SUMMARY
2-Cys peroxiredoxins (Prxs) play two different roles depending on the physiological status of the cell. They are thioredoxin-dependent peroxidases under low oxidative stress, and ATP-independent chaperones upon exposure to high peroxides concentrations. These alternative functions have been associated with changes in the oligomerization state from low (LMW) to high (HMW) molecular weight species. Here we present the structures of Schistosoma mansoni PrxI in both states: the LMW decamer and the HMW 20-mer, formed by two stacked decamers. The latter is the first structure of a 2-Cys Prx chaperonic form. Comparison of the structures sheds light on the mechanism by which chemical stressors, such as high H2O2 concentration and acidic pH, are sensed and translated into a functional switch in this protein family. We also propose a model to account for the in vivo formation of long filaments of stacked Prx rings.
INTRODUCTION
Oxidative stress is a potential hazard for protein folding. Under redox stress conditions, ATP-dependent foldases are no longer sufficient to cope with protein misfolding and cells employ ATP-independent holdases to counteract the new redox milieu (Kumsta and Jacob, 2009). Holdases are a particular branch of molecular chaperones, which prevent protein aggregation by placing client proteins in quarantine. Within this group, redox-regulated chaperones have a fascinating mechanism of action: they sense the redox level of the environment and translate this redox information into quaternary structure changes and functional activation. As an example, upon cysteine residues oxidation, heat shock protein 33 (HSP33) forms dimers and exposes hydrophobic patches at the N-terminus to bind non-native proteins (Winter et al., 2005).
Among the redox-regulated chaperones, peroxiredoxins (Prxs) have recently become the object of increasing attention, due to their ubiquitous distribution and implication in several human pathological conditions (Hofmann et al., 2002; Wood et al., 2003; Flohé et al., 2003; Sayed et al., 2006). Additionally, Prxs are moonlighting proteins: at high H2O2 concentrations they act as holdases, while in the absence of oxidative stress they are thioredoxin (Trx)-dependent peroxidases (Jang et al., 2004).
Typical 2-Cys Prxs are the largest and most widely distributed subfamily of Prx proteins (Soito et al., 2011). Their catalytic properties as antioxidant scavengers rely on the peroxidatic Cys (Cysp), a redox-active cysteine located in the first turn of helix α2 in the N-terminal region of the enzyme. Cysp reduces H2O2 by releasing H2O and forming a sulfenic acid derivative, Cysp-SOH. Cysp-SOH can be converted to a disulfide by the resolving cysteine (Cysr) located at the C-terminus of the other subunit in the obligate homodimer (Wood et al., 2003; Winterbourn and Hampton, 2008; Neuman et al., 2009). Oxidation of Cysp unwinds the first turn of the α2 helix, thus allowing Cysr to attack Cysp. The resulting disulfide bridge is reduced by Trx and/or glutathione, thus completing the enzymatic cycle (Wood et al., 2003; Neumann et al., 2009). The peroxidase activity of 2-Cys Prxs has been shown to be mediated by low-molecular weight (LMW) species. These can be dimers or (do)-decamers, made up of either six or five dimers arranged in a ring-like structure with pseudo six- or five-fold symmetry, respectively. In particular, the (do)-decamers are most abundant in the presence of reduced cysteines, whereas disulfide-bond formation shifts the equilibrium towards the dimers (Barranco-Medina et al., 2009).
At high H2O2 concentrations, before resolution by Cysr, Cysp-SOH may react with additional H2O2 forming sulfinic acid, Cysp-SO2H, and/or sulfonic acid, Cysp-SO3H. This overoxidation is associated with a change in function resulting in the loss of peroxidase activity and onset of chaperone activity. This change in biochemical function is coupled to a shift in the oligomerization equilibrium towards higher-order oligomers (HMW; Kumsta and Jacob, 2009; Jang et al., 2004; Barranco-Medina et al., 2009; Moon et al., 2005). In particular, the chaperone activity of yeast cytosolic PrxI has been attributed to different HMW species whose structures, determined by electron microscopy, consist either of rings with 5-fold symmetry or spherically-shaped particles (Jang et al., 2004). Moreover, overoxidation of yeast PrxI to Cysp-SO3H shifts the equilibrium towards a HMW form of about 530 kDa, which is proposed to be a double (do)-decamer that is responsible for the chaperone function (Lim et al., 2008). HMW species of several other 2-Cys Prxs have been shown by electron microscopy to be constituted of two or more stacked (do)-decameric rings (Wood et al., 2004; Gourlay et al., 2003). Further, in cultured epithelial cells under high oxidative stress, long filaments of stacked Prx rings have been detected and shown to alert cells of perturbations in peroxide homeostasis, thus determining cell cycle arrest (Gourlay et al., 2003; Phalen et al., 2006).
In vivo, formation of HMW forms is reversible, due to the presence of sulfiredoxin (Srx). Srx specifically reduces Cysp-SO2H, restoring the peroxidase activity and favouring the dissociation of HMW complexes (Lowther and Haynes, 2011). Recently, the complex between human PrxI and Srx has been solved by X-ray crystallography (Jönsson et al., 2008). This structure shows that Cysp of the Prx dimer is exposed on the protein outer surface and the C-terminus is completely unwound and lodged in a hydrophobic groove of Srx. Even though this 3D-complex has provided clues to understand the mechanism of recognition between 2-Cys Prxs and Srx, the principal substrate of Srx under physiological conditions is likely to be the HMW species (Lowther and Haynes, 2011).
At variance with the well characterized peroxidase activity of Prx, neither the atomic structures of HMW forms or the mechanism by which the quaternary assembly is modified and the holdase ability acquired have been elucidated yet. Here we present two distinct 3D crystal structures of cytosolic PrxI from the human parasite Schistosoma mansoni (GenBank: Thioredoxin peroxidase I, AF301002): the LMW decamer (herein called SmPrxI-LMW) and the HMW double decamer assembly (called SmPrxI-HMW). The latter represents the first crystallographic structure of a HMW assembly of a typical 2-Cys Prx.
S. mansoni is a causative agent of schistosomiasis, a debilitating parasitic disease affecting approximately 200 million people in tropical areas. The worm lacks catalase, and its glutathione peroxidases are not efficient scavengers of hydroperoxides; thus the oxidative stress generated by the host’s immune response is essentially counteracted by Prxs (Sayed et al., 2006; Sayed and Williams, 2004). As previously reported for other typical 2-Cys Prxs (Kumsta and Jacob, 2009; Jang et al., 2004), our experiments demonstrate that SmPrxI-LMW acts as a peroxidase, while SmPrxI-HMW is endowed with holdase activity. Given the high sequence and structural conservation of Prx proteins (Wood et al., 2003), the structure of SmPrxI-HMW may represent a general organization of 2-Cys Prx HMW species. Based on structural comparisons between SmPrxI-LMW and -HMW, we propose a mechanism explaining how chemical modifications of Cysp trigger the formation of HMW assemblies. Moreover, we propose a model of how the long filaments of stacked rings (Phalen et al., 2006) may be built.
RESULTS
Quaternary Assembly of SmPrxI as a Function of pH and Oxidation State
Size Exclusion Chromatography (SEC) experiments were carried out to assess the quaternary states of SmPrxI under different redox conditions and at different pH values. We analysed three samples: i) reduced SmPrxI at pH 7.4; ii) reduced SmPrxI at pH 4.2; iii) overoxidized SmPrxI-SO2H/SO3H at pH 7.4. The overoxidized SmPrxI derivatives were obtained by Trx-dependent reduction of the Cysp-Cysr disulfide bonded derivative in the presence of 50 mM H2O2, following the previously reported procedure (Sayed et al; 2006; Sayed and Williams, 2004).
