Abstract
Transcription factor-induced lineage reprogramming or transdifferentiation experiments are essential for understanding the plasticity of differentiated cells. These experiments helped to define the specific role of transcription factors in conferring cell identity and played a key role in the development of the regenerative medicine field. We here investigated the acquisition of DNA methylation changes during C/EBPα-induced pre-B cell to macrophage transdifferentiation. Unexpectedly, cell lineage conversion occurred without significant changes in DNA methylation not only in key B cell- and macrophage-specific genes but also throughout the entire set of genes differentially methylated between the two parental cell types. In contrast, active and repressive histone modification marks changed according to the expression levels of these genes. We also demonstrated that C/EBPα and RNA Pol II are associated with the methylated promoters of macrophage-specific genes in reprogrammed macrophages without inducing methylation changes. Our findings not only provide insights about the extent and hierarchy of epigenetic events in pre-B cell to macrophage transdifferentiation but also show an important difference to reprogramming towards pluripotency where promoter DNA demethylation plays a pivotal role.
INTRODUCTION
The development of transcription factor-mediated cell transdifferentiation strategies have played a fundamental role to define the specific contribution of transcription factors within a cell type, provide key information about gene regulatory networks and facilitate new approaches for the generation of custom designed cell types in regenerative medicine (1).
The instructive role of transcription factors to specify cell lineage was first demonstrated when Weintraub's team reported that forced expression of MyoD in fibroblasts induces myotube formation (2). Since then, other factors have proven their ability to induce transdifferentiation or lineage reprogramming. For instance, GATA1 converts monocytes into erythroid-megakaryocytic cells or eosinophils (3) and PU.1 converts erythroid-megakaryocytic cells into myeloid cells (4). The most dramatic example of transcription factor-mediated cell reprogramming can be considered the generation of induced pluripotent stem cells (iPSCs) from somatic cells by ectopic expression of the transcription factors OCT4, SOX2, KLF4 and MYC (5–9).
An excellent model for transcription factor-induced lineage reprogramming is based on the ectopic expression of C/EBPα in primary lymphoid cells. Enforced expression of C/EBPα by retroviral infection converts around 60% committed B-cell progenitors into functional macrophages (10). Transdifferentiation is initiated by the coordinated inhibition of Pax5 activity, resulting in downregulation of CD19 and other B cell-specific genes, and the synergistic action between C/EBPα and PU.1 that results in the activation of macrophage-specific genes. Recently, a system based on a leukaemic pre-B cell line, HAFTL, stably infected with an inducible form of C/EBPα was developed. In this system, B cells undergo 100% conversion into macrophages within 2–3 days after β-estradiol addition (11). The efficiency, tightness, rapidity, stromal independence and availability of unlimited cells make the system unique for investigating the molecular basis of C/EBPα-dependent B cells to macrophage transdifferentiation. Given the leukaemic nature of this cell line, this model allows to test the interaction of additional alterations that may counteract or accelerate lineage reprogramming, in a context similar to several pathological processes.
Transdifferentiation by forced expression of a transcription factor involves direct regulation of its target genes in cooperation with elements of the epigenetic machinery within chromatin. Most studies addressing epigenetic changes associated with cell reprogramming have focused on reprogramming towards pluripotency where DNA methylation changes play a pivotal role (12). The role of DNA methylation is best known in the context of promoters, particularly those containing CpG islands, where this modification is generally associated with gene repression (13). In reprogramming towards pluripotency, DNA demethylation is essential to restore DNA methylation and expression patterns similar to those present in embryonic stem (ES) cells (9,14). In this process, activation-induced cytidine deaminase (AID) has been demonstrated to be required for promoter demethylation and induction of genes like OCT4 and NANOG (15). However, there is emerging evidence for substantial differences in DNA methylation between ES cells and iPSCs (16–19). Recent data indicate that transcription factor-mediated reprogramming to iPSCs result in megabase-scale regions of aberrant methylation (20). These findings highlight the importance of investigating the specific contribution of different transcription factors in the establishment of epigenetic patterns in reprogramming experiments.
In the present study, we have investigated the acquisition of DNA methylation changes during C/EBPα-dependent B cell to macrophage transdifferentiation. Unexpectedly, we found that C/EBPα-mediated transdifferentiation of B cell into functional macrophages occurs without detectable DNA methylation changes at the promoter of key differentiation genes. High-throughput analysis and the use of two different model systems confirmed that this process takes place in the absence of significant DNA methylation changes. Remarkably, C/EBPα activates gene expression by binding to hypermethylated promoters and inducing changes in histone modifications of macrophage associated genes. Our data challenge the current perception of the significance of DNA methylation changes in cell lineage reprogramming and suggests a hierarchy of epigenetic changes necessary to ensure functional C/EBPα-induced B cell to macrophage reprogramming.
MATERIALS AND METHODS
Cells
Normal primary macrophages and pre-B cells were derived from bone marrow as described elsewhere (10). HAFTL cells are a foetal-liver-derived, Ha-ras-oncogene-transformed mouse pre-B cell line (21) and were provided by Dr B. Birnstein. RAW 264.7 cells are a mouse leukaemic monocyte macrophage cell line (22) and were provided by Dr D. Cox. C10 cells were previously developed (11) and consist of a clone derived from HAFTL cells infected with a β-estradiol-inducible viral system containing a fusion between C/EBPα and the oestrogen hormone-binding domain (C/EBPαER).
Induction of reprogramming assays
C10 cells were grown in RPMI 1640 without phenol red (Lonza) supplemented with 10% charcoal/dextran-treated FBS (Hyclone) and 50 µM 2-mercaptoethanol (GIBCO), incubated at 37°C in 5% CO2. They were induced by the addition of 100 nM β-estradiol (Calbiochem) and grown with 10 nM IL-3 and CSF-1 (Peprotech). Control cells were treated with 0.1% ethanol (solvent). Reprogramming of primary pre-B cells was carried out by transducing C/EBPα cloned in pMIG retrovirus vector as previously reported (10). Cell-surface antigens were stained with directly conjugated antibodies against Mac-1 and CD19 (BD PharMingen). Cells were analyzed with a FACSCanto flow cytometer (BD Biosciences, San Diego, CA, USA), via FlowJo software (Tree Star, Ashland, OR, USA). FACS was also used to measure forward and side scatter. For pulse-induction transdifferentiation assays cells were washed thoroughly after 48 h and incubated with 10 µM of the β-estradiol antagonist ICI 182780 (Tocris Bioscience).
DNA methylation analysis
DNA methylation status was determined by sequencing bisulfite-modified genomic DNA as described by Herman et al. (23). For quantitative purposes, each amplicon was cloned and between 15 and 20 clones were sequenced as described in Javierre et al. (24). For each gene, primers were designed using the Methyl Primer Express® v1.0 program (Applied Biosystems). Both promoter CpG islands-containing genes and genes with low CpG density promoters were analyzed. Classification was performed following algorithms by Wu et al. (25). For promoter CpG island-containing genes, CpG island shores were also analyzed (26). The primer sequences, product lengths and annealing temperatures used for bisulfite sequencing reactions are shown in Supplementary Table S5.
MethylCAP-chip
MethylCAP, a technique based on capture of methylated DNA with the MBD domain of MeCP2, was performed as described (27,28) using MethylCAP kit (Diagenode). The MBD-bound and input fractions were then analyzed by qPCR with primers of genes previously analyzed (like Cebpa, Dock8, Ebf1 and Pax5) to confirm enrichment of methylated genes. Whole-genome amplification was then performed with both input and immunoprecipitated fractions using the GenomePlex WGA amplification Kit (Sigma). Further confirmation of the enrichment of methylated in immunoprecipitated fractions was again performed by qPCR of the aforementioned genes. Three independent experiments were performed for hybridization on mouse 385K promoter microarrays (Nimblegen). Ten different samples were analyzed: five were related to primary cells (pre-B cells, pre-B cells infected with the C/EBPα retrovirus and induced with β-estradiol for 0, 48 and 120 h, and macrophages) and five corresponding to the cell line-based system (HAFTL B cells, C10 cells at 0, 48 and 120 h after estradiol induction and RAW macrophages). In each case, competitive hybridization was performed, in which input and the MBD-bound fractions were, respectively, labelled with Cy3 and Cy5. For each microarray hybridization, we used the raw feature intensities to form log ratios and denoted these with M, as done by others (29).
