Abstract
The conjugation of siRNA to molecules, which can be internalized into the cell via natural transport mechanisms, can result in the enhancement of siRNA cellular uptake. Herein, the carrier-free cellular uptake of nuclease-resistant anti-MDR1 siRNA equipped with lipophilic residues (cholesterol, lithocholic acid, oleyl alcohol and litocholic acid oleylamide) attached to the 5′-end of the sense strand via oligomethylene linker of various length was investigated. A convenient combination of H-phosphonate and phosphoramidite methods was developed for the synthesis of 5′-lipophilic conjugates of siRNAs. It was found that lipophilic siRNA are able to effectively penetrate into HEK293, HepG2 and KB-8-5 cancer cells when used in a micromolar concentration range. The efficiency of the uptake is dependent upon the type of lipophilic moiety, the length of the linker between the moiety and the siRNA and cell type. Among all the conjugates tested, the cholesterol-conjugated siRNAs with linkers containing from 6 to 10 carbon atoms demonstrate the optimal uptake and gene silencing properties: the shortening of the linker reduces the efficiency of the cellular uptake of siRNA conjugates, whereas the lengthening of the linker facilitates the uptake but retards the gene silencing effect and decreases the efficiency of the silencing.
INTRODUCTION
Small interfering RNAs (siRNAs) (1–3) have broad application within molecular biology and experimental pharmacology, being widely used for the control of gene expression (4–6). Currently, siRNAs are used successfully for the validation of potent drug targets for anti-cancer therapy (7,8). A factor that significantly limits their biomedical application, are the challenges associated with the inefficient delivery of siRNAs to target cells and tissues. Various approaches have been developed in an attempt to overcome this problem. These different approaches can be assigned to one of two major groups: viral (9,10) and non-viral (11–13) methods. Viral-based RNAi provides an efficient and long-lasting silencing in cultured cells and in laboratory animals systems; however, immunogenicity, in the case of adenoviral vectors, is a factor that limits their biomedical application (14). Furthermore, the potential tumorogenicity as a result of the integration into the host genome, in the case of lenti- and retroviral vectors is an additional limiting factor (15–17). Non-viral approaches include the following groups of methods: firstly, high-pressure intravenous injections (18–20); secondly, the delivery of siRNA in the complexes with cationic lipids, polymers and different types of particles; thirdly, the covalent conjugation of siRNAs with different carrier molecules (11,21,22). The first approach can be applied only to laboratory animals, since it often results in organ damage and immune activation. The second group of methods is a member of a quickly developing field of science; however, the toxicity of lipids and polymers (23) and the insufficient transfection efficacy in vivo (11), severely limits the application of available up-to-date formulations. The conjugation of siRNA to the molecules, which can be internalized into the cell by natural transport mechanisms, is an approach that shows considerable promise in the attempt to overcome the problem of toxicity and target delivery (22,24). Steroids and other hydrophobic lipid groups can be attached to siRNA, thereby extending the siRNA circulation time and enhancing the direct cellular uptake (25–27). The potential of cholesterol (26,27), α-tocopherol (28), aptamers (29–31), antibodies (32–34) and cell-penetrating peptides (35–38) in the alteration of the bioavailability and distribution of siRNAs has been described; however, the silencing efficacy of different conjugates varies substantially and the optimization of the composition and structure of the conjugates is required.
Within this study, we investigated the carrier-free cellular accumulation and silencing activity of various lipophilic conjugates of the nuclease-resistant anti-MDR1 siRNA. The following lipophilic moieties: cholesterol, oleyl alcohol, lithocholic acid and oleylamide of lithocholic acid, were attached to the 5′-end of the sense strand of siRNA directly or via aliphatic amino-propyl-, -hexyl-, -octyl-, -decyl- and -dodecyl- linker. It was ascertained that the efficiency of cellular accumulation is dependent upon the type of lipophilic residues, the type of the target cells and the length of the linker between siRNA and lipophilic residue.
MATERIALS AND METHODS
General remarks
RNA phosphoramidites, 2′-O-methylphosphoramidites and other reagents for the oligonucleotide synthesis were obtained from Glen Research (USA). 3-Aminopropan-1-ol, 6-aminohexan-1-ol, cholesterol, cholesteryl chloroformate and lithocholic acid were purchased from Sigma-Aldrich (USA), oleylamine and oleyl alcohol were supplied from Acros (Belgium) and 8-aminooctan-1-ol, 10-aminodecan-1-ol, 12-aminododecan-1-ol were acquired from TCI (Belgium). Other chemicals were supplied by Merck (Germany) and Fluka (Switzerland). Solvents were supplied from Panreac (Spain).
Column chromatography was performed with Silica gel 60 Å 230–400 mesh (Sigma), and thin-layer chromatography (TLC) was performed on Silica gel 60 F254 aluminum sheets (Merck) in CH3OH/CH2Cl2 5/95. 1H and 31P NMR spectra were recorded on a Bruker AV-300 spectrometer with tetramethylsilane as an internal standard, or 85% phosphoric acid as an external standard, respectively. RNA synthesis (0.4 µmol scale) was performed on the automatic ASM-800 DNA/RNA synthesizer (Biosset, Russia). Preparative reverse phase high performance liquid chromatography (RP-HPLC) was performed on an Alliance chromatographic system (Waters, USA) equipped with XBridge C18 column, 2.5 μm, 4.6 × 50 mm (Waters, USA) at 50°C. The flow rate was 1 ml/min (buffer A, 0.05 M NaClO4; buffer B, 0.05 M NaClO4/90% CH3CN); the gradient was as follows: (i) 0.00 min, 0.0% buffer B; 35.00 min, 90.0% buffer B for the conjugated siRNAs; (ii) 0.00 min, 0.0% buffer B; 35.00 min, 20.0% buffer B for the non-conjugated siRNAs. The oligonucleotide-containing fractions were evaporated in Speedvac concentrator (SVC-100 H, Savant, USA) and precipitated as Na+ salts. The preparative PAGE was performed using denaturing 12% polyacrylamide gel (acrylamide:N,N′-methylenebisacrylamide (30:0.5), 8 M urea, 89 mМ Tris–borate, рН 8.3, 2 mМ Na2EDTA, 20 V/cm). After electrophoretic separation, oligonucleotide material was visualized by UV shadowing, extracted from the gel with 0.3 M NaOAc (pH 5.2) containing 0.1% sodium dodecyl sulfate (SDS), followed by precipitation with ethanol. LC-ESI and MALDI-TOF mass-spectra were obtained with an ESI MS/MSD XCT (Agilent Technologies, USA) and a autoflex III (Bruker Daltonics, Germany) mass spectrometer.
Synthesis
Cholesterol-3-(carboxyaminopropan-3-ol) (2), cholesterol-3-(carboxyaminohexan-6-ol) (3), cholesterol-3-(carboxyaminooctan-8-ol) (4), cholesterol-3-(carboxyaminodecan-10-ol) (5) and cholesterol-3-(carboxyaminododecan-12-ol) (6) were synthesized by analogy with (39). Cholesteryl chloroformate (1, 0.5 g, 1.1 mmol) and dry triethanolamine (TEA, 0.15 ml, 1.1 mmol) were dissolved in dry dichloromethane (4 ml) and added to anhydrous aminoalcohol (1.1 mmol). The mixture was stirred at room temperature for 2 h (TLC, AcOEt/hexane 50/50), following this dichloromethane (20 ml) was added, and the resulting solution was then washed with saturated aqueous (sat. aq.) NaHCO3 (20 ml × 2) and water (20 ml), dried over Na2SO4, and then filtered and evaporated under reduced pressure. The residue was dissolved in AcOEt/hexane (10/90) and purified via silica gel column chromatography (elution with AcOEt/hexane 10/90 to 50/50). The characteristics of 2 to 6 are presented in the Supplementary Materials.
3α-(Acetoxy)-5β-cholanic acid (8)
Lithocholic acid (7, 0.5 g, 1.3 mmol) was dissolved in dry pyridine (10 ml) under argon and cooled to 0°C. 4-Dimethylaminopyridine (0.02 g, 0.13 mmol) and acetyl chloride (1.1 ml, 16 mmol) were added to the solution. After the mixture was stirred at room temperature for 1 h (under TLC control), water (3 ml) was added and the solution was evaporated under reduced pressure. Dichloromethane (15 ml) was used to dissolve the residue, which was subsequently washed with sat. aq. NaCl (20 ml) and water (15 ml). The organic phase was dried over Na2SO4, then filtered and evaporated to dryness. The crude product was co-evaporated with toluene (10 ml × 2), ethanol (10 ml × 2), acetonitrile (10 ml × 2) and dichloromethane (10 ml × 2) in order to remove the traces of pyridine and purified by silica gel chromatography (elution with CH3OH/CH2Cl2 0/100 to 5/95). The characteristics of 8 are presented in Supplementary Methods.
3α-(Acetoxy)-5β-cholanic acid pentafluorophenyl ester (9) was synthesized by analogy with (40). Compound 8 (0.4 g, 1.0 mmol) was dissolved in dry dichloromethane (13 ml) under argon and cooled to 0°C. N,N-Dicyclohexylcarbodiimide (0.25 g, 1.2 mmol) was added to the solution. The mixture was stirred for 30 min at 0°C. Following this, pentafluorophenol (0.2 g, 1.1 mmol) in dichloromethane (1 ml) was added and the stirring was then continued at room temperature for an additional 20 h (under TLC control). The precipitated N,N-dicyclohexylurea was filtered off and washed with cold dichloromethane; combined filtrates were evaporated under reduced pressure. The oily residue obtained was then diluted with dichloromethane (15 ml) and washed with sat. aq. NaCl (20 ml) and water (15 ml). The organic phase was dried over Na2SO4, filtered and evaporated until complete dryness had been achieved. Compound 9 (0.55 g, 98%) thus obtained was directly used for the next step without purification. Rf (TLC, CH3OH/CH2Cl2 5/95): 0.8.
