip5 mRNA would play an important role in this process. We discovered that this UTR is well conserved among the mammals and contains a stem-loop structure that is flanked by putative seed sites for two miRNAs. We followed a rationale outlined in a recent review to experimentally validate predicted miRNA targets (Kuhn et al. 2008). This scheme requires the simultaneous satisfaction of four criteria: (1) Computational prediction of miRNA-mRNA seed pairs, (2) ΔG analysis of the 3′-UTR for the given mRNA to verify that miRNAs target accessible regions, (3) Co-expression of both miRNA and mRNA in vivo, and (4) A functional assay to demonstrate regulation. The data obtained herein support the concept that two miRNAs as well as a stem-loop structure in the 3′-UTR of mammalian Zip5 mRNAs function in the translational control of ZIP5 in response to zinc.
Materials and methods
Animal care and use
Experiments involving mice were performed in accordance with the guidelines from the National Institutes of Health for the care and use of animals and were approved by the Institutional Animal Care and Use Committee. Mouse diets were purchased from Dyets Inc. (Dyets.com) and were identical except for zinc levels, which were as follows: the zinc-deficient (ZnD) diet contained 1 ppm zinc plus deionized water supplied in a zinc-free acid-washed bottle; the zinc-adequate (ZnA) diet contained 30 ppm zinc with tap water. The effects of zinc deficiency during pregnancy were monitored as described in detail previously (Dufner-Beattie et al. 2003, 2004, 2007). The embryonic visceral yolk sacs (VYS) were harvested from day 14 pregnant dams fed either a ZnA or ZnD diet beginning on day 8 of pregnancy. Tissues were either snap frozen in liquid nitrogen for later extraction of total RNA or immediately processed to isolate soluble and polysomal ribonucleoprotein (RNP) fractions.
RNA extraction
RNA was isolated using the Trizol method (Life technologies). In brief, tissues or cells were snap frozen in liquid nitrogen and pulverized using a pestle and mortar pre-equilibrated in liquid nitrogen. Frozen-powdered samples were homogenized in Trizol (1 ml/100 mg tissue) using a Polytron, and processed according to manufacturer’s instructions (Life technologies).
Quantitative PCR
The NCode™ VILO™ miRNA cDNA synthesis Kit (Life technologies) was used to polyadenylate small RNA and synthesize cDNA using total RNA input (0.5 μg per sample) according to manufacturer’s recommendations. The NCode™ EXPRESS SYBR® Green ER™ miRNA qRT-PCR Kit (Life technologies) was used in qPCR analysis (primers listed in Supplemental Table 1) on a real-time Rotor-Gene™ 3000 system (Qiagen) to analyze miRNA expression levels. The NCode miRNA qRT-PCR kit has the ability to distinguish small RNAs with single nucleotide differences. SuperScript® III Reverse Transcriptase (Life technologies) was used to synthesize cDNAs from total RNA (1 μg per sample) according to manufacturer’s recommendations. Rotor-Gene™ SYBR® Green RT-PCR kit (Qiagen) was used in a real time Rotor-Gene™ 3000 system according to manufacturer’s recommendations (Qiagen) to analyze mRNA expression levels (primers listed in Supplemental Table 1). The ΔΔCt method was used to interpret qPCR results. For all qPCR data, each bar in a graph represents the mean with standard deviation from three independent qPCR runs and each run has triplicates. Two to three independent cDNA syntheses were used for the separate runs. When necessary, data were normalized to beta-actin or the renilla co-transfected control. Student’s t test was used to determine significance (P < 0.05).
Computational prediction of miRNAs and RNA secondary structures
The following algorithms are publicly available and continuously updated. RNA secondary structure predictions were derived from the publicly accessible mFOLD algorithm (http://mfold.bioinfo.rpi.edu/zukerm/). Three experimentally validated algorithms that predict miRNA-mRNA target pairs can be found at the following URLs along with links to pilot and other representative publications: miRanda (miRBase) http://microrna.sanger.ac.uk/, TargetScan http://www.targetscan.org/, and PicTar http://pictar.mdc-berlin.de/.
Generation of Zip5 3′-UTR luciferase reporter constructs
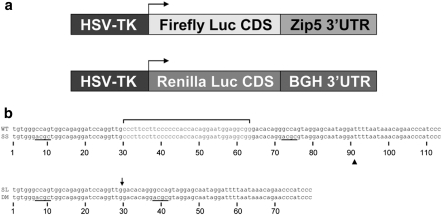
The firefly luciferase coding sequence and the Renilla luciferase coding sequence were amplified by PCR using oligonucleotides indicated in Supplemental Table 2. The PCR products were subcloned into pcDNA3.1 Hygro(+) vector (Promega) using the engineered NheI site at the 5′ end and BamHI site at the 3′ end. The CMV promoter was then replaced with the HSV-TK promoter as follows: An approximately 0.9 kb BglII-NheI fragment encompassing the CMV promoter was excised from the vector and replaced with a 1 kb BglII-NheI fragment containing the HSV-TK promoter from the pRL-TK vector (Promega). The full-length wild-type Zip5 3′-UTR and mutant 3′-UTRs (see Fig. 6b) were generated by annealing oligonucleotides with an engineered NotI site at the 5′ end and an ApaI site at the 3′ end. These UTRs were subcloned downstream of the stop codon in firefly luciferase. Renilla luciferase was subcloned into pcDNA3.1 Hygro(+) with the same restriction sites and promoter replacement indicated above. The bovine growth hormone polyadenylation signal 190 bases downstream of the stop codon served as the 3′-UTR for renilla luciferase. The renilla luciferase construct served as a normalization control in transfection assays. All constructs were sequence verified.
Fig. 6.
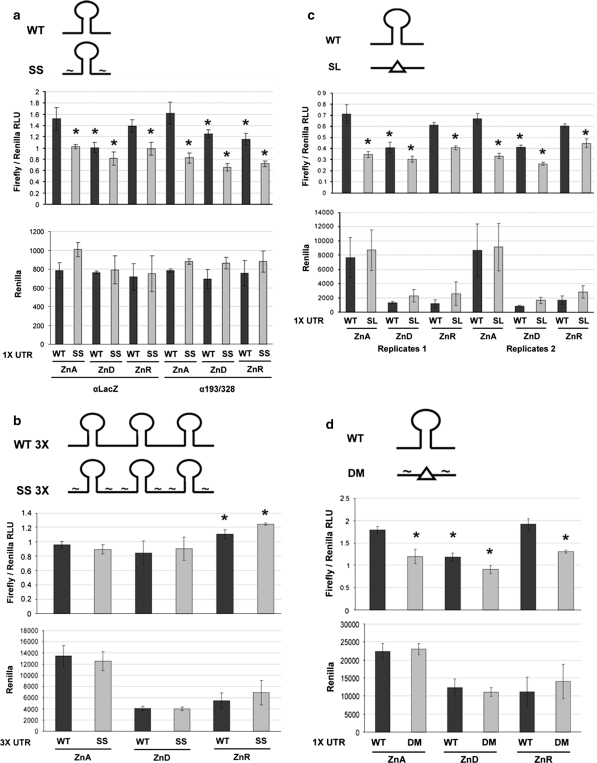
Diagram and nucleotide sequences of the luciferase-Zip5 3′-UTR wild-type and mutant reporter constructs. a Diagram of the structure of the firefly and renilla luciferase reporter constructs. Expression of the firefly and renilla coding sequences was driven by the HSV-TK promoter. The bovine growth hormone polyadenylation signal and site served as the 3′-UTR for the renilla luciferase construct. b Wild-type and mutant Zip5 3′-UTR sequences used in these studies: The 34 nucleotide stem-loop; (sequence in red). The mutated seed sites; (black bars) in the seed site mutant (SS) and the double mutant (DM). The stem-loop mutant (SL) with the deletion site indicated by an arrow. The double mutant (DM) with deletion of the stem-loop and with scrambled seed sites. WT and SS concatamers were constructed by adding two tandem repeats of WT or SS fragments (sequence indicated by triangle) and the third UTR repeat in the concatamers included the polyadenylation signal and site (see Fig. 7)
Generation of miR-193a and miR-328 antagomiR expression vectors
AntogomiR constructs were generated using the BLOCK-iT™ Pol II miR RNAi expression vector kit with EmGFP according to manufacturer’s instructions (Life technologies). The antagomiRs are cloned into an intronic site in the expression vector. Expression of EmGFP can only occur if the intron is correctly processed, thereby ensuring expression of the siRNA or antagomiRs. A control vector encoding EmGFP and a siRNA targeting LacZ was supplied by the manufacturer. The oligonucleotides used to generate antagomiRs are given in Supplemental Table 2. For each miRNA target, bases 2–22 of the given miRNA were used to generate a complementary antagomiR. These can then be processed out of a precursor stem-loop using endogenous cellular small RNA processing pathways. As indicated in Supplemental Table 2, the nucleotides in bold uppercase are the sense sequences of bases 1–8 and 11–21 of the underlined antagomiR that aid in stem-loop formation. The antisense strand is a reverse complement of the sense oligonucleotide except for 5′ overhangs. Other bases are suggested by the manufacturer for cloning and a terminal loop for antagomiR processing which also contains an MscI cut site that can be used during sequence confirmation to overcome strong secondary structure formation. All constructs were sequence verified.