Microarray data analysis
Normalization
For each probe, the log2 ratio of the MBD-bound and the input (designated as M) was computed. To allow array comparison we performed a vsn normalization of the probes (30) using the limma package (31). No relevant changes were observed if different normalization methods (such as ‘median-centring and quantile normalization’) were used.
Gene selection by probes
Probes were first mapped to the mm9 version of the Mus musculus genome; those probes that were not mapped near a transcriptions starting site (TSS) were discarded. This region near the TSS was defined as the −2000 to +500 bp region (32) however more constrained definitions of a TSS are possible (33). For each gene an M value was computed as the average value for all the probes mapped to its TSS. Only those genes with at least three probes mapped to their TSS were considered in the genome wide analysis.
Gene selection by differential methylation
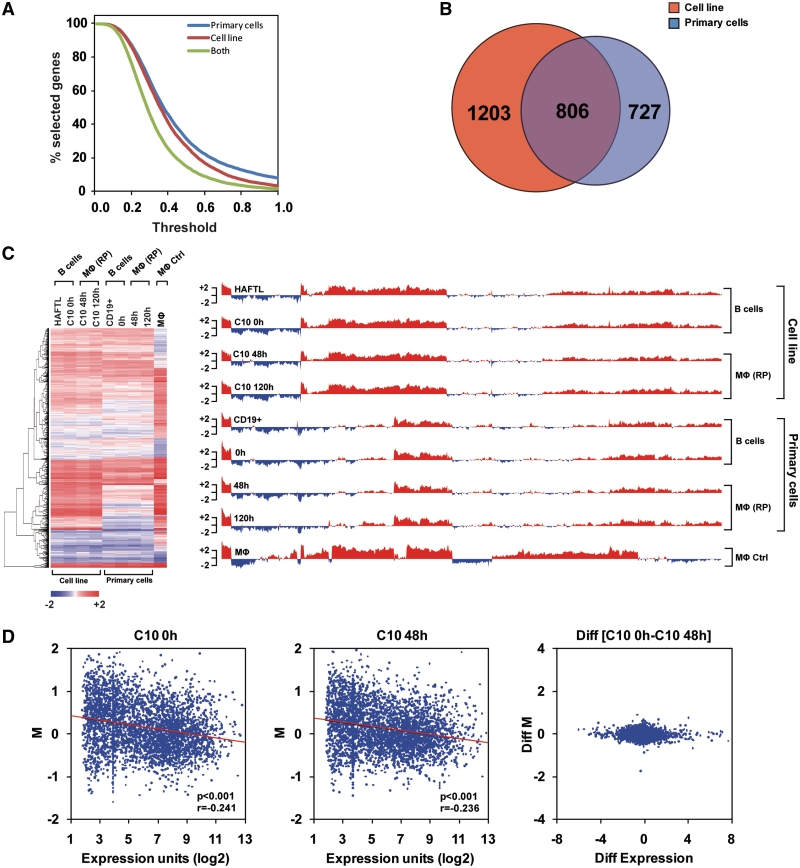
We considered for each gene the difference of the minimum and maximum value within primary cells (PC) and cell lines (CL), denotes, respectively, by PCmaxM and CLmaxM. For a given threshold (th) a gene was selected in PC (CL) experiment if PCmaxM < th (CLmaxM < th). We computed the number of selected genes for different thresholds (Figure 2A). For any given threshold most genes considered differentially methylated in PC were differentially methylated in CL. To set a restrictive definition of differential methylation a threshold of 0.6 was selected. However for different values of threshold we observed similar results, so our conclusions are fairly robust. Number of differentially (diff) methylated genes in PC (diffPC) and CL (diffCL) are 2009 and 1533, respectively. The intersection of diffPC and diffCL is 806 genes.
Figure 2.
High-throughput DNA methylation analysis shows no significant changes during C/EBPα-mediated transdifferentiation. (A) Plot showing the proportion of genes that are differentially methylated depending on the selection threshold used. Data corresponds to the C10-based system, primary cells and the intersection of both sets of data. (B) Venn diagram showing the overlap of differentially methylated genes between the primary cells and C10-based cell system. (C) Heatmap and barplot representing M (log ratio raw intensities between immunoprecipitated DNA and input in MethylCAP experiments) for C10 cells at 0, 48 and 120 h after β-estradiol addition, primary pre-B cells, infected with C/EBPα retrovirus and β-estradiol induced for 0, 48 and 120 h, as well as HAFTL pre-B cells and primary macrophages. (D) Plots representing M versus Affymetrix expression data (11) for C10 at 0 and 48 h after β-estradiol addition (left and centre panels). The difference between two different situations: diff(C10 48 h–C10 0 h) for both M and expression is also represented (right panel).
Comparison of DNA methylation data with Affymetrix data
To compare DNA methylation level (M values) versus Affymetrix expression data (11) we first performed normalization of the raw Affymetrix expression data by Robust Multi-chip Analysis (RMA) (34) via the Affy package. Normalized log2 scaled values were then averaged, and ratios were calculated to isolate probes showing an at least 2-fold change.
Quantitative RT-PCR expression analyses
Total RNA was isolated by using TRIzol Reagent (Invitrogen Co.) according to the manufacturer’s recommendations, and cDNA was produced with the SuperScript II Reverse Transcriptase (Invitrogen Co.). Quantitative Real-Time PCR (Q-RT-PCR) was performed on a LightCycler 480 II System using LightCycler 480 SYBR Green Mix (Roche). Reactions were carried out in triplicates and Q-RT-PCR data was analyzed using the Standard Curve Method. Primer sequences are shown in Supplementary Table S5.
Immunoprecipitation and chromatin immunoprecipitation assays
Immunoprecipitation was performed by standard procedures in HAFTL, RAW and C10 cells at 0, 48 and 120 h after β-estradiol treatment. Cellular extracts were prepared in 50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 150 mM NaCl, 1% Triton–X-100 and protease cocktail inhibitors (Complete, Roche Molecular Biochemicals).
For chromatin immunoprecipitation (ChIP) assays, C10 cells treated with β-estradiol for 0, 48 and 120 h, HAFTL, and RAW cells were crosslinked with 1% formaldehyde and subjected to immunoprecipitation after sonication. ChIP experiments were performed as described (35). The following antibodies were used: Anti-H3 (Abcam, ab 1791), Anti-AcH3 (K9/K14) (Diagenode, pAb-005-044), Anti-3meK4H3 (Millipore, 17-614), Anti-3meK27H3 (Millipore, 07-449), Anti-C/EBPα (14AA) (Santa Cruz Biotechnology, sc-61), Anti-p300 (N-15) (Santa Cruz Biotechnology, sc-584), Anti-p300 (C-20) (Santa Cruz Biotechnology, sc-585) and Anti-RNA polymerase II CTD repeat YSPTSPS (phospho S5) (Abcam, ab5131). Analysis was performed by real-time quantitative PCR. Data are represented as the ratio between the ‘bound’ fraction of the histone modification antibody to that of the C-t histone H3 (total) antibody for histone marks, and as the ratio between the bound fraction over the input for transcription factors. Primer sequences are shown in Supplementary Table S5. In some cases, the methylation of DNA isolated from ChIP experiments was analyzed. In those cases, DNA was pooled from a minimum of five experiments and subjected to bisulfite modification, as described earlier.
