Abstract
The mom gene of bacteriophage Mu encodes an enzyme that converts adenine to N6-(1-acetamido)-adenine in the phage DNA and thereby protects the viral genome from cleavage by a wide variety of restriction endonucleases. Mu-like prophage sequences present in Haemophilus influenzae Rd (FluMu), Neisseria meningitidis type A strain Z2491 (Pnme1) and H. influenzae biotype aegyptius ATCC 11116 do not possess a Mom-encoding gene. Instead, at the position occupied by mom in Mu they carry an unrelated gene that encodes a protein with homology to DNA adenine N6-methyltransferases (hin1523, nma1821, hia5, respectively). Products of the hin1523, hia5 and nma1821 genes modify adenine residues to N6-methyladenine, both in vitro and in vivo. All of these enzymes catalyzed extensive DNA methylation; most notably the Hia5 protein caused the methylation of 61% of the adenines in λ DNA. Kinetic analysis of oligonucleotide methylation suggests that all adenine residues in DNA, with the possible exception of poly(A)-tracts, constitute substrates for the Hia5 and Hin1523 enzymes. Their potential ‘sequence specificity’ could be summarized as AB or BA (where B = C, G or T). Plasmid DNA isolated from Escherichia coli cells overexpressing these novel DNA methyltransferases was resistant to cleavage by many restriction enzymes sensitive to adenine methylation.
INTRODUCTION
Bacterial restriction–modification (R-M) systems function as a genetic immune system that cleaves foreign DNA, e.g. phage or plasmid DNA entering the host cell (1). Typically, R-M systems comprise enzymes responsible for two opposing activities: a DNA methyltransferase (MTase) and the restriction endonuclease (REase), which recognize the same sequence. A MTase introduces a specific methylation that protects the DNA against the REase cleavage.
Phage and plasmids employ various strategies to avoid restriction, such as modification of the phage genome, transient occlusion of restriction sites, subversion of host R-M activities and direct inhibition of restriction enzymes (2). Several phage genomes encode enzymes that modify nucleosides in DNA leading to generalized protection of phage DNA within bacterial hosts that carry R-M systems (2–5).
The bacteriophage Mu mom gene encodes a protein responsible for the dA′x DNA modification (6). The Mom protein modifies ~15% of DNA adenine residues in loosely defined target sequences 5′-(C or G)-A-(C or G)-N-(C or T)-3′ (7). Mass spectrometry analyses have suggested that the modified deoxyribonucleoside dA′x corresponds to α-N-(9 -β-d-2′-deoxyribofuranosylpurin-6-yl)-glycinamide (8). This unusual modification of DNA is not required for Mu lytic or lysogenic growth and is generally dispensable for phage growth (9). However, the bacteriophage Mu dA′x DNA modification protects the viral genome against cleavage by a wide variety of REases (10). Thus, it serves as a protective measure against nucleolytic attack when the Mu genome infects a cell possessing a DNA ‘host specificity’ different from that of the bacterium in which the phage replicated.
The expression of mom is harmful to the host, so it is strictly controlled and is a late function in the Mu growth cycle, when the host cell is already destined for death (11). The mom gene is subject to a series of unusual regulatory controls including the action of the phage-encoded Com protein (‘zinc finger-like’ translational regulator) (12). The com and mom genes constitute a single operon located at the right end of the Mu phage genome and the shared com–mom gene promoter is positively regulated by the zinc-binding protein Com (13,14).
Other Mu-like phages, such as SP18 (15), often encode homologs of Mom and its regulatory proteins at corresponding positions in their genome, although there are exceptions to this rule. For example, the transposable Mu-like phage B3 of Pseudomonas aeruginosa encodes a Com homolog (ORF47), but instead of a homolog of Mom it has a DNA adenine MTase (16). The activity of this ORF48 protein has yet to be demonstrated experimentally.
The determination of the genome sequences for Haemophilus influenzae Rd and Neisseria meningitidis type A strain Z2491 in 2002 led to the identification of the Mu-like prophages FluMu and Pnm1, respectively (17). A genomic island containing genes related to phage Mu was also discovered in the H. influenzae biogroup aegyptius Brazilian purpuric fever (BPF) strain F3031 by McGillivary et al. (18). Homologs of the com gene were readily identified in all of these prophages. However, at the position corresponding to the mom gene within the operons containing the com-like genes, the DNA sequences and the encoded proteins showed no similarity to the mom gene or the Mom protein, respectively. For example, downstream of the FluMu HI1522.1 gene (whose product is 44% identical to Mu Com), in the position analogous to mom, is the HI1523 gene, whose product appears to be unrelated to Mom. The gene nma1821 encoded by prophage Pnm1 (17), and ORF44 from the genomic island of H. influenzae biogroup aegyptius BPF strain F3031 (18) are also located in the same locus as Mu mom, and their products show 47 and 70% identity to the HI1523 protein, respectively. It has been reported that the FluMu HI1523 protein weakly resembles the Streptococcus sanguis DNA m6A MTase M.StsI (17), but this assertion was based on similarity that is statistically insignificant (a region of 56 residues with only 33% amino acid identity within a protein of 281 residues).
Further, bioinformatic analyses performed by us identified sequence similarity between HI1523 and NMA1821, and a family of DNA adenine-N6 (m6A) MTases. All known DNA m6A MTases belong to the Rossmann-fold MTase superfamily that is structurally and evolutionarily unrelated to the Mom protein that is a member of the acyltransferase superfamily (19). Therefore, we speculated that the HI1523 gene product and its homologs from other prophages modify adenine bases in the DNA, perhaps utilizing a mechanism different from that of the Mom protein. The aim of this study was to test this hypothesis by functionally characterizing these putative MTases.
MATERIALS AND METHODS
Enzymes, chemicals and oligonucleotides
REases, T4 DNA ligase, DNA polymerases, DNaseI, calf intestine alkaline phosphatase (CIP), DNA size standards, unlabeled N6-methyladenine (m6A), N4-methylcytosine (m4C), C5-methylcytosine (m5C) and S-adenosyl-l-methionine (AdoMet) were obtained from MBI Fermentas (Vilnius, Lithuania); M.HaeIII, CviQI, TfiI and NlaIII REases were from New England Biolabs (Ipswich, MA, USA). Snake venom phosphodiesterase I, P1 nuclease and deoxynucleosides were from Sigma. TLC PEI Cellulose F glass plates were from Merck. Chromatography resin was from Sigma (Ni-nitrilotriacetic acid-agarose, Ni-NTA). Synthetic deoxyoligonucleotides (sequences listed in Supplementary Tables S1 and S2) were synthesized by the Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland) and Sigma-Aldrich (Poland). [methyl-3H]-AdoMet (2.68 TBq/mmol) was from NEN Life Science Products (Boston, USA). Kits for plasmid preparation, genomic DNA isolation and DNA purification were from A&A Biotechnology (Gdynia, Poland). All enzymes were used following the manufacturer's recommendations.
Bacterial strains, plasmids, media and growth conditions
Escherichia coli strain Top10 (Invitrogen) F− mcrA Δ(mrr-hsdMRS-mcrBC) φ80lacZ ΔM15 ΔlacX74 deoR recA1 araD139 Δ(araA-leu)7697 galU galK rspL endA1 nupG was used for cloning experiments and strain ER2566 (New England Biolabs, Ipswich, MA, USA) F− λ− fhuA2 (lon) ompT lacZ::T7 geneI gal sulA11 Δ(mcrC-mrr)114::IS10 R(mcr-73::mini-Tn10)2 R(zgb-210::Tn10)1 (Tets) endA1 (Dcm) was used for recombinant protein overproduction. Escherichia coli strains were cultured under standard conditions in Luria-Bertani (LB) medium (20). When required, media were supplemented with antibiotics at the following final concentrations: ampicillin (Ap)—100 µg ml−1; kanamycin (Kn)—50 µg ml−1; tetracycline (Tc)—10 µg ml−1. To repress T7 RNA polymerase expression in ER2566 strains before induction, glucose was added to cultures at a final concentration of 1.0%.
