Abstract
We describe an inexpensive and efficient method for generating functional pools of Dicer-substrate small interfering RNAs (siRNAs) in a single reaction tube. The method exploits a highly active form of the enzyme Dicer from Giardia lamblia, which is capable of accurately processing double-stranded RNA (dsRNA) into 25–27 nt RNA pools during in vitro transcription. The small RNAs produced function as substrates of human Dicer in vitro and induce gene silencing with potency equivalent to traditional siRNAs when introduced into mammalian cells. The overall reaction is simple, can be carried out in any laboratory with access to a PCR machine, and is amenable to high-throughput processes.
INTRODUCTION
The use of small interfering RNAs (siRNAs) to knockdown specific gene sequences is a powerful tool for reverse-genetic experiments in a variety of mammalian cell types (1). However, siRNAs are costly to synthesize, and potent siRNA sequences, with minimal off-target effects, are often best identified experimentally. To overcome these limitations, several groups have developed enzymatic methods for generating pools of siRNAs against a chosen target gene (2,3). siRNA pools are believed to be less susceptible to off-target effects, as no single siRNA sequence is present in high abundance (4). Typically, protocols for generating siRNA pools involve transcription of the target gene into forward and reverse strand RNAs, followed by purification and annealing to form dsRNA. Alternatively, purified single stranded RNA can be treated with an RNA dependent RNA polymerase to generate long dsRNA (5). Long dsRNAs are then cleaved into small RNA fragments using either recombinant human Dicer or bacterial RNase III enzymes. These methods have been used by many groups for generating functional pools of small RNAs (6–10). However, the established protocols employ multiple discrete steps and are relatively time-consuming.
We have developed a simplified method for generating siRNA pools that can be carried out in a single reaction tube. The method takes advantage of a Dicer homolog from the protozoan Giardia lamblia, which cleaves long dsRNA substrates into small RNAs 25-27 nt in length (11). Because Giardia Dicer is highly active in a wide range of reaction conditions, it can be included in the dsRNA transcription reaction. DNA templates for the transcription can be taken directly from PCRs without further clean-up. Therefore, the entire reaction, starting with the cDNA clone of a target gene and gene-specific PCR primers, and ending with a pool of siRNAs against the target, can be carried out with very little manipulation of the sample. The small RNAs produced are substrates for human Dicer, which cleaves them into siRNAs 20–23 nt in length, and promote sequence specific silencing of target genes when introduced into mammalian cells.
METHODS AND MATERIALS
Protein expression and purification
Giardia Dicer (XP_001705536) was produced and purified from a baculovirus expression system. The plasmid pFB-GiDcr, which contains the Giardia Dicer coding region cloned into pFastBac HTA (Invitrogen), was used to generate baculovirus using the Bac-to-Bac method (Invitrogen). For protein expression, 250 ml of Sf9 cells (1 × 106 cells per ml) were grown at 27°C in ESF 921 media (Expression Systems, Woodland, CA, USA) and infected with 0.5 ml of amplified baculovirus bearing the Giardia Dicer gene. Forty-eight to seventy-two hours after infection, cells were pelleted by centrifugation and resuspended in 5 ml of a lysis buffer composed of 300 mM NaCl, 10 mM imidazole, 0.5 mM Tris(2-carboxyethyl)phosphine (TCEP), 0.5% Triton X-100 and 50 mM sodium phosphate, pH 8. Resuspended cells were lysed with seven strokes of a Dounce tissue grinder and then centrifuged to pellet insoluble material. The soluble cell lysate was then applied to 3 ml of HIS-Select nickel chelate resin (Sigma) in batch. After a 30 min incubation period, the HIS-Select resin was pelleted by centrifugation and the unbound supernatant fluid was discarded. The resin was washed by resuspending in 50 ml of a wash buffer containing 300 mM NaCl, 20 mM imidazole, 0.5 mM TCEP and 50 mM sodium phosphate, pH 8 and then pelleted again by centrifugation. The wash step was repeated a total of five times. The protein was then eluted from the resin in 9 ml of an elution buffer composed of: 300 mM NaCl, 300 mM imidazole, 0.5 mM TCEP, 50 mM sodium phosphate, pH 8. Eluted protein was dialyzed overnight against 0.1 M NaCl, 10% glycerol, 1 mM TCEP and 40 mM Tris, pH 8. The dialyzed protein was concentrated to 0.5 mg/ml. During the final concentration steps the protein was exchanged into a protein storage buffer composed of 50% glycerol, 0.1 M NaCl, 1 mM TCEP and 40 mM Tris, pH 8. All purification steps were carried out on ice or at 4°C. The final purified protein was stored at −80°C at 0.5 mg/ml in the storage buffer.
