Abstract
Translation of the TnaC nascent peptide inhibits ribosomal activity in the presence of l-tryptophan, inducing expression of the tnaCAB operon in Escherichia coli. Using chemical methylation, this work reveals how interactions between TnaC and the ribosome are affected by mutations in both molecules. The presence of the TnaC-tRNAPro peptidyl-tRNA within the ribosome protects the 23S rRNA nucleotide U2609 against chemical methylation. Such protection was not observed in mutant ribosomes containing changes in 23S rRNA nucleotides of the A748–A752 region. Nucleotides A752 and U2609 establish a base-pair interaction. Most replacements of either A752 or U2609 affected Trp induction of a TnaC-regulated LacZ reporter. However, the single change A752G, or the dual replacements A752G and U2609C, maintained Trp induction. Replacements at the conserved TnaC residues W12 and D16 also abolished the protection of U2609 by TnaC-tRNAPro against chemical methylation. These data indicate that the TnaC nascent peptide in the ribosome exit tunnel interacts with the U2609 nucleotide when the ribosome is Trp responsive. This interaction is affected by mutational changes in exit tunnel nucleotides of 23S rRNA, as well as in conserved TnaC residues, suggesting that they affect the structure of the exit tunnel and/or the nascent peptide configuration in the tunnel.
INTRODUCTION
Expression of the E. coli tryptophanase operon, tnaCAB, is induced in response to l-tryptophan (Trp) by the action of the TnaC nascent peptide. TnaC stalls the ribosomes that have produced it in response to high Trp; the stalled ribosomes inhibit transcription attenuation, resulting in increased operon expression (1). The tna operon consists of a leader regulatory region, which includes tnaC, and two downstream structural genes, tnaA and tnaB. tnaA encodes tryptophanase, an enzyme involved in the degradation of Trp to obtain indole, energy and ammonia (2). In addition to its roles in biosynthetic pathways, the indole molecule is used in bacteria as a signal in regulating biofilm formation and quorum sensing (3). tnaB encodes a Trp permease for Trp transport into the cell (2). tnaC specifies the 24-residue TnaC regulatory leader peptide. Immediately downstream of tnaC there is a non-coding segment which contains Rho-dependent terminator sequences (2).
Transcription initiation of the tna operon is regulated by catabolite repression (4). Despite transcription activation by CAP/cAMP, transcription of the tna operon is terminated prematurely at the Rho-dependent termination sequences when low concentrations of Trp are present in the growth media (1). However, in the presence of high Trp, transcription of the tna operon is not attenuated at those sites and mRNA containing tnaA and tnaB is produced (1). Hydrolysis of the terminal TnaC-tRNAPro peptidyl-tRNA by the action of RF-2 protein during translation termination is inhibited by Trp (5,6), causing the translating ribosome to transiently stall at the tnaC stop codon. The stalled ribosome masks the binding sequences for the Rho termination factor; the absence of interaction of Rho with the nascent mRNA allows transcription to continue into the tnaA and tnaB structural genes (6,7).
Analyses of the primary structure of the TnaC peptide from many bacterial species have revealed that the Trp residue at the 12th position (W12), an aspartic acid residue at the 16th position (D16), and a proline residue at the last position (P24) of E. coli TnaC are highly conserved (8,9). These conserved TnaC residues are essential for TnaC-mediated Trp induction (8). Changing these amino acid residues abolishes Trp induction in vivo, the ability of Trp to inhibit the hydrolysis induced by RF-2, and Trp-inhibition of TnaC-tRNAPro cleavage induced by puromycin (8,10). The relative position of these conserved residues in the TnaC peptide is important as well: insertion or deletion of single amino acids between the W12 and the P24 residues abolishes Trp induction (11). These data indicate that the nature and positions of the conserved TnaC residues are important for the nascent peptide's regulatory activity at the level of translation.
Nascent peptides mediating ribosome stalling are widespread in the microbial world (12). Some notable examples are SecM from the secMA operon of E. coli (13), MifM that regulates expression of the yidC2 gene of Bacillus subtilis (14), ErmCL from the erm operon of erythromycin-resistant bacteria (15), and the evolutionary conserved fungal arginine attenuator peptide (AAP) (16,17). As is the case for TnaC, these regulatory peptides contain amino acid residues whose nature and relative positions are essential for stalling activity (13–15,18).
Changes in the large subunit 23S rRNA sequence or in ribosomal protein L22 affect Trp induction. Insertion of an additional adenine nucleotide in the G745–A752 region (designated +A751ins), or the substitutions U2609C, A752C and A752U in the 23S rRNA, abolish the action of Trp to induce TnaC-mediated ribosome stalling (19). Replacements of the K90 residue of ribosomal protein L22 also affect Trp induction (19). These ribosomal components are located in the narrowest region of the ribosome exit tunnel (20). Cryo-electron microscopy (EM) structures of ribosomes containing TnaC-tRNAPro molecules suggest that W12 and D16 of TnaC are in close proximity to the K90 and R92 residues of ribosomal protein L22, and to 23S rRNA nucleotides A751–A752 (21). Cross-linking analysis confirms that the W12 residue of TnaC is in close proximity to the G745–A752 region of the 23S rRNA (19). The cryo-EM structure also indicates that the P24 residue of the TnaC peptide is close to the peptidyl transferase center (PTC) nucleotide U2585, and adjacent to the nucleotides G2583 and U2584; mutations in the two latter positions are tolerated but affect Trp induction (21,22).
The evidence suggests that essential TnaC residues interact with components of the ribosomal exit tunnel, and that these interactions induce structural changes that are transferred from the TnaC-exit tunnel contact points to the PTC, resulting in inhibition of peptidyltransferase activity (11,23). The cryo-EM model suggests three possible routes where structural changes could be induced and transferred from the ribosomal exit tunnel to the PTC. In one possible route, the structural changes are transmitted through ribosomal protein L4 and the A2058–2059:2060–2062:2503:2451 23S rRNA nucleotides (21). However, changes at most of these positions do not affect Trp induction (19,24), although they affect the action of the SecM and ErmCL nascent peptides (24). A second possible route considers transmission of structural changes through the nascent TnaC peptide chain. Finally, in the third proposed route, transmission of structural changes occurs through the interactions observed between L22 and the A751–A752:U2609:U1781–U1782:U2586–U2585 23S rRNA nucleotides (21). This last route contains mostly those nucleotides in which changes are known from experimental data to affect Trp induction.
