Abstract
Independently folded domains in RNAs frequently adopt identical tertiary structures regardless of whether they are in isolation or are part of larger RNA molecules. This is exemplified by the P15 domain in the RNA subunit (RPR) of the universally conserved endoribonuclease P, which is involved in the processing of tRNA precursors. One of its domains, encompassing the P15 loop, binds to the 3′-end of tRNA precursors resulting in the formation of the RCCA–RNase P RNA interaction (interacting residues underlined) in the bacterial RPR–substrate complex. The function of this interaction was hypothesized to anchor the substrate, expose the cleavage site and result in re-coordination of Mg2+ at the cleavage site. Here we show that small model-RNA molecules (~30 nt) carrying the P15-loop mediated cleavage at the canonical RNase P cleavage site with significantly reduced rates compared to cleavage with full-size RPR. These data provide further experimental evidence for our model that the P15 domain contributes to both substrate binding and catalysis. Our data raises intriguing evolutionary possibilities for ‘RNA-mediated’ cleavage of RNA.
INTRODUCTION
Like proteins, RNAs are composed of different domains and these can have different functions, for example one domain can bind a small ligand or cofactor while another constitutes the active site (1; see also Refs 2 and 3). When separated, the domains can fold in a similar way compared to the fold they have in the full-length RNA molecule. This is exemplified by the P4-P6 domain of the Tetrahymena group I intron (4–6; for a review see, e.g. Ref. 7) and the group II intron where domain five retains its catalytic activity when separated from the full-length RNA (8–12; for a review see, e.g. Ref. 13). This is also true for the catalytic RNA component (RPR) of the universally conserved endoribonuclease P, which is involved in the processing of tRNA precursors [Figure 1 (14–21)].
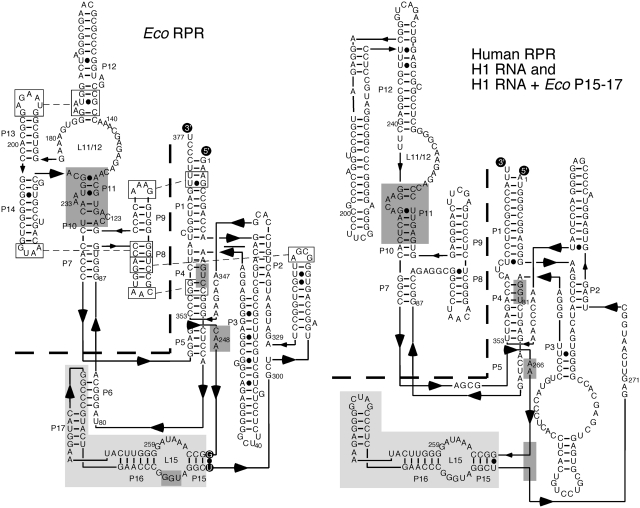
Figure 1.
Predicted secondary structures of wild-type Eco RPR and human RPR, H1 RNA (58). The specificity (S) and catalytic (C) domains are separated with the dashed line and the Eco RPR P15–P17 domain is highlighted in light grey. The Eco RPR P15–P17 domain was introduced into H1 RNA generating H1 RNA + Eco P15–P17 as indicated at the position marked with a grey box. Eco RPRΔP15–P17 carries a deletion of P15–P17 except the GU pair marked with black circles (22). The highlighted regions in dark grey mark regions in RPR (including P15–P17) known to be important for binding of the substrate and that show structural differences, for details see main text. P refers to helices in Eco RPR and H1 RNA.
We recently provided experimental evidence that two eukaryotic, human and Giardia lamblia, RPRs mediate cleavage in the absence of proteins. However, the rates were dramatically lower compared to the bacterial RPR. Human RPR (H1 RNA) displays a rate 106–107-fold lower compared to Escherichia coli RPR (M1 RNA or Eco RPR; 22). Although the core RPR structure is conserved, there are important structural differences between human and bacterial RPRs (23). For example, the P15–P17 domain with its internal loop, the P15 loop, is missing in H1 RNA (Figure 1). The P15 loop constitutes the binding site for the 3′-end (the 3′-RCCA motif; interacting residues underlined) of the precursor tRNA (pre-tRNA) substrate (24). Deleting this region in Eco RPR results in a 103–104-fold reduction in the rate of cleavage (22). To increase the cleavage activity of H1 RNA, we introduced Eco RPR's P15–P17 domain into H1 RNA. Here we report that although such a chimeric RPR did not improve the cleavage activity, small RNAs containing only the P15 loop were able to mediate cleavage of both model-RNA hairpin substrates and full-length tRNA precursors. This surprising finding provides new insight into RPR-mediated cleavage.
MATERIALS AND METHODS
Preparation of RPR, small model RNAs containing the P15 loop, Eco RPR domain constructs and substrates
Eco RPR was prepared as run-off transcripts using T7 DNA-dependent RNA polymerase as described (Ref. 22 and references therein). The pATSerUG substrate derivatives were either purchased from Dharmacon USA (Lafayette, CO) or prepared as run-off transcript using T7 DNA-dependent RNA polymerase (22). The tRNA precursors pSu1 and pSu3 were generated as run-off transcript following standard procedures. The substrates, pATSerUG and pSu1, were 5′-end labelled using γ-32P-ATP as described (22) while pSu3 and also pATSerUG (where the latter was used in the TLC assays, see below) were internally labeled with α-32P-UTP and α-32P-GTP, respectively (final specific activity ≥ 5 Ci/mmol).
The P15–P17 and P15–P15.1 RNA constructs were generated from PCR templates using the following primers: P15–P17 F-primer and P15–P17 R-primer, P15–P15.1 F-primer and P15–P15.1 R-primer (Supplementary Table S1). The PCR templates were designed to carry the T7 Promoter sequence at the 5′-end as described (22). The P15 PCR-product (P15 RNA) was cloned into a plasmid, subsequently linearized and used as a transcription template for T7 DNA-dependent RNA polymerase. The chemically synthesized (unmodified and modified) versions of the P15 RNA were purchased from Dharmacon USA (Lafayette, CO, USA). The transcribed constructs and the chemically synthesized constructs were gel purified as described (25). For preparation of the H1P15–P17 RNA and Eco RPR domain constructs, see Supplementary Information.