The H. influenzae biotype aegyptius strain ATCC 11116 (kind gift from Promega to A.P.) and H. influenzae Rd (21) were grown at 37°C in 5% CO2 on brain heart infusion (BHI) medium supplemented with both nicotinamide-adenine dinucleotide and hemin at 2 µg ml−1 (sBHI) or on sBHI medium supplemented with mitomycin C at 35 ng ml−1.
The hia5 gene from H. influenzae biotype aegyptius ATCC 11116 (a homolog of ORF44 from H. influenzae biogroup aegyptius BPF strain F3031, AAV37176), the hin1523 gene from H. influenzae Rd (NP_439673) and the nma1821 gene (YP_002343106) from N. meningitidis type A strain Z2491 were cloned from the respective genomic DNAs. The following plasmid vectors were used in this work: pBluescript KS (Apr, Stratagene, USA) in cloning experiments, and pET28a and pET30a (Knr, Invitrogen, USA) in recombinant protein expression experiments. For the determination of DNA MTase specificity, phage λ DNA (Fermentas, Lithuania) and plasmid pAltOKI (Tcr) were used. pAltOKI is a derivative of the vector pAlterEx2 (Promega, Madison, USA) obtained by insertion of a synthetic deoxyoligonucleotide duplex (oligonucleotides Pst1TGCA and Pvu1AT, Supplementary Table S1) between the PstI and PvuI sites.
All routine DNA manipulations were carried out according to standard protocols (20). DNA sequencing was performed at the Institute of Biochemistry and Biophysics, Polish Academy of Sciences (Warsaw, Poland).
Cloning and sequence analysis of the hia5 gene
The genomic sequence of H. influenzae biotype aegyptius ATCC 11116 had not been determined. Assuming similarity between H. influenzae biotype aegyptius strains F3031 (18) and ATCC 11116, we designed primers for PCR (8Hf and 8Hr, Supplementary Table S1) based on the sequences flanking ORF44 (GenBank Acc. No. AY647244). The amplified PCR product was cloned into the SmaI site of the vector pBluescriptKS to produce plasmid pHia5KS. The cloned DNA fragment was sequenced and its length was found to be 1332 bp. This sequence has been submitted to the GenBank database under accession number JF268249.
For recombinant expression, the hia5 gene was amplified by PCR from plasmid pHia5KS DNA using primers 8HfNde and 8HXho (Supplementary Table S1). The obtained fragment (0.85 kb) was double digested with NdeI and XhoI, gel purified, and cloned into NdeI/XhoI-digested expression vector pET30a to add a C-terminal His6-tag to the encoded protein. To confirm that no mutations had been introduced during the cloning process, the insert of the resulting plasmid pHia5ET was sequenced.
Cloning nma1821 (ORF NMA1821) and hin1523 (ORF HI1523)
The procedure for cloning the hia5 gene was also employed to clone the nma1821 gene from N. meningitidis type A strain Z2491 (primers NmeNde and NmeXho) into expression vector pET30a, and the hin1523 gene from H. influenzae Rd (primers HindNco and HindXho, Supplementary Table S1) into expression vector pET28a. Therefore, C-terminal His6-tags were added to the encoded Nme1821 and Hin1523 proteins. Subsequently, the efficiency of recombinant Hin1523 protein isolation was found to be lower than for the other two recombinant proteins. Therefore, we subcloned the hin1523 gene on an NcoI/XhoI fragment into expression vector pET30a to add His6-tag at the N-terminus. Thus, the final recombinant Hin1523 protein carried His6-tags on both termini. The resulting plasmids pNmeET and pHinET were verified by sequencing.
Bioinformatic analyses
Sequence searches against the non-redundant protein sequence (nr) database to identify evolutionarily related proteins were performed using PSI-BLAST (22). Expectation (E) values of <10−3 were considered to be indication of homology. A multiple sequence alignment of selected homologs of Hia5, Nme1821 and Hin1523 proteins was made using PROMALS (23). Additional similarity searches and the alignment of the Hia5 family with sequences of proteins with known structure were done using the GeneSilico metaserver (http://genesilico.pl/meta2) (24).
Site-directed mutagenesis
The substitution of codons for aspartate with those for alanine within the cloned genes was carried out by PCR according to the ExSite protocol (Stratagene, USA). Oligonucleotide primers (Supplementary Table S1) were used to introduce the desired changes into the coding sequences of recombinant plasmids. The obtained clones were named pHia5D194AET, pNme1821D191AET and pHin1523D194AET.
Expression in E. coli and protein purification
All His6-tagged recombinant enzymes were expressed in E. coli strain ER2566. An overnight culture was diluted (1/100) into 500 ml of fresh LB medium supplemented with Kn and incubated at 37°C with shaking to an OD600 of 1.0. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and the incubation continued at 20°C for another 4 h. The culture was then centrifuged to harvest the cells and the bacterial pellet resuspended in buffer A (50 mM HEPES buffer, pH 8.0; 300 mM NaCl) supplemented with 1 mM PMSF, 10 mM 2-mercaptoethanol, 10% glycerol and 0.5% Triton X-100. After sonication to lyse the cells, the debris was removed by centrifugation at 40 000g for 1 h and the supernatant applied to a 6-ml Ni-NTA Agarose (Sigma) column pre-equilibrated with buffer A containing 20 mM imidazole. The column was washed with buffer A supplemented with 50 mM imidazole and then 70 mM imidazole. The recombinant protein was eluted from the column with buffer A containing 150 mM imidazole. The purity of the isolated protein was determined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE).
DNA methylation assay
The DNA modification activities of the purified recombinant enzymes were assayed in a 30 -μl reaction with 1 μg of λ DNA dam− dcm− at 37°C for 1 h in buffer M (10 mM Tris-HCl, pH 7.5, 10 mM EDTA, 5 mM 2-mercaptoethanol, 1 mg ml−1 bovine serum albumin (BSA)) and 2 μM [3H]AdoMet. The reaction was terminated by the addition of 0.5% SDS and the mixture was spotted onto 20 mm × 20 mm DE81 filter paper discs (Whatman, Brentford, UK). The filters were air-dried and then washed three times (10 min each) in a large volume of 50 mM KH2PO4, once in pure water, and once in 70% ethanol. Incorporation of the 3H-labeled methyl group was measured using a liquid scintillation counter (Wallace, Pharmacia).
For kinetic analysis, oligonucleotide duplexes were used as substrates in methylation assays to assess the sequence specificity of the enzymes. The complementary oligonucleotides (45 μM) were mixed, heated at 90°C for 5 min and then slowly cooled to 25°C to allow duplex formation (Supplementary Table S2). The reaction mixtures contained 1 µM oligo duplex, 6 μM [3H]AdoMet, 100 μM unlabeled AdoMet and 106 nM recombinant protein in buffer M. Therefore, the AdoMet concentration exceeds more than 2-fold of the concentration of potential substrate adenine residues in the TA50 duplex (10 times in the CA10 and GA10 duplexes, 5 times in the AA21 duplex, 106 times in Cm2A1, 35 times in Cm2A3). Reactions were incubated at 37°C, aliquots removed at specified time periods, mixed with 0.5% SDS and processed as described above. Each methylation experiment was carried out at least in triplicate. Initial velocities of reactions were calculated from the slope of reaction progress curves.
Thin-layer chromatography to identify the methylated base
The identity of the base methylated by the studied MTases was determined as described previously (25) using λ DNA dam− dcm− and [methyl-3H]AdoMet.
REase cleavage of DNA methylated by the Hia5, Nme1821 and Hin1523 enzymes
The DNAs of plasmids pHia5ET, pNmeET and pHinET isolated from IPTG-induced and uninduced bacterial cultures were used as substrates for cleavage by selected restriction enzymes. Typically, 1 µg of DNA was digested with 10 U of REase for 4 h in a 20 µl reaction in appropriate buffer conditions, as suggested by the manufacturer. For enzymes with only one recognition site in the plasmid DNA, additional digestion with an enzyme not sensitive to adenine methylation was carried out to linearize the DNA. Sequences recognized by REases HindIII, PstI, EcoRI and Eco32I were absent from pHia5ET, so in these cases, plasmid pAltOKI was introduced into strain ER2566(pHia5ET), and the plasmid preparation isolated from the double transformants was examined as described above. Digestion products were separated by electrophoresis on 0.8% agarose gels, which were then stained with ethidium bromide and photographed on a UV transilluminator.