Reduced SmPrxI at pH 7.4 eluted as one homogeneous peak centred around 250 kDa, compatible with a decameric assembly, and at pH 4.2 as a peak at 500 kDa (Figure 1A), indicating the presence of a 20-mer HMW species. The sample containing SmPrxI-SO2H/SO3H at pH 7.4 eluted with almost indistinguishable profiles as a peak with intermediate MW, suggesting the presence of HMW and LMW species in rapid equilibrium relative to the time of the experiment. Each fraction was collected and dialysed in order to assay the peroxidase and chaperone enzymatic activities.
Figure 1. Quaternary Assembly and Chaperone Activities of SmPrxI.
(A) SEC of SmPrxI was performed using a TSK G3000PWXL column equilibrated with a mix of PBS and Na-citrate. Three different protein samples were fractionated on the column: (▲) reduced SmPrxI in PBS/Na-citrate pH 7.4; (□) reduced SmPrxI in PBS/Na-citrate pH 4.2; (●) SmPrxI-SO2H/SO3H in PBS/Na-citrate pH 7.4. Horse heart cytochrome c was used as an internal reference (indicated by the arrow at 12.4 kDa).
(B) The chaperone activity of reduced and overoxidized SmPrxI was measured by the temperature-induced aggregation of citrate synthase (CS) from porcine heart (Sigma-Aldrich) at 43 °C in 50 mM Hepes pH 7.0: CS 1 μM (control) (■); CS 1 μM + reduced SmPrxI-monomer 10 μM (○); CS 1 μM + SmPrxI-SO2H/SO3H-monomer 0.5 μM (▲), 1 μM (□), 2 μM (●).
(C) The chaperone activity of reduced SmPrxI in 50mM Na-citrate pH 4.2 was measured using the same procedure as in Panel B: CS 0.8 μM (control) (▲); CS 0.8 μM + SmPrxI-SO2H/SO3H-monomer 10 μM (+); CS 0.8 μM + reduced SmPrxI-monomer 10 μM (×), 20 μM (□); 50 μM (●).
Peroxidase and Chaperone Activities
Reduced SmPrxI at pH 7.4 displays peroxidase activity with apparent steady state parameters KM(H2O2)=12 μM and kcat=1.1 s−1, at fixed concentrations of E. coli Trx (1.6 μM) and Trx-reductase (200 nM). The holdase activity of this form is negligible at pH 7.0 (Figure 1B), even in the presence of 10-fold excess of reduced SmPrxI. It was not possible to increase the molar ratio over 10:1 SmPrxI-monomer:citrate synthase (CS) due to the precipitation of reduced SmPrxI in the assay conditions (i.e., absence of salts and 43°C). These conditions are required to allow measurements of chaperone activity, since CS precipitation was prevented by even very small amounts of salts.
At pH 4.2, under reducing conditions, no peroxidase activity of SmPrxI could be detected, but a holdase activity, weaker than that observed for overoxidized Prx at pH 7.0, was present (Figure 1C). The apparent stoichiometry under these conditions is higher than for the overoxidized SmPrxI, approaching 20:1 SmPrxI-monomer:CS, possibly due to a reduction in affinity (Figure 1C).
Overoxidized SmPrxI-SO2H/SO3H at pH 7.0 was shown to be active as a holdase chaperone at stoichiometric concentrations of the SmPrxI-monomer with CS (Figure 1B). In the same conditions, peroxidase activity was not detected, as previously reported (Sayed et al, 2006).
At pH 4.2 SmPrxI-SO2H/SO3H was shown to be active as a holdase, even though to a lesser extent than at pH 7.0 (Figure 1C). The molar ratio SmPrxI-monomer:CS could not be increased over 10:1 due to the low solubility of SmPrxI-SO2H/SO2H in the assay conditions (see above).
Even though the holdase assay is semi-quantitative, it is possible to roughly estimate the relative activities of SmPrxI under the different conditions tested. Taking the ratio SmPrxI-monomer:CS 10:1 as reference condition, the protection activity of the different SmPrxI forms against CS precipitation is the following: 100% for overoxidized SmPrxI at pH 7; 50% for reduced SmPrxI at pH 4.2; 20% for overoxidized SmPrxI at pH 4.2; close to 0% for reduced SmPrxI at pH 7. This under estimates the actual holdase activity of overoxidized SmPrxI at pH 7, which reaches its asymptote at a lower SmPrxI-monomer:CS ratio than 10:1. Moreover, overoxidized SmPrxI increases the lag time of CS precipitation at pH 7, whereas at pH 4.2 the lag time varies scarcely, if at all.
Crystal Structures of SmPrxI
Following the results of SEC experiments, we set up crystallization trials in the experimental conditions required to stabilize either the LMW or the HMW species. We obtained two structures by molecular replacement: a decameric assembly (SmPrxI-LMW), under conditions similar to those yielding the SEC fraction showing peroxidase activity (i.e., reducing environment and pH 7.6); a double decamer (SmPrxI-HMW), under the same conditions yielding the single SEC peak with chaperone activity (i.e., reducing environment and pH 4.2).
SmPrxI-LMW: the Single Decamer
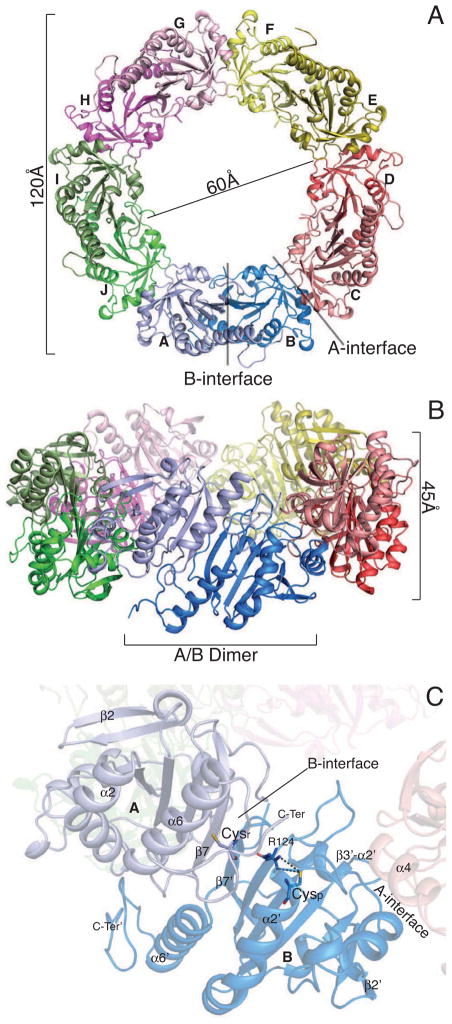
The 3.0 Å resolution electron density map of reduced SmPrxI-LMW allowed us to fit almost all 185 amino acids for each of the 10 polypeptide chains, which were identified with letters from A to J (see Tables 1 and S1). The overall structure of SmPrxI-LMW (Figure 2A–B) consists of a decameric ring with 52 point group symmetry and is superimposable with that of homologous proteins crystallised in the reduced form (Hall et al., 2009). The ring is 4.5 nm thick and has outer and inner diameters of 12 and 6 nm, respectively.
Table 1.
Data Collection and Refinement Statistics
| Data processing
| ||
|---|---|---|
| SmPrxI-LMW | SmPrxI-HMW | |
| Crystallization conditions | 20% PEG 3350, 20mM DTT, 0.2M potassium citrate, pH 7.6 | 20 % PEG 8K, 0.2 M sodium chloride, 0.1 M phosphate-citrate buffer, pH 4.2 |
| PDB code | 3ZTL | 3ZVJ |
| Space group | P1 | P21 |
| Unit cell dimensions (Å) | a= 49.51 | a= 126.91 |
| b= 116.90 | b= 204.94 | |
| c= 117.35 | c= 126.84 | |
| α= 69.12°, β= 92.00°, γ =86.93° | α= γ =90.00°, β= 114.60° | |
| Resolution range (Å) | 43.0-3.0 (3.16-3.0) | 30.0-3.0 (3.11-3.0) |
| <I/σ(I)> | 9.0 (3.2) | 6.1 (2.8) |
| % Completeness | 98.9 (98.7) | 96.0 (91.6) |
| Multiplicity | 4.0 (4.0) | 3.6 (2.6) |
| Rmerge raw data (%) | 0.15 (0.47) | 0.23 (0.40) |
| Rmerge detwinned data | No twinning was detected | 0.13 |
| Twinning fraction | No twinning was detected | 0.5 |
| No. molecules/asymmetric unit | 10 | 20 |
|
Refinement statistics
| ||
| Resolution range (Å) | 29.8–3.0 | 29.8–3.0 |
| No. reflections | 46759 | 112406 |
| Rfactor/Rfree | 0.21/0.26 | 0.27/0.29 |
| No. atoms | 14405 | 26995 |
| Mean B-factor | 27.1 | 69.6 |
| RMS bond lengths deviations | 0.0040 Å | 0.0040 Å |
| RMS bond angles deviations | 0.93° | 0.72° |
Values in the high resolution shell are shown in parentheses
Figure 2. Crystal Structure of SmPrxI-LMW.
(A) Ribbon representation of SmPrxI-LMW structure (top view). The pentameric arrangement of the dimers is highlighted. The internal and external diameters of the decamer are shown, together with the B-interface (A/B dimer) and A-interface (B/C monomers). Dimers are indicated by five different colours. Within each dimer, the two monomers have different shades. See also Figure S1.
(B) Ribbon representation of SmPrxI-LMW structure (front view). The A/B dimer and the thickness of the decamer are indicated. See also Figure S2.
(C) Overall fold of one SmPrxI-LMW dimer. The catalytically active Cysp (Cys48), within the first turn of the α2 helix, and Cysr (Cys169), in the C-terminal tail, together with Arg124 (responsible for lowering the pKa of Cysp) are displayed as sticks. The active site is in the fully-folded (FF) conformation (see text). Structure elements that undergo conformational changes between SmPrxI-LMW and SmPrxI-HMW (i.e., α2 and α6 helices, β2 strand, C-terminus), together with the A-interface (helix α4 and β3–α2 loop) and B-interface (β7 strands), are indicated. See also Figure S3.
Each subunit shows the same secondary structure arrangement occurring in all typical 2-Cys Prxs solved to date (Wood et al., 2003). The overall topology (Figure S1) consists of a modified Trx fold with a seven-stranded β-sheet (β1–β7) sandwiched by: two α-helices (α1 and α5) and one short β-hairpin on the internal face of the decameric ring; four α-helices (α2, α3, α4 and α6) on the external face (Figure 2A–B). The two protomers of each obligate homodimer are correlated by a pseudo 2-fold axis perpendicular to the plane containing the extended β-sheet (Figure 2C) that, in turn, forms an angle of 27° with respect to the pseudo 5-fold symmetry axis. Both the monomer-monomer interface within each dimer and the dimer-dimer interface (called B- and A-interfaces, respectively; Aran et al., 2009) resemble those observed in the structures of other typical 2-Cys Prx (Aran et al., 2009; Barranco-Medina et al., 2008; Nelson et al., 2011).
The intra-dimeric B-interface (Figures 2B-C and S2) is symmetrical and is contributed by: i) the antiparallel β7-β7′ strand interaction from each of the two subunits, through which the continuous β-sheet of the dimer is formed (the apex indicates structural elements from the interacting monomer); ii) on the internal face of the ring, the N-terminal region of one subunit interacts with the β–hairpin and part of the α5′ helix of the other subunit; iii) on the external face of the ring, the β7-α6 loop interacts with the α6′ helix, and the interacting α2 helix and C-terminal region of the other subunit comprise the active site.
The active-site (Figure 2C) is in the fully-folded (FF) conformation (Wood et al., 2003): residues 47–50, including the reduced Cysp (Cys48), form the first turn of the α2 helix, and the C-terminal tail (residues 165–185), containing Cysr (Cys169), is folded in a hairpin-like structure.
The sulphur atom of Cysp is engaged in an electrostatic interaction with the NH2 atom of Arg124 (3.6 Å), but it is too distant from the sulphur atom of Cysr to form a disulfide bridge with it (~13 Å in the A/B dimer). This implies that, during the catalytic cycle, a structural rearrangement must occur to allow the formation of a disulfide bridge between Cysr and Cysp (Wood et al., 2003).
The inter-dimeric A-interface is also symmetrical, and involves contacts between: i) the α4 helix of one subunit and α4′ helix and β3′-α2′ loop of the adjacent subunit, on the external side of the decameric ring; and ii) the β5-α5 loop of one subunit and the short β-hairpin of the other subunit on the ring internal side (Figures 2B–C and S3).
The Structure of SmPrxI-HMW: Two Stacked Decamers
Reduced SmPrxI-HMW crystallized at pH 4.2 is a 20-mer arranged in two stacked decamers (Figure 3). Since both decamers forming the SmPrxI-HMW have a 52 point group symmetry, they can stack one on top of the other without a preferential orientation. The 3 Å resolution electron density map was clearly visible for residues 3–46 and 50–165 for all 20 subunits, while density for residues 47–49 is missing in some subunits (Table S1). Only a few residues of the C-terminal tail could be fitted in the electron density map and they did not assume regular secondary structures.
Figure 3. Crystal Structure of SmPrxI-HMW.
(A) Overall surface of SmPrx-HMW. The HMW species is constituted by two decamers (grey and orange, respectively) locked one above the other along the five-fold symmetry axis.
(B) The upper and lower decamer of SmPrxI-HMW (view from top) appear to be rotated by 18° around the five-fold symmetry axis. To highlight this feature, the Cα atoms of residue Phe161 of adjacent monomers within each of the two decamers are joined by grey and orange lines, respectively.
(C) Secondary structure elements forming the pawls allowing the locking of the cogwheel rings at the R-interface, namely the α2 and α6 helices and the β2 strand from the B monomer (upper ring) and K and M monomers (lower ring), are shown in dark grey and orange, respectively. The thickness of the double-decamer is also indicated. See also Figure S4.
(D) Magnification of the contacts between the secondary structure elements of the two stacked decamers at the R-interface. The α2 and α6 helices create a negatively charged region due to the orientation of their carbonyl groups. The positively charged residues, Lys164 and His165 at the end of α6, stabilize this architecture. The β2 strands are joined by polar contacts between Glu21 and Lys23 of chain B and Glu21 of chain M. Structural elements of the B monomer are indicated with apexes.
(E) The cogwheel model. Schematic representation of the exploded linearized rings of SmPrxI-HMW. The secondary structure elements (α2, α6 and β2) implicated in the self-assembly of the HMW species are shown.
