Abstract
Translation initiation factor eIF3 acts as the key orchestrator of the canonical initiation pathway in eukaryotes, yet its structure is greatly unexplored. We report the 2.2 Å resolution crystal structure of the complex between the yeast seven-bladed β-propeller eIF3i/TIF34 and a C-terminal α-helix of eIF3b/PRT1, which reveals universally conserved interactions. Mutating these interactions displays severe growth defects and eliminates association of eIF3i/TIF34 and strikingly also eIF3g/TIF35 with eIF3 and 40S subunits in vivo. Unexpectedly, 40S-association of the remaining eIF3 subcomplex and eIF5 is likewise destabilized resulting in formation of aberrant pre-initiation complexes (PICs) containing eIF2 and eIF1, which critically compromises scanning arrest on mRNA at its AUG start codon suggesting that the contacts between mRNA and ribosomal decoding site are impaired. Remarkably, overexpression of eIF3g/TIF35 suppresses the leaky scanning and growth defects most probably by preventing these aberrant PICs to form. Leaky scanning is also partially suppressed by eIF1, one of the key regulators of AUG recognition, and its mutant sui1G107R but the mechanism differs. We conclude that the C-terminus of eIF3b/PRT1 orchestrates co-operative recruitment of eIF3i/TIF34 and eIF3g/TIF35 to the 40S subunit for a stable and proper assembly of 48S pre-initiation complexes necessary for stringent AUG recognition on mRNAs.
INTRODUCTION
Canonical translation initiation ensures timely and spatially coordinated formation of the trimeric complex between the 40S small ribosomal subunit (40S subunit), initiator Met-tRNAiMet and an mRNA at its extreme 5′ end, and concludes with the assembly of an elongation-competent 80S ribosome at the authentic AUG start codon. The entire process is orchestrated by numerous eukaryotic initiation factors (eIFs) with eIF3 representing the most intricate factor (1). The multiple essential roles of eIF3 during initiation include stabilization of eIF2/GTP/Met-tRNAiMet ternary complex (TC) binding to 40S subunits, recruitment of 5′-7mG capped mRNAs to 40S subunits, assistance in scanning of the 5′ untranslated region (5′ UTR) of the mRNA, and finally in aiding AUG initiation codon recognition (2).
This crucial involvement of eIF3 in nearly every step of translation initiation is also reflected in its structural complexity. Mammalian eIF3 consists of at least 13 subunits (eIF3a-m) assembled into a 750 kDa particle. In budding yeast, eIF3 comprises five essential core subunits (a/TIF32, b/PRT1, c/NIP1, i/TIF34, g/TIF35) and one loosely associated, non-essential subunit j/HCR1, all of which have corresponding orthologs in mammals (1). At least in yeast, this core eIF3 complex associates with eIFs 1, 5 and the TC in vivo to form the so-called multifactor complex (MFC). Surprisingly, detailed biochemical analysis carried out with purified eIF3 subcomplexes identified that the trimeric complex of a/TIF32, b/PRT1 and c/NIP1 promoted TC and mRNA recruitment to the 40S subunit and even stimulated translation in vitro on a model mRNA as efficiently as the wild-type (wt) five subunit complex (3). Hence given the fact that i/TIF34 and g/TIF35 subunits are otherwise essential for cell proliferation and their individual depletions result in a typical polysome run-off (4), these results perhaps indicate that their contributions to general translation initiation have a more stimulatory character. Indeed, we have recently shown that both small eIF3 subunits augment various aspects of linear scanning and, as such, may have differential effects on efficiency of translation of coding mRNAs containing short, less structured versus long, highly structured 5′ UTRs (5). Interestingly, in vitro reconstitution of human eIF3 also supported the dispensability of highly conserved eIF3g and eIF3i in stimulation of canonical eIF3 functions. Instead, non-conserved eIF3e, eIF3f and eIF3h in complex with eIF3a, eIF3b, and eIF3c were proposed to be the functionally indispensable subunits of mammalian eIF3 (6). In contrast, however, two other groups reported purification of a human eIF3 subcomplex closely resembling the yeast core eIF3 complex (7,8). Hence more experiments are required to clarify these discrepancies.
The structural complexity of eIF3 is well illustrated by its elaborate subunit–subunit interaction web, which has been mapped in great detail for yeast eIF3 and its associated eIFs (9). The b/PRT1 subunit serves as the major scaffolding subunit of eIF3 and associates with the other core subunits in both yeast and mammals (7–10). The b/PRT1 N-terminal domain (NTD) contains a conserved RNA recognition motif (RRM) (11,12), which provides an interaction surface for the C-terminal half of a/TIF32 and the NTD of j/HCR1 (12,13), followed by a middle domain predicted to fold into two β-propeller structures (14), the second of which contains a binding site for c/NIP1. Finally, the extreme C-terminal domain (CTD) of the b/PRT1 scaffold is required for association of i/TIF34 and g/TIF35 subunits (10). Whereas i/TIF34 is predicted to adopt a seven-bladed β-propeller structure made up of seven WD-40 repeats with unknown binding sites for b/PRT1 and g/TIF35, the latter subunit interacts with i/TIF34 and b/PRT1 through its NTD containing a predicted Zn-finger domain (10). The g/TIF35-CTD then adopts an RRM fold that is not involved in any subunit–subunit interactions (9,10). Better understanding of all functions of individual interactions among eIF3 subunits clearly requires more detailed information at the molecular level; however, no atomic structures of any yeast eIF3 subunits have been determined to date.
Here we report the 2.2 Å resolution crystal structure of the i/TIF34 subunit in complex with the minimal CTD of b/PRT1 (654–700), the boundaries of which we defined by solution NMR spectroscopy. Mutating the conserved residues mediating the b/PRT1—i/TIF34 contact results in lethality or severe growth phenotypes owing to the loss of the i/TIF34-g/TIF35 mini-module from the rest of eIF3 and from pre-initiation complexes (PICs). Since binding of the remaining a/TIF32-b/PRT1-c/NIP1 subcomplex and eIF5 is also unexpectedly destabilized, aberrant PICs containing eIFs 2 and 1 accumulate and dramatically increase leaky scanning over the AUG start codon in the manner suppressible by overexpression of g/TIF35 and eIF1. Hence we propose that stable association of the i/TIF34–g/TIF35 mini-module with the rest of eIF3 via b/PRT1 significantly stabilizes binding of eIF3 and eIF5 to the nascent pre-initiation complexes in vivo. This way these critical interactions serve to prevent accumulation of mis-assembled PICs on mRNAs and thus ensure stringent selection of the AUG start codon.
MATERIALS AND METHODS
Protein expression and purification
DNA fragments encoding yeast i/TIF34 and b/PRT1(630–724) were prepared by PCR from pGEX–TIF34 or pGEX–PRT1, respectively (10), using appropriate primers and subcloned into pET28a vector (Novagen) with a Tobacco Etch Virus (TEV) protease cleavage site instead of the original thrombin cleavage site. A DNA fragment encoding b/PRT1(654–700) was prepared by PCR from b/PRT1(630–724) plasmid DNA and subcloned into a modified pET28a vector (Novagen, containing an N-terminal His6-tag fused to a lipoyl domain (12,15) followed by a TEV cleavage site and the standard pET28a multiple cloning site). Point mutations in b/PRT1(630–724) were introduced following the QuickChange protocol (Stratagene). Proteins were expressed in Escherichia coli Rosetta(DE3) cells (Novagen) in LB rich or M9 minimal media supplemented with 15NH4Cl or 15NH4Cl and 13C-glucose. Protein expression was induced by addition of 1 mM IPTG at OD600 of ~0.8 and further incubation for 14 h at 16° C. All proteins were purified using two 5 mL HiTrap chelating columns (HiTrap, GE Healthcare) in series and charged with nickel sulfate under standard conditions (lysis and loading buffer A: 20 mM HEPES pH 7.5, 500 mM NaCl, 30 mM imidazole, 10% glycerol (w/v), 5 mM 2-mercaptoethanol; elution buffer B: same as buffer A with 300 mM imidazole). For b/PRT1 peptides this was followed by TEV protease cleavage for His6-tag or His6-tag lipoyl fusion removal (dialysis buffer for TEV cleavage at room temperature: 20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 5 mM 2-mercaptoethanol), while i/TIF34 was left uncleaved. TEV cleavage reaction mixtures were then reloaded onto HiTrap chelating columns to remove the His6-TEV protease, the His6-tag or His6-tag lipoyl fusion as well as minor contaminating proteins using buffers A and B (see above). After purification, the proteins were dialyzed 3 times against 2 L storage/NMR buffer (50 mM sodium phosphate buffer pH 7.5, 150 mM NaCl, 5 mM 2-mercaptoethanol).
Purified His6–i/TIF34 and b/PRT1 proteins were assembled into complexes at a 1:1.2 molar ratio, concentrated using Vivaspin 20 mL centrifugal devices with 10 000 MWCO (Sartorius) and passed over a Sephacryl S-200 gel filtration column (GE Healthcare) using storage/NMR buffer as running buffer. Fractions corresponding to the 1:1 complex were pooled, concentrated to 7–10 mg/ml and either used directly for NMR experiments or dialyzed against 2 times 2 L crystallization buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM 2-mercaptoethanol) using Slide-A-Lyzer 0.5 mL dialysis cassettes with 10 000 MWCO (Thermo Scientific).
NMR spectroscopy
NMR experiments were performed on Bruker DMX600 or AVANCE800 spectrometers equipped with cryoprobes. Partial protein backbone assignments of 13C/15N-labeled b/PRT1(630–724) in complex with unlabeled His6–i/TIF34 were achieved by means of through-bond heteronuclear scalar correlations with standard pulse sequences from the Bruker pulse sequence library. Chemical shift patterns upon binding of 15N-labeled b/PRT1(630–724) or b/PRT1(654–700) to His6–i/TIF34 were compared using standard 15N-HSQC and 15N-TROSY pulse sequences. All NMR samples were prepared in NMR buffer (see above) with addition of 10% D2O (v/v) at protein concentrations of 100–200 µM. All spectra were recorded at 20° C.