The fact that some elements of the ribosome exit tunnel are important for the function of the nascent TnaC peptide suggests that they may interact with this regulatory peptide. The proximity of these elements to the nascent peptide observed in the cryo-EM structure are also consistent with this idea (21). However, changes in these elements could also affect the structure of the ribosome exit tunnel in a manner that indirectly affects interactions between the exit tunnel and TnaC. In this study, we show that the presence of the nascent TnaC peptide within the ribosome induces protection against chemical methylation of exit tunnel 23S rRNA nucleotide U2609. We observed that mutational changes in the nucleotides constituting the G745–A752 region of the 23S rRNA, and in conserved TnaC residues, that abolish TnaC-mediated regulation also reduced the methylation protection of U2609 conferred by wild-type nascent TnaC. These results indicate that changes in the G745–A752 and U2609 regions greatly reduced the capacity of Trp and TnaC to inhibit ribosome function. The proximity of these regions of the ribosome to TnaC suggests that functional interactions are impaired by these mutational changes.
MATERIALS AND METHODS
Bacterial strains, plasmids and mutagenesis procedures
The E. coli K-12 strains, and plasmids containing selected genes used in this study, are listed in Table 1. Strains with replacements of the 23S rRNA gene were generated using plasmids pNK (19), and pK4–16 (Selwyn Quan and Catherine Squires, personal communication), which contain the rrnB operon. Replacements of tnaC sequences were generated in the pGF2500 plasmid that contains the tna promoter, a wild-type tnaC gene, the tna intercistronic region and a rpoBC terminator (5). Mutations in these genes were made using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies). Complementary primers were designed with the desired replacements flanked by ~10–15 nucleotides of the wild-type sequences on each side of the change. The mutagenesis reactions were performed as recommended by the manufacturer in 50 μl final volume with 10–100 ng of plasmid and 10 pmol of each complementary primer. The plasmids that contained the desired replacements were confirmed by sequencing using the following primer: (5′-ACGGAATTCCTTGCCGAGTTTGACTC-3′) which is complementary to the 3′-end of the rpoBC region.
Table 1.
Escherichia coli bacterial strains and plasmids used in this work
| Strains or plasmids | Relevant genotype | References |
|---|---|---|
| Strains | ||
| SR-14 | Δ7 rrn Δ(lacZYA) ΔrecA λ tnap tnaC(tnaA’-‘lacZYA) (prrnC-sacB, ptRNA67) | (19) |
| SQ351 | MG1655 Δ7 rrn Δ(lacZYA) (pKK3535, ptRNA67) | (19) |
| AW122 | Derived from SQ351 (prrnC-sacB, ptRNA67) | This work |
| AW182 | MG1655 Δ7 rrn Δ(lacZYA) att7::tnaptnaC(tnaA’-‘lacZYA) (prrnC-sacB, ptRNA67) | This work |
| AW216 | MG1655 Δ7 rrn Δ(lacZYA) att7::tnaptnaC(tnaA’-‘lacZYA) (pNK, ptRNA67) | This work |
| AW218 | MG1655 Δ7 rrn Δ(lacZYA) att7::tnaptnaC(tnaA’-‘lacZYA) (pKKU2609C, ptRNA67) | This work |
| AW221 | MG1655 Δ7 rrn Δ(lacZYA) att7::tnaptnaC(W12R)(tnaA’-‘lacZYA) (pNK, ptRNA67) | This work |
| AW227 | MG1655 Δ7 rrn Δ(lacZYA) att7::tnaptnaC(tnaA’-‘lacZYA) (pNH153, ptRNA67) | This work |
| AW326 | MG1655 Δ7 rrn Δ(lacZYA) att7::tnaptnaC(K18A)(tnaA’-‘lacZYA) (pNK, ptRNA67) | This work |
| AW600 | MG1655 Δ7 rrn Δ(lacZYA) att7::tnaptnaC(D16A)(tnaA’-‘lacZYA) (pNK, ptRNA67) | This work |
| Plasmids | ||
| pNK | Wild-type rrnB operon; Ampr, derived from ColE1 | (13) |
| pNH153 | Derived from pNK; has an insertion at position 751 in the 23S rRNA gene | (13) |
| pKKU2609C | Derived from pNK; has a T-to-C replacement at position 2609 in the 23S rRNA gene | (33) |
| pNKA752C | Derived from pNK; has a A-to-C replacement at position 752 in the 23S rRNA gene | (36) |
| pK4–16 | Wild-type rrnB operon; Kmr, derived from SC101 | Quan and Squires, personal communication |
| prrnC-sacB | Wild-type rrnC gene and a sacB gene, derived from SC101 | (45) |
| pKK3535 | Wild-type rrnB operon, derived from pBR322 | (37) |
| ptRNA67 | tRNA encoding plasmid | (45) |
| pGF2500 | Wild-type tnaC gene with the rpoBC terminator, derived from pUC18 | (5) |
| pAW137 | Has the tnaptnaC(ΔN2-H22) with BsaI–XhoI–BsaI linker-tna-'-'lacZYA cloning reporter gene, derived from pACYC184 | This work |
Creation of the tnaA’-‘lacZ reporter gene at the att7 site and tnaA’-‘lacZ induction experiments
A DNA fragment containing the tnap tnaC(tnaA’-‘lacZY) reporter gene was amplified from the chromosome of the SVS1144 strain (1) using the primers 5′-CTGGTCGACGCTTCTGTATTGGTAAGTAACCGCGC-3′ and 5′-CTAGTCGACGCTTAAGCGACTTCATTCACCTGACG-3′. These primers amplify a DNA fragment containing 262 bp upstream of the start codon of tnaC through 2 bp downstream of the start codon of lacY. These elements are flanked by SalI sites on both ends of the PCR product. This DNA fragment was cloned in the SalI site of pACYC184 plasmid. Inverse PCR products obtained with the primers 5′-CTGCTCGAGGGTCTCACGCCCTTGAATTGCCCTTCTGTAGC-3′ and 5′-CTACTCGAGGGTCTCACATAATGCACTTATCCTCGCAAGAC-3′ containing XhoI restriction sites were digested with XhoI enzyme and ligated. This procedure eliminates the tnaC region that specifies the second through the 22nd codons. The plasmid now contains a tnaC gene with an internal deletion marked with a BsaI–XhoI–BsaI cloning site, which allows direct insertion of synthetic oligonucleotides containing wild-type and mutated tnaC coding sequences [tnaC(ΔN2-H22) BsaI–XhoI–BsaI linker-tnaA’]. To avoid potential complications arising from the inverse PCR, the new cloning construct was moved into pRS552 plasmid as a BamHI fragment (1), to generate the tnaC(ΔN2-H22)-tnaA’-‘lacZYA reporter gene that later was transferred back into pACYC184, generating plasmid pAW137. Annealed oligos were used to insert either wild-type or mutant tnaC sequence into pAW137. Five microliters of each 100 µM complementary oligos were mixed in a microcentrifuge tube, incubated in boiling water for 2 min and allowed to cool to room temperature. Annealed oligos were ligated to BsaI digested pAW137, creating derivatives containing wild-type or mutant TnaC sequences. The SalI DNA fragments from these pAW137 derivatives were cloned into the XhoI site of pGRG36 plasmid (39). pGRG36 encodes the Tn7 transposition machinery which allows site-specific transposition at the att7 locus of E. coli (39). Two different orientations of the wild-type or mutant tnaC-tnaA’-’lacZYA reporter genes result from this cloning method. Strains containing one of each orientation were obtained for every construct. The plasmids derived from pGRG36 were transformed into chemical-competent AW122 cells. Transformants were selected on Luria-Bertani (LB) plates containing 100 μg/ml ampicillin at 32°C. The transposition protocol to move tnaC-tnaA’-‘lacZYA reporters to the chromosomal att7 site was carried out as described (39). Transposition was verified by replica plating colonies on LB plates containing, or lacking, 100 μg/ml ampicillin. The sequence of the region from tnap through the tnaA’-‘lacZ junction of the att7 integrants was confirmed by using PCR to amplify the att7 locus using the primers 5′-GCGGCGACAACAGTTGCGACGGTGGTACG-3′ and 5′-GCGGTTTTCTCCGGCGCGTAAAAATGCGCTCAGG-3′ followed by sequencing of the resulting fragment. To analyze the expression of the tnaA’-‘lacZ reporter gene we performed β-galactosidase (β-gal) assays as previously (39). β-gal activity is reported in Miller units.