Assay conditions and determination of the kinetic constants under single-turnover conditions
The cleavage reactions were performed in buffer C [50 mM 4-morpholineethanesulfonic acid (MES), 0.8 M NH4OAc pH 6.0] at 37°C in the presence of 160 mM Mg(OAc)2, or as indicated. In all reactions, before mixing substrate with the catalytic RNA, the latter were pre-incubated at 37°C in buffer C and 160 mM Mg(OAc)2 for 10 min; the substrates were subsequently added and incubated as indicated. The reactions were performed under single-turnover conditions at concentrations of substrate and catalytic RNA as indicated in the figure.
For the CP RNA, the kinetic constants kobs and kobs/Ksto (= kcat/KM) were determined under saturating single-turnover conditions in buffer C (160 mM Mg2+ pH 6.0) as previously described (26–28). At this pH, the chemistry of cleavage of pATSerUG by full-length Eco RPR is suggested to be rate limiting (29). Briefly, kobs and kobs/Ksto were calculated [by linear regression from Eadie–Hofstee plots (30,31)] by determination of the initial rates at different CP RNA concentrations, ranging from 0.1 to 37.5 µM. The final concentration of substrate was ≤20 nM. The 5′-leader product was quantitated and used for calculating the extent of cleavage. The rate constants kobs and kobs/Ksto are defined on the basis of the following simplified reaction scheme.
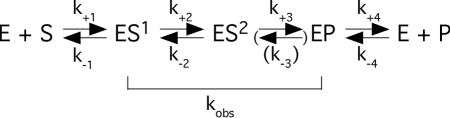
In Scheme 1, kobs reflects the rate of cleavage as indicated while k+1 equals the rate constant kobs/Ksto (= kcat/Km). On the basis of previous reports (27,28,32,33) it is likely that k−1 >> kobs. Hence, Ksto ~ Kd under the reaction conditions used in the present study. The rate constant k−3 and the corresponding arrow are put in parenthesis since there is no precedent for the reverse reaction, i.e. ligation.
Analysis of the 5′-end of the 5′-matured cleavage product
The cleavage site was inferred by comparing the mobility of the 5′-cleavage fragments generated by using the different catalytic RNAs. The presence of pGp at the 5′-end of the large cleavage product was verified by two-dimensional thin-layer chromatography (TLC) as described (34), using α-32P-GTP internally labeled pATSerUG (specific activity at least 5 Ci/mmol) as substrate (22). As a control, we used pATSerUG cleaved with wild-type Eco RPR. Incubation of the substrate alone did not result in the occurrence of pGp at the 5′-end of the large cleavage product (22).
RESULTS
The domain of Eco RPR that interacts with the 3′-end of the substrate mediates cleavage on its own
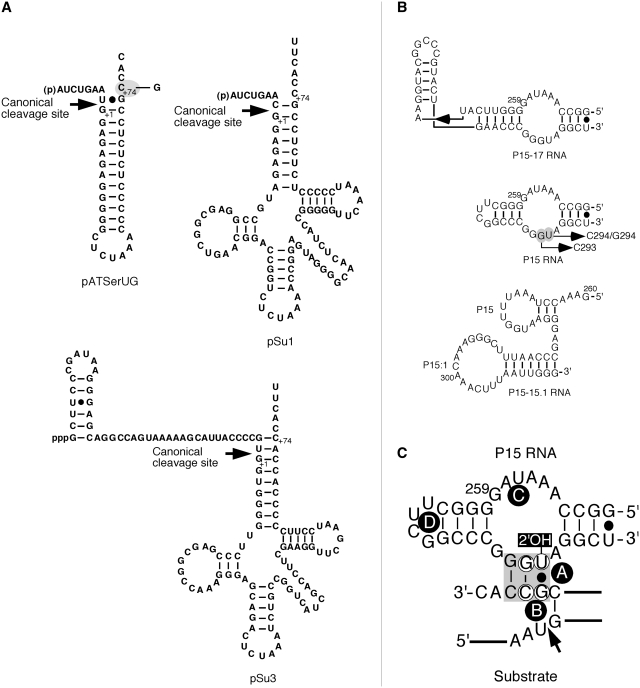
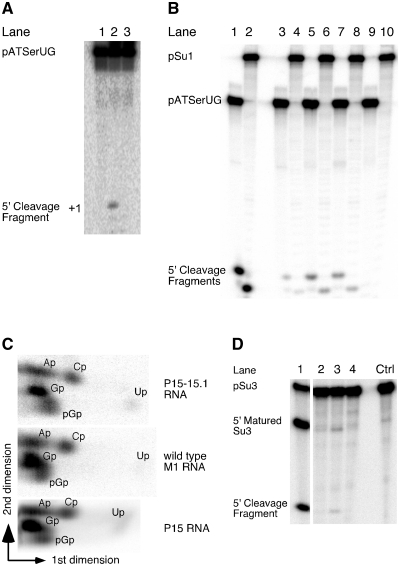
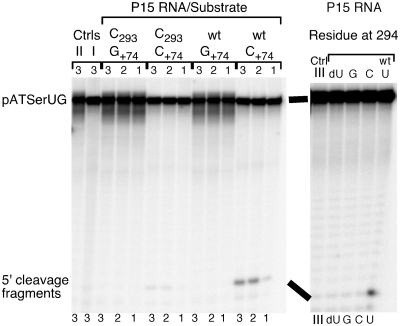
Given the low cleavage activity for H1 RNA, our initial aim was to investigate whether it is possible to increase the H1 RNA activity by the introduction of missing domains, in particular the Eco RPR's P15–P17 domain (Figure 1). As substrate we used the well-characterized model hairpin loop substrate pATSerUG (Figure 2A). Consistent with our previous data, H1 RNA mediated cleavage of the model substrate pATSerUG with a low level (14; Figure 3A). Introduction of the Eco RPR P15–P17 domain (Figure 1) into H1 RNA (or addition of the P15–P17 RNA in trans, data not shown) did not improve the rate of cleavage of pATSerUG (Figure 3A) under conditions where it is possible to monitor weak RPR cleavage activity using the model substrate pATSerUG (see ‘Materials and Methods’ section). For the H1P15–P17 RNA construct (Figure 3A; cf. lane 3) no cleavage was detected, which might be due to the folding of the RNA but this was not further investigated. However, surprisingly, the P15–P17 RNA alone promoted cleavage at a low rate (Figures 2B and 3B; cf. lane 5). On the basis of the mobility of the 5′-cleavage fragment, we inferred that pATSerUG was cleaved at the canonical cleavage site +1. To define the region in the P15–P17 RNA responsible for mediating the cleavage, we used a second small RNA representing only the P15 stem, the P15 loop and part of the P16 stem (Figure 2B). The solution structure of this RNA, referred to as P15 RNA, has been determined by NMR spectroscopy (35). The P15 RNA also cleaved pATSerUG at a low rate at the same position as wild-type Eco RPR and P15–P17 RNA did (cf. lanes 1, 3 and 5 Figure 3B). We confirmed by TLC using α-32P-GTP internally labeled pATSerUG that cleavage mediated by the P15–P17 RNA and the P15 RNA generated pGp (36), the hallmark of RNase P-mediated cleavage (Figure 3C; data not shown for the P15–P17 RNA; spontaneous or metal(II)-ion induced cleavage would not have resulted in the appearance of pGp, see e.g. Ref. 22). Hence, this demonstrates the presence of 5′-phosphate and 3′-hydroxyl terminus after cleavage using these short truncated RNAs. We also tested cleavage of the full-length tRNA precursors pSu1 and pSu3 (Figure 2A) and as shown in Figure 3B (cf. lanes 4 and 6) both P15 RNA and P15–P17 RNA promoted cleavage of pSu1 with a low rate. The mobility of the 5′-cleavage fragments suggests that both these RNAs cleaved pSu1 at the canonical cleavage site. In the case of pSu3, we also detected weak cleavage however this was only observed for the P15–P17 RNA (Figure 3D). A likely reason for the weak cleavage of pSu3, compared to pATSerUG and pSu1, is that A+73 and C+74 are less accessible to pairing with residues ‘C293’ and ‘U294’ in the P15 loop since they are paired with U−1 and G−2, respectively [Figure 2A and C; see also below (18,19)].