The effect of in vitro enzyme activity toward GATC sites was determined using a protection assay in which λ DNA dam− dcm− was incubated with the recombinant proteins and then the DNA was challenged with REases. A typical assay reaction was a 100 μl mixture containing 80 μM AdoMet, 10 µg DNA and purified recombinant protein (~0.9 µg) in buffer M. The reaction mixture was incubated at 37°C for 60 min, extracted with phenol/chloroform and the DNA was precipitated with ethanol. The DNA was then cleaved separately with REases MboI, DpnI and Bsp143I in the appropriate buffers at 37°C for 4 h. In control reactions λ DNA dam− dcm− or λ DNA dam+ dcm+ were used to assess the activity of each enzyme. Plasmid or λ DNA without added endonuclease was used as negative controls.
HPLC DNA methylation assay
The substrate λ DNA dam− dcm− (10 µg) was methylated as described above for 3 h. Following phenol/chloroform extraction and DNA precipitation, samples were incubated with 1 U DNaseI in 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 mM NaCl and 0.1 mg/ml BSA, for 1 h at 37°C in a total volume of 30 µl. The DNaseI was then heat-inactivated and the mixtures were treated with P1 nuclease (2.5 U) and CIP (1 U) overnight at 37°C. A volume of 20 µl of each mixture was applied to a dC18 Atlantis column (3.0 × 150 mm, 5 µM particle size, Waters Associates Inc.) equilibrated with buffer E (50 mM KH2PO4 adjusted to pH 5.5 with KOH). High performance liquid chromatography (HPLC) runs were performed at a flow rate of 0.9 ml min−1 at ambient temperature using an isocratic mixture of buffer E and methanol (85 : 15). The column eluate was monitored by measuring absorbance at 260 nm. The obtained chromatograms were analyzed with the Empower™ program (Waters Associates Inc.). All samples were analyzed in quadruplicate. The same procedure was used for quantification of the base composition of chromosomal DNA isolated from the H. influenzae biotype aegyptius ATCC 11116 strain (5 µg). As controls, λ DNA dam− dcm− and λ DNA dam+ dcm+ were analyzed in the same way.
In order to calibrate the retention times of different nucleosides, six standards containing 400 pmol of the nucleosides dC, dT, dA, and dG, dmA and dmC (obtained by treating dm5CTP and dm6ATP with alkaline phosphatase under the same conditions as the DNA samples) were also analyzed (10 replicates of each). The average HPLC peak areas of the samples were compared with those of the standards and the mole percents calculated.
Determination of the genomic DNA methylation status of H. influenzae biotype aegyptius ATCC 11116 and H. influenzae Rd by restriction digestion
Genomic DNA was isolated from cells from the cultures of both H. influenzae strains that were either untreated or treated with mitomycin C, using a Genome-DNA kit (A&A Biotechnology). DNA (1 μg) was incubated with 10 U of the selected REases at 37°C for 4 h. The extent of cleavage was assessed by agarose gel electrophoresis.
RESULTS
Identification of Mu-like sequences in H. influenzae biotype aegyptius strain ATCC 11116
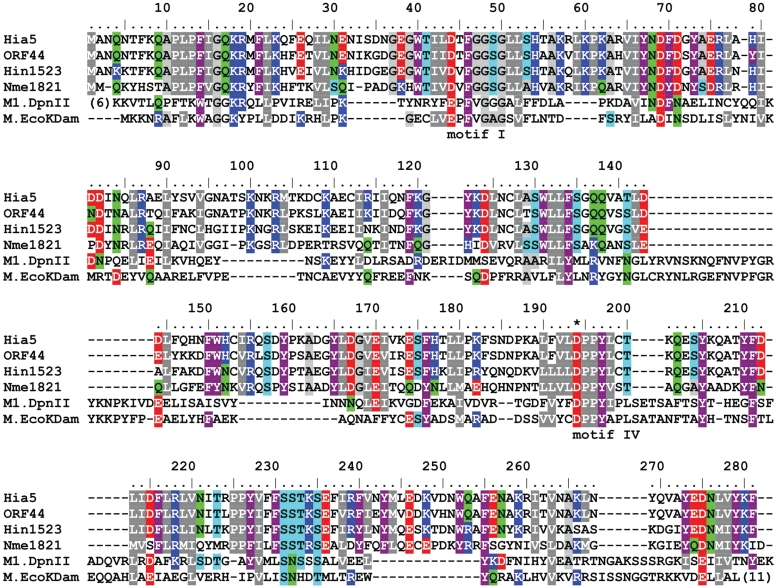
The 1332 bp sequenced fragment of H. influenzae biotype aegytptius strain ATCC11116 (for cloning see ‘Materials and Methods’ section) showed 86% identity to the DNA sequence between nucleotides 34 230 and 35 558 of the genomic island identified in the H. influenzae biogroup aegyptius BPF strain F3031 (Acc no. AY647244) (Supplementary Figure S1). Inspection of the sequence of this 1332-bp fragment revealed two open reading frames (ORFs), named comHia and hia5, located between nucleotides 232–348 and 413–1255, respectively (Supplementary Figure S2). The comHia translation product was 98% identical to the protein encoded by ORF43 of H. influenzae biotype aegytptius strain F3031 (located on the genomic island) (18) and also shared sequence similarity with Mu Com (49% identical over its entire length of 39 residues). Moreover, the putative ComHia protein contained four cysteines (positions 9, 12, 29 and 32) in analogous positions to Mu Com (Supplementary Figure S3), where these residues (Cys6, 9, 26 and 29) are required to form the zinc finger (12). This similarity suggested that ComHia performs the same function as Mu Com. The predicted protein product of the hia5 gene was 281 amino acids long and shared 80% identity with ORF44 of the H. influenzae biotype aegytptius strain F3031 genomic island, 69% with Hin1523 and 43% with Nme1821 (Figure 1).
Figure 1.
Sequence alignment of the Hia5, ORF44, Hin1523, Nme1821 proteins and two DNA MTases with known crystal structures: M1.DpnII (2dpm) and M.EcoKDam (2g1p). The number of terminal residues omitted for clarity is shown in parentheses (). Conserved motifs are indicated beneath the alignment and conserved residues are highlighted by shading (aliphatic in grey, aromatic in purple, positively charged in blue, negatively charged in red, with amide groups in green, with hydroxyl groups in cyan). The catalytic aspartate residue substituted by alanine is indicated by an asterisk.
Protein expression and purification
Recombinant Hia5, Nme1821and Hin1523 proteins were expressed in E. coli ER2566 cells carrying the plasmid constructs pHia5ET, pNmeET and pHinET, respectively. These His-tagged proteins were purified using metal affinity chromatography (‘Materials and Methods’ section). The apparent molecular masses of the recombinant proteins (34 kDa for Hia5, 33.6 kDa for Nme1821 and 38 kDa for Hin1523; Supplementary Figure S4) were in good agreement with those predicted from the nucleotide sequences.
Analysis of the amino acid sequences of Hia5, Nme1821 and Hin1523 proteins
The deduced amino acid sequences of the proteins Hia5, Nme1821 and Hin1523 were used as queries in PSI-BLAST searches of the National Center for Biotechnology Information (NCBI) nr database. The highest similarity scores over the full length of the sequences were with a large number of uncharacterized proteins annotated as putative MTases, followed by bona fide adenine MTases with lower, but still significant scores, regardless of the relatively low-level sequence identity (e.g. MboIA with the e-value 4e-7 and 15% identity, reported in a third iteration when using Hia5 as the query). Interestingly, the ORF48 of P. aeruginosa phage B3, which encodes a DNA adenine MTase homolog (16), was not detected among the 1000 most similar sequences. The homology between the Hia5 family and Dam-related MTases was further supported by the protein fold-recognition methods run via the GeneSilico metaserver (24). For example, using the HHsearch algorithm (26), E. coli Dam (M.EcoKDam), Streptococcus pneumoniae DpnM (M1.DpnII), and T4 Dam (M.EcoT4Dam) were identified with 100% probability as the closest homologs of Hia5 with known structure, and all consensus predictors indicated this prediction as the most probable (data not shown).