Puromycin assay
Stalled ribosome complexes were isolated using pGF2500 variants as previously indicated (19). Ten microliters of isolated stalled complexes dissolved in buffer A (35 mM Tris–acetate, pH 8.0, 10 mM magnesium acetate, 175 mM potassium acetate, 10 mM ammonium acetate and 1 mM DTT) were mixed with 1 µl of water or 1 µl of 20 mM Trp. The mixtures then were mixed with 1 µl of water or 1 µl of 0.2 mM puromycin, and these mixtures were then incubated for 10 min at room temperature. The reactions were stopped by adding an equal volume of loading buffer [100 mM Tris–HCl, pH 6.8, 24% (v/v) glycerol, 8% sodium dodecyl sulfate, 4% (v/v) β-mercaptoethanol and 0.4 mg/ml bromophenol blue]. The products of this reaction were resolved using 10% Tris–tricine polyacrylamide gels. The gels were dried by vacuum and then exposed to X-ray films.
Methylation protection and primer extension assays
Fifty microliters of either 50S ribosomal subunits (40 A260), obtained as previously indicated (19), or isolated stalled complexes dissolved in buffer A were mixed with either 2 μl of dimethyl sulfate (DMS, 1:6 dilution in ethanol) or 50 μl of 100 mg/ml 1-cyclohexyl-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT) solution. Mixtures containing DMS or CMCT were incubated at room temperature for 10 or 30 min, respectively. DMS reactions were stopped by adding 25 μl of a solution containing 1.4 M of β-mercaptoethanol and 1 M Tris–HCl (pH 8.0). The final mixtures were diluted by adding 10 mM EDTA in Diethyl pyrocarbonate (DEPC) treated water. The methylated rRNA was obtained by standard phenol chloroform extractions. Integrity of the extracted rRNAs was verified on 2% agarose gels. RNA was quantified using UV spectroscopy at A260. Primer extension analysis were performed to detect the methylation of nucleotides in the 23S rRNA as indicated previously (19).
RESULTS
Nucleotide changes in the ribosome exit tunnel that affects TnaC/Trp inhibition of the ribosome function
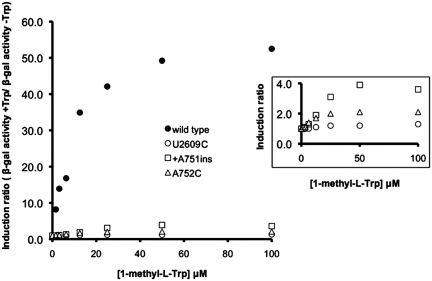
Mutations in 23S rRNA nucleotides that constitute the ribosome exit tunnel reduce TnaC-mediated operon induction in response to Trp (19,22). We used bacterial strains in which the seven rRNA operons were deleted from their chromosomal locations to analyze how A752C, U2609C and +A751 insertion mutations in the 23S rRNA affected the inhibition of ribosome function by Trp (19). These strains contained a homogeneous population of mutant ribosomes (Supplementary Figure S1). To determine the effect of ribosomal mutations on TnaC-mediated regulation in response to Trp and Trp analogs, we used bacteria that contained a tnaC tnaA’-‘lacZ reporter gene (19). We tested the effects of these ribosomal mutations on the expression of the reporter gene in vivo using several concentrations of 1-methyl-l-Trp (1MT). This Trp analog induces operon expression but is not cleaved by tryptophanase and thus is a more efficient inducer in vivo than Trp (42). The results are summarized in Figure 1 (primary data are given in Supplementary Table S1). Based on the induction curve, 40 µM of 1MT would be sufficient for maximal expression of the reporter gene in cells containing wild-type ribosomes (Figure 1, closed circles). For cells containing ribosomes with the +A751ins mutation, even at 100 µM 1MT operon induction was 12-fold lower than cells containing wild-type ribosomes (Figure 1, compare open squares with closed circles). Similar results were observed with cells containing A752C mutant ribosomes, in which 100 µM 1MT induced 25-fold less operon expression than in wild-type cells (Figure 1, compare open triangles with closed circles). These results indicated that +A751ins or A752C ribosomes allowed at most, slight induction of the tna reporter operon at high concentrations of 1MT (Figure 1, inset). Finally, we observed in cells containing ribosomes with the U2609C replacement that expression of the reporter gene was not induced by the addition of any amount of 1MT tested (Figure 1, open circles and data not shown). This result indicated that the U2609C replacement completely abolished TnaC-mediated regulation in response to 1MT. In summary, we observed differences in the way that the +A751ins, A752C and the U2609C mutations affected TnaC-mediated regulation.
Figure 1.