Figure 2.
(A) Secondary structures of pATSerUG, pSu1 and pSu3. The canonical RNase P cleavage sites are marked with arrows. The 3′-CCA residues in pATSerUG correspond to positions +74, +75 and +76 in full-length tRNA precursors and consequently the numbering of these positions follows the numbering in the tRNA precursors. The residue C+74 in pATSerUG was mutated to G as indicated. pSu1 and pSu3 correspond to the precursors to tRNASerSu1 and tRNATyrSu3, respectively. (B) The secondary structures of P15–P17 RNA, P15 RNA, and P15–P15.1 RNA. These RNAs were generated as outlined in ‘Materials and Methods’ section. P15–P17 RNA and P15 RNA were based on Eco RPR (Figure 1) while the P15–P15.1 RNA was based on M. hyopneumoniae RPR (31). The highlight residue marked in grey corresponds to G293 and was changed to C as indicated. (C) Model illustrating the interaction between the P15 RNA and the RCC-motif of the substrate (grey area), the RCCA–RPR interaction (interacting residues underlined). The residues in the P15 RNA that correspond to residues C293 and U294 in Eco RPR are encircled. These residues were replaced, C293 with G and U294 with G or C. The encircled residues in the substrate correspond to G+73 and C+74 in the substrate, and C+74 was substituted with G. The 2′-OH of U294 is highlighted and was replaced with 2′-H. The arrow marks the cleavage site and A–D (encircled in black) corresponds to Mg2+ ions that have been identified in the P15 RNA and in the substrate (19).
Figure 3.
(A) Cleavage of pATSerUG with H1 RNA (lane 2) and H1P15–P17 RNA (lane 3). The concentrations of substrates and catalytic RNA were: ≤20 nM for the substrates, 4.8 µM for H1 RNA and 4.3 µM for H1P15–P17 RNA (Eco RPR P15–P17 inserted into H1 RNA; see Figure 1). Lane 1 pATSerUG incubated under the same conditions without RPR. Reaction times were 24 h in all cases. (B) Cleavage of pATSerUG and pSu1 with different RNAs. Lane 1, pATSerUG incubated with wild-type Eco RPR; lane 2, pSu1 incubated with wild-type Eco RPR; lane 3, pATSerUG incubated with P15 RNA; lane 4, pSu1 incubated with P15 RNA; lane 5, pATSerUG incubated with P15–P17 RNA; lane 6, pSu1 incubated with P15–P17 RNA; lane 7, pATSerUG incubated with P15–P15.1 RNA; lane 8, pSu1 incubated with P15–P15.1 RNA and lanes 9 and 10, incubation of pATSerUG and pSu1 alone, respectively. The concentrations of substrates and catalytic RNA were: ≤20 nM for the substrates, 0.16 µM for wild-type Eco RPR, 24 µM for P15–P17 RNA, 39 µM for P15 RNA and 23 µM for P15–P15.1 RNA. Reaction times were 20.5 h in all cases except for wild-type Eco RPR (10 sec). All reactions were performed at 160 mM Mg2+ in reaction buffer C at 37°C. (C) Two-dimensional TLC demonstrating the presence of pGp at the 5′-end of the 5′-matured cleavage product after cleavage of [α-32P]GTP internally labeled pATSerUG as indicated (14). (D) Cleavage of [α-32P]UTP internally labeled pSu3 (final specific activity ≥5 Ci/mmol) in reaction buffer C (see ‘Materials and Methods’ section) with different catalytic RNAs: lane 1 wild-type Eco RPR, lane 2 P15 RNA, lane 3 P15–P17 RNA and lane 4 P15–P15.1 RNA. The reaction time was 22 h in all cases except for wild-type Eco RPR (10 sec). The concentrations of: wild-type Eco RPR 0.16 µM, P15 RNA 39 µM, P15–P17 RNA 24 µM and P15–P15.1 23 µM. Negative control (Ctrl) incubation of pSu3 alone in reaction buffer C for 22 h. For further details see ‘Materials and Methods’ section.