3α-(Acetoxy)-5β-cholanic acid 3-hydroxypropylamide (10) and 3α-(acetoxy)-5β-cholanic acid 6-hydroxyhexylamide (11). Aminoalcohol (1.3 mmol) was co-evaporated with dry pyridine (3 ml × 3), dissolved in dry dichloromethane (1 ml) and added to the mixture of compound 6 (0.5 g, 0.9 mmol) and TEA (three equiv) in dry dichloromethane (20 ml) cooled to 0°C. The reaction mixture was stirred at room temperature under argon for 40 min (under TLC control) and washed with sat. aq. NaCl (50 ml × 2) and water (50 ml). The organic phase was dried over Na2SO4. Following this, it was filtered and evaporated to dryness. The crude product was then purified via silica gel column chromatography (elution with CH3OH/hexane/CH2Cl2 0/50/50 to 8/46/46) in order to afford the compounds 10 and 11 as white solids. The characteristics of 10 and 11 are presented in the Supplementary Materials.
3α-(Hydroxy)-5β-cholanic acid pentafluorophenyl ester (12) was synthesized by analogy with (41). Lithocholic acid (7) (0.5 g, 1.35 mmol) was dissolved in dry dichloromethane (13 ml) and then cooled to 0°C. Following this N,N-dicyclohexylcarbodiimide (0.34 g, 1.65 mmol) was added to the solution. After stirring for 30 min at 0°C, pentafluorophenol (0.27 g, 1.5 mmol) in dichloromethane (1 ml) was added; the stirring was then continued at room temperature under argon for an additional 20 h (under TLC control). The precipitated N,N-dicyclohexylurea was filtered off and washed with cold dichloromethane. Combined filtrates were then evaporated under reduced pressure. The oily residue obtained was then diluted with dichloromethane (15 ml) and washed with sat. aq. NaCl (20 ml) and water (15 ml). The organic phase was dried over Na2SO4, filtered and evaporated to dryness. The obtained compound 12 (0.7 g, 96%), was directly used for the next step without purification. Rf (TLC, CH3OH/CH2Cl2 5/95): 0.8.
3α-(Hydroxy)-5β-cholanic acid oleylamide (13). Compound 12 (0.7 g, 1.3 mmol) was dissolved in dry dichloromethane (30 ml) and then cooled to 0°C. A cold mixture of oleylamine (0.65 ml, 2.0 mmol) and TEA (0.55 ml, 4.0 mmol) in dichloromethane (2 ml) was added to the resulting solution. The reaction mixture was stirred at room temperature for an additional 1 h (under TLC control) and washed with sat. aq. NaCl (50 ml × 2) and water (50 ml). The organic phase was dried over Na2SO4 and filtered. In order to afford the compound 13 as a white solid, the crude product was purified via silica gel column chromatography (elution with CH3OH/hexane/CH2Cl2 0/50/50 to 4/48/48) following the evaporation of solvents. The characteristics of 13 are presented in the Supplementary Materials.
3α-(Succinimidyloxycarbonyl)-5β-cholanic acid oleylamide (14) was synthesized by analogy with (40). The amide 13 (0.45 g, 0.72 mmol) was dissolved in dry dichloromethane (6 ml). N,N′-disuccinimidyl carbonate (0.55 g, 2.1 mmol), dry TEA (0.5 ml) and dry acetonitrile (3 ml) were added to the solution. The reaction mixture was stirred at room temperature under argon for 24 h (under TLC control) and was then evaporated until dryness had been achieved. The residue was dissolved in dichloromethane (20 ml), washed with sat. aq. NaHCO3 (20 ml × 2), dried over Na2SO4, filtered and then evaporated under reduced pressure. Compound 14 (0.54 g, 98%) was obtained as a colorless powder, which was directly used for the next step in the absence of further purification. Rf (TLC, CH3OH/CH2Cl2 5/95): 0.2.
3α-(Carboxyaminopropan-3-ol)-5β-cholanic acid oleylamide (15) and 3α-(carboxyaminohexan-6-ol)-5β-cholanic acid oleylamide (16) were synthesized via analogy with (40). Aminoalcohol (0.7 mmol) was dried via co-evaporation with dry pyridine (3 ml × 3), dissolved in dry pyridine (0.8 ml) and subsequently added to the solution of compound 14 (0.54 g, 0.7 mmol) in dry dichloromethane (4 ml). The resulting mixture was then cooled to 0°C. Following stirring at room temperature under argon for 20 h (under TLC control), the reaction mixture was evaporated to dryness, dissolved in dichloromethane (20 ml), washed with sat. aq. NaHCO3 (20 ml × 2) and water (20 ml); dried over Na2SO4 and then evaporated under reduced pressure. The residue was purified via silica gel column chromatography (elution with CH3OH/CH2Cl2 0/100 to 4/96) to afford 15 and 16. The characteristics of 15 and 16 are shown in the Supplementary Materials.
Synthesis of lipophilic H-phosphonates (17–28)
H-Phosphonates of lipophilic compounds were synthesized via analogy with (42). Imidazole (1 g, 14.5 mmol) was dissolved in dry acetonitrile (20 ml) and then cooled to 0°C. TEA (2 ml, 14.0 mmol) and PCl3 (0.4 ml, 4.5 mmol) were added to this solution and subsequently stirred for 25 min at 0°C. Lipophylic compound (1 mmol) was dissolved separately in dry dichloromethane (10 ml) and added drop-wise into the mixture for 40 min under argon. The reaction mixture was then stirred for an additional 1 h at room temperature (under TLC control) followed by the addition of water (3 ml) and subsequent evaporation. The residue was dissolved in dichloromethane (30 ml), washed with water (20 ml × 2), dried over Na2SO4, filtered and then evaporated under reduced pressure. The crude product was dissolved in CH3OH/CH2Cl2 (10/90) and was then subjected to silica gel chromatography (elution with CH3OH/CH2Cl2 10/90 to 70/30 with 0.1% TEA), in order to afford the corresponding H-phosphonate. The characteristics of H-phosphonates (17–28) are shown in the Supplementary Materials.
Synthesis of siRNA
Oligoribonucleotides were synthesized on an automatic ASM-800 synthesizer at 0.4 μmol scale using solid phase phosphoramidite synthesis protocols (43) optimized for the instrument, with a 10 min coupling step for 2′-O-TBDMS-protected phosphoramidites (0.1 M in acetonitrile), 6 min coupling step for 2′-O-methylated phosphoramidites (0.05 M in acetonitrile) and 5-ethylthio-H-tetrazole (0.25 M in acetonitrile) as an activating agent. A mixture of acetic anhydride with 2,6-lutidine in THF and N-methylimidazole in THF were utilized as capping reagents. The oxidizing agent was 0.02 M iodine in pyridine/water/THF (1/9/90). Dichloroacetic acid (3%) in dichloromethane was used as a detritylating reagent. The 3′-end of the antisense strand of siRNA used in the uptake assay was labeled by fluorescein (FITC) as described in (44).
Synthesis of lipophilic conjugates of oligonucleotides
Conjugates were prepared via analogy with (42). Each H-phosphonate of the lipophilic compound (17–28) (4.5 µmol) was dissolved in dichloromethane (150 µl). The 5′-O-detritylated, support-bound oligonucleotide (15 mg, 0.45 µmol) was then transferred from the synthesis column to a 0.6 ml screw top glass vial and subsequently blended with a solution of the corresponding H-phosphonate (50 µl), followed by treatment with pivaloyl chloride (1.3 µl, 12.5 µmol in 150 µl pyridine/acetonitrile, 2/1) for 5 min. In order to remove an uncoupled lipophilic compound, the support-bound conjugate was washed with acetonitrile. In order to increase the yield of the conjugates the condensation was repeated twice. The conjugate-containing support was then washed with acetonitrile, treated with I2 (0.2 ml of 0.1 M solution in pyridine/water, 98/2) for 20 min, washed with acetonitrile, acetone and diethyl ether, and then subsequently dried.
Deprotection, purification and annealing of siRNAs and lipophilic analogs
The oligoribonucleotides (Table 1) and their conjugates (Table 2) were cleaved from the support and deprotected by 40% methylamine in water at 65°C for 15 min. 2′-O-Silyl groups were removed upon treatment with a mixture of NMP/TEA·3HF/TEA (150/100/75) at 65°C for 1.5 h. Deprotected oligoribonucleotides and conjugates were isolated by RP-HPLC and precipitated with ethanol as Na+ salts. Alternatively, they were isolated via preparative electrophoresis in 12% polyacrylamide/8 M urea gel, followed by elution from the gel with 0.3 M NaOAc (pH 5.2)/0.1% SDS solution and precipitated with ethanol as Na+ salts. The purified oligoribonucleotides were characterized by electrophoretic mobility in 12% dPAAG, MALDI-TOF-MS and LC-ESI-MS (Table 2). siRNA duplexes were annealed at 10 µM in 30 mM HEPES-KOH, pH 7.4, 100 mM potassium acetate, 2 mM magnesium acetate and stored at −20°C.
Table 1.