Mutations of 23S rRNA nucleotides that affect tna operon expression. Bacterial cells expressing the indicated 23S rRNA alleles were used to analyze expression of β-gal from a tnaC-tnaA’-‘lacZ protein fusion. Bacterial cultures were grown in minimal medium containing 0.2% glycerol, 0.05% acid-hydrolyzed casein, 0.01% vitamin B1 and variable amounts of 1 MT as an inducer. The figure on the right shows an amplification of the plots between the induction values zero to four.
Nucleotide residues of the ribosome exit tunnel that are protected from methylation by the presence of nascent TnaC
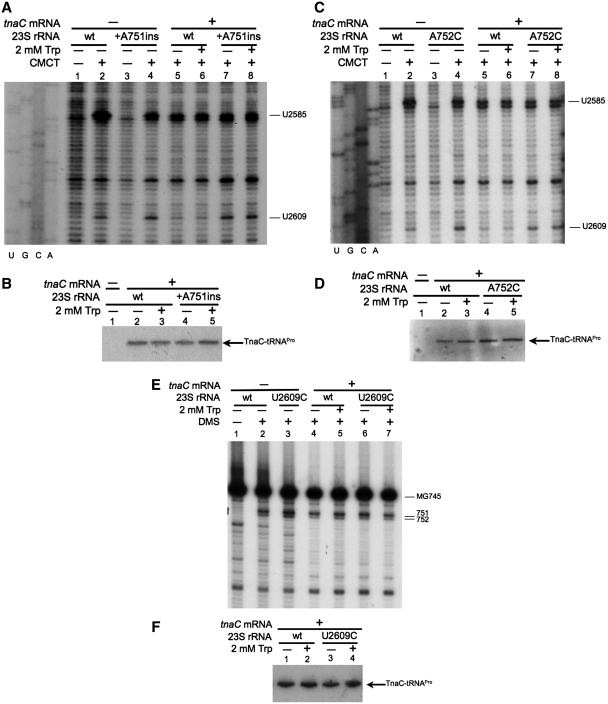
The action of Trp on tna operon expression requires the presence of the TnaC nascent peptide within the ribosome (5). We performed methylation protection assays to determine if the nascent TnaC peptide protects 23S rRNA nucleotides A751, A752 and U2609 from chemical methylation (see ‘Materials and Methods’ section). These nucleotide residues are water accessible in vacant ribosomes (ribosomes not engaged in polypeptide synthesis), and can therefore be methylated by alkylating agents (41). We also examine methylation of U2585, which forms part of the PTC (25). The alkylating agent CMCT-induced methylation of U2585 and U2609 in vacant wild-type and +A751ins mutant ribosomes (Figure 2A, compare lane 2 with lane 1, or lane 4 with lane 3). We observed that the U2585 methylation level was slightly less (30 ± 5%, n = 4) in vacant +A751ins ribosomes than vacant wild-type ribosomes (Figure 2A, compare lane 4 with lane 2). We also observed that the methylation level of U2609 was slightly higher (25 ± 3%, n = 4) in vacant +A751ins ribosomes than in wild-type ribosomes (Figure 2A, compare lane 4 with lane 2). The A752C mutant ribosomes, like the +A751ins ribosomes, differed from the wild-type. Vacant A752C ribosomes showed slightly reduced methylation (32 ± 5%, n = 4) of U2585 and an increased methylation (50 ± 3%, n = 4) of U2609 compared to wild-type ribosomes (Figure 2C, compare lane 4 with lane 2). Trp did not affect methylation levels in vacant ribosomes (Supplementary Figure S2). These observations suggested that there were differences in the architecture of the ribosome exit tunnel between vacant +A751ins or A752C mutant ribosomes and wild-type ribosomes.
Figure 2.
23S rRNA nucleotides that are protected by the TnaC nascent peptide. (A, C and E) Methylation protection assays were performed with ribosomes containing the indicated 23S rRNA alleles. Ribosomes translating (+) or not (−) messengers containing the tnaC gene sequences were analyzed in a buffer containing (+) or not (−) Trp. The ribosomes were exposed (+) or not (−) to the indicated alkylating agents. These assays were performed with [32P]-labeled oligonucleotides complementary to nucleotides 2654–2674 of 23S rRNA for (A and C), and complementary to nucleotides 821–838 of 23S rRNA for (E). Results obtained with ribosomes translating tnaC sequences in absences of the alkylating agents are shown in Supplementary Figure S3. Nucleotides methylated are indicated. cDNA synthesis on 23S rRNA template obtained from wild-type ribosomes is usually stopped by the naturally methylated G745 nucleotide (MG745) (E) (34). (B, D and F) Northern blot assays performed with the ribosomes indicated above. The ribosomal components were resolved in 10% denaturing Tris–tricine polyacrylamide gels. The presence of the TnaC-tRNAPro in the complexes was determined using a [32P]-labeled oligonucleotide complementary to the anti-codon region of the tRNAPro1 (35).
The methylation levels of U2585 and U2609 were reduced when the wild-type ribosomes contained TnaC-tRNAPro. This reduction occurred independent of the presence of Trp (Figure 2A, compare lanes 5 or 6 with lane 2). The methylation of the U2609 nucleotide was substantially reduced (70 ± 3%, n = 4) (Figure 2A, compare lanes 5 or 6 with lane 2) when TnaC-tRNAPro was in the ribosome. These results indicated that the presence of the TnaC-tRNAPro within the wild-type ribosome reduced the accessibility of the U2585 and U2609 nucleotides with a greater effect on U2609 accessibility. In contrast, we did not observe any change in the methylation level of U2585 or U2609 in +A751ins (Figure 2A, compare lanes 7 or 8 with lane 4) or A752C (Figure 2C, compare lanes 7 or 8 with lanes 5 or 6) mutant ribosomes containing TnaC-tRNAPro. These results indicate that TnaC-tRNAPro did not protect the U2585 and U2609 nucleotides in the +A751ins and A752C mutant ribosomes. These differences did not reflect changes in the capacity of mutant ribosomes to synthesize TnaC-tRNAPro as determined by Northern blots (Figure 2B and D).
Finally, we analyzed the U2609C mutant ribosomes. We focused our efforts on determining the DMS accessibility of the A751 and A752 nucleotides because changes in these nucleotides affected the CMCT accessibility of the U2609 nucleotide (see above). The water soluble alkylating agent DMS methylates A751 and A752 in vacant wild-type and U2609C mutant ribosomes (Figure 2E, compare lanes 2 or 3 with lane 1) (41). The methylation levels of A751 and A752 nucleotides were not affected in wild-type or U2609 mutant ribosomes containing a TnaC-tRNAPro molecule, in either the presence or absence of Trp (Figure 2E, compare lanes 4 and 5 with lane 2, or lanes 6 and 7 with lane 3). These results indicate that the presence of TnaC-tRNAPro did not affect the DMS accessibility of these two nucleotides.