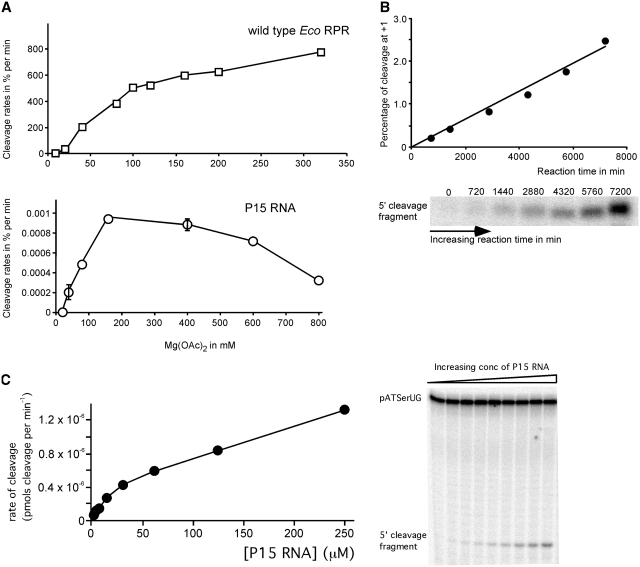
Comparing the Mg2+ profiles in cleavage of pATSerUG revealed that wild-type Eco RPR and the P15 RNA have similar Mg2+ requirements with a plateau around 160 mM. However, at higher [Mg2+], the rate of cleavage for the P15 RNA was lower compared to cleavage at 160 mM (Figure 4A). Moreover, the percentage of cleavage of pATSerUG with P15 RNA increased linearly over time (Figure 4B) and with increasing concentration of ribozyme (Figure 4C; complete cleavage in any of our catalytic RNA substrate combinations was not observed and maximum fraction of substrate converted into product was ~10%). From the data shown in Figure 2C, it is evident that the rate does not plateau at the highest concentration of P15 RNA tested. Therefore, the data could not be fit to obtain the kobs value. The absence of saturable behavior might be due to substrate binding being rate limiting and/or P15 RNA folding being sub-optimal. We know from our earlier NMR spectroscopy studies that the P15 RNA folds into different structures in solution however we do not know which of the structures are catalytically active (35). Although, the kobs value for H1 RNA is 2.6 × 10−6 min−1 (22), i.e. 106–107-fold lower compared to Eco RPR (Supplementary Table S2). On the basis of that observation, the percentage of cleavage of pATSerUG by P15 RNA is similar to that observed for H1 RNA under these conditions; therefore, we estimate that the rate of cleavage for P15 RNA is roughly in the same range as for H1 RNA. Moreover, we also note that the P15–P17 RNA appears to be more active than the P15 RNA and this might be due to the increased flexibility of the P15 RNA structure. It is also conceivable, but not mutually exclusive, that the larger P15–P17 RNA interacts more productively with the 5′-leader of the substrate.
Figure 4.
(A) Cleavage of pATSerUG with wild-type Eco RPR and P15 RNA expressed as a percentage of cleavage per min as a function of Mg2+. Concentration of: substrate ≤20 nM, wild-type Eco RPR 3.2 µM and P15 RNA 39.1 µM. Data are the average of two independent experiments. Bars indicate the experimental range. (B) Cleavage of pATSerUG in percentage as a function of time and accumulation of the 5′-cleavage fragment over time as indicated. Same concentrations of P15 RNA and substrate as in Figure 3 were used. (C) A typical experiment illustrating cleavage of pATSerUG with P15 RNA as a function of increasing concentration of P15 RNA. The concentration of substrate was ≤20 nM and the reaction time after mixing of substrate and P15 RNA was 25 h.
Structural changes in the P15 RNA and in the 3′-end of the substrate influence cleavage
The P15–P17 RNA and P15 RNA were generated using T7 DNA-dependent RNA polymerase. To eliminate the possibility that the RNA preparations contained trace amounts of Eco RPR, dot blot analysis and Reverse Transcription Polymerase Chain Reaction (RT-PCR) were performed (22). These two assays did not reveal any traces of Eco RPR in our small RNA preparations. Moreover, a chemically synthesized P15 RNA showed very similar cleavage properties as our original P15 RNA (not shown). In addition, substitution of residues participating in the RCCA–RNase P RNA, or RCCA–RPR, interaction (either in the P15 RNA or in the substrate and interacting residues underlined; Figure 2A and B) reduced cleavage compared to the ‘P15 RNA/wild-type pATSerUG’ situation (Figure 5; note that the P15 RNA variants were chemically synthesized). These results parallel our previous data using full-length Eco RPR and emphasizes the importance of the structural architecture of the RCCA–RPR interaction for catalysis (see also below; 24,29,37–41). From Figure 5 it is also apparent that the 2′-OH of U294 (Figure 2C) plays an important role for catalysis since replacement of this 2′-OH with a 2′-H resulted in a loss of activity under these conditions.
Figure 5.
Cleavage of different pATSerUG derivatives with chemically synthesized variants of the P15 RNA at 160 mM Mg2+ as indicated. The experiments were performed under single-turnover conditions at pH 6.0 and at 37°C as described in ‘Materials and Methods’ section. The concentrations of substrates and P15 RNA variants were ≤20 nM and 39.1 µM, respectively. In cleavage of the +74 variants the reaction times were 4 h (lanes labeled 1), 24.5 h (lanes labeled 2) and 26 h (lanes labeled 3). C+74 (wild-type pATSerUG) and G+74 refer to the identity of the residue at position 74 in the substrate while C293 refers to the identity of the residue at position 293 (wild-type: G293) in the P15 RNA (for comparison we use Eco RPR numbering; Figure 1A). In the right panel pATSerUG was cleaved with chemically synthesized P15 RNA variants carrying substitutions at position 294 (Figure 1C). A reaction time of 18 h was used while assaying the different P15 variants. Controls (Ctrl) incubation of substrate in reaction buffer C without the P15 RNA; Ctrl I pATSerUG (26 h); Ctrl II pATSerUG(G+74) (26 h); Ctrl III pATSerUG (18 h).
Taken together, these data eliminate the presence of a contaminating activity. Hence, together with the data discussed above, we conclude that the region of RPR interacting with the 3′-end of the substrate when separated from the full-length RPR can promote cleavage at the canonical RNase P cleavage site.
