Abstract
Anti-miRs are oligonucleotide inhibitors complementary to miRNAs that have been used extensively as tools to gain understanding of specific miRNA functions and as potential therapeutics. We showed previously that peptide nucleic acid (PNA) anti-miRs containing a few attached Lys residues were potent miRNA inhibitors. Using miR-122 as an example, we report here the PNA sequence and attached amino acid requirements for efficient miRNA targeting and show that anti-miR activity is enhanced substantially by the presence of a terminal-free thiol group, such as a Cys residue, primarily due to better cellular uptake. We show that anti-miR activity of a Cys-containing PNA is achieved by cell uptake through both clathrin-dependent and independent routes. With the aid of two PNA analogues having intrinsic fluorescence, thiazole orange (TO)-PNA and [bis-o-(aminoethoxy)phenyl]pyrrolocytosine (BoPhpC)-PNA, we explored the subcellular localization of PNA anti-miRs and our data suggest that anti-miR targeting of miR-122 may take place in or associated with endosomal compartments. Our findings are valuable for further design of PNAs and other oligonucleotides as potent anti-miR agents.
INTRODUCTION
MicroRNAs (miRNAs) are endogenous small non-coding RNAs that regulate gene expression post-transcriptionally. Many miRNAs have been described in different species and with functions associated with a wide range of cellular processes and human diseases (1,2). Although alternative miRNA biogenesis pathways have been uncovered recently (3–5), the principal biosynthetic route involves processing in the nucleus of an initial primary miRNA transcript (pri-miRNA) by an RNase III type enzyme, Drosha, resulting in a ~70–80 nt long stem-loop precursor-miRNA (pre-miRNA). The pre-miRNA is then exported to the cytosol where another RNase III type enzyme, Dicer, removes the loop to give an imperfectly paired double-stranded mature miRNA of ~22 nt long. One miRNA strand is then selected by a ribonucleoprotein complex necessary for miRNA activity, miRISC (6). Upon loading in miRISC, miRNAs bind to the 3′-untranslated region (UTR) of their mRNA targets and repress their expression by inducing mRNA degradation and/or by preventing mRNA translation (7–9). MiRNA targets are recognized by the miRNA mainly through the miRNA ‘seed’ sequence comprising nucleotides 2–8 at the 5′-end of the miRNA (10). miRNA loading into miRISC pre-organizes the seed sequence for target recognition (11,12). Apart from these nucleotides, there is no need for extensive complementarity between the miRNA and its mRNA target, which thus allows one miRNA to be able to regulate many mRNAs (10).
Although the cellular site of mRNA repression by miRNAs is unclear, it is believed that miRNAs become associated with certain cytoplasmic foci and endosomal compartments. Cytosolic foci known as P-bodies contain not only miRNAs and their target mRNAs but also other key components of miRISC such as Argonaute proteins (Ago) and Trinucleotide-Repeat-Containing-Gene-6 (TNRC6) (13). Similarly, GW-Bodies (bodies enriched in GW182, a protein that binds Ago), which were found to be independent of P-bodies, also contain the same miRISC components, often associated with multivesicular body (MVB)-like membranes (14). MiRNAs have also been found in exosomes (14,15) and more recently also in the nucleus (16) and mitochondria (17).
Oligonucleotide analogues (ON) complementary to miRNAs can inhibit their function and are known as anti-miRs. Anti-miRs have been used extensively as tools to gain understanding of specific miRNA functions and as potential therapeutics (18–23). The first anti-miR chemistries were based on 2′-O-methyl ribonucleosides (18,24–27). Later, other anti-miR chemistries became more promising, such as 2′-O-methoxyethyl (MOE) (19,28) and 2′-fluoro/2′-methoxyethyl mixmers (2′F/MOE) (29), charge neutral ONs, such as Peptide Nucleic Acids (PNA) (22,30–32) and high-affinity Locked Nucleic Acid (LNA) in mixmers with DNA (21,23,33,34), OMe (30,32,34) or MOE (28,29). To date only an LNA/DNA mixmer has advanced to clinical trials (Santaris Pharma News release 23 September 2010).
Further development of anti-miRs as reagents and therapeutics is likely to benefit from an improved understanding of the anti-miR biology: the chemical structure requirements for anti-miR targeting as well as mechanistic aspects, such as cell uptake and subcellular localization. To this end, we have chosen to study miR-122, which is a 23-mer-long, liver-specific miRNA that has generated considerable interest as a therapeutic target (23). Inhibition of miR-122 has been shown to lead to lowering of plasma cholesterol levels in mice and primates (18,19,21). Furthermore, miR-122 is required for hepatitis C virus (HCV) replication (26), such that treatment with an LNA/DNA anti-miR leads to suppression of HCV viremia in primates (23).
We and others have shown that PNAs are highly effective as anti-miRs (22,30–32). We reported that a 23-mer PNA fully complementary to miR-122, containing three Lys residues at the C-terminus and one Lys at the N-terminus as well as a Cys residue for post-synthetic modifications (Cys-K-PNA23mer-K3) effectively inhibited miR-122 in a human hepatic cell line (Huh7) and in primary rat hepatocytes without the use of any transfection agents, as seen by up-regulation of miR-122 mRNA targets (30). More recently, we showed that the same PNA anti-miR compared as well in inhibitory activity in a miR-122-sensitive luciferase reporter system in Huh7 cells as an LNA/OMe anti-miR containing phosphorothioate (PS) linkages, each delivered in the absence of transfection agents (32). We also showed efficient inhibition of miR-155 by a 23-mer PNA anti-miR having the structure K-PNA23mer-K3 in B-cells and in mice, again without use of transfection agents (22).
In our new studies, we have explored various aspects of targeting of miR-122 by cationic PNA anti-miRs. We first addressed the PNA sequence and attached amino acid requirements for efficient miRNA targeting and show that activity of the PNA anti-miR is enhanced substantially by the presence of a free thiol group. We demonstrate that this is due primarily to better cellular uptake, an unanticipated discovery. We also investigated the mechanisms of cellular uptake for Cys-K-PNA-K3 and shed light on a possible subcellular compartment where the targeting of miR-122 by PNA anti-miRs might occur. Our data should prove useful for further design of PNAs and other ONs as potent anti-miR agents.
MATERIALS AND METHODS
PNA synthesis
PNA ONs were synthesised as previously described (22) and their sequences are shown in Table 1, with the exceptions of Met-K-PNA23mer-K3, Ser-K-PNA23mer-K3 and MPA-K-PNA23mer-K3. These three PNA ONs had the same sequence as Cys-K-PNA23mer-K3 but the terminal Cys residue was replaced by methionine, serine or 3-mercaptopropionic acid, respectively. RNA oligonucleotide having the same sequence as miR-122 (5′-UGGAGUGUGACAAUGGUGUUUGU-3′) was purchased from Dharmacon. TO-PNAs were synthesized on 2 µmol scale on Merck NovaSyn TGR resin preloaded with the C-terminal Lys (80 µmol/g) by using an Intavis ResPep parallel synthesizer and 1 ml polyethylene syringe reactors. For Fmoc deprotection the resin was treated with a solution of piperidine/DMF (1:4, 200 µl) for 2 min followed by draining of the reactor. The procedure was repeated and the resin washed with DMF (3 × 200 µl). For couplings, stock solutions were prepared: PNA monomers in NMP (0.2 M), Fmoc-Lys(Boc) in NMP (0.3 M), Fmoc-TO-monomer in NMP (0.2 M), HCTU in NMP (0.3 M), NMM in DMF (4 M). Solutions were combined for preactivation (6 eq Lys, 5.4 eq HCTU, 18 eq NMM or 4 eq PNA monomer, 3.6 eq HCTU, 12 eq NMM) and after 2 min transferred to the syringe reactor. The reactor was drained after 30 min. Double coupling and triple coupling was used when necessary. The resin was washed with DMF (3 × 200 µl). For capping, the resin was treated with DMF/acetic anhydride/2,6-lutidine (89:5:6, 200 µl) for 2 min, and washed with DMF (3 × 200 µl). For final cleavage the resin was washed with CH2Cl2 (10 × 200 µl), and dried. The resin was treated for 1 h with a solution of L-cysteine methyl ester hydrochloride (7.5 mg) in TFA/m-cresol/water (93:5:2, 200 µl). After draining, the resin was washed with TFA (2 × 200 µl). Work-up and purification was performed as previously described (35). Cys-K-(TO)PNA-K3 and K-(TO)PNA-K3 were synthesized at 40 µmol scale as previously described (22) but with manual coupling of the TO-PNA monomer. BoPhpC-PNA was synthesized purified and characterized as previously described (36). Briefly, PNAs were synthesized using an Applied Biosystems 433A peptide synthesizer and standard Fmoc chemistry on a 10 µmol scale and on a Rink amide resin. Crude oligomers were purified by RP-HPLC and their identities confirmed by HRMS (ESI-TOF). For PNA Cys-alkylation, 100 nmoles PNA ONs dissolved in water was incubated for 10 min at room temperature with 10 equivalents N-ethylmaleimide (NEM). The reaction was monitored by MALDI-TOF mass spectrometry. Upon completion, the reaction was purified by RP-HPLC, the product lyophilized and re-suspended in water.