Exit tunnel nucleotide interactions that are important for TnaC-tRNAPro stalling activity
X-ray crystal structures of the E. coli 50S ribosomal subunits have shown that the 23S rRNA A752 and U2609 nucleotides form a base-pair interaction (26). To understand if this interaction is important for TnaC-mediated regulation we produced bacterial strains containing directed replacements at either or both positions in the 23S rRNA. These strains also contained the tnaA’-‘lacZ Trp-inducible reporter gene (19). We determined the effects of these 23S rRNA mutations on the expression of this reporter gene by measuring β-gal activity in cells grown with or without Trp (Table 2). Strain with wild-type ribosomes (A752/U2609) exhibited high expression levels of the tnaA’-‘lacZ reporter gene in the presence but not the absence of Trp. Substitutions of A752 with uridine or cytosine substantially reduced induction of the reporter gene (Table 2, combinations A752U/U2609 and A752C/U2609). However, the A752G mutation did not affect the induction of the reporter (Table 2, nucleotide combination A752G/U2609). These results indicated that a purine nucleotide, A or G, was required at nucleotide 752 to enable TnaC function. Substitutions at position U2609 with A or C also abolished reporter gene induction by Trp in this analysis (Table 2, nucleotide combinations A752/U2609C and A752/U2609A). Finally, while most combinations of nucleotide replacements at both positions gave uninducible phenotypes (Table 2, nucleotide combinations A752U/U2609A, A752U/U2609C, A752C/U2609A, A752C/U2609C), the combination A752G/U2609C in 23S rRNA conferred an inducible phenotype similar to the wild-type combination A752/U2609, whereas A752/U2609C did not (Table 2).
Table 2.
A752 and U2609 nucleotide changes that affect tnaA’-‘lacZ expression in the bacterial cell
| 23S rRNAs | β-gal activity (Miller units)a ± SD |
Induction ration (+Trp/−Trp)b | |
|---|---|---|---|
| −Trp | +Trp | ||
| A752/U2609 (wt) | 120 ± 10 | 4500 ± 20 | 37.5 |
| A/U combination | |||
| A752/U2609A | 110 ± 12 | 100 ± 10 | 1.0 |
| A752U/U2609 | 130 ± 15 | 140 ± 20 | 1.1 |
| A752U/U2609A | 90 ± 10 | 200 ± 13 | 2.2 |
| G/C combination | |||
| A752/U2609C | 75 ± 10 | 85 ± 6 | 1.1 |
| A752G/U2609 | 150 ± 20 | 3500 ± 22 | 23.3 |
| A752G/U2609C | 120 ± 15 | 4200 ± 20 | 35.0 |
| A752C/U2609 | 120 ± 10 | 160 ± 10 | 1.3 |
| Other combinations | |||
| A752C/U2609A | 130 ± 9 | 140 ± 8 | 1.1 |
| A752C/U2609C | 110 ± 12 | 140 ± 10 | 1.3 |
| A752U/U2609C | 90 ± 9 | 100 ± 8 | 1.1 |
aCultures of SR-14 derived strains obtained by replacement of the prrnC-sacB plasmid by pK4–16 variants were grown in minimal medium plus 0.2% glycerol, 0.05% acid-hydrolyzed casein, 0.01% vitamin B1 and 50 µg/ml kanamycin with (+Trp) or without (−Trp) 100 µg/ml Trp. β-gal assays were performed in four independent experiments.
bRatio of values for cultures grown with Trp (+Trp) and those grown without Trp (−Trp).
TnaC residues that protect the U2609 nucleotide
The presence of TnaC-tRNAPro within the wild-type ribosome reduced CMCT accessibility of the U2609 nucleotide (Figure 2A). These results suggested that either components of the TnaC peptide may be in close proximity to U2609, or interactions of TnaC-tRNAPro with other regions of the ribosome induced structural changes affecting this nucleotide. The model of TnaC-tRNAPro bound to a ribosome suggested by cryo-EM structures indicates that the TnaC residues W12, D16 and K18 are in the vicinity of the A751, A752 and U2609 (21). We determined expression levels in vivo of the tnaA’-‘lacZ reporter construct containing W12R, D16A or K18A substitutions in TnaC to establish their importance for TnaC-mediated regulation (Table 3). The replacements W12R and D16A substantially reduced induction of the reporter gene in response to Trp (Table 3) (8). In contrast, the TnaC K18A mutation retained regulatory capacity (Table 3). A K18E mutation also did not interfere with regulatory function (data not shown). These results indicated that residues W12 and D16, but not K18, were important for Trp induction.
Table 3.
TnaC residue changes that affect tnaA’-‘lacZ expression in bacterial cell
| TnaC peptide | β-gal activity (Miller units)a ± SD |
Induction ratio (+Trp/−Trp)b | |
|---|---|---|---|
| −Trp | +Trp | ||
| wt | 30 ± 1 | 717 ± 85 | 23.9 |
| W12R | 51 ± 3 | 42 ± 2 | 0.8 |
| D16A | 30 ± 1 | 29 ± 0 | 1.0 |
| K18A | 23 ± 1 | 985 ± 20 | 42.8 |
aCultures of E. coli bacterial strains AW216 (Wt), AW221 (W12R), AW326 (K18A) and AW600 (D16A) were grown in minimal medium plus 0.2% glycerol, 0.05% acid-hydrolyzed casein, 0.01% vitamin B1 and 100 µg/ml ampicillin with (+Trp) or without (−Trp) 100 µg/ml Trp. β-gal assays were performed in three independent experiments.
bRatio of values for cultures grown with Trp (+Trp) and those grown without Trp (−Trp).