The P15 domain of a type B bacterial RPR mediates cleavage
On the basis of secondary structure, bacterial RPR can be divided into two main classes, type A (Ancestral) and type B (Bacillus like). One distinct structural difference is the structure of the P15–P17 domain, which in type B RPR is replaced with a P15 hairpin loop (Figure 2B). Type B RPR has also a second hairpin loop, P15.1 that is missing in type A [Figure 2B; (42)]. In spite of the difference in structure, the type B P15 hairpin loop interacts with the 3′-RCCA-motif of pre-tRNAs in a similar way as the type A relative (37). To investigate whether the P15 domain of type B promotes cleavage, we generated the P15–P15.1 RNA [based on Mycoplasma hyopneumoniae RPR, Hyo P RNA, Figure 2B (43)]. This RNA-mediated cleavage of pATSerUG and pSu1 at the expected positions (Figure 3B) with rates comparable to those observed for P15–P17 and P15 RNA (see above). The presence of pGp after cleavage of internally labeled pATSerUG was confirmed by TLC (Figure 3C). We also investigated whether an RNA representing just the P15 hairpin loop promoted cleavage but no activity was detected under these conditions (data not shown) indicating an important role for P15.1, perhaps in stabilizing the folding of the P15 loop. We conclude that a small RNA representing the P15–P15.1 domain of a type B RPR also mediates cleavage.
Importance of the P15–P17 domain in cleavage promoted by the Eco RPR C domain
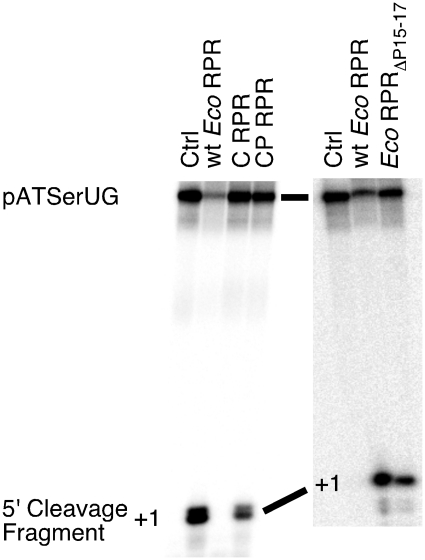
RNase P RNA consists of the catalytic (C) and the specificity (S) domains [Figure 1 (15,16)] where the C domain alone mediates cleavage in the absence and in the presence of the RNase P protein with significantly reduced activity (44,45). To investigate the role of the P15–P17 domain in relationship with the C domain, we tested constructs with and without the P15–P17 domain (Figure 1 and Supplementary Figure S1). Figure 6 shows that the C domain lacking P15–P17 (C construct) did not promote any detectable cleavage of pATSerUG under these conditions at 160 mM Mg2+ while its presence did (CP construct). The kobs for the CP construct was similar to the kobs previously determined for Eco RPRΔP15–P17 RNA (Supplementary Table S2) and in keeping with rates determined for cleavage using a C domain derived from a type B RPR (45). Note also that deleting P15–P17 in Eco RPR result in a catalyst with significantly reduced activity (Figure 6) in keeping with our previous findings (22; Supplementary Table S2). These data show that the C domain of type A mediates cleavage in the absence of protein and further corroborate the importance of the P15–P17 domain and its role for efficient catalysis. In this context, we note that the S domain is almost completely missing in some archaeal RPRs and these RPRs do mediate cleavage in the absence of proteins (46).
Figure 6.
Cleavage activities of pATSerUG for the Eco RPR C domain with and without the P15–P17 domain (left panel), and for wt Eco RPR and Eco RPRΔP15–P17 (right panel). Amount of RNA added: C construct 7 µM and CP construct 5.5 µM. For wt Eco RPR and Eco RPRΔP15–P17, we used 0.8 µM and 2.8 µM, respectively. Irrespective of RPR construct the concentration of γ-32P 5′-end labeled pATSerUG was ≤20 nM. Time of incubations were: 4 h for the Eco RPR C-domain variants, 0.5 min for wt Eco RPR and 4 h for Eco RPRΔP15–P17. Ctrl (control): incubation of pATSerUG alone for 4 h in the reaction buffer C.
DISCUSSION
The conserved P15 loop in bacterial RPR plays an important role in substrate binding and catalysis (18–21). Here we showed that the P15 loop RNA mediates cleavage of various substrates when separated from the full-length RPR. In the group II intron system the most conserved region, domain V (D5) plays a key role for catalytic activity and as part of small RNA molecules it catalyzes hydrolysis of the exon–intron junction when added in trans (47–49). Hence, this suggests similarities comparing RPR and the group II intron RNA and emphasizes that RNAs with complex structure are composed of individually functional domains.
Deleting the P15-loop region in a full-length RPR does not abolish but reduces cleavage activity significantly as in the case of e.g. human RNase P RNA, H1 RNA (Supplementary Table S2 and Figure 6; 22). Moreover, compared to Eco RPR, the rate of cleavage for P15 RNA and H1 RNA are significantly reduced with rates a few orders of magnitude higher than the rate constant for spontaneous hydrolysis (22,50), supporting the notion that cleavage depends on several determinants in the substrate. In keeping with this expectation is the finding that deleting the S domain resulting in the CP construct (this report) or the P15–P17 domain caused an ~4000-fold lower rate while no detectable cleavage was observed at pH 6 and 160 mM Mg2+ when the S domain and P15–P17 were deleted (Figure 6 and Supplementary Table S2). Consequently, dependent on the number of determinants in the substrate (18,26), or determinant binding sites in RPR, the efficiency of cleavage is affected to a varying extent. This idea helps rationalize the ability of the P15 RNA to promote cleavage since it can interact productively with the 3′-end of the substrate, i.e. formation of the RCCA–RPR interaction (Figure 2C). Also, compared to Eco RPR, H1 RNA shows structural differences in domains that are important substrate interaction sites that likely rationalize why human RPR is such a poor catalyst. Apart from the P15–P17 domain, other known regions that play a role in substrate binding are structurally altered; for example the binding sites for the T loop and the −1 residue in the substrate (18; Figure 1). However, addition of the P15–P17 domain to H1 RNA did not give any detectable activity (or improvement as in the case when the P15–P17 was added in trans). At present, we have no explanation for this observation, but we speculate that one possibility is folding of the RNA constructs such that substrate binding or catalysis is affected. Moreover, these constructs might interact differently with the T loop in the model substrate pATSerUG, which is known to influence the cleavage efficiency (26–28). Also in the H1 RNA case, even with the P15–P17 domain present we do not know whether the P6 domain is present or not in our H1 RNAP15–P17 construct and this might also be a factor to consider.