Western blotting
Cellular extracts and samples from immunoprecipitation experiments were electrophoresed and western blotted according to standard procedures. The blots were developed with the ECL detection kit (Amersham Pharmacia Biotech). Anti-C/EBPα, and anti-p300 antibodies were those listed earlier.
RESULTS
Key differentiation genes exhibit a differential DNA methylation status in pre-B and macrophage cells
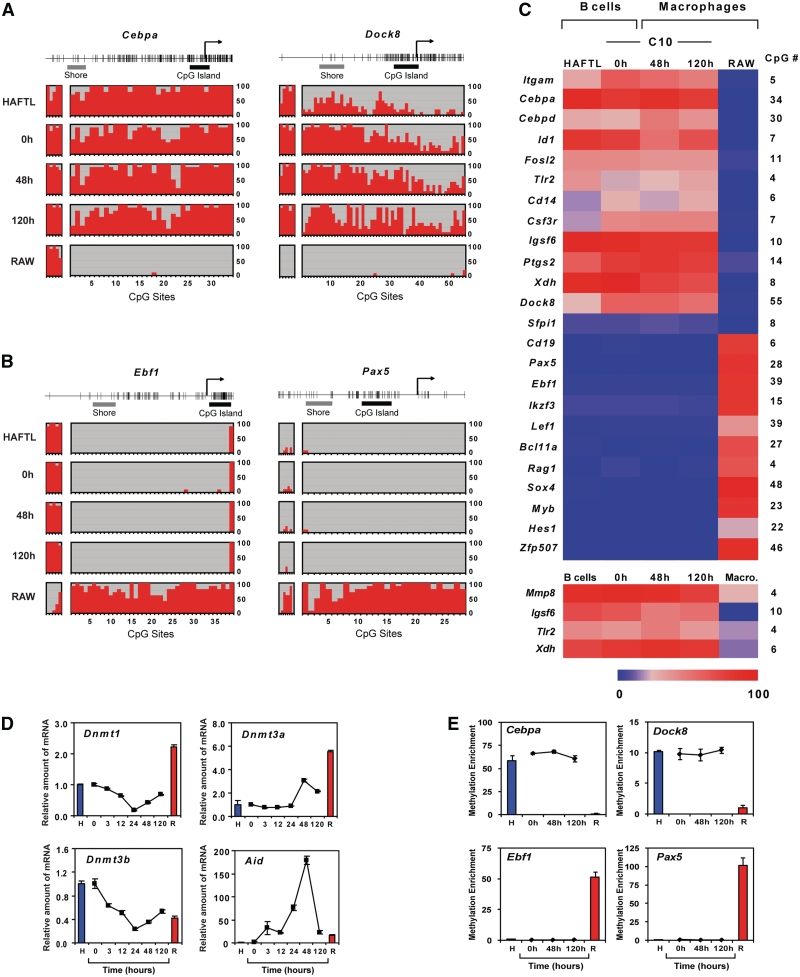
To investigate the existence of DNA methylation changes in association with C/EBPα-mediated B cell to macrophage transdifferentiation, we first mapped the DNA methylation status of 45 key genes relevant to B cell and macrophage function (Supplementary Table S1) to select those with significant differences between B cells and macrophages. For that, we examined both primary cells and cell lines: mouse primary bone marrow derived pre-B cells and macrophages and, alternatively, HAFTL B cells and RAW 264.7 macrophages. The gene selection included bona fide markers of cell identity for the two cell types, as well as genes with known expression changes occurring during B cell to macrophage reprogramming (11). We performed bisulfite genomic sequencing in a 300- to 450-bp region around the transcription start site (TSS) of the 45 genes, 30 of which contained CpG islands and 15 contained promoters with low CpG density (Supplementary Table S1). For 24 of the 45 genes, we found differential DNA methylation at CpG sites near the TSS when comparing HAFTL B cells and RAW macrophages (Supplementary Table S1). Cd19, Pax5, Ikzf3 and other B cell-specific genes, were methylated in RAW macrophages but not in HAFTL B cells. Conversely, genes like Itgam, Cebpa, Dock8 and other macrophage-associated genes, were methylated in HAFTL cells but not in RAW cells (Supplementary Table S1). Other genes, like Tcf3 (E2a), Mef2c, Fmr1 and Foxo1 (among the B cell-specific set), or Cebpb, Evi2a and Emb (among the macrophage-specific group) showed no differences (Supplementary Table S1). Both HAFTL and the C/EBPα-inducible pre-B cell line C10 had virtually identical DNA methylation patterns for all studied genes (not shown), indicating that HAFTL cells remain an appropriate reference for C10 cells.
In contrast to the cell lines studied, primary B cells and macrophages were devoid of DNA methylation for the majority of genes listed earlier (Supplementary Table S1). Only 5 of the 45 analyzed genes exhibited relevant levels of DNA methylation, and of those only 4 (Mmp8, Igsf6, Tlr2 and Xdh) had a differential methylation status. The higher levels of promoter methylation observed for HAFTL and RAW cells with respect to primary B cells and macrophages are consistent with previous findings by others. As mentioned, HAFTL and RAW cells are both leukaemogenic (21,22). Data generated by multiple groups have shown that a large number of promoters can become de novo methylated at early stages in tumourigenesis (33,36,37). Many of these DNA methylation events occur at the promoter of genes that are repressed in the normal primary tissue, including macrophage-specific genes that are repressed in primary B cells, and B-cell-specific genes repressed in macrophages. It has also been reported that extended in vitro proliferation results in hypermethylation of differentiation genes in a pattern reminiscent of that reported in primary tumours (38). We therefore decided to monitor epigenetic changes in C/EBPα-mediated B cell to macrophage transdifferentiation in both systems.
B cell to macrophage transdifferentiation occurs in the absence of DNA methylation changes at key differentiation genes
To explore DNA methylation changes during B cell to macrophage transdifferentiation we first analyzed C/EBPαER-expressing B C10 cells at three times during reprogramming, by focusing on the 24 genes that display differential DNA methylation between HAFTL B cells and RAW macrophages (Supplementary Tables S1 and Figure S1). We tested cells at 0 h, immediately after the addition of β-estradiol; at 48 h, when the phenotype has already become that of a macrophage-like cell and reversion does not occur following β-estradiol depletion; and at 120 h, when the macrophage-like phenotype has had time to fully stabilize. Unexpectedly, the DNA methylation patterns for these genes in C10 cells did not change during transdifferentiation and was identical for the 24 genes analyzed in C10-derived functional macrophages and uninduced C10 cells or HAFTL B cells (Figure 1A–C). Thus, macrophage-associated genes remained methylated in C10-derived macrophages and conserved an identical methylation pattern to that observed in HAFTL and unreprogrammed C10 B cells (Figure 1A and C). Conversely, B cell-associated genes that are not expressed either in RAW cells or in reprogrammed cells, were only methylated in RAW cells, and reprogrammed macrophages retained the unmethylated status present in HAFTL B cells (Figure 1B and C). Similar results were obtained in transdifferentiation experiments with primary cells since there were no changes in DNA methylation during reprogramming for the few candidate genes displaying differences between the two parental primary cell types (Figure 1C and Supplementary Figure S2B).
Figure 1.