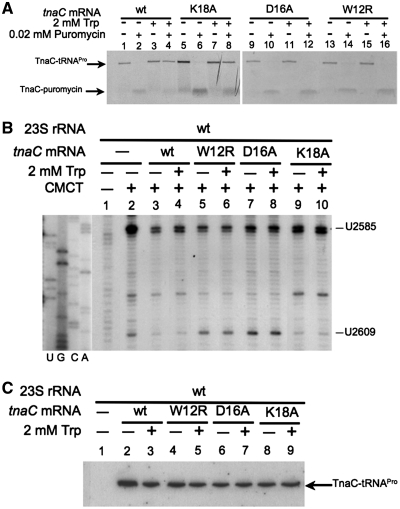
We examined the impact of Trp on the function of ribosome complexes containing wild-type, W12R, D16A and K18A TnaC nascent peptides using puromycin, an aminoacyl-tRNA analog that is an A-site substrate at the PTC. Puromycin was added to wild-type ribosomes containing wild-type or mutated TnaC peptides (Figure 3A). The addition of Trp inhibited puromycin activity on ribosome containing either wild-type or K18A nascent TnaC peptides (Figure 3A, compare lane 4 with lane 2, or lane 8 with lane 6). However, the addition of Trp did not inhibit puromycin activity on ribosomes containing either the W12R or D16A mutant TnaC peptides (Figure 3A, compare lane 12 with lane 10, or lane 16 with lane 14). These results indicated that the replacements W12R and D16A, but not K18A, affected the capacity of TnaC to inhibit peptidyltransferase activity in response to Trp, consistent with their regulatory phenotypes in vivo.
Figure 3.
Nascent TnaC peptide residues involved in the protection of the U2609 nucleotide. (A) Isolated ribosome complexes containing the indicated tnaC mRNAs were tested with (+) or without (−) puromycin in the presence (+) or absence (−) of Trp. The final products of each reaction were resolved on 10% Tris–tricine polyacrylamide gels. The TnaC-tRNAPro and TnaC-puromycin molecules position are indicated with arrows. (B) Methylation protection assays performed with wild-type ribosomes containing the indicated tnaC mRNAs. The experiments were carried out as indicated in Figure 2 using the alkylating agent CMCT. Nucleotides methylated by the presence of CMCT are indicated. (C) Northern blot assays performed with the ribosome complexes indicated above. The TnaC-tRNAPro in the ribosome complexes was detected as indicated in Figure 2.
Finally, we analyzed the effects of W12R, D16A and K18A TnaC peptides on the protection of the U2609 nucleotide from methylation by CMCT (Figure 3B). The presence of wild-type TnaC-tRNAPro substantially reduced the methylation level of the U2609 nucleotide that was observed in vacant wild-type ribosomes (Figure 3B, compare lanes 3 and 4 with lane 2), as also observed in the experiments shown in Figure 2. In contrast, in ribosome complexes containing W12R or D16A TnaC-tRNAPro the methylation level of U2609 was not affected (Figure 3B, compare lanes 5 and 6, or lanes 7 and 8 with lane 2). These results indicated that W12R and D16A mutations reduced the protection of U2609 conferred by the presence of the wild-type TnaC-tRNAPro within the ribosome. However, the methylation of U2609 within ribosomes containing K18A TnaC-tRNAPro was similar to that observed with ribosomes containing wild-type TnaC-tRNAPro (Figure 3B, compare lanes 9 and 10 with lane 2 or lanes 3 and 4). These effects on U2609 methylation were independent of the presence of Trp (Figure 3B, compare − lanes with + lanes). These results indicated that the K18A replacement did not affect TnaC-tRNAPro-mediated protection of U2609 from methylation.
DISCUSSION
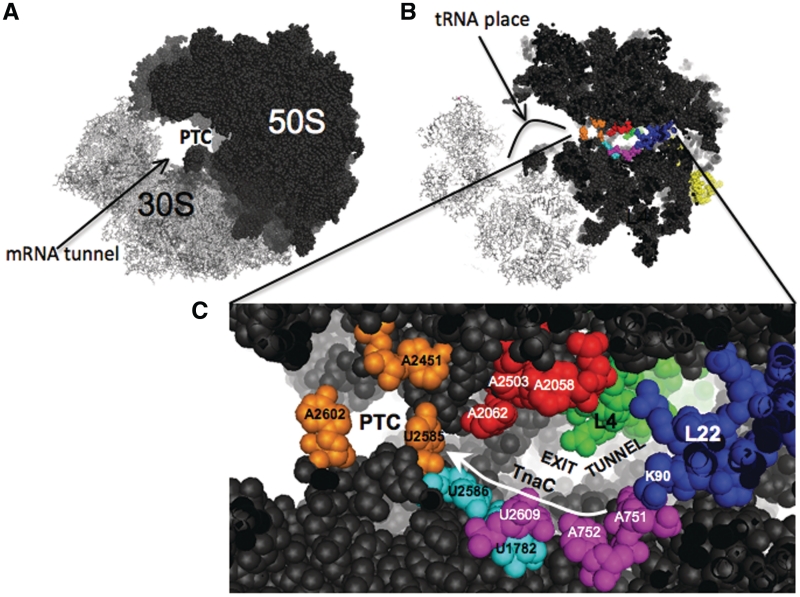
The data presented here show that, in ribosomes that can respond to functional TnaC, functional TnaC-tRNAPro in the ribosome exit tunnel protects U2609 of the 23S rRNA from methylation by CMCT (Figure 2). These results are consistent with the structural model obtained from cryo-EM data, where it has been observed that U2609 conformation is affected by the presence of the TnaC peptide (21). The cryo-EM model suggests that the changes in the conformation of U2609 are not the result of major structural changes in the exit tunnel (21). These data suggest that interaction between U2609 and TnaC residues is a major reason for the difference in methylation sensitivity of U2609 when comparing ribosomes containing either no peptide or non-functional TnaC peptide to ribosomes containing functional TnaC peptide. The cryo-EM model further suggests that the K18 residue of TnaC might be involved in positioning U2609 (21). However, our mutagenesis analyses did not support this view. Replacing the K18 residue by alanine, a small non-charged amino acid, did not affect either Trp induction in vivo (Table 3) or inhibition of TnaC-tRNAPro cleavage by puromycin (Figure 3A). Also, the protection of U2609 was not affected by the K18A TnaC mutation (Figure 3B). The TnaC mutations W12R and D16A, which eliminate Trp-mediated ribosome stalling (Table 3 and Figure 3A), abolished the protection from methylation of U2609 (Figure 3B). Therefore, these essential residues of the TnaC peptide may interact with U2609 directly. Alternatively, W12 and D16 interactions with other elements of the ribosome may relay structural changes through the TnaC peptide to establish a position for U2609 that protects this nucleotide from methylation (21,27). Cryo-EM structures of eukaryotic ribosomes containing either of the regulatory peptides CMV or AAP suggest that amino acids that are important for stalling interact with the A751 and U2609 nucleotides (28). The essential residues for stalling Ser-12 of CMV and the Asp-12 of AAP seem to be in the proximity of A751 (18,28,29). Meanwhile, the important residues Lys-18 of CMV and Trp-19 of AAP are close to U2609 (18,28,29). These positions correspond with the conserved residues W12 and Ile-19 of TnaC (8). Furthermore, comparison of the cryo-EM structures of ribosomes containing each of these regulatory peptides reveal that they interact in similar manner with the A751–752 and U2609 region (43). These observations suggest that the interactions between the region constituted by A751–A752 and U2609 and residues of regulatory peptides are essential for stalling in prokaryotic and eukaryotic systems. This region of the ribosomal exit tunnel might be a common anchor-place for regulatory peptides (Figure 4). The ErmCL peptide seems to be the exception as the action of this regulatory peptide is not affected by mutations in the A751 and U2609 nucleotides (24). The ErmCL peptide is shorter and might not reach these nucleotides, in fact ErmCL is anchored to the A2058 nucleotide by erythromycin (Figure 4) (15).