In agreement with their overall sequence similarity to known DNA adenine MTases, the Hia5, Nme1821 and Hin1523 sequences were found to contain the characteristic motif DPPY, also called motif IV (Figure 1), which is typical for MTases that modify amino groups in various substrates. The importance of this motif in the catalytic activity of DNA m6A MTases has been confirmed experimentally (27). In addition to the conserved DPPY motif, these enzymes possessed a well-conserved FxGxG motif, also called motif I, which is involved in AdoMet binding (Figure 1). However, the region corresponding to the target recognition domain in Hia5 (residues ~80–170) appeared highly divergent compared with those of known DNA MTase structures, which hampered the modeling of a Hia5–DNA complex and detailed prediction of the protein–DNA interactions.
Identification of the base methylated by the Hia5, Nme1821 and Hin1523 enzymes
In in vitro assays, recombinant Hia5, Nme1821 and Hin1523 were shown to incorporate the [3H] methyl group from AdoMet into the substrate DNA. However, variants in which the DPPY motif had been altered to APPY by site-directed mutagenesis were inactive (Table 1). This result confirmed that these proteins exhibit DNA MTase activity. Of the three enzymes tested, Hin1523 displayed the lowest activity. The chemical identity of the methylated base produced by the DNA MTase activity of these enzymes was identified using thin-layer chromatography. Tritium label was present only in the position corresponding to the m6dA standard (Table 2), which implied that all of the analyzed MTases modify adenine at the N6 position to form m6A.
Table 1.
Efficiency of in vitro methylation of phage λ DNA by M.HaeIII, Hia5, Nme1821 and Hin1523 enzymes and their altered catalytic motif IV (APPY) variants
| Sample | Methylation level (dpm) |
|---|---|
| Negative control: no enzyme | 130 |
| Negative control: enzyme heated to 80°C for 15 min before reaction | 150 |
| M.HaeIII | 19 000 |
| Hia5 | 18 000 |
| Hia5D194A | 226 |
| Nme1821 | 17 000 |
| Nme1821D191A | 231 |
| Hin1523 | 11 000 |
| Hin1523D194A | 195 |
Table 2.
Identification, by thin-layer chromatography, of the base methylated by the Hia5, Hin1523 and Nme1821 enzymes
| Enzyme |
3H incorporation (dpm) |
|||
|---|---|---|---|---|
| Solvent Ga |
Solvent Db |
|||
| m6dA | m4dC + m5dC | m6dA + m4dC | m5dC | |
| Hia5 | 5820 | 27 | 5700 | 22 |
| Nme1821 | 5670 | 20 | 5595 | 20 |
| Hin1523 | 5033 | 17 | 5001 | 25 |
am4dC and m5dC comigrate in solvent G. bm6dA and m4dC comigrate in solvent D.
Assessment of the sequence specificity of the Hia5, Nme1821 and Hin1523 enzymes using the endonuclease protection assay
To define the sequence specificity of the three novel MTases, we used a panel of adenine methylation-sensitive endonucleases in a REase digestion assay. It was anticipated that the presence of methylated adenine, introduced by the studied proteins, within a REase recognition site, would interfere with cleavage and therefore change the pattern of cleaved fragments.
First, we tested plasmids isolated from the E. coli ER2566 strain carrying the gene encoding the Hia5 protein for susceptibility to cleavage by enzymes sensitive or insensitive to methylation of adenine residues. Plasmid DNAs were prepared from the bacterial host, either induced for 4 h with 1 mM IPTG (pHia5ETi DNA) or not induced (pHia5ET DNA), and then subjected to digestion by REases. According to REBASE (28), 30 of the REases tested are blocked by adenine methylation at their cleavage site. Another 18 REases, insensitive to m6A, were used as controls to confirm the susceptibility of the substrate DNAs to digestion. The results of digestions by different restriction enzymes as an indicator of modification introduced by the Hia5 enzyme are presented in Table 3 (Supplementary Figure S5).
Table 3.
Summary of the sensitivity of different REases to DNA modifications introduced by the Hia5 enzyme
| REase | Recognition site | Number of sites in pHia5ET | Sensitivity to m6A | Cleavage |
|---|---|---|---|---|
| BglII | AGaTCT | 1 | S | N |
| Bsh1285I | CGRYCG R = A/G | 5 | S | N |
| Bsu15I | ATCGAT | 2 | S | N |
| CviQI | GTAC | 3 | S | N |
| DraI | TTTAAA | 2 | S | N |
| Eco32I | GATATC | 0 (1)b | S | N |
| EcoRI | GAATTC | 0 (1)b | S | N |
| HincII | GTYRAC | 1 (GTTAAC) | S | N |
| HindIII | AaGCTT | 0 (1)b | S | N |
| HinfI | GANTC | 19 | S | N |
| HphI | GGTGA/TCACC | 16 | S | N |
| KspAI | GTTAAC | 1 | S | N |
| MboIa | GATC | 24 | S | N |
| MluI | db ACGCGT | 1 | S | N |
| MlyI | GAGTC/GACTC | 9 | S | N |
| MunI | CAATTG | 1 | S | N |
| NlaIII | CATG | 24 | S | N |
| PaeI | GCATGC | 1 | S | N |
| PstI | CTGCAG | 0 (2)b | S | N |
| PvuII | dbCAGCTG | 3 | S | N |
| SspI | aATATT | 3 | S | N |
| VspI | ATTAAT | 5 | S | N |
| XbaI | TCTaGA | 1 | S | N |
| XhoI | CTCGAG | 1 | S | N |
| TaiI | ACGT | 16 | S | N |
| TaqI | TCGA | 16 | S | N |
| TfiI | GAWTC | 10 | S | N |
| AluI | AGCT | 24 | S | P, L |
| TasI | AATT | 28 | S | P, L |
| Tru1I | TTAA | 29 | S | P, L |
| CaiI | CAGNNNCTG | 1 | U | N |
| CseI | GACGC | 12 | U | N |
| Eam1104I | GAAGAG | 3 | U | N |
| Eco147I | AGGCCT | 1 | U | N |
| Eco47III | AGCGCT | 3 | U | N |
| Eco91I | GGTNACC | 1 | U | N |
| LweI | GCATC | 24 | U | N |
| Mph1103I | ATGCAT | 3 | U | N |
| NdeI | CaTATG | 1 | U | N |
| PfeI | GAWTC | 10 | U | N |
| PscI | ACATGT | 1 | U | N |
| SchI | GAGTC/GACTC | 9 | U | N |
| XmiI | GTMKAC | 1 | U | N |
| XapI | RAATTY | 6 | U | N |
| AdeI | CACNNNGTG | 1 | U | N |
| BpiI | GAAGAC | 5 | U | N |
| Bsp119I | TTCGAA | 1 | U | N |
| BcnI | CCSGG | 12 | (−) | Y |
| BglI | GGCN5GCC | 1 | (−) | Y |
| Bme1390I | CCNGG | 21 | (−) | Y |
| Bpu1102I | GCTNAGC | 2 | U | Y |
| Bsh1236I | CGCG | 37 | (−) | Y |
| Bsp143Ia | GATC | 24 | Not | Y |
| Bsp120I | GGGCCC | 1 | (−) | Y |
| Bsp68I | TCGCGA | 1 | Not | Y |
| BspLI | GGNNCC | 20 | (−) | Y |
| BspOI | GCTAGC | 0 (2)b | U | Y |
| Bst1107I | GTATAC | 1 | U | Y |
| BsuRI | GGCC | 22 | (−) | Y |
| Csp6I | GTAC | 3 | U | Y |
| DpnIa | Gm6ATC | 24 | Not | Y |
| Eco130I | CCWWGG | 1 | (−) | Y |
| Eco88I | CYCGRG | 2 | (−) | Y |
| EcoO109I | RGGNCCY | 3 | (−) | Y |
| FspBI | CTAG | 6 | U | Y |
| Hin1I | GRCGYC | 5 | (−) | Y |
| Hin6I | GCGC | 32 | (−) | Y |
| HpyF10VI | GCN7GC | 41 | (−) | Y |
| MbiI | GAGCGG/CCGCTC | 4 | U | Y |
| MspI | CCGG | 29 | (−) | Y |
| NheI | GCTAGC | 0 (2)b | U | Y |
| PvuI | CGATCG | 1 | Not | Y |
| SmaI | CCCGGG | 1 | (−) | Y |
aTested in λ DNA dam- in vitro methylated with the Hia5 enzyme. bnumber of recognition sites in plasmid pAltOKI.