Sequences of oligoribonucleotides
| Designation | Sequencea (5′–3′) |
|---|---|
| ON1 (sense strand of siMDR) | GGCUUGACAAGUUGUAUAUGG |
| ON2 (antisense strand of siMDR) | AUAUACAACUUGUCAAGCCAA |
| ON3 (sense strand of siScr) | CAAGUCUCGUAUGUAGUGGUU |
| ON4 (antisense strand of siScr) | CCACUACAUACGAGACUUGUU |
a2′-О-methyl-modified nucleotides are underlined.
Table 2.
5′-lipophilic conjugates of the sense strands of siRNAs
| No. | Conjugate | Yielda (%) | Mass found | Mass calculated |
|---|---|---|---|---|
| 29 | Ch0-ON1 | 8/88b | 7286.3c | 7285.8 |
| 30 | Ch3-ON1 | 9/89b | 7386.5c | 7386.8 |
| 31 | Ch6-ON1 | 7/87.5b | 7428.7c | 7428.8 |
| 32 | Ch8-ON1 | 9.5/89.3d | 7456.5e | 7456.8 |
| 33 | Ch10-ON1 | 10/89.5d | 7485.3e | 7484.8 |
| 34 | Ch12-ON1 | 9.5/89.3d | 7513.2e | 7512.8 |
| 35 | Lt3-ON1 | 8.5/88.4b | 7332.8c | 7332.7 |
| 36 | Lt6-ON1 | 11/90b | 7374.8e | 7374.7 |
| 37 | Ol-ON1 | 7/87.5b | 7167.2e | 7167.7 |
| 38 | OlLt0-ON1 | 7/87.5d | 7524.7c | 7525.2 |
| 39 | OlLt3-ON1 | 9/89d | 7625.7c | 7626.2 |
| 40 | OlLt6-ON1 | 8.5/88.4d | 7667.7c | 7668.2 |
| 41 | Ch0-ON3 | 11/90b | 7208.5e | 7208.7 |
| 42 | Ch3-ON3 | 9/89b | 7308.9e | 7309.7 |
| 43 | Ch6-ON3 | 10/89.5b | 7351.2e | 7351.7 |
| 44 | Ch8-ON3 | 10/89.5d | 7380.1e | 7379.7 |
| 45 | Ch10-ON3 | 11/90d | 7407.8e | 7407.7 |
| 46 | Ch12-ON3 | 10.5/89.8d | 7436.2e | 7435.7 |
| 47 | Lt3-ON3 | 11/90b | 7255.1e | 7255.6 |
| 48 | Lt6-ON3 | 11/90b | 7297.4e | 7297.6 |
| 49 | OlLt0-ON3 | 9/89d | 7448.4e | 7448.1 |
| 50 | OlLt3-ON3 | 9/89d | 7549.2e | 7549.1 |
| 51 | OlLt6-ON3 | 9/89d | 7590.5e | 7591.1 |
aOverall yield/coupling yield (per support-bound starting nucleoside) after isolation.
bOverall yield/coupling yield (per support-bound starting nucleoside) after isolation by RP-HPLC
cLC-ESI-MS (ESI MS/MSD XCT, Agilent Technologies, USA).
dOverall yield/coupling yield (per support-bound starting nucleoside) after isolation by PAGE.
eMALDI-TOF-MS (autoflex III, Bruker Daltonics, Germany).
Cell cultures
Human HEK293 and HepG2 cell lines were obtained from the Institute of Cytology RAS, St. Petersburg, Russia. Multiple drug resistant human cell line KB-8-5 growing in the presence of 300 nM vinblastine was generously provided by Prof. M. Gottesman (National Institutes of Health, USA). The cells were grown in Dulbecco modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 µg/ml streptomycin and 0.25 µg/ml amphotericin at 37°C in a humidified atmosphere containing 5% CO2/95% air.
Cellular accumulation assay
One day before the experiment KB-8-5, HepG2 and HEK 293 cells in the exponential phase of growth were plated in 24-well plates at a density of 1.7 × 105 cells/well. The growth medium was then replaced by fresh DMEM (200 µl/well) without FBS. Each of the FITC-labeled lipophilic siRNAs was added to the cells in 50 µl of Opti-Mem to give the final concentration varying from 0.2 to 5 µM. Four hours post-transfection cells were trypsinized and fixed in 2% formaldehyde in phosphate-buffered saline (PBS). In the case of KB-8-5 cells, the accumulation of conjugates was estimated 1, 2, 4, 8 and 24 h post-transfection. Cells were analyzed using Cytomics FC 500 (Beckman Coulter, USA) flow cytometer (excitation wave length 488 nm, emission 530 ± 30 nm). Of cells, 10 000 from each sample were analyzed. The percentage of FITC positive cells with green fluorescence exceeded the level of the auto-fluorescence of untreated cells (here and after referred to as ‘transfected cells’). The mean values of cell fluorescence intensity normalized to this value of untreated cells were used for data presentation.
Confocal microscopy
In order to prove the intracellular localization of the lipophilic siRNAs, the confocal fluorescent microscopy was utilized. KB-8-5 cells in the exponential phase of growth were plated on the glass coverslips in 24-well plates at a density of 1.3 × 105 cells/well one day before the experiment. FITC-labeled cholesterol-containing siRNAs were then added to the cells to final concentration 2 µM. Following 8 h of incubation the cells were washed with PBS, fixed in 4% formaldehyde/PBS at 37°C for 20 min, permeabilized in 0.5% saponin/PBS for 10 min and stained with Rhodamine Phalloidin (Millipore, USA) and dissolved in PBS for 15 min for cytoskeleton (ß-actin) visualization. The cells on the coverslips were then washed with PBS and mounted on the object-plate in DAPI/Antifade solution (Millipore, USA). The analysis of lipophilic siRNA localization was performed using a confocal fluorescent microscope LSM 510 Meta (Carl Zeiss, Germany) at 100× magnification using optical filters BP 420–480 nm, BP 505–530 nm and LP 560 nm.
MTT Assay
In order to determine the number of living cells a colorimetric assay was used, based on the reduction of the dye 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) in mitochondria (45). Exponentially growing KB-8-5 cells were plated in 96-well plates (2.5 × 103 cells/well) in a vinblastine-free medium one day before the experiment. Following this lipophilic conjugates of siRNA (1–5 µM) were added to the cells. After 24 h of incubation, the vinblastine was added to the cells to the final concentration of 300 nM. After 4, 5 and 6 days of incubation the MTT assay was then performed according to the procedure described in (45). The relative number of living cells normalized to the number of living cells in control was used for data presentation.
Western blotting
The level of P-glycoprotein (P-gp) in KB-8-5 cells was evaluated in a vinblastine-free medium. The cells in the exponential phase of growth were plated in 48-well plates (7 × 103–104 cells/well). After 24 h had elapsed, cholesterol-containing siRNAs (5 µM) were added to the cells as previously described. Alternatively, the cells were transfected with non-modified siRNA (0.2 µM) using Lipofectamine 2000 (Invitrogene, Carlsbad, CA) (0.8 µl per well) according to the manufacturer's protocol. The cells were then cultured for 3–8 days. During this incubation period cells were re-plated after 4 and 5 days of incubation into 48-well plates (2 × 104 cells/well) for the time points: 6 and 8 days, respectively. The cells were lysed in 40 µl of sample buffer (Sigma-Aldrich, USA) at the time points 3, 4, 5, 6 and 8 days of incubation. Of each sample, 20 µl was loaded on a 10% SDS/polyacrylamide gel and then separated at 60 mA for 1 h. The proteins were transferred from PAAG to PVDF membrane (Millipore, USA) using SemiPhor (Hoefer, USA) and the membrane was then blocked overnight in 0.5% non-fat dried milk in 0.05 M Tris–HCl, 0.15 M NaCl, 0.1% Tween-20 pH 7.5. The membranes were incubated with monoclonal anti-P-gp and anti-β-actin antibodies (Sigma-Aldrich, USA) at 1:3000 and 1:6000 dilutions, respectively for 1 h. After the membranes were washed in buffer B, they were subsequently incubated for 30 min with secondary rabbit anti-mouse antibodies conjugated with alkaline phosphatase (Invitrogen, USA). Chromogenic detection was then performed using Western Blue Stabilized Substrate for alkaline phosphatase (Promega, USA). The reaction was stopped via rinsing with water. The human β-actin protein was used as an internal control. Data were analyzed using GelPro 4.0. software (Media Cybernetics, USA).
RESULTS
The sequence of siRNA (Table 1) targeted to 557–577 nt of the human MDR1 mRNA (here and after siMDR) was selected in our previous study (46). It was demonstrated that siMDR has the ability to inhibit the MDR1 expression both at the level of mRNA and P-gp. Furtermore, siMDR is capable of reversing the multiple drug resistance of cancer cells. siRNA that has no significant homology to any known mRNA sequences from mouse, rat or human was used as a negative control (here and after referred to as ‘siScr’, Table 1). These siRNAs contained 2′-O-methyl modifications in CpA, UpA and UpG motives, earlier identified as the primary sites of nuclease attack (47). 5′-Hydroxyl-position of the sense strands of siRNAs was used for the introduction of the lipophilic moieties since the modification of this position is better tolerated by RNAi machinery than the modification of the antisense or both strands (25,48,49).