Table 1.
Sequences of PNAs used in this study. Indicated in the grey box is the miR-122 ‘seed’ sequence-targeting region
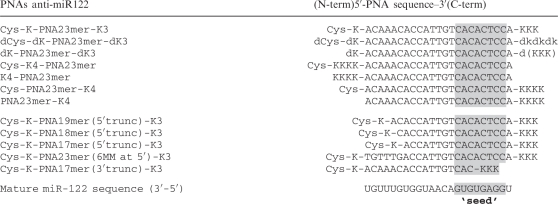 |
Note that while PNA sequences are written from 5′/N-term to 3′/C-term, the mature miR-122 sequence is indicated from 3′ to 5′ to show base pairing between the miRNA and the anti-miRs.
Cell culture and transfections
Huh7 cells and HEK293ET cells were maintained in Dulbecco's modified eagle medium containing 10% foetal bovine serum and Penicillin/Streptomycin antibiotics (Full Media: DMEM/FBS/PenStrep) at 37°C/5% CO2. Lipofections were carried out for 4 h using Lipofectamine 2000 following the manufacturer's protocols in 150 µl final volume of transfection. Following lipofection, cells were washed with PBS and media was replaced by Full Media. All PNA ONs were delivered in the absence of transfection agents in serum-free media (opti-MEM) unless stated otherwise, as described previously (32).
Luciferase assays
MiR-122 dual luciferase reporter system and luciferase assays was described previously (32). Unless stated otherwise, Huh7 cells were lipofected with 100 ng miR-122 dual luciferase plasmid and the next day washed and incubated with PNA anti-miRs for 4 h in serum-free media (opti-MEM). After PNA incubation, cells were washed and grown in Full Media until luciferase measurements were carried out. All experiments were carried out in triplicate.
Assays with endocytosis inhibitors
Huh7 cells were plated and lipofected with miR-122 dual-luciferase reporter construct as described previously (32). Cells were washed once with PBS and were incubated for 2 h with 5 µM Cys-K-PNA23mer-K3 in 100 µl opti-MEM or 100 µl of either 30 µM chlorpromazine (CPZ), 2.5 mM methyl-β-cyclodextrin (MBCD), 0.45 M sucrose or 2.5 µg/ml Latrunculin B. All the inhibitor solutions were freshly made up in opti-MEM just before use. For specific inhibition of clathrin-mediated endocytosis (CME), Huh7 cells were co-lipofected with miR-122 dual luciferase reporter construct and with 100 ng AP180-C construct (37) and PNA incubation was carried out the next day in opti-MEM. For AP180-C plus MBCD treatment, Huh7 cells were pre-incubated for 1 h in opti-MEM or opti-MEM containing 2.5 mM MBCD before addition of PNA and cell incubation with PNA was carried out for 1 h. After PNA incubations, cells were washed twice with PBS and 200 µl Full Media was added. 2 h later media was again replaced by 200 µl Full Media. Luciferase expression was measured 48 h after PNA incubation.
Assays with chloroquine and calcium
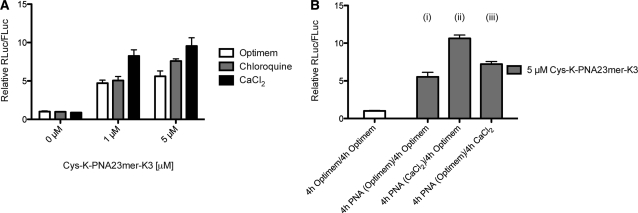
Huh7 cells expressing miR-122 dual luciferase reporter construct were incubated for 4 h with 1 or 5 µM Cys-K-PNA23mer-K3 in 100 µl opti-MEM in the presence or absence of 6 mM CaCl2 or 100 µM chloroquine. After PNA incubation media was replaced by Full Media. Luciferase expression was carried out 48 h after PNA incubation. For Figure 5B, PNA-treated cells were washed twice with 150 µl PBS and media was replaced by 100 µl opti-MEM in the presence or absence of 6 mM CaCl2 but without PNA present. Four hours later, cells were washed and media was replaced by Full Media and Luciferase expression measured after 48 h.
Figure 5.
(A) Effect of 100 µM chloroquine or 6 mM CaCl2 on Cys-K-PNA23mer-K3 anti-miR activity as seen by luciferase assay. Huh7 cells were incubated with the indicated amounts of PNA in the presence or absence of chloroquine or CaCl2. (B) Effect of CaCl2 on Cys-K-PNA23mer-K3 anti-miR activity due to increase of endocytotic uptake or enhanced endosomal escape (see text for details) as seen by luciferase assay.
Cell viability assays
Cell viability assays were carried out using CellTiter 96 AQueous One Solution Cell Proliferation Assay (Promega; cat number 63582) following the manufacturer’s protocol (see Supplementary Methods). The assay is based on the MTS tetrazolium reagent.
FACS analysis
Huh7 or HEK293ET cells were incubated with 3 µM TO-PNA anti-miRs in opti-MEM for 4 h, after which media was replaced by Full Media (see Supplementary Methods). After 24 h of PNA incubation, FACS analysis was performed in a FACSCalibur analyzer measuring 10000 gated events per sample. TO-PNA fluorescence was recorded by excitation at 488 nm (argon laser) and detection at 530 nm. Data was processed using FlowJo version 9.3.2.
Confocal microscopy
Huh7 cells were plated, plasmid transfected and incubated with TO-PNA or BoPhpC-PNA as described in Supplementary Methods. Before visualization by confocal microscopy, cells were washed and fixed with 4% EM-grade paraformaldehyde (PFA; Electron Microscopy Sciences). We obtained similar results when performing live imaging (data not shown). For transferrin co-localization studies, Alexa 594-conjugated human transferrin (Invitrogen; T-13343) at 50 µg/ml was mixed with Cys-K(TO) PNA-K3 at 3 µM in opti-MEM clear media (no phenol red). Cells were incubated with 200 µl transferrin/TO-PNA solution for 1.5 h and then washed 3 times with 200 µl PBS, fixed, nuclei stained and visualized by confocal microscopy. Additional detailed protocols in Supplementary Methods.
Cell fractionation and Immuno-precipitation
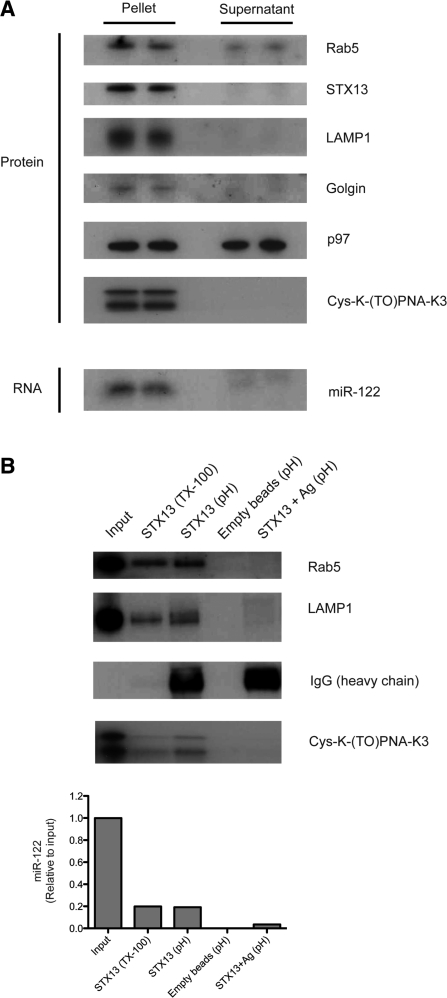
Cell fractionation and immunoprecipitation (IP) experiments were based on Steuble et al. (38) with some modifications (see Supplementary Methods). For Figure 7A, the membrane fraction was resuspended in 12 ml HB, while for IP (Figure 7B) it was suspended in 3.5 ml HB. For STX13 plus antigen IP experiment (Figure 7B, lane 5), beads were incubated with 5 µg pure STX13 protein (Synaptic Systems; 110-13 P) in 500 µl final volume HB for 30 min at 4°C prior to incubation with membrane fraction sample.
Figure 7.