Y: complete cleavage; N: no digestion; P: partial digestion, L: plasmid linearized; S: sensitive to m6A; U: unknown sensitivity to m6A; Not: not sensitive to m6A; (−): absence of adenine residues in recognition sequence; db: methylation of both strands required to block cleavage; A: the enzyme will not cut if the marked adenine is methylated; a: the enzyme is not sensitive to methylation of the marked adenine; A: there are no data on the sensitivity of the enzyme to methylation of the unmarked adenine.
Seven REases that do not have adenine in their target sequence, and eight others with only cytosine or guanine residues in the defined part of degenerate sequences (e.g. Bme1390I CCNGG or BglI GGCN5GCC) cleaved all DNA substrates tested. Likewise, three other REases (Bsp68I, Bsp143I and PvuI) that are known to be insensitive to adenine methylation in their cognate sequences, produced the expected restriction pattern. In contrast, all 30 REases with previously identified sensitivity to the presence m6A in their recognition sequences failed to cleave pHia5ETi DNA, or partial cleavage was observed with a large proportion of DNA fragments corresponding to linearized plasmid. The pHia5ET plasmid DNA isolated from non-induced cells was cleaved by all restriction enzymes tested.
The E. coli ER2566 strain contains EcoKDam MTase that modifies the adenine residue in the sequence GATC. To determine whether GATC sequences also represent a substrate for Hia5 modification, λ DNA dam− dcm− was methylated in vitro using Hia5. The status of this methylation was then tested by incubating the treated DNA with an excess of the following REases: DpnI (requires adenine methylation of GATC sites for cleavage), MboI (inhibited by m6A methylation) and Bsp143I (cleaves both methylated and unmethylated GATC sites, but not methylated sites when the methylation is on the C). The extent of digestion of Hia5-modified λ DNA by these REases showed that virtually all GATC sites were methylated (Supplementary Figure S6).
Experiments analogous to those described above were performed for the Nme1821 and Hin1523 enzymes (Supplementary Tables S3 and S4). The cleavage pattern for Nme1821-methylated DNA was very similar to that for Hia5, whereas in the case of Hin1523, the pHinETi plasmid DNA was more sensitive to REase cleavage than the pNmeETi or pHia5ETi DNAs.
Quantitative determination of m6A in DNA methylated by Hia5 or Hin1523
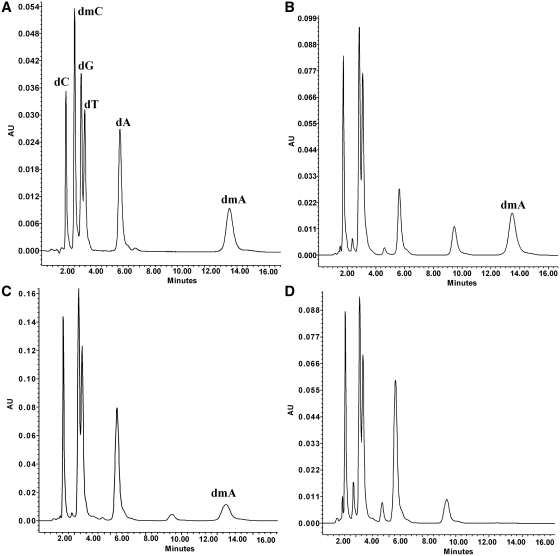
The results described above indicated that m6A was introduced in many nucleotide contexts. An HPLC DNA methylation assay was conducted to evaluate the extent of adenine modification resulting from the activity of the Hia5 and Hin1523 enzymes. The results of this analysis (Figure 2, for control DNAs see Supplementary Figure S7) clearly showed that the tested enzymes catalyzed extensive methylation of adenine residues in the substrate λ DNA. In contrast, identical analysis of λ DNA treated with the Hia5D194A variant did not reveal any dmA content, which confirmed the loss of DNA MTase activity in the Hia5D194A protein (Figure 2).
Figure 2.
HPLC profiles of deoxynucleosides in the standard mixture (A), and the enzymatic digests of Hia5-methylated λ DNA (B), Hin1523-methylated λ DNA (C) and Hia5D194A-methylated λ DNA (D). In digests of Hia5- and Hin1523-methylated λ DNA, in addition to major deoxynucleoside components, a peak was eluted at 13.7 min, corresponding to that of dmA. In the Hia5 and Hin1523 HPLC profiles and also those of Hia5D194A and λ DNA dam+ dcm+ (Supplementary Figure S7), we detected an unknown peak eluting at 9.5 min, but were unable to assign it to any obvious chemical species.
The HPLC assay results also allowed calculation of the quantity of adenine residues transformed into m6A (Table 4) by the studied enzymes: Hia5 methylated >60% of adenine residues in λ DNA and Hin1523 methylated 29.6%.
Table 4.
Percentages of the base content of λ DNA (control or methylated with Hia5, Hia5D194A or Hin1523 proteins) and total DNA of H. influenzae biotype aegyptius ATCC 11116 as determined by the HPLC DNA methylation assay
| DNA | dC | dmC | dG | dT | dA | dmA | dmA/ΣA | %GC |
|---|---|---|---|---|---|---|---|---|
| λ dam− dcm− | 23.60 | 0.00 | 26.10 | 25.30 | 25.00 | 0.00 | 0.0 | 49.70 |
| λ dam+ dcm+ | 23.60 | 0.04 | 25.88 | 24.58 | 25.78 | 0.12 | 0.46 | 49.52 |
| λ methylated by Hia5 | 23.90 | 0.00 | 25.40 | 24.00 | 10.20 | 16.50 | 61.79 | 49.30 |
| λ methylated by Hia5D194A | 25.00 | 0.00 | 25.10 | 24.60 | 25.20 | 0.00 | 0.00 | 50.20 |
| λ methylated by Hin1523 | 23.40 | 0.00 | 24.90 | 24.70 | 19.00 | 8.00 | 29.63 | 48.30 |
| Total DNA of H. influenzae biotype aegyptius ATCC 11116 | 21.00 | 3.70 | 19.20 | 26.80 | 28.80 | 0.50 | 1.70 | 43.90 |
ΣA = dA + dmA. Values presented are the means obtained using several independent DNA preparations.
As summarized in Table 4, the %GC content measured for λ DNA that had been in vitro methylated with the tested enzymes was in good agreement with reference data, i.e. 49.8% GC. The lower level of m6A in λ DNA dam+ dam+, than expected from the number of dam target sites (0.46 versus 0.92%), probably reflected undermethylation of the substrate GATC sites by M.EcoKDam. It has previously been demonstrated that only ~50% of Dam sites in λ DNA are methylated, presumably because the MTase does not have the opportunity to fully methylate the DNA before it is packaged into the phage head (29,30).
Sequence preferences of MTases
The observed high level of adenine methylation strongly suggested that the Hia5 and Hin1523 enzymes have minimal sequence specificity. Therefore, instead of using the classical approach to identify multiple methylated sequences followed by the search for consensus between them we have used a set of oligonucleotide duplexes as the substrates. Each of the duplexes contained repetitions of a dinucleotide in which an adenine was accompanied by another nucleotide (CA, GA, TA and AA). Examples of methylation reaction are shown in Supplementary Figure S8. Increase of radiolabel incorporation was linear for ~30 min.