Defining the minimal i/TIF34-binding domain of b/PRT1 by NMR spectroscopy
Residues 641–724 of b/PRT1 were previously shown to be required for i/TIF34-binding in vitro (9,10). Crystallization of proteins and their complexes is often hindered by flexible, unstructured tails and this prompted us to further define the precise boundaries of the interaction site by NMR spectroscopy (16,17). To achieve this, we expressed unlabeled i/TIF34 and a 13C,15N-labeled C-terminal fragment of b/PRT1 spanning residues 630–724 and recorded 15N-HSQC spectra of the peptide both in the free and i/TIF34-bound form. The spectrum of the free 10 kDa peptide displays severe resonance overlap in the amide region and mainly random coil chemical shifts indicative of no intrinsic secondary structure as also confirmed by CD spectroscopy (data not shown and Supplementary Figure S1A). Upon binding of i/TIF34 to the b/PRT1 peptide, a 50 kDa complex is formed with much slower tumbling times of the bound peptide in solution resulting in shorter transverse relaxation times and concomitant broadening of amide signals from b/PRT1 residues participating in the interaction with i/TIF34. On the other hand, a significant portion of the sharper random coil crosspeaks of b/PRT1 remains unchanged indicating that they are not part of the i/TIF34 binding interface. For several of these random coil resonances much longer transverse relaxation times allowed us to obtain backbone assignments using standard triple resonance experiments. We unambiguously assigned a four amino acid stretch spanning residues Q651-M654 from the N-terminus as well as the entire extreme C-terminus starting at residue A701 of b/PRT1, while the middle segment spanning residues 655–700 could not be assigned due to short transverse relaxation times from an interaction with i/TIF34. We therefore designed and expressed a 15N-labeled peptide comprising residues 654–700 of the C-terminus of b/PRT1. This shorter peptide displays the same broad, shifted resonances when bound to i/TIF34 as the longer 15N-labeled peptide comprising residues 630–724, including a downfield shifted Trp-NεH crosspeak of W674, but lacks the sharp random coil Trp-NεH crosspeak (W644) from the N-terminal half of the longer peptide (Supplementary Figure S1B). This unambiguously defines that the minimum i/TIF34 binding site of b/PRT1 comprises residues 654–700. This conclusion is also confirmed by isothermal titration calorimetry (ITC), which displays very similar affinities for the long b/PRT1(630–724) and the minimal C-terminal peptide of b/PRT1(654–700) with KD-values of 81 nM and 160 nM, respectively (Supplementary Figure S1C). Thus optimized complex of yeast full-length i/TIF34 and the 654–700 fragment of b/PRT1, lacking b/PRT1 random coil segments, was subjected to crystallization and after optimization yielded well diffracting crystals suitable for structure determination of the complex.
Crystallization
Crystallization screening by sitting drop vapor diffusion was performed with 200 nl drops (1:1 ratio of protein solution to mother liquor) against 1600 commercial conditions at room temperature. Crystals were obtained in 0.1 M Tris–HCl pH 8.8, 0.1 M Li2SO4, 28% PEG 4000, 2–8% 1,5-diAminoPentane dihydrochloride. Heavy atom derivatives were obtained by soaking crystals in saturated solutions of K2PtCl4 (potassium tetrachloroplatinate (II) and C2H5HgSC6H4CO2Na (sodium ethyl-mercuri-thiosalicylate, thiomersal) for 2–5 min.
Diffraction data collection and structure determination
Native diffraction dataset was collected from a single crystal at 100K at the Diamond synchrotron facility (UK) on beamline I03. Data of heavy atom derivatives were collected using a MAR-DTB image-plate detector and rotating anode X-ray generator. The data were indexed with MOSFLM (18) and further processed using SCALA (19) and TRUNCATE (20) from CCP4 package (21). Experimental phases were obtained by MIR, combining Pt and Hg derivatives, scaling them to the native dataset and further processing with SHARP (22). Although the phase information from the Pt derivative was weak beyond (and at) 3.8 Å resolution, adding it significantly improved the experimental map. PHYRE server (23) homology modeling solution of i/TIF34 was used as a search model in Phaser (24) to find second molecule in the asymmetric unit. Map was improved with solvent flattening in Parrot (25). Major part of the model was built with Buccaneer (26), with remaining parts hand-built using Coot (27). Iterative rounds of model building in Coot, refinement with Refmac (28) and validation using MolProbity server (29) were ultimately performed without non-crystallographic symmetry (NCS). Initially, NCS was applied during the first rounds of refinement but no significant improvement was observed. Therefore we decided not to use NCS in the refinement, especially since one part of the protein (the flexible loop) was different between the two molecules. The final structure consists of four protein chains: B and D—i/TIF34, E and F—b/PRT1(654–700). Chains B and D have TEV protease cleavage site, and therefore start at the residue Glu-9 leaving methionine as the first amino acid. Chain B contains residues −9 to 259 and 273 to 342, chain D −9 to 342, chains E and F 661 to 699. Data collection and refinement statistics are summarized in Supplementary Table S1.
Yeast strains, plasmids, and biochemical methods
Lists of strains and plasmids used in this study (Supplementary Tables S3–S5), details of their construction, as well as description of all well-established biochemical assays used throughout the study can be found in the Supplementary Data.
RESULTS
The overall crystal structure of i/TIF34–b/ PRT1(654–700) complex
Using solution NMR spectroscopy, we defined the minimal C-terminal i/TIF34-binding site of b/PRT1(654–700) lacking any b/PRT1 random coil segments, which could hinder crystallization (16,17) (Figure 1A, Supplementary Figures S1 and S2A, ‘Material and Methods’ section). The crystallographic structure of thus optimized yeast i/TIF34–b/PRT1(654–700) complex was determined at 2.2 Å resolution by MIR heavy atom phasing (data statistics in Supplementary Table S1). Two complexes were found in the asymmetric unit and both of them displayed excellent electron density (Supplementary Figures S3 and S4) except for a few terminal residues of both the b/PRT1 and i/TIF34 molecules and a specific loop region spanning well-conserved residues 260–272 in one of the two i/TIF34 molecules (discussed in more detail later).
Figure 1.
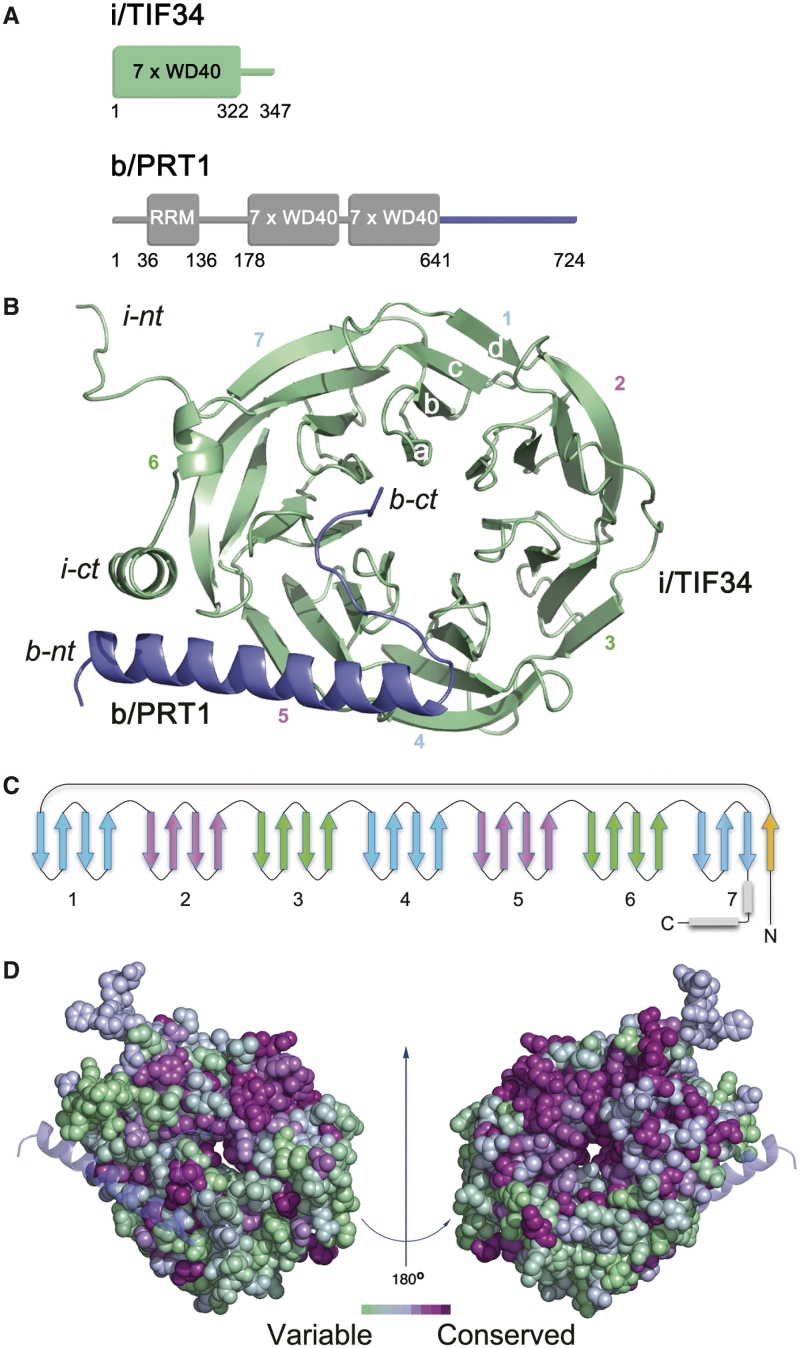
Structure of i/TIF34–b/PRT1 complex. (A) Schematic drawing of the predicted protein domains of yeast i/TIF34 (top) and b/PRT1 subunits of eIF3 (bottom). The folded domains and their boundaries are indicated. Predicted unstructured regions are shown as lines. (B) Overview of the structure: cartoon representation of i/TIF34 (green) and b/PRT1(654–700) (blue). WD-40 blades 1–7, β-strands a–d of blade 1 and N-terminus (nt) and C-terminus (ct) are labeled. The complex is shown from the bottom side of the β-propeller, where loops occur between strands a–b and c–d. (C) Topology diagram of the i/TIF34 fold. (D) Space-filling view of the i/TIF34 in complex with b/PRT1 (bottom view, left and top view, right), colored according to sequence conservation. A gradient of green to purple indicates the degree of phylogenetic conservation, with variable shown as green and most conserved as dark purple. The conservation heat plot of the i/TIF34 surface was generated by ConSurf using multiple alignment of Human, Drosophila, Arabidopsis and Saccharomyces cerevisiae i/TIF34 protein homologues. All structural figures were generated using PyMOL (http://www.pymol.org).
The i/TIF34 subunit adopts a seven-bladed β-propeller fold with a short, bent α-helix at the C-terminus (Figure 1B). As commonly seen in β-propeller structures (30), one β-sheet of the last blade 7 is formed by N-terminal residues 1–7, while the subsequent amino acid residues consecutively form blades 1–7 (Figure 1C). Phylogenetic analysis shows that the most conserved residues comprise blades 1, 2, 6 and 7 (Figure 1D and Supplementary Figure S2A) and that several loops between the blades also include highly conserved residues. The β-propeller structure usually serves as a platform for protein–protein interactions (31). Interacting protein partners often bind to the top or bottom of the β-propeller structure, but binding along the groove between blades has also been observed (32–34). The b/PRT1(654–700) fragment binds to the bottom side of the β-propeller along the loops of blades 5 and 6 of i/TIF34 (Figure 1B). The b/PRT1 residues 663 to 689 form a long α-helix while the extended C-terminus (residues 690–699) advances towards the central cavity of the i/TIF34 β-propeller and is held mostly by contacts via main chain atoms (Figure 1B). These interactions are independent of the contacts between the two complexes within the asymmetric unit (Supplementary Figures S3 and S4).