(A) Representative cell fractionation experiment: protein analysis by western blot showing enrichment of markers for membrane-bound compartments in the ‘pellet’ fraction as compared to the ‘supernatant’ (cytosolic) fraction. Cys-K-(TO)PNA-K3 was detected by north-western blot approach (same gel as western Blot). RNA analysis for miR-122 detection by northern blot (same samples as protein gels shown above). (B)Representative experiment of immuno-precipitation for Syntaxin 13 (STX13)-positive compartments. Top panel: western blot and north-western blot for detection of endosomal markers and Cys-K-(TO)PNA-K3, respectively. Input: ~20% IP. Bottom panel: RT–qPCR for miR-122 detection in RNA extracted from samples treated as in B top panel. Input: ~65% IP. Ag: STX13 antigen. TX-100: TritonX-100 elution (mild detergent). pH: pH shock elution.
RNA extraction and protein extraction
RNA was extracted using TRIzol LS (Invitrogen) following the manufacturer’s protocols. The obtained RNA pellet was re-suspended in water and was re-precipitated as described previously (39). Quantification was carried out using a NanoDrop 2000 spectrophotometer (Thermo Scientific). For protein extraction, 200 µl sample obtained after cellular fractionation or IP were thoroughly mixed with 600 µl methanol (MeOH) and 100 µl chloroform. Then 600 µl water was added and mixed. Samples were centrifuged for 5 min at room temperature at 13 000 rpm for phase separation. The upper phase was discarded. 600 µl MeOH was added to the remaining phases, mixed and centrifuged for 15 min at room temperature at 13 000 rpm. The supernatant was discarded and the pellet was air-dried. Samples obtained after IP experiments were re-suspended in 35 µl 4× NuPAGE LDS sample buffer (Invitrogen) and were not quantified. Samples obtained after cell fractionation were re-suspended in 1% SDS and quantified using a QuantiPRO BCA assay kit (Sigma) following the manufacturer's protocol.
Western blot and antibodies
Western blots were carried out using standard procedures (see Supplementary Methods). Primary antibodies used: anti-Rab5 (Sc-46692; Santa Cruz Biotechnology) used at 1:2000 dilution, anti-Lamp1 (H4A3; Developmental Studies Hybridoma Bank) used at 1:10000, anti-Golgin (A-21270; Molecular Probes/Invitrogen) used at 1:1000 dilution, anti-p97 (MA1-21412; Pierce/Thermo Scientific) used at 1:2000 dilution, anti-STX13 (110132; Synaptic Systems) used at 1:10000 dilution. For IP experiments, IgG heavy chain was detected when the membrane was incubated with anti-STX13 (cross-reaction). All secondary antibodies were ZyMax IgG (H+L) HRP Conjugated (Invitrogen) and were used at 1:3000 dilution. All antibodies were diluted in PBS/0.1% Tween20/5% Milk.
North-western blot
Proteins were extracted and electrophoresed in protein gels as described above (and Supplementary Methods). After gel transfer, the lower portion of the membrane (below 17 KDa in size) was cut and incubated in UltraHyb Oligo hybridization buffer (Ambion/Applied Biosystems; AM8663) for 30 min at 42°C. Then, 250 pmol of an RNA oligonucleotide having the same sequence as miR-122 (see above) was 5′-end-radiolabeled using [γ-32P]ATP and added to the membrane-containing hybridization buffer. The membrane was left hybridizing with the radiolabeled probe overnight at 42°C and the next day washed as previously described (39) and exposed to X-ray films.
Northern blot
Northern blots were carried out as previously described (30,39) with one modification: 2.5 µg of RNA was dissolved in 8 M urea/20% formamide loading dye and samples were loaded in 15% TBE-Urea pre-cast gels (Invitrogen) and ran for 65 min at 180 V.
miR-122 reverse transcription quantitative real-time PCR
Quantification of miR-122 by quantitative real-time PCR (RT–qPCR) was carried out essentially as described previously (30) with some modifications. Absolute miR-122 quantification method was carried out using a calibration curve that was prepared by performing serial dilutions of a single stranded RNA oligonucleotide having the same sequence as miR-122. A 5 µl sample was used for cDNA synthesis. Then 9 µl cDNA taken immediately from the cDNA reaction was combined with 11 µl qPCR Master mix for qPCR step.
RESULTS
PNA anti-miR and attached amino acid requirements for efficient miR-122 inhibition in cells
We described recently a convenient reporter system for assessing the potency of anti-miRs against miR-122 (32), which is based on a dual-luciferase miRNA sensor assay (20). Briefly, a miR-122 recognition site, perfectly complementary to the full mature miR-122 sequence was inserted at the 3′-untranslated region (UTR) of Renilla Luciferase (RLuc) to bring RLuc under the control of miR-122 (repressed luciferase expression). In this construct, a second luciferase gene is present (Firefly; FLuc) that is not under miR-122 regulation and which serves as an internal control. When this dual-luciferase construct is introduced into miR-122 positive cells (e.g. Huh7 cells), RLuc is repressed. Upon addition of an anti-miR, de-repression of RLuc is observed as a consequence of anti-miR binding to miR-122, leading to an increase in relative RLuc/FLuc ratio (32). Such an approach has become widely used to measure anti-miR activity (20,21,32–34,40).
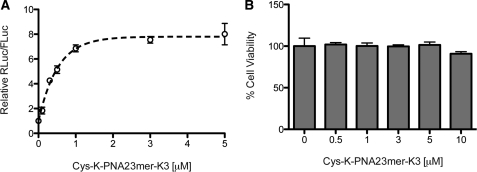
Dual-luciferase reporter Huh7 cells were incubated with increasing amounts of Cys-K-PNA23mer-K3 anti-miR for 4 h in serum-free media in the absence of a transfection agent, followed by cell growth in Full Media for 2 days in the absence of added PNA. This showed dose-dependent RLuc de-repression (miR-122 inhibition) by the PNA anti-miR (Figure 1A). MiR-122 inhibition was seen at sub-micromolar concentrations, as reported recently (32), with saturation above 1 µM anti-miR concentration. Importantly, Cys-K-PNA23mer-K3 did not show any statistically significant decrease in cell viability up to 10 µM (one-way ANOVA with Dunnet's multiple comparison test) (Figure 1B).
Figure 1.
(A) Dose-dependent up-regulation of luciferase expression in Huh7 cells treated with Cys-K-PNA23mer-K3 in the absence of transfection agents. (B) Toxicity assay for Huh7 cells treated as in (A).
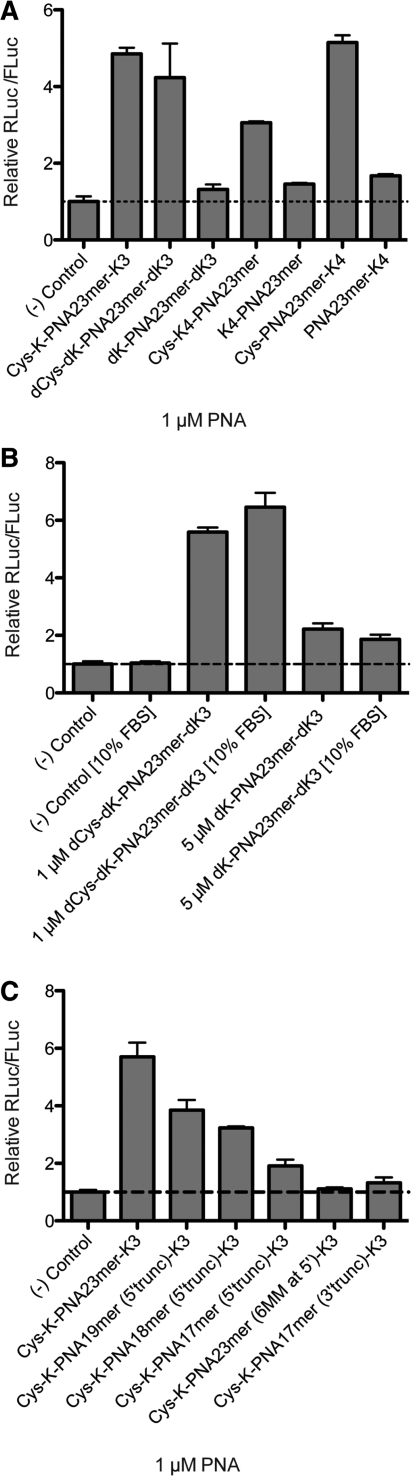
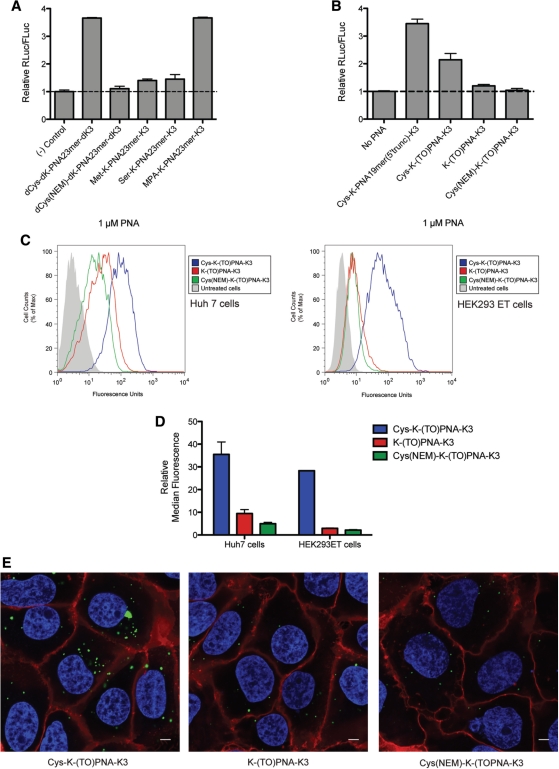
We then asked whether the presence of the N-terminal Cys residue, the positioning of the Lys residues or use of D-amino acids have an effect on anti-miR activity. Dual-luciferase reporter Huh7 cells were treated as before with different cationic PNA anti-miRs (Table 1) at 1 µM, a concentration just below anti-miR activity saturation to ensure maximal dynamic range. Whereas use of all D-amino acids (dCys-dK-PNA23mer-dK3) led to only a mild reduction in miR-122 inhibition (Figure 2A), additional removal of the terminal Cys (dK-PNA23mer-dK3) reduced activity dramatically. Positioning of the Lys residues at the N-terminus of the PNA anti-miR also showed a detrimental effect (Cys-K4-PNA23mer) as compared to the PNA anti-miR containing all four Lys residues at the C-terminus (Cys-PNA23mer-K4). Similarly, upon removal of the terminal Cys, the activity in each case was substantially reduced (K4-PNA23mer and PNA23mer-K4) (Figure 2A). Comparable activity levels were obtained for experiments with PNA anti-miRs with all D-amino acids when pre-incubated with 10% foetal bovine serum (FBS) for 2 h and then added to reporter Huh7 cells for 4 h also in 10% FBS (Figure 2B), suggesting that the cellular uptake of these PNA anti-miRs is not affected by serum proteins. In the case of anti-miR lacking the terminal Cys (dK-PNA23mer-dK3) a higher concentration was used (5 µM) to obtain a better activity comparison between untreated [(-) control] and dK-PNA23mer-dK3 treated cells, but no difference was also seen at 1 µM (data not shown).