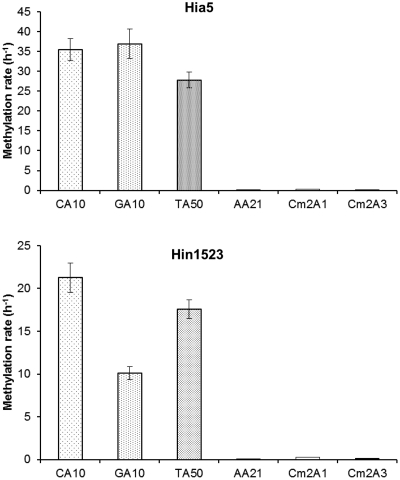
Substrates with repetitions of the dinucleotides CA and GA (CA10, GA10, respectively) were methylated with comparable rates by Hia5, and TA was methylated slightly slower (Figure 3). The dinucleotide CA was methylated by the Hin1523 enzyme with the highest rate, TA was methylated slightly slower and GA two times slower than two others (Figure 3). The poly-dA/poly-dT duplex was not methylated by either the Hia5 or Hin1523 enzymes. To exclude the nature of the sequence (tract character) and potential duplex instability as the cause of this negative result, we tested substrates containing one or three adenines flanked by m6As i.e. m6A A m6A (Cm2A1) and m6A AAA m6A (Cm2A3). These duplexes were also not methylated by Hia5 and Hin1523.
Figure 3.
Comparison of initial velocities of methylation reactions of oligonucleotide duplexes (Supplementary Table S2) carried out by the Hia5 and Hin1523 enzymes.
Hia5, Nme1821 and Hin1523 enzymes used as tools to test the sensitivity of REases to adenine methylation in their recognition sequences
Due to the extensive methylation of adenines introduced by the activity of Hia5, Nme1821 and Hin1523, these enzymes might be useful as tools in testing the sensitivity of REases to adenine methylation in their recognition sequences. We examined the activity of 23 commercially available REases, for which sensitivity to m6A had not been established, following methylation of their target sequences by Hia5, Hin1523 or Nme1821 (Table 3 for Hia5, Supplementary Table S3 for Nme1821 and Supplementary Table S4 for Hin1523). Sixteen of the tested REases were unable to cleave plasmid pHia5ETi DNA isolated from the strain overexpressing the Hia5 protein. These results were particularly valuable for nine REases that possess only one adenine residue in their recognition sequence (AdeI, CaiI, CseI, Eco147I, Eco47III, Eco91I, LweI, PfeI and SchI), which had to be the target for Hia5 methylation. It has previously been shown that NdeI is insensitive to methylation of the first adenine within its recognition sequence (CATATG), but its sensitivity to methylation of the second adenine was unknown. NdeI did not cut Hia5-methylated DNA, so it may be suggested that the Hia5 enzyme modified the second adenine within the CATATG sequence and as a result, NdeI lost its ability to recognize this target. Seven REases with unknown sensitivity to m6A that did not cleave DNA methylated by Hia5 contain more than one adenine residue in their recognition sequence and it is not certain which position interfered with cleavage when methylated. Another seven REases digested Hia5 methylated DNA to produce the expected restriction pattern. Since our results demonstrated that Hia5 can modify adenines in many nucleotide contexts, it may be concluded that these endonucleases are indeed insensitive to adenine methylation. Lastly, cleavage by CviQI was blocked when the adenine in its recognition site GTAC was methylated. However, REase Csp6I, an isoschizomer of CviQI, was not sensitive to adenine methylation in target sites.
m6A status in H. influenzae genomic DNA
Genomic DNAs of H. influenzae biotype aegyptius ATCC 11116 and H. influenzae Rd isolated from regular cultures and cultures treated with mitomycin C were incubated with an excess of REases and then the extent of cleavage was assessed by agarose gel electrophoresis. Treatment of bacterial cultures with mitomycin C had no apparent effect on the restriction patterns of the genomic DNAs (data not shown). Each DNA was found to be resistant to digestion with MboI, while at the same time cleaved by DpnI, which was as expected, because these bacterial strains contain Dam activity and therefore, GATC sites were modified. Bsp143II did not digest the genomic DNA of H. influenzae biotype aegyptius ATCC 11116 on account of the presence of M.HaeII activity (31) and HindIII did not digest the genomic DNA of H. influenzae Rd due to M.HindIII activity in this strain (32). The genomic DNAs were sensitive to all other enzymes used in this experiment (e.g. HinfI, CviQI and Csp6I) (Supplementary Figure S9—data shown only for genomic DNA of H. influenzae biotype aegyptius ATCC 11116 isolated from a regular culture).
The results described above are in good agreement with the HPLC analysis of the m6A content in the H. influenzae biotype aegyptius ATCC 11116 chromosomal DNA, which was calculated to be 1.7%. The m6A detected in this experiment was probably the result of the Dam MTase activity (Table 4, Supplementary Figure S7). Taken together, these data indicate that the Hia5 and Hin1523 enzymes are normally inactive in the respective Haemophilus strains, even in cultures treated with mitomycin C.
DISCUSSION
Since the annotation of the Mu genome in 2002 (17), Mu-like transposable phage family members have been identified during the sequencing of several bacterial genomes. Comparison of these Mu-like phages, prophages and phage-related elements indicates that their genome organizations are similar.
The 1332-bp fragment amplified from genomic DNA of the H. influenzae ATCC 11116 strain analyzed in this study was highly similar (86% identity) to a segment of the H. influenzae F3031 genomic island containing genes related to phage Mu (18). This high level of sequence similarity indicated that this ATCC 11116 genomic fragment represents part of a complete or defective Mu-like prophage present in this bacterial host.
Mu-like prophages often carry genes homologous to those of phage Mu and also other apparently unrelated genes that appear to perform analogous functions (17). Two of ORFs (nma1821, hin1523) characterized in this study are present at the right end of two Mu-like prophage genomes (17), and the third (hia5) is predicted to be a part of a Mu-like sequence. All three localize in the prophage genomes at positions syntenic to the mom gene of Mu. The putative protein products of the hia5, hin1523 and nma1821 genes also share sequence similarity, but are not homologous to the Mom protein of Mu—the modifying enzyme that converts adenine residues in DNA to acetamidoadenine. In this study, we cloned, expressed and characterized the hia5, hin1523 and nma1821 genes and identified their products as novel DNA MTases with extremely relaxed specificity.
According to the nature of the reactions they catalyze, DNA MTases can be divided into three separate groups, generating m6A, m4C or m5C. All known DNA MTases have conserved motifs, of which I–VIII and X are common to most subfamilies (33). X-ray crystallographic analyses have revealed that the region containing the conserved motifs corresponds to the structurally and evolutionarily conserved catalytic domain, in which motifs X and I–III are involved in AdoMet binding and motifs IV–VIII form the active site. The common AdoMet binding region is conserved across the whole MTase superfamily, while the ‘catalytic motifs’, especially motif IV, exhibit subfamily-specific patterns (33,34).
The enzymes that were the subject of this study belong to a previously uncharacterized subfamily of DNA adenine MTases. Despite their relatively distant sequence similarity to characterized MTases, they possess the characteristic signature motifs, including the catalytic motif IV (DPPY), which is typical for MTases that modify amino groups in various substrates, including RNAs and proteins. We have shown that in vitro these enzymes catalyzed the transfer of methyl groups from an AdoMet donor to the substrate DNA, and that they modified only adenine residues, converting them into m6A. To further confirm that the Hia5, Hin1523 and Nme1821 proteins act as adenine MTases, we altered their predicted catalytic sites by site-directed mutagenesis. Substitution of aspartate in the DPPY motifs with alanine (point substitution D194A for Hia5 and Hin1523, and D191A for Nme1821) completely abolished the adenine MTase activity of these enzymes.