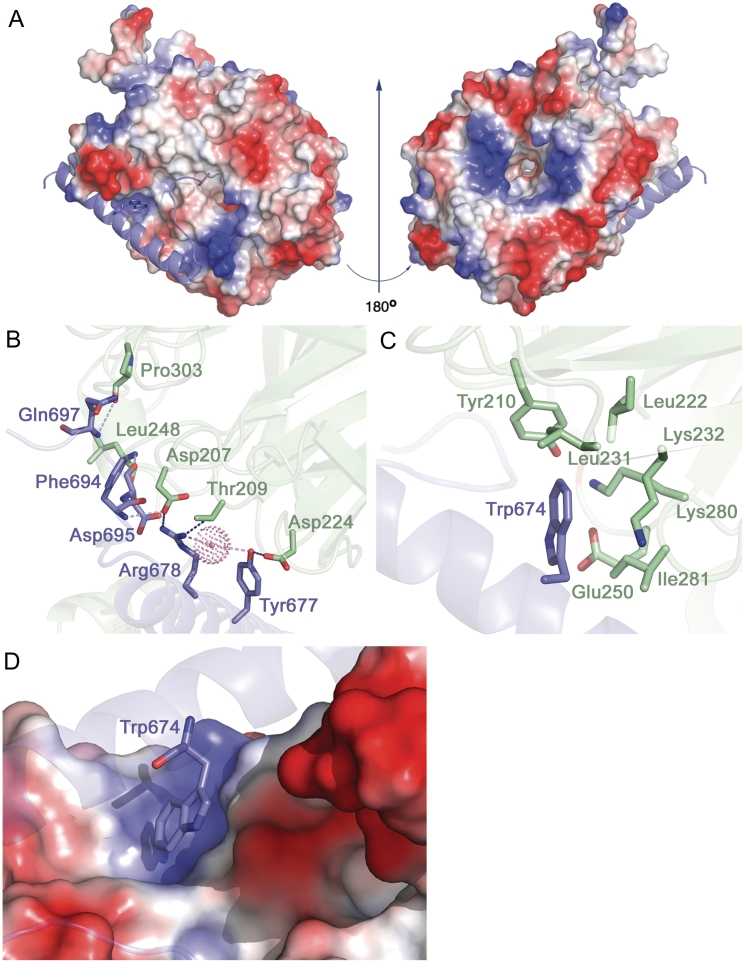
The b/PRT1-binding interface of i/TIF34 comprises residues from β-sheets and loops in blades 5 and 6 (Figure 1B). The total buried surface area of the b/PRT1–i/TIF34 interface is 1028.7 Å2 and 1054.6 Å2 (complex I and complex II, respectively, in Supplementary Figure S4) as calculated using PISA (35). The b/PRT1-interacting surface of i/TIF34 bears a few important charged regions (Figure 2A; left). The remaining surface of this side of the β-propeller shows dominantly negative charge, while patches of positive charge dominate around the central cavity on the top side of the β-propeller (formed by blades 2–5, Figure 2A; right). This charge distribution is conserved suggesting distinct interfaces for the interactions with other components of the translational machinery (Figures 1D, 2A and Supplementary Figure S2A).
Figure 2.
Molecular details of i/TIF34 and b/PRT1 interactions. (A) Electrostatic potential (±5kT/e) of the solvent-accessible surface of i/TIF34 in complex with b/PRT1(654–700) rendered on the molecular surface of the complex. A gradient of blue to red shows positive to negative charge, respectively, as calculated using PyMOL built-in APBS tools (55). PQR file for analysis of Poisson–Boltzmann electrostatics calculations was generated using PDB2PQR tool (56) and further used for APBS. b/PRT1(654–700) is shown as a cartoon in blue. View from the b/PRT1 binding site is shown on the left; the ‘reverse’ side of i/TIF34 shown on the right has negative charge clustering at the highly conserved blade 1 (upper part), blades 4 and 5 (the bottom part) and positive charge around the central cavity. (B–C) The i/TIF34-b/PRT1 binding interface involves highly conserved amino acids from both proteins. i/TIF34 is shown in green, b/PRT1 in blue. (B) Y677 and R678 from b/PRT1 form H-bonds (blue) with D207, T209 and D224 from i/TIF34 (only interacting side chains are shown). Other residues making contacts (light blue) via main chain atoms are also shown in sticks. One water molecule (shown in dots) is in close proximity (light pink) to R678 NH1 (2.6 Å) and Y677 OH (3.3 Å). (C) b/PRT1 W674 is surrounded by hydrophobic and charged amino acids (only side chains are shown), which form a shallow pocket. (D) Surface representation of the i/TIF34 hydrophobic pocket, which accommodates b/PRT1 W674. This interaction serves as a ‘lock’ for the i/TIF34 and b/PRT1 interaction interface.
Seven residues of the b/PRT1(654–700) fragment are in close proximity (<3.5 Å apart) for direct contact with eight residues of i/TIF34. In both proteins, the interacting residues involve highly conserved amino acids (Figure 1D and Supplementary Figure S2A; and also listed in Supplementary Table S2). At the b/PRT1–i/TIF34 interface hydrogen bonds are formed between the most conserved residues Y677-D224 and R678-D207/T209 (Figure 2B and Supplementary Figure S2B), implying that these residues are crucial for b/PRT1–i/TIF34 complex formation. The second important feature of the complex interface is the insertion of the aromatic ring of b/PRT1 W674 into a pocket of i/TIF34 formed by hydrophobic amino acids (L231 and I281 as well as the conserved L222) and the side chains of polar amino acids (Y210 and K280 as well as the conserved K232 and E250) (Figure 2C, D and Supplementary Figure S2B). The nearest i/TIF34 residues are within 3.8–4.8 Å of W674. W674 probably makes a π–cation interaction with K280 (36) and additional hydrophobic interactions with the other ‘pocket’ residues (Figure 2D). Phylogenetic investigation shows that in other organisms including human, frog and Drosophila, Phe or Tyr is found instead of Trp and K280 replaced by either Leu or Val, implying conservation of the hydrophobic nature of this interaction (Supplementary Figure S2A). In addition to the specific side chain contacts along the α-helix of b/PRT1, there is a series of interactions between main chain atoms of the extended C-terminal part of b/PRT1 starting with N690 and i/TIF34 residues in blades 5 and 6 (Supplementary Table S2). Interestingly, none of the i/TIF34 residues directly interacting with b/PRT1 appeared in previous mutational studies by Asano and co-workers (10). For direct comparison see Supplementary Figure S5 and the corresponding text.
The major difference between the two i/TIF34–b/PRT1(654–700) complexes in the asymmetric unit lies within the loop made of residues 260–272 in blade 6 of i/TIF34 on the opposite side of the b/PRT1 binding interface. One of the complexes has well-defined electron density surrounding these residues (complex II in Supplementary Figure S6A), whereas the other molecule lacks defined electron density around this area (position of the residues omitted from the PDB file are indicated by the red arrow in Supplementary Figure S6A). The well-defined loop in complex II contacts another symmetry-related i/TIF34 molecule (residues 80–87 and 130–139; not shown) and forms a salt bridge between E270 and K91 in the same chain of the symmetry-related complex fixing the loop in a detectable state. The flexible loop residues are predominantly negatively charged (Supplementary Figure S6B) and residues 260–279 display high sequence conservation from yeast to human (Supplementary Figure S2A) suggesting an important interaction module with either the 40S subunits or other partners within the eIF3 complex.
Disrupting the conserved b/PRT1–i/TIF34 interaction results in lethality or severe growth phenotypes
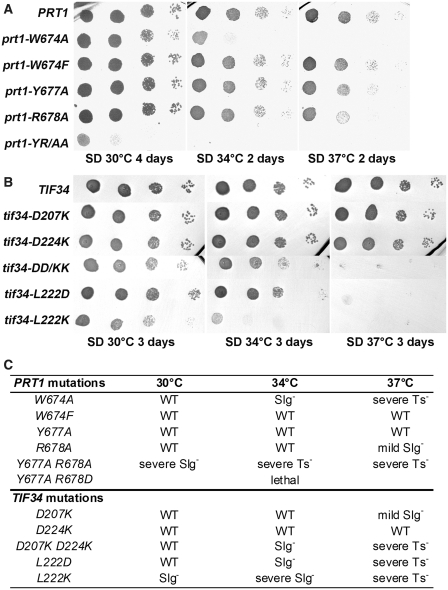
To confirm the critical aspects of the b/PRT1–i/TIF34 contacts revealed by the structure analysis and to investigate the functional consequences of their loss in living cells, we first substituted Y677 and R678 (alone or in combination), or W674 of b/PRT1 with Ala, Asp or Phe residues, respectively, and tested them for growth phenotypes. The corresponding mutations were generated in a plasmid copy of PRT1-His, encoding His8-tagged b/PRT1, and introduced into a prt1Δ strain harboring wt PRT1 on a URA3 plasmid, which was subsequently evicted by counter-selection on medium containing 5-fluoroorotic acid (5-FOA). Whereas the double Y677A R678D mutation is lethal and the Y677A R678A (dubbed YR/AA herein) double mutant imparts a very severe slow growth (Slg–) phenotype, individual Y677A and R678A substitutions show little to no effect on growth rates (Figure 3A). Severe Slg− and temperature sensitive (Ts−) phenotypes are also found associated with the W674A mutation. Strikingly, the ‘phylogenetic correction’ substitution in W674F (see above) displays wt-like behavior under all tested conditions (Figure 3A; summarized in 3C). Importantly, in vitro electrophoretic mobility shift assays performed with recombinant purified i/TIF34 and b/PRT1-CTD(630–724) variants confirm these in vivo results: b/prt1-W674F mutant protein competes well with wt b/PRT1-CTD for i/TIF34 binding, whereas b/prt1-W674A and the YR/AA double mutant are not able to compete for i/TIF34 binding at all (Supplementary Figure S7). These results confirm that an aromatic side chain (Phe or Trp) is required to fill the hydrophobic pocket on i/TIF34 and indicate that the contacts of Y677 and R678 with D207 and D224, respectively, are redundant (see more biochemical evidence further below).
Figure 3.
Phenotypic and biochemical analysis of i/TIF34 and b/PRT1 mutations that disrupt subunit interactions. (A) The prt1-W674A and -YR/AA but not -W674F mutations produce severe slow growth and temperature sensitive phenotypes. The YAH06 (prt1Δ) strain was transformed with the corresponding plasmids carrying individual mutant alleles and the resident pCR52 (PRT1,URA3) covering plasmid was evicted on 5-FOA. The resulting strains were then spotted in four serial 10-fold dilutions on SD medium and incubated at 30, 34 and 37°C. (B) The tif34-DD/KK, -L222D and L222K mutations produce severe slow growth and temperature sensitive phenotypes. The H450 (tif34Δ) strain was transformed with the corresponding plasmids carrying individual mutant alleles and the resident YEp-i/TIF34-U (TIF34, URA3) covering plasmid was evicted on 5-FOA. The resulting strains were then spotted in four serial 10-fold dilutions on SD medium and incubated at 30, 34 and 37°C. (C) Summary of phenotypes of mutations analyzed in this study.
In a similar fashion, specific substitutions of D207 and D224 (alone or in combination), or L222 of i/TIF34, generated in a plasmid copy of TIF34-HA encoding HA-tagged i/TIF34, were tested for growth defects in a tif34Δ strain. In agreement with the aforementioned results, individual D207K and D224K substitutions produce no effects; however, the combined D207K D224K (dubbed DD/KK herein) mutant displays a severe Ts− phenotype (Figure 3B). Finally, both charge substitutions of L222 in L222D or L222K, which probably drastically disturb the L222-containing hydrophobic pocket or perhaps even the entire local structure around blades 5 and 6 (Figure 2C and D), also impart severe Ts− and Slg− phenotypes (Figure 3B; summarized in 3C).
Impairment of the contact between b/PRT1 and i/TIF34 eliminates association of i/TIF34 and g/TIF35 from the rest of eIF3 in vivo
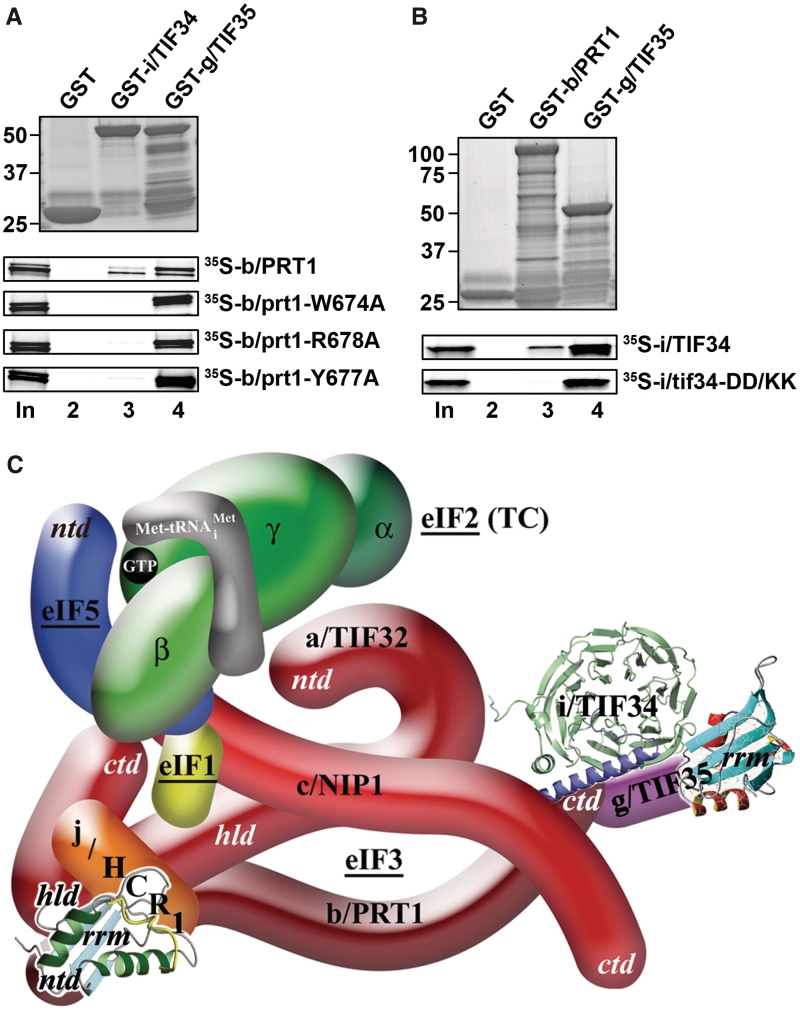
To further test whether the mutations under study impair a direct contact between b/PRT1 and i/TIF34 proteins we introduced three single-Ala-substitution mutations into full length b/PRT1, synthesized and radiolabeled the resulting mutant proteins in vitro and tested their binding affinities towards i/TIF34 and g/TIF35 fused with the GST moiety. As shown in Figure 4A, all three completely eliminate binding of b/PRT1 specifically to i/TIF34 but not to g/TIF35. Similarly, both single D/K substitutions and the DD/KK double mutation of i/TIF34, the latter of which was chosen for further analysis, abolished binding of [35S]-labeled i/TIF34 to GST-b/PRT1 but not to GST-g/TIF35 (Figure 4B and data not shown).
Figure 4.
The tif34-DD/KK mutation impairs the direct interaction between i/TIF34 and b/PRT1 in vitro and the revised 3D model of eIF3 in the MFC. (A) The prt1-W674A, -Y677A, and -R678A mutations impair the direct interaction between b/PRT1 and i/TIF34 in vitro. Full-length i/TIF34 (lane 3) and g/TIF35 (lane 4) fused to GST, and GST alone (lane 2), were tested for binding to 35S-labeled wt b/PRT1 and its mutant derivatives; 10% of input amounts added to each reaction is shown in lane 1 (In). (B) Full-length b/PRT1 (lane 3) and g/TIF35 (lane 4) fused to GST, and GST alone (lane 2), were tested for binding to 35S-labeled wt i/TIF34 and the DD/KK mutant derivative. (C) A revised 3D model of eIF3 and its associated eIFs in the MFC (based on the data from (9); ntd, N-terminal domain; ctd, C-terminal domain; hld, HCR1-like domain; rrm, RNA recognition motif; TC, ternary complex). The NMR structure of the interaction between the RRM of human eIF3b (green and light blue) and the N-terminal peptide of human eIF3j (yellow) (12), the NMR structure of the C-terminal RRM of human eIF3g (red and sky-blue) (5), and the X-ray structure of the yeast i/TIF34–b/PRT1 complex (this study), were used to replace the original schematic representations of the corresponding molecules.
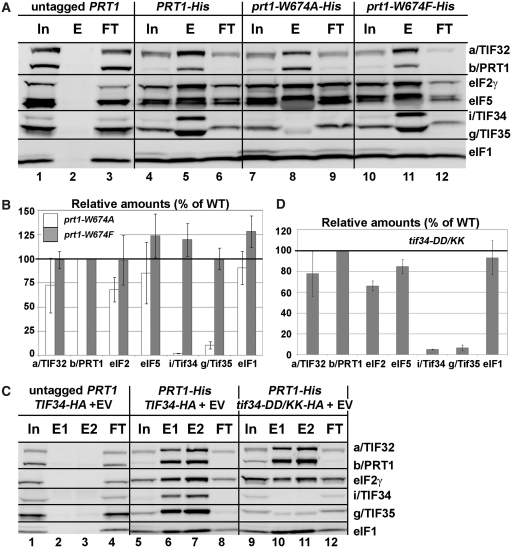
Next we wished to demonstrate the effect of our mutations on the association of i/TIF34 with the rest of eIF3 in vivo. Towards this end we analyzed formation of the entire eIF3-containing MFC (see our model in Figure 4C) in yeast cells by Ni2+-chelation chromatography using His8-tagged PRT1 as bait. As reported previously (13), a fraction of a/TIF32, j/HCR1, eIF2, eIF5 and eIF1 co-purified specifically with wt b/PRT1-His but not with its untagged version (Figure 5A, lanes 4–6 versus 1–3). The prt1-W674A mutation of one of the two contacts between i/TIF34 and b/PRT1 severely diminishes (by >90%) association of i/TIF34, and in contrast to the above in vitro experiments, also that of g/TIF35 with the MFC, whereas prt1-W674F shows no effects (Figures 5A and B, lanes 7–9 versus 10–12). This concurs well with our genetic data (Figure 3A). In addition, the overall integrity of the MFC also seems to be modestly affected. Similarly, the tif34-DD/KK mutation of the other contact also severely reduces binding of i/TIF34 and g/TIF35 to the purified b/PRT1-His complex (Figures 5C and D, lanes 9–12 versus 5–8). Importantly, overexpressing His8-tagged TIF35 as bait in the background of tif34-DD/KK further supported these novel observations as g/TIF35-His practically failed to pull down any of the MFC components with the exception of i/TIF34 (Supplementary Figure S8A). The fact that the DD/KK mutation does not affect the mutual interaction between i/TIF34 and g/TIF35 in vivo is in perfect agreement with our in vitro binding data (Figure 4B). Together these results strongly suggest that binding of i/TIF34 with b/PRT1, in addition to the direct g/TIF35–b/PRT1 interaction, is a necessary prerequisite for stable eIF3-association of g/TIF35 in vivo indicating that the observed growth phenotypes are a direct consequence of the loss of the essential i/TIF34–g/TIF35 mini-module from the rest of eIF3. Hence the fact that the individual prt1-Y677A, prt1-R678A, tif34-D207K and tif34-D224K mutations diminish the b/PRT1–i/TIF34 interaction in vitro (Figure 4) yet produce no significant growth phenotypes (Figure 3) can be explained by proposing that their impact in vivo is largely overcome by a stabilization effect of simultaneous binding of g/TIF35 to i/TIF34 and b/PRT1 in the context of the entire eIF3 complex, as was observed (Supplementary Figure S8B and C). In other words, their aforementioned redundancy is dependent on the presence of g/TIF35 in vivo. Indeed, this effect is not powerful enough in the case of more deleterious double mutations. Since we had confirmation that genetic and molecular defects of both prt1-W674A and tif34-DD/KK mutations have the same nature, we decided to focus our further analysis on the latter.
Figure 5.