Synthesis of lipophilic siRNAs
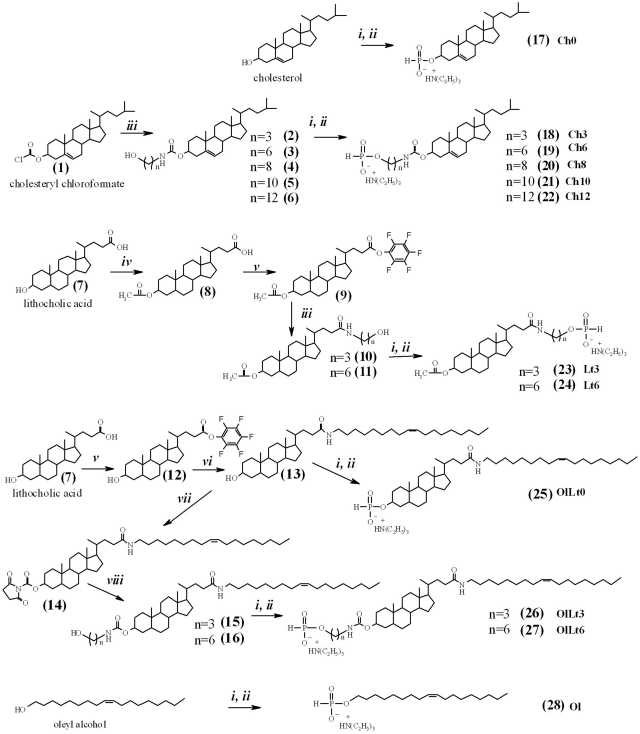
A combination of H-phosphonate and phosphoramidite methods was applied to the synthesis of 5′-lipophilic conjugates of siRNAs. The modified lipophilic synthons were then synthesized, beginning with cholesterol, cholesteryl chloroformate (1) and lithocholic acid (7) (Figure 1). Lithocholic acid was selected due to the fact that both carboxylic and hydroxyl groups are presented in one molecule and can be used for attachment to siRNAs or esterification with fatty acids. The lipophilicity of the lithocholic acid was enhanced via introduction of the oleylamine residue giving compounds 13, 15 and 16. The lipophilic compounds 1, 7 and their derivatives bearing linkers of various lengths were transformed into the corresponding H-phosphonates via interaction with phosphorus triimidazolide followed by hydrolysis (Figure 1). This reaction gives H-phosphonates 17–28 with high yield (41–94%). Structures of H-phosphonates and their precursors were confirmed by NMR and MS methods (see Supplementary Materials for details).
Figure 1.
Synthesis of lipophilic H-phosphonates 17–28. Reagents: (i) PCl3, imidazole, TEA; (ii) H2O; (iii) aminopropanol, aminohexanol, aminooctanol, aminodecanol or aminododecanol, TEA; (iv) acetyl chloride, DMAP; (v) DCC, pentafluorophenol; (vi) oleylamide, TEA; (vii) N,N′-disuccinimidyl carbonate, TEA; (viii) aminopropanol or aminohexanol.
The sense and antisense strands of siRNAs were synthesized according to the phosphoramidite method. Support-bound 5′-hydroxyl-containing sense strands of siRNA were used for the condensation with H-phosphonates 17–28 in the presence of pivaloyl chloride. The lipophilic conjugates obtained were fully deprotected according to the standard procedures, using methylamine and TEA·3HF (43).
Isolation of the oligoribonucleotides conjugated with cholesterol (29–34, 41–46), lithocholic acid (35, 36, 47, 48) and oleyl alcohol (37) from reaction mixtures was accomplished via reverse-phase HPLC. With regards to the conventional oligoribonucleotides, the retention time increased from 5 min to 15–23 min for lipophilic oligoribonucleotides depending on the nature of the lipophilic component and the linker length (Supplementary Figure S1). Purification of the oligoribonucleotides conjugated with the litocholic acid oleylamide (38–40, 49–51) via RP-HPLC resulted in the loss of oligonucleotide material because of their high lipophilicity. Preparative PAGE was used followed by conjugates precipitation as Na+ salts, in order to overcome these obstacles. The overall yield of the lipophilic oligoribonucleotides purified by HPLC or PAGE methods was 8-12% (per support-bound starting nucleoside) (Table 2).
All purified lipophilic oligoribonucleotides were analyzed by LC-ESI and MALDI-TOF mass spectrometry. The calculated molecular weights were in agreement with the measured values, thereby confirming the structures of the isolated products (Table 1).
Carrier-free cellular uptake of lipophilic siRNAs
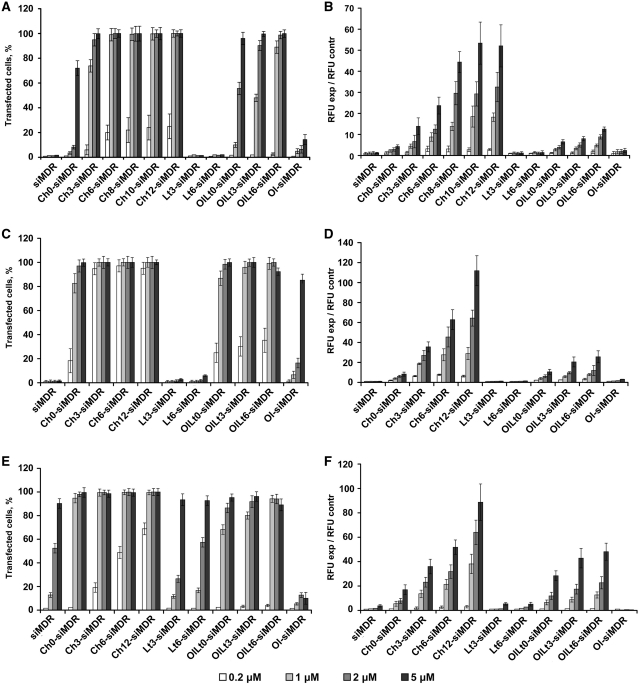
The accumulation of the FITC-labeled siRNA and its lipophilic conjugates into KB-8-5, HepG2 and HEK 293 cells was measured using flow cytometry after 4 h of incubation with the corresponding siRNA. Two parameters reflecting the efficiency of the process were used for the comparison: the transfection efficiency (here and after referred to as ‘TE’) estimated as a percentage of cells with green fluorescence exceeding the maximum level of cell auto-fluorescence and the normalized mean value of the intensity of the green fluorescence of the cell measured in relative fluorescent units (here and after referred to as “RFU”) normalized to that of untreated cells. The data show that the efficacy of cellular accumulation is dependent upon the type of cell line; however, all conjugates of siRNA with cholesterol or with oleylamide of lithocholic acid accumulated in all cells under study, with a higher degree of effectiveness than other conjugates (Figure 1). The 100% transfection of KB-8-5 cells was observed after the incubation of the cells with these conjugates at 2 and 5 μM concentrations (Figure 2A); conversely, the same level of transfection was achieved at the concentrations of the conjugates 0.2 and 1 μM, respectively for HepG2 and HEK293 cells (Figure 2C and E). Non-modified siRNA and conjugates of siRNA and lithocholic acid did not penetrate in KB-8-5 and HepG2 cells (Figure 2A and C), whereas the TE of HEK293 with these conjugates reached 90% at 5 μM concentration (Figure 2E). The accumulation of the siRNA conjugated with oleic alcohol in the cells was insignificant. The comparison of mean values of the cell fluorescence intensity revealed a substantial preference of cholesterol-conjugated siRNA in comparison with other conjugates (Figure 2B, D and F). The cholesterol residue conjugated to siRNA via aminohexyl and aminododecyl linkers enabled a 4–5-fold higher accumulation of siRNA in KB-8-5 cells at a 5 μM concentration of the conjugates, in comparison with that of oleylamide of lithocholic acid conjugated to siRNA (Figure 2B). Similar results were obtained in HepG2 cells. The differences between cellular accumulation of the conjugates in HEK293 cells were less pronounced, but still substantial (Figure 2).
Figure 2.
Cellular accumulation of FITC-labeled lipophilic siRNAs. The percentage of FITC-positive cells in the population (%) (A, C and E) and the normalized mean value of the cell fluorescence (RFU exp/RFU contr) (B, D and F) after incubation of KB-8-5 (A and B), HepG2 (C and D) and HEK293 (E and F) cells in the presence of corresponding siRNAs are shown. Data were obtained via flow cytometry. Fifteen thousand events were counted in each sample. Mean values (±SD) from three independent experiments are presented.
It was discovered that the length of the linker between the siRNA and lipophilic residue significantly influences the cellular accumulation. The conjugates containing linkers of 6-12 carbon atoms accumulated in all tested cells more efficiently than conjugates in which the lipophilic residue was attached to siRNA directly or via a shorter linker (Figure 2). The estimation of TE at the lower concentrations of the conjugates (0.2–1 μM) revealed that the percentage of transfected cells increased with the increase of the linker length (Figure 2A, C and E). TE of HEK293 cells was 0, 20, 50 and 70% for the siRNA with cholesterol tethered directly (without linker) or via the aminopropyl, aminohexyl and aminododecyl linkers, respectively (Figure 2E). This tendency can evidently be observed via comparison of the mean fluorescence values (Figure 2B, D and F). For instance, the lengthening of the linker from 0 to 12 carbon atoms increased the mean fluorescence intensity from 5 to 110 RFU for KB-8-5 cells, from 10 to 117 RFU for HepG2 cells and from 20 to 90 RFU for HEK293 cells, when the conjugates were used at concentration 5 μM (Figure 2B, D and F).