Figure 2.
(A) Effect of Cys, D-amino acids and positioning of Lys residues on Cys-K-PNA23mer-K3 anti-miR activity as seen by luciferase assay. Huh7 cells were treated with 1 µM PNAs. (B) Luciferase assay of Huh7 cells treated with dPNAs at the indicated concentrations in the presence or absence of 10% FBS. (C) Effect of truncations and mismatches on Cys-K-PNA-K3 anti-miR activity as seen by luciferase assay. Huh7 cells were treated with 1 µM PNAs.
The activity of Cys-K-PNA-K3 was reduced as the length of the ON was shortened (Figure 2C). However, even a 19-mer PNA anti-miR containing a 4-nucleotide truncation at the 5′-end of the anti-miR [Cys-K-PNA19mer (5′trunc)-K3] was still able to inhibit miR-122 significantly. Interestingly, a 6-nucleotide truncation at the 5′-end of the anti-miR [Cys-K-PNA17mer (5′trunc)-K3] was better tolerated than an equivalent 6 nucleotides mismatch in that position [Cys-K-PNA23mer (6MM at 5′)-K3]. Also, a 6-nt truncation at the 5′-end of the anti-miR [Cys-K-PNA17mer (5′trunc)-K3] was better tolerated than a 6-nt truncation at 3′-end of the anti-miR [Cys-K-PNA17-mer (3′trunc)-K3]. This highlights the importance of targeting the ‘seed’ sequence of the miRNA (Table 1). All the PNA anti-miRs showed similar melting temperatures when hybridized against a fully complementary RNA strand containing the same sequence as miR-122 (Supplementary Table S1). This suggests that there are factors involved in efficient targeting of miRNAs beyond the RNA binding affinity of the anti-miR.
Thiol-dependent enhanced cellular uptake for cationic PNA anti-miRs
We further investigated the function of the terminal Cys residue in PNA anti-miRs and asked if the enhanced anti-miR activity is due to the presence of a free thiol group. PNA23-mer anti-miR was alkylated at the Cys residue using N-ethylmaleimide (NEM) and the anti-miR added to dual luciferase reporter Huh7 cells at 1 µM concentration for 4 h as before. Alkylation (dCys(NEM)-dK-PNA23mer-dK3) led to a dramatic loss in anti-miR activity (Figure 3A), similar to that of dK-PNA23mer-dK3 anti-miR lacking a Cys residue (Figure 2A). Similarly, replacement of the Cys by Met, which contains a methylated thiol group (Met-K-PNA23mer-K3), or by Ser (Ser-K-PNA23mer-K3) resulted in each case in a substantial reduction in anti-miR activity (Figure 3A). In contrast, activity was completely recovered when the Cys was replaced by 3-mercaptopropionic acid (MPA-K-PNA23mer-K3). These data strongly suggest that the enhanced PNA anti-miR activity for Cys-containing PNA anti-miRs is due to the presence of a terminal-free thiol group.
Figure 3.
(A) Effect of Cys modification or replacement within Cys-K-PNA23mer-K3 on anti-miR activity as seen by luciferase assay. Huh7 cells were treated with 1 µM PNAs. (B) Luciferase activity assay for 1 µM TO-PNAs as compared to a sequence-equivalent unmodified PNA anti-miR. (C) Representative FACS histogram for Huh7 (left panel) or HEK293ET (right panel) cells treated with TO-PNAs at 3 µM. (D) Median fluorescence relative to untreated cells observed for Huh7 cells and HEK293ET cells treated as in (C) by FACS analysis; shown are results of three independent experiments. (E) Confocal microscopy of Huh7 cells treated with TO-PNAs at 3 µM. Green, TO-PNAs; blue, Hoechst staining; red, Cell Mask plasma membrane staining. Scale bar corresponds to 5 µm. All three images were taken with the same confocal microscopy settings.
We asked whether the activity increase obtained with a terminal free thiol group was due to enhanced cellular uptake or improved miRNA targeting following cell internalization. For these studies, we made use of a fluorescent PNA in which a PNA nucleobase is replaced by thiazole orange (TO) (41–43) (Supplementary Figure 1A). The TO dye is able to behave as universal base (41). Moreover, the fluorescence emission of the TO-PNA monomer in the context of a PNA ON is enhanced upon binding to a complementary DNA or RNA strand in vitro (41–43) and in cells (43). We carried out a scan of K-PNA19mer-K3 anti-miR by replacement of individual PNA units at different sequence positions by the TO-PNA monomer and by measurement of the fluorescence differences when the PNA is single-stranded (ss-PNA) or in a duplex with a complementary miR-122 RNA (PNA–RNA duplex) at 25 or 37°C (Supplementary Figure S1B and C). A 19-mer anti-miR containing a TO-PNA monomer at the second to last position at the 5′-end [PNA 17, K-(TO)PNA-K3] was amongst the best TO-PNA anti-miRs in terms of fluorescence enhancement upon RNA binding. The K-(TO)PNA-K3 anti-miR also showed enhanced fluorescence by confocal microscopy as compared to the other TO-PNA anti-miRs (data not shown). Further, the K-(TO)PNA-K3 was one of the most active TO-PNA anti-miRs as seen by luciferase up-regulation measurements (data not shown). Importantly, the Cys-K-(TO)PNA-K3 maintained much of the anti-miR activity as compared to Cys-K-PNA19mer-K3 (Figure 3B). Removal of the terminal Cys residue [K-(TO)PNA-K3] or alkylation of the terminal Cys [Cys(NEM)-K-(TO)PNA-K3] led to a substantial loss in anti-miR activity, just as for unmodified PNA anti-miRs (Figure 3B, compared with Figures 2A and 3A).
When Huh7 cells were incubated with 3 µM Cys-K-(TO)PNA-K3, with the anti-miR lacking Cys [K-(TO)PNA-K3] or with the alkylated Cys anti-miR [Cys(NEM)-K-(TO)PNA-K3] under standard 4 h/24 h incubation conditions, FACS analysis showed that the relative median fluorescence was 35.5 ± 5.4 for Huh7 cells treated with Cys-K-(TO)PNA-K3, but only 9.4 ± 1.7 and 4.9 ± 0.5 for cells treated with K-(TO)PNA-K3 or Cys(NEM)-K-(TO)PNA-K3, respectively (Figure 3C, left panel and 3D). This suggested enhanced uptake of the Cys-containing PNA. However, under these conditions, the median fluorescence is dependent on cellular uptake of the anti-miR and dependent on binding of the TO-anti-miR to miR-122. We therefore carried out the same experiment on HEK293ET cells that do not express the RNA target miR-122. We found that the relative median fluorescence was also much higher for HEK293ET cells after treatment with Cys-K-(TO)PNA-K3 (28.3 ± 0.1) as compared to K-(TO)PNA-K3 (2.9 ± 0.2) or Cys(NEM)-K-(TO)PNA-K3 (2.1 ± 0.2) (Figure 3C, right panel and 3D). Similarly, the relative median fluorescence was higher for Cys-K-(TO)PNA-K3 than for K-(TO)PNA-K3 in undifferentiated (myoblast) or differentiated (myotubes) H2K mdx mice-derived muscle cells (data not shown). We also carried out confocal microscopy on Huh7 cells treated with the TO-PNA anti-miRs to verify that the observed fluorescence by FACS was detected inside the cells and not bound externally to the plasma membrane. Figure 3E shows the fluorescence inside the cell in vesicular shapes in the cytosol but fluorescence was not observed in the nucleus or plasma membrane. In agreement with FACS, cells treated with Cys-K-(TO)PNA-K3 were more fluorescent than cells treated with K-(TO)PNA-K3 or Cys(NEM)-K-(TO)PNA-K3. Confocal microscopy on HEK293ET cells also showed Cys-K-(TO)PNA-K3 anti-miR fluorescence only inside cells (data not shown). Overall, the data are strongly consistent with the view that the terminal Cys residue enhances PNA anti-miR cell uptake. Moreover, this seems to be a general feature for several cell lines.
Endocytosis of Cys-K-PNA23mer-K3
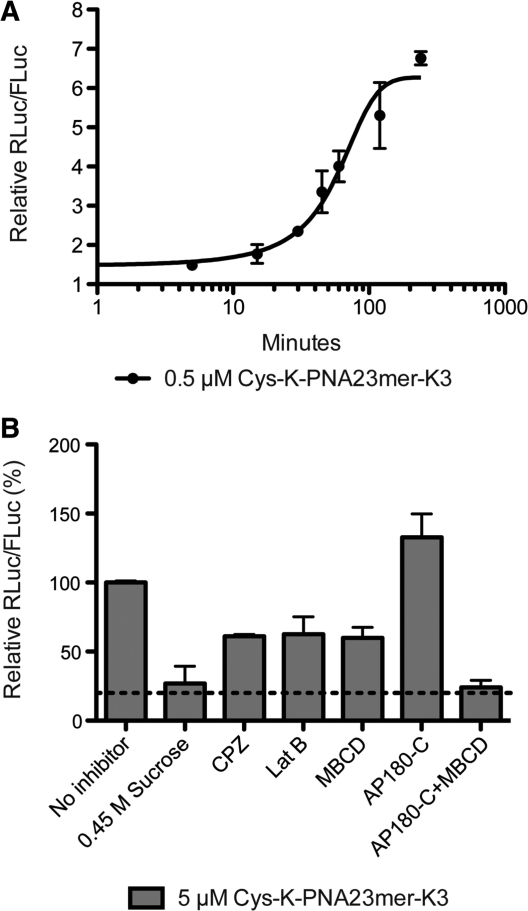
Dual-luciferase reporter Huh7 cells were incubated with 0.5 µM Cys-K-PNA23mer-K3 (a concentration at which PNA activity is sub-saturating, Figure 1A) for various times up to 240 min in serum-free media. After PNA incubations, cells were washed thoroughly to remove undelivered PNA, media replaced by Full Media and Luciferase expression measured after 48 h as before. The activity data plotted against time fit to a sigmoidal-shaped curve (Figure 4A). A low level of activity (RLuc/FLuc 1.5–2.0) is obtained within 5 min of PNA anti-miR incubation, which is followed by a rapid increase after about 15 min and reached a plateau by 100 min incubation time. We ruled out the possibility that the low-level activity at early time-points is due to Cys-K-PNA23-mer-K3 remaining bound to the plasma membrane after washing and internalized during subsequent cell growth, because in a control experiment where cells were incubated with Cys-K-PNA23-mer-K3 for 15 min, washed thoroughly and further incubated for 1 h with excess of scrambled Cys-K-PNA23-mer-K3 control anti-miR as competitor, no decrease in Cys-K-PNA23-mer-K3 activity was observed (Supplementary Figure S2).
Figure 4.
(A) Kinetics of uptake for Cys-K-PNA23mer-K3 anti-miR. Huh7 cells were treated with 0.5 µM PNA for the indicated time. Cells were then washed and luciferase expression was measured 48 h after PNA incubation. (B) Effect of endocytosis inhibitors on Cys-K-PNA23mer-K3 uptake as seen by luciferase assay. CPZ: chlorpromazine; Lat B: Latrunculin B; MBCD: Methyl-β-cyclodextrin; AP180-C: dominant-negative form of AP180.
We then asked whether an endocytotic mechanism was involved in Cys-K-PNA23-mer-K3 uptake. To do this, dual-luciferase reporter Huh7 cells were incubated for 2 h with 5 µM Cys-K-PNA23-mer-K3 (saturating conditions of activity) in media containing one of a number of endocytosis inhibitors (Figure 4B). Use of hypertonic media (0.45 M sucrose) completely abolished the PNA anti-miR activity. However incubation in the presence of 30 µM chlorpromazine (CPZ), 5 µg/ml Latrunculin B (LatB) or 2.5 mM methyl-β-cyclodextrin (MBCD) only partially reduced Cys-K-PNA23mer-K3 anti-miR activity (~40% reduction). Similar results were obtained in each case when the cells were pre-incubated with inhibitor for 1 h followed by incubation with PNA anti-miR for 1 h in the presence of inhibitor (data not shown). A total of 0.45 M sucrose is a general inhibitor of endocytosis and mainly blocks CME, but it has also been shown to block fluid-phase endocytosis in fibroblasts (44) and to inhibit a clathrin-independent mechanism of receptor-mediated endocytosis in ependymoma cells, possibly by disturbing actin filaments (45). High sucrose was shown to reduce uptake of another cationic PNA (K8-PNA-K3) in HeLa cells (46). CPZ is thought to inhibit CME as it blocks uptake of transferrin (a typical marker for CME) (47). LatB prevents actin polymerization involved in CME but mainly in clathrin-independent endocytotic mechanisms (48). Finally, MBCD removes cholesterol from the cell plasma membrane also affecting mainly clathrin-independent endocytosis (CIE) (48). A similar reduction in Cys-K-PNA23mer-K3 activity in the presence of MBCD was seen when the cells were incubated with the PNA anti-miR for 15 min (early PNA anti-miR uptake time point) (Supplementary Figure S3A) suggesting that there is no difference in cell entry mechanism for Cys-K-PNA23mer-K3 at either early or later time points.
To further inhibit CME specifically, we used a plasmid construct expressing the C-terminus of the clathrin-assembly protein AP180 (AP180-C; (37)) that has been validated as a dominant negative approach for inhibition of CME (37,49). Huh7 cells containing co-transfected miR-122 dual luciferase reporter and AP180-C plasmids and incubated with Cys-K-PNA23mer-K3 anti-miR as before showed slightly improved activity (Figure 4B). This suggests that an alternative clathrin-independent endocytotic uptake mechanism is able to compensate for this loss of CME (48) and also gives rise to enhanced uptake (50,51). When over-expression of AP180-C during cell incubation with Cys-K-PNA23mer-K3 was combined with MBCD treatment, to inhibit general CIE, activity was completely abolished, which suggests complete loss of productive cellular uptake (Figure 4B). Such cell incubation conditions did not affect cell viability (Supplementary Figure 3B). Overall the inhibitor data suggest that Cys-K-PNA23-mer-K3 is internalized in Huh7 cells by CME but can also enter productively to reach miR-122 by a clathrin-independent but actin- and cholesterol-dependent mechanism of endocytosis.
Endosome trapping does not significantly affect Cys-K-PNA23mer-K3 anti-miR activity
We asked whether following PNA internalization, endosome trapping would limit the efficiency of Cys-K-PNA-K3 anti-miR. Dual-luciferase reporter Huh7 cells were incubated with 1 or 5 µM Cys-K-PNA23-mer-K3 for 4 h in the presence or absence of 100 µM chloroquine or 6 mM CaCl2, conditions reported to increase endosomal release and nuclear splicing inhibitory activity of cationic PNA (46,52) (Figure 5A). Chloroquine had only a small effect on Cys-K-PNA23mer-K3 anti-miR activity, whereas calcium ion addition led to a very significant increase in Cys-K-PNA23mer-K3 anti-miR activity. Since an increase in cytosolic calcium ion concentration has been shown to stimulate CME and CIE in pancreatic β-cells (53) and to accelerate endocytosis of vSNARES (54), we asked if the calcium ion–induced increase in Cys-K-PNA23-mer-K3 anti-miR activity is due to enhanced cellular uptake of the PNA. Thus, dual-luciferase reporter Huh7 cells were incubated for (i) 4 h with 5 µM Cys-K-PNA23-mer-K3 followed by a further 4 h in the absence of PNA, or (ii) 4 h with PNA and 6 mM calcium ions followed by a further 4 h incubation in 6 mM calcium ions in the absence of PNA, or (iii) 4 h with PNA alone followed by incubation with 6 mM calcium for 4 h in the absence of PNA (Figure 5B). Careful cell washings were carried out between incubations to prevent carry-over. The contribution to Cys-K-PNA23-mer-K3 anti-miR activity in the presence of calcium ions due to endosomal release (condition iii) was small and similar to the effect of chloroquine (Figure 5A; 5 µM PNA), whereas the activity stimulation was much larger under condition ii, when calcium ions were present at the start. These results suggest that endosomal trapping does not significantly affect Cys-K-PNA23-mer-K3 anti-miR activity and that the predominant effect of calcium ions in Huh7 cells is to stimulate PNA endocytosis. Note that we were able to verify that for Cys-K-(TO)PNA-K3 chloroquine treatment of Huh7 cells results in a pattern of fluorescence consistent with partial endosomal release, but we were unable to carry out the same verification for calcium ions as we found the TO fluorescence to be quenched inside Huh7 cells in the presence of calcium ions (data not shown).
Subcellular localization of PNA anti-miRs
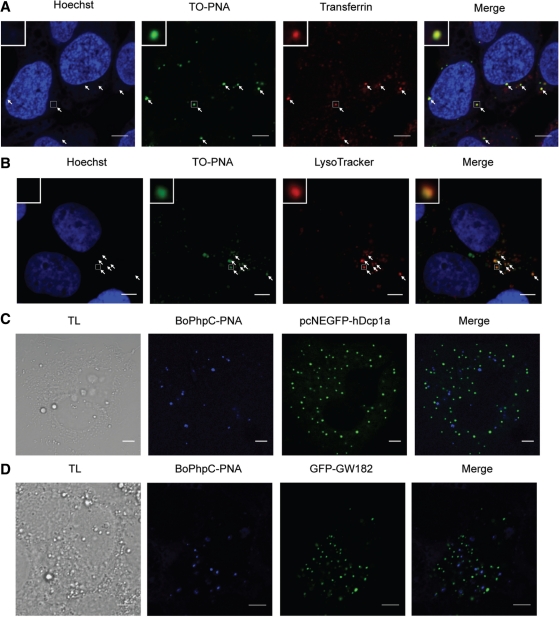
MiRNAs were found associated with endosomal compartments as well as cytoplasmic foci known as P-bodies and GW-bodies (13,14,55). We therefore asked whether targeting of miRNA by PNA anti-miR might occur within these compartments. To test if Cys-K-(TO)PNA-K3 is found in early endosomes, Huh7 cells were co-incubated with Cys-K-(TO)PNA-K3 and fluorescently labelled transferrin for 1.5 h. Cells were thoroughly washed, fixed, nuclei were stained and cells were visualized by confocal microscopy. Cys-K-(TO)PNA-K3 was found to co-localize with transferrin in small vesicles likely to be early endosomes and thus consistent with CME (Figure 6A). In a second experiment, Huh7 cells were incubated with Cys-K-(TO)PNA-K3 for 4 h. Twenty-four hours later, lysosomes and nuclei were stained, and cells were fixed and visualized by confocal microscopy (Figure 6B). The results showed that the PNA was co-localized with lysosomes, implying it had travelled through the complete endocytotic pathway.
Figure 6.
(A) Confocal microscopy of Huh7 cells co-incubated with 3 µM Cys-K-(TO)PNA-K3 and 50 µg/mL Alexa 594-conjugated human transferrin for 1.5 h. (B) Confocal microscopy of Huh7 cells treated with 3 µM Cys-K-(TO)PNA-K3 observed at 24 h after PNA treatment; lysosomes were stained using LysoTracker Red. (C) Representative image of confocal microscopy of Huh7 cells expressing P-body marker (pcNEGFP-hDcp1a) treated with 3 µM BoPhpC-PNA. (D) Representative image of confocal microscopy of Huh7 cells expressing GW-body marker (GFP-GW182) treated with 3 µM BoPhpC-PNA.
Since P-bodies and GW-bodies were suggested to associate with endosomal compartments (55), we investigated whether PNAs colocalize with these cytoplasmic structures. Due to fluorescence compatibility issues between TO-PNAs and markers for P-bodies and GW-Bodies, we utilized an alternative fluorescent PNA unit, [bis-o-(aminoethoxy)phenyl]pyrrolocytosine (BoPhpC) (36) (Supplementary Figure S4A) that has been used for PNA localization in cells (56). BoPhpC-PNA anti-miR exhibited similar activity as K-(TO)PNA-K3 in the miR-122 dual luciferase reporter system (Supplementary Figure S4B). BoPhpC-PNA was able to enter Huh7 cells in the absence of transfection agents and co-localized with K-(TO)PNA-K3 (Supplementary Figure S4C). Confocal microscopy showed that BoPhpC-PNA was also found in early and late endosomes as seen by co-localization with fluorescently labelled transferrin and LysoTracker, respectively (Supplementary Figure S4D and E). Huh7 cells expressing pcNEGFP-hDcp1a (a plasmid expressing the human decapping enzyme 1 as a P-body marker (57)) or GFP-GW182 (58) were incubated with BoPhpC-PNA for 1 - 4 h and confocal microscopy was carried out at 2, 4, 6, 8, 10 and 24 h after PNA incubation. No co-localization with P-bodies or GW-Bodies was detected under these conditions (Figure 6C and D).
We were unable to test whether PNA anti-miR co-localizes with miR-122 by in situ hybridization for miRNA detection, since the miRNA is not detected by this technique in the presence of a high-affinity anti-miR (33). Instead, we carried out cellular fractionation on Huh7 cells treated with Cys-K-(TO)PNA-K3 48 h after PNA incubation. Nuclei and cell debris were first separated from cytoplasm by centrifugation, which was then divided into membrane-bound compartments (‘pellet’) and the remaining cytosol (‘supernatant’) by ultracentrifugation. Proteins and RNA were extracted from each fraction and analyzed by western blot and northern blot, respectively. PNA ONs may be recovered by standard protein extraction and detected by electrophoresis as discrete bands in standard protein gels after western blotting by use of a radiolabeled RNA complementary probe (north-western blot), similarly to a previous report (59). In western blotting, there was enrichment in the ‘pellet’ fraction of markers for membrane-bound compartments including Rab5 (early endosomes), Syntaxin13 (STX13; recycling endosomes), Lysosomal-associated membrane protein 1 (LAMP1; late endosomes/lysosomes) and Golgin (Golgi apparatus) (Figure 7A). As a control, we detected p97, a cytosolic AAA-ATPase that associates with endoplasmic reticulum (ER) for ER-associated protein degradation (ERAD) (60), which is expected to be present in both the ‘pellet’ and ‘supernatant’ fractions. Cys-K-(TO)PNA-K3 was also detected in the ‘pellet’ fraction as seen by the north-western blotting approach. Note that Cys-K-(TO)PNA-K3 was detected as two bands instead of one, similarly to the reported detection of a PNA ON conjugated to a cationic cell penetrating peptide, whereas a charge neutral PNA ON of the same sequence was detected as a single band (59). Finally, northern blotting showed enrichment of mature miR-122 in the ‘pellet’ fraction as compared to the ‘supernatant’, in agreement with mature miRNAs being found within endosomal compartments (55). The same results were obtained for cellular fractionation of Huh7 cells treated with K-(TO)PNA-K3 (without Cys) (data not shown).
We have been unable to date to fully separate different endosomal compartments using continuous or discontinuous opti-prep gradients (data not shown). Instead we carried out IP to discriminate between endosomal and other membrane-bound compartments, such as Golgi or endoplasmic reticulum. Protein A coated beads were incubated with a STX13 anti-serum and then the anti-STX13-containing beads were incubated with the ‘pellet’ fraction to recover endosomal vesicles (mainly recycling but also early and late endosomes). Elution of the STX13-positive compartments was carried out either by treatment with a mild detergent (Triton X-100) to elute the internal components of the vesicles and part of trans-membrane proteins, or by a pH shock to break the bead-antibody bond to recover IgG and STX13-positive components. Endosomal markers Rab5 and Lamp1 were detected when the samples were eluted with either Triton X-100 treatment or pH shock (Figure 7B). Pre-treatment of anti-STX13-containing beads with STX13 antigen mostly abolished the detection of these markers. As expected, IgG was detected only under pH shock conditions. STX13 could not be detected, since it cross-reacted with the IgG present in the samples. Both Cys-K-PNA(TO)-K3 and miR-122 were present in STX13-positive compartments as seen by north-western blotting and by RT–qPCR, respectively (Figure 7B). Northern blot for miR-122 detection could not be carried out due to the small amounts of RNA obtained after the IP. Overall these results are fully consistent with the view that PNA anti-miRs passage through the endocytotic pathway and target miR-122 in STX13-positive endosomal compartments.
DISCUSSION
Lys-functionalized PNA anti-miRs are versatile tools to inhibit miRNAs inside cells without the need for added transfection agent (22,30,32,39). However, information has been lacking to date on the PNA and amino acid sequence requirements for anti-miR activity. Our studies on PNA anti-miR truncations (Figure 2C), agrees with a previous study where truncations of a full-length 22-mer MOE PS anti-miR at the 5′-end were better tolerated than truncations at the 3′-end (28). Nevertheless, a large drop in activity still occurred upon 5′-truncation by 3 nucleotides of MOE PS anti-miRs (28), whereas inhibition of miR-122 remained strong for the 5′-truncation of 4 nucleotides from the PNA anti-miR (19-mer Figure 2C) and was reduced drastically only for a 6-nt truncation (17-mer). In contrast, LNA-containing anti-miRs do not need to target the full-length miRNA to maintain strong anti-miR activity (21,23,33) and ‘tiny’ all-LNA 8mers targeting only the seed sequence of miR-21 or miR-122 have been shown to be very effective in cells and in mice (40). Thus, it is clear that the difference in length requirements for anti-miR potency is chemistry dependent. Based on the available literature and the data presented here, it is reasonable to suppose that the stronger the RNA-binding chemistry, the greater the ability to tolerate anti-miR truncation whilst still maintaining anti-miR potency. However, all PNA anti-miRs tested here showed high melting temperatures with complementary RNA in vitro (Supplementary Table S1), whilst their anti-miR potencies were different (Figure 2C). Thus other miRNA targeting factors beyond the affinity of the anti-miR must play a strong role in potency determination, as has been suggested previously (28,29,32).
Our data suggest that targeting the ‘seed’ sequence of the miRNA is crucial for PNA anti-miR activity (Figure 2C), in agreement with other reports (28,40). MiRNA loading into miRISC pre-organises the seed sequence for target recognition (11,12). Further, the 5′-end of an siRNA-like duplex or a 5′-phosphorylated 21-base DNA guide strand loaded into Ago serves as the nucleation site for mRNA recognition and binding (61–63). It seems likely that an anti-miR first interacts with the miRNA seed sequence followed by a ‘zipping-up’ mechanism towards the 3′-end of the miRNA to form a stable miRNA:anti-miR duplex. This would imply that the targeting by anti-miRs occurs once the miRNA has been loaded in miRISC (24,25,28,40). Since a 6-nt truncation at the 5′-end of the PNA anti-miR was better tolerated than an equivalent 6-nt mismatch (Figure 2C), even though both these PNAs showed almost the same melting temperature (Supplementary Table S1), it is likely that the mismatches destabilize the duplex in the context of miRISC.
Positioning the 4 Lys residues at the N-terminus of the PNA (Cys-K4-PNA23mer) resulted in a substantial drop in anti-miR activity compared to location at the C-terminus of the PNA (Cys-PNA23mer-K4) (Figure 2A). However, substitution of all L-amino acids by D-amino acids (dCys-dK-PNA23mer-dK3) did not significantly affect the anti-miR activity. This is consistent with our recent demonstration that a PNA anti-miR containing 4 × D-Lys could inhibit miR-155 in cells and in vivo and partially recapitulated the gene expression profile of miR-155 knockout mice at doses that did not cause toxicity (22). However, the most striking observation was that the presence of a terminal Cys (in either L or D configuration) dramatically enhanced PNA anti-miR cellular activity (Figure 2A). Interestingly, this was not due to the particular structure of Cys but due to the presence of a free thiol group (Figure 3A), leading to enhanced cellular uptake of the PNA anti-miRs (Figure 3C–E) in Huh7 liver cells, HEK293ET cells (Figure 3C and D) and H2K mdx mice-derived muscle cells (data not shown). We therefore believe that the most likely explanation for the enhanced anti-miR activity observed for thiol-containing PNAs is due to improved bulk PNA uptake.
Aubry et al. (64) showed for Cys disulfide-conjugated cell penetrating peptides (CPPs) that thiol/disulfide exchange reactions may occur at the cell surface to enhance cell entry and they reported higher cell association for peptides containing Cys residues, either oxidized or reduced, as compared to their Cys-free versions, leading to enhanced internalization. They were also able to internalize a Protein Kinase C peptide inhibitor into CHO K1 cells by addition of an activated Cys residue. We found also that a disulfide-conjugated Cys-K-PNA23-mer-K3 dimer was able to inhibit miR-122 in Huh7 cells as well as the monomeric version (data not shown), which is consistent with a redox involvement at the plasma membrane. We have not investigated specifically whether the enhanced cellular uptake for Cys-K-PNA-K3 is due to enhanced cell association. However, when we used confocal microscopy at different time-points we could only observe TO-PNAs inside the cell and not associated with the plasma membrane (Figure 3E and data not shown). Nevertheless, since Cys-K-PNA-K3 is both cationic and thiol-containing, and since cationic CPPs are known to associate with cells through negatively charged heparan sulfate-containing glycosylaminoglycans (65), improved cell attachment cannot be ruled out as an explanation for the enhanced cellular uptake observed.
The presence of a thiol group in peptides has been shown to correlate with enhanced cellular uptake. The MPG series of peptides have been used to complex DNA and siRNA and these contain a C-terminal cysteamide residue that is a pre-requisite for efficient delivery into cells (66,67). Similarly, internalization of the CPP Pep-1, used for delivery of protein cargoes, was inhibited in the absence of its C-terminal cysteamide (68). The precise role of the cysteamide in MPG and Pep-1 has not been clearly demonstrated, although it is thought that it plays a role in stabilising the nanoparticle complex or in membrane anchoring (66–68). However, our results would indicate that the free thiol may also play a more direct role in cellular delivery for these nanostructures.
Sulfur replacement of non-bridging oxygen atoms in ONs have also led to enhanced activity and cell delivery in the absence of transfection agents. For example, lipid-conjugated thio-phosphoramidates exhibited higher anti-telomerase activity as compared to lipid-conjugated phosphoramidate ONs in cancer cells, thought to be due to enhanced delivery (69). Similarly, DNA PS ONs were better taken up by mouse B-cells than their PO versions, which correlated with better cell-surface binding of the PS ON (70). We showed recently better anti-miR activity for an LNA/OMe PS anti-miR than an LNA/OMe PO anti-miR delivered in the absence of transfection agents, suggesting better cellular uptake (32). Our results here showing a steep increase in Cys-K-PNA23-mer-K3 anti-miR activity at lower concentrations that reaches a plateau beyond 1 µM PNA (Figure 1A) are similar to the uptake curves reported by Zhao et al. (70), who argued that DNA PS ONs may congregate at high-affinity binding sites on cells. No convincing evidence has yet emerged for specific receptor proteins for binding of ONs or PNA, but our results suggest that proteins capable of interacting either with free thiols or with phosphorothioates may help to enhance cell uptake, and this subject deserves to be explored further.
The extent to which these thiol/sulfur-mediated enhanced uptake mechanisms are relevant in vivo still remains to be addressed. We have carried out a preliminary study on targeting miR-122 in mice by dCys-dK-PNA23mer-dK3 and dK-PNA23mer-dK3. After a short-term i.p. dosage protocol of 3 doses on alternating days at 40 mg/kg PNA, both PNAs gave rise to a 20% reduction in cholesterol levels in plasma 10 days after the last injection as compared to PBS-treated mice (data not shown), showing that PNA anti-miRs are active in mouse liver. The reason for the lack of phenotypic activity difference between these two PNAs is not yet clear. One possibility is that, in vivo, the presence of a reactive thiol group on the PNA compromises the enhancement of cell uptake seen in cell culture, through sequestration by a thiol-reactive protein perhaps. We are currently investigating long-term dosage protocols as well as the possibility of delivery of dCys-dK-PNA23mer-dK3 as disulfide-conjugated dimers.
In our cell studies, FACS analysis and confocal microscopy (Figures 3 and 6) were greatly aided by use of two novel fluorescent PNA residues. Intrinsic PNA fluorescence has advantages over an attached fluorescent tag to avoid substantial compromise of activity or alteration of delivery properties or cell localization (71–74), as well as eliminating the danger of tag removal during cell delivery (75). Here, both TO-PNA and BoPhpC-PNA allowed monitoring of PNA cell delivery and trafficking without substantial compromise of PNA activity (Figure 3 and 6 and Supplementary Figure S4). A BoPhpC-PNA was shown previously to inhibit mutant huntingtin protein in GM04281 cells (56). Our studies are the first to use this fluorescent analogue in efficient inhibition of miRNA (Supplementary Figure S4). TO-PNAs have recently been shown to be useful tools for imaging of influenza H1N1 mRNA in living cells (43). In our work, we report the first intracellular functional activity for TO-PNAs, in this case for miR-122 targeting (Figure 3B). TO-PNA has an additional advantage in that binding to a complementary RNA strand restricts depletion of the TO excited state by hampering twisting motions, resulting in enhanced emitted fluorescence (43). We confirmed this property for in vitro RNA binding (Supplementary Figure S1). In cells, an increased fluorescence emission was detected for Huh7 cells that express the target miR-122 as compared to HEK293ET cells that do not express the target miR-122 (Figure 3C and D). To rule out better TO-PNA uptake in Huh7 cells over HEK293ET cells, we thought to treat Huh7 cells with a scrambled TO-PNA anti-miR. However, the precise positioning of the TO-PNA monomer within the anti-miR sequence affects the fluorescence emission properties significantly (Supplementary Figure S1), so such an approach cannot be used. Although we cannot rule out completely cell-specific uptake differences, since TO-PNA anti-miRs were shown to inhibit miR-122 in Huh7 cells (Figure 3B), it seems likely that the enhanced median fluorescence detected for TO-PNAs in Huh7 cells is due to TO-PNAs binding to the miR-122 RNA target.
The mechanism of cellular uptake for Cys-containing cationic PNA ONs was investigated. Cys-K-(TO)PNA-K3 was found to be co-localized with transferrin, which indicates that the bulk PNA prefers a CME route of entry (Figure 6A). However, the use of a functional assay is paramount to determine the pathways of the active fraction of PNA that can reach the desired RNA target. Anti-miR activity results showed clear evidence for both clathrin-dependent and independent uptake pathways for the active fraction of the PNA, since activity was not fully blocked by CPZ, LatB or MBCD and completely blocked only when both CME and CIE were simultaneously inhibited (Figure 4B). Interestingly, upon specific inhibition of CME through the use of the dominant-negative form of AP180 (AP180-C) (37), activity of Cys-K-PNA23mer-K3 was enhanced a little, suggesting that the CIE pathway may lead to slightly more productive PNA delivery. However, in general both types of pathway led to anti-miR inhibitory activity. In the absence of endocytosis inhibitors, the kinetics for productive uptake of Cys-K-PNA23mer-K3 showed a fast but mild uptake at early time points (within 5 min) that rapidly increased and reached a plateau by 100 min incubation times (Figure 4A). One possibility is that the rapid increase in uptake is due to endosomal recycling and the plateau stage corresponds to saturation of the endocytotic machinery. Consistent with this model, our data suggest that Cys-K-PNA23mer-K3 internalization at early or late time points occurs via the same endocytotic mechanism (Supplementary Figure S3A).
Our data suggest that the mechanisms of cellular targeting of miRNA are different from that of mRNA and pre-mRNA. Koller et al. suggested recently (76) that RNase-H active MOE/DNA PS gapmers targeted to mRNA and delivered without transfection agents did not co-localize with P-bodies and that the bulk of the ON was found in lysosomes after 24 h. However, they concluded that two endocytotic pathways explained their antisense activity data best. The non-productive pathway led to lysosomes, while the productive uptake involved a novel pathway that was Adaptor Protein Complex 2 (AP2)-dependent, but clathrin- and caveolae-independent. For miRNA targeting, activity is clearly achievable unaided by transfection agents through established endocytotic pathways and there is no evidence for an independent productive pathway. In the case of pre-mRNA targeting, we and others have found that the nuclear-active fraction of cationic PNA for splicing redirection is internalized predominantly by CME (50,51,77) and endosome trapping significantly limits antisense activity (46,50,52). For miRNA targeting, the choloroquine experiment showed that promotion of endosomal release into the cytosol did not significantly affect Cys-K-PNA23mer-K3 anti-miR activity (Figure 5). Although both Cys-K-(TO)PNA-K3 and BoPhpC-PNA were found in early and late endosomes (Figure 6A and B and Supplementary Figure S4), suggestive of endosome trapping, enough PNA is able to reach and inhibit miR-122 (Figure 3B and Supplementary Figure S4B). Moreover, we have found that both the PNA anti-miR and also miR-122 are associated with endosomal compartments (Figure 7, see below). This provides strong evidence that targeting of miRNAs by anti-miRs occurs within or associated with endosomal compartments and does not require PNA release into the cytosol. Note also that we showed recently that LNA/OMe PS and other modified ONs are also highly active inhibitors of miR-122 in cells at sub-micromolar concentrations in the absence of transfection agents (32; Threlfall et al., manuscript submitted for publication).
How therefore is an anti-miR able to access the miRNA while travelling through the endosomal pathway? MiRNAs loaded into miRISC are thought to traffic to P-bodies following mRNA binding, whereupon Cap and polyA tails of the repressed mRNA are removed (55). The complex is then thought to move to GW-bodies, where miRISC may disassemble. P-bodies and GW-bodies were shown to often associate with MVBs that may fuse with the plasma membrane and release miRNAs and mRNAs into exosomes or, alternatively, traffic to lysosomes for cargo degradation. BoPhpC-PNA was not seen to co-localize with P-bodies or GW-bodies (Figure 6C and D), similarly to a report that antagomiRs did not co-localize with P-bodies in mouse liver (we note however use of a marker for GW-bodies, GFP-GW182) (27). Obad et al. detected tiny-LNA anti-miR-21 delivered without transfection agents as discrete punctate signals in the cytoplasm of HEK293 cells that, in some cases, co-localized with FLAG-tagged Ago2 (40). Our cell fractionation studies showed that both miR-122 and TO-PNAs were enriched in fractions containing membrane-bound compartments together with markers for early, recycling and late endosomes as well as markers for Golgi and ER (Figure 7A). Both miR-122 and Cys-K-(TO)PNA-K3 anti-miR were detected during immunoprecipitation identification of STX13-positive compartments (Figure 7B), indicative that targeting of miRNAs with anti-miRs may take place in recycling endosomes. Therefore, anti-miRs are perhaps able to access the miRNA either by fusion of anti-miR-containing endosomal vesicles with miRNA-containing endosomal compartments or by leakage of enough anti-miR into the compartment where miRNA passages, possibly at the endosome recycling step. However, a full understanding of the trafficking and sub-cellular encounter between PNA anti-miR and its complementary miRNA will require detailed further investigation and needs to take account not only of localization of bulk fluorescent anti-miR but also to address the issue of the active targeting fraction, which is currently hard to distinguish biophysically.
Our studies have addressed a number of important aspects of PNA anti-miR design for efficient targeting in cultured cells and have pointed to the mechanisms of cell uptake leading to encounter of the target miRNA. To date, only the outcome for the miRNA upon targeting by anti-miRs of different chemistries has been addressed in depth (27,29,39). There is still much to learn on the mechanisms, localization and structural requirements for efficient miRNA inhibition in cells and in vivo by anti-miRs. We hope that our studies on PNA are a useful contribution along this pathway of understanding, and may help towards design of better therapeutic candidates.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Methods, Supplementary Table 1, Supplementary Figures 1–4.
FUNDING
Medical Research Council (MRC Unit programme U105178803); Cesar Milstein Scholarship from the Darwin Trust of Edinburgh, Scotland (to A.G.T.); NSERC Canada (to R.H.E.H.). Funding for open access charge: MRC.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Alexander Ludwig (MRC-LMB, Cambridge UK) for technical advice and reagents for cell fractionation and immuno-precipitation studies, Harvey McMahon and Leon Lagnado (MRC-LMB, Cambridge UK) for AP180-C construct, Edward K.L. Chan (University of Florida, Health Science Center; Gainesville, FL; USA) and Olivier Voinnet (IBMP-CNRS UPR 2357, Strasbourg; France) for GFP-GW182 construct, Jens Lykke-Andersen (MCD Biology; University of Colorado, Boulder; USA) for pcNEGFP-hDcp1a construct and David Loakes and members of the Gait group for helpful discussions.
REFERENCES
- 1.Bushati N, Cohen SM. MicroRNA functions. Annu. Rev. Cell Dev. Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 2.Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao W, Cui Q. An analysis of human microRNA and disease associations. PLoS one. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chong MMW, Zhang G, Cheloufi S, Neubert TA, Hannon GJ, Littman DR. Canonical and alternate functions of the microRNA biogenesis machinery. Genes Dev. 2010;24:1951–1960. doi: 10.1101/gad.1953310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheloufi S, Dos Santos CO, Chong MMW, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465:584–589. doi: 10.1038/nature09092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Siomi H, Siomi MC. Posttranscriptional regulation of microRNA biogenesis in animals. Mol. Cell. 2010;38:323–332. doi: 10.1016/j.molcel.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 7.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 8.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 2009;21:452–460. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lambert NJ, Gu SG, Zