SUMMARY
Diurnal variation in nitrogen homeostasis is observed across phylogeny. But whether these are endogenous rhythms, and if so, molecular mechanisms that link nitrogen homeostasis to the circadian clock remain unknown. Here, we provide evidence that a clock-dependent peripheral oscillator, Krüppel-like factor15 transcriptionally coordinates rhythmic expression of multiple enzymes involved in mammalian nitrogen homeostasis. In particular, Krüppel-like factor15-deficient mice exhibit no discernable amino acid rhythm, and the rhythmicity of ammonia to urea detoxification is impaired. Of the external cues, feeding plays a dominant role in modulating Krüppel-like factor15 rhythm and nitrogen homeostasis. Further, when all behavioral, environmental and dietary cues were controlled in humans, nitrogen homeostasis still expressed endogenous circadian rhythmicity. Thus, in mammals, nitrogen homeostasis exhibits circadian rhythmicity, and is orchestrated by Krüppel-like factor15.
INTRODUCTION
Despite its abundance in the earth’s atmosphere, mammals cannot freely assimilate nitrogen, and are dependent on ingestion of amino acids. Nitrogen fixation is an elementary biological process through which microorganisms that exist in the roots of leguminous plants convert atmospheric nitrogen to ammonia. Thus, plants serve as the major source of amino acids (AAs) for mammalian organisms; accordingly amino acids in organisms are termed essential (diet-dependent) or non-essential (synthesized from other essential amino acids in vivo). In addition to serving as building blocks of proteins, AAs are critical for diverse biological functions, including gluconeogenesis, hormone synthesis, nutrient signaling, neurotransmission and embryonic stem-cell growth (Wang et al., 2009; Wu, 2009). Following utilization of AAs, organisms also face the burden of detoxifying the by-products, i.e., converting ammonia to urea (Morris, 2002). The importance of this homeostatic process is perhaps best demonstrated in congenital disorders of amino acid (AA) metabolism/ammonia detoxification that often present with dysfunction of multiple organ systems – particularly cognitive impairment (Gropman et al., 2007). Recent studies have also shed light on a potentially direct pathogenic role for amino acids. Interestingly, supplementation of essential AAs significantly reduced survival in Drosophila (Grandison et al., 2009), while supplementation of branched chain amino acids (BCAA) enhanced survival in mice (D'Antona et al., 2010). Thus, organisms face a delicate task of imbibing and metabolizing amino acids, and imbalance alters survival.
The behavior, activity and survival of organisms are influenced by the 24-hour rotation of the earth on its axis (Foster and Roenneberg, 2008). Studies over the last two decades have identified components of the endogenous core clock machinery (CCM) that govern 24-hour rhythms even in the absence of external cues (e.g., light) (Reppert and Weaver, 2002). The CCM exists in the central circadian pacemaker in the hypothalamus, the suprachiasmatic nucleus, as well as in most peripheral cells, and consists of a series of positive and negative feedback loops generated by the basic-helix-loop-helix family of transcription factors Clock and Bmal1, the Period and Cryptochrome families and the nuclear receptors Reverbα and Rorα (Ko and Takahashi, 2006). The CCM in the suprachiasmatic nucleus is entrained by cues such as light exposure and acts via neural and endrocrine signals to appropriately synchronize the CCM in peripheral tissues (Dibner et al., 2010). However, circadian timing in peripheral tissues many also be affected by other factors, such as rhythmic feeding and temperature (Dibner et al., 2010). Multiple lines of evidence identify the CCM as centrally involved in regulating metabolic homeostasis – particularly with respect to glucose and lipid levels (Duez and Staels, 2008; Green et al., 2008; Lamia et al., 2008; Le Martelot et al., 2009; Marcheva et al., 2010; Rudic et al., 2004; Turek et al., 2005; Yin et al., 2007). A relationship of the CCM to AA utilization and excretion (i.e. nitrogen homeostasis) remains unknown.
Interestingly, several aspects of nitrogen flux across several phylogenies in nature occur in a diurnal fashion. The first process of “nitrogen fixation” in diazotrophs is highly diurnal due to the oscillatory pattern of nitrogenase activity (Balandreau et al., 1974; Wheeler, 1969). As a consequence, AA concentration in the leaves of plants exhibit diurnal variation (Bauer et al., 1977). Intriguingly, studies from humans and rodents over five decades ago reported diurnal variation in plasma levels of AAs that is modulated by changes in diet (Feigin et al., 1969; Feigin et al., 1967; Fernstrom et al., 1979). Further, recent unbiased metabolite analyses in yeast and mice have identified that AAs and urea-cycle intermediates exhibit oscillatory behavior (Minami et al., 2009; Tu et al., 2007). In addition, screening of the hepatic proteome revealed rhythmicity of AA metabolic enzymes and urea cycle enzymes (Reddy et al., 2006). However, the molecular mechanisms that control these diurnal rhythms, and whether nitrogen homeostasis exhibits true endogenous circadian rhythmicity, i.e., oscillation in the absence of external cues, remain unknown.
Krüppel-like factors belong to the zinc-finger family of transcription factors, and are implicated in coordinating numerous biological processes from pluripotency to carcinogenesis (McConnell and Yang, 2010). Our laboratory previously demonstrated that Krüppel-like factor 15 (Klf15) regulates glucose homeostasis through effects on AA metabolism (Gray et al., 2007). Subsequent genome-wide microarray analyses of muscle and liver tissues identified additional targets of Klf15 as being involved in AA metabolism. Thus, we hypothesized that Klf15 may be involved in regulating the rhythmic utilization of amino acids. In the present study we demonstrate that nitrogen homeostasis exhibits 24 hour periodicity in mice and humans. Further, we identify Klf15 as a clock-dependent peripheral regulator of rhythmic amino acid utilization and excretion of amino acids.
RESULTS
Klf15 expression exhibits 24 hour periodicity
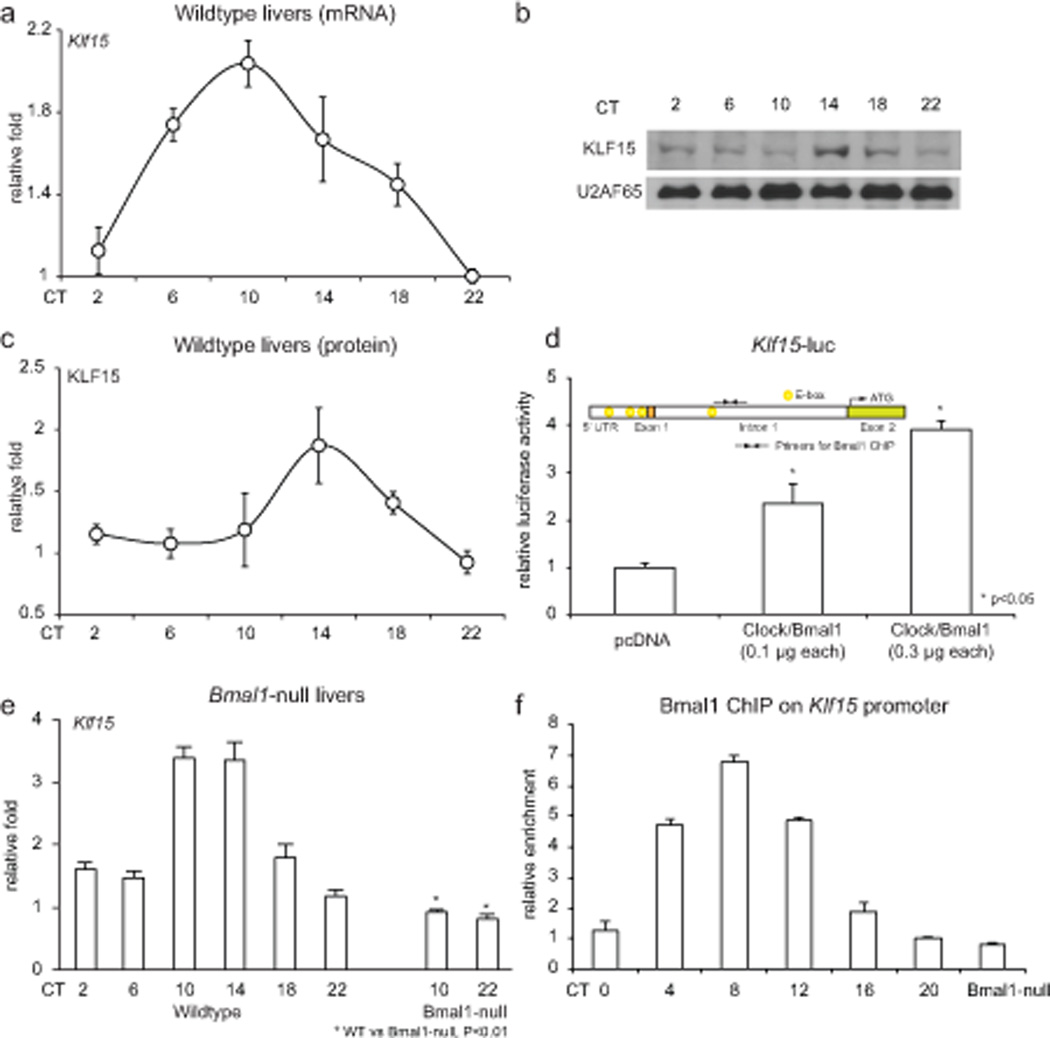
As a first step, we examined whether Klf15 expression itself was rhythmic. WT mice were sacrificed every 4 hours under light/dark conditions (L/D) or constant dark (D/D) conditions for 24-hours. Klf15 expression exhibits rhythmic oscillation in several peripheral organs including liver and skeletal muscle under both conditions (Fig. 1a–c & Supplemental Fig. 1a,b), confirming the notion that Klf15 expression is rhythmic. As next steps, we examined whether the CCM mediates Klf15 rhythmicity in peripheral organs, and how this occurs. Examination of the regulatory region of Klf15 identified four “E-box” binding motifs (Fig. 1d inset), and CLOCK/BMAL1 induce Klf15 in a dose dependent manner in hepatocyte cell lines (Fig. 1d). Consistent with this observation, Klf15 rhythmic variation was abrogated in Bmal1-null livers (Fig. 1e). Further, Klf15 rhythmicity was also abrogated in livers of several CCM-mutant mouse lines including Per2/Cry1 KO, Per1/2 KO and Reverbα KO (Supplemental Fig. 1c, d). Finally, chromatin immunoprecipitation (ChIP) revealed rhythmic occupancy of Bmal1 on the Klf15 promoter (Fig. 1f). These data support a direct role for the CCM in orchestrating the 24 hour periodicity of Klf15.
Figure 1. Klf15 exhibits 24-hour periodicity and is driven by the core clock machinery.
(a) Klf15 mRNA accumulation from WT mice livers (n=5 per time point). (b) Representative KLF15 and U2AF65 protein expression from WT and Klf15-null liver nuclei. (c) KLF15 protein densitometry from three replicates. (d) Klf15-luciferase is induced on a dose dependent fashion by CLOCK/BMAL1, and inset illustrates four “E Box” motifs in the Klf15 promoter (−5 kb). (e) Klf15 mRNA accumulation in WT and Bmal1 KO livers. (g) (h) Rhythmic binding of BMAL1 on the Klf15 promoter (n=3 per time point). Data presented as mean ± SEM.
Nitrogen homeostasis exhibits 24 hour periodicity in mice
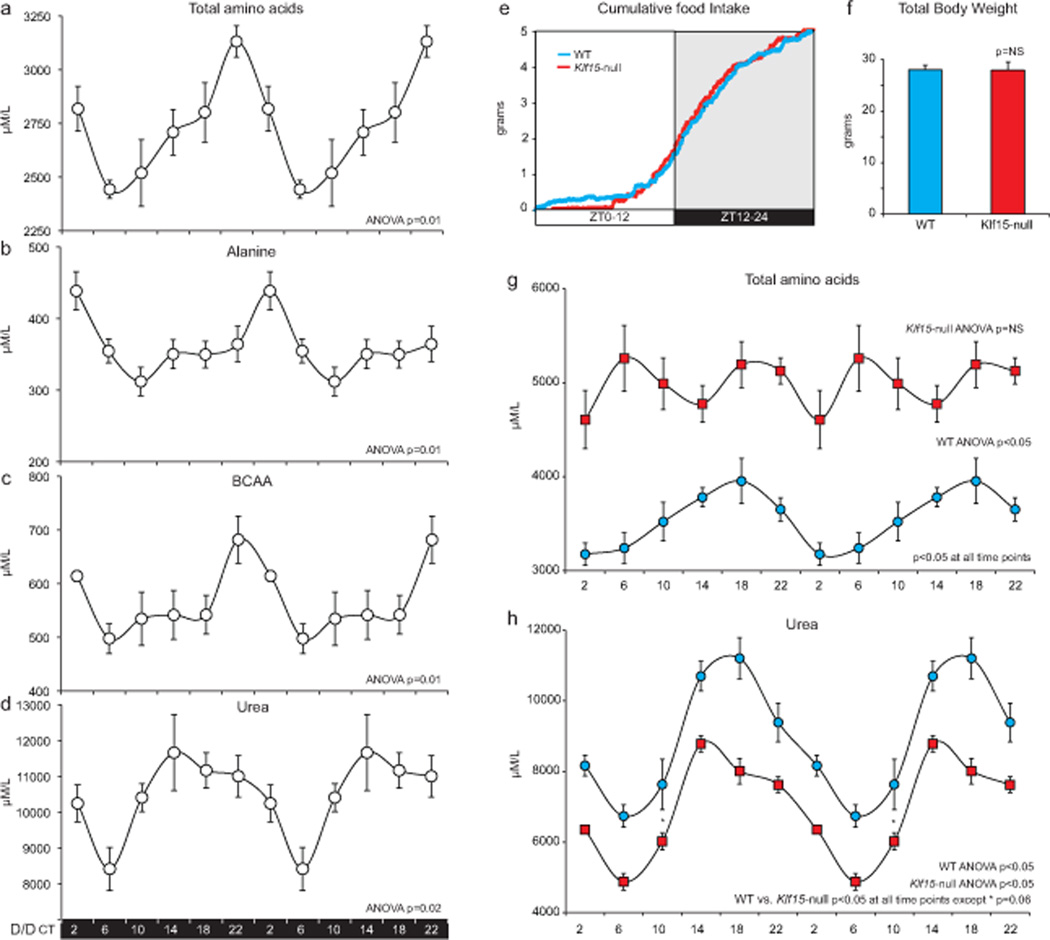
In addition to daily rhythms in Klf15, we next examined if AA utilization and excretion also oscillate. Wild-type (WT) mice were placed in constant darkness for 38 hours (D/D), after which plasma was collected every four hours for the following 24-hours. Interestingly, the total AA pool, as well as major circulatory amino acids (AAs), e.g., alanine and branched chain amino acids (BCAA) exhibited 24 hour rhythms in constant darkness (Fig. 2a–c). Further, the detoxified excretory product of nitrogeneous waste i.e., urea also oscillates with similar 24 hour periodicity (Fig. 2d). Of the twenty AA’s, fourteen AA’s were rhythmic under D/D as detailed in Supplemental Table 1. Thus, nitrogen homeostasis in mice exists with a 24 hour periodicity even under constant environmental conditions.
Figure 2. Nitrogen homeostasis exhibits 24-hour periodicity, driven by Klf15, in mice.
(a–d) Plasma total amino acid pool, alanine, BCAA and urea measured every four hours over a circadian period after placing mice in constant darkness for 38 hours (n=5 per time point). The data are double-plotted, and ANOVA was used to determine rhythmicity. (e) Cumulative food intake measured every 5 minutes in WT and Klf15-null mice (n=4 per group). (f) Total body weights of WT and Klf15-null mice (n=4 per group). (g, h) Plasma total AA pool, urea from WT and Klf15-null mice measured every four hours under L/D and double plotted to illustrate rhythmicity (n=5 per group per time point). Data presented as mean ± SEM.
Klf15 regulates rhythmic AA utilization
Next, the effect of Klf15 proficiency and deficiency on nitrogen homeostasis was assessed under L/D over a 24-hour period. Because mammals are dependent on diet for essential AAs, we first assessed the cumulative and total food intake over 24-hours. The cumulative food intake (measured every five minutes, Fig. 2e) and aggregate food consumed over 24-hours (Supplemental Fig. 2a) were nearly identical in WT and Klf15-null mice. Consistent with this observation, the total body weights of Klf15-null mice were similar to their WT counterparts (Fig. 2f). Further, the free running period under constant dark and constant light conditions were similar between WT and Klf15-null mice (Supplemental Fig. 2b,c). Interestingly, the hepatic expression of oscillation of several components of the CCM was preserved in the Klf15-deficient state (Supplemental Fig. 2d). However, a marked alteration in both the absolute levels and rhythmicity of the total AA pool and urea were observed in Klf15-null mice (Fig. 3g, h). A detailed analysis of each individual AA is provided in Supplemental Table 1. Collectively, these observations identify Klf15 as an essential regulator of rhythmic nitrogen homeostasis.
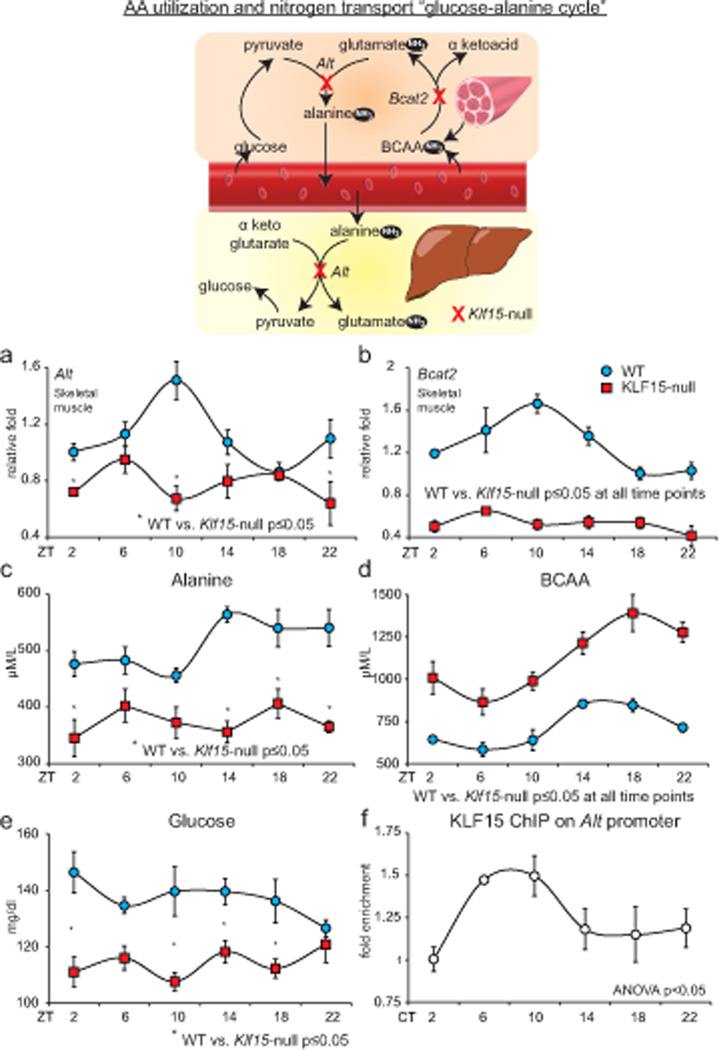
Figure 3. Klf15 regulates rhythmic amino acid utilization.
Schematic illustrates the inter-organ transport, utilization of amino acids “the glucose-alanine cycle.” (a, b) Skeletal muscle Alt and Bcat2 expression in WT and KLF15-null mice (n=4 per group per time point). (c, d) Plasma alanine and BCAA in WT and Klf15-null mice (n=5 per group per time point) (e) Plasma glucose in WT and Klf15-null mice (n=5 per group per time point). (f) ChIP for KLF15 on Alt promoter (n=3 per time point). (# p<0.05 at all time points between WT and Klf15-null). Data presented as mean ± SEM.
We next sought to elucidate the molecular basis for the non-rhythmic nitrogen homeostasis in the Klf15 deficient state. In mammals, the liver and skeletal muscles are centrally involved in coordinating nitrogen homeostasis. During the daily feed/fast rhythms, the glucose-alanine cycle serves two main purposes – (a) to supply carbon skeletons to the liver to sustain glucose levels, and (b) to facilitate transport and elimination of nitrogenous waste (Felig, 1975). During the fed state, when glucose is freely available, skeletal muscle oxidizes glucose to generate pyruvate that is transaminated by alanine transaminase (Alt) to produce alanine (Fig. 3 schematic) (Felig, 1975). Further, in the post-absorptive state, carbons generated from breakdown of muscle AAs are the principal precursors for hepatic gluconeogenesis. Since branched chain amino acids (BCAA) compose 35% of the essential AAs in muscle (Harper et al., 1984), they are major donors of carbon and dispose their nitrogen through the glucose-alanine cycle. The skeletal muscle mitochondrial branched chain transaminase (Bcat2) is the first step in BCAA catabolism and converts BCAA to glutamate and α-keto acids (Harper et al., 1984). The glutamate is subsequently utilized by Alt in skeletal muscle to synthesize alanine. The alanine spills into the circulation, is absorbed by the liver, and ultimately donates its carbons for gluconeogenesis and its nitrogen for urea synthesis (Fig. 3 schematic). Intriguingly, the expression patterns of Alt, Bcat2 in skeletal muscle and Alt in liver of WT mice were found to exhibit robust diurnal rhythms – an effect that was abrogated in both tissues with Klf15-deficiency (Fig. 3a,b and Supplemental Fig. 3a). Consistent with this defect, skeletal muscle and liver concentrations of alanine and glutamate were reduced, and BCAA was significantly increased (Supplemental Fig. 3a). Consequently, plasma alanine was reduced without rhythmicity (Fig. 3c), whereas, plasma BCAA was persistently increased with abnormal rhythmicity in Klf15-null mice (Fig. 3d). An important consequence of impaired alanine availability was persistently low glucose in Klf15-null mice (Fig. 3e). This occurred despite compensatory adaptive changes in several hepatic gluconeogenic enzymes (Glut2, Pepck, Pfkfbp2 and G6PC, Supplemental Fig. 3b), insulin and glucagon (Supplemental Fig. 3b). Next, to examine if Alt and Bcat2 are direct transcriptional targets for Klf15, we utilized gain of function studies and ChIP. Adenoviral overexpression of Klf15 induced Alt in hepatocytes and Alt, Bcat2 in skeletal myotubes (Supplemental Fig. 3c). Further, AA analysis of cell culture supernatant from Klf15 overexpressing hepatocytes revealed reduced alanine and increased glutamate concentrations (Supplemental Fig. 3c). Examination of the Alt promoter region identified conserved consensus DNA binding sites for Krüppel-like factors - C(A/T)CCC (Miller and Bieker, 1993)(Supplemental Fig. 3d). Finally, ChIP analysis of WT livers over a circadian period identified rhythmic enrichment of KLF15 on the Alt promoter (Fig. 3f and Supplemental Fig. 3d).
Klf15 regulates rhythmic urea synthesis
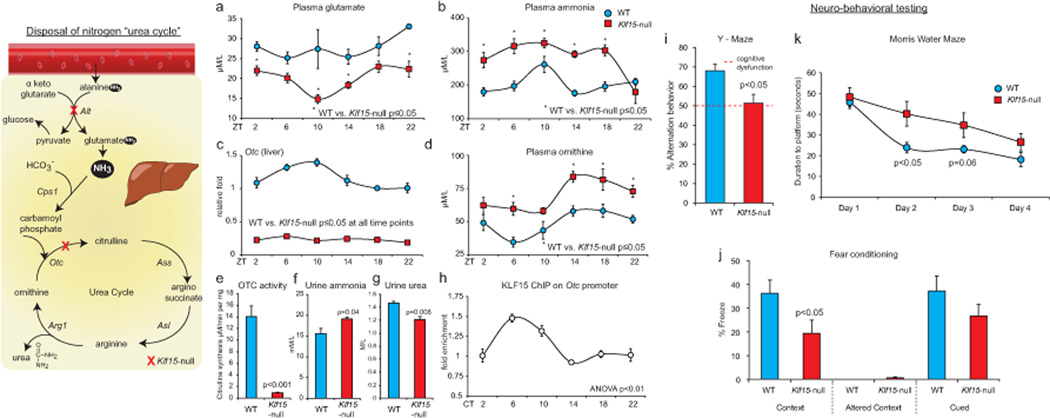
The final common pathway in liver for detoxification of nitrogenous waste occurs through glutamate, the major nitrogen donor for ammonia (Brosnan and Brosnan, 2009). Surprisingly, despite reduced plasma glutamate in Klf15-null mice (Fig. 4a), there was increased accumulation of plasma ammonia (Fig. 4b). This led us to examine expression of the hepatic urea cycle enzymes. The urea cycle (Fig. 4 schematic) is responsible for detoxification of ammonia to urea, and occurs predominantly in the mammalian liver (Morris, 2002). Interestingly, the expression of ornithine transcarbamylase (Otc), a hepatic mitochondrial urea cycle enzyme was markedly reduced and devoid of rhythmicity in Klf15-null mice (Fig. 4c). In contrast, the 24 hour periodicity of expression of other urea cycle enzymes (Cps1, Asl, Ass and Arg1) was either unchanged or increased with Klf15-deficiency (Supplemental Fig. 4a). Importantly, OTC enzymatic activity from KLF15-null hepatic mitochondrial extracts was reduced to ~6% of their WT counterparts (Fig. 4e). Consistent with this defect, plasma/tissue concentrations of ornithine were markedly increased (Fig. 4d and Supplemental Fig. 4b). Further, urine analysis revealed elevated levels of ammonia and reduced urea, supportive of impaired hepatic ureagenesis (Fig. 4f, g). Next, to examine if Otc is a transcriptional target for Klf15, we performed adenoviral overexpression in hepatocytes and ChIP. Klf15 overexpression induced Otc expression, and analysis of hepatocyte cell culture supernatant revealed reduced ornithine concentration (Supplemental Fig. 4c). Further, ChIP identified rhythmic enrichment of KLF15 on a conserved region of the Otc promoter (Supplemental Fig. 4c and Fig. 4h). Next, to determine if the observed hyperammonemia was associated with cognitive dysfunction we performed extensive neurocognitive behavioral testing. Klf15-null mice exhibited significant dysfunction of short-term memory as evidenced by reduced percentage of alternation behavior in the Y-maze test (Fig. 4i). To test for memory more specifically we determined contextual (hippocampal) and cued (amygdala) fear-based memory. Klf15-null mice exhibit reduced freezing in the context in which they had been previously received an aversive stimulus suggestive of impaired hippocampal-based memory (Fig. 4j). However, no significant difference was noted for cued fear conditioning (Fig. 4j). Finally, in the Morris water maze, a selective test for hippocampal function, Klf15-null mice exhibit an initial significant impairment in learning but no differences in retention suggesting that Klf15-null mice exhibit delay in learning but ability to ultimately acquire and retain the task (Fig. 4k). In summary, these data suggest that impaired detoxification of ammonia impairs selective aspects of cognitive function in Klf15-null mice.
Figure 4. Klf15 regulates rhythmic nitrogeneous waste excretion.
Schematic illustrates the excretion of nitrogenous waste products, i.e., “the urea cycle.” (a, b) Plasma glutamate, ammonia in WT and Klf15-null mice (n=5 per group per time point). (c) Otc expression in WT and Klf15-null livers (n=4 per group per time point). (d) Plasma ornithine in WT and Klf15-null mice (n=5 per group per time point). (e) OTC enzymatic activity measured from liver mitochondrial extracts from WT and KLF15-null mice (n=4 per group). (f, g) Urinary levels of urea and ammonia in WT and KLF15-null mice (n=5 per group). (h) ChIP for KLF15 on the Otc promoter (n=3 per time point). (# p<0.05 at all time points between WT and Klf15-null). (i–k)Results of neurobehavioral testing for (i) Y-maze, a test of working memory (n=3 per group) (j) Fear conditioning during contextual changes (hippocampal function) and altered cues (amygdalar function) (n=8 per group) and (k) Morris water maze test, a test of hippocampal function (n=8 per group). Data presented as mean ± SEM.
Feeding regulates nitrogen homeostasis and the peripheral core clock machinery
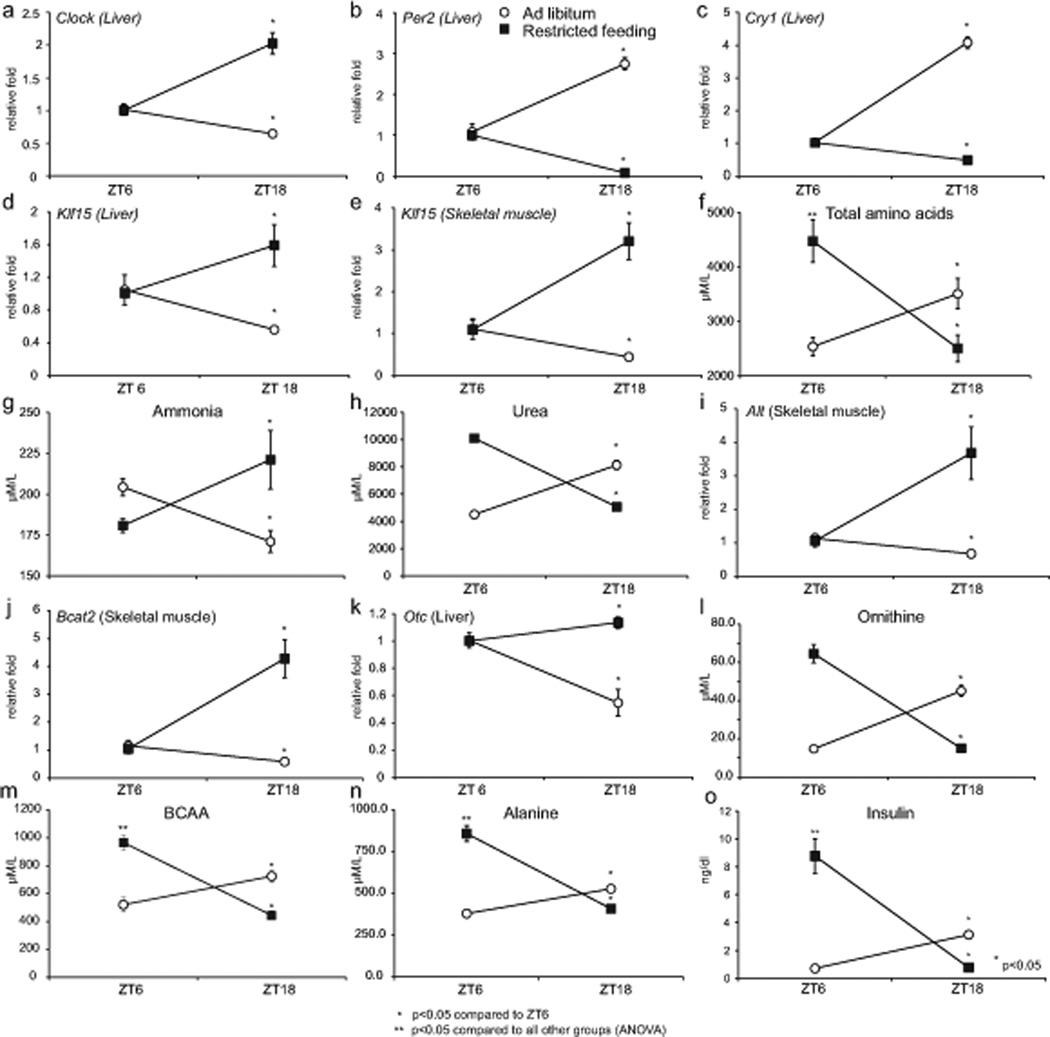
Previous studies have identified that rhythmic feeding is a dominant input that modulates CCM expression in peripheral organs (Damiola et al., 2000). Thus, we reasoned that rhythmic feeding could alter Klf15 diurnal expression pattern and nitrogen homeostasis. To test this hypothesis, mice were fed for a six-hour period during the light phase (ZT3-ZT9) or ad libitum for a period of one-month under L/D conditions. Animals were then harvested at two time points corresponding to the zenith and nadir of the total AA pool under L/D (Fig. 2g). Consistent with previous studies (Damiola et al., 2000), restricting food intake to the light phase altered expression of several components of the CCM in liver including Clock, Per2, Cry1, Reverbα and Dbp (Fig. 5a–c and Supplemental Fig. 5a). Interestingly, restricted feeding also altered expression of Klf15 in liver and skeletal muscle, the total AA pool (including alanine and BCAA), ammonia and urea (Fig. 5d–h). Of the fifteen AAs that exhibit rhythmic oscillation in LD, the zenith and nadir of thirteen were shifted significantly by daytime feeding (Supplemental Table 1). Further, examination of enzymes in AA metabolism revealed Alt and Bcat2 to be altered in skeletal muscle, but not in the liver (Fig. 5i, j and Supplemental Fig. 5b). Of the nitrogen disposal machinery in the liver, Otc expression was reversed, and corroborated by changes in plasma ornithine (Fig. 5k,l and Supplemental Fig. 5b). In an additional set of experiments, mice were harvested at all time points of a 24-hour period following restricted feeding. Consistent with our aforementioned experiments, we identified full reversal in gene expression of Bmal1, Klf15 and Otc in the liver (Supplemental Fig. 5c). Thus, feeding rhythms play a key role in modulating Klf15 and rhythmic nitrogen homeostasis.
Figure 5. Feeding alters Klf15 expression and nitrogen homeostatsis.
Following ad-libitum or feeding restricted to the light-phase (ZT3-ZT9) for one month: (a, b & c) liver expression of Clock, Per2 and Cry1, (d, e) liver and skeletal muscle expression of Klf15, (f, g & h) total plasma AA, ammonia and urea concentrations (I, j) skeletal muscle Alt and Bcat2 expression (k) liver expression of Otc (l, m, n & o) plasma ornithine, alanine, BCAA, insulin (n=5 per group per time point). Data presented as mean ± SEM.
Adaptation to high protein diet in WT and Klf15-null mice
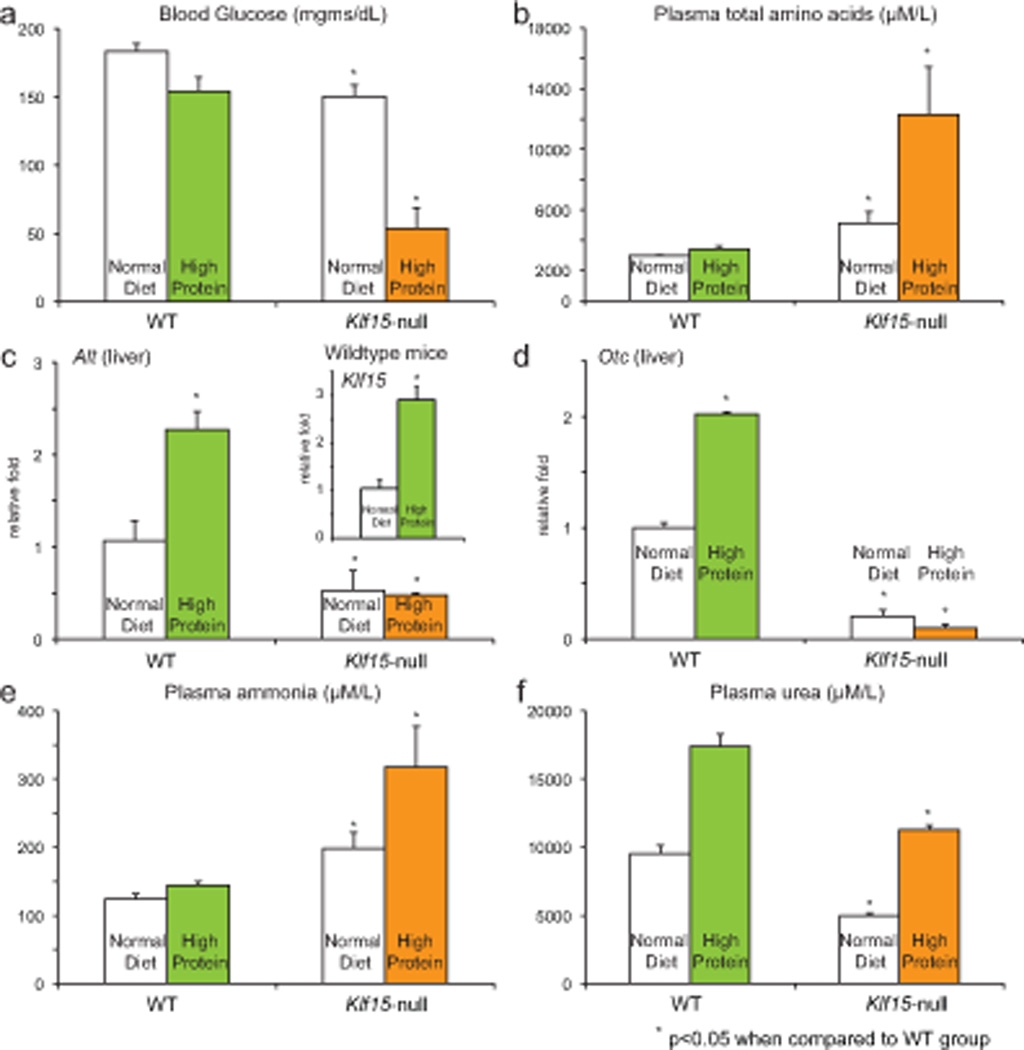
Our study suggests an important role for Klf15 in regulating rhythmic flow of nitrogen by orchestrating rhythmic variation in expression of amino acid utilization and excretion enzymes across multiple organs. To further substantiate the importance of these regulatory effects in vivo, we challenged WT and Klf15-null mice to a high protein diet (70% protein as casein) or normal diet (18% protein) for one week. In WT mice, despite the low levels of carbohydrate in the high protein diet (18.7 %, normal diet has 62.3 % carbohydrate), the mice were able to maintain euglycemia (Fig. 6a) by extracting carbon skeletons from amino acids, i.e., gluconeogenesis through greater Klf15 and Alt expression (Fig. 6b, c). In addition, ammonia levels were also maintained at physiological levels by increasing ureagenesis (Fig. 6e, f) accompanied by a significant increase in hepatic Otc expression (Fig. 6d). In sharp contrast, Klf15-null mice exhibit near fatal hypoglycemia on high protein diet (one of five mice died at one week of high protein diet due to severe hypoglycemia), and demonstrated significant accumulation of amino acids in plasma, indicative of impaired utilization of amino acids (Fig. 6a–c). In addition, and consistent with a significant defect in hepatic Otc function (Fig. 6d), Klf15-null mice exhibited marked hyperammonemia with impaired ureagenesis (Fig. 6e, f). This global failure of nitrogen flux, and inability to adapt to high protein diet further illustrates the role of Klf15 in controlling utilization and excretion of nitrogen in mammalian organisms.
Figure 6. WT and Klf15-null adaptation to high protein diet.
Following one week of high protein diet (70% casein) or normal diet (18% protein) WT and Klf15-null (a) blood glucose (n=5 per group), (b) plasma amino acids, (c, d) liver expression of Klf15, Alt, Otc (d) plasma ammonia and (e) plasma urea. Data presented as mean ± SEM.
Nitrogen homeostasis is circadian in humans
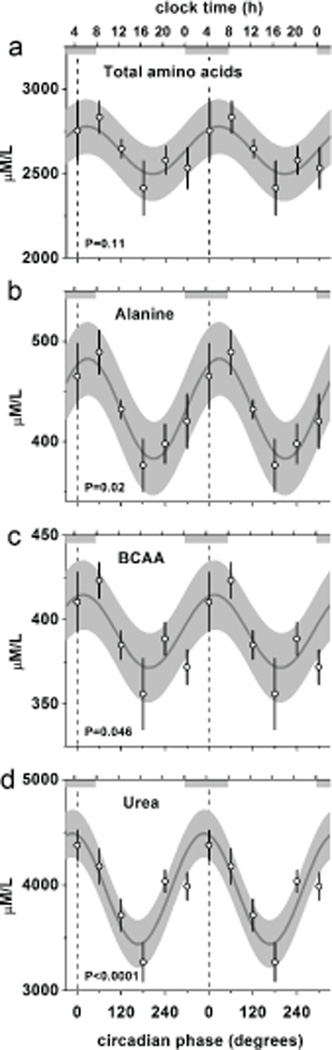
Studies from the 1960’s conducted for six continuous days clearly established that AAs exhibit diurnal rhythm in humans (Feigin et al., 1967). Our studies in mice suggested that such rhythms could occur in the absence of external cues. However, since food intake exhibits endogenous circadian rhythmicity in mice, we cannot determine if nitrogen homeostasis is driven simply by the feeding/fasting cycle or by an endogenous circadian rhythm independent of dietary cues. Thus, to determine if the circadian system per se influences nitrogen homeostasis in humans independent of environmental, behavioral and dietary influences, we scheduled individuals to live on 28-hour ‘days’ for 7 cycles, across 196 h, in persistent dim light as previously described (Scheer et al., 2009). This forced desynchrony protocol dissociates the behavioral sleep/wake and fasting/feeding cycle (imposed 28-h cycle) from the circadian system (endogenous ~24-h cycle) and thus allows the determination of circadian system influences not confounded by the influence of changes in behavior (including food intake) and environment (including light). In this way, blood samples were obtained in the fasted state (~12 hours since the last meal) at different internal circadian phases. Using this gold-standard method in human circadian research, we discovered that several AAs including alanine and BCAA exhibit robust endogenous circadian rhythms (Fig. 7b, c). Of the 20 AAs, six exhibited endogenous circadian rhythmicity in humans (Fig. 7 & Supplemental Fig 6), and the total amino acid pool demonstrated a trend towards rhythmicity (Fig. 7a). Finally, plasma urea and ornithine also oscillated with endogenous ~24-hour periodicity (Fig. 7d & Supplemental Fig. 6).
Figure 7. Human endogeneous circadian nitrogen homeostasis.
Fasting total plasma amino acid pool, alanine, BCAA and urea exhibit endogenous circadian rhythmicity in humans during the forced desynchrony protocol. The cosine models (black lines) and 95% confidence intervals (gray areas) are based on mixed model analyses and use precise circadian phase data. To show that these models adequately fit the actual data, we also plot the proportional changes across 60 circadian degree windows per individual multiplied by the group average with SEM error bars (open circles with error bars). Data are double plotted to aid visualization of rhythmicity. Lower X-axis, circadian phase in degrees, with fitted core body temperature minimum assigned 0-degrees, and 360 degrees equal to the individual circadian period (group average 24.09h); top X-axis, corresponding average clocktime for these individuals; vertical dotted lines, core body temperature minimum; horizontal gray bars, corresponding average habitual sleep episode in the two weeks prior to admission.
DISCUSSION
Using complementary approaches in mice and humans we demonstrate that nitrogen homeostasis is a conserved endogenous circadian process in mammals. Our initial studies in mice were done under the classical conditions to ascertain rhythmicity, i.e., constant darkness. In D/D we observed that a 24-hour rhythmicity existed for total amino acid pool, ammonia and urea (Fig. 2a–d). Furthermore, in our human study we dissociated the influence of the circadian timing system from that of all environmental, behavioral and dietary influences on nitrogen homeostasis by placing humans in persistent dim light for 196 hours, and altered the habitual feed/fasting and sleeping rhythms to 28 hour cycles. Despite these alterations and while fasted, nitrogen homeostasis proceeded with a 24-hour rhythmicity (Fig. 7), convincing of an endogenous circadian driving force behind these rhythms.
The teleological role of circadian rhythms is postulated to coordinate behavioral rhythms (activity, feeding, sleeping) to the anticipated energetic needs of an organism (Green et al., 2008). Of these functions, glucose homeostasis is paramount due to the obligate use of glucose by the brain and limited ability of the mammalian liver to store glycogen. Therefore, within a few hours after a meal, AAs derived from skeletal muscle provide the carbon skeleton for hepatic gluconeogenesis (Felig, 1975). However, as a consequence of utilizing carbons from AAs, mammalian organisms face the burden of eliminating the toxic nitrogenous waste. Our study identifies Klf15 as a clock-driven peripheral circadian factor essential for coordinating delivery of the carbon skeletons required for glucose production and elimination of ammonia to urea. The importance of this regulation is underscored by the fact that failure to faithfully coordinate rhythmic utilization and excretion of amino acids can lead to severe pathophysiological consequences. Indeed, failure of rhythmic amino acid utilization leads to persistently low glucose with impaired circadian glucose homeostasis (Fig. 3), whereas failure of rhythmic excretion causes hyperammonemia and attendant cognitive dysfunction (Fig. 4).
Our study also links the circadian clock to nitrogen homeostasis through a Klf15-dependent mechanism (graphical abstract). Klf15 rhythm was disrupted in several circadian clock mutant mouse lines, and our data supports a direct clock-dependent transcriptional basis for this regulation (Fig. 1e, Supplemental Fig. 1c,d). This was also confirmed in an independent genome wide ChIP-sequencing study that identified rhythmic BMAL1 binding to the Klf15 promoter (Rey et al., 2011). Feeding rhythms have been identified to play an important role in entraining the peripheral clock (Damiola et al., 2000). Further, depleting food intake led to failure of rhythmic variation of transcripts in the liver, whereas, restricted feeding in a clock mutant induced oscillation of several transcripts in the liver (Vollmers et al., 2009). Since nitrogen utilization and disposal largely occurs in peripheral organs, we examined the role of restricted feeding, and identified that feeding plays an important role in rhythmic nitrogen homeostasis by altering expression of Klf15, and other amino acid utilization enzymes (Fig. 5). The essential role of Klf15 in mediating amino acid rhythms was also evident from the observation that normal feeding/activity rhythms and near-normal hepatic clock gene expression rhythms (Supplemental Fig. 2) in the Klf15-null mice were not sufficient to maintain amino acid rhythms. Further, administration of a high protein diet was also not sufficient to overcome the enzymatic deficiencies caused by the absence of Klf15 (Fig. 6). Thus, our studies suggest a central role for Klf15 in transcriptionally coupling rhythmic variation in amino acid metabolic enzymes to the circadian clock and feeding rhythms. There are several important limitations to our work. First, because Klf15 is under control of Bmal1 (Fig. 1), our observations suggest that nitrogen homeostasis may also be under the control of the circadian clock. However, studies assessing nitrogen homeostasis in Clock-mutant or Bmal1 null mice are likely to be confounded by abnormal feeding rhythms, altered food intake, and body composition (Turek et al., 2005; Shi et al., 2010). Further, since nitrogen homeostasis occurs across multiple organs – compound tissue specific deletion models in Clock mutants will be needed to investigate this issue. While such studies are decidedly non-trivial, they are clearly of interest and will provide a more comprehensive understanding of the link between the circadian clock and rhythmic nitrogen homeostasis in mammalian organisms.
Finally, because AAs can activate many signaling pathways and are precursors for hormones/nitrogenous substances (Wu, 2009), dysregulation of rhythmic utilization and excretion may play a pathogenic role in the onset of common disease states. For example, restricted feeding also resulted in a marked elevation of the total AA pool, BCAA and alanine (Fig. 5f, m & n). Interestingly, this was associated with a significant increase in plasma insulin (Fig. 5o and Supplemental Fig. 7), and is consistent with the well-known relationship between shift work and insulin resistance (Scheer et al., 2009). Indeed, studies have identified a pathogenic role for altered BCAA levels in the development of insulin resistance (Newgard et al., 2009). More recently, Shah et al. identified a significant relationship between BCAA levels, urea cycle metabolites in patients with coronary artery disease (Shah et al., 2010). Finally, Klf15-null mice develop cardiac and vascular myopathy characterized by heart failure and aneurismal aortic dilatation following exposure to angiotensin II infusion (Haldar et al., 2010). These findings support a potential link between impaired nitrogen homeostasis and susceptibility to common disease states. Thus, examination of rhythmic changes in nitrogen homeostasis may have wide clinical implications for diagnosis, prognosis and therapy.
EXPERIMENTAL PROCEDURES
Mice and dietary perturbations
Wildtype male mice on C57BL6/J background were purchased from The Jackson Laboratory (Bar Harbor, ME), and acclimatized to our facility for 3–4 weeks. Generation of systemic Klf15-null mice was previously described (Fisch et al., 2007), and Klf15-null mice have been backcrossed into the C57BL6/J background for over 10 generations. WT and Klf15-null mice were housed under strict light/dark conditions (6 AM lights on and 6PM lights off), and had free access to standard chow/water. For L/D experiments, mice were euthanized with CO2 inhalation or isoflurane every four hours for 24 hours. For D/D experiments, mice were placed in complete darkness for 38 hours (starting at the end of light phase ZT12) followed by harvest every four hours over a 24-hour period. Studies on the Bmal1-null, Per1/2 double KO, Per2/Cry1 KO and Reverba were as previously described (Hogenesch et al., 2000; Oster et al., 2002; Preitner et al., 2002; Zheng et al., 2001). For restricted feeding studies, mice had free access to food (ad libitum group) or only during the light phase ZT3-ZT9 for one month. High protein diet feeding was conducted for one week: the control diet for this study consisted of 18.1% protein, 62.3% carbohydrate and 6.2% fat (TD110483, Harlan laboratories), and high protein diet was 70% casein, 18.7% carbohydrate and 6.2% fat (TD 03367, Harlan laboratories).
Physiological studies
Wheel-running behavior was monitored in mice using Clocklab software (Actimetrics). Animals were housed in light-dark until stably entrained, then wheel running was recorded for three weeks in L/D, three weeks in D/D, and 17 days in L/L. Food intake analysis was measured using the Dietmax system (Accuscan Instruments, Columbus, OH) every five minutes at the Cincinnati mouse metabolic phenotyping core (MMPC).
Tissue harvest and biochemical analysis
Prior to euthanasia, glucose was measured from tail blood using glucometer (Accu-Check, Roche Diagnostics). Following this, mice were euthanized and blood collected from the inferior vena cava with a heparin-coated syringe. For constant dark experiments, following euthanasia the optic nerves were severed before proceeding to organ harvest. Organs were washed in cold phosphate buffered saline, and flash frozen in liquid nitrogen. Plasma was separated by centrifugation and frozen in aliquots for analysis. Plasma insulin, glucagon were measured using radio-immuno assay, and amino acids, ammonia, urea and ornithine were measured using high performance liquid chromatography at the Vanderbilt MMPC. Tissue amino acids, ammonia, urea and ornithine were extracted by homogenizing weighed tissue pieces in 10% 5-sulphosalicylic acid dissolved in distilled water. Briefly, amino acid analysis was performed using a Biochrom 30 analyzer, a PC controlled automatic liquid chromatograph with a post column detection system. Prepared samples are injected into a column of cation exchange resin and separated by buffers of varying pH and ionic strength. They are then reacted with ninhydrin at 1350 C and the absorbance maxima read at 440 and 570nm. The retention time of the peak identifies the amino acid, and the area under the peak indicates the quantity present. Samples were prepared by deproteinizing with 10% SSA (5-Sulfosalicylic Acid) and centrifugation. The resulting supernatant was then added to an equal quantity of Lithium citrate loading buffer, which lowers the pH prior to introduction to the cation exchange column. A known quantity of Norleucine may also be added to act as an internal standard. The PC presents both a detailed chromatogram of each sample and the amount of each amino acid based on comparison to a standard mixture of acidic and basic amino acids. These results were confirmed by manual calculations based on the area of each peak.
OTC activity analysis
Hepatic mitochondrial extracts were prepared, and OTC activity measured as previously described (Lee and Nussbaum, 1989; Ye et al., 1996).
Cell culture studies
For adenoviral overexpression studies, Hepa1–6 cells or AML12 mouse hepatocyte cell lines and differentiated C2C12 cells were used. For AA analysis, adenoviral overexpression was performed for 48 hours in serum-free media. The media was removed, spun to remove debris, deproteinized and analyzed for AA, ornithine concentration. The promoter region of Klf15 (-5kb before the translation start site in the second exon) was cloned into PGL3 reporter vector (Promega, Madison, WI). Transient transfection studies were conducted in HepG2 cells using Fugene HD (Roche, Indianapolis, IN).
RNA isolation and Real-time PCR analysis
RNA was isolated from frozen liver and skeletal muscle samples by homogenization in Trizol reagent by following manufacturer’s instructions (Invitrogen, Carlsbad, CA). RNA was reverse transcribed following DNAse treatment. Real-time PCR was performed using standard or LNA based Taqman approach with primers/probes designed and validated from the Universal Probe Library (Roche, Indianapolis, IN). The results were normalized to Beta actin or Gapdh.
Western Immunoblotting
Liver samples were homogenized in buffer containing 50mM Tris, 150mM NaCl, 1mM EDTA, 1% Triton X-100, 0.5% Sodium deoxycholate and 1% SDS supplemented with protease and phosphatase inhibitors (Roche, Indianapolis, IN). Nuclear lysates were prepared using the NE-PER kit following manufacturer’s instructions (Thermo Scientific, Rockford, IL). A goat polyclonal antibody against KLF15 was used (ab2647, Abcam, Cambridge, MA).
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed from mouse livers as previously described (Tuteja et al., 2009)(Ripperger and Schibler, 2006). Briefly, wildtype mouse livers were harvested every four hours and fixed with 11% formaldehyde for 10 minutes. Following this, chromatin was prepared and sonicated using Bioruptor (Diagnode, Sparta, NJ). The sonicated chromatin was flash frozen in liquid nitrogen and stored at −80F for subsequent analysis. Immunoprecipitation was conducted using Dynabeads (Invitrogen, Carlsbad, CA) bound to BMAL1 or KLF15 antibody. The promoter regions of putative KLF15 target genes were manually examined for conserved regions using Kalign (EMBL-EBI). Real-time PCR analysis was performed using following primers Alt (F – aactagctgtcccgtctcca and R – ctctgatgagccactgcaag), Otc (F- acctgggctcagttagggtag R- cgtcatgatttgtaatgacctaaga), 28S (F- ctgggtataggggcgaaagac R- ggccccaagacctctaatcat), non-target region (F- cctctgtgcctgtgaagga R- catcagtgtcccctgacaga). The relative abundance was normalized to abundance of 28S between the input and immunoprecipitated samples as previously described (Tuteja et al., 2009).
Neurobehavioral analysis
Described under the supplemental methods section.
Human study
We studied 10 adult participants [5 female; mean age 25.5 years (range 19–41 years); mean body mass index 25.1 kg/m2 (20–28 kg/m2)], as previously published (Scheer et al., 2009). The forced desynchrony protocol consisted of 7 recurring 28-h sleep–wake cycles under dim light conditions (±1.8 lux) to minimize any influence of light on the circadian system. During this period a fasting plasma sample was collected from each participant every 28 h across the full 196 h of the forced desynchrony, with each sample collected approximately 12 hours since the last meal (within 1 hour of awakening and prior to breakfast). By analyzing the plasma samples every 28 h throughout the forced desynchrony protocol, samples were distributed across the circadian cycle, allowing assessment of the effects of the circadian system, independent of the effects of the behavioral cycles of sleep/fasting and wake/eating (Scheer et al., 2009). A sufficient number of samples across the entire circadian cycle was available for analysis in 8 of these 10 participants.
Statistical analysis
All data are presented as mean ± SEM. For circadian mouse studies, the statistical significance between time points to assess rhythmicity was performed using analysis of variance (ANOVA). The statistical difference between two individual groups was assessed using the Student’s t-test. P<0.05 was considered significant. For the human forced desynchrony protocol, the effect of the endogenous circadian rhythm was analyzed using cosinor analyses including the circadian (fundamental ~24-h) rhythmicity and a linear component (hours since the start of the forced desynchrony protocol) and mixed model analysis of variance with restricted maximum likelihood (REML) estimates of the variance components (JMP, SAS Institute) (Nelson et al., 1979). For analytes without significant effect of the linear component, a simple cosine model was used. Continuously recorded core body temperature was used as individualized circadian phase marker in the human study (Scheer et al., 2009).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Drs. Alfred F. Connors Jr, David S. Rosenbaum, Jonathan S. Stamler, Douglas T. Hess and Satish C. Kalhan for support and suggestions, to Dr. Ueli Schibler for reagents, to Drs. John Le Lay and Klaus H. Kaestner for providing chromatin immunoprecipitation protocol, to Ken Grimes and Wanda Snead at the Vanderbilt MMPC, to Dana Lee at the Cincinnati MMPC, to Jenny Marks at BWH, to Diana Awad Scrocco for proof-reading, to Mike Mustar for artistic illustrations, CWRU rodent behavioral core and to members of the Jain laboratory for their assistance. Funding sources: NIH grants HL094660 (D.J.), R01-HL09480601 and P30-HL101299 (F.A.J.L.S.), K24-HL76446 (S.A.S.), HL072952 (S.M.H.), HL097023 (G.H.M.), HL075427, HL076754, HL084154, HL086548, HL097595 (M.K.J.), DK059630 (Cincinnati MMPC), DK59637 (Vanderbilt MMPC),SNF grant 31003A/131086 (U.A.), and M01-RR02635 (BWH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Balandreau JP, Millier CR, Dommergues YR. Diurnal variations of nitrogenase activity in the field. Appl Microbiol. 1974;27:662–665. doi: 10.1128/am.27.4.662-665.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A, Urquhart AA, Joy KW. Amino Acid Metabolism of Pea Leaves: Diurnal Changes and Amino Acid Synthesis from N-Nitrate. Plant Physiol. 1977;59:915–919. doi: 10.1104/pp.59.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosnan ME, Brosnan JT. Hepatic glutamate metabolism: a tale of 2 hepatocytes. Am J Clin Nutr. 2009;90:857S–861S. doi: 10.3945/ajcn.2009.27462Z. [DOI] [PubMed] [Google Scholar]
- D'Antona G, Ragni M, Cardile A, Tedesco L, Dossena M, Bruttini F, Caliaro F, Corsetti G, Bottinelli R, Carruba MO, et al. Branched-chain amino acid supplementation promotes survival and supports cardiac and skeletal muscle mitochondrial biogenesis in middle-aged mice. Cell metabolism. 2010;12:362–372. doi: 10.1016/j.cmet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & development. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annu Rev Physiol. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Duez H, Staels B. The nuclear receptors Rev-erbs and RORs integrate circadian rhythms and metabolism. Diab Vasc Dis Res. 2008;5:82–88. doi: 10.3132/dvdr.2008.0014. [DOI] [PubMed] [Google Scholar]
- Feigin RD, Dangerfield HG, Beisel WR. Circadian periodicity of blood amino acids in normal and adrenalectomized mice. Nature. 1969;221:94–95. doi: 10.1038/221094a0. [DOI] [PubMed] [Google Scholar]
- Feigin RD, Klainer AS, Beisel WR. Circadian periodicity of bloof amino-acids in adult men. Nature. 1967;215:512–514. doi: 10.1038/215512b0. [DOI] [PubMed] [Google Scholar]
- Felig P. Amino acid metabolism in man. Annu Rev Biochem. 1975;44:933–955. doi: 10.1146/annurev.bi.44.070175.004441. [DOI] [PubMed] [Google Scholar]
- Fernstrom JD, Wurtman RJ, Hammarstrom-Wiklund B, Rand WM, Munro HN, Davidson CS. Diurnal variations in plasma neutral amino acid concentrations among patients with cirrhosis: effect of dietary protein. Am J Clin Nutr. 1979;32:1923–1933. doi: 10.1093/ajcn/32.9.1923. [DOI] [PubMed] [Google Scholar]
- Fisch S, Gray S, Heymans S, Haldar SM, Wang B, Pfister O, Cui L, Kumar A, Lin Z, Sen-Banerjee S, et al. Kruppel-like factor 15 is a regulator of cardiomyocyte hypertrophy. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7074–7079. doi: 10.1073/pnas.0701981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster RG, Roenneberg T. Human responses to the geophysical daily, annual and lunar cycles. Curr Biol. 2008;18:R784–R794. doi: 10.1016/j.cub.2008.07.003. [DOI] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Wang B, Orihuela Y, Hong EG, Fisch S, Haldar S, Cline GW, Kim JK, Peroni OD, Kahn BB, et al. Regulation of gluconeogenesis by Kruppel-like factor 15. Cell metabolism. 2007;5:305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gropman AL, Summar M, Leonard JV. Neurological implications of urea cycle disorders. J Inherit Metab Dis. 2007;30:865–879. doi: 10.1007/s10545-007-0709-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldar SM, Lu Y, Jeyaraj D, Kawanami D, Cui Y, Eapen SJ, Hao C, Li Y, Doughman YQ, Watanabe M, et al. Klf15 deficiency is a molecular link between heart failure and aortic aneurysm formation. Sci Transl Med. 2010;2:26ra26. doi: 10.1126/scitranslmed.3000502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper AE, Miller RH, Block KP. Branched-chain amino acid metabolism. Annu Rev Nutr. 1984;4:409–454. doi: 10.1146/annurev.nu.04.070184.002205. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu YZ, Moran SM, Shimomura K, Radcliffe LA, Takahashi JS, Bradfield CA. The basic helix-loop-helix-PAS protein MOP9 is a brain-specific heterodimeric partner of circadian and hypoxia factors. J Neurosci. 2000;20:RC83. doi: 10.1523/JNEUROSCI.20-13-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum Mol Genet. 2006;15(Spec No 2):R271–R277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Sasso GL, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JT, Nussbaum RL. An arginine to glutamine mutation in residue 109 of human ornithine transcarbamylase completely abolishes enzymatic activity in Cos1 cells. The Journal of clinical investigation. 1989;84:1762–1766. doi: 10.1172/JCI114360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, et al. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell BB, Yang VW. Mammalian Kruppel-like factors in health and diseases. Physiol Rev. 2010;90:1337–1381. doi: 10.1152/physrev.00058.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Kruppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, Kasukawa T, Kakazu Y, Iigo M, Sugimoto M, Ikeda S, Yasui A, van der Horst GT, Soga T, Ueda HR. Measurement of internal body time by blood metabolomics. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:9890–9895. doi: 10.1073/pnas.0900617106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SM., Jr Regulation of enzymes of the urea cycle and arginine metabolism. Annu Rev Nutr. 2002;22:87–105. doi: 10.1146/annurev.nutr.22.110801.140547. [DOI] [PubMed] [Google Scholar]
- Nelson W, Tong YL, Lee JK, Halberg F. Methods for cosinor-rhythmometry. Chronobiologia. 1979;6:305–323. [PubMed] [Google Scholar]
- Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell metabolism. 2009;9:311–326. doi: 10.1016/j.cmet.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster H, Yasui A, van der Horst GT, Albrecht U. Disruption of mCry2 restores circadian rhythmicity in mPer2 mutant mice. Genes & development. 2002;16:2633–2638. doi: 10.1101/gad.233702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Reddy AB, Karp NA, Maywood ES, Sage EA, Deery M, O'Neill JS, Wong GK, Chesham J, Odell M, Lilley KS, et al. Circadian orchestration of the hepatic proteome. Curr Biol. 2006;16:1107–1115. doi: 10.1016/j.cub.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F. Genome-Wide and Phase-Specific DNA-Binding Rhythms of BMAL1 Control Circadian Output Functions in Mouse Liver. PLoS Biol. 2011;9:e1000595. doi: 10.1371/journal.pbio.1000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rudic RD, McNamara P, Curtis AM, Boston RC, Panda S, Hogenesch JB, Fitzgerald GA. BMAL1 and CLOCK, two essential components of the circadian clock, are involved in glucose homeostasis. PLoS Biol. 2004;2:e377. doi: 10.1371/journal.pbio.0020377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheer FA, Hilton MF, Mantzoros CS, Shea SA. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Bain JR, Muehlbauer MJ, Stevens RD, Crosslin DR, Haynes C, Dungan J, Newby LK, Hauser ER, Ginsburg GS, et al. Association of a peripheral blood metabolic profile with coronary artery disease and risk of subsequent cardiovascular events. Circ Cardiovasc Genet. 2010;3:207–214. doi: 10.1161/CIRCGENETICS.109.852814. [DOI] [PubMed] [Google Scholar]
- Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian clock gene Bmal1 is not essential; functional replacement with its paralog, Bmal2. Curr Biol. 2010;20:316–321. doi: 10.1016/j.cub.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu BP, Mohler RE, Liu JC, Dombek KM, Young ET, Synovec RE, McKnight SL. Cyclic changes in metabolic state during the life of a yeast cell. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16886–16891. doi: 10.1073/pnas.0708365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, et al. Obesity and metabolic syndrome in circadian Clock mutant mice. Science (New York, NY. 2005;308:1043–1045. doi: 10.1126/science.1108750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja G, White P, Schug J, Kaestner KH. Extracting transcription factor targets from ChIP-Seq data. Nucleic Acids Res. 2009;37:e113. doi: 10.1093/nar/gkp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science (New York, NY. 2009;325:435–439. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler CT. Diurnal Fluctuation in Nitrogen Fixation in Nodules of Alnus Glutinosa and Myrica Gale. New Phytologist. 1969;68 675-&. [Google Scholar]
- Wu G. Amino acids: metabolism, functions, and nutrition. Amino Acids. 2009;37:1–17. doi: 10.1007/s00726-009-0269-0. [DOI] [PubMed] [Google Scholar]
- Ye X, Robinson MB, Batshaw ML, Furth EE, Smith I, Wilson JM. Prolonged metabolic correction in adult ornithine transcarbamylase-deficient mice with adenoviral vectors. The Journal of biological chemistry. 1996;271:3639–3646. doi: 10.1074/jbc.271.7.3639. [DOI] [PubMed] [Google Scholar]
- Yin L, Wu N, Curtin JC, Qatanani M, Szwergold NR, Reid RA, Waitt GM, Parks DJ, Pearce KH, Wisely GB, et al. Rev-erbalpha, a heme sensor that coordinates metabolic and circadian pathways. Science (New York, NY. 2007;318:1786–1789. doi: 10.1126/science.1150179. [DOI] [PubMed] [Google Scholar]
- Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, et al. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.