Kinetics of accumulation of lipophilic siRNAs in KB-8-5 cells
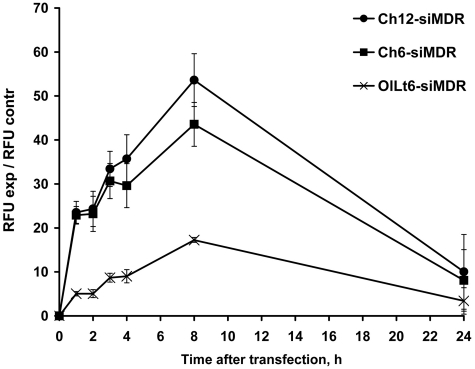
The time course of the accumulation of lipophilic siRNAs in KB-8-5 cells was monitored for 1–24 h. FITC-labeled conjugates Ch6-siMDR, Ch12-siMDR and OlLt6-siMDR were selected for these experiments since they demonstrated an efficient accumulation in all cell types (Figure 2). The maximal mean fluorescence of the cells was observed at 8 h for all tested conjugates (Figure 3). Cellular accumulation of cholesterol-containing siRNAs was 2.5–3-fold higher than this value for OlLt6-siMDR. After 24 h of incubation with the lipophilic siRNAs the mean values of the cell fluorescence was substantially reduced (Figure 3).
Figure 3.
The time course of lipophilic siRNA accumulation in KB-8-5 cells. The incubation time after carrier-free transfection of cholesterol-conjugated siMDR with hexyl (Ch6-siMDR) or dodecyl (Ch12-siMDR) amino-linkers and siMDR conjugated to oleylamide of lithocholic acid moiety (OlLt6-siMDR) varied from 1 to 24 h. The efficacy of cellular uptake was estimated as the mean value of the cell fluorescence intensity (RFU). Data were obtained via flow cytometry, in each sample 10 000 events were counted. Mean values (±SD) from three independent experiments are presented.
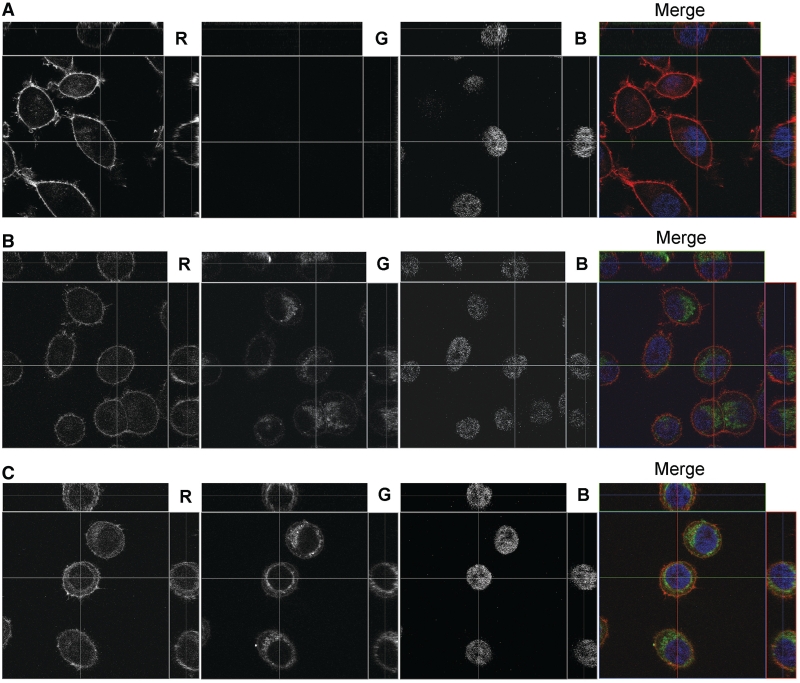
The localization of FITC-labeled Ch6-siMDR and Ch12-siMDR in KB-8-5 cells was studied by confocal fluorescent microscopy (Figure 4) in order to prove the accumulation of the lipophilic siRNAs inside the cells. Non-modified siRNA did not penetrate into the cells (Figure 4A), whereas cholesterol-containing siRNAs accumulated inside the cells effectively after 8 h of incubation (Figure 4B and C). The green signal corresponding to FITC-labeled siRNAs is located in the cytoplasm of the cells as small discrete granules, and the granules are concentrated around the nuclei. Consequently, cholesterol-conjugated siRNA do accumulate in the cell cytoplasm.
Figure 4.
Accumulation of the cholesterol-contaning siRNAs in KB-8-5 cells. The carrier-free transfection of non-modified siMDR (A), Ch6-siMDR (B) Ch12-siMDR (C) was analyzed by confocal fluorescent microscopy at 100× magnification after 8 h of incubation. Three-channel (RGB) pictures were obtained using staining by Rhodamine Phalloidin (R), binding to the actin filaments; FITC (G), attached to cholesterol-modified and non-modified siRNAs and DAPI (B), intercalating in DNA.
Kinetics of MDR1 gene silencing by cholesterol-containing siMDR
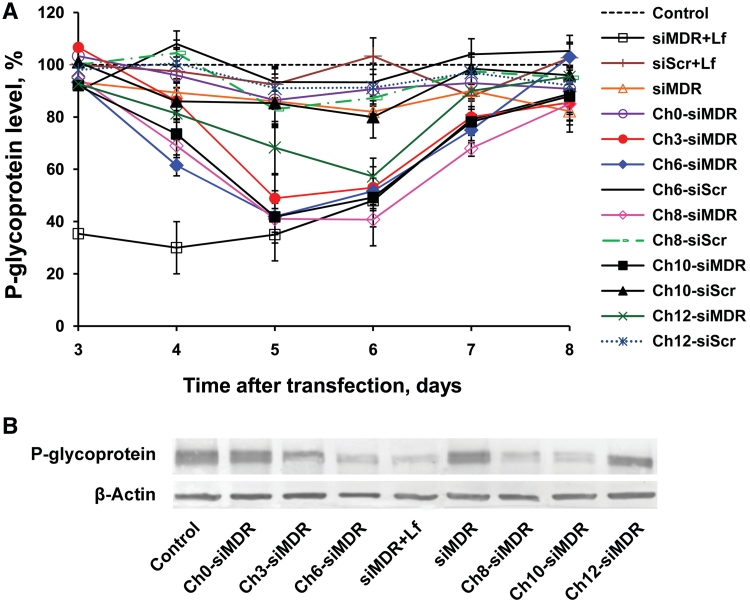
The silencing activity of the cholesterol-conjugated siMDR was investigated using drug resistant KB-8-5 cells. These cells are capable of growing in the presence of 300 nM vinblastine, due to the over-expression of the MDR1 gene. The level of the MDR1 gene product–P-gp was estimated by western blotting after 3–8 days of incubation of cells with 5 µM conjugates in the absence of cytostatic in order to avoid the negative selection of living cells. The start time point (3 days) was chosen taking into account the half-life time of P-gp: 48–72 h (50). The level of β-actin in the same samples was used as an internal standard. Scrambled siRNA (siScr) and its lipophilic analogs were used as controls of specificity. The levels of P-gp were normalized to the levels of β-actin for data presentation (Figure 5A and B). It was found that the efficiency of the MDR1 gene silencing increased when the linker length augmented from 0 to 6-10 carbon atoms and correlated with the efficacy of cellular accumulation of the lipophilic siRNAs. The further increase of linker length, to aminododecyl, resulted in a decrease of the silencing activity and retardation of P-gp level reduction (Figure 5A). The 40% P-gp suppression was observed at Day 4 after addition of Ch6-siMDR to the cells, the action of Ch8-, Ch10- and Ch12-siMDR was less pronounced (30, 25 and 18% reduction of P-gp level, respectively) and was detected at the same time point, the levels of the P-gp in the cells incubated with Ch0- and Ch3-siMDR did not differ from the controls. The maximum levels of P-gp silencing were detected in the cells incubated with cholesterol-containing siMDRs with linkers of 3–10 carbon atoms for 5 days (60% for Ch6-, Ch8- and Ch10-siMDRs and 50% for Ch3-siMDR) (Figure 5). It is noteworthy that there is no statistically relevant difference between the silencing activities of conjugates containing linkers with 3-10 carbon atoms; whereas Ch3-siMDR has a tendency to be less efficient than others. Ch12-siMDR was reliably less efficient than Ch6-, Ch8- and Ch10-siMDRs: this conjugate induced only 30% reduction of the P-gp level after 4 days of incubation (P < 0.02, Ch12-siMDR versus Ch6-, Ch8- or Ch10-siMDR). At Day 6 of incubation the silencing activities of Ch6-, Ch8- and Ch10-siMDRs were similar (Figure 5A). In contrast, the silencing activity of Ch12-siMDR increased to 57% and did not differ to a reliable degree from the activity of Ch3-, Ch6- and Ch10-siMDRs at Day 6, the activity of Ch8-siMDR was slightly higher (P < 0.05, Ch8-siMDR versus Ch12-siMDR). The silencing effect decreased at days 7–8 and did not exceed 15–20% for these conjugates (Figure 5A). Ch0-siMDR was inactive. Hence, the linker length between cholesterol residue and siRNA is critical for the biological activity of the conjugates.
Figure 5.
Silencing of P-gp expression in KB-8-5 cells by non-modified or cholesterol-containing siRNAs. (A) Kinetics of P-gp level suppression following incubation with cholesterol-modified siRNAs or transfection of siRNA with Lipofectamine 2000. The data were obtained via western blotting carried out after 3–8 days of incubation followed by data quantitative analysis by GelPro 4.0. program (Media Cybernetics, USA). The human β-actin protein was used as an internal standard. Data were normalized to the ratio of the P-gp/β-actin levels in untreated cells (control). Mean values (±SD) from three independent experiments are shown. (B) Levels of P-gp in KB-8-5 cells at Day 5 post-transfection. An untreated sample of KB-8-5 cells was taken as a control.