Transdifferentiation of B cells to macrophages occurs without DNA methylation changes in key genes. (A) Selected examples of bisulfite genomic sequencing at the promoter CpG islands and CpG island shores of two macrophage-specific genes in parental HAFTL B cells, RAW macrophages, and C/EBPαER-expressing B C10 cells (derived from HAFTL) at 0 h (after C/EBPα induction) and reprogrammed into functional macrophages (48 and 120 h). Percentage of DNA methylation is represented by red bars and CpG site number included in the analysis is indicated at the bottom. The two analyzed regions are CpG island (right panel, black bar) and CpG island shore (left panel, grey bar). (B) Selected examples of bisulfite genomic sequencing at the promoter CpG islands and CpG island shores of two B cell-specific genes in parental HAFTL and RAW cells and C10 cells at 0 h and fully reprogrammed into functional macrophages (48 and 120 h). The two analyzed regions are CpG islands (right panel, black bar) and CpG island shores (left panel, grey bar). (C) Heatmap of DNA methylation data for 13 macrophage-specific genes (top) and 11 B cell-specific genes (bottom) in HAFTL and RAW cells and C10 cells at three times during β-estradiol-induced reprogramming (0, 48 and 120 h). Heatmap of DNA methylation data including the four macrophage-specific genes that are differentially methylated between primary B cells and macrophages are also included at the bottom. The number of CpG sites analyzed for each gene is indicated on the right of the heatmap. Values are in Supplementary Table S2. (D) qRT-PCR kinetics of expression of DNA methyltransferases (Dnmt1, Dnmt3a and Dnmt3b) and activation induced deaminase (Aid). H and R, respectively, designate HAFTL and RAW cells. (E) qPCR analysis of MBD-bound (methylated) fractions obtained from MethylCAP experiments with HAFTL and RAW cells and C10 cells at 0, 48 and 120 h for two macrophage-specific and two B-cell-specific genes.
To check whether DNA methylation changes may be acquired over longer time periods, after the cell population had divided at least once (11) we also analyzed the DNA methylation patterns of some of the genes at 7, 10, 15 and 30 days after addition of β-estradiol to C10 cells, much time after the phenotype is no longer reversible. Neither acquisition nor loss of DNA methylation was observed for any of the genes (Supplementary Figure S3).
The lack of significant observable changes may be associated to absence of changes in expression levels of DNA methyltransferases (DNMTs) or activation induced cytidine deaminase (AID). To test this possibility, we measured by qRT-PCR the levels of the three bona fide DNMTs, DNMT1, DNMT3a and DNMT3b, and of AID. While Dnmt1 and Dnmt3b became first downregulated and then slightly upregulated again, Dnmt3a and Aid became upregulated after 48 h before dropping at 120 h (Figure 1D).
It is generally assumed that transcriptionally relevant CpG methylation occurs in promoters and most methylation studies have therefore focused on CpG island-containing promoters (13). However, it has recently been reported that DNA methylation changes at CpG island shores, CpG containing regions adjacent to CpG islands up to 2 kb distant, can also correlate with changes in gene expression (26). We therefore analyzed the DNA methylation status of CpG island shores for some of the CpG island-containing promoters included in our study. We found that the methylation status at CpG island shores did not strictly correlate with expression status in HAFTL or RAW cells. For instance, the CpG island shore in Cebpa was methylated both in HAFTL and RAW cells (Figure 1A). In other cases, like Dock8 or Pax5, their CpG island shores exhibit identical methylation patterns to their adjacent CpG islands in HAFTL and RAW cells (Figure 1A and B). Importantly however no changes in the CpG island shores were observed during transdifferentiation in any of the genes analyzed (Figure 1A and B). We also checked the DNA methylation status of the gene bodies, intragenic sequences >500 bp downstream of the transcription start sites, which has also been shown to be correlated with gene activity (39,40). Similarly to the results obtained for CpG islands and CpG island shores no changes were observed during transdifferentiation for the genes studied (Supplementary Figure S4).
The absence of DNA methylation changes was unexpected considering that many lineage-specific genes exhibit 10- to 100-fold expression changes during the transdifferentiation process (11). It was also surprising that activated macrophage-specific genes retain hypermethylated promoters, given the well established relationship between promoter CpG methylation and gene repression and that promoter hypermethylation of some of the genes studied here, like Cebpa or Cebpd, are associated with loss of expression (41,42).
To investigate whether DNA methylation influences the expression status of these genes in the cell types studied, we cultured HAFTL, C10 and RAW cells in the presence of the DNA demethylating agent 5-azadC to measure the effect of pharmacologically induced DNA demethylation on the expression levels of these genes. We found increased expression of all methylated genes, suggesting that DNA methylation indeed helps to maintain their lineage-specific repressed status in each corresponding situation (Supplementary Figure S5). However the levels obtained following pharmacological demethylation did not reach the ones present in the cell type where these genes are normally expressed indicating the existence of additional factors regulating their expression (Supplementary Figure S5B).
High-throughput analysis confirms absence of significant changes in DNA methylation during pre-B cell to macrophage transdifferentiation
To check the generality of our conclusions regarding the absence of DNA methylation changes, we performed high-throughput methylation analysis in C/EBPα mediated transdifferentiation in both the C10- and primary cell-based systems. To this end, we performed methylCAP (27,28) associated with hybridization on mouse 385K promoter microarrays for the 10 different samples previously studied. Preliminary analysis of methylCAP samples by qPCR of previously studied lineage-specific genes confirmed enrichment of methylated DNA in the MBD-bound fraction as well as absence of changes in those genes during reprogramming (Figure 1E). Following hybridization, we mapped probes to the genome and selected those within the −2000 to +500 bp region around the transcription start site (32). A comparison of the DNA methylation data (obtained as the log ratio of the raw intensities, and designated as M) for pre-B cells and macrophages revealed that, at any given threshold, a high proportion of the genes that are differentially methylated between HAFTL pre-B cells and RAW macrophages are also differentially methylated between primary pre-B cells and macrophages (Figure 2A), highlighting the similarities between C10 cells and primary pre-B cells. To unify comparisons of potential acquisition of DNA methylation changes of both C10 cells and primary cells we performed all comparisons with respect to primary macrophages (although comparisons of C10 cells against RAW macrophages were also made and yielded similar results, Supplementary Figure S6). We confirmed that candidate genes previously characterized by bisulfite sequencing were also differentially methylated between control pre-B cells and macrophages and showed no changes in methylation during transdifferentiation (Supplementary Figure S6A). Analysis of the data showed that 1533 genes are differentially methylated between HAFTL and macrophages, whereas 2009 are differentially methylated between primary pre-B cells and macrophages (both at a threshold of 0.6) (Supplementary Table S3). A total of 806 genes differentially methylated between primary B cells and macrophages were common between the two systems (Figure 2B).
Our analysis revealed that neither C10 cells nor primary B cells undergo any significant DNA methylation changes during C/EBPα-mediated transdifferentiation. Hierarchical clustering of the two sets of samples reinforced the notion that reprogrammed macrophages (at 48 and 120 h) in both primary cells and the C10-based system have virtually identical DNA methylation patterns to primary pre-B cells and HAFTL cells, respectively, and have not undergone changes that would make their DNA methylation profiles more similar to the ones present in control macrophages (primary macrophages and RAW cells). This is illustrated by the heatmap and barplot generated after hierarchical clustering analysis (Figure 2C), where only differentially methylated genes were considered in each case. Among the genes that are differentially methylated between B cells and macrophages and that do not undergo methylation changes during transdifferentiation there are important regulators of cell cycle, differentiation and identity (Supplementary Table S4). For instance, these lists include Hox (Hoxb5, Hoxb8, Hoxc9, Hoxd10, Hoxd11, Hoxd12) and Fox (Foxc1, Foxc2, Foxd3, Foxf1a, Foxg1, Foxd1, Foxq1) genes, which play crucial roles in differentiation and development, interleukins (Il11, Il12b, Il17a, Il18r1, etc) as well as macrophage- and lymphoid-specific genes. We performed t-test to compare the M values of cells at 48 and 120 h with respect to B cells in both primary and cell lines and therefore to investigate the potential existence of small sets of genes that may undergo DNA methylation changes during transdifferentiation, however not significant results were produced (all genes had an FDR > 0.1), confirming our previous conclusion.