The interface between the two decameric rings stacked on top of each other in SmPrxI-HMW is R (ring). The R-interface (Figures 3C–D and S4) comprises two regions: (i) the α2 and α6 helices of subunit B (upper ring) are in contact with the equivalent helices of subunit K (lower ring); and (ii) strand β2 of subunit B interacts with the equivalent strand β2 of subunit M (lower ring). If we compare each single decamer to a cogwheel, the secondary structure elements present in these regions may resemble pawls through which the two rings are interlocked (Figure 3D–E). The observed joints are made possible by a rotation of 18° around the five-fold symmetry axes of one decamer with respect to the other (Figure 3B). The four helices α2 and α6 of the adjacent B and K monomers are oriented in such a way that all of their carbonyl groups, and hence the helix dipoles, point towards the R-interface. This creates a negatively charged area centred on residues Lys164 and His165, located at the end of α5 helix of both B and K subunits (Figure 3D). The dipoles of the peptide bonds are expected to raise the pKa of His165 at the end of the α6-helix. Thus, the positively charged side-chains of Lys164 and His165 are expected to stabilize the negative charge of the helix dipoles (Sali et al., 1988), thus contributing to the stabilization of the stacked decamers. On the same pawl, Glu21 and Lys23, belonging to the β2 strands, stabilize the interface between monomers B and M with two polar contacts (Figure 3D).
Comparison Between SmPrxI-LMW and SmPrxI-HMW
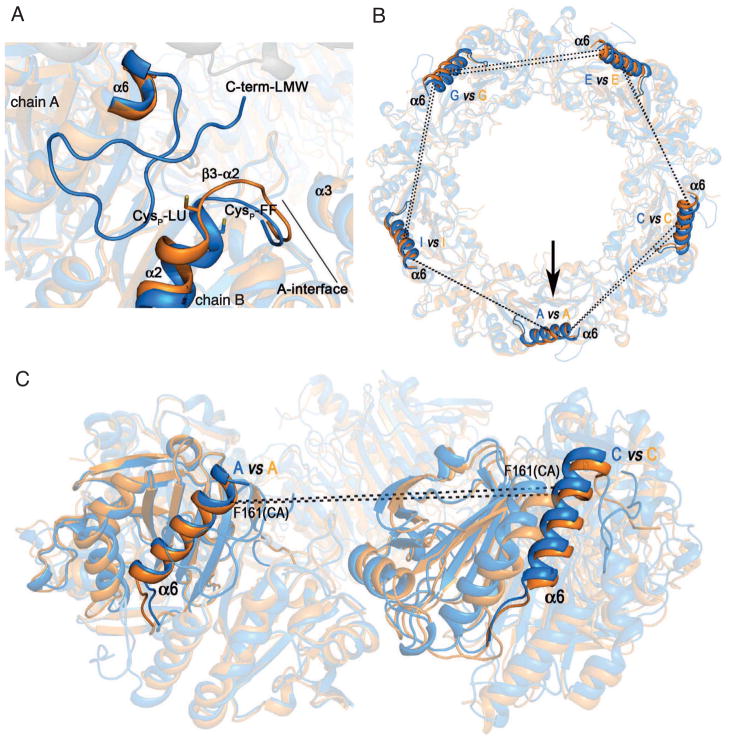
The monomers constituting SmPrxI-HMW have the same overall fold as those in SmPrxI-LMW. Optimal superposition of the Cα atoms of residues in the common regions of LMW and HMW monomers (i.e., 3–46, 50–165, see Table S1) provided root mean square deviation (RMSD) values ranging between 0.8 and 1.0 Å (Table S2). The highest differences occur in the active site. The FF conformation observed in SmPrxI-LMW becomes locally unfolded (LU) in SmPrxI-HMW (Figure 4A), and electron density for a large number of C-terminal residues is missing in the electron density map (Table S1), similarly to what previously reported for other Prx structures in the dimeric form (Wood et al., 2003; Lowther and Haynes, 2011; Jönsson et al., 2008). Furthermore, unwinding of the first turn of the α2 helix in SmPrxI-HMW leads to exposure of Cysp (Figure 4A and Movies S1 and S2).
Figure 4. Tertiary and Quaternary Structural Changes Between SmPrxI-LMW and SmPrxI-HMW.
(A) Superposition of the A/B dimers of SmPrxI-LMW (blue) and SmPrxI-HMW (orange) highlighting the changes in the active sites. The first turn of the α2 helix and the β3-α2 loop in the fully folded (FF) conformation of SmPrxI-LMW and in the locally unfolded (LU) conformation of SmPrxI-HMW are displayed as cartoons. The side-chains of Cysp of both structures are displayed as sticks. The C-terminal extension is visible in the electron density map only in SmPrxI-LMW.
(B) Changes in the quaternary structure between adjacent dimers of SmPrxI-LMW and SmPrxI-HMW decamers (view from the top). Distances between the Cα atoms of Phe161 in monomers A, C, E, G and I are shown as dashed lines after optimal superposition of the A/B dimers (highlighted by an arrow) of SmPrxI-LMW (cyan) and SmPrxI-HMW (orange). The displacement of the α6 helices increases with distance from the best-matched dimers.
(C) Changes in the quaternary structure between two adjacent dimers (view from the side of the ring). Only the A/B and C/D dimers of both SmPrxI-LMW (blue) and SmPrxI-HMW (orange) are displayed. The distances between the Cα atom of Phe161 within the α6 helix of monomer A and the equivalent atom of monomer C in SmPrxI-LMW and SmPrxI-HMW are shown as dashed lines, as a marker of the quaternary changes between the two structures.
The homodimer represents both the minimal structural unit of Prxs and the smallest unit that is populated during the catalytic cycle. Superposition of the SmPrxI-LMW and SmPrxI-HMW dimers in the conserved regions provided RMSD values ranging between 0.8 and 1.0 Å, very similar to those obtained for the monomers (Table S2). The main change associated with the structural transition from LMW to HMW form occurs at the B-interface. Indeed, the pattern of contacts between the α2 helix of one monomer and the C-terminus of the adjacent monomer at the B-interface is almost completely different between the two forms (Figure S2 and Movies S1 and S2). The two structural features distinctive of HMW dimers (i.e., unfolded C-terminus and unwound α2 helix) are strongly correlated, since a folded C-terminus would clash with the unwound α2′ helix at the B interface.
The HMW and LMW decamers differ because of both tertiary and quaternary structural changes. The RMSD values calculated after optimal superposition of the LMW and HMW forms in the common decamer regions are, on average, 1.1Å, a value that is slightly but significantly higher than the average of those calculated after optimal superpositions of both dimers and monomers (Table S2). The local unfolding of the β3-α2 loop (Figure 4A) results in a pressure on the α3 helix of the adjacent dimer, which, in turn, determines a slight increase of the A-interface in the region near Cysp. However, the A-interface is perturbed to a lesser extent than the B-interface (Figure S3).
The quaternary structural changes affecting the relative position of the dimers within LMW and HMW decamers can be visualized using as a marker the distance between residues belonging to the α6 helix, whose conformation does not change in the two forms. Figure 4C shows the α6 helices of monomers A (within the A/B dimer) and C (within the adjacent C/D dimer) after optimal superposition of the A/B dimers belonging to SmPrxI-LMW and SmPrxI-HMW, respectively (highlighted by an arrow in Figure 4B). In SmPrxI-HMW, the distance between the Cα atoms of the Phe161 residue within the α6 helix of any monomer and the equivalent residue in the adjacent dimer is on average shorter by 1.5 Å than in SmPrxI-LMW (Figure 4C). In view of the strong geometrical constraints imposed by the closed-ring arrangement of both SmPrxI-LMW and SmPrxI-HMW decamers, and the observed structural conservation of the A-interfaces, we believe that the structural changes highlighted in Figures 4B–C are significant.