The comparison of P-gp levels after carrier-free uptake of the conjugates with P-gp levels observed after Lipofectamine 2000 mediated delivery of siRNA revealed the difference in the kinetics of the silencing process: in the first case the level of P-gp at Day 3 of incubation of the cells with the conjugates virtually did not change in comparison with the controls (Figure 5A), whereas in the second case (Lipofectamine 2000 mediated transfection) 65% reduction of the P-gp level was observed. At Days 5 and 6, the inhibitory effects of conventional siMDR (200 nM) transfected with Lipofectamine 2000 was similar to the effects of Ch3-, Ch6-, Ch8-, Ch10- and Ch12-siMDRs (5 µM) entering into the cells in a carrier-free mode.
Reversion of multiple drug resistance phenotype of cancer cells
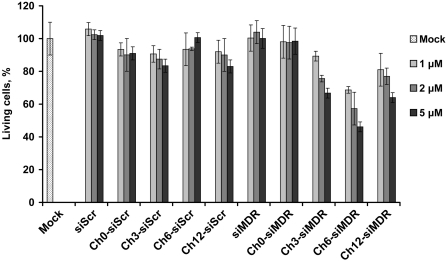
The ability of cholesterol-containing siMDRs to reverse MDR phenotype of KB-8-5 cells was subjected to comparison. After 6 days of cell incubation in the presence of lipophilic siRNAs (1–5 µM) and 300 nM vinblastine, a number of living cells was assayed via an MTT-test. siScr and its lipophilic analogs were used as controls. The obtained results (Figure 6) demonstrate that control siRNAs (Ch0-, Ch3-, Ch6-, Ch8-, Ch10- and Ch12-siScr) did not affect the resistance of the cell to the cytostatic. Incubation of the cells with non-modified siMDR or with Ch0-siMDR also did not alter the cells viability, thereby verifying that the cholesterol-containing siRNAs are not toxic for the cells. Quite conversely, the number of living cells incubated with Ch6-siMDR (5 μM) decreased to 45% as compared to the controls, whereas Ch3-siMDR and Ch12-siMDR (5 μM) were less active: ~65–70% of the cells survived in their presence. The results on the number of living cells at Day 6 of incubation obtained in these experiments correlates well with the data on P-gp suppression levels at Day 6 of incubation (Figures 5 and 6).
Figure 6.
Restoration of KB-8-5 cells sensitivity to vinblastine. The number of living cells (%) in the presence of 300 nM vinblastine was estimated after 6 days of incubation with cholesterol-conjugated siRNAs in the absence of transfection agent. siScr containing the same type of modification were used as controls. Untreated cells are denoted as Mock.
DISCUSSION
MDR1 gene encodes P-gp, the transmembrane pump that effluxes a number of various compounds including cytostatics out of the cells (51,52). Over-expression of this gene is one of the main origins for multiple drug resistance of the cells impeding the chemotherapy of oncological diseases (53). Temporal silencing of MDR1 gene expression by siRNAs decreases the P-gp level and reverses the MDR phenotype. Efficient gene silencing can be achieved only in the case of the efficient delivery of siRNA into the cytoplasm of the cells. However, the negatively charged cellular membrane organized in lipid bilayer is the main barrier on the way of negatively charged siRNA to the cellular targets. Conjugation of siRNAs to lipophilic molecules, such as cholesterol, bile and fatty acids, is a promising approach to solving the problem of siRNA delivery in vivo (26,27). These modifications improved the bio-distribution of siRNAs (26,37) and significantly increased their cellular uptake (25,27,35). The presence of the cholesterol in siRNA did not however guarantee efficient cellular uptake and gene silencing without the aid of the transfection agent (54). Moreover, the data on the efficiency of carrier-free uptake and the biological effects of lipophilic siRNAs vary significantly in different reports where different synthetic methods for the introduction of lipophilic moieties resulted in the obtaining of the conjugates with different structures (26,37,54).
In this study, we posited a new synthetic scheme for the preparation of lipophilic siRNAs and investigated the influence of the type of lipophilic residue and the length of the aliphatic amino-linker between the lipophilic residue and the siRNA on the efficiency of the carrier-free cellular accumulation and the silencing activity of the conjugates. There exist two approaches to the synthesis of lipophilic conjugates of oligonucleotides, namely the synthesis in solution (41) and the solid phase synthesis (25,27,39,55–58). At the time of writing, the common method for the chemical modification of siRNA at the 5′-end is the use of lipophilic compounds as phosphoramidite synthons in solid phase chemical synthesis by the phosphoramidite method (25,27). The H-phosphonate method is rarely used for these purposes (55,57,58) in spite of the fact that H-phosphonates have the advantages of stability and readily available compounds. The developed approach gives high coupling yields (87.5–90%) (Table 2, Supplementary Figure S2) in the condensation of lipophilic H-phosphonates with 5′-hydroxyl of polymer-bound oligoribonucleotide in the presence of pivaloyl chloride.
The results obtained demonstrate that the efficiency of the carrier-free cellular accumulation of lipophilic siRNAs is dependent upon the type of lipophilic residues, the type of the target cells and the length of the linker connecting the lipophilic residue and siRNA strand. The most efficient accumulation of siRNAs was observed in HepG2 cells, while the accumulation in the drug-resistant cell line KB-8-5 was the least efficient. Among the lipophilic siRNAs tested, the cholesterol-containing siRNAs displayed the best TE in all the cells that were tested. Incubation of the cells with cholesterol-containing siRNAs (1–5 µM) in a serum-free medium leads to the transfection of 100% of cells in the population, siRNAs containing oleylamide of lithocholic acid accumulated a slightly less acutely; whereas the two other types of siRNA, containing both lithocholic acid and oleic alcohol residues displayed very poor cellular accumulation properties. Thus indicating that cholesterol is the promising lipophilic molecule provided carrier-free transport of modified siRNA into cells.
Cholesterol is an important biogenic molecule, which can be internalized into and effluxed out of the cell via natural cellular mechanisms (59,60). It is well known that the majority of cholesterol is transported throughout the blood stream as cholesteryl esters in the form of lipid-protein particles known as low-density lipoproteins (LDL) (61,62). Apolipoprotein B organizes the particle and mediates the specific binding of LDL to cell-surface receptor proteins (61). Hence the internalization of the cholesterol into cells occurs via receptor-mediated endocytosis. It was suggested that LDL receptors could facilitate the cellular uptake of the cholesterol-contaning siRNAs in vivo via receptor-mediated endocytosis (27). Our data correlates well with the data reported in (27), in which was shown that the efficiency of the interaction of lipophilic siRNA with LDL-particles depends on the hydrophobicity of the lipophilic residue and cholesterol-conjugated siRNAs associate with LDL very efficiently. This fact may explain the superior cellular accumulation of cholesterol-conjugated siRNAs in hepatocytes (HepG2) and embryonic kidney cells (HEK293) expressing a high level of LDL receptors (63–65) (Figure 1C–F). However, it does not explain the accumulation of non-modified siRNA and conjugate of siRNA and lithocholic acid into HEK293 cells, neither does it explain the significantly poor accumulation of the same siRNAs in HepG2 cells (Figure 1C and E). Furthermore, the efficient uptake of lipophilic-siRNA in serum-free condition and the reduction of their accumulation in the presence of serum (data not shown) do not support the participation of LDL receptors in this process.
An alternative hypothesis concerning the mechanism of carrier-free uptake of lipophilic siRNA by the cells puts forward the transmembrane protein SID-1, which was shown to be essential for the systemic RNAi (66) and promotes the transport of lipophilic siRNA conjugates into the cells (27). It is assumed, that SID-1 forms a channel for dsRNA diffusion or mediates dsRNA uptake indirectly. However, the participation of SID-1 also does not explain the obtained data, since the level of SID-1 in kidney cells (HEK293) is very lower whereas the expression of this protein in liver (HepG2) is high (27).
The adsorption of the lipophilic conjugates on the cell surface and their subsequent transport into the cells via the mechanism of adsorption endocytosis/pinocytosis could be the alternative option for their cellular uptake. In this case, two main factors could determine the efficacy of adsorption of the conjugates on the cell surface: the hydrophobicity of the conjugates and the distance between negatively charged cellular membrane and anionic siRNA. In light of this fact, more hydrophobic siRNAs containing oleylamide of lithocholic acid accumulate in the cells considerably better than less hydrophobic siRNAs containing lithocholic acid (Figure 2). Conversely, the long aliphatic linker in the conjugate structure provides an optimal distance between the cellular membrane and siRNA, along with an increase of the hydrophobicity of the conjugates. As a result, lipophilic siRNAs with amino-linkers of 6-12 carbon atoms exibit the best cellular accumulation (Figures 1–3). The substantial difference between the efficiency of the cellular accumulation of cholesterol-conjugated siRNAs and siRNAs equipped with the lithocholic acid suggests also the role of specific interactions with the cellular membrane (inherent in the binding with the proteins), since the structure of the moieties is similar, with the exception of the presence of the only free hydroxyl group in position 3 of lithocholic acid (Figure 1).