Cell culture
The rat extraembryonic endoderm XEN cell line was previously described (Soares et al. 1987). Cells were maintained at 37°C in a humidified 5% CO2 incubator in Dulbecco’s modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS), 100 units penicillin/ml, and 100 μg streptomycin/ml. To generate ZnD conditions in culture, heat-inactivated fetal bovine serum (FBS) was treated with Chelex 100 resin, as described (Kim et al. 2004). Essential metals except zinc were repleted by the DMEM which was adjusted to 10% Chelex-treated FBS. Cells were cultured in ZnA or ZnD medium for 24 to 48 h before harvest.
Luciferase assays
XEN cells were seeded at low density (1.5 × 104 cells per well) in gelatin-treated 24 well plates and allowed to attach in ZnA medium. Cells were then kept in ZnA medium or switched to ZnD medium and transfected using Lipofectamine2000 (Life technologies) at a 1:1.4 ratio of DNA to Lipofectamine2000. Cells were co-transfected with 150 ng/well each of a Zip5 3′-UTR luciferase reporter and a renilla luciferase reporter control plus 600 ng/well of anti-LacZ siRNA/EmGFP expression vector to monitor transfection efficiency (Life technologies). These transfected XEN cells were cultured for 16 h, fresh medium was added and cells were harvested 8 h later. Some ZnD transfected cells were repleted with zinc (40 μM ZnCl2) during the last 6–8 h before harvest.
To examine the potential roles of miR-328 and miR-193a, XEN cells were plated as above and transfected in ZnA medium with 600 ng total/well of antagomiR/EmGFP expression vectors for miR-328 and miR-193a (1:1) or with the control vector anti-LacZ siRNA/EmGFP. Transfected cells were given fresh ZnA medium 12 h after transfection and cultured for an additional 30 h. At that time, the transfected cells were switched to ZnD medium or kept in ZnA medium and transfected with a Zip5 3′-UTR luciferase reporter and a renilla luciferase reporter control plus 600 ng total/well of either antagomiR/EmGFP expression vectors for miR-328 and miR-193a (1:1) or the control vector anti-LacZ siRNA/EmGFP. These cells were given fresh medium 16 h after the second transfection and then harvested 8 h later. Some ZnD transfected cells were repleted with zinc (40 μM ZnCl2) during the last 6–8 h before harvest.
Harvested transfected cells were lysed at room temperature with 1× passive lysis buffer (Promega) by moderate shaking for 30 min at 25°C then assayed (or stored at −80°C until assayed) using the dual luciferase reporter system on a Turnerbiosystems Veritas microplate luminometer according to the manufacturer’s specifications (Promega). All firefly luciferase values were normalized to the co-transfected renilla luciferase values and were expressed as relative light units (RLUs). Data represent the average of 3–4 replicate wells and are expressed as the mean ± standard deviation (SD). All experiments were repeated independently to confirm validity of results. Student’s t tests were performed to determine statistical significance between normalized luciferase means. All means were compared relative to the respective wild-type ZnA sample set for that experiment. Significance was set to P < 0.05.
Preparation of soluble and polysomal RNP fractions
S100 soluble cytoplasmic RNP fractions and polysomal RNP pellets were prepared essentially as described (Dignam et al. 1983; Leibold and Munro 1988). Fresh VYS tissue (100 mg) or XEN cells (100 mg) were homogenized, using a polytron, in ice-cold homogenization buffer (40 mM Hepes, 5 mM MgCl2, 125 mM KCl, pH 7.4, with or without 400 units/ml RNAse Block (Agilent)). Homogenates were centrifuged at 4°C for 5 min at 500×g to pellet nuclei and the recovered supernatant was then centrifuged for 10 min at 8,000×g to remove mitochondria. That supernatant was recovered and centrifuged at 100,000×g for 30 min at 4°C. The high speed supernatent was considered the soluble fraction and the pellet was considered the polysomal RNP pellet. The polysomal RNP pellet was resuspended in homogenization buffer by brief sonication. Protein content was determined by BCA. All extracts were adjusted to 20% glycerol and stored at −80°C or used immediately.
RNA was extracted from the soluble and polysomal RNP fractions by the addition of a volume of Trizol (Life technologies) equal to that of the recovered soluble fraction. Samples were homogenized and RNA isolated according to the manufacturer’s instructions. Soluble and polysomal RNAs were resuspended in the same volume such that the relative abundance of RNAs in the respective fractions could be represented (Weaver et al. 2007). RNAs (4 μg of polysomal and a volume equivalent of soluble fraction) were resolved in pre-run 12.5% polyacrylamide/8.3 M urea/1 × TBE gels at 18 V/cm. Gels were stained with ethidium bromide to visualize fractionation and integrity of total RNA.
T1 RNAse footprinting
The full length 113 nucleotide mouse Zip5 3′-UTR was generated by annealing and ligating synthetic overlapping oligonucleotides with free 3′A residues into the pGEMT vector (mouse; Fig. 1). A sequence verified clone was linearized with Nco I and SP6 RNA polymerase was used to synthesize a 32P body-labeled sense-strand RNA or a non-radioactive sense-strand RNA competitor. The RNA synthesis reactions (30 μl) were as follows: Linear DNA template (1 μg) in transcription buffer (Agilent) containing 10 mM DTT, 0.5 mM each of ATP, GTP, UTP and 0.5 mM CTP for non-radioactive RNA competitor or 110 μCi of α32P-CTP (800 Ci/mmol) for radioactive RNA. SP6 RNA polymerase (10 U) was added and the reaction mixture was incubated at 37°C for 1 h. The DNA template was removed by the addition of 4 units of RNAse-free DNAseI (NEB) for 15 min at 37°C. The synthesized RNAs were precipitated by the addition of 1.5 μg of heat-denatured, fragmented salmon sperm DNA (ssDNA), 0.3 M ammonium acetate, and 2.5 volumes of cold ethanol. RNA pellets were washed twice with 70% ethanol, briefly dried, and diluted into 50% glycerol. ssDNA was used to prevent non-specific binding and served as a carrier that did not interfere with the T1 RNAse digestion. Reactions with ssDNA omitted generated similar T1 digests but had variability in loading due to non-quantitative transfer.
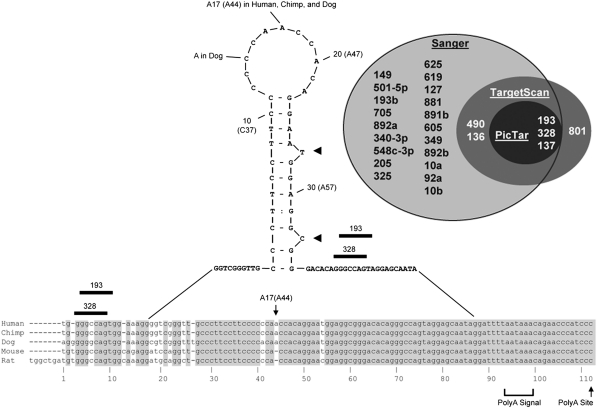
Fig. 1.
Alignment of Slc39a5 (Zip5) 3′-UTRs across five mammalian species showing an mFOLD (Zuker 2003) predicted stable stem-loop structure and Venn-diagram of candidate miRNAs predicted by three algorithms to target this UTR. The full-length sequences of Zip5 3′ UTRs from human, chimp, dog, mouse, and rat were aligned and annotated with empirical information from available Zip5 cDNAs (Dufner-Beattie et al. 2004). The polyadenylation signal (polyA signal) and the polyadenylation site (polyA site) are indicated. Yellow indicates nucleotides conserved in all species shown; green and cyan indicate nucleotides conserved in two or more species shown. The most stable stem-loop predicted by mFOLD (Zuker 2003) is shown. Stem-loop residue A17 is A44 in the full length UTR sequence. Two conserved single nucleotide bulges are indicated by heavy arrowheads. miR-328 and miR-193 seed sites are indicated with black bars. Venn-diagram: light gray shaded region, miRNAs predicted by the miRBase algorithm (Sanger); dark gray shaded region, miRNAs predicted by TargetScan; black shaded region, miRNAs predicted by PicTar. Only miR-328, miR-193, and miR-137 are predicted by all three algorithms to target the 3′-UTR of Zip5 mRNA
T1 digests (20 μl) did not contain RNAse inhibitors but the reactions contained 30 fmol (20,000 dpm) labeled UTR RNA, 6 mM Hepes, 0.75 mM MgCl2, 170 mM KCl and 17.5% glycerol, pH 7.4, ±1 μg of soluble or polysomal protein, ±100-fold molar excess cold competitor, and ±1 unit of T1 RNAse (Ambion). Protein binding and cold competitor reactions were incubated for 30 min at 25°C before the addition of T1 RNAse. T1 digestions were carried out for up to 45 min at 25°C. Reactions were quenched by addition of formamide to a final concentration of 50% and EDTA to a final concentration of 9.5 mM, denatured at 95°C for 5 min, snap-chilled on ice for several minutes and resolved in pre-run 12.5% polyacrylamide/8.3 M urea/1 × TBE gels at 18 V/cm. Gels were then visualized by autoradiography using Biomax film (Kodak) according to manufacturer’s recommendations.
RNA electrophoretic mobility shift assays (EMSAs)
32P-labeled or unlabeled probes corresponding to nucleotides 22 through 84 of the Zip5 3′-UTR (mouse; Fig. 1) were generated from cloned double-stranded oligonucleotides, as detailed above. Reaction conditions were identical to those in the T1 RNAse footprinting reactions described above except that they also contained excess polyI:C (0.5 μg/μl) or heparin (0.35 mg/ml) (both from Sigma) and 8 units of RNAse Block. The soluble or the polysomal RNP fractions (1 μg protein) were allowed to bind at 25°C for 1 h before complexes were resolved in pre-run 4% polyacrylamide gels containing 25 mM Tris, 192 mM Glycine, pH 8.3, at 8 V/cm. Gels were dried and visualized by autoradiography using Biomax film (Kodak) according to manufacturer’s recommendations.
Results
Conservation of the Slc39a5 (Zip5) mRNA 3′-untranslated region among several mammals, and prediction of miRNAs that may target this region
Alignment of the sequences of the Zip5 3′-UTR from several mammals reveals a high degree of sequence conservation throughout the ~113 nucleotide region (Fig. 1). Human and chimp sequences are identical. Mouse and rat sequences have only four nucleotide differences with the exception of seven nucleotides unique to the rat at the 5′ end of the UTR. Mouse and human sequences have only eight nucleotide differences. The mFOLD algorithm predicts secondary structures within the Zip5 3′-UTR. Among the group of predicted structures, a 34 nucleotide stem-loop structure that is highly conserved and which has the highest predicted stability (−13.06 kcal/mol) is shown in Fig. 1. Two stem bulges and a terminal loop are predicted for this putative structure in each of these mammals (Fig. 1). The predicted stem-loop has very little differences between the five species, including, most notably, an additional base (A17) in the terminal loop of three species (human, chimp, dog).
We next determined if miRNAs are predicted to target the Slc39a5 (Zip5) 3′-UTR (Fig. 1). The publicly available miRanda (miRBase) algorithm renders a list of approximately two dozen putative miRNAs that potentially target the 3′-UTR of Zip5 mRNA. We narrowed the possible candidates by employing two other algorithms that also predict miRNA-mRNA seed pairs, namely TargetScan (Friedman et al. 2009; Grimson et al. 2007) and PicTar (Grun et al. 2005; Krek et al. 2005; Lall et al. 2006). All three algorithms predict miRNA-mRNA interactions based on 5′ miRNA seed sites in the 3′-UTRs of mRNAs; however, their weighting and threshold values for other criteria including site conservation or combinatorial action differ and thus their outputs differ.
Three conserved miRNAs, miR-328, miR-137 and miR-193a-3p (also referred to as miR-193 or miR-193a; for uniformity, miR-193a will be used throughout this paper), were predicted by all three algorithms to target the 3′-UTR of the mammalian Zip5 mRNAs (Fig. 1). miR-193b shares the same seed site sequence as miR-193a and therefore was also examined. We found that miR-137 is not expressed in any tissues that express detectable levels of Zip5 and is therefore not considered further (data not shown). It is noteworthy that PicTar predicts Zip5 as the number one target for miR-193a and the number four target for miR-328 in mammals. These results satisfied the first criterion to validate miRNA targets.
T1 RNAse protection and EMSA assays suggest that the Zip5 3′-UTR is structured and can be specifically and constitutively bound by polysome-associated protein(s)
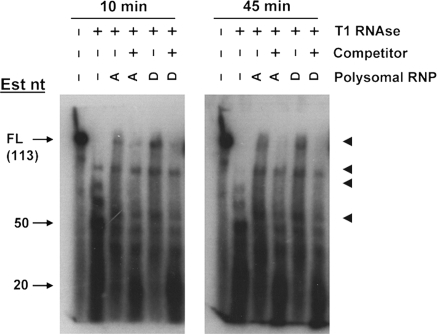
We performed T1 RNAse footprinting (Fig. 2) with or without polysomal RNPs isolated from day 14 embryonic VYS harvested from pregnant mice fed a ZnA or ZnD diet (Dalton et al. 1996, 2003, 2004). Results of the T1 digest in the absence of proteins indicate that the Zip5 3′-UTR is highly structured. T1 cleaves 3′ of single-stranded guanosine residues in RNA. The longest stretch of sequence in the Zip5 3′-UTR without guanosines is 21 nucleotides (Fig. 1). Therefore, the presence of T1 digestion products larger than 21 nucleotides out to 45 min of T1 digestion in the absence of polysomal RNP fraction (Fig. 2) suggests that the Zip5 3′-UTR adopts a stable structure. The predicted structure (Fig. 1) does not contain single-stranded guanosines in the terminal loop. The T1 RNAse footprinting of the labeled UTR preincubated with polysomal RNP fraction from either ZnA or ZnD VYS resulted in protection of some apparently full-length Zip5 3′-UTR and yielded RNA fragments larger than those found in the digests without polysomal RNP fraction (Fig. 2). Inclusion of a 100-fold molar excess of competitor in the binding reaction eliminated protection of the full-length UTR by polysomal RNPs suggesting that the binding is specific. This UTR RNA with scrambled miRNA seed sites (see Fig. 6) yielded T1 footprints indistinguishable from those of the native UTR (data not shown) suggesting that they did not contribute significantly to the binding in this assay.
Fig. 2.
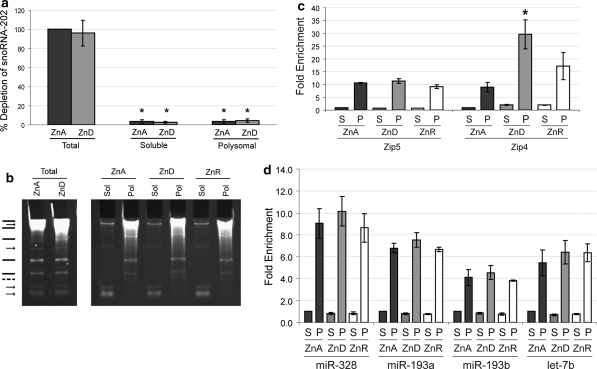
T1 RNAse footprinting of the full length Zip5 3′-UTR with or without visceral yolk sac polysomal ribonucleoprotein fractions. Uniformly labeled full-length Zip5 3′-UTR RNA (113 nucleotides; Fig. 1) was synthesized in vitro, incubated with or without VYS polysomal RNPs (Polysomal RNPs) and then digested with T1 RNAse (±T1 RNAse) for 10 min or 45 min as indicated. Polysomal RNPs were prepared from day 14 embryonic VYS taken from pregnant dams fed a ZnA (A) or ZnD (D) diet beginning on day 8 of pregnancy. Where indicated, the reactions also included a 100-fold molar excess of unlabeled competitor (±competitor). Reaction products were size fractionated and visualized by autoradiography. Approximate sizes of products are indicated (Est nt) and dark arrowheads demarcate four major reaction products
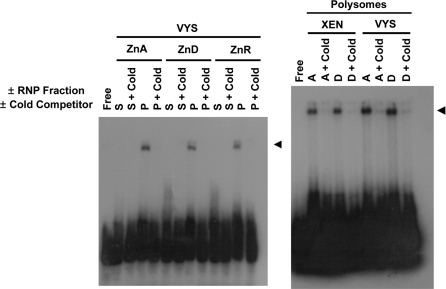
To further examine specificity of protein interactions with the Zip5 3′-UTR, we performed native RNA EMSA with a labeled UTR fragment encompassing the stem-loop structure as well as the downstream miRNA seed sites (nucleotides 22–84), but not the polyadenylation signal (Fig. 1). Binding reactions were performed with or without either the soluble or the polysomal RNP fraction, and with or without a 100-fold molar excess of cold competitor (unlabeled RNA of the same sequence). The mobility of the Zip5 3′-UTR stem-loop probe was shifted when the polysomal RNP fractions were included in the binding reactions but not when the soluble RNP fractions were included (Fig. 3; left panel). As described later in the Results section, enrichment for polysomal and concomitant depletion of nuclear transcripts (see Fig. 5) from these fractions suggests that the polysomal RNP fractions are highly enriched. Additionally, the mobility shifts were eliminated by inclusion of excess cold competitor (Fig. 3) suggesting that the binding is specific. The mobility shift obtained using the rat extraembryonic XEN cell line polysomal RNP fraction was indistinguishable from that obtained with the VYS polysomal RNP fraction. Indistinguishable mobility shifts were obtained with high salt extracts of the polysomal RNP fractions in the binding reactions (data not shown). Thus, the Zip5 3′-UTR must efficiently compete for binding with polysome-associated protein(s). We also noted that the EMSA shifts were resistant to 400 mM KCl which is indicative of high-affinity binding with the Zip5 3′-UTR stem-loop structure (data not shown).
Fig. 3.
EMSA using the Zip5 3′-UTR stem-loop RNA and soluble or polysomal ribonucleoprotein fractions from visceral yolk sac or XEN cells. A uniformly labeled Zip5 3′-UTR sequence, corresponding to nucleotides 22 through 84 of the Zip5 3′-UTR (mouse; Fig. 1), was incubated with or without either the soluble (S) or the polysomal (P) RNP fraction isolated from embryonic VYS or XEN cells. VYS were harvested on day 14 from pregnant dams fed a zinc adequate (ZnA; A) or zinc deficient (ZnD; D) diet beginning on day 8 of pregnancy. XEN cells were cultured in ZnA (A) or ZnD (D) media for 48 h. Where indicated, a 100-fold molar excess of unlabeled competitor (cold) was included in the reaction. Reaction products were size fractionated and visualized by autoradiography
Fig. 5.
Quantification of miR-328, miR-193a, miR-193b, snoRNA 202 and miR-let-7b in the soluble and the polysomal RNP fractions from the VYS. Embryonic visceral yolk sacs (VYS) were harvested from pregnant dams fed a ZnA or ZnD diet or fed a ZnD diet and then given an oral gavage of zinc (ZnR). RNAs were extracted and resuspended from the soluble and the polysomal RNP fractions such that the relative abundance of RNA in the respective fractions was represented (Weaver et al. 2007). a Total RNA, polysomal RNA or soluble RNA were reverse transcribed and amplified for the predominantly nucleolar snoRNA-202. Values are reported as a percent depletion relative to the total ZnA 2−ΔΔCt value. b Total, polysomal (Pol) and soluble (Sol) RNAs were resolved under denaturing conditions and detected by ethidium bromide staining. Arrows indicate RNAs that consistently partition into the soluble RNP fraction. Lines indicate RNAs that consistently partition into the polysomal RNP fraction (the dashed line represents a faint band in total RNA that partitions into the polysomal RNP fraction). c Zip5 and Zip4 mRNAs were amplified in soluble (S) and polysomal (P) RNAs to confirm effective fractionation of the cell lysates and the expected effects of zinc on the abundance of these mRNAs in the VYS. Values are reported as fold enrichment relative ZnA. An asterisk indicates a P value <0.05 as determined by Student’s t test. d Quantification of miR-328, miR-193a, miR-193b, and let-7b abundance in the soluble and polysomal RNP fractions. For each miRNA, the ZnA soluble RNP fraction 2−ΔΔCt value was set to 1 and all other 2−ΔΔCt values were reported as fold enrichment relative to that value. All values shown in this figure represent the mean of three independent qPCR runs (where each run was done in triplicate) ± SD of the three runs. Student’s t test was used to determine significance
The EMSA results further revealed that the VYS and XEN polysome-induced mobility shifts were not significantly affected by whether the polysomal RNPs came from ZnA or ZnD tissues or cells. Taken together, the T1 RNAse footprinting and the EMSA results demonstrate that a protein(s) or RNP complex associated with the polysome fraction can bind with high affinity and specificity to the Zip5 3′-UTR, most likely binding to the stem-loop structure. However, the binding alone does not appear to be regulated by zinc availability.
Two overlapping miR-328 and miR-193a seed-sites are within predicted accessible regions of the 3′-UTR of Zip5
The majority of experimentally confirmed miRNA-mRNA interactions target putatively accessible regions in the 3′-UTR of the mRNA of interest (Kuhn et al. 2008). Thus, a criterion we considered was whether these predicted miRNAs may target accessible regions of the 3′-UTR of Zip5. As shown in Fig. 1, each of the miRNAs was predicted to target putatively accessible regions of the 3′-UTR of Zip5 mRNA. Additionally, miR-328 and miR-193a were predicted to target accessible loops even in other predicted secondary structures of lower predicted stability (data not shown).
As is often the case for mRNAs regulated by miRNAs, two putative seed-pairing sites for miR-328 and miR-193a were noted in the 3′-UTR of Zip5 and in both cases the seed-paring sites for these miRNAs overlapped with a one nucleotide shift. The downstream sites were in an optimal location in the UTR since it has previously been shown that >15 nucleotides downstream of the stop codon is a favorable context for the majority of miRNA seed sites (Grimson et al. 2007). The upstream overlapping seed-sites for miR-328 and miR-193a were predicted to be in a less favorable location within 4–5 nucleotides downstream of the stop codon, yet these sites were conserved in humans, chimpanzees, rats and mice. In summary, miR-328 and miR-193a putatively target evolutionarily-conserved and redundant seed-sites in accessible regions of the thermodynamically-predicted lowest folding energy state for the Zip5 3′-UTR. This satisfies the second criterion to validate miRNA targets.
miR-328, miR-193a and miR-193b are detected in tissues which actively express and regulate ZIP5 in response to zinc
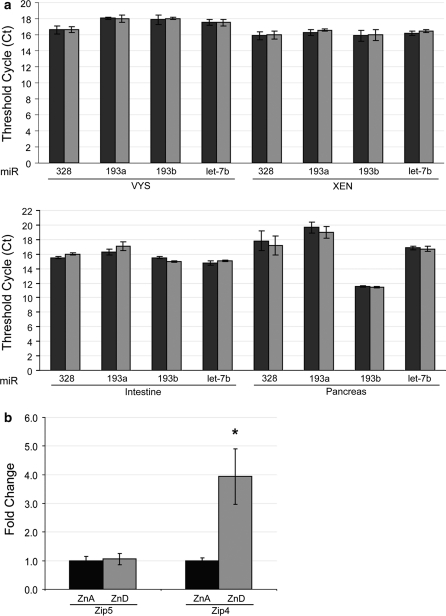
To examine the expression levels of miR-328, miR-193a and miR-193b, we used a sensitive and specific real time quantitative PCR method able to distinguish mature miRNAs differing by a single nucleotide. First, we examined steady state levels of miR-328, miR-193a, miR-193b and let-7b (not predicted to target the Zip5 3′-UTR) in total RNAs from VYS, intestine and pancreas of mice fed a ZnA diet or a ZnD diet and from rat XEN cells cultured in ZnA or ZnD media. Compared to let-7b, a miRNA known to associate with polysomes (Maroney et al. 2006), the three miRNAs of interest had comparable expression levels in total cellular RNA (Fig. 4a). Raw Ct values are plotted for the part a results to show absolute abundance and not relative abundance. No zinc-dependent changes in abundance were observed (Fig. 4a). Because VYS was used for subsequent analyses, the relative abundance of Zip5 and Zip4 mRNAs was also quantified in the ZnA and ZnD VYS RNA samples (Fig. 4b) since the Zip4 mRNA is stabilized and accumulates during zinc deficiency in the mouse VYS while Zip5 mRNA abundance remains constant (Dufner-Beattie et al. 2003, 2004; Weaver et al. 2007). miR-328, miR-193a and miR-193b were each expressed with comparable abundance in the mouse VYS, intestine and pancreas, tissues known to endogenously express and regulate Zip5 which thereby satisfies the third criterion to validate miRNA targets.
Fig. 4.
Quantification of miR-328, miR-193a, miR-193b, Zip4 and Zip5 mRNA abundance in total RNAs from ZnA and ZnD visceral yolk sac, intestine, pancreas and XEN cells. Embryonic visceral yolk sacs (VYS), intestine and pancreas were harvested on day 14 from pregnant dams fed a ZnA or ZnD diet beginning on day 8 of pregnancy. XEN cells were cultured in ZnA or ZnD medium for 48 h. Total RNA was reverse transcribed and probed via quantitative PCR for miR-328, miR-193a, miR-193b, let-7b miRNA abundance. VYS was also probed for Zip4 and Zip5 mRNA abundance. a miRNA abundance was measured in ZnA (black bars) and ZnD (gray bars) VYS, intestine, pancreas and XEN cells. Raw Ct values are shown. Let-7b (not predicted to target Zip5) is shown as a miRNA known to be associated with polysomes (Maroney et al. 2006). Each bar represents the mean of three independent qPCR runs (where each run was done in triplicate) ± SD Student’s t test was used to determine significance. b Zip4 and Zip5 mRNA abundance in VYS. The ΔΔCt method was used to quantify qPCR results. Zip5 and Zip4 mRNA abundance was normalized to beta-actin mRNA and are presented as fold change relative to their respective ZnA values. Each bar represents the mean of three independent qPCR runs (where each run was done in triplicate) ± SD. An asterisk indicates a P value <0.05 as determined by Student’s t test
Association of miR-328, miR-193a and miR-193b with the polysomal ribonucleoprotein fraction
The association of these small RNAs with polysomes was examined. We reasoned that if these miRNAs target Zip5, they should be enriched in the polysomal RNP fraction but might shuttle between the soluble and the polysomal RNP fractions in a zinc-responsive manner. RNA extracts were prepared from both the soluble and the polysomal RNP fractions of VYS taken from animals that were ZnA, ZnD or zinc-repleted by an oral gavage of zinc. The nucleolar snoRNA-202 was almost entirely depleted from the soluble and the polysomal RNP fractions (Fig. 5a) suggesting complete removal of large organelles such as nuclei. Total RNAs from ZnA and ZnD VYS were resolved on a denaturing gel (Fig. 5b, left panel) as were RNAs from the soluble and the polysomal RNP fractions from ZnA, ZnD, and zinc repleted VYS (Fig. 5b, right panel). These RNAs demonstrated distinct and repeatable banding patterns, confirming effective fractionation of the lysates. Furthermore, both Zip5 and Zip4 mRNAs were significantly enriched in the polysomal RNP fractions as noted previously (Weaver et al. 2007). The induction of Zip4 mRNA in the ZnD samples confirmed the ZnD state and the intermediate value for the 5 h gavage sample confirmed those VYS were in the adaptive transition returning to ZnA levels. We then examined the fold enrichment of the various miRNAs in the soluble and the polysomal RNP fractions relative to their respective abundance in the ZnA soluble fraction. Each of these miRNAs were each at least 4-fold enriched in the polysomal RNP fraction (Fig. 5d) and miR-328 was 10-fold enriched in that fraction. There were no significant zinc-dependent changes in the association of these miRNAs with the polysomal RNP fraction.
The Zip5 3′-UTR enhances translation when zinc is available and this function depends on the stem-loop as well as the miRNA seed sites
To study the functional significance of the Zip5 3′-UTR stem-loop and miRNA seeds-sites, we employed a rat extraembryonic endoderm XEN cell line previously shown to express extraembryonic endoderm markers (Soares et al. 1987). Extraembryonic endoderm is known to express and regulate ZIP5 in response to zinc (Dufner-Beattie et al. 2004; Weaver et al. 2007), but the rat XEN cells did not contain significant levels of Zip5 mRNA (data not shown). However, the XEN cells do express miR-328, miR-193a and miR-193b (Fig. 4) and do exhibit specific Zip5 3′-UTR-protein interactions indistinguishable from VYS (Fig. 3). To determine the structure–function relationships in the Zip5 3′-UTR, we engineered expression vectors in which the firefly luciferase coding sequence has the full-length mouse Zip5 3′-UTR cloned downstream of its stop codon (Figs. 6a, 7). In addition to the full-length wild-type mouse Zip5 3′-UTR, this UTR with the miRNA seed-sites scrambled and/or the stem-loop deleted were examined. The nucleotide sequences for the various UTR structures are shown with detail in Fig. 6b and diagrammatically in Fig. 7 for each given experiment. The firefly luciferase-UTR vectors were transfected into XEN cells along with a renilla luciferase expression vector internal control and the effects of zinc were assessed (Fig. 7). Raw renilla luciferase values are shown since zinc deficiency impacts global translation and firefly luciferase values are normalized to renilla luciferase in these experiments.
Fig. 7.
Functional analyses of the Zip5 3′-UTR in transfected XEN cells. XEN cells were cultured in ZnA or ZnD media for 30–48 h and then transfected with the luciferase Zip5 3′-UTR reporter constructs. The UTR structure is diagramed above each panel (see Fig. 6 for sequence details). Some ZnD transfected cells had 40 μM zinc added during the last 6–8 h of culture (ZnR). All firefly luciferase values were normalized to co-transfected renilla luciferase values. Each bar represents the mean of 3–4 replicas ± SD. An asterisk indicates a P value <0.05. All sample means were compared to the respective wild-type ZnA mean for the given experiment. a The wild-type (WT) and seed site (SS) mutant UTR vectors were also co-transfected with either an antagomiR vector expressing GFP and siRNA targeting LacZ (αLacZ) or antagomiR vectors expressing GFP and siRNAs targeting miR-193a and miR-328 (α193/328), as described in detail in the “Materials and methods” section. b Concatamerized wild-type Zip5 3′-UTR (WT 3×) and concatamerized Zip5 3′-UTR with scrambled seed sites (SS 3×) were transfected with a GFP expression vector as carrier. c The full length wild-type Zip5 3′-UTR (WT) or the Zip5 3′-UTR with deletion of the stem-loop (SL) were transfected with a GFP expression vector as carrier. Two independent experiments are shown. d The full length wild-type Zip5 3′-UTR (WT) or the Zip5 3′-UTR with both scrambled seed sites and deletion of the putative stem-loop (DM) were transfected with a GFP expression vector as carrier
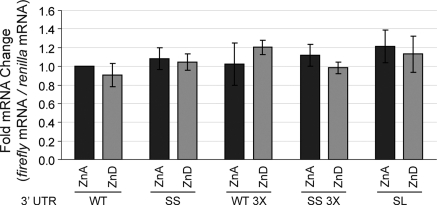
The full-length wild-type Zip5 3′-UTR (Fig. 6b; WT) and a mutant with scrambled miRNA seed sites (Fig. 6b; SS) were examined (Fig. 7a). Disruption of the miRNA seed sites alone significantly reduced expression of firefly luciferase vector when zinc was adequate in the culture medium. The level of expression of the mutant with scrambled miRNA seed sites in ZnA medium was indistinguishable from that of the wild-type 3′-UTR reporter in ZnD medium. Zinc deficiency caused about a 40% reduction in the activity of the wild-type UTR expression vector, but did not significantly further reduce the level of expression of the mutant with scrambled seed sites (Fig. 7a). Restoring zinc in the culture medium restored expression of the vector with the wild-type Zip5 3′-UTR but had little effect on the expression of the vector with scrambled miRNA seed sites. Quantitative real time PCR analysis demonstrated that changes in firefly and renilla luciferase activity in the transfected cells did not reflect changes in the abundance of these mRNAs (Fig. 8) but rather reflected changes in the accumulation of these proteins, suggestive of translation regulation.
Fig. 8.
Examination of steady state levels for firefly and renilla encoding mRNAs in transiently transfected XEN cells. XEN cells were cultured and transfected as described in the legend to Fig. 7. The following constructs were examined: The wild-type Zip5 3′-UTR (WT); this UTR with scrambled seed sites (SS); this UTR bearing a deletion of the stem-loop (SL); a concatamer of wild-type Zip5 3′-UTR (WT 3×) and a concatamer of this UTR with scrambled seed sites (SS 3×) (see Fig. 6 for details). Total RNA from ZnA (dark gray bars) or ZnD (light gray bars) transfected XEN cells was reverse transcribed and amplified via qPCR to measure relative abundance of firefly and renilla luciferase mRNAs. The ΔΔCt method was used to interpret qPCR results. All firefly 2−ΔΔCt values were first normalized to renilla 2−ΔΔCt values. The WT ZnA normalized value was set to 1 and the 2−ΔΔCt values for all other samples were reported as relative fold changes to the WT 1 × ZnA value. Each bar represents the mean of three independent qPCR runs (where each run was done in triplicate) ± SD. Student’s t test was used to determine significance
The roles of miR-328 and miR-193a in the regulation of ZIP5 translation was further examined by co-expressing antagomiRs to these miRNAs in the transfections in order to disrupt miRNA function(s). AntagomiRs were transfected 48 h before the luciferase and renilla reporter vectors and the transfected cells were cultured as above. Co-transfection of the anatogomiRs did not affect the activity of the wild-type 3′-UTR when zinc was adequate or further repress its expression when zinc was deficient (Fig. 7a). However, zinc-responsive upregulation for the wild-type 3′-UTR reporter by zinc repletion was attenuated (Fig. 7a). The antagomiRs were not as robust as the seed site mutations and were not able to delineate the individual effects of the two miRNAs. This may reflect the incomplete loss of function of these apparently abundant miRNAs.
Other studies have shown that concatamerized UTRs often can enhance reporter gene regulation. Concatamerization of the wild-type or the seed site mutant 3′-UTRs was therefore examined (Fig. 7b). The expression of the firefly luciferase vector with three tandem copies of the wild-type UTR (with only the last copy containing the polyA signal) was resistant to the effects of zinc deficiency and the basal expression of firefly luciferase was unaffected by the seed site mutations. However, both UTR constructs retained some responsiveness to zinc repletion in the culture medium. These results suggest that sequences in the Zip5 3′-UTR in addition to the miRNA seed sites also contribute to translational control of Zip5 mRNA.
The role of the stem-loop structure was therefore examined next (Fig. 7c). Compared with the wild-type UTR, deletion of the stem-loop significantly reduced basal expression of firefly luciferase in ZnA medium, attenuated its further repression by zinc deficiency and its induction by zinc repletion. These results were similar to what was observed when the seed sites in this UTR were mutated (Fig. 7a) and these data indicate that the Zip5 3′-UTR stem-loop also contributes to translational activation of ZIP5 in response to zinc availability. As noted above, these changes in firefly and renilla luciferase expression in the transfected cells did not reflect changes in the abundance of these mRNAs (Fig. 8).
Finally, we examined the effects of mutating both the stem-loop and the miRNA seed sites on the expression of firefly luciferase (Figs. 6b (DM), 7d). The double mutant UTR yielded results similar to those obtained with either the stem-loop or the seed site mutation alone. Taken together, these studies of the Zip5 3′-UTR suggest that both the stem-loop structure and the redundant miRNA seed sites participate in the regulation of Zip5 mRNA translation in response to zinc. These structures do not appear to function synergistically or additively.
Discussion
We previously reported that the adaptive response to dietary zinc in mice involves the rapid post-transcriptional regulation of Zip5 in the intestine and pancreas, but also in the visceral yolk sac during pregnancy (Dufner-Beattie et al. 2004; Weaver et al. 2007). Those studies suggested that Zip5 is not regulated by futile protein degradation during zinc deficiency but instead Zip5 mRNA remains polysome-associated and can be rapidly translated when adequate zinc is restored. Zip5 protein is localized to the basolateral membranes of intestinal enterocytes, visceral yolk sac endoderm, and pancreatic acinar cells (Dufner-Beattie et al. 2004; Weaver et al. 2007) and appears to be unique thus far among the Slc30a and Slc39a families of zinc transporters with regard to positive translational regulation in response to zinc. We hypothesized that Zip5 expression is regulated by a translational stall mechanism that is zinc-responsive (Weaver et al. 2007). Herein, we sought to determine the molecular mechanisms of this zinc-responsive translational regulation.
The role of the iron-responsive element (IRE) stem-loop structure in translational control of the ferritin heavy and light chain mRNAs is well documented (Leibold et al. 1990; Walden et al. 2006). The lowest free energy fold for the 28 nucleotide IRE has a ΔG value of −7.32 kcal/mol. The ferritin heavy and light chain mRNAs have IREs in their 5′ UTRs which provides a binding site for IRP leading to blocked translation of these mRNAs during iron deficiency (Leibold et al. 1990; Walden et al. 2006). Other mRNAs contain IREs and are also regulated by iron availability either at the level of mRNA stability or translation. In contrast, the lowest predicted free energy fold for the 34 nucleotide Zip5 stem-loop (ΔG = −13.06 kcal/mol) is somewhat more stable than that of the IRE and an in vitro protein binding assay with a Zip5 3′-UTR probe indicated constitutive binding that is not zinc-responsive.
The Zip5 3′-UTR stem-loop appears to function as a positive regulator of ZIP5 translation in response to zinc. The stem-loop sequence is unique in the mouse and human genomes. Thus, its function in control of translation may be unique to Zip5 mRNA. However, the 18 bases (bases 15–32 of the Zip5 stem-loop) of the terminal loop and 3′ stem (CACCACAGGAATGGAGGC) is exactly repeated in a very large intron (50,000 nucleotides long) in the mouse Runx1 transcription factor gene but was not found in the human Runx1 gene. Runx1 is involved in myeloid cell lineage differentiation and hematopoiesis, a zinc-dependent process (Elagib et al. 2003; Nagamura-Inoue et al. 2001), but the significance of this sequence in mouse Runx1, if any, is unknown. Further studies are required to elucidate the molecular mechanisms involved in the positive control of Zip5 mRNA translation by its 3′-UTR stem-loop structure.
MicroRNAs have been implicated in many post-transcriptional regulatory mechanisms, including P-body localization of mRNAs, mRNA degradation, translation initiation, translation stall, and even translation upregulation (Fabian et al. 2010). Most numerous are reports of mechanisms resulting in either inhibition of cap-dependent initiation of translation or mRNA deadenylation and decay (Fabian et al. 2010). However, miRNA-mediated upregulation of tumor necrosis factor α mRNA translation, an AU-rich element containing mRNA, and other target mRNAs has been reported during cell cycle arrest (Vasudevan et al. 2007, 2008; Vasudevan and Steitz 2007). Translational control often involves the 3′-UTR of an mRNA and the Zip5 3′-UTR is highly conserved across multiple mammalian species and contains redundant seed sites for two miRNAs (miR-328 and miR-193a). Redundant miRNA seed sites are a common feature of mRNA targets. None of the other Slc30a (Znts) or Slc39a (Zips) zinc transporters are predicted targets of miR-328 or miR-193a. These miRNAs fulfill several criteria which support the concept that they target Zip5 mRNA. Computational prediction identifies these miRNA-mRNA seed pairs in the conserved Zip5 3′-UTR and ΔG analysis of the 3′-UTR suggests that these miRNAs target accessible regions. Furthermore, these miRNAs are co-expressed in tissues which express abundant Zip5 mRNA in vivo, and they are, at least in part, associated with polysomes in these tissues. Finally, transfection experiments revealed that these miRNA seed sites are important for active translation of Zip5 mRNA when zinc is replete, and antagomiRs against these miRNAs attenuated translation of a firefly luciferase reporter gene. Taken together, these studies provide strong evidence that miR-328 and miR-193a participate in the regulation of Zip5 mRNA translation when zinc is replete. The mechanism of this regulation is not entirely understood but an mRNA degradation mechanism appears unlikely under these conditions. We previously reported that Zip5 mRNA abundance is unchanged in vivo with zinc availability in wild-type animals (Weaver et al. 2007) and the transfection data reported herein do not appear to reflect changes in firefly luciferase mRNA levels. Inhibition of translation initiation or a P-body localization mechanism seem unlikely given that Zip5 mRNA remains polysome-associated in vivo (Weaver et al. 2007).
It should also be noted that ZIP5 protein is destabilized in vivo during zinc deficiency whereby it is internalized and degraded (Weaver et al. 2007). Clearly this mechanism contributes significantly to the dramatic loss of ZIP5 during zinc deficiency, and may explain why the Zip5 3′-UTR alone was insufficient to mediate a complete loss of the firefly luciferase protein during zinc deficiency in our transfection experiments. Therefore, our transfection studies can only assay the translational control component of the overall Zip5 regulatory mechanism.
The results reported herein suggest that the Zip5 3′-UTR stem-loop and redundant miRNA seed sites function in a non-additive, non-synergistic manner. They appear to function in the same pathway. Thus, the Zip5 3′-UTR functions as an integrated, zinc-responsive translational regulatory element. To our knowledge, this represents the first demonstration of a zinc-responsive translational regulatory element and clarifies our previous findings that ZIP5 protein levels but not mRNA levels are zinc-responsive (Dufner-Beattie et al. 2004; Weaver et al. 2007). Recently, miR-398 in Arabidopsis thaliana was reported to regulate both cytosolic Cu/Zn superoxide dismutase and Cox5b-1 (a cytochrome c oxidase subunit) by causing the degradation of these mRNAs in response to copper limitation (Yamasaki et al. 2007). In addition, the plastocyanin mRNA was similarly down-regulated by miR-397, miR-408, and miR-857 (bdel-Ghany and Pilon 2008). Those findings provide precedent for miRNA-mediated gene regulation in metal homeostasis and further suggest that more than one mechanism of action for miRNAs in metal homeostasis exists.