Recombinant human Dicer (NM_030621) was expressed and purified as described previously (11,12). Purified human Dicer can be stored in the same storage buffer as Giardia Dicer (above). T7 RNA polymerase was expressed in Escherichia coli strain BL21(DE3) and purified as described (13). E. coli RNase III was purchased from New England Biolabs (M0245S).
Transcription/dicing reaction
Templates for in vitro transcription were generated by amplifying a ~500 base-pair segment of the desired target gene in a PCR (30 cycles) using Taq DNA polymerase (New England Biolabs, M0273S) or Pfu Turbo DNA polymerase (Agilent, 600254-52). PCR primers contained the promoter sequence for T7 RNA polymerase (TAATACGACTCACTATAG) on their 5′-ends, allowing amplified DNA to be transcribed in both directions in the subsequent transcription/dicing reaction.
A 5 µl aliquot of the PCR was used directly, without purification, in a 100 µl transcription/dicing reaction containing: 10 mM DTT, 10 mM MgCl2, 2 mM ATP, 2 mM CTP, 2 mM GTP, 2 mM UTP, 1 µg/µl T7 RNA polymerase, 20 ng/µl Giardia Dicer, and 40 mM Tris, pH 8. Transcription/dicing reactions were incubated at 37°C for 2–3 h. The 20 ng/µl of purified Giardia Dicer could be replaced with 8 µl of the commercially available ‘PowerCut’ Giardia Dicer (Fisher Scientific, F602S). However, the PowerCut enzyme had significantly less activity than our purified Giardia Dicer and thus reactions using PowerCut Dicer required a much longer incubation time (16 h at 37°C). The DNA template and any remaining single-stranded RNA were then degraded by adding 1 µl DNase I (New England Biolabs, M03035S) and 1 µl RNase T1 (Roche, 10-109-193-001) and incubating an additional 15–30 min at 37°C. Enzymes were removed by extracting with 100 µl phenol/chloroform/isoamyl alcohol (25:24:1, pH 6.7). The aqueous layer was transferred to a new tube where RNA was precipitated by adding 10 µl of 3 M sodium acetate (pH 5.2) and 250 µl of 100% ethanol. After centrifugation, the pelleted RNA was washed with 1 ml of 70% ethanol and then dissolved in 50 µl of water. At this stage of the preparation the small RNAs could be transfected directly into mammalian cells. However, residual unincorporated nucleotides were often still present and interfered with accurate RNA concentration measurement. Unincorporated nucleotides were removed by passing the sample through an illustra MicroSpin G-25 column (GE Healthcare Biosciences). RNA concentration was determined by UV spectroscopy.
Small RNA pools generated by human Dicer or bacterial RNase III were prepared as described previously (2,3).
In vitro human Dicer cleavage assays
A radiolabeled small RNA pool against firefly luciferase was generated by including α-32P-UTP in a transcription/dicing reaction. Small RNAs were purified on a native polyacrylamide gel (14%) in 0.5x TBE buffer. Purified small RNAs were incubated with 3.3 µg of purified human Dicer in a buffer solution containing 150 mM NaCl, 20 mM Tris pH 8.0, 2 mM MgCl2, 0.5 mM TCEP and incubated at 37°C for ≥90 min. Diced products were precipitated, resolved on by denaturing PAGE (14%) and imaged by phosphorimaging.