Another important issue coming from the obtained data is retardation in the development of the silencing effect induced by cholesterol-conjugated siRNA following carrier-free cellular accumulation in comparison with that of siRNA delivered into the cells by Lipofectamine 2000 (Figure 4A). It is known that intracellular trafficking of siRNA begins in endocytic vesicles, which subsequently turns into early endosomes, late endosomes (pH 5–6) and lysosome (pH 4.5) (67). siRNA must escape from the endosomes into the cytosol, where it can be incorporated into RISC, since siRNA molecule is extremely unstable in lysosomes. The endosomal escape is one of the key rate-limiting steps in the delivery process. The problem of endosomal trapping has been intensively discussed in literature, with respect to the conjugates of antisense oligonucleotides and cell-penetrating peptides (68–70), when it was found that conjugates containing only stable chemical bonds accumulated readily in the cells, but displays no biological activity. Cholesterol-containing siRNA do escape from the endosomes since high levels of the target gene silencing are observed, but the process of their release into the cytoplasm seems to be rather slow and less efficient than for siRNA/Lipofestamine complexes: maximal level of P-gp silencing was detected at Day 5 for Ch6-, Ch8-, Ch10-siRNA and at Day 2 for siRNA/Lipofectamine 2000 complexes (Figure 5). Hence, the conclusion can be drawn that lipophilic siRNAs are trapped on the endosomes for some time. The obtained data show that the increase of the length of the aliphatic amino-linker between the cholesterol residue and siRNA above 10 carbon atoms impedes the endosomal release of lipophilic siRNA, retards the development of the silencing effect and decreased the efficiency of the silencing: the conjugate with dodecyl amino-linker was less active than the conjugates with the linkers of 6-10 carbon atoms (Figure 6).
The results consistent with the kinetics of P-gp suppression were obtained in the experiments on KB-8-5 cell viability. This cell line is characterized by the over-expression of the MDR1 gene. As a result of this, the cells have the ability to grow in the presence of 300 nM vinblastine. Silencing of the MDR1 gene by siRNAs brings about the death of KB-8-5 cells, at concentrations of vinblastine that were previously tolerated. The pronounced reduction of the number of living cells was observed at Day 6 of cells incubation with cholesterol-containing siMDR (Figure 6). This effect was similar to the effect observed at Day 4 after cell transfection with siMDR/Lipophectamine 2000 (54).
Lipofectamine 2000 contains a fusogenic lipid DOPE (dioleylphosphatidyl-ethanolamine) responsible for endosomal release of siRNA by facilitating the interactions between the lipoplexes and the endosomal membranes (67,71), but this formulation is toxic for cells and is not stable in the presence of blood plasma, and is subsequently is incompatible with regards to therapeutic usage. Unlike Lipophectamine and other commercially available cationic lipid formulations, the conjugates of siRNA with cholesterol that were subjected to study are stable and non-toxic for cells. Furthermore, unlike the conjugates of antisense oligonucleotides, they do release from the endosome with a moderate delay and display a substantial level of biological activity.
Thus, the structure of lipophilic siRNA was optimized by variation of the type of lipophilic moiety and length of the linker between this moiety and siRNA. The designed anti-MDR1 siRNAs (Ch6-, Ch8- and Ch10-siMDR) were found to penetrate efficiently into the cells in a carrier-free mode, to silence the expression of P-gp and to restore the sensitivity of drug resistant cancer cells to vinblastine. Designed lipoophilic siRNAs are promising agents for in vivo applications. For this reason, it is necessary to further investigate the optimization of their structure, in order to facilitate their endosomal release.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods and Supplementary Figures S1 and S2.
FUNDING
The Russian Academy of Science under the programs ‘Molecular and Cell Biology’ (21.1); ‘Science to Medicine’ (37); President’ program for support of leading scientific schools (SS-7101.2010.4); Russian Foundation for Basic Research (11-04-01017-a and 11-04-12095-ofi-m-2011); Ministry of Science and Education of the Russian Federation (14.740.11.1058) and Siberian Branch of Russian Academy of Sciences (41). Funding for open access charge: Siberian Branch of Russian Academy of Sciences (Grant number 41).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Dr V. Koval for MS analysis, Ms A. Iglina for her assistance in the synthesis of the conjugates, Dr R. Serikov for RP-HPLC chromatography of the conjugates and A.V. Vladimirova for the cell maintenance.
REFERENCES
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 3.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 4.Chen SH, Zhaori G. Potential clinical applications of siRNA technique: benefits and limitations. Eur. J. Clin. Invest. 2010;41:221–232. doi: 10.1111/j.1365-2362.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- 5.Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3:318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- 6.Lares MR, Rossi JJ, Ouellet DL. RNAi and small interfering RNAs in human disease therapeutic applications. Trends Biotechnol. 2010;28:570–579. doi: 10.1016/j.tibtech.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jankovic R, Radulovic S, Brankovic-Magic M. siRNA and miRNA for the treatment of cancer. J. BUON. 2009;14:S43–S49. [PubMed] [Google Scholar]
- 8.Pirollo KF, Chang EH. Targeted delivery of small interfering RNA: approaching effective cancer therapies. Cancer Res. 2008;68:1247–1250. doi: 10.1158/0008-5472.CAN-07-5810. [DOI] [PubMed] [Google Scholar]
- 9.Couto LB, High KA. Viral vector-mediated RNA interference. Curr. Opin. Pharmacol. 2010;10:534–542. doi: 10.1016/j.coph.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Mowa MB, Crowther C, Arbuthnot P. Therapeutic potential of adenoviral vectors for delivery of expressed RNAi activators. Expert Opin. Drug Deliv. 2010;7:1373–1385. doi: 10.1517/17425247.2010.533655. [DOI] [PubMed] [Google Scholar]
- 11.Leng Q, Woodle MC, Lu PY, Mixson AJ. Advances in systemic siRNA delivery. Drugs Future. 2009;34:721. doi: 10.1358/dof.2009.034.09.1413267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posadas I, Guerra FJ, Cena V. Nonviral vectors for the delivery of small interfering RNAs to the CNS. Nanomedicine. 2010;5:1219–1236. doi: 10.2217/nnm.10.105. [DOI] [PubMed] [Google Scholar]
- 13.Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA in vivo. FEBS J. 2010;277:4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- 14.Bessis N, GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther. 2004;11:S10–S17. doi: 10.1038/sj.gt.3302364. [DOI] [PubMed] [Google Scholar]
- 15.Check E. A tragic setback. Nature. 2002;420:116–118. doi: 10.1038/420116a. [DOI] [PubMed] [Google Scholar]
- 16.Kaiser J. Gene therapy. Seeking the cause of induced leukemias in X-SCID trial. Science. 2003;299:495. doi: 10.1126/science.299.5606.495. [DOI] [PubMed] [Google Scholar]
- 17.Sinn PL, Sauter SL, McCray PB., Jr Gene therapy progress and prospects: development of improved lentiviral and retroviral vectors–design, biosafety, and production. Gene Ther. 2005;12:1089–1098. doi: 10.1038/sj.gt.3302570. [DOI] [PubMed] [Google Scholar]
- 18.De Souza AT, Dai X, Spencer AG, Reppen T, Menzie A, Roesch PL, He Y, Caguyong MJ, Bloomer S, Herweijer H, et al. Transcriptional and phenotypic comparisons of Ppara knockout and siRNA knockdown mice. Nucleic Acids Res. 2006;34:4486–4494. doi: 10.1093/nar/gkl609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song E, Lee SK, Wang J, Ince N, Ouyang N, Min J, Chen J, Shankar P, Lieberman J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- 20.Zender L, Kubicka S. Suppression of apoptosis in the liver by systemic and local delivery of small-interfering RNAs. Methods Mol. Biol. 2007;361:217–226. doi: 10.1385/1-59745-208-4:217. [DOI] [PubMed] [Google Scholar]
- 21.De Paula D, Bentley MV, Mahato RI. Hydrophobization and bioconjugation for enhanced siRNA delivery and targeting. RNA. 2007;13:431–456. doi: 10.1261/rna.459807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeong JH, Mok H, Oh YK, Park TG. siRNA conjugate delivery systems. Bioconjug. Chem. 2009;20:5–14. doi: 10.1021/bc800278e. [DOI] [PubMed] [Google Scholar]
- 23.Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control Release. 2006;114:100–109. doi: 10.1016/j.jconrel.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 24.Thomas M, Kularatne SA, Qi L, Kleindl P, Leamon CP, Hansen MJ, Low PS. Ligand-targeted delivery of small interfering RNAs to malignant cells and tissues. Ann. NY Acad. Sci. 2009;1175:32–39. doi: 10.1111/j.1749-6632.2009.04977.x. [DOI] [PubMed] [Google Scholar]