Figure 4.
Regions of the ribosomal exit tunnel essential for stalling. (A) Lateral vision of the 70S ribosome of E. coli (20). (B) Sagittal plane section of the 70S ribosome (20). (C) Visual amplification of the PTC and the first part of the exit tunnel. Nucleotides in orange constitute the PTC region; these nucleotides are involved in the peptidyl transferase and hydrolysis of peptidyl-tRNAs during translation (38). Nucleotides in red are essential for stalling induced by the nascent peptides ErmCL and SecM (13,24). Nucleotides in pink and the amino acid residue K90 of the ribosomal protein L22 are essential for stalling induced by SecM and TnaC (13,19). Nucleotides in cyan connect the nucleotides U2585 and U2609. White arrow indicates possible structural relay from the exit tunnel to the PTC produced by the TnaC nascent peptide.
X-ray structures of the E. coli 50S ribosomal subunit have shown that the A751 and A752 help form the exit tunnel. The exit tunnel structure is presumably stabilized by base-pairing, base-stacking interactions, and by the presence of the extended loop of ribosomal protein L22 (20,44). Furthermore, A752 has base-pairing interactions with U2609 (26). A752-U2609 base-pairing interaction is also important for antibiotic binding (26). Changes in either A752 or U2609 resulted in reduced regulation by TnaC in response to Trp (Table 2). These changes could eliminate base-pairing interactions between nucleotides at these two positions. In this regard, the A752G single substitution and the A752G/U2609C double substitution allowed TnaC-mediated regulation (Table 2), suggesting that the G replacement at the 752 position might generate G:U base-pairing interaction with the U2609 or G:C interaction with C2609 nucleotide retaining the contacts between these two nucleotide positions. Therefore, it seems that the base pairing between the 752 and the 2609 nucleotide positions are required for TnaC function (Figure 4).
In vacant ribosomes lacking a nascent peptide in the exit tunnel, the +A751ins and the A752C mutations affected the conformation of the U2609 nucleotide as well as that of the U2585 nucleotide, a residue located in the PTC, as assessed by their sensitivity to chemical methylation (Figure 2A and B). The +A751ins also affects the function of other regulatory peptides such as SecM (13). These results indicate that these mutations generate perturbations in the structure of the exit tunnel. These perturbations might be transferred from U2609 to U2585 through the nucleotides U1782 and U2586 (Figure 4, cyan nucleotides). Similar results have been observed in erythromycin-resistant ribosomes containing mutations in the extended loop of the ribosomal protein L22 (32). Also, mutations in the loop of L22 affect the function of the regulatory nascent ErmCL peptide (15) as well as the nascent TnaC peptide (19). Therefore, perturbations in the shape of the exit tunnel induced by changes in the G745–A752 nucleotide region, as well as the loop of L22, may affect the way that nascent peptides interact with the ribosome. This is consistent with the idea that the shape of the exit tunnel is a determining factor for the function of regulatory nascent peptides (30,31).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table S1 and Supplementary Figures S1–S3.
FUNDING
The National Institutes of Health USA Foundation (R01 GM47498 to M.S.S.), Robert A. Welch Foundation (A-1310 to M.J.B.); Research Experiences for undergraduates program sponsored by University of Alabama in Huntsville and Alabama Space Grant Consortium (to S.M.); University of Alabama in Huntsville startup funds (to L.R.C.V.). Funding for open access charge: University of Alabama in Huntsville startup funds (to L.R.C.V.).
Conflict of interest statement. None declared.
Supplementary Material
AKNOWLEDGEMENTS
We thank Catherine Squires and Selwyn Quan for the pK4–16 plasmid and Jacqueline Moreno for the generation of the bacterial cells containing the pK4–16 mutant plasmids used in this study. We thank Charles Yanofsky for his valuable help during the elaboration of this article. L.R.C.V. dedicates this article to the memory of Dr Gopy Podila, who supported with enthusiasm his career as a professor.
REFERENCES
- 1.Stewart V, Yanofsky C. Evidence for transcription antitermination control of tryptophanase operon expression in Escherichia coli K-12. J. Bacteriol. 1985;164:731–740. doi: 10.1128/jb.164.2.731-740.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deeley MC, Yanofsky C. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J. Bacteriol. 1981;147:787–796. doi: 10.1128/jb.147.3.787-796.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Stationary-phase quorum-sensing signals affect autoinducer-2 and gene expression in Escherichia coli. Appl. Environ. Microbiol. 2004;70:2038–2043. doi: 10.1128/AEM.70.4.2038-2043.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deeley MC, Yanofsky C. Transcription initiation at the tryptophanase promoter of Escherichia coli K-12. J. Bacteriol. 1982;151:942–951. doi: 10.1128/jb.151.2.942-951.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong F, Yanofsky C. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 2001;276:1974–1983. doi: 10.1074/jbc.M008892200. [DOI] [PubMed] [Google Scholar]
- 6.Gong F, Ito K, Nakamura Y, Yanofsky C. The mechanism of tryptophan induction of tryptophanase operon expression: tryptophan inhibits release factor-mediated cleavage of TnaC-peptidyl-tRNA(Pro) Proc. Natl Acad. Sci. USA. 2001;98:8997–9001. doi: 10.1073/pnas.171299298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Konan KV, Yanofsky C. Rho-dependent transcription termination in the tna operon of Escherichia coli: roles of the boxA sequence and the rut site. J. Bacteriol. 2000;182:3981–3988. doi: 10.1128/jb.182.14.3981-3988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cruz-Vera LR, Yanofsky C. Conserved residues Asp16 and Pro24 of TnaC-tRNAPro participate in tryptophan induction of tna operon expression. J. Bacteriol. 2008;190:4791–4797. doi: 10.1128/JB.00290-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trabuco LG, Harrison CB, Schreiner E, Schulten K. Recognition of the regulatory nascent chain TnaC by the ribosome. Structure. 2010;18:627–637. doi: 10.1016/j.str.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gollnick P, Yanofsky C. tRNA(Trp) translation of leader peptide codon 12 and other factors that regulate expression of the tryptophanase operon. J. Bacteriol. 1990;172:3100–3107. doi: 10.1128/jb.172.6.3100-3107.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 12.Cruz-Vera LR, Sachs MS, Squires CL, Yanofsky C. Nascent polypeptide sequences that influence ribosome function. Curr. Opin. Microbiol. 2011;14:160–166. doi: 10.1016/j.mib.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 13.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 14.Chiba S, Lamsa A, Pogliano K. A ribosome-nascent chain sensor of membrane protein biogenesis in Bacillus subtilis. EMBO J. 2009;28:3461–3475. doi: 10.1038/emboj.2009.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 16.Fang P, Spevak CC, Wu C, Sachs MS. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl Acad. Sci. USA. 2004;101:4059–4064. doi: 10.1073/pnas.0400554101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hood HM, Spevak CC, Sachs MS. Evolutionary changes in the fungal carbamoyl-phosphate synthetase small subunit gene and its associated upstream open reading frame. Fungal Genet. Biol. 2007;44:93–104. doi: 10.1016/j.fgb.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Spevak CC, Ivanov IP, Sachs MS. Sequence requirements for ribosome stalling by the arginine attenuator peptide. J. Biol. Chem. 2010;285:40933–40942. doi: 10.1074/jbc.M110.164152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 20.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 21.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, et al. Structural insight into nascent polypeptide chain-mediated translational stalling. Science. 2009;326:1412–1415. doi: 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang R, Cruz-Vera LR, Yanofsky C. 23S rRNA nucleotides in the peptidyl transferase center are essential for tryptophanase operon induction. J. Bacteriol. 2009;191:3445–3450. doi: 10.1128/JB.00096-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sachs MS, Geballe AP. Biochemistry. Sense and sensitivity–controlling the ribosome. Science. 2002;297:1820–1821. doi: 10.1126/science.1076865. [DOI] [PubMed] [Google Scholar]
- 24.Vazquez-Laslop N, Ramu H, Klepacki D, Kannan K, Mankin AS. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A° resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 26.Dunkle JA, Xiong L, Mankin AS, Cate JH. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc. Natl Acad. Sci. USA. 2010;107:17152–17157. doi: 10.1073/pnas.1007988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yap MN, Bernstein HD. The plasticity of a translation arrest motif yields insights into nascent polypeptide recognition inside the ribosome tunnel. Mol. Cell. 2009;34:201–211. doi: 10.1016/j.molcel.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhushan S, Meyer H, Starosta AL, Becker T, Mielke T, Berninghausen O, Sattler M, Wilson DN, Beckmann R. Structural basis for translational stalling by human cytomegalovirus and fungal arginine attenuator peptide. Mol. Cell. 2010;40:138–146. doi: 10.1016/j.molcel.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 29.Degnin CR, Schleiss MR, Cao J, Geballe AP. Translational inhibition mediated by a short upstream open reading frame in the human cytomegalovirus gpUL4 (gp48) transcript. J. Virol. 1993;67:5514–5521. doi: 10.1128/jvi.67.9.5514-5521.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiba S, Kanamori T, Ueda T, Akiyama Y, Pogliano K, Ito K. Recruitment of a species-specific translational arrest module to monitor different cellular processes. Proc. Natl Acad. Sci. USA. 2011;108:6073–6078. doi: 10.1073/pnas.1018343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vazquez-Laslop N, Mankin AS. Picky nascent peptides do not talk to foreign ribosomes. Proc. Natl Acad. Sci. USA. 2011;108:5931–5932. doi: 10.1073/pnas.1103011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gregory ST, Dahlberg AE. Erythromycin resistance mutations in ribosomal proteins L22 and L4 perturb the higher order structure of 23 S ribosomal RNA. J. Mol. Biol. 1999;289:827–834. doi: 10.1006/jmbi.1999.2839. [DOI] [PubMed] [Google Scholar]
- 33.Garza-Ramos G, Xiong L, Zhong P, Mankin A. Binding site of macrolide antibiotics on the ribosome: new resistance mutation identifies a specific interaction of ketolides with rRNA. J. Bacteriol. 2001;183:6898–6907. doi: 10.1128/JB.183.23.6898-6907.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gustafsson C, Persson BC. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cruz-Vera LR, Gong M, Yanofsky C. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc. Natl Acad. Sci. USA. 2006;103:3598–3603. doi: 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cruz-Vera LR, New A, Squires C, Yanofsky C. Ribosomal features essential for tna operon induction: tryptophan binding at the peptidyl transferase center. J. Bacteriol. 2007;189:3140–3146. doi: 10.1128/JB.01869-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 38.Youngman EM, Brunelle JL, Kochaniak AB, Green R. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell. 2004;117:589–599. doi: 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 39.McKenzie GJ, Craig NL. Fast, easy and efficient: site-specific insertion of transgenes into enterobacterial chromosomes using Tn7 without need for selection of the insertion event. BMC Microbiol. 2006;6:39. doi: 10.1186/1471-2180-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JH. Experiments in Molecular Genetics. NY: Cold Spring Harbor; 1972. [Google Scholar]
- 41.Moazed D, Noller HF. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell. 1989;57:585–597. doi: 10.1016/0092-8674(89)90128-1. [DOI] [PubMed] [Google Scholar]
- 42.Yanofsky C, Horn V, Gollnick P. Physiological studies of tryptophan transport and tryptophanase operon induction in Escherichia coli. J. Bacteriol. 1991;173:6009–6017. doi: 10.1128/jb.173.19.6009-6017.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wilson DN, Beckmann R. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr. Opin. Struct. Biol. 2011;21:274–282. doi: 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Lebars I, Yoshizawa S, Stenholm AR, Guittet E, Douthwaite S, Fourmy D. Structure of 23S rRNA hairpin 35 and its interaction with the tylosin-resistance methyltransferase RlmAII. EMBO J. 2003;22:183–192. doi: 10.1093/emboj/cdg022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaporojets D, French S, Squires CL. Products transcribed from rearranged rrn genes of Escherichia coli can assemble to form functional ribosomes. J. Bacteriol. 2003;185:6921–6927. doi: 10.1128/JB.185.23.6921-6927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.