Cell culture, transfection and luciferase assays
HeLa, HEK 293 FT and NIH 3T3 cells were grown in DMEM (Gibco) supplemented with 10% fetal bovine serum (Lonza) and antibiotic-antimycotic (Gibco) at 37°C under a 5% CO2 atmosphere. The day before transfection, cells were plated onto 24-well dishes to ~60% confluency. Plasmids and siRNA were transfected into cells using Lipofectamine 2000 (Invitrogen), according to the manufacture's instructions. Small RNA pools targeting firefly luciferase (derived from nucleotides 347-868 of pIS0, Addgene plasmid 12178,) were transfected with 83 ng of pIS0 (14), and 17 ng of pIS2 (Addgene plasmid 12177), which encodes Renilla luciferase (15) in each well of a 24-well dish. Luciferase activity from cell lysates was measured 24 h post-transfection using the Dual Luciferase Assay system (Promega) on a 20/20 n luminometer (Turner Biosystems).
PME-1 knock-down
HEK 293 cells were plated in 6-well dishes and transfected with 100 pmol (33 nM, final concentration) of a small RNA pool targeting nucleotides 420–967 of the human PME-1 gene (NM_016147) using Lipofectamine 2000 (Invitrogen). 100 pmol of a synthetic siRNA targeting nucleotides 153–171 was transfected as a positive control for PME-1 knock-down. Untransfected cells and cells transfected with 100 pmol of a small RNA pool targeting Renilla luciferase served as negative controls. 48 h after transfection, cells were washed and resuspended in 500 µl of DPBS (Gibco). Cells were disrupted by sonication and denatured with 4× SDS/PAGE loading buffer (reducing). Proteins in the lysate were separated by SDS/PAGE, and analyzed by western blotting using standard methods. PME-1 antibodies were purchased from Millipore (07-095). Blots were probed following the manufacturers’ instructions, and were visualized and quantified using the Odyssey Imaging System (Li-Cor).
RESULTS AND DISCUSSION
Giardia Dicer can generate small RNAs during in vitro transcription
Previous methods for generating siRNA pools involve multiple discrete steps including transcription, RNA purification, annealing and long dsRNA cleavage steps. However, in living cells all of these processes occur simultaneously and thus, in principle, the entire reaction should be able to be carried out in vitro in a single reaction tube. To explore this idea we tested different RNases for the ability to efficiently cleave a 500 bp dsRNA into siRNAs during in vitro transcription by T7 RNA polymerase.
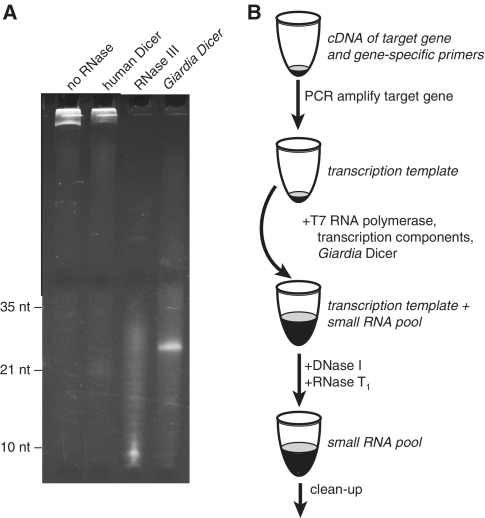
Recombinant human Dicer has been used to successfully generate siRNA pools from purified long dsRNA (2). However, we found human Dicer is unable to match the pace of transcription in vitro, even when high enzyme concentrations are used, and converts only a small fraction of the dsRNA into siRNAs (Figure 1A). This is most likely due to the enzyme's poor catalytic efficiency and susceptibility to product inhibition (16,17). Bacterial RNase III has also been used to generate siRNA pools (3) and, in contrast to human Dicer, this enzyme does efficiently convert dsRNA into small RNAs during in vitro transcription. However, RNase III lacks the precision of Dicer and generates a pool of small RNAs ranging from 30 nt down to less than 10 nucleotides in length, with the most abundant species being about 9 nt. In titration experiments we were unable the find a concentration of RNase III that could reliably convert a long dsRNA into a ~21 nt pool during in vitro transcription (Supplementary Figure S1).