- 25.Lorenz C, Hadwiger P, John M, Vornlocher HP, Unverzagt C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 26.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 27.Wolfrum C, Shi S, Jayaprakash KN, Jayaraman M, Wang G, Pandey RK, Rajeev KG, Nakayama T, Charrise K, Ndungo EM, et al. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 28.Nishina K, Unno T, Uno Y, Kubodera T, Kanouchi T, Mizusawa H, Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of alpha-tocopherol. Mol. Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 29.Chu TC, Twu KY, Ellington AD, Levy M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicke BJ, Stephens AW. Escort aptamers: a delivery service for diagnosis and therapy. J. Clin. Invest. 2000;106:923–928. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNamara JO, II, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E, Sullenger BA, Giangrande PH. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- 32.Dassie JP, Liu XY, Thomas GS, Whitaker RM, Thiel KW, Stockdale KR, Meyerholz DK, McCaffrey AP, McNamara JO, II, Giangrande PH. Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors. Nat. Biotechnol. 2009;27:839–849. doi: 10.1038/nbt.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 34.Xia CF, Boado RJ, Pardridge WM. Antibody-mediated targeting of siRNA via the human insulin receptor using avidin-biotin technology. Mol. Pharm. 2009;6:747–751. doi: 10.1021/mp800194y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu YL, Ali A, Chu CY, Cao H, Rana TM. Visualizing a correlation between siRNA localization, cellular uptake, and RNAi in living cells. Chem. Biol. 2004;11:1165–1175. doi: 10.1016/j.chembiol.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 36.Endoh T, Ohtsuki T. Cellular siRNA delivery using cell-penetrating peptides modified for endosomal escape. Adv. Drug Deliv. Rev. 2009;61:704–709. doi: 10.1016/j.addr.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 37.Moschos SA, Jones SW, Perry MM, Williams AE, Erjefalt JS, Turner JJ, Barnes PJ, Sproat BS, Gait MJ, Lindsay MA. Lung delivery studies using siRNA conjugated to TAT(48-60) and penetratin reveal peptide induced reduction in gene expression and induction of innate immunity. Bioconjug. Chem. 2007;18:1450–1459. doi: 10.1021/bc070077d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muratovska A, Eccles MR. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- 39.MacKellar C, Graham D, Will DW, Burgess S, Brown T. Synthesis and physical properties of anti-HIV antisense oligonucleotides bearing terminal lipophilic groups. Nucleic Acids Res. 1992;20:3411–3417. doi: 10.1093/nar/20.13.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manoharan M, Kesavan V, Rajeev KG. 2005 US Patent 20050107325 A1. [Google Scholar]
- 41.Rump ET, de Vrueh RL, Sliedregt LA, Biessen EA, van Berkel TJ, Bijsterbosch MK. Preparation of conjugates of oligodeoxynucleotides and lipid structures and their interaction with low-density lipoprotein. Bioconjugate Chem. 1998;9:341–349. doi: 10.1021/bc970176z. [DOI] [PubMed] [Google Scholar]
- 42.Sergeeva ZA, Lokhov SG, Ven'yaminova AG. Oligo(2′-O-methylribonucleotides) and their derivatives. II. Synthesis and properties of oligo(2'-O-methylribonucleotides) modified with N-(2-hydroxyethyl)phenazinium and steroid groups at the 5'-terminus. Bioorgan. Khim. 1996;22:916–922. [Google Scholar]
- 43.Bellon L. Oligoribonucleotides with 2'-O-(tert-butyldimethylsilyl) groups. Curr. Protoc. Nucleic Acid Chem. 2001 doi: 10.1002/0471142700.nc0306s01. doi: 10.1002/0471142700.nc0306s01. [DOI] [PubMed] [Google Scholar]
- 44.Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4542. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a tetrazolium-based semiautomated colorimetric assay: assessment of radiosensitivity. Cancer Res. 1987;47:943–946. [PubMed] [Google Scholar]
- 46.Logashenko EB, Vladimirova AV, Repkova MN, Venyaminova AG, Chernolovskaya EL, Vlassov VV. Silencing of MDR 1 gene in cancer cells by siRNA. Nucleosides Nucleotides Nucleic Acids. 2004;23:861–866. doi: 10.1081/NCN-200026032. [DOI] [PubMed] [Google Scholar]
- 47.Volkov AA, Kruglova NS, Meschaninova MI, Venyaminova AG, Zenkova MA, Vlassov VV, Chernolovskaya EL. Selective protection of nuclease-sensitive sites in siRNA prolongs silencing effect. Oligonucleotides. 2009;19:191–202. doi: 10.1089/oli.2008.0162. [DOI] [PubMed] [Google Scholar]
- 48.Chiu YL, Rana TM. siRNA function in RNAi: a 115 chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Fougerolles A, Vornlocher HP, Maraganore J, Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2003;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richert ND, Aldwin L, Nitecki D, Gottesman MM, Pastan I. Stability and covalent modification of P-glycoprotein in multidrug-resistant KB cells. Biochemistry. 1988;27:7607–7613. doi: 10.1021/bi00420a006. [DOI] [PubMed] [Google Scholar]
- 51.Anthony V, Skach WR. Molecular mechanism of P-glycoprotein assembly into cellular membranes. Curr. Protein Pept. Sci. 2002;3:485–501. doi: 10.2174/1389203023380503. [DOI] [PubMed] [Google Scholar]
- 52.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim. Biophys. Acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 53.Gottesman MM, Pastan I. Clinical trials of agents that reverse multidrug-resistance. J. Clin. Oncol. 1989;7:409–411. doi: 10.1200/JCO.1989.7.4.409. [DOI] [PubMed] [Google Scholar]
- 54.Kruglova NS, Meshchaninova MI, Ven'iaminova AG, Zenkova MA, Vlasov VV, Chernolovskaia EL. Cholesterol-modified anti-MDR1 small interfering RNA: uptake and biological activity. Mol. Biol. 2010;44:284–293. [PubMed] [Google Scholar]
- 55.Krieg AM, Tonkinson J, Matson S, Zhao Q, Saxon M, Zhang LM, Bhanja U, Yakubov L, Stein CA. Modification of antisense phosphodiester oligodeoxynucleotides by a 5′-cholesteryl moiety increases cellular association and improves efficacy. PNAS. 1993;90:1048–1052. doi: 10.1073/pnas.90.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Manoharan M, Tivel KL, Cook PD. Lipidic nucleic acids. Tetrahedron Lett. 1995;36:3651–3654. [Google Scholar]
- 57.Stein CA, Pal R, DeVico AL, Hoke G, Mumbauer S, Kinstler O, Sarngadharan MG, Letsinger RL. Mode of action of 5 -linked cholesteryl phosphorothioate oligodeoxynucleotides in inhibiting syncytia formation and infection by HIV-1 and HIV-2 in vitro. Biochemistry. 1991;30:2439–2444. doi: 10.1021/bi00223a020. [DOI] [PubMed] [Google Scholar]
- 58.Venyaminova AG, Sergeyeva ZA, Bashirova VZ. H-Phosphonates of hydroxysterols as new key compounds for the synthesis of sterol derivatives of mono-, oligonucleotides and their analogues. Bioorgan. Khim. 1991;17:1294–1296. [Google Scholar]
- 59.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 60.Plosch T, Kosters A, Groen AK, Kuipers F. The ABC of hepatic and intestinal cholesterol transport. Handb. Exp. Pharmacol. 2005:465–482. doi: 10.1007/3-540-27661-0_17. [DOI] [PubMed] [Google Scholar]
- 61.Burnett JR, Barrett PH. Apolipoprotein B metabolism: tracer kinetics, models, and metabolic studies. Crit. Rev. Clin. Lab. Sci. 2002;39:89–137. doi: 10.1080/10408360208951113. [DOI] [PubMed] [Google Scholar]
- 62.Deckelbaum RJ, Shipley GG, Small DM. Structure and interactions of lipids in human plasma low density lipoproteins. J. Biol. Chem. 1977;252:744–754. [PubMed] [Google Scholar]
- 63.Havekes LM, De Wit ECM, Princen HMG. Cellular free cholesterol in Hep G2 cells is only partially available for down-regulation of low-density-lipoprotein receptor activity. Biochem. J. 1987;247:739–746. doi: 10.1042/bj2470739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kambouris AM, Roach PD, Calvert GD, Nestel PJ. Retroendocytosis of high density lipoproteins by the human hepatoma cell line, HepG2. Arteriosclerosis. 1990;10:582–590. doi: 10.1161/01.atv.10.4.582. [DOI] [PubMed] [Google Scholar]
- 65.Marsche G, Frank S, Raynes JG, Kozarsky KF, Sattler W, Malle E. The lipidation status of acute-phase protein serum amyloid A determines cholesterol mobilization via scavenger receptor class B, type I. Biochem. J. 2007;402:117–124. doi: 10.1042/BJ20061406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feinberg EH, Hunter CP. Transport of dsRNA into cells by the transmembrane protein SID-1. Science. 2003;301:1545–1547. doi: 10.1126/science.1087117. [DOI] [PubMed] [Google Scholar]
- 67.Dominska M, Dykxhoorn DM. Breaking down the barriers: siRNA delivery and endosome escape. J. Cell Sci. 2010;123:1183–1189. doi: 10.1242/jcs.066399. [DOI] [PubMed] [Google Scholar]
- 68.Turner JJ, Arzumanov AA, Gait MJ. Synthesis, cellular uptake and HIV-1 Tat-dependent trans-activation inhibition activity of oligonucleotide analogues disulphide-conjugated to cell-penetrating peptides. Nucleic Acids Res. 2005;33:27–42. doi: 10.1093/nar/gki142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Turner JJ, Fabani M, Arzumanov AA, Ivanova G, Gait MJ. Targeting the HIV-1 RNA leader sequence with synthetic oligonucleotides and siRNA: chemistry and cell delivery. Biochim. Biophys. Acta. 2006;1758:290–300. doi: 10.1016/j.bbamem.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 70.Turner JJ, Ivanova GD, Verbeure B, Williams D, Arzumanov AA, Abes S, Lebleu B, Gait MJ. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res. 2005;33:6837–6849. doi: 10.1093/nar/gki991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farhood H, Serbina N, Huang L. The role of dioleoyl phosphatidylethanolamine in cationic liposome mediated gene transfer. Biochim. Biophys. Acta. 1995;1235:289–295. doi: 10.1016/0005-2736(95)80016-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.