The current findings provide insights into the control of zinc-responsive translational regulation. We report the first demonstration of a conserved RNA stem-loop, interacting with two miRNAs in this regulatory mechanism. We speculate that the complex interaction of an RNA secondary structure with miRNPs serves to fine-tune zinc homeostasis simultaneously in multiple tissues in a cell-autonomous manner. In conclusion, the Zip5 3′-UTR functions as an integrated zinc-responsive element to control Zip5 translation in response to changes in zinc availability.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
This work was funded, in part, by a National Institutes of Health grant to GKA (DK63975). BPW was supported, in part, by a KU Medical Center Biomedical Research Training Program Scholarship and by an ARRA supplement award (DK63975-8S1). We thank Jim Geiser in the lab for excellent technical assistance, Dr. Christer Hogstrand (King’s College London) for helpful discussions in initiating our miRNA studies, and Dr. Yi M. Weaver (CU Boulder) for helpful discussions in data analysis. We also thank Dr. Mohammad Rumi and Dr. Michael Soares (KUMC) for the generous gift of XEN cells.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Benjamin P. Weaver, Email: Benjamin.Weaver@Colorado.edu
Glen K. Andrews, Email: gandrews@kumc.edu
References
- Afonyushkin T, Vecerek B, Moll I, Blasi U, Kaberdin VR. Both RNase E and RNase III control the stability of sodB mRNA upon translational inhibition by the small regulatory RNA RyhB. Nucleic Acids Res. 2005;33:1678–1689. doi: 10.1093/nar/gki313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard P, Yang Q, Marzluff WF, Clarke HJ. The stem-loop binding protein regulates translation of histone mRNA during mammalian oogenesis. Dev Biol. 2005;286:195–206. doi: 10.1016/j.ydbio.2005.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altuvia S, Zhang AX, Argaman L, Tiwari A, Storz G. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 1998;17:6069–6075. doi: 10.1093/emboj/17.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrick BA, Lee AL, Grendell RL, Derynck R. Inhibition of translation of transforming growth factor-β3 mRNA by its 5′ untranslated region. Mol Cell Biol. 1991;11:4306–4313. doi: 10.1128/mcb.11.9.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashizuka M, Fukuda T, Nakamura T, Shirasuna K, Iwai K, Izumi H, Kohno K, Kuwano M, Uchiumi T. Novel translational control through an iron-responsive element by interaction of multifunctional protein YB-1 and IRP2. Mol Cell Biol. 2002;22:6375–6383. doi: 10.1128/MCB.22.18.6375-6383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- bdel-Ghany SE, Pilon M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J Biol Chem. 2008;283:15932–15945. doi: 10.1074/jbc.M801406200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceman S, Nelson R, Warren ST. Identification of mouse YB1/p50 as a component of the FMRP-associated mRNP particle. Biochem Biophys Res Commun. 2000;279:904–908. doi: 10.1006/bbrc.2000.4035. [DOI] [PubMed] [Google Scholar]
- Dalton T, Fu K, Palmiter RD, Andrews GK. Transgenic mice that overexpress metallothionein-I resist dietary zinc deficiency. J Nutr. 1996;126:825–833. doi: 10.1093/jn/126.4.825. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufner-Beattie J, Wang F, Kuo YM, Gitschier J, Eide D, Andrews GK. The acrodermatitis enteropathica gene ZIP4 encodes a tissue-specific, zinc-regulated zinc transporter in mice. J Biol Chem. 2003;278:33474–33481. doi: 10.1074/jbc.M305000200. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Kuo YM, Gitschier J, Andrews GK. The adaptive response to dietary zinc in mice involves the differential cellular localization and zinc-regulation of the zinc transporters ZIP4 and ZIP5. J Biol Chem. 2004;279:49082–49090. doi: 10.1074/jbc.M409962200. [DOI] [PubMed] [Google Scholar]
- Dufner-Beattie J, Weaver BP, Geiser J, Bilgen M, Larson M, Xu W, Andrews GK. The mouse acrodermatitis enteropathica gene Slc39a4 (Zip4) is essential for early development and heterozygosity causes hypersensitivity to zinc deficiency. Hum Mol Genet. 2007;16:1391–1399. doi: 10.1093/hmg/ddm088. [DOI] [PubMed] [Google Scholar]
- Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray NK, Pantopoulos K, Dandekar T, Ackrell BAC, Hentze MW. Translational regulation of mammalian and Drosophila citric acid cycle enzymes via iron-responsive elements. Proc Natl Acad Sci USA. 1996;93:4925–4930. doi: 10.1073/pnas.93.10.4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N. microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol. 2005;1:e13. doi: 10.1371/journal.pcbi.0010013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess MA, Duncan RF. Sequence and structure determinants of Drosophila Hsp70 mRNA translation: 5′-UTR secondary structure specifically inhibits heat shock protein mRNA translation. Nucleic Acids Res. 1996;24:2441–2449. doi: 10.1093/nar/24.12.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- Kim BE, Wang FD, Dufner-Beattie J, Andrews GK, Eide DJ, Petris MJ. Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J Biol Chem. 2004;279:4523–4530. doi: 10.1074/jbc.M310799200. [DOI] [PubMed] [Google Scholar]
- Kong YW, Cannell IG, de Moor CH, Hill K, Garside PG, Hamilton TL, Meijer HA, Dobbyn HC, Stoneley M, Spriggs KA, Willis AE, Bushell M. The mechanism of micro-RNA-mediated translation repression is determined by the promoter of the target gene. Proc Natl Acad Sci USA. 2008;105:8866–8871. doi: 10.1073/pnas.0800650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, Macmenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37:495–500 [DOI] [PubMed]
- Kuhn DE, Martin MM, Feldman DS, Terry AV, Jr, Nuovo GJ, Elton TS. Experimental validation of miRNA targets. Methods. 2008;44:47–54. doi: 10.1016/j.ymeth.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P, Kao HL, Gunsalus KC, Pachter L, Piano F, Rajewsky N. A genome-wide map of conserved microRNA targets in C. elegans. Curr. Biol. 2006;16:460–471. doi: 10.1016/j.cub.2006.01.050. [DOI] [PubMed] [Google Scholar]
- Leibold EA, Guo B. Iron-dependent regulation of ferritin and transferrin receptor expression by the iron-responsive element binding protein. Annu Rev Nutr. 1992;12:345–368. doi: 10.1146/annurev.nu.12.070192.002021. [DOI] [PubMed] [Google Scholar]
- Leibold EA, Munro HN. Cytoplasmic protein binds in vitro to a highly conserved sequence in the 5′ untranslated region of ferritin heavy- and light-subunit mRNAs. Proc Natl Acad Sci USA. 1988;85:2171–2175. doi: 10.1073/pnas.85.7.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibold EA, Laudano A, Yu Y. Structural requirements of iron-responsive elements for binding of the protein involved in both transferrin receptor and ferritin mRNA post-transcriptional regulation. Nucleic Acids Res. 1990;18:1819–1824. doi: 10.1093/nar/18.7.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Yu Y, Fisher J, Nilsen TW. Evidence that microRNAs are associated with translating messenger RNAs in human cells. Nat Struct Mol Biol. 2006;13:1102–1107. doi: 10.1038/nsmb1174. [DOI] [PubMed] [Google Scholar]
- Mullner EW, Neupert B, Kuhn LC. A specific mRNA binding factor regulates the iron-dependent stability of cytoplasmic transferrin receptor mRNA. Cell. 1989;58:373–382. doi: 10.1016/0092-8674(89)90851-9. [DOI] [PubMed] [Google Scholar]
- Munro HN, Aziz N, Leibold EA, Murray M, Rogers J, Vass JK, White K. The ferritin genes: structure, expression, and regulation. Ann N Y Acad Sci. 1988;526:113–123. doi: 10.1111/j.1749-6632.1988.tb55497.x. [DOI] [PubMed] [Google Scholar]
- Muralidharan B, Bakthavachalu B, Pathak A, Seshadri V. A minimal element in 5′UTR of insulin mRNA mediates its translational regulation by glucose. FEBS Lett. 2007;581:4103–4108. doi: 10.1016/j.febslet.2007.07.050. [DOI] [PubMed] [Google Scholar]
- Nagamura-Inoue T, Tamura T, Ozato K. Transcription factors that regulate growth and differentiation of myeloid cells. Int Rev Immunol. 2001;20:83–105. doi: 10.3109/08830180109056724. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Soares MJ, Schaberg KD, Pinal CS, De SK, Bhatia P, Andrews GK. Establishment of a rat placental cell line expressing characteristics of extraembryonic membranes. Dev Biol. 1987;124:134–144. doi: 10.1016/0012-1606(87)90466-0. [DOI] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Steitz JA. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell. 2007;128:1105–1118. doi: 10.1016/j.cell.2007.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545–1549. doi: 10.4161/cc.7.11.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walden WE, Selezneva AI, Dupuy J, Volbeda A, Fontecilla-Camps JC, Theil EC, Volz K. Structure of dual function iron regulatory protein 1 complexed with ferritin IRE-RNA. Science. 2006;314:1903–1908. doi: 10.1126/science.1133116. [DOI] [PubMed] [Google Scholar]
- Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553–560. doi: 10.1016/j.molcel.2006.03.034. [DOI] [PubMed] [Google Scholar]
- Weaver BP, Dufner-Beattie J, Kambe T, Andrews GK. Novel zinc-responsive post-transcriptional mechanisms reciprocally regulate expression of the mouse Slc39a4 and Slc39a5 zinc transporters (Zip4 and Zip5) Biol Chem. 2007;388:1301–1312. doi: 10.1515/BC.2007.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H, bdel-Ghany SE, Cohu CM, Kobayashi Y, Shikanai T, Pilon M. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J Biol Chem. 2007;282:16369–16378. doi: 10.1074/jbc.M700138200. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.