Disrupting the b/PRT1–i/TIF34 interaction eliminates association of the i/TIF34-g/TIF35 mini-module from the MFC in vivo. (A and B) WCEs prepared from YAH06 (prt1Δ) bearing untagged b/PRT1 (lanes 1–3), 8xHis-tagged b/PRT1 (lanes 4–6), and two of its mutant derivatives (lanes 7–9 and 10–12) were incubated with Ni2+ agarose and the bound proteins were eluted and subjected to western blot analysis with the antibodies indicated in each row. (In) lanes contained 5% of the input WCEs; (E) lanes contained 100% of eluate from the resin; (FT) lanes contained 5% of the flow through. (B) The Western signals for indicated proteins in the E fractions of the wt PRT1-His and its mutants were quantified, normalized for the amounts of the wt b/PRT1 in these fractions and plotted in the histogram as percentages of the corresponding values calculated for the wt b/PRT1. (C and D) WCEs were prepared from YAH12 (prt1Δ tif34Δ) bearing untagged PRT1 and wt TIF34 (lanes 1–4) and from YAH11 (prt1Δ tif34Δ) bearing 8xHis-tagged PRT1 and either wt TIF34 plus empty vector (lanes 5–8) or mutant tif34-DD/KK plus empty vector (lanes 9–12) and analyzed analogously to (A and B).
Disruption of the b/PRT1–i/TIF34 interaction prevents 40S-binding of the i/TIF34–g/TIF35 mini-module and selectively affects stability of pre-initiation complexes in vivo
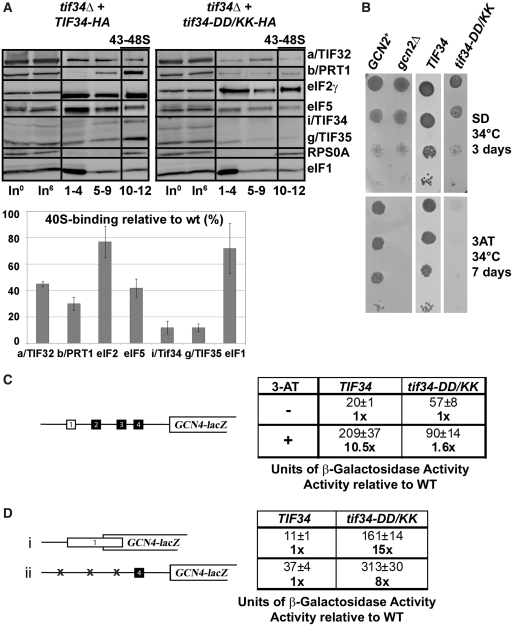
We showed previously that a partial eIF3 subcomplex containing a/TIF32 and c/NIP1, and eIF5, but lacking the b/PRT1, i/TIF34 and g/TIF35 subunits, still interacted with the 40S ribosomes in vivo (albeit with ~2-fold reduced affinity compared to wt 5-subunit eIF3), whereas the b/PRT1–i/TIF34–g/TIF35 subcomplex lacking the N-terminal RRM of b/PRT1 did not (13,37). Hence given that mutating the contact residues between b/PRT1 and i/TIF34 strongly impairs association of i/TIF34 and g/TIF35 with eIF3 in the MFC (Figure 5), it could be consequently expected that the ribosome occupancy of only i/TIF34 and g/TIF35 gets impaired. To examine this, we measured binding of selected eIF3 subunits and other MFC components to 40S subunits by formaldehyde cross-linking followed by high velocity sedimentation in sucrose gradients, as this method provides the best available approximation of the native composition of 43S/48S pre-initiation complexes in vivo (38). In accord with our prediction, we observed a relative ~90% decrease in the amounts of i/TIF34 and g/TIF35 associated with 40S subunits in whole-cell extracts (WCEs) obtained from the tif34-DD/KK cells grown at the semipermissive temperature (Figure 6A, ‘43–48S’ lanes). Surprisingly, rather significant reductions in 40S-binding (~40%) were also observed for other eIF3 subunits and eIF5, whereas binding of eIF2 (TC) and eIF1 was reduced only marginally (~20%). The lack of an increase in abundance of unbound eIF3 and eIF5 factors in the tif34-DD/KK gradients, which might have been expected given the pronounced loss of these proteins from the PICs, is attributed to an increased instability of eIFs defective to properly bind to 40S relative to those tightly bound in PICs during sedimentation. This phenomenon was also observed by us and others in the past (39–41). Quantification of input lanes ruled out a possibility of an increased proteolysis of eIF3 and eIF5 in the living cells as well as in the WCEs even after prolonged incubation on ice (Figure 6A). We conclude that the physical detachment of i/TIF34 and g/TIF35 from the rest of eIF3 not only prevents their binding to the 40S ribosome but also significantly weakens 40S-association of the remaining a/TIF32-b/PRT1-c/NIP1 subcomplex and eIF5. Remarkably, these findings thus may suggest that the mutant cells contain a subpopulation of PICs containing only eIF1 and the TC (see further below).
Figure 6.
Disruption of the b/PRT1–i/TIF34 interaction prevents 40S-binding of the i/TIF34–g/TIF35 mini-module and dramatically increases leaky scanning over the AUG start site producing a severe Gcn- phenotype. (A) Physical detachment of i/TIF34 and g/TIF35 from the rest of eIF3 selectively affects stability of pre-initiation complexes in vivo. Transformants of H450 (tif34Δ) bearing wt or mutant i/TIF34-HA were grown in SD medium at 34 C to an OD600 of approximately 1.5 and cross-linked with 2% HCHO prior to harvesting. WCEs were prepared, separated on a 7.5–30% sucrose gradient by centrifugation at 41 000 rpm for 5 h and subjected to western blot analysis (note that the anti-RPS0A antibodies were generated in this study; see Supplementary Data). Fractions 1–4, 5–9, and 10–12 (43–48S) were pooled; lanes ‘In0’ and ‘In6’ show samples of the input WCEs (5%) that were processed immediately before (h0) or after (h6) incubation for 6 h on ice, mimicking the duration of the HCHO fractionation experiment to document the stability of the factors of interest in WCEs. Proportions of the 40S-bound proteins relative to the amount of 40S subunits were calculated using NIH ImageJ from three independent experiments. The resulting values obtained with the wt strain were set to 100% and those obtained with mutant strains were expressed as percentages of the wt (SDs are given). This experiment was repeated seven times with similar results. (B) tif34-DD/KK imparts the Gcn- phenotype. H417 (GCN2), H418 (gcn2Δ) and H450 (tif34Δ) bearing wt or mutant i/TIF34-HA were spotted in four serial 10-fold dilutions on SD (upper panel) or SD containing 30 mM 3-AT (lower panel) and then incubated at 34°C for 3 and 7 days, respectively. (C) tif34-DD/KK severely prevents derepression of GCN4-lacZ upon starvation. The H450 strains as in panel A were transformed with the GCN4-lacZ reporter p180 and grown in SD medium at 34°C to an OD600 of approximately 1. The β-galactosidase activities were measured in the WCEs and expressed in units of nmol of O-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per mg of protein. To induce GCN4-lacZ expression, strains were grown in minimal medium to an OD600 approximately 0.5 and then treated with 10 mM 3-AT for 6 h. The table gives mean values and standard deviations obtained from at least six independent measurements with three independent transformants, and activities with 3-AT-induction relative to those without induction. (D) Detachment of the i/TIF34–g/TIF35 mini-module from eIF3 provokes unusually severe leaky scanning defect. The H450 strains as in (A) were transformed with the GCN4-lacZ reporter plasmids pM226 (i) and plig102-3 (ii) and analyzed as in (C). The table gives activities in mutant relative to wt cells.
Detachment of the i/TIF34–g/TIF35 mini-module from eIF3 dramatically increases leaky scanning over the AUG start site producing a severe Gcn− phenotype
To elucidate the functional consequences of this PIC-assembly defect, we employed the translational control mechanism of GCN4, which depends on four short upstream open reading frames (uORFs) found in its mRNA leader and has been adapted to serve as an experimental tool for monitoring various translational steps [reviewed in (42)]. The expression of GCN4, a transcriptional activator of many biosynthetic genes, is delicately regulated in a nutrient-dependent manner by the GCN2 kinase. Under nutrient-replete conditions the kinase is inactive and the GCN4 expression is repressed. Upon amino-acid starvation, GCN2 becomes activated and derepresses GCN4 synthesis by reducing the steady state levels of the TC. Mutants defective in the TC formation and/or its recruitment to the 40S subunit mimic starvation conditions and constitutively derepress GCN4 even under nutrient-replete conditions, producing the Gcd− phenotype. Conversely, mutants that fail to derepress GCN4 under starvation conditions provoke the Gcn− phenotype, which signals defects in the steps following assembly of 48S PICs, such as processivity of scanning, AUG recognition or subunit joining (42).
In accord with the nearly wt levels of 40S-bound TC (Figure 6A), tif34-DD/KK did not display the Gcd− phenotype; however, it did impart the severe Gcn− phenotype characterized by a failure to grow in the presence of 3-aminotriazole (3-AT) at 34°C (Figure 6B, column 4; 3-AT is an inhibitor of histidine biosynthetic genes.) Using the wt GCN4-lacZ reporter plasmid we indeed confirmed almost an absolute failure to derepress GCN4 in response to 3-AT in the DD/KK cells (Figure 6C). The fact that the tif34-DD/KK cells sport the Gcn− phenotype as severe as the deletion of GCN2 by itself (Figure 6B, column 2) suggests that the DD/KK mutation deregulates GCN4 translational control by a dramatic impairment of one or more initiation steps following the TC recruitment.
To investigate this, we decided to employ the GCN4-lacZ reporter constructs with specific modifications in the GCN4 mRNA leader, which have been successfully used in the past to reveal malfunctioning in scanning processivity, scanning rates, stringency of AUG selection or in subunit joining (12,39,40,43). Defects in recognition of the AUG start codon resulting in so-called leaky scanning can be identified with two GCN4-lacZ constructs: one where the first uORF (uORF1) is elongated and overlaps the beginning of GCN4 (Figure 6D, constructs i); the other where only the last uORF (uORF4), non-permissive for reinitiation (44,45), is preserved in the mRNA leader of GCN4-lacZ (Figure 6D, construct ii). Only those ribosomes that skip (leaky scan) AUGs of the corresponding uORFs in both constructs may initiate on GCN4-lacZ, thereby producing an increase in β-galactosidase activity. Accordingly, both tif34-DD/KK and prt1-W674A show an unusually robust increase (up to 15-fold) of expression from both of these constructs (Figure 6D and Supplementary Figure S8D). Surprisingly, further analysis of other potential defects described above did not reveal any clearly distinguishable defects (see ‘Discussion’ section). Taken together these results strongly suggest that stable association of the i/TIF34–g/TIF35 mini-module with the rest of eIF3 stabilizes binding of eIF3 and eIF5 to the PICs to ensure their proper assembly required for stringent AUG recognition.
Increased gene dosage of TIF35 suppresses growth and leaky scanning defects of the i/tif34-DD/KK mutant by preventing formation of aberrant PICs
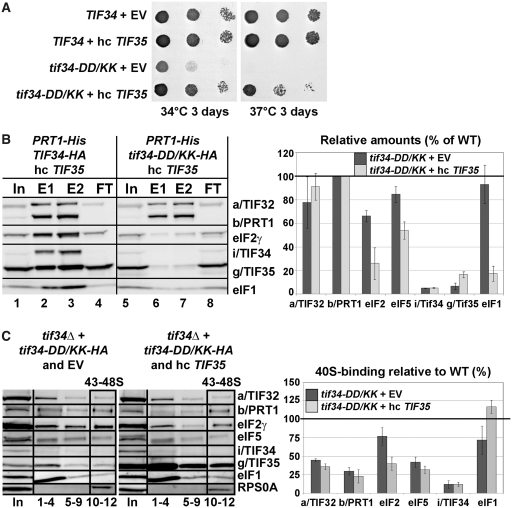
Given the close interdependence of both small eIF3 subunits in terms of their stability and incorporation into the eIF3 complex it may be possible to suppress some of the observed phenotypes of tif34-DD/KK by increasing the gene dosage of TIF35. Accordingly, overexpressing TIF35 significantly suppressed its growth deficiency (Figure 7A), unexpectedly, however, it did not strengthen association of i/TIF34 and g/TIF35 with b/PRT1-His in the MFC (Figure 7B, lanes 5–8 versus 1–4 and versus Figure 5C, lanes 9–12; these experiments were carried out at the same time and are summarized in the quantification plot of Figure 7B). Instead, it repeatedly decreased the amounts of eIF5, and mainly of eIF2 and eIF1 co-purifying with the rest of eIF3 suggesting that the increased dosage of g/TIF35 destabilized formation of the MFC in vivo. Sequestration of eIF2 and eIF1 in a separate complex by excess of g/TIF35 in cells could explain this unexpected phenomenon; however, was not observed (Supplementary Figure S8A). Hence, the molecular details of this mechanism are at present unknown to us. Most importantly, whereas it had no impact on the amounts of the 40S-bound eIF3 subunits and eIF5 when compared to wt and i/tif34-DD/KK cells bearing an empty vector, high dosage of g/TIF35 significantly decreased the amounts of eIF2 (~2-fold) and evened the eIF1 amounts to wt levels (Figure 7C, compare 43–48S fractions and the quantification plot summarizing results from Figures 6A and 7C). (Increased amounts of g/TIF35 in the ribosomal fractions are caused by trailing of the large excess of this protein through sucrose gradients and thus are not significant.) Importantly, the fact that the eIF2 levels actually matched those of eIF3 and eIF5 suggests that destabilization of the MFC in turn prevents formation of the aberrant TC-containing PICs. We therefore propose that the presence of aberrant PICs in the cell interferes with the canonical initiation process and significantly contributes to the observed growth defects associated with the loss of the i/TIF34–g/TIF35 mini-module from the rest of eIF3.
Figure 7.
Increased gene dosage of TIF35 partially suppresses growth defects of tif34-DD/KK mutant by preventing formation of the aberrant PICs. (A) High copy expression of TIF35 partially suppresses growth phenotypes of DD/KK. The H450 strains as in Figure 6A were transformed with empty vector or hc TIF35 and spotted in three serial 10-fold dilutions on SD medium and incubated at 34°C and 37°C for 3 days. (B) High dosage of g/TIF35 destabilizes formation of the MFC in vivo. The YAH12 (prt1Δ tif34Δ) strains as in Figure 5C were transformed with hc TIF35 and subjected to Ni2+-chelation chromatography as described in Figure 5A. The histogram shown on the right combines data from Figure 5C and D and this panel; the data were obtained in parallel experiments carried out at the same time. (C) High dosage of g/TIF35 prevents formation of aberrant TC-containing PICs in vivo. The H450 transformants as in (A) were subjected to formaldehyde cross-linking as described in Figure 6A. Proportions of the 40S-bound proteins relative to the amount of 40S subunits are shown in the histogram on the right. The resulting values obtained with the wt strain were set to 100% and those obtained with the DD/KK strain transformed with empty vector or high copy TIF35 were expressed as percentages of the former (SDs are given).
Hence it is likely that the robustly increased tendency to leaky scan initiating AUGs in tif34-DD/KK (Figure 6D) comes from malfunctioning of these erroneous PICs during the AUG recognition process. In strong support of this assumption, high dosage g/TIF35 fully suppressed the severe leaky scanning defect of the DD/KK mutant as well as its Gcn− phenotype (Figure 8A and B). These findings also argue that this unusually severe leaky scanning defect is actually the primary cause of the Gcn− phenotype in the mutant cells. To our knowledge, this type of a direct causal dependence has not been observed before.
Figure 8.
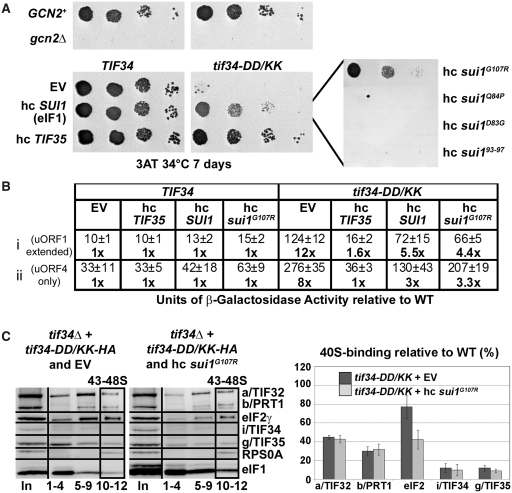
Increased gene dosage of TIF35, SUI1 (eIF1), and its mutant allele sui1G107R suppresses the Gcn− phenotype of tif34-DD/KK as well as its severe leaky scanning defect; and high copy sui1G107R disrupts aberrant PICs in vivo. (A) The H450 strains as in Figure 6A were transformed with empty vector, hc SUI1 (eIF1) or its mutant alleles, and with hc TIF35, respectively, spotted in four serial 10-fold dilutions on 3-AT containing SD media and tested for growth at 34°C for 7 days. (B) The strains as in (A) were further transformed with constructs shown in Figure 6D and analyzed as described in there. (C) High dosage of sui1G107R disrupts the aberrant TC-containing PICs in vivo. The H450 transformants as in panel A were subjected to formaldehyde cross-linking as described in Figure 6A. Proportions of the 40S-bound proteins relative to the amount of 40S subunits are shown in the histogram on the right. The resulting values obtained with the wt strain were set to 100% and those obtained with the DD/KK strain transformed with empty vector or hc sui1G107R were expressed as percentages of the former (SDs are given).
Increased gene dosage of SUI1 (eIF1) selectively suppresses the Gcn− and leaky scanning phenotypes of the tif34-DD/KK mutant
Initiation factors eIF1, eIF1A and eIF5 are considered to be the key controllers of the start selection process in eukaryotes (46). To examine their effect in the background of our leaky scanning DD/KK mutant, we overexpressed them individually in the DD/KK mutant strain while scoring for suppression of its Gcn− phenotype on 3-AT plates. Interestingly, overexpressing eIF1 but not eIF1A and eIF5 markedly suppressed the Gcn− but not the Slg− phenotype of DD/KK (Figures 8A and data not shown). To test whether this pronounced effect requires stable PIC association of eIF1, we examined eIF1 mutants with either severely weakened (sui1Q84P, sui1D83G, sui193-97) or wt (sui1G107R) PIC affinity (41,47) for their ability to suppress the Gcn− phenotype. Out of these mutants, only high copy (hc) sui1G107R with wt PIC affinity was capable of nearly wt Gcn− phenotype suppression (Figure 8A, right-hand panel). Hence, in order to suppress the DD/KK mutant, eIF1 must be capable of stable 40S-binding. Accordingly, high dosage of wt eIF1 as well as sui1G107R also considerably suppressed the leaky scanning defect (Figure 8B). As expected, overexpressing eIF1A and eIF5 had no effect (data not shown). These experiments thus further support the idea that the leaky scanning defect is the main contributor to the failure of mutant cells to induce GCN4 expression under starvation conditions.
In contrast to increased dosage of g/TIF35, which destabilized formation of the MFC in vivo (Figure 7B), Ni2+-chelation experiments from WCE of the tif34-DD/KK cells overexpressing either wt eIF1 or sui1G107R repeatedly showed no further destabilization of the MFC integrity in the mutant cells (data not shown). However, we did observe a clear negative effect of overexpressing sui1G107R on the amount of TC associated with 40S subunits in the DD/KK mutant in vivo reminiscent of that displayed by hc TIF35 (Figure 8C compare with Figure 7C). Interestingly, no similar TC reduction was detected in cells overexpressing wt eIF1 (data not shown).
DISCUSSION
In this study we show that residues 654–700 of b/PRT1 are sufficient for i/TIF34 binding and present, to our knowledge, the first atomic-resolution structure of the interaction between two essential core eIF3 subunits. Our structure reveals that the two major contacts between i/TIF34 and b/PRT1(654–700) occur on the bottom side of the i/TIF34 β-propeller through conserved residues in blades 5 and 6. Disrupting the first contact between D207 and D224 of i/TIF34 and the corresponding Y677 and R678 of b/PRT1 produces lethal or Ts− phenotypes and severely diminishes MFC- and 40S-association of i/TIF34, and also that of g/TIF35, in vivo (Figures 3–6). Importantly, it also destabilizes binding of the rest of eIF3 and eIF5 to the 40S subunit (Figure 6). Likewise, disrupting the second contact between W674 of b/PRT1 and the hydrophobic pocket of i/TIF34 confers essentially the same phenotypes (Figures 3–5, Supplementary Figure S8D, and data not shown). Together these results clearly indicate that both contacts between b/PRT1 and i/TIF34 critically stabilize association of the i/TIF34–g/TIF35 mini-module with the rest of eIF3, in close co-operation with those contacts that g/TIF35 makes with both subunits. This in turn ensures efficient loading of these two small eIF3 subunits onto the 40S where they are subsequently required for proper assembly of the 48S PICs.
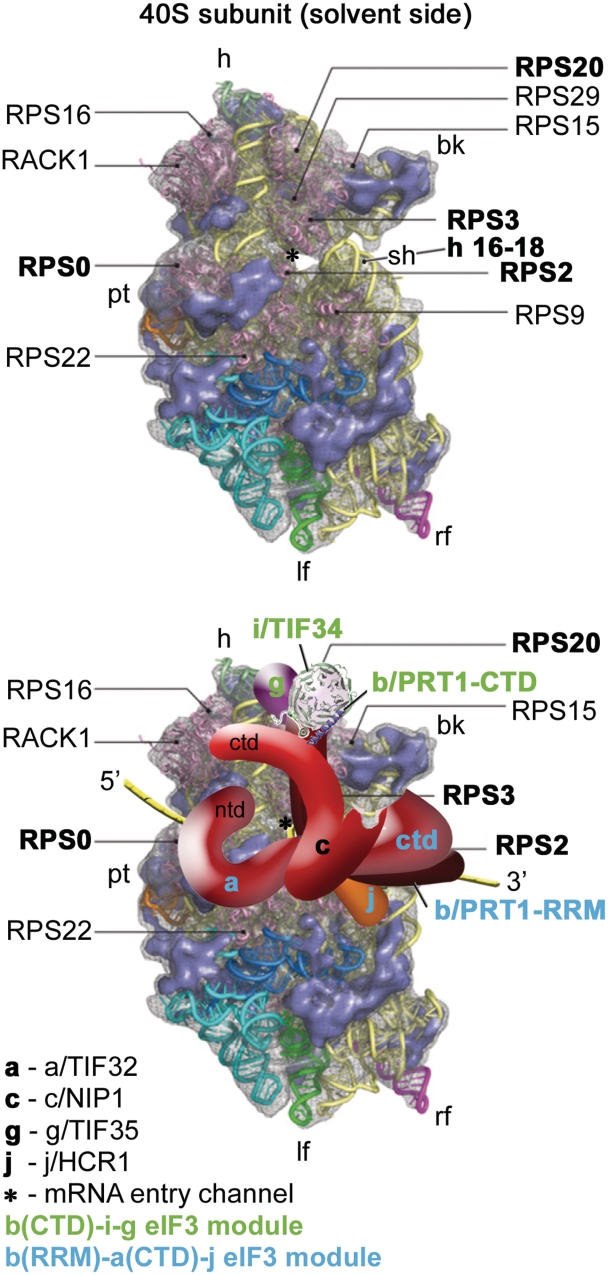
Several important intermolecular bridges between yeast eIF3 and the solvent-exposed side of the 40S ribosome were previously identified, including those between the NTD of a/TIF32 and the small ribosomal protein RPS0A, the a/TIF32-CTD and helices 16–18 of 18S rRNA and RPS2 and RPS3, and the CTD of j/HCR1 and RPS2 (12,37,44,48). Note that both RPS2 and 3 are situated near the mRNA entry pore (Figure 9, upper panel). Besides them, the CTD of c/NIP1 and the b/PRT1-RRM were also shown to critically contribute to the eIF3 affinity for the 40S subunit; however, their binding partners remain to be identified (12,37). Whether or not the remaining eIF3 subunits in i/TIF34 and g/TIF35 likewise participate in this functionally crucial eIF3-ribosome binding activity remained unclear until now. On the one hand a partial subcomplex composed of i/TIF34, g/TIF35 and b/PRT1 lacking its N-terminal RRM showed zero 40S-binding affinity in vivo (9). On the other hand, another subcomplex comprising c/NIP1, the critical N-terminal half of a/TIF32, and eIF5 showed a substantial affinity for the 40S subunits in vivo, though not as strong as that of the wt 6-subunit eIF3 (37). Based on these findings and the data presented here, we propose that whereas the major and essential driving force of the 40S-binding affinity of yeast eIF3 lies in the three largest subunits, as proposed earlier (37), i/TIF34 and g/TIF35 provide complementary 40S-binding activity that is required for stabilization of the entire 48S PICs. It is therefore conceivable that the proper establishment of all intermolecular bridges between eIF3 and the 40S ribosome is needed to ensure precise positioning of eIF3 on the small subunit and thereby flawless functioning of eIF3 not only in formation of 43S and 48S PICs, but also in the subsequent initiation steps.
Figure 9.
A model of two eIF3 modules bound to the opposite termini of the scaffold b/PRT1 subunit situated near the mRNA entry channel of the 40S subunit. (Upper panel) The Cryo-EM reconstruction of the 40S subunit is shown from the solvent side with ribosomal RNA represented as tubes. Ribosomal proteins, with known bacterial homologs and placement, are shown as pink cartoons and labeled (adapted from (57)). Positions of RPS0, 2, 3 and 20 and 18S rRNA helices 16–18 are highlighted in bold. The mRNA entry channel is designated by an asterisk. (Lower panel) Hypothetical location of S. cerevisiae eIF3 on the back side of the 40S subunit based on the data presented in this study and elsewhere, including the interactions between RPS0 and a/TIF32-NTD; RPS2 and j/HCR1; RPS2 and 3 and a/TIF32-CTD; helices 16-18 of 18S rRNA and a/TIF32-CTD; and RPS3 and 20 and g/TIF35 (see text for details). The schematic representations of b/PRT1-CTD and i/TIF34 were replaced with the X-ray structure as in Figure 4C. Two eIF3 modules represented by the b/PRT1-CTD–i/TIF34–g/TIF35 and the b/PRT1-RRM–a/TIF32-CTD–j/HCR1 are color-coded in green and blue, respectively. The yellow lines represent mRNA.
This last notion resonates with our findings that the DD/KK mutation produces severe Gcn− phenotype owing to the robustly increased skipping of the AUG start codon (Figure 6). It is assumed that this so-called leaky scanning phenotype results from an inability of the 48S PIC to switch from the open/scanning-conducive conformation of the 40S head region to the closed/scanning-arrested one that occurs upon AUG recognition and is strictly regulated by eIFs 1 and 1A (reviewed in (46)). The conformational change upon scanning arrest is characterized by dissolution of the contact between RPS3 and helix 16 of 18S rRNA and reformation of the helix18–helix34-RPS3 connection designated as the latch on the mRNA entry channel (49). Interestingly, the leaky scanning phenotype was also observed with mutations disrupting the web of interactions among the a/TIF32-CTD, the b/PRT1-RRM and j/HCR1 located most likely below the mRNA entry channel (12,48) (Figure 9, lower panel). Likewise, g/TIF35 was recently found to interact with RPS3 and RPS20, two subunits located above the mRNA entry channel (5) (Figure 9). This advocates that both termini of b/PRT1 and their associated eIF3 subunits can influence the precision and/or timing of this critical conformational transition upon AUG start codon recognition (see our model in Figure 9, lower panel). We envisage three functional consequences of our mutants that could explain the leaky scanning as well as growth defects.
First, the lack of i/TIF34 and g/TIF35 may change the overall conformation and/or orientation of the rest eIF3 on the ribosome in a way that interferes with dynamics of the latch closing upon start codon recognition, allowing the 40S ribosome to skip the authentic AUG codon and continue scanning downstream. The aforementioned fact that g/TIF35 interacts with RPS20 and mainly with RPS3, which is one of the main components of the ‘latch mechanism’ (5), is consistent with this hypothesis. If true, the scaffold b/PRT1 subunit would serve to connect two eIF3 modules at each of its termini as indicated in Figure 9 (a/TIF32-CTD–j/HCR1 at the N-terminal RRM, and i/TIF34–g/TIF35 at the C-terminal α-helix) that would work together and with c/NIP1 (39) and other eIFs (46) to fine-tune the AUG selection process.
The second contributor to the leaky scanning phenotype might originate from defects in the interaction of the PICs with mRNA in the background of the disrupted interactions between eIFs. It is evident now that eIF3 is crucial for productive mRNA recruitment (50,51) and there is also evidence that eIF3 directly interacts with mRNA (5,44,52). This might suggest that changes in how eIF3 interacts with the PICs could negatively affect the way the mRNA interacts with the mRNA binding channel having an ultimate impact on fidelity of start codon recognition.
The third contribution could arise from our finding that breaking the contact between b/PRT1 and i/TIF34 subunits leads to accumulation of erroneous PICs containing the TC and eIF1 (Figure 6A), the amounts of which can be reduced by overexpressing TIF35 (Figure 7C) or sui1G107R (Figure 8C). The fact that high dosage of g/TIF35 suppresses the Gcn− and partially also the Slg− phenotypes of tif34-DD/KK but paradoxically does not better the assembly of the MFC and 48S PICs indicates that the presence of these aberrant complexes has a dominant negative effect on the initiation process reducing the overall fitness of mutant cells. Given the lack of any precedent for these repeatedly observable effects, we can only hypothesize that such forms of aberrant PICs would originate from simultaneous binding of all eIFs associated around eIF3 in the MFC to the ribosome followed by rapid dissociation of the unstably bound eIF3 (since lacking the i/TIF34–g/TIF35 mini-module) and eIF5 (eIF5 is known to bind to eIF3 very tightly (3)). It is assumed that the MFC has to undergo a relatively extensive rearrangement when it associates with the 40S subunit (37), as eIFs 1, 2, and most probably also 5 occur on the 40S interface side, whereas eIF3 binds to the solvent-exposed side [reviewed in (2)]. Hence we can stipulate that eIF3 and eIF5 preferentially fall off during this rearrangement period. We further stipulate that the resulting TC-eIF1-rich PICs would have the TC locked in a conformation conducive to scanning but incompatible with initiation (the so-called the Pout conformation of the TC), as recently described for some eIF1A mutants (53), and as such they would be skipping the AUG start codons with dramatically increased frequency, as observed with our mutants. How to explain the paradox mentioned above? We propose that the increased dosage of g/TIF35 completely eliminates the MFC-driven PIC assembly pathway that is considered to be the most efficient way of the PIC formation (1). As a consequence, PICs in the mutant cells overexpressing g/TIF35 must form solely by a less efficient yet still fully functional stochastic association of individual eIFs with the 40S ribosome, reminiscent of the bacterial initiation reaction, with no need for ribosomal rearrangement.
The fact that high dosage of sui1G107R also reduced amounts of TC-containing PICs in tif34-DD/KK (Figure 8C) but, in contrast to high copy TIF35, did not further destabilize the MFC in the mutant cells suggests a different molecular mechanism of suppression. In the light of earlier observations with reconstituted translational systems showing that mammalian eIF1 disrupts aberrantly formed PICs in vitro (54), we could speculate that an increased dosage of eIF1 disrupts the aberrant PICs directly also in vivo. In support, we found that the sui1G107R mutant, which in contrast to other sui1 mutants decreases the fidelity of start codon recognition without increasing the rate of eIF1 release from the PICs, suppressed the Gcn− and leaky scanning phenotypes of the DD/KK mutant like wt eIF1 (Figure 8A and B). These results imply that the eIF1 effect in DD/KK cells requires its stable physical presence on the 40S ribosome, where it could directly manipulate the aberrant PICs. However, at odds with this scenario, overexpression of wt eIF1 had no impact on the amounts of the 40S-bound TC in tif34-DD/KK. Hence at present we cannot offer any satisfactory explanation for this rather intriguing observation.
The aforementioned paradox that overexpressing TIF35 significantly suppresses the growth defect of the tif34-DD/KK mutant without rescuing the 40S association of eIF3 and eIF5 may also evoke a notion that the yeast cells can grow at wt rates with only a relatively small fraction (~40%) of the normal amounts of eIF3, eIF2 and eIF5 bound to 40S PICs. While we indeed cannot rule out this possibility, we think that in reality the 40S-binding of all factors in the living cells is most likely not as dramatically affected as we see in our cross-linking experiments, where the effect of a weaker 40S-association might get magnified by a mechanical breakage of cells and all other in vitro manipulations.
Finally, it should also be noted that we recently generated and analyzed two specific mutations in i/TIF34 and g/TIF35, neither of which had any effects on eIF3 and PIC assembly, that produced a severe slow scanning defect and significantly reduced processivity of scanning through stable secondary structures (5). Neither of these scanning phenotypes was, however, found associated with the W674A and DD/KK mutations analyzed here. This is not surprising, since the latter mutations affect translation by destabilizing the MFC and PIC formation, while the former mutations, which have no assembly defects, primarily disrupt eIF3 functions downstream of the 48S PIC assembly. This is consistent with the dual role of eIF3 as a central hub required for mRNA binding to the 43S PIC (48,50,51) as well as a crucial player in scanning and AUG recognition (5,12,39). It will now be intriguing to investigate the binding determinants of g/TIF35 in a partial subcomplex with i/TIF34 and b/PRT1-CTD and their interactions with the 40S subunit using the powerful combination of structural, genetic and biochemical approaches in order to continue replacing the hic sunt leones on our illustrative model of eIF3 with real structures as presented in Figure 4C.
ACCESSION CODES
Structure factors and coordinates for the i/TIF34-b/PRT1(654-700) complex have been deposited in the Protein Data Bank under accession code 3zwl.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–9, Supplementary Methods, Supplementary References [58–64].
FUNDING
The Wellcome Trust grant (090812/B/09/Z); Howard Hughes Medical Institute, and Inst. Research Concept (AV0Z50200510 to L.S.V.); the Medical Research Council and HFSP (RGP0024/2008 to P.J.L.); EMBO Long-term fellowship (to D.D.). Funding for open access charge: The Wellcome Trust grant (090812/B/09/Z).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Jon Lorsch, Monica Liu and Phil Evans for critical reading of the manuscript and to an unknown reviewer for his insightful comments. We are indebted to Phil Evans and Gerhard Reitmayr for help with diffraction data analysis and Python scripting. We also gratefully acknowledge members of Lukavsky, Valášek, Neuhaus, Nagai and Krásný laboratories for numerous discussions.
REFERENCES
- 1.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CUT, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Phan L, Schoenfeld LW, Valášek L, Nielsen KH, Hinnebusch AG. A subcomplex of three eIF3 subunits binds eIF1 and eIF5 and stimulates ribosome binding of mRNA and tRNAiMet. EMBO J. 2001;20:2954–2965. doi: 10.1093/emboj/20.11.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naranda T, Kainuma M, McMillan SE, Hershey JWB. The 39-kilodalton subunit of eukaryotic translation initiation factor 3 is essential for the complex's integrity and for cell viability in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:145–153. doi: 10.1128/mcb.17.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cuchalová L, Kouba T, Herrmannová A, Danyi I, Chiu W-l, Valášek L. The RNA recognition motif of eukaryotic translation initiation factor 3g (eIF3g) is required for resumption of scanning of posttermination ribosomes for reinitiation on GCN4 and together with eIF3i stimulates linear scanning. Mol. Cell. Biol. 2010;30:4671–4686. doi: 10.1128/MCB.00430-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser CS, Lee JY, Mayeur GL, Bushell M, Doudna JA, Hershey JW. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40S ribosomal subunits in vitro. J. Biol. Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- 8.Zhou M, Sandercock AM, Fraser CS, Ridlova G, Stephens E, Schenauer MR, Yokoi-Fong T, Barsky D, Leary JA, Hershey JW, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl Acad. Sci. USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valášek L, Nielsen KH, Hinnebusch AG. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asano K, Phan L, Anderson J, Hinnebusch AG. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:18573–18585. doi: 10.1074/jbc.273.29.18573. [DOI] [PubMed] [Google Scholar]
- 11.ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J. Biol. Chem. 2007;282:8165–8174. doi: 10.1074/jbc.M610860200. [DOI] [PubMed] [Google Scholar]
- 12.ElAntak L, Wagner S, Herrmannová A, Karásková M, Rutkai E, Lukavsky PJ, Valášek L. The indispensable N-terminal half of eIF3j co-operates with its structurally conserved binding partner eIF3b-RRM and eIF1A in stringent AUG selection. J. Mol. Biol. 2010;396:1097–1116. doi: 10.1016/j.jmb.2009.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valášek L, Phan L, Schoenfeld LW, Valášková V, Hinnebusch AG. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recognition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. 2005;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- 15.Dodd RB, Allen MD, Brown SE, Sanderson CM, Duncan LM, Lehner PJ, Bycroft M, Read RJ. Solution structure of the Kaposi's sarcoma-associated Herpesvirus K3 N-terminal domain reveals a novel E2-binding C4HC3-type RING domain. J. Biol. Chem. 2004;279:53840–53847. doi: 10.1074/jbc.M409662200. [DOI] [PubMed] [Google Scholar]
- 16.Muchmore SW, Sattler M, Liang H, Meadows RP, Harlan JE, Yoon HS, Nettesheim D, Chang BS, Thompson CB, Wong S-L, et al. X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature. 1996;381:335–341. doi: 10.1038/381335a0. [DOI] [PubMed] [Google Scholar]
- 17.Szymczyna BR, Taurog RE, Young MJ, Snyder JC, Johnson JE, Williamson JR. Synergy of NMR, computation, and X-ray crystallography for structural biology. Structure. 2009;17:499–507. doi: 10.1016/j.str.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leslie A. The integration of macromolecular diffraction data. Acta Cryst.Section D. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 19.Evans P. Scaling and assessment of data quality. Acta Cryst. Section D. 2006;62:72–82. doi: 10.1107/S0907444905036693. [DOI] [PubMed] [Google Scholar]
- 20.French S, Wilson K. On the treatment of negative intensity observations. Acta Cryst Section A. 1978;34:517–525. [Google Scholar]
- 21.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 22.de La Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 23.Kelley LA, Sternberg MJE. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protocols. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 24.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Cryst. Section D. 2005;61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 25.Zhang KY, Cowtan K, Main P. Combining constraints for electron-density modification. Methods Enzymol. 1997;277:53–64. doi: 10.1016/s0076-6879(97)77006-x. [DOI] [PubMed] [Google Scholar]
- 26.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Cryst. Section D. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. Section D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. Section D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coyle SM, Gilbert WV, Doudna JA. Direct Link between RACK1 function and localization at the ribosome in vivo. Mol. Cell. Biol. 2009;29:1626–1634. doi: 10.1128/MCB.01718-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, Roberts R. Human genome and diseases: WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jennings BH, Pickles LM, Wainwright SM, Roe SM, Pearl LH, Ish-Horowicz D. Molecular recognition of transcriptional repressor motifs by the WD domain of the groucho/TLE corepressor. Mol. Cell. 2006;22:645–655. doi: 10.1016/j.molcel.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the Î2-propeller of clathrin. Proc. Natl Acad. Sci. USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 36.Gallivan JP, Dougherty DA. Cation-pi interactions in structural biology. Proc. Natl Acad. Sci. USA. 1999;96:9459–9464. doi: 10.1073/pnas.96.17.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Valášek L, Mathew A, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. The yeast eIF3 subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Valášek L, Szamecz B, Hinnebusch AG, Nielsen KH. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- 39.Valášek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J. 2007;26:1602–1614. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nanda JS, Cheung Y-N, Takacs JE, Martin-Marcos P, Saini AK, Hinnebusch AG, Lorsch JR. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 2009;394:268–285. doi: 10.1016/j.jmb.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 43.Nielsen KH, Szamecz B, Valasek L, Jivotoskaya A, Shin BS, Hinnebusch AG. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valášek L. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev. 2008;22:2414–2425. doi: 10.1101/gad.480508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Munzarová V, Pánek J, Gunišová S, Dányi I, Szamecz B, Valášek LS. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet. 2011;7:e1002137. doi: 10.1371/journal.pgen.1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell SF, Lorsch JR. Should I stay or should i go? Eukaryotic translation initiation factors 1 and 1a control start codon recognition. J. Biol. Chem. 2008;283:27345–27349. doi: 10.1074/jbc.R800031200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiu W-L, Wagner S, Herrmannova A, Burela L, Zhang F, Saini AK, Valasek L, Hinnebusch AG. The C-terminal region of eukaryotic translation initiation factor 3a (eIF3a) promotes mRNA recruitment, scanning, and, together with eIF3j and the eIF3b RNA recognition motif, selection of AUG start codons. Mol. Cell. Biol. 2010;30:4415–4434. doi: 10.1128/MCB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 50.Mitchell SF, Walker SE, Algire MA, Park E-H, Hinnebusch AG, Lorsch JR. The 5′-7-Methylguanosine Cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol. Cell. 2010;39:950–962. doi: 10.1016/j.molcel.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jivotovskaya A, Valášek L, Hinnebusch AG, Nielsen KH. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CUT, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saini AK, Nanda JS, Lorsch JR, Hinnebusch AG. Regulatory elements in eIF1A control the fidelity of start codon selection by modulating tRNAiMet binding to the ribosome. Genes Dev. 2010;24:97–110. doi: 10.1101/gad.1871910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 55.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc. Natl Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35:W522–W525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Taylor DJ, Devkota B, Huang AD, Topf M, Narayanan E, Sali A, Harvey SC, Frank J. Comprehensive molecular structure of the eukaryotic ribosome. Structure. 2009;17:1591–1604. doi: 10.1016/j.str.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuzmic P. DynaFit--a software package for enzymology. Methods in Enzymology. 2009;467:247–280. doi: 10.1016/S0076-6879(09)67010-5. [DOI] [PubMed] [Google Scholar]
- 59.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: Application to HIV proteinase. Analyt. Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 60.Nielsen KH, Valášek L. In vivo deletion analysis of the architecture of a multi-protein complex of translation initiation factors. Methods Enzymol. 2007;431:15–32. doi: 10.1016/S0076-6879(07)31002-1. [DOI] [PubMed] [Google Scholar]
- 61.Grant CM, Miller PF, Hinnebusch AG. Requirements for intercistronic distance and level of eIF-2 activity in reinitiation on GCN4 mRNA varies with the downstream cistron. Mol. Cell. Biol. 1994;14:2616–2628. doi: 10.1128/mcb.14.4.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cigan AM, Foiani M, Hannig EM, Hinnebusch AG. Complex formation by positive and negative translational regulators of GCN4. Mol. Cell. Biol. 1991;11:3217–3228. doi: 10.1128/mcb.11.6.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 64.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.