We have proposed that the function of the RCCA–RPR interaction is to anchor the substrate, expose the cleavage site, and as a consequence result in re-coordination of Mg2+ ion(s) (24). Our present findings that small RNAs representing the domain that interacts with the 3′-terminal RCCA motif in the substrate mediates cleavage support that the P15 loop contributes to both substrate binding and catalysis. Specifically, the P15 RNA is expected to bind 6 Mg2+ (51) and one is positioned close to the 2′-OH of U294 [A; Figure 2C (25)]. Most likely and consistent with our previous data using full-size Eco RPR (19,25), 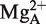 stabilizes the interaction between residue +73 in the substrate and U294, which has been suggested to influence positioning of
stabilizes the interaction between residue +73 in the substrate and U294, which has been suggested to influence positioning of 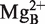 that generates the nucleophile (41). Interestingly, substituting the 2′-OH with 2′-H at position U294 (Figure 2C) resulted in loss of activity indicating an important role for the 2′-OH at this position (Figure 5). Hence, in this model the 2′-OH of U294 is likely to play a role for binding of
that generates the nucleophile (41). Interestingly, substituting the 2′-OH with 2′-H at position U294 (Figure 2C) resulted in loss of activity indicating an important role for the 2′-OH at this position (Figure 5). Hence, in this model the 2′-OH of U294 is likely to play a role for binding of 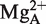 . However, in the crystal structure of RNase P in complex with tRNA we note that no metal(II)-ions are detected in the vicinity U294 (21). This might be due to the fact that this structure represents the post-cleavage stage and/or due to the resolution (4.1 Å).
. However, in the crystal structure of RNase P in complex with tRNA we note that no metal(II)-ions are detected in the vicinity U294 (21). This might be due to the fact that this structure represents the post-cleavage stage and/or due to the resolution (4.1 Å).
Both the P15 RNA, when separated from the rest of the RPR, and H1 RNA mediate cleavage (this report, 22). The former lacks all regions except the one that interacts with the 3′-RCCA-motif while this region is missing in H1 RNA (Figures 1 and 2). These two catalysts nevertheless cleave the same substrates resulting in products with the same ends, 5′-phosphate and 3′-hydroxyl. One question is therefore how this can be rationalized. Available data suggest that chemical groups in the substrate affect positioning of Mg2+ in the vicinity of the cleavage site as well as the rate of cleavage (18,19 and references therein). Hence, we argue that it is conceivable that 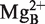 (see Figure 2C and also Refs 19 and 41) is associated with the substrate and when the P15 RNA (or H1 RNA)-substrate complex is formed
(see Figure 2C and also Refs 19 and 41) is associated with the substrate and when the P15 RNA (or H1 RNA)-substrate complex is formed 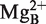 is repositioned to ensure that the nucleophilic attack occurs from the correct orientation in relation to the scissile phosphorous center to generate a 5′-phosphate and a 3′-hydroxyl as cleavage products. This model is also applicable to when other RPRs are used and is consistent with that Mg2+ are positioned near the 5′-end of the tRNA in the crystal structure of RNase P in complex with tRNA (21). In this context note that changing the structural topography of the RCCA–RPR interaction influences catalysis (see above; 24,29,37–41). We therefore hypothesized that this affects positioning of Mg2+ at and near the cleavage site, and cleavage efficiency (29). This becomes more apparent in the P15 ‘RNA-mediated’ reaction than in the Eco RPR case likely due to that cleavage by P15 RNA depends on the RCCA–RPR interaction while in the case of full-length Eco RPR several determinants are present (see above). Therefore, this is a likely reason to why the C293 variant did not rescue cleavage of pATSerUG(G+74) (Figure 5).
is repositioned to ensure that the nucleophilic attack occurs from the correct orientation in relation to the scissile phosphorous center to generate a 5′-phosphate and a 3′-hydroxyl as cleavage products. This model is also applicable to when other RPRs are used and is consistent with that Mg2+ are positioned near the 5′-end of the tRNA in the crystal structure of RNase P in complex with tRNA (21). In this context note that changing the structural topography of the RCCA–RPR interaction influences catalysis (see above; 24,29,37–41). We therefore hypothesized that this affects positioning of Mg2+ at and near the cleavage site, and cleavage efficiency (29). This becomes more apparent in the P15 ‘RNA-mediated’ reaction than in the Eco RPR case likely due to that cleavage by P15 RNA depends on the RCCA–RPR interaction while in the case of full-length Eco RPR several determinants are present (see above). Therefore, this is a likely reason to why the C293 variant did not rescue cleavage of pATSerUG(G+74) (Figure 5).
Functional RNAs are composed of domains (1) and it has been suggested that the C domain is the ancestral part of RPR (52). The P15 loop constitutes an autonomous metal ion-binding domain (25). Together with our present findings, this observation suggests the possibility that the P15 loop is an ancient part of RPR. However, according to Sun and Caetano-Anollés (53) the P15 domain was recruited at a later stage in evolution. The driving force could have been the appearance of more complex RNA molecules, such as hairpin loop RNAs, that needed to be processed and/ or the need for faster reactions or multiple turnover reactions (it has not yet been demonstrated that P15 RNA or H1 RNA performs multiple turnover). In the hammerhead ribozyme the presence of a peripheral domain gives a significant rate-enhancement that is due to stabilization of the active conformation (54,55). Note also that the ancient part of pre-tRNA is considered to be the top domain, i.e. the acceptor stem, T stem and T loop (56,57), and it has been suggested that the top domain of tRNA and the C domain coevolved (53). Moreover, the rate of cleavage for H1 RNA and other eukaryotic RPRs, which have lost the P15 loop (23), are significantly reduced (22). It is therefore possible that the presence of protein(s) is required for efficient cleavage and multiple turnovers when the number of substrate determinants are reduced. Hence, the protein might have compensated for the loss of elements such as the P15 loop. Consistent with this postulate, we recently showed that an archaeal RPR in complex with two of its proteins displayed substrate recognition properties that coincided with those of the Eco RPR rather than the archaeal RPR (27). Moreover Lai et al. (46) recently identified archaeal RPRs with significantly reduced S domains and given the importance of the bacterial S domain for substrate binding (21,26–28) it is likely that proteins compensate for the loss of the S domain in these archael RPRs (see also Ref. 33). In this context, it is also important to note that metal(II)-ion induced cleavage generates products with 5′-OH and 2′,3′-cyclic phosphate at their ends. Therefore, it is plausible (not mutually exclusive) that through evolution there was a selection pressure for evolving RNA-motifs capable of mediating RNA cleavage that yields product termini with 5′-phosphate and 3′-OH.
In conclusion, given that human RNase P RNA and that the P15 loop as part of small RNAs mediate cleavage, opens for new ways to study and understand RNA based catalysis and its evolution. In particular the catalytic P15 RNA will be useful to identify chemical groups needed to mediate cleavage of its substrate.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1 and 2, Supplementary Figure 1 and outline of construction of the H1P15–P17 RNA, C- and CP constructs.
FUNDING
The Swedish Research Council (to L.A.K. and to Uppsala RNA Research Center in the form of Linné support); the Foundation for Strategic Research. Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank our colleagues for discussions and critical reading of the article in particular, Drs F. Darfeuille, S. DasGupta and S. G. Svärd and Ms T. Bergfors is acknowledged for critical reading.
REFERENCES
- 1.Masquida B, Westhof E. A modular and hierarchial approach for all-atom RNA modeling. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2006. pp. 659–681. [Google Scholar]
- 2.Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: building blocks of a molecular biomolecule. Q. Rev. Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- 3.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 5.Murphy FL, Cech TR. An independently folding domain of RNA tertiary structure withion the Tetrahymna ribozyme. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- 6.Cech TR, Damberger SH, Gutell RR. Representation of the secondary and tertiary structure of group I introns. Nat. Struct. Biol. 1994;1:273–280. doi: 10.1038/nsb0594-273. [DOI] [PubMed] [Google Scholar]
- 7.Hoegland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D. In: The RNA World. 3rd edn. Gesteland RF, Cech TR, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2006. pp. 133–205. [Google Scholar]
- 8.Michel F, Jacquier A, Dujon B. Comparison of fungal mitochondrial introns reveal extensive homologies in RNA secondary structure. Biochimie. 1982;64:857–861. doi: 10.1016/s0300-9084(82)80349-0. [DOI] [PubMed] [Google Scholar]
- 9.Michel F, Dujon B. Conservation of RNA secondary structure in two intron families including mitochondrial-, chloroplast-, and nuclear-encoded members. EMBO J. 1983;2:33–38. doi: 10.1002/j.1460-2075.1983.tb01376.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmelzer C, Schmidt C, Schweyen RJ. Identification of splicing signals in introns of yeast mitochondrial split genes: mutational alterations in intron bI1 and secondary structures in related introns. Nucleic Acids Res. 1982;10:6797–6808. doi: 10.1093/nar/10.21.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmelzer C, Schmidt C, May K, Schweyen RJ. Determination of functional domains in intron bI1 of yeast mitochondrial RNA by studies of mitochondrial mutations and a nuclear suppressor. EMBO J. 1983;2:2047–2052. doi: 10.1002/j.1460-2075.1983.tb01698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costa M, Michel F, Westhof E. A three-dimensional perspective on exon binding by a group II self-splicing intron. EMBO J. 2000;19:5007–5018. doi: 10.1093/emboj/19.18.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyle AM, Fedorova O, Waldsich C. Folding of group II introns: a model system for large, multidomain RNAs? Trends Biol. Sci. 2007;32:138–145. doi: 10.1016/j.tibs.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Guerrier-Takada C, Altman S. Reconstitution of enzymatic activity from fragments of M1 RNA. Proc. Natl Acad. Sci. USA. 1992;89:1266–1270. doi: 10.1073/pnas.89.4.1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan T. Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry. 1995;34:902–909. doi: 10.1021/bi00003a024. [DOI] [PubMed] [Google Scholar]
- 16.Loria A, Pan T. Domain structure of the ribozyme from eubacterial ribonuclease P. RNA. 1996;2:551–563. [PMC free article] [PubMed] [Google Scholar]
- 17.Kazantsev AV, Pace NR. Bacterial RNase P: a new view of an ancient enzyme. Nat. Rev. Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- 18.Kirsebom LA. RNase P RNA mediated cleavage: Substrate recognition and catalysis. Biochimie. 2007;89:1183–1194. doi: 10.1016/j.biochi.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Kirsebom LA, Trobro S. RNase P RNA-mediated cleavage. IUBMB Life. 2009;61:189–200. doi: 10.1002/iub.160. [DOI] [PubMed] [Google Scholar]
- 20.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584:287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reiter NJ, Osterman A, Torres-Larios A, Swinger KK, Pan T, Mondragón A. Structure of a bacterial ribonuclease P holoenzyme in complex with tRNA. Nature. 2010;468:784–789. doi: 10.1038/nature09516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikovska E, Svärd SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marquez SM, Chen JL, Evans D, Pace NR. Structure and function of eukaryotic ribonuclease P RNA. Mol. Cell. 2006;24:445–456. doi: 10.1016/j.molcel.2006.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirsebom LA, Svärd SG. Base pairing between Escherichia coli RNase P RNA and its substrate. EMBO J. 1994;13:4870–4876. doi: 10.1002/j.1460-2075.1994.tb06814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kufel J, Kirsebom LA. The P15-loop of Escherichia coli RNase P RNA is an autonomous divalent metal ion binding domain. RNA. 1998;4:777–788. doi: 10.1017/s1355838298970923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brännvall M, Kikovska E, Wu S, Kirsebom LA. Evidence for induced fit in bacterial RNase P RNA-mediated cleavage. J. Mol. Biol. 2007;372:1149–1164. doi: 10.1016/j.jmb.2007.07.030. [DOI] [PubMed] [Google Scholar]
- 27.Sinapah S, Wu S, Chen Y, Pettersson BMF, Gopalan V, Kirsebom LA. Cleavage of model substrates by archaeal RNase P: role of protein cofactors in cleavage-site selection. Nucleic Acids Res. 2011;39:1105–1116. doi: 10.1093/nar/gkq732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu S, Chen Y, Lindell M, Mao G, Kirsebom LA. Functional coupling between a distal interaction and the cleavage site in bacterial RNase P RNA mediated cleavage. J. Mol. Biol. 2011 doi: 10.1016/j.jmb.2011.05.049. doi:10.1016/j.jmb.2011.05.049. [DOI] [PubMed] [Google Scholar]
- 29.Brännvall M, Kikovska E, Kirsebom LA. Cross talk in RNase P RNA mediated cleavage. Nucleic Acids Res. 2004;32:5418–5429. doi: 10.1093/nar/gkh883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hofstee BHJ. On the evaluation of the constants Vm and KM in enzyme reactions. Science. 1952;116:329–331. doi: 10.1126/science.116.3013.329. [DOI] [PubMed] [Google Scholar]
- 31.Dowd JE, Riggs DS. A comparison of estimates of Michaelis-Menten Kinetic constants from various linear transformations. J. Biol. Chem. 1965;240:863–869. [PubMed] [Google Scholar]
- 32.Stage-Zimmermann TK, Uhlenbeck OC. Hammerhead ribozyme kinetics. RNA. 1998;4:875–889. doi: 10.1017/s1355838298980876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen W-Y, Pulukkunat DK, Cho I-M, Tsai H-Y, Gopalan V. Dissecting functional cooperation among protein subunits in archaeal RNase P, a catalytic ribonucleoprotein complex. Nucleic Acids Res. 2010;38:8316–8327. doi: 10.1093/nar/gkq668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guerrier-Takada C, van Belkum A, Pleij CWA, Altman S. Novel reactions of RNase P with a tRNA-like structure in turnip yellow mosaic virus. Cell. 1988;53:267–272. doi: 10.1016/0092-8674(88)90388-1. [DOI] [PubMed] [Google Scholar]
- 35.Glemarec C, Kufel J, Földesi A, Maltseva T, Sandström A, Kirsebom LA, Chattopadhyaya J. The NMR structure of P15 RNA domain of Escherichia coli RNase P RNA using its non-uniformly deuterium labeled counterpart [the 'NMR-window' concept] Nucleic Acids Res. 1996;24:2002–2035. doi: 10.1093/nar/24.11.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson HD, Altman S, Smith JD. Purification and properties of a specific Escherichia coli ribonuclease which cleaves a tyrosine transfer ribonucleic acid precursor. J. Biol. Chem. 1972;247:5243–5251. [PubMed] [Google Scholar]
- 37.Svärd SG, Kagardt U, Kirsebom LA. Phylogenetic comparative mutational analysis of the base-pairing between RNase P RNA and its substrate. RNA. 1996;2:463–472. [PMC free article] [PubMed] [Google Scholar]
- 38.Brännvall M, Kirsebom LA. Metal ion cooperativity in ribozyme cleavage of RNA. Proc. Natl Acad. Sci. USA. 2001;98:12943–12947. doi: 10.1073/pnas.221456598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brännvall M, Kirsebom LA. Complexity in orchestration of chemical groups near different cleavage sites in RNase P RNA mediated cleavage. J. Mol. Biol. 2005;351:251–257. doi: 10.1016/j.jmb.2005.06.031. [DOI] [PubMed] [Google Scholar]
- 40.Brännvall M, Pettersson BMF, Kirsebom LA. Importance of the +73/294 interaction in Escherichia coli RNase P RNA substrate complexes for cleavage and metal ion coordination. J. Mol. Biol. 2003;325:697–709. doi: 10.1016/s0022-2836(02)01195-6. [DOI] [PubMed] [Google Scholar]
- 41.Kikovska E, Mikkelsen N-E, Kirsebom LA. The naturally trans acting ribozyme RNase P RNA has leadzyme properties. Nucleic Acids Res. 2005;33:6920–6930. doi: 10.1093/nar/gki993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haas ES, Brown JW. Evolutionary variation in bacterial RNase P RNAs. Nucleic Acids Res. 1998;26:4093–4099. doi: 10.1093/nar/26.18.4093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svärd SG, Mattsson JG, Johansson K-E, Kirsebom LA. Cloning and characterization of two mycoplasmal RNase P RNA genes. Mol. Microbiol. 1994;11:849–859. doi: 10.1111/j.1365-2958.1994.tb00363.x. [DOI] [PubMed] [Google Scholar]
- 44.Green CJ, Rivera-León R, Vold BS. The catalytic core of RNase P. Nucleic Acids Res. 1996;24:1497–1503. doi: 10.1093/nar/24.8.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loria A, Pan T. The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry. 1999;38:8612–8620. doi: 10.1021/bi990691f. [DOI] [PubMed] [Google Scholar]
- 46.Lai LB, Chan PP, Cozen AE, Bernick DL, Brown JW, Gopalan V, Lowe TM. Discovery of a minimal form of RNase P in Pyrobaculum. Proc. Natl Acad. Sci. USA. 2010;107:22493–22498. doi: 10.1073/pnas.1013969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin K, Pyle AM. Branch-point attack in group II introns is a highly reversible transesterfication, providing a potential proofreading mechanism for 5'-splice site selection. RNA. 1995;1:391–406. [PMC free article] [PubMed] [Google Scholar]
- 48.Ferré-D'Amaré AR, Zhou K, Doudna JA. A general module for RNA crystallization. J. Mol. Biol. 1998;279:621–631. doi: 10.1006/jmbi.1998.1789. [DOI] [PubMed] [Google Scholar]
- 49.Pyle AM. The tertiary structure of group II introns: implications for biological function and evolution. Cri. Rev. Biochem. Mol. Biol. 2010;45:215–232. doi: 10.3109/10409231003796523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Emilsson GM, Nakamura S, Roth A, Breaker RR. Ribozyme speed limits. RNA. 2003;9:907–918. doi: 10.1261/rna.5680603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beebe JA, Kurz JC, Fierke CA. Magnesium ions are required by Bacillus subtilis ribonuclease P RNA for both binding and cleaving precursor tRNAAsp. Biochemistry. 1996;35:10493–10505. doi: 10.1021/bi960870m. [DOI] [PubMed] [Google Scholar]
- 52.Altman S, Kirsebom LA. Ribonuclease P. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1999. pp. 351–380. [Google Scholar]
- 53.Sun F-J, Caetano-Anollés G. The ancient history of the structure of ribonuclease P and the early origins of archaea. BMC Bioinformatics. 2010;11:153. doi: 10.1186/1471-2105-11-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson JA, Uhlenbeck OC. When to believe what you see. Mol. Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Maizels N, Weiner AM. Phylogeny from function: evidence from the molecular fossil record that tRNA originated in replication, not translation. Proc. Natl Acad. Sci. USA. 1994;91:6729–6734. doi: 10.1073/pnas.91.15.6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun F-J, Caetano-Anollés G. The origin and evolution of tRNA inferred from phylogenetic analysis of structure. J. Mol. Evol. 2008;66:21–35. doi: 10.1007/s00239-007-9050-8. [DOI] [PubMed] [Google Scholar]
- 58.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.