To define the target sequences of these novel MTases, we used an endonuclease protection assay. All 30 REases with previously established sensitivity to m6A in their recognition sequences were unable to cleave DNAs isolated from strains expressing the tested genes or only a partial cleavage was observed. On the contrary, the control REases, which are insensitive to m6A, were able to cleave the DNAs to produce the expected restriction patterns. These results were the first indication of the massive DNA methylation catalyzed by the Hia5, Hin1523 and Nme1821 enzymes. Corroborating evidence was obtained using an HPLC DNA methylation assay to evaluate the base composition of λ DNA methylated with Hia5 in vitro. This analysis revealed that as much as 61% of the adenine residues were converted to m6A (29.6% by Hin1523). In other words more than every second adenine in λ DNA was modified by the Hia5 enzyme, which strongly suggested that a predicted sequence specificity cannot be longer than a dinucleotide. To directly test this assumption, we used substrates with repetitions of the dinucleotides CA, GA, TA and AA (poly-A tract). The first two were methylated by the Hia5 enzyme with comparable rates and TA was methylated slightly slower. This slight difference could be associated with the nature of the substrate. Adenines are present in both the upper and lower strands of the TA50 duplex, unlike in the CA10 and GA10 duplexes, where they are present only in one strand. It is possible that only one of the two diagonally arranged adenines in the TA duplex is methylated efficiently, while the other may be methylated at a lower rate.
Hin1523 also methylated the aforementioned duplexes, although the rate of CA10 methylation was the fastest. The TA50 duplex was modified by Hin1523 slightly slower similar to the Hia5 enzyme. Weak discrimination against the GA10 duplex observed for the Hin1523 does not exclude GA dinucleotides as substrates, since the GA10 duplex methylation rate was only two times lower than the rates measured for the CA10 and TA50 duplexes.
The incorporation signal obtained for the AA21 substrate (poly(A)/poly(T) tract) was at the background level. Adenines occurring in the Cm2A1 and Cm2A3 duplexes also appeared to be poor substrates for both the Hia5 and Hin1523 enzymes, suggesting that internal adenines within poly(A)-stretches are effectively not methylated.
Our results suggest that all adenine residues in double-stranded DNA, with the exception of poly(A)-tracts, constitute potential substrates for the Hia5 and Hin1523 enzymes, and that these enzymes are indeed sequence non-specific. Formally, their potential ‘sequence specificity’ could be summarized as AB or BA (where B = C, G or T). 71% of adenines in λ DNA occur in AB (BA) sequences. Sixty-one percent level of Hia5 λ DNA methylation in HPLC assay is close to this value.
To our knowledge, this is the first example of an exocyclic amino MTase with such extraordinary sequence promiscuity. High levels of adenine residue methylation in vivo and in vitro in a heterologous host (E. coli) have been described for M.CviQII that modifies the dinucleotide sequence AG (35), but this was demonstrated only by a restriction protection assay and the extent of M.CviQII modification has never been quantified.
REases are commonly used as molecular biology tools and their sensitivity to methylation has to be considered when digesting DNA, because cleavage may be blocked or impaired when a particular base in the recognition site is methylated. Our results suggest that the novel MTases characterized in this study can be very useful in verifying the adenine methylation sensitivity of many REases and in revealing such behavior in those endonucleases with unknown sensitivity to this DNA modification. Indeed, we demonstrated that 16 REases with unknown sensitivity to m6A in their cognate sequences were in fact sensitive to this modification. The knowledge whether a particular REase is sensitive to a nucleotide modification in its recognition sequence may be utilized to analyze patterns of regional methylation or to establish the methylation status of genomic DNA.
In eukaryotic cells, nuclear DNA is subject to enzymatic methylation resulting in the formation of m5C residues, mainly in CG and CNG sequences. In plants and animals, this DNA methylation is species-, tissue- and even organelle-specific. It changes with age and is regulated by hormones. On the other hand, genome methylation can control hormonal signaling (36).
It has long been known that m6A is present in the DNA of algae and their viruses, fungi and protists. Furthermore, and in spite of the common opinion that m6A is not found in the DNA of higher eukaryotes, this modified base has been detected in plastid, mitochondrial and nuclear plant DNA and in mosquito DNA (37). Accumulating evidence suggests that m6A affects the regulation of gene expression in mammalian cell (38–40). The use of the novel MTases identified in this work might allow us to study the effects of methylation on eukaryotic DNA–protein interactions and, in consequence, its impact on transcription in mammalian or plant cells.
Sequence searches with the Hia5 sequence against a non-redundant database (NCBI) revealed many closely related proteins, suggesting that there may exist more DNA : m6A MTases with similar properties, i.e. extremely relaxed substrate specificity. All identified Hia5 homologs were present in bacterial parasites, pathogens or commensal strains in mammals, including humans (data not shown). Thus, the contribution of Mu-like sequences to the virulence of their bacterial hosts should be considered.
Despite hosting Mu-like prophages encoding these novel MTases, we did not observe additional methylation of DNA during the normal growth of H. influenzae Rd (lysogenic for FluMu) and H. influenzae biotype aegyptius ATCC 11116, or after induction with mitomycin C, beyond the native methylation resulting from the activities of M.HinDam and M.HindIII, and M.HaeII, respectively. We were also unable to isolate FluMu phage particles (data not shown). Similarly, McGillivary et al. (18) failed to purify phage particles from the H. influenzae biotype aegyptius F3031. The expression of the mom gene is under strict control mediated by the Com protein (11). Possible Com (or Com homolog) binding sites and associated regulatory elements have been identified at the 5′-end of the hin1523 gene (17). This suggests that the Mom protein of Mu and these novel MTases could be regulated by a similar mechanism and may explain why we were unable to detect in vivo MTase activity under laboratory conditions; the hin1523 gene presumably remains repressed until the prophage enters the lytic cycle.
Saariaho et al. (41) revealed that FluMu encodes a functional transposase and contains critical transposase binding sites, but they were also unable to obtain FluMu virus plaques. Other attempts to induce Mu-like prophage excision and cell lysis using the DNA-damaging agents mitomycin C and UV irradiation have also been unsuccessful (42). Under certain conditions it is likely that the host may benefit from the presence of prophages (in particular those unable to enter a lytic cycle) by conferring immunity to superinfection by related phages (43).
Although the biological role of these newly characterized MTases for Mu-like phages and their hosts remains unknown, the discovery of a group of DNA:m6A MTases with extremely relaxed substrate specificity has significance due to their potential as tools in molecular biology research, particularly in the study of the poorly characterized role of adenine methylation in eukaryotic DNA.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–9.
FUNDING
Ministry of Science and Higher Education (2P04B00827 to M.R., NN301032634 to A.P.); 7th EU Framework Programme (HEALTH-PROT, GA No 229676 to J.M.B.). Funding for open access charge: Institute of Microbiology University of Warsaw.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Krzysztof Skowronek for useful comments and critical reading of the manuscript. Adam Jagielski is gratefully acknowledged for his help with the HPLC DNA methylation assays.