Superposition of SmPrxI-LMW to each decamer of SmPrxI-HMW revealed that unfolding of the C-terminal region downstream of the α6 helix of each subunit is required to allow the stacking of the decamers, as it removes a protrusion on the ring surface that otherwise would clash with the other ring at the R-interface (Figure S4 and Movie S1).
In summary, the quaternary changes at the R-interface appear to be related to: i) tertiary changes (unfolding of the C-terminus and unwinding of the first turn of α2 helix), allowing the direct ‘vertical’ interaction between monomers B and K (Figure 3C–E); ii) propagation of tertiary changes ‘horizontally’ through the A-interfaces within the single ring. As an example, the β3-α2 loop (residues 45–50, Figure 4A) of monomer B protrudes towards the α3 helix of monomer C; this in turn moves as a rigid body (see RMSD values in Table S2) making the α6 helix act as a pawl for interlocking the decamer cogwheels (Figure 4B–C).
DISCUSSION
Prxs sense the redox environment by changing the oxidation state of the catalytic cysteine thiol to a sulfinic/sulfonic acid (-SO2H/-SO3H). This chemical modification is at the basis of the moonlighting behaviour of Prxs, in that it induces the formation of HMW complexes, which are competent for the molecular chaperone activity and are implicated in cell signalling (Jang et al., 2004; Phalen et al., 2006; Hall et al., 2009). Neither the 3D structure of these HMW species nor their mechanism of assembly have been reported to date. Comparison of the structures of SmPrxI-LMW (the decameric ring) and SmPrxI-HMW (the double decamer) here presented reveal the quaternary transitions by which Prxs sense and respond to chemical stressors (high oxidant concentration and acidic pH), switching to HMW complexes endowed with chaperone activity. Moreover, they suggest where unfolded proteins may be bound by the HMW forms, and how these may pile up to form long filaments.
The following events are involved in the activation of SmPrxI and of typical 2-Cys Prxs: (i) overoxidation of the sulphur atom of Cysp, which causes destabilization of the α2 helix; (ii) concerted tertiary variations, i.e. unwinding of the first turn of the α2 helix; unfolding of the C-terminal tail; protrusion of the β3-α2 loop towards the nearby dimer; and (iii) quaternary structural changes leading to the formation of the HMW species (Movies S1 and S2).
The first important questions to address are why both oxidative and acidic stimuli should cause the same tertiary and quaternary structural changes in SmPrxI, and whether SmPrxI-HMW crystallized at acidic pH is representative of the structure acquired by the same protein or other 2-Cys Prxs when exposed to oxidative stress. It is known that under oxidative stress SmPrxI, as well as other eukaryotic 2-Cys Prx, switches its structure and its function (from LMW/peroxidase to HMW/chaperone), both in vitro and in vivo (Kumsta and Jacob, 2009; Jang et al., 2004). SEC experiments here presented indicate that the HMW species is populated both at pH 4.2 and after H2O2 treatment at physiological pH. We propose that overoxidation and protonation of Cysp, which is at the beginning of helix α2, trigger unwinding of this helix through two different mechanisms. In the case of overoxidation, the increased volume and hydrophilicity of Cysp-SO2H/SO3H are likely to favour its removal from the compact hydrophobic pocket formed by Tyr40, Pro41, Ala42, Thr45 and Pro49 that, together with Arg124, surround the reduced Cysp in the LMW species. Once Cysp is removed from its pocket, the first turn of the α2 helix can unwind and the β3-α2 loop assume a LU conformation (Figure 4A). This is favoured by the presence of a proline residue adjacent to Cysp (Pro49), highly conserved in typical 2-Cys Prxs (Hofmann et al., 2002), which reduces the propensity of the first turn of the α2 helix to retain its secondary structure. In the case of acidic pH, a very similar structural change would be caused by the loss of the ionic bond between Cysp and Arg124. Indeed in all typical 2-Cys Prxs, Cysp is in contact with a conserved arginine residue, homologous to Arg124 of SmPrxI, which lowers the pKa of the catalytic residue from 8.5 to 5–6 (Winterbourn and Hampton, 2008; Neumann et al., 2009). Accordingly, in SmPrxI, one Cys residue, likely to be Cysp, has a pKa=6.0 (Figure S5). At pH 4.2 this residue is fully protonated, and the ionic bond with Arg124 cannot be formed. We propose that the versatile chemical nature of the pocket containing Cysp evolved to trigger the same local unfolding of the active site in response to stimuli as different as overoxidation and acidification.
Unwinding of the first turn of helix α2 has been previously observed in the structure of the dimeric form of human PrxI (HsPrxI) bound to human sulfiredoxin (HsSrx), the enzyme that restores the active form of HsPrxI by reducing the overoxidized Cysp (Jönsson et al., 2008). As a result, Cysp is exposed towards the Srx active site, similarly to what observed for the Cysp to Asp mutant in a different structure (Jönsson et al., 2008; Jönsson et al., 2009). The similarity between the A/B dimer of SmPrxI-HMW and the isolated dimer of HsPrxI bound to Srx, in particular in the β3-α2 loop and α2 helix region, is highlighted in Figure 5A. These observations suggest that unwinding of the first turn of the α2 helix and the increased accessibility of Cysp to solvent and/or interacting protein may be a general structural mechanism by which 2-Cys Prxs respond to chemical stressors.
Figure 5. Comparison With Other 2-Cys Prxs.
(A) Comparison between the tertiary structures of the A/B dimer of SmPrxI-HMW (orange) and human PrxI (HsPrxI, magenta) in complex with human HsSrx (green; PDB ID: 2RII; Jönsson et al., 2008). SmPrxI-LMW (blue) and the hydrophobic surface of the HsSrx region involved in HsPrxI recognition are also shown (Jönsson et al., 2008 and 2009). The β3-α2 loops of SmPrxI-HMW and HsPrxI have a very similar conformation, which is different from that adopted by the same loop in SmPrxI-LMW, as highlighted by the relative position of the homologous residues Phe46 in SmPrxI and Phe50 in HsPrxI.
(B) Superposition between two adjacent dimers of SmPrxI-HMW (orange cartoons) and two copies of the oxidized dimer of Rattus norvegicus HBP23 (RnPrx, violet cartoons; PDB ID: 1QQ2). Equivalent aromatic residues contributing to the A-interface of SmPrxI-HMW and to the modelled interface in RnPrx are displayed as sticks.
The next point to address concerns the relationships between the tertiary structural change just described and the other observed changes in tertiary structure (i.e., the unfolding of the C-terminal tail and the concomitant protrusion of the β3-α2 loop towards the nearby dimer), and between these tertiary events and the change in the quaternary assembly from LMW to HMW. Superposition of SmPrxI-LMW and SmPrxI–HMW showed that the C-terminal region cannot assume the FF hairpin-like conformation observed in the LMW form when the first turn of α2 helix is unwound. As a consequence, unwinding of this helix necessarily triggers a conformational change of the C-terminus, and vice versa. Accordingly, in the structure of HsPrxI in complex with HsSrx, where helix α2 is unwound, the C-terminal arm of HsPrxI is unfolded and embraces HsSrx (Figure 5A; Jönsson et al., 2008; Jönsson et al., 2009).