Our data not only suggest that DNA methylation changes are dispensable during C/EBPα-mediated reprogramming but also point out a change in the relationship between DNA methylation and expression levels in C/EBPα-mediated transdifferentiation. To test this hypothesis we compared high-throughput methylation data of C10 cells at 0 and 48 h after estradiol addition with their corresponding Affymetrix expression data (11) (accession number GSE17316). Scatter plots where DNA methylation is represented against expression data indicate the existence of a general inverse correlation between methylation and expression levels (Figure 2D, left panel, P < 0.001, r = −0.241; centre panel, P < 0.001, r = −0.236). However, when looking at the transition between C10 cells at 0 (functional pre-B cells) and 48 h (functional macrophages) (represented as diff [C10 48 h − C10 0 h]), we observed that during transdifferentiation many genes undergo changes in expression without a change in DNA methylation and Pearson correlation (P < 0.01) showed that no relation between transcription changes and DNA methylation changes exists during this process (Figure 2D, right). Identical results were obtained by looking at the genes obtained from the separate analysis of cell lines (Supplementary Figure S6C).
Histone H3 modifications accompany gene expression changes during B cell to macrophage transdifferentiation
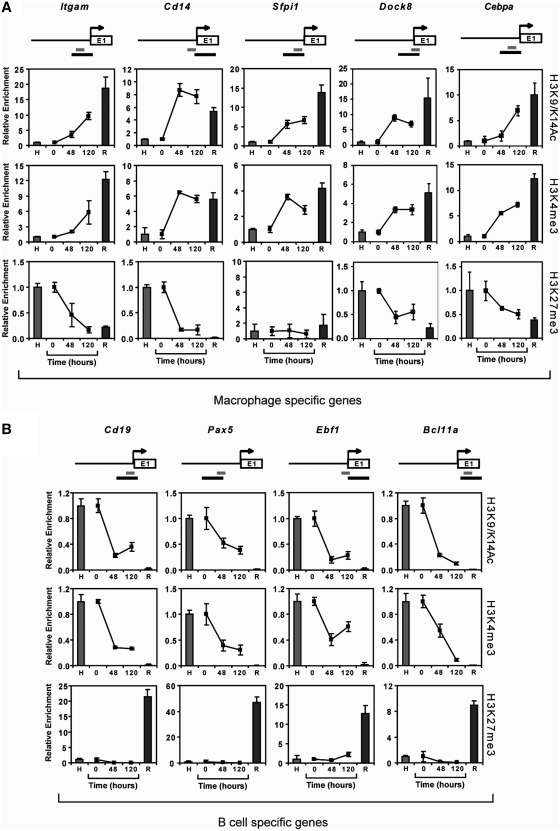
The lack of DNA methylation changes during B cell to macrophage transdifferentiation in genes that undergo expression changes suggests that alternative epigenetic factors, like histone modifications, have a stronger association with the expression status of these genes, at least in our model system. To address this issue we chose five macrophage-specific genes (Itgam, Sfpi1, Cd14 and Dock8) and four B cell-specific genes (Cd19, Pax5, Ebf1 and Bcl11a), for which we found differential promoter DNA methylation between HAFTL and RAW cells (Supplementary Table S1). These genes undergo changes in transcription as expected during transdifferentiation (Supplementary Figure S7; (10,11). We then performed ChIP assays for three histone H3 modifications well-characterized for their localization at promoters and functional association with gene expression status: histone H3 K4 trimethylation (H3K4me3) and acetylation (H3K9acK14ac), associated with active transcription, and histone H3 K27 trimethylation (H3K27me3), associated with gene repression (43–45). We analyzed the profile of these histone modification marks at the promoter of the two groups of genes studied in a region near the TSS overlapping with the region analyzed for DNA methylation (Figure 3, top).
Figure 3.
Changes in the histone modification profile accompany changes in the expression status of key genes. Quantitative ChIP assays show changes in the levels of H3K9acK14ac and H3K4me3, as active marks, and H3K27me3, as a repressive mark, in both macrophage-specific genes (A) and B cell-specific genes (B). HAFTL pre-B cells (H) and RAW macrophages (R) are used as controls and C10 cells at 0 h and fully reprogrammed into functional macrophages (48 and 120 h). In all cases, data is normalized with respect to the values in HAFTL cells. On top of each gene, a scheme depicting the sequence covered for DNA methylation analysis (black bar) and ChIP analysis (grey bar), in relation with the transcription start site (arrow) is shown.
We observed that active histone modifications, histone H3 K9/K14 acetylation and K4 trimethylation, correlated with an increase in gene expression of macrophage-specific genes (Figure 3A). For B cell-associated genes, H3 K9/K14 acetylation and K4 trimethylation decreased in parallel with their expression during transdifferentiation (Figure 3B). Likewise, there was a correlation between gene expression and histone H3 K4 trimethylation and K9/K14 acetylation in pre-B cells and macrophages. For H3K27me3, we observed the opposite behaviour, as it was absent at the promoter of B-cell-specific genes in pre-B cells and present in macrophages. In macrophage-associated genes H3K27me3 was present in HAFTL and absent in RAW cells. Interestingly, we did not observe an increase in H3K27me3 H3 in B cell-associated genes during reprogramming (Figure 3B, bottom). However, for macrophage-specific genes a loss of H3K27me3 did occur in parallel with an increase in gene expression (Figure 3A, bottom), except for Sfpi1, which is expressed in both HAFTL and RAW cells.
We then wondered whether changes in histone modifications persist if the induction of CEBP/a is turned off at 48 h or, in other words, if histone modifications return to the pre-B cell state in the absence of the inducer. As described in our previous study (11), transdifferentiation is no longer reversible after 48 h. We therefore performed pulse-induction transdifferentiation assays cells where β-estradiol was washed out at 48 and investigated the histone H3 modification profile at 120 h, when the phenotype should have stabilized. No differences were found with respect to standard trandifferentiation experiments (Supplementary Figure S8), supporting the notion of the stability of this histone modification profile associated with altered DNA methylation patterns in transdifferentiated macrophages.
C/EBPα binds the methylated promoter of macrophage-specific genes and recruits p300 regardless of their DNA methylation status
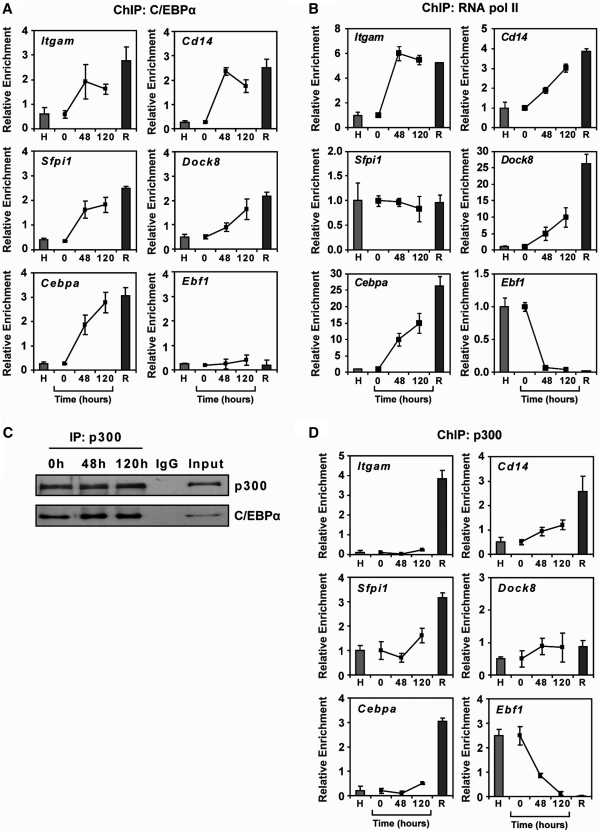
C/EBPα-mediated transdifferentiation involves expression changes in several thousand genes (11). A significant proportion of these changes is likely caused by direct C/EBPα binding and regulation, although the full range of target genes is currently unknown. Some of the macrophage-specific genes included in our study, like Cd14, Sfpi1 or Cepba, are known direct C/EBPα targets (46–48) and sequence inspection of genes like Itgam or Dock8 showed the presence of C/EBPα binding sites (49) near their respective transcription start sites. We used ChIP assays to investigate C/EBPα binding at the Cd14, Itgam, Sfpi1, Dock8 and Cebpa promoters in C10 cells during transdifferentiation. We also tested the promoter of the pre-B cell-specific gene Ebf1. Our analysis showed specific binding of C/EBPα to all the above macrophage-specific genes in reprogrammed cells (48 and 120 h) but not to Ebf1 (Figure 4A). C/EBPα association appeared to occur in reprogrammed macrophages, in which these promoters are methylated, at comparable levels to the ones observed in RAW macrophages, where these promoters are unmethylated. Interestingly, methylation at these promoters does not interfere with the association of the active form of the RNA Pol II, as demonstrated by ChIP assays (Figure 4B).