Figure 1.
Single pot reaction to generate small RNA pools. (A) Three ribonucleases were tested for the ability to generate siRNA pools during in vitro transcription. Reactions contained either 275 ng/ul human Dicer, 0.01 unit/µl ‘ShortCut’ RNase III, or 10 ng/µl Giardia Dicer. RNA products were resolved by denaturing PAGE and stained with ethidium bromide. (B) Schematic of the optimized method for producing small RNAs using Giardia Dicer.
We next examined the ability of Dicer from the protozoan Giardia lamblia to process long dsRNA into siRNAs during in vitro transcription. The Giardia enzyme is a small and simplified form of Dicer that is missing many of the accessory domains found in Dicer enzymes from higher eukaryotes. We choose this enzyme because it is easily expressed and purified in recombinant form (Supplementary Figure S2), is much less susceptible to product inhibition than human Dicer (11), and accurately cleaves dsRNA into small RNAs of defined length (18). Indeed, unlike the human enzyme, Giardia Dicer was able to keep pace with transcription in vitro and converted essentially all of the transcribed dsRNA into small RNAs (Figure 1A). Furthermore, in contrast to bacterial RNase III, the products of Giardia Dicer fell within a narrow size range, with the vast majority of all the small RNAs produced about 26 nt in length. We also found that, like T7 RNA polymerase (19), Giardia Dicer is not inhibited by components from PCR and thus DNA templates for transcription/dicing reactions may be taken directly from PCRs without an impact on small RNA yield. The result is a simple procedure that starts with PCR of a segment of a target gene and leads to remarkably high yields of small RNAs 25–27 nt in length in just a few hours with very little manual intervention (Figure 1B). At the end of the reaction the DNA template can be degraded by adding DNase I. For some templates we found that unprocessed single-stranded RNA accumulated during transcription. These RNAs can be degraded by the addition of RNase T1 at the end of the reaction. Treatment with RNase T1 does not significantly impact gene-silencing potency of the small RNAs (data not shown). In our hands, a 100 µl reaction containing 3–5 µl of a standard PCR and 2 µg of recombinant Giardia Dicer, was sufficient to generate 20–100 µg of small products in just a few hours.
Giardia Dicer products are substrates for human Dicer
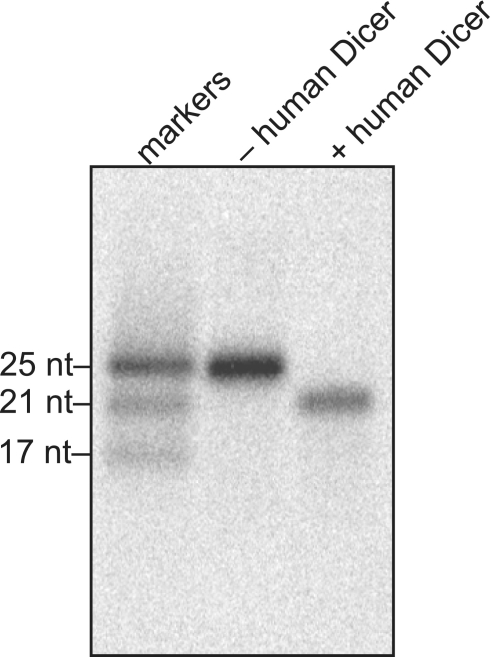
The size of small RNAs generated by Giardia Dicer is 25–27 nt (11), which is 4–6 nt longer than traditional 21-nt siRNAs. In this respect, Giardia products resemble commercially available synthetic 27 nucleotide RNAs named ‘Dicer-substrate siRNAs’ (DsiRNAs) (20). We therefore suspected that the small RNAs generated by Giardia Dicer would serve as effective substrates for cleavage by human Dicer. To test this idea directly, we introduced an internal 32P-label into the RNA during the co-transcriptional dicing reaction and used the purified, radiolabeled small RNAs as substrates in a cleavage reaction with recombinant human Dicer (Figure 2). As predicted, human Dicer efficiently cleaved the small RNAs generated by Giardia Dicer into proper siRNAs, about 21 nt in length. Therefore, upon introduction into mammalian cells Giardia Dicer products could enter the RNAi pathway through the action of endogenous human Dicer. Also, like commercially available DsiRNAs, products of Giardia Dicer should be short enough to avoid the interferon response (20).
Figure 2.
Giardia Dicer products are substrates for human Dicer. Internally radiolabeled products of Giardia Dicer were incubated with or without recombinant human Dicer, and then analyzed by denaturing PAGE.
Giardia Dicer products silence luciferase reporters in mammalian cells
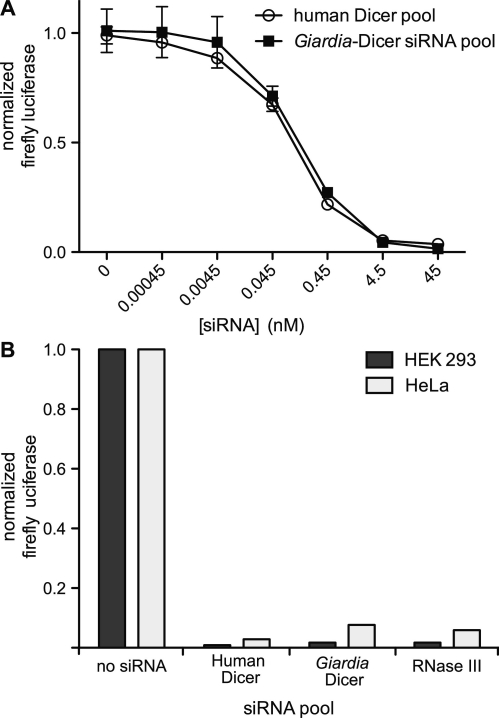
We next tested the ability of Giardia Dicer siRNA pools to specifically silence a reporter gene when transfected into mammalian cells. Giardia Dicer was used to generate small RNAs against a 500 bp segment of the firefly luciferase gene. Various concentrations of the small RNAs were transfected with fixed amounts of firefly and Renilla luciferase reporter plasmids into HEK 293 cells. Renilla luciferase was included to normalize for transfection efficiency and also served as an indicator of non-specific gene silencing. Transfecting 4.5 nM Giardia Dicer small RNAs silenced more than 95% of the co-transfected firefly reporter compared to the no siRNA control (Figure 3A). Increasing the concentration of small RNAs to 45 nM silenced >98% of the firefly luciferase, with no appreciable effect on expression of Renilla. A pool of siRNAs generated by recombinant human Dicer, using the method of Myers et al. (2), produced a nearly identical dose response (Figure 3A), revealing that small RNA pools generated by Giardia Dicer have very similar silencing potencies to siRNA pools made by human Dicer. This observation is in contrast to the previous report that 27-nt DsiRNAs are more efficient at inducing RNAi than 21 nt siRNAs (20). Likewise, in a separate experiment, small RNAs generated by human Dicer, Giardia Dicer or RNase III gave rise to similar levels of luciferase silencing when transfected into either HeLa or HEK 293 cells (Figure 3B). Efficient silencing of firefly luciferase in NIH 3T3 cells (data not shown) also demonstrated that Giardia Dicer products can induce RNAi of a reporter gene in multiple mammalian cells types. The combined results suggest that the small RNAs produced by Giardia Dicer are functionally equivalent to small silencing RNAs generated using previously described methods.
Figure 3.
Silencing of a luciferase reporter by Giardia Dicer products. (A) HEK293 FT cells were transfected with firefly and Renilla luciferase reporter plasmids and increasing concentrations of siRNA pools targeting firefly luciferase. Small RNA pools generated by either Giardia or human Dicer enzymes were tested. Relative firefly luciferase activity levels (normalized to Renilla levels) are plotted is as a function of siRNA concentration. Data points are averages of three independent measurements ± standard deviation. (B) HEK 293 or HeLa cells were transfected with firefly and Renilla luciferase reporters and siRNA pools (200 ng/ml; 12–15 nM final concentration) targeting firefly luciferase that had been generated by human Dicer, Giardia Dicer, or bacterial RNase III. Relative firefly activity levels are shown.
Giardia Dicer products silence endogenous targets in mammalian cells
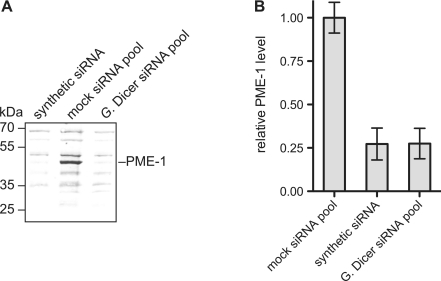
Because Giardia Dicer siRNAs silence a reporter gene as efficiently as other siRNA pools, we expected that they should also be able to silence an endogenous mammalian gene. To test this idea we used Giardia Dicer to generate a pool of small RNAs targeting protein phosphatase methylesterase-1 (PME-1). Small RNAs were transfected into HEK 293 cells (33 nM final concentration). After 48 h the cells were lysed and PME-1 levels were measured by quantitative western blotting (Figure 4). Small RNA pools against Renilla luciferase were transfected separately as a negative control. The PME-1 small RNAs reduced the amount of PME-1 in the crude cell lysate to 27% the level of the negative control. Transfecting a validated synthetic siRNA targeting PME-1 (21) at the same concentration reduced the level of PME-1 by a similar amount (Figure 4). These data demonstrate that, like conventional siRNAs, small RNAs generated using Giardia Dicer in transcription/dicing reactions can be used to knock-down endogenous genes in mammalian cells.
Figure 4.
Silencing of an endogenous gene by Giardia Dicer products. (A) Western blot for PME-1 levels in HEK 293 cells transfected with a small RNA pool targeting PME-1. A validated synthetic siRNA targeting PME-1 was used as a positive control for silencing and transfection efficiency. (B) Results from three independent PME-1 silencing experiments. Arithmetic mean and standard deviation are shown.
CONCLUSION
We have tested the transcription/dicing reaction on a variety of template sequences and found it to be a reproducible and robust method for generating small RNA pools (Supplementary Figure S3). However, it should be noted that in some cases we observed the accumulation of long, Dicer-resistant single stranded RNA species, thought to arise from preferential transcription from one DNA strand, which can promote toxicity or gene-silencing artifacts when transfected into mammalian cells. In these cases, it is important to include the RNase T1 digestion at the end of the transcription/dicing reaction to degrade any unprocessed single stranded RNA. We also found that older preparations of Giardia Dicer had reduced enzymatic activity, which can result in incomplete processing of long dsRNA when used in the transcription/dicing reaction. We found that Giardia Dicer stored in 50% glycerol at −80°C remains active for 4–6 months (storing at −20°C reduced its lifetime). Similarly, the commercially available ‘PowerCut’ Giardia Dicer had lower dicing activity than enzyme freshly prepared in house (data not shown). In cases of low dicing activity, longer reaction times (overnight at 37°C) often led to more complete dsRNA cleavage. Furthermore, it is always beneficial to confirm complete processing of long RNAs in transcription/dicing reactions by denaturing PAGE in order to avoid artifacts in downstream RNAi assays.
In summary, we have developed a simple and rapid method for generating small RNA pools against any target gene. The strengths of the procedure include low cost of materials, technical simplicity and speed. We imagine the method will be particularly beneficial in situations in which several RNAi experiments would be informative but allocated resources are limiting. Furthermore, because the entire reaction can be carried out in a single reaction tube this method could be adapted to a high-throughput format and used for inexpensive large-scale RNAi screens.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
National Institutes of Health (grant number R01GM086701; the Pew Scholars Program in Biomedical Sciences and a Blasker award from the San Diego Foundation (to I.J.M); Graduate Research Fellowship from the National Science Foundation (to A.J.P.); University of California work-study program (to K.Z.G.). Funding for open access charge: NIH R01GM086701.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 2.Myers JW, Jones JT, Meyer T, Ferrell JE., Jr Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- 3.Yang D, Buchholz F, Huang Z, Goga A, Chen CY, Brodsky FM, Bishop JM. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl Acad. Sci. USA. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Myers JW, Chi JT, Gong D, Schaner ME, Brown PO, Ferrell JE. Minimizing off-target effects by using diced siRNAs for RNA interference. J. RNAi Gene Silencing. 2006;2:181–194. [PMC free article] [PubMed] [Google Scholar]
- 5.Aalto AP, Sarin LP, van Dijk AA, Saarma M, Poranen MM, Arumae U, Bamford DH. Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage phi6 RNA-dependent RNA polymerase. RNA. 2007;13:422–429. doi: 10.1261/rna.348307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandman O, Liou J, Park WS, Meyer T. STIM2 is a feedback regulator that stabilizes basal cytosolic and endoplasmic reticulum Ca2+ levels. Cell. 2007;131:1327–1339. doi: 10.1016/j.cell.2007.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwata A, Christianson JC, Bucci M, Ellerby LM, Nukina N, Forno LS, Kopito RR. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proc. Natl Acad. Sci. USA. 2005;102:13135–13140. doi: 10.1073/pnas.0505801102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rojas M, Arias CF, Lopez S. Protein kinase R is responsible for the phosphorylation of eIF2alpha in rotavirus infection. J. Virol. 2010;84:10457–10466. doi: 10.1128/JVI.00625-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazzio TG, Panning B. Condensin complexes regulate mitotic progression and interphase chromatin structure in embryonic stem cells. J. Cell Biol. 2010;188:491–503. doi: 10.1083/jcb.200908026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kakinuma N, Roy BC, Zhu Y, Wang Y, Kiyama R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J. Cell Biol. 2008;181:537–549. doi: 10.1083/jcb.200707022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macrae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- 12.De N, Macrae IJ. Purification and assembly of human Argonaute, Dicer, and TRBP complexes. Methods Mol. Biol. 2011;725:107–119. doi: 10.1007/978-1-61779-046-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zawadzki V, Gross HJ. Rapid and simple purification of T7 RNA polymerase. Nucleic Acids Res. 1991;19:1948. doi: 10.1093/nar/19.8.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 15.Farh KK, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP. The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W. Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J. 2002;21:5875–5885. doi: 10.1093/emboj/cdf582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 19.Batey RT, Kieft JS. Improved native affinity purification of RNA. RNA. 2007;13:1384–1389. doi: 10.1261/rna.528007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- 21.Bachovchin DA, Mohr JT, Speers AE, Wang C, Berlin JM, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Schurer SC, et al. Academic cross-fertilization by public screening yields a remarkable class of protein phosphatase methylesterase-1 inhibitors. Proc. Natl Acad. Sci. USA. 2011;108:6811–6816. doi: 10.1073/pnas.1015248108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.