Unfolding of the C terminus, in turn, results in: (i) changes in the surface of the ring decamer at the R-interface, allowing its interaction with another Prx decamer (Figures 3C–D and S4A); (ii) recognition of proteins with sulfiredoxin activity that restore the peroxidase activity through reduction of Cysp-SO2H and disassembly of the HMW species (Jönsson et al., 2008; Jönsson et al., 2009); (iii) recognition and binding of unfolded proteins.
We sequentially ordered the events above as local-tertiary-quaternary, but they might occur in a concerted way: on the one hand, oxidation/protonation of Cysp is driven by a large free energy change; on the other hand, the aggregation of FF dimers to the LMW or of LU dimers to HMW occurs at relatively low concentrations (see Experimental procedures), therefore it must be associated with a negative free energy. Consequently, both local (oxidation/protonation) and quaternary (aggregation) events are likely to contribute to the stabilization of the tertiary structural changes.
Tertiary changes analogous to those observed in SmPrxI-HMW, are known to take place when Cysp is disulfide bridged to Cysr (located in the C-terminal region) during the enzymatic cycle of Prxs. However, when this disulfide bridge is formed, the dimeric form of the protein is stabilized, while HMW states are not populated. Superposition of two copies of the eukaryotic Rattus norvegicus PrxI (RnPrx; Hirotsu et al., 1999), another 2-Cys Prx, to two adjacent dimers of SmPrxI-HMW (Figure 5B) shows that the presence of the disulfide bridge in RnPrx: (i) constrains the loop β3-α2 in a different orientation from that observed in SmPrxI-HMW, resulting in a perturbation of the contacts between the α3 helices that is likely to hamper formation of the A-interface; and (ii) prevents the complete unfolding of the C-terminus. These features are likely to block the propagation of the local unfolding of α2 helix and C-terminus, both horizontally through the A-interface and vertically at the R-interface, in the disulfide forms of Prx dimers.
The structure of the SmPrxI-HMW chaperone here presented resembles that of chaperonins such as HSP60 and its bacterial homologue GroEL. These are large complexes of two heptamer rings stacked on each other, enclosing a cavity where the correct folding of misfolded proteins takes place. The observed similarity is in agreement with the results of a recent study proposing that the most likely origin of the chaperone fold is the Prx-like fold (Dekker et al., 2011).
It has been hypothesized that the HMW species of Prxs bind unfolded substrates via unfolded hydrophobic regions, in analogy with other stress-regulated chaperones (Kumsta and Jacob, 2009). Although the binding site for unfolded polypeptides in the structure of SmPrxI-HMW cannot be identified with certainty, we propose a role for the C-terminus. This is unstructured and disordered in SmPrxI-HMW, but not in SmPrxI-LMW, which has no holdase activity. Additionally, the C-terminus of HsPrxI has been shown to interact with hydrophobic patches on the surface of partner proteins (Jönsson et al., 2008). Moreover, the presence of 20 unfolded C-termini in the 20-mer HMW, five at each rim and ten radiating from the equatorial portion of the structure, is consistent with the stoichiometry of the SmPrxI-monomer:CS ratio observed in our experiments.
Multi-ring stacking of 2-Cys Prxs has been observed both in vivo and in vitro by electron microscopy (Gourlay et al., 2003; Phalen et al., 2006; Wood et al., 2002; Harris et al., 2001), which has been correlated with cellular signalling. All Prx decamers intrinsically possess the structural information to lock with one another. Interlocking of SmPrxI decamers is mediated by two series of five pawls represented by the conserved α2 and α6 helices and β2 strands (Wood et al., 2003). Due to the pseudo 2-fold symmetric structure of the dimers, the pawls lean out above and below the plane of the ring. Like SmPrxI-LMW, SmPrxI-HMW (Figure 3C–E) has ten pawls available for interlocking with other decamers, leading to the formation of multi-ring structures. In these high-order assemblies the α2 and α6 helices longitudinally span through the rings and a discontinuous β-sheet, comprising the whole central β–sheet of the dimer, wraps the supramolecular structure in a left-handed helicoidal embrace (Figure 6). The external and internal diameters of this filament are in good agreement with those calculated by electron microscopy (Wood et al., 2002; Harris et al., 2001).
Figure 6. Theoretical Model of the Long Filament Structure.

Surface representation of the modelled filamentous-like structure of SmPrxI. This model has been built by joining two copies of the SmPrxI-HMW structure after optimal superposition of the lower decamer (chains K–T) of the first copy to the upper decamer (chains A–J) of a third copy of the molecule and of the upper decamer of the second copy (chains A–J) to the lower decamer (chains K–T) of the third copy. The α-helices (cyan) belonging to different decamers, which vertically span the filament, and a discontinuous β-sheet (red), which wraps the structure in a helicoidal embrace, are highlighted.
Pathophysiological role
The symptoms of schistosomiasis are due to egg secretion by the female worm residing in the blood stream. To leave the human body and close the parasite life cycle, the eggs both induce inflammation to penetrate the host tissues and, at the same time, prevent the formation of large granuloma that might hamper their escape (Green and Colley, 1981; Stadecker, 1992). We suggest that SmPrxI, one of the few proteins secreted by the eggs (Cass et al., 2007; Williams et al., 2001), is required to maintain this delicate equilibrium. The pH at a site of inflammation is below 6.0 (Poon et al., 2002); in addition, the granuloma contains immune cells that generate oxidative stress (neutrophils, eosinophils). Under these conditions SmPrxI might be in the stress-induced chaperone form here reported and may help controlling excessive precipitation of proteins associated with granuloma formation.
EXPERIMENTAL PROCEDURES
Protein Preparation
Recombinant 6-histidine-tagged SmPrxI, cloned from adult S. mansoni cDNA, was expressed in Escherichia coli and purified as previously described (Sayed et al., 2006; Sayed and Williams, 2004).
To obtain the overoxidized derivative, recombinant 1 μM SmPrxI was incubated in 50 mM potassium phosphate, 2 mM EDTA pH 7.0 with 1 mM NADPH, 200 nM E. coli Trx-reductase, 4 μM E. coli Trx, and 50 mM H2O2 at 25°C for 3 hours in aerobic conditions. The mixture was then dialyzed by ultrafiltration using concentrator tubes (Amicon) with a 50 kDa cut-off, concentrated to 1 mg/mL, and used for further experiments.
Size Exclusion Chromatography
Size exclusion chromatography (SEC) was performed on HPLC (LabService Inc.) at 25°C with a flow rate of 0.5 mL/min using a Tosoh TSK G3000PWxl column equilibrated with phosphate buffered saline (PBS) and 100 mM sodium-phosphate/sodium-citrate either at pH 7.4 or at pH 4.2. Molecular weight standards were DNA-binding protein (DPS) from Listeria innocua (230 kDa), ferritin from Pyrococcus furiosus (500 kDa) and cytochrome C from horse heart (12.4 kDa). Three different protein samples were run on the column: 1.5 mg/mL of reduced SmPrxI in PBS/Na-citrate pH7.4; 1.3 mg/mL of reduced SmPrxI in PBS/Na-citrate pH4.2; 1.0 mg/mL of SmPrxI-SO2H/SO3H in PBS/Na-citrate pH7.4. Each SEC run was repeated three times.