Figure 4.
C/EBPa associates with P300 and with macrophage-and B cell-specific genes during transdifferentiation. (A) Quantitative ChIP assays showing the binding of C/EBPα to the promoter of five macrophage-specific genes. Ebf1 is used as an example of B-cell specific genes. HAFTL pre-B cells (H) and RAW macrophages (R) are used as controls. (B) Quantitative ChIP assays showing the binding of RNApol II to the promoter of five macrophage-specific genes. Ebf1 is used as an example of B-cell specific genes. HAFTL pre-B cells (H) and RAW macrophages (R) are shown as controls. (C) C/EBPα and p300 are associated in C10 cells, at 0, 48 and 120 h following transdifferentiation induction, as demonstrated by immunoprecipitation. (D) Quantitative ChIP assays showing the binding of p300 to the promoter of five macrophage-specific genes. Ebf1 is used as an example of B-cell-specific genes. HAFTL pre-B cells (H) and RAW macrophages (R) are used as controls.
In addition, it has been shown that C/EBPα, as well as other C/EBP family members, can recruit co-activators such as p300, a bona fide histone acetyltransferase (50,51). Therefore, we investigated whether C/EBPα can directly associate with, and recruit p300 to the promoter of some of the genes studied. To this end, we performed immunoprecipitation experiments in C10 cells at 0, 48 and 120 h following estradiol addition. We confirmed that p300 associates with C/EBPα in C10 cells (Figure 4C). We then performed ChIP experiments with p300 in C10 cells following reprogramming induction, in parallel with HAFTL B cells and macrophages. We found increased and specific binding of p300 to the promoter of some of the macrophage-specific genes during transdifferentiation (Figure 4D), although the profile of association was slightly different to the one observed for C/EBPα. In fact, we observed that the B cell-specific gene Ebf1 shows a gradual decrease in p300 association (Figure 4C), suggesting that additional factors are involved in eliciting changes in the recruitment of p300 histone acetyltransferase and other histone modifying enzymes.
Uncoupling between DNA methylation and histone H3 modifications and C/EBPα binding during B cell to macrophage transdifferentiation
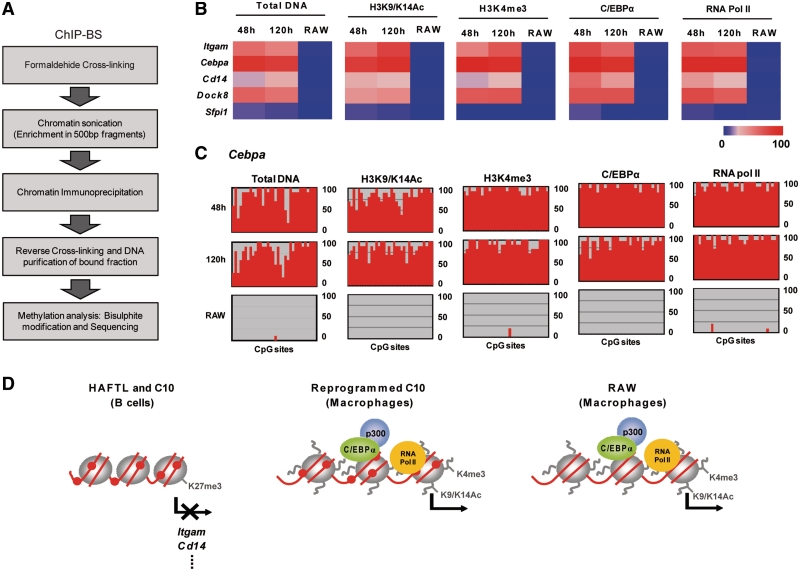
The above results suggest that C/EBPα and RNA Pol II associate the promoters of macrophages-specific genes without sensing or inducing a modification of the DNA methylation status. They also suggest that during C/EBPα-mediated B cell to macrophage transdifferentiation the DNA methylation status of the genes whose expression changes are at least partially uncoupled from their histone modification status. We therefore decided to assess whether histone H3 K9/K14 acetylation and K4 trimethylation, as well as C/EBPα and RNA Pol II binding, were associated with macrophage-specific genes that remain methylated state in reprogrammed C10 macrophages. To this end, we performed bisulfite sequencing of DNA isolated from chromatin immunoprecipitation experiments using H3K4me3, H3K9acK14ac and C/EBPα antibodies (Figure 5A). Comparison of the DNA methylation status of the DNA associated with H3K4me3, H3K9acK14ac, C/EBPα and RNA Pol II in four macrophage-associated genes (Itgam, Cebpa, Cd14 and Dock8) in both reprogrammed macrophages and RAW cells showed that the DNA methylation status of these genes and the two histone H3 modifications and C/EBPα and Pol II binding are uncoupled (Figure 5B). The absence of methylation changes at any CpG sites in the DNA fraction associated with C/EBPα, RNA Pol II or H3K4me3, H3K9acK14ac was confirmed by looking at the methylation profiles throughout the entire sequence (an example is shown in Figure 5C).
Figure 5.
Combination of ChIP analysis and bisulfite sequencing of B cell-specific and macrophage genes shows uncoupling of histone modification marks and DNA methylation status for reprogrammed C10 cells. (A) Scheme depicting the rationale for ChIP-BS experiments, where DNA methylation status of the DNA fraction bound to a specific histone modification or transcription factors is determined. (B) Five heatmaps show the DNA methylation status (as assessed by bisulfite genomic sequencing of multiple clones) of five macrophage-specific genes associated with H3K9acK14ac, H3K4me3, C/EBPα and RNA pol II in C10 reprogrammed macrophages (at 48 and 120 h) and RAW macrophages. (C) An example of the results obtained from ChIP-BS experiments where the profiles of the Cebpa promoter CpG island associated with H3K9acK14ac, H3K4me3, C/EBPα and RNA pol II are compared for reprogrammed macrophages and RAW macrophages. (D) Schematic representation focusing on the different epigenetic profiles observed for the two different sets of macrophages: those resulting from C/EBPα-dependent transdifferentiation of C10 cells and RAW cells. Histone octamers are represented by grey circles. DNA is represented as a red line in which only methylated CpG dinucleotides are shown (as red circles). Histone tails are lines protruding from octamers, where H3K9acK14ac, and H3K4me3 are indicated. Loss of these two marks is indicated by absence of the protruding lines. The arrows indicate transcription of macrophage-specific genes.