REFERENCES
- 1.Bertani G, Weigle JJ. Host controlled variation in bacterial viruses. J. Bacteriol. 1953;65:113–121. doi: 10.1128/jb.65.2.113-121.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tock MR, Dryden DT. The biology of restriction and anti-restriction. Curr. Opin. Microbiol. 2005;8:466–472. doi: 10.1016/j.mib.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins BM. Plasmid promiscuity: meeting the challenge of DNA immigration control. Environ. Microbiol. 2002;4:495–500. doi: 10.1046/j.1462-2920.2002.00332.x. [DOI] [PubMed] [Google Scholar]
- 4.Bickle TA, Kruger DH. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kruger DH, Bickle TA. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol. Rev. 1983;47:345–360. doi: 10.1128/mr.47.3.345-360.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hattman S. Unusual modification of bacteriophage Mu DNA. J. Virol. 1979;32:468–475. doi: 10.1128/jvi.32.2.468-475.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hattman S. Specificity of the bacteriophage Mu mom+ -controlled DNA modification. J. Virol. 1980;34:277–279. doi: 10.1128/jvi.34.1.277-279.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swinton D, Hattman S, Crain PF, Cheng CS, Smith DL, McCloskey JA. Purification and characterization of the unusual deoxynucleoside, alpha-N-(9-beta-D-2'-deoxyribofuranosylpurin-6-yl)glycinamide, specified by the phage Mu modification function. Proc. Natl Acad. Sci. USA. 1983;80:7400–7404. doi: 10.1073/pnas.80.24.7400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toussaint A. The DNA modification function of temperate phage Mu-1. Virology. 1976;70:17–27. doi: 10.1016/0042-6822(76)90232-4. [DOI] [PubMed] [Google Scholar]
- 10.Kahmann R. Methylation regulates the expression of a DNA-modification function encoded by bacteriophage Mu. Cold Spring Harb. Symp. Quant. Biol. 1983;47(Pt 2):639–646. doi: 10.1101/sqb.1983.047.01.075. [DOI] [PubMed] [Google Scholar]
- 11.Hattman S. Unusual transcriptional and translational regulation of the bacteriophage Mu mom operon. Pharmacol. Ther. 1999;84:367–388. doi: 10.1016/s0163-7258(99)00042-x. [DOI] [PubMed] [Google Scholar]
- 12.Witkowski RT, Hattman S, Newman L, Clark K, Tierney DL, Penner-Hahn J, McLendon G. The zinc coordination site of the bacteriophage Mu translational activator protein, Com. J. Mol. Biol. 1995;247:753–764. doi: 10.1006/jmbi.1995.0178. [DOI] [PubMed] [Google Scholar]
- 13.Hattman S, Ives J, Wall L, Maric S. The bacteriophage Mu com gene appears to specify a translation factor required for mom gene expression. Gene. 1987;55:345–351. doi: 10.1016/0378-1119(87)90295-2. [DOI] [PubMed] [Google Scholar]
- 14.Wulczyn FG, Bolker M, Kahmann R. Translation of the bacteriophage Mu mom gene is positively regulated by the phage com gene product. Cell. 1989;57:1201–1210. doi: 10.1016/0092-8674(89)90057-3. [DOI] [PubMed] [Google Scholar]
- 15.Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, Nakayama K, Hayashi T. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009;5:e1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braid MD, Silhavy JL, Kitts CL, Cano RJ, Howe MM. Complete genomic sequence of bacteriophage B3, a Mu-like phage of Pseudomonas aeruginosa. J. Bacteriol. 2004;186:6560–6574. doi: 10.1128/JB.186.19.6560-6574.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgan GJ, Hatfull GF, Casjens S, Hendrix RW. Bacteriophage Mu genome sequence: analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 2002;317:337–359. doi: 10.1006/jmbi.2002.5437. [DOI] [PubMed] [Google Scholar]
- 18.McGillivary G, Tomaras AP, Rhodes ER, Actis LA. Cloning and sequencing of a genomic island found in the Brazilian purpuric fever clone of Haemophilus influenzae biogroup aegyptius. Infect. Immun. 2005;73:1927–1938. doi: 10.1128/IAI.73.4.1927-1938.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaminska KH, Bujnicki JM. Bacteriophage Mu Mom protein responsible for DNA modification is a new member of the acyltransferase superfamily. Cell Cycle. 2008;7:120–121. doi: 10.4161/cc.7.1.5158. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 21.Glover SW, Piekarowicz A. Host specificity of DNA in Haemophilus influenzae: restriction and modification in strain Rd. Biochem. Biophys. Res. Commun. 1972;46:1610–1617. doi: 10.1016/0006-291x(72)90793-0. [DOI] [PubMed] [Google Scholar]
- 22.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 24.Kurowski MA, Bujnicki JM. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 2003;31:3305–3307. doi: 10.1093/nar/gkg557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piekarowicz A, Golaszewska M, Sunday AO, Siwinska M, Stein DC. The HaeIV restriction modification system of Haemophilus aegyptius is encoded by a single polypeptide. J. Mol. Biol. 1999;293:1055–1065. doi: 10.1006/jmbi.1999.3198. [DOI] [PubMed] [Google Scholar]
- 26.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 27.Bheemanaik S, Reddy YV, Rao DN. Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem. J. 2006;399:177–190. doi: 10.1042/BJ20060854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE–a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2010;38:D234–D236. doi: 10.1093/nar/gkp874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ringquist S, Smith CL. The Escherichia coli chromosome contains specific, unmethylated dam and dcm sites. Proc. Natl Acad. Sci. USA. 1992;89:4539–4543. doi: 10.1073/pnas.89.10.4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hattman S. Plasmid-controlled variation in the content of methylated bases in bacteriophage lambda deoxyribonucleic acid. J. Virol. 1972;10:356–361. doi: 10.1128/jvi.10.3.356-361.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roberts RJ, Breitmeyer JB, Tabachnik NF, Myers PA. A second specific endonuclease from Haemophilus aegyptius. J. Mol. Biol. 1975;91:121–123. doi: 10.1016/0022-2836(75)90375-7. [DOI] [PubMed] [Google Scholar]
- 32.Roy PH, Smith HO. DNA methylases of Hemophilus influenzae Rd. II. Partial recognition site base sequences. J. Mol. Biol. 1973;81:445–459. doi: 10.1016/0022-2836(73)90516-0. [DOI] [PubMed] [Google Scholar]
- 33.Malone T, Blumenthal RM, Cheng X. Structure-guided analysis reveals nine sequence motifs conserved among DNA amino-methyltransferases, and suggests a catalytic mechanism for these enzymes. J. Mol. Biol. 1995;253:618–632. doi: 10.1006/jmbi.1995.0577. [DOI] [PubMed] [Google Scholar]
- 34.Klimasauskas S, Timinskas A, Menkevicius S, Butkiene D, Butkus V, Janulaitis A. Sequence motifs characteristic of DNA [cytosine-N4] methyltransferases: similarity to adenine and cytosine-C5 DNA-methylases. Nucleic Acids Res. 1989;17:9823–9832. doi: 10.1093/nar/17.23.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan SH, Zhu Z, Dunigan DD, Van Etten JL, Xu SY. Cloning of Nt.CviQII nicking endonuclease and its cognate methyltransferase: M.CviQII methylates AG sequences. Protein Expr. Purif. 2006;49:138–150. doi: 10.1016/j.pep.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Vanyushin BF. Enzymatic DNA methylation is an epigenetic control for genetic functions of the cell. Biochemistry. 2005;70:488–499. doi: 10.1007/s10541-005-0143-y. [DOI] [PubMed] [Google Scholar]
- 37.Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: the other methylated base of DNA. Bioessays. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tronche F, Rollier A, Bach I, Weiss MC, Yaniv M. The rat albumin promoter: cooperation with upstream elements is required when binding of APF/HNF1 to the proximal element is partially impaired by mutation or bacterial methylation. Mol. Cell Biol. 1989;9:4759–4766. doi: 10.1128/mcb.9.11.4759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knebel D, Doerfler W. N6-methyldeoxyadenosine residues at specific sites decrease the activity of the E1A promoter of adenovirus type 12 DNA. J. Mol. Biol. 1986;189:371–375. doi: 10.1016/0022-2836(86)90518-8. [DOI] [PubMed] [Google Scholar]
- 40.Truss M, Bartsch J, Chalepakis G, Beato M. Artificial steroid hormone response element generated by dam-methylation. Nucleic Acids Res. 1992;20:1483–1486. doi: 10.1093/nar/20.7.1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saariaho AH, Lamberg A, Elo S, Savilahti H. Functional comparison of the transposition core machineries of phage Mu and Haemophilus influenzae Mu-like prophage Hin-Mu reveals interchangeable components. Virology. 2005;331:6–19. doi: 10.1016/j.virol.2004.09.041. [DOI] [PubMed] [Google Scholar]
- 42.Evans TJ, Coulthurst SJ, Komitopoulou E, Salmond GP. Two mobile Pectobacterium atrosepticum prophages modulate virulence. FEMS Microbiol. Lett. 2010;304:195–202. doi: 10.1111/j.1574-6968.2010.01901.x. [DOI] [PubMed] [Google Scholar]
- 43.Snyder L. Phage-exclusion enzymes: a bonanza of biochemical and cell biology reagents? Mol. Microbiol. 1995;15:415–420. doi: 10.1111/j.1365-2958.1995.tb02255.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.