Assays of Chaperone and Peroxidase Activities
The elution fractions of reduced SmPrxI and SmPrxI-SO2H/SO3H were dialyzed against 50 mM Hepes pH 7.0 and then concentrated and assayed for the peroxidase and holdase activities. Trx-dependent peroxidase activity of Prxs linked to NADPH oxidation was monitored by the decrease in absorbance at 340 nm as previously described (Sayed et al., 2006). The holdase activity was measured by using 0.8–1 μM porcine heart CS (Jang et al., 2004; Cremers et al., 2010). Turbidity, induced at 43°C, in the presence or absence of SmPrxI samples was monitored at 360 nm in a spectrophotometer (Hewlett Packard, USA) equipped with a thermostatic cell holder both at pH 7.4 and 4.2. Each experiment was repeated three times.
Crystallization of SmPrxI-HMW and SmPrxI-LMW
The reduced form of SmPrxI was obtained by treating a solution of the enzyme (7 mg/mL) in PBS with 5 mM DTT. Crystallization trials were set up using the vapour diffusion method by both the hanging and sitting drop procedures. 1 μL of the reservoir solution (100 mM potassium citrate, 20% PEG 3350 and 20 mM DTT) was mixed with an equal volume of the protein solution. In the case of SmPrxI-HMW, reducing agents were not used. SmPrxI-HMW (7 mg/mL) was crystallized in 100 mM PBS/citrate buffer pH4.2, 200 mM NaCl and 20% PEG 8000. Both forms of the crystals were cryoprotected by adding 30% PEG 200 to the mother liquor and flash cooling in liquid nitrogen.
X-ray Data Collection, Processing and Refinement
Diffraction patterns of both SmPrxI-LMW and SmPrxI-HMW were collected at 100K, λ=0.918Å at the beamline ID14.1 of BESSYII (Berlin, Germany). For SmPrxI-LMW, data processing and reduction were performed using MOSFLM and SCALA in the CCP4 suite (Battye et al., 2011; Evans, 2011; Winn et al., 2011). The crystal belonged to space group P1. The unit cell contained 10 monomers per asymmetric unit with an estimated solvent content of 52%. Molecular replacement was performed with MolRep (Vagin and Teplyakov, 2010), using human PrxIV (PDB code 2pn8) as search model. Model building and refinement were performed using Coot (Emsley et al., 2010) and REFMAC5 (Murshudov et al., 2011) respectively. Data processing and reduction of SmPrxI-HMW crystal were performed using HKL2000 (Otwinowski and Moinor, 1997). PHENIX/Xtriage (Adams et al., 2010) revealed that the diffraction pattern belongs to space group P21 with pseudo-merohedral twinning (Yeates, 1997). The unit cell contained 20 monomers per asymmetric units with an estimated solvent content of 59.3%. Molecular replacement was performed using SmPrxI-LMW as search model. Model building and refinement were performed using Coot (Emsley et al., 2010) and PHENIX/Refine (Adams et al., 2010), respectively. The latest step of refinement revealed a twinning fraction of 0.5. NCS averaging on both structures was performed as a control and no differences were observed in the statistics and maps. Prior to deposition in the PDB, both structures were validated using MolProbity (Davis et al., 2004). In SmPrxI-LMW: 96.8% of residues are in the favoured regions, 3.2% in the allowed regions. In SmPrxI-HMW: 92.4% in favoured regions, 7.6% in allowed regions. A summary of data collection and refinement statistics are presented in Table 1. Figures were produced with PyMOL (Schrödinger LLC) and CCP4MG (Potterton et al., 2004).
Structure Analyses
Secondary structure assignment has been performed with DSSP (Kabsch and Sander, 1983) and solvent accessible surface area calculations with Naccess (http://www.bioinf.manchester.ac.uk/naccess/). Structure visualization and analyses were performed using the programs InsightII (Accelrys Inc.), PyMOL (Schrödinger LLC) and in house-built programs. The distance thresholds used for inter-atomic contacts were 4.0 and 3.6 Å for polar and hydrophobic atoms, respectively.
Supplementary Material
Acknowledgments
Supported by grants from the Fondazione Roma (project “Rational Approach to the Specific Inhibition of Plasmodium falciparum and Schistosoma mansoni”); Sapienza University of Rome “Ricerche Universitarie” 2009 e 2010; MIUR of Italy “FIRB/Proteomica 2007-protRBRN07BMCT”; National Institute of Allergy and Infectious Diseases Grant AI065622; European Community’s Seventh Framework Programme (FP7/2007-2013) under grant agreement n.°226716.
We acknowledge the Helmholtz-Zentrum Berlin - Electron storage ring BESSY II for provision of synchrotron radiation at beamline ID14.1 and would like to thank Drs. Manfred Weiss and Uwe Mueller for assistance. We are grateful to Prof. L. Mazzarella, Drs. L. Savino and G. Avella for stimulating discussions on crystallographic data; Dr. P. Ceci for providing protein markers used in SEC experiments, and Dr. G. Giardina for morphing and movies.
ABBREVIATIONS
- Prx
Peroxiredoxin
- Trx
Thioredoxin
- Trx-reductase
thioredoxin reductase
- CS
citrate synthase
- LMW
low molecular weight
- HMW
high molecular weight
- PBS
phosphate buffer saline
- DTT
dithiothreitol
- SEC
size exclusion chromatography
- EDTA
Ethylenediaminetetraacetic acid
- PEG
polyethylene glycol
- FF
fully folded
- LU
locally unfolded
Footnotes
ACCESSION NUMBERS
Coordinates of SmPrxI-LMW and SmPrxI-HMW, together with structure factors, have been deposited in the Protein Data Bank under accession numbers 3ZTL and 3ZVJ, respectively.