DISCUSSION
Our results provide evidence that C/EBPα-induced B cell to macrophage transdifferentiation occurs without substantial DNA methylation changes. In contrast, C/EBPα is able to induce changes in active and repressive histone H3 modifications at the promoters of key differentiation genes. The first implication of our findings is that DNA methylation differences between control and reprogrammed macrophages, that retain the methylation profile of pre-B cells, do not seem to be functionally relevant, since both sets of macrophages are functionally indistinguishable (10,11). This finding challenges our general perception about the role of promoter methylation in gene expression. Thus, among the genes studied in detail herein, promoter DNA methylation-dependent gene silencing has been demonstrated for Pax5, Ebf1, Bcl11a or Cebpa for normal and leukaemic haematopoietic cells (41,42). Together with our own results with DNA demethylating agents these data at first sight indicate that DNA methylation influences their expression. However, bisulfite genomic sequencing of macrophage-specific genes and high-throughput DNA methylation analysis in reprogrammed macrophages clearly shows that some of these genes remain highly methylated at their promoters in a situation where high expression levels can also be detected (11). The result is independent of the model system (primary B cells, HAFTL-based cell line) used to test C/EBPα-induced transdifferentiation, although the number of hypermethylated genes is larger in the C10 system given its leukaemic nature. However, a high proportion of genes that are differentially methylated between pre-B cells and macrophages in primary cells are also represented in the C10-based system, highlighting the similarities between the two models. Our results suggest that additional factors, such as active histone modifications, are able to overcome the repressive effect of DNA methylation in macrophage-specific genes in reprogrammed macrophages.
Our data contrast with the results obtained for transcription factor-mediated reprogramming systems for the generation of iPSCs. Several studies have recently addressed the issue of DNA methylation changes during reprogramming towards pluripotency of somatic cell types, using nuclear transfer or OCT4, SOX2, KLF4 and MYC expression (9,14,16–19,52). Studies comparing the functional and molecular properties of resulting iPSCs and ES cells showed that their expression and DNA methylation patterns were highly similar (9,14). For instance, transcription factor-induced reprogramming of fibroblasts into iPSCs resulted in demethylation of the Nanog and Oct4 promoters to a similar extent as present in ES cells, in contrast to their highly methylated status in fibroblasts (9). In contrast, cells trapped in partially reprogrammed pluripotent states failed in activating pluripotency genes that were hypermethylated in the cell type of origin (14). In all the systems studied, DNA demethylation appears to play a critical role during reprogramming although the extent of ressemblance in methylation patterns between iPSCs and ES cells is variable depending on the strategy followed in the generation of iPSCs. Among different strategies, transcription factor-dependent reprogramming differs not only in the dynamics of demethylation with respect to nuclear transfer methods (14) but also in its extent (18,19). In fact, detailed transcriptional and epigenetic comparison between transcription factor-obtained iPSCs and ES cells have shown that iPSCs retain residual DNA methylation signatures characteristic of the somatic tissues of origin (16,19). In line with this, our model system, in which cell lineage conversion is achieved by the ectopic expression of a single transcription factor, represents the most extreme example of residual DNA methylation characteristic of the cell type of origin. In fact, our analysis reveals the absence of substantial promoter CpG methylation changes. Moreover, detailed analysis of selected genes indicates that neither promoters (both CpG island-containing or with low CpG density) nor CpG island shores of any of the genes studied undergo DNA methylation changes. However, the genome-wide assays used here would not detect single CpG demethylation events that might be occurring within a sea of methylated residues. It also remains an open possibility that alternative pathways to demethylation for macrophage-specific genes may take place, such as oxidation of 5-methylcytosine (5meC) by the recently described TET family of 5meC hydroxylases (53,54). In fact we have obtained recent evidence that supports this possibility (our unpublished data). Finally, it is possible that other transcription factor-induced cell lineage reprogramming systems will show different outcomes with respect to the type and extent of DNA methylation changes.
Another implication of the absence of DNA methylation changes in our system is the ability of C/EBPα to efficiently induce changes in the expression of its target genes without altering or sensing the DNA methylation status at their promoters (CpG island shores and gene bodies). Previous reports have shown that CpG methylation can directly interfere with transcription factor binding. For instance, the transcription factor UBF, a single methylated CpG site inhibits binding to nucleosomal rDNA (55). The explanation is that CpG methylation either directly interferes with transcription factor binding or affects the local chromatin structure, thus indirectly interfering with binding. In view of these data the ability of C/EBPα to bind to highly methylated macrophage-specific promoters is remarkable although this might be explained by the fact that its canonical binding site does not contain any CG (49).
The absence of DNA methylation changes at C/EBPα-target genes suggests that C/EBPα is neither able to directly recruit DNMTs (or demethylating activities like AID) nor facilitates the recruitment of these enzymes, at least during the time period that leads to the generation of functional macrophages. In contrast, C/EBPα can associate with p300 [(50,51) and this study], as well as other histone modification enzymes, including CREB-binding protein (CBP) and histone deacetylase 1 (HDAC1) (56,57). It is possible that the association with these histone modification enzymes mediates the recruitment of additional enzymes, such as histone H3 K4 or K27 methyltransferases.
In contrast to the apparent absence of changes in DNA methylation, histone H3 K9/K14 acetylation and K4 and K27 trimethylation were found to accompany changes in expression during reprogramming of the genes studied. The only exception is H3 K27me3 in the B-cell-specific genes Cd19, Pax5, Ebf1 and Bcl11a. Multiple reports have demonstrated that H3 K27me3 and DNA methylation are often associated (36,58). Binding of EZH2-containing Polycomb-group (PcG) complex, that has H3 K27me3 catalytic activity, shows correlation with genes that become hypermethylated in cancer (33,36,37). Since B-cell-specific genes do not become hypermethylated during B cell to macrophage reprogramming, we can speculate whether the absence of both marks at the promoters of these genes is associated. Interestingly, the requirement of PcG complexes for efficient reprogramming towards pluripotency, where DNA methylation changes are essential, has recently been demonstrated (59).
A remarkable implication of our findings is that DNA methylation and activating histone H3 modifications are apparently uncoupled in our system. For both B cell-specific and macrophage-associated genes, we find that two different DNA methylation states for reprogrammed macrophages and the RAW macrophage cell line occur in association with the same histone H3 modification and expression pattern (Figure 5C). In these two observed situations, histone modifications better correlate with gene expression than DNA methylation in reprogrammed macrophages. Our data support the notion of a hierarchy between different epigenetic marks at the promoter of macrophage associated genes where H3K9acK14ac and H3H3 K24me3 overimpose their functional effects over the DNA methylation status of these genes, particularly in the case of macrophage-specific genes that remain hypermethylated and where the high levels of expression are associated with active histone H3 marks.
In summary, our findings not only provide insights about the extent and hierarchy of epigenetic events during B cell to macrophage reprogramming but also highlight the importance in the development of improved reprogramming strategies, such as alternative combinations of factors for the faithful generation of desired new cell types.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–S8 and Supplementary Tables S1–S5.
FUNDING
Spanish Ministry of Science and Innovation (MICINN) [grants PI081346 (FIS) to E.B.; CSD2006-49 (CONSOLIDER) to E.B. and T.G.; 2009SGR184 (AGAUR) to E.B.; HEROIC to T.G.; Ramon y Cajal Programme to M.P.; German Academic Exchange Service to L.B.; ICREA to T.G.; BFU2008-0410 (MICINN) to M.P.; Marie Curie International Re-integration Grant (MIRG-CT-2007-208119) (FP7-PEOPLE) to M.P.; Swedish Research Council to V.R.; Stockholm County to J.T.; Karolinska Institutet to D.G.-C. and J.T.; Virtual Physiological Human Network of Excellence (VPH-NoE, Grant Agreement No.: 223 920) to D.G.-C. and J.T. and European Science Foundation LESC/EMRC FFG Reference 2669 to D.G.-C.]. Funding for open access charge: IDIBELL and MICINN.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Paul Wade for helpful comments on the manuscript, Marina Corominas and Luisa de Andres for excellent technical assistance.