Supplemental Information includes five figures, two tables, two movies and can be found online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aran M, Ferrero DS, Pagano E, Wolosiuk RA. Typical 2-Cys peroxiredoxins – modulation by covalent transformations and noncovalent interactions. FEBS J. 2009;276:2478–2493. doi: 10.1111/j.1742-4658.2009.06984.x. [DOI] [PubMed] [Google Scholar]
- Barranco-Medina S, Kakorin S, Lázaro JJ, Dietz KJ. Thermodynamics of the dimer-decamer transition of reduced human and plant 2-cys peroxiredoxin. Biochemistry. 2008;47:7196–7204. doi: 10.1021/bi8002956. [DOI] [PubMed] [Google Scholar]
- Barranco-Medina S, Lázaro JJ, Dietz KJ. The oligomeric conformation of peroxiredoxins links redox state to function. FEBS Lett. 2009;583:1809–1816. doi: 10.1016/j.febslet.2009.05.029. [DOI] [PubMed] [Google Scholar]
- Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass CL, Johnson JR, Califf LL, Xu T, Hernandez HJ, Stadecker MJ, Yates JR, 3rd, Williams DL. Proteomic analysis of Schistosoma mansoni egg secretions. Mol Biochem Parasitol. 2007;155:84–93. doi: 10.1016/j.molbiopara.2007.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremers CM, Reichmann D, Hausmann J, Ilbert M, Jakob U. Unfolding of metastable linker region is at the core of Hsp33 activation as a redox-regulated chaperone. J Biol Chem. 2010;285:11243–11251. doi: 10.1074/jbc.M109.084350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32(Web Server issue):W615–W619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker C, Willison KR, Taylor WR. On the evolutionary origin of the chaperonins. Proteins. 2011;79:1172–1192. doi: 10.1002/prot.22952. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:2126–2132. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PR. An introduction to data reduction: space-group determination, scaling and intensity statistics. Acta Crystallogr D Biol Crystallogr. 2011;67:282–292. doi: 10.1107/S090744491003982X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flohé L, Budde H, Hofmann B. Peroxiredoxins in antioxidant defense and redox regulation. BioFactors. 2003;19:3–10. doi: 10.1002/biof.5520190102. [DOI] [PubMed] [Google Scholar]
- Gourlay LJ, Bhella D, Kelly SM, Price NC, Lindsay JG. Structure-Function Analysis of Recombinant Substrate Protein 22kDa (SP-22) J Biol Chem. 2003;278:32631–32637. doi: 10.1074/jbc.M303862200. [DOI] [PubMed] [Google Scholar]
- Green WF, Colley DG. Modulation of Schistosoma mansoni egg-induced granuloma formation: I-J restriction of T cell-mediated suppression in a chronic parasitic infection. Proc Natl Acad Sci. 1981;78:1152–1156. doi: 10.1073/pnas.78.2.1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Karplus PA, Poole LB. Typical 2-Cys peroxiredoxins – structures, mechanisms and functions. FEBS J. 2009;276:2469–2477. doi: 10.1111/j.1742-4658.2009.06985.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JR, Schröder E, Isupov MN, Scheffler D, Kristensen P, Littlechild JA, Vagin AA, Meissner U. Comparison of the decameric structure of peroxiredoxin-II by transmission electron microscopy and X-ray crystallography. Biochim Biophys Acta. 2001;547:221–34. doi: 10.1016/s0167-4838(01)00184-4. [DOI] [PubMed] [Google Scholar]
- Hirotsu S, Abe Y, Okada K, Nagahara N, Hori H, Nishino T, Hakoshima T. Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23kDa/proliferation-associated gene product. Proc Natl Acad Sci USA. 1999;96:12333–12338. doi: 10.1073/pnas.96.22.12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann B, Hecht HJ, Flohé L. Peroxiredoxins. Biol Chem. 2002;383:347–364. doi: 10.1515/BC.2002.040. [DOI] [PubMed] [Google Scholar]
- Jang HH, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Jönsson TJ, Johnson LC, Lowther WT. Structure of the sulphiredoxin-peroxiredoxin complex reveals an essential repair embrice. Nature. 2008;451:98–101. doi: 10.1038/nature06415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jönsson TJ, Johnson LC, Lowther WT. Protein engineering of the quaternary sulfiredoxin. peroxiredoxin enzyme substrate complex reveals the molecular basis for cysteine sulfinic acid phosphorylation. J Biol Chem. 2009;284:33305–33310. doi: 10.1074/jbc.M109.036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kumsta C, Jakob U. Redox-regulated chaperones. Biochemistry. 2009;48:4666–4676. doi: 10.1021/bi9003556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim JC, Choi HI, Park YS, Nam HW, Woo HA, Kwon KS, Kim YS, Rhee SG, Kim K, Chae HZ. Irreversible oxidation of the active-site cysteine of peroxiredoxin to cysteine sulfonic acid for enhanced molecular chaperone activity. J Biol Chem. 2008;283:28873–80. doi: 10.1074/jbc.M804087200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowther WT, Haynes AC. Reduction of cysteine sulfinic acid in eukaryotic, typical 2-Cys Peroxiredoxins by Sulfiredoxin. Antioxid Redox Signal. 2011;15:99–109. doi: 10.1089/ars.2010.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JC, et al. Oxidative stress-dependent structural and functional switching of a human 2-Cys peroxiredoxin isotype II that enhances HeLa cell resistance to H2O2-induced cell death. J Biol Chem. 2005;280:28775–28784. doi: 10.1074/jbc.M505362200. [DOI] [PubMed] [Google Scholar]
- Murshudov GN, Skubák P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KJ, Knutson ST, Soito L, Klomsiri C, Poole LB, Fetrow JS. Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Proteins. 2011;79:947–964. doi: 10.1002/prot.22936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CA, Cao J, Manevich Y. Peroxiredoxin I and its role in cell signaling. Cell Cycle. 2009;8:4072–4078. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor Z. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Phalen TJ, Weirather K, Deming PB, Anathy V, Howe AK, van der Vliet A, Jönsson TJ, Poole LB, Heintz NH. Oxidation state governs structural transitions in peroxiredoxin II that correlate with cell cycle arrest and recovery. J Cell Biol. 2006;175:779–789. doi: 10.1083/jcb.200606005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon S, Rybchyn MS, Easterbrook-Smith SB, Carver JA, Pankhurst GJ, Wilson MR. Mildly acidic pH activates the extracellular molecular chaperone clusterin. J Biol Chem. 2002;277:39532–39540. doi: 10.1074/jbc.M204855200. [DOI] [PubMed] [Google Scholar]
- Potterton L, McNicholas S, Krissinel E, Gruber J, Cowtan K, Emsley P, Murshudov GN, Cohen S, Perrakis A, Noble M. Developments in the CCP4 molecular-graphics project. Acta Crystallogr D Biol Crystallogr. 2004;60:2288–2294. doi: 10.1107/S0907444904023716. [DOI] [PubMed] [Google Scholar]
- Sali D, Bycroft M, Fersht AR. Stabilization of protein structure by interaction of alpha-helix dipole with a charged side chain. Nature. 1988;335:740–743. doi: 10.1038/335740a0. [DOI] [PubMed] [Google Scholar]
- Sayed AA, Williams DL. Biochemical characterization of 2-Cys peroxiredoxins from Schistosoma mansoni. J Biol Chem. 2004;279:26159–66. doi: 10.1074/jbc.M401748200. [DOI] [PubMed] [Google Scholar]
- Sayed AA, Cook SK, Williams DL. Redox balance mechanisms in Schistosoma mansoni rely on peroxiredoxins and albumin and implicate peroxiredoxins as novel drug targets. J Biol Chem. 2006;281:17001–17010. doi: 10.1074/jbc.M512601200. [DOI] [PubMed] [Google Scholar]
- Soito L, Williamson C, Knutson ST, Fetrow JS, Poole LB, Nelson KJ. PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 2011;39(Database issue):D332–D337. doi: 10.1093/nar/gkq1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadecker MJ. The role of T-cell mediated suppression in the immunomodulation of schistosomiasis. Parasitol Today. 1992;8:199–204. doi: 10.1016/0169-4758(92)90264-3. [DOI] [PubMed] [Google Scholar]
- Vagin A, Teplyakov A. Molecular Replacement with MOLREP. Acta Crystallogr D Biol Crytallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- Williams DL, Asahi H, Botkin DJ, Stadecker MJ. Schistosome infection stimulates host CD4(+) T helper cell and B-cell responses against a novel egg antigen, thioredoxin peroxidase. Infect Immun. 2001;69:1134–41. doi: 10.1128/IAI.69.2.1134-1141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J, Linke K, Jatzek A, Jakob U. Severe oxidative stress causes inactivation of DnaK and activation of the redox-regulated chaperone Hsp33. Mol Cell. 2005;17:381–392. doi: 10.1016/j.molcel.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Winterbourn CC, Hampton MB. Thiol chemistry and specificity in redox signaling. Free Radic Biol Med. 2008;45:549–561. doi: 10.1016/j.freeradbiomed.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Poole LB, Hantgan RR, Karplus PA. Dimers to Doughnuts: Redox-Sensitive Oligomerization of 2-Cys Peroxiredoxins. Biochemistry. 2002;41:5493–5504. doi: 10.1021/bi012173m. [DOI] [PubMed] [Google Scholar]
- Wood ZA, Schröder E, Robin Harris J, Poole LB. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci. 2003;28:32–40. doi: 10.1016/s0968-0004(02)00003-8. [DOI] [PubMed] [Google Scholar]
- Yeates TO. Detecting and overcoming crystal twinning. Methods Enzymol. 1997;276:344–358. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.