REFERENCES
- 1.Graf T, Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- 2.Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- 3.Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 1995;9:1250–1262. doi: 10.1101/gad.9.10.1250. [DOI] [PubMed] [Google Scholar]
- 4.Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412. doi: 10.1101/gad.12.15.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 6.Wernig M, Meissner A, Foreman R, Brambrink T, Ku M, Hochedlinger K, Bernstein BE, Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 8.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, Creyghton MP, Steine EJ, Cassady JP, Foreman R, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maherali N, Sridharan R, Xie W, Utikal J, Eminli S, Arnold K, Stadtfeld M, Yachechko R, Tchieu J, Jaenisch R, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Xie H, Ye M, Feng R, Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- 11.Bussmann LH, Schubert A, Vu Manh TP, De Andres L, Desbordes SC, Parra M, Zimmermann T, Rapino F, Rodriguez-Ubreva J, Ballestar E, et al. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 13.Illingworth RS, Bird AP. CpG islands—‘a rough guide’. FEBS Lett. 2009;583:1713–1720. doi: 10.1016/j.febslet.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 14.Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doi A, Park IH, Wen B, Murakami P, Aryee MJ, Irizarry R, Herb B, Ladd-Acosta C, Rho J, Loewer S, et al. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009;41:1350–1353. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng J, Shoemaker R, Xie B, Gore A, LeProust EM, Antosiewicz-Bourget J, Egli D, Maherali N, Park IH, Yu J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat. Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, et al. Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467:285–290. doi: 10.1038/nature09342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polo JM, Liu S, Figueroa ME, Kulalert W, Eminli S, Tan KY, Apostolou E, Stadtfeld M, Li Y, Shioda T, et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010;28:848–855. doi: 10.1038/nbt.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lister R, Pelizzola M, Kida YS, Hawkins RD, Nery JR, Hon G, Antosiewicz-Bourget J, O'Malley R, Castanon R, Klugman S, et al. Hotspots of aberrant epigenomic reprogramming in human induced pluripotent stem cells. Nature. 2011;471:68–73. doi: 10.1038/nature09798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alessandrini A, Pierce JH, Baltimore D, Desiderio SV. Continuing rearrangement of immunoglobulin and T-cell receptor genes in a Ha-ras-transformed lymphoid progenitor cell line. Proc. Natl Acad. Sci. USA. 1987;84:1799–1803. doi: 10.1073/pnas.84.7.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raschke WC, Baird S, Ralph P, Nakoinz I. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell. 1978;15:261–267. doi: 10.1016/0092-8674(78)90101-0. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc. Natl Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–179. doi: 10.1101/gr.100289.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP. Redefining CpG islands using hidden Markov models. Biostatistics. 2010;11:499–514. doi: 10.1093/biostatistics/kxq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG. Whole-genome DNA methylation profiling using MethylCap-seq. Methods. 2010;52:232–236. doi: 10.1016/j.ymeth.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 28.Bock C, Tomazou EM, Brinkman AB, Muller F, Simmer F, Gu H, Jager N, Gnirke A, Stunnenberg HG, Meissner A. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat. Biotechnol. 2010;28:1106–1114. doi: 10.1038/nbt.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber W, von Heydebreck A, Sueltmann H, Poustka A, Vingron M. Parameter estimation for the calibration and variance stabilization of microarray data. Stat. Appl. Genet. Mol. Biol. 2003;2 doi: 10.2202/1544-6115.1008. Article3. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 32.Palmke N, Santacruz D, Walter J. Comprehensive analysis of DNA-methylation in mammalian tissues using MeDIP-chip. Methods. 2011;53:175–184. doi: 10.1016/j.ymeth.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 33.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schubeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat. Genet. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 34.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 35.Ballestar E, Paz MF, Valle L, Wei S, Fraga MF, Espada J, Cigudosa JC, Huang TH, Esteller M. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 2003;22:6335–6345. doi: 10.1093/emboj/cdg604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 37.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, et al. Epigenetic stem cell signature in cancer. Nat. Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 38.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat. Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc. Natl Acad. Sci. U.S.A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tada Y, Brena RM, Hackanson B, Morrison C, Otterson GA, Plass C. Epigenetic modulation of tumor suppressor CCAAT/enhancer binding protein alpha activity in lung cancer. J. Natl Cancer Inst. 2006;98:396–406. doi: 10.1093/jnci/djj093. [DOI] [PubMed] [Google Scholar]
- 42.Agrawal S, Hofmann WK, Tidow N, Ehrich M, van den Boom D, Koschmieder S, Berdel WE, Serve H, Muller-Tidow C. The C/EBPdelta tumor suppressor is silenced by hypermethylation in acute myeloid leukemia. Blood. 2007;109:3895–3905. doi: 10.1182/blood-2006-08-040147. [DOI] [PubMed] [Google Scholar]
- 43.Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 44.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc. Natl Acad. Sci. USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kummalue T, Friedman AD. Cross-talk between regulators of myeloid development: C/EBPalpha binds and activates the promoter of the PU.1 gene. J. Leukoc. Biol. 2003;74:464–470. doi: 10.1189/jlb.1202622. [DOI] [PubMed] [Google Scholar]
- 47.Legraverend C, Antonson P, Flodby P, Xanthopoulos KG. High level activity of the mouse CCAAT/enhancer binding protein (C/EBP alpha) gene promoter involves autoregulation and several ubiquitous transcription factors. Nucleic Acids Res. 1993;21:1735–1742. doi: 10.1093/nar/21.8.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Z, Hetherington CJ, Zhang DE. CCAAT/enhancer-binding protein activates the CD14 promoter and mediates transforming growth factor beta signaling in monocyte development. J. Biol. Chem. 1999;274:23242–23248. doi: 10.1074/jbc.274.33.23242. [DOI] [PubMed] [Google Scholar]
- 49.Akira S, Isshiki H, Sugita T, Tanabe O, Kinoshita S, Nishio Y, Nakajima T, Hirano T, Kishimoto T. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–1906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oelgeschlager M, Janknecht R, Krieg J, Schreek S, Luscher B. Interaction of the co-activator CBP with Myb proteins: effects on Myb-specific transactivation and on the cooperativity with NF-M. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 51.Mink S, Haenig B, Klempnauer KH. Interaction and functional collaboration of p300 and C/EBPbeta. Mol. Cell. Biol. 1997;17:6609–6617. doi: 10.1128/mcb.17.11.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ito S, D'Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol. Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 56.Kovacs KA, Steinmann M, Magistretti PJ, Halfon O, Cardinaux JR. CCAAT/enhancer-binding protein family members recruit the coactivator CREB-binding protein and trigger its phosphorylation. J. Biol. Chem. 2003;278:36959–36965. doi: 10.1074/jbc.M303147200. [DOI] [PubMed] [Google Scholar]
- 57.Wiper-Bergeron N, Wu D, Pope L, Schild-Poulter C, Hache RJ. Stimulation of preadipocyte differentiation by steroid through targeting of an HDAC1 complex. EMBO J. 2003;22:2135–2145. doi: 10.1093/emboj/cdg218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat. Rev. Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 59.Pereira CF, Piccolo FM, Tsubouchi T, Sauer S, Ryan NK, Bruno L, Landeira D, Santos J, Banito A, Gil J, et al. ESCs require PRC2 to direct the successful reprogramming of differentiated cells toward pluripotency. Cell Stem Cell. 2010;6:547–556. doi: 10.1016/j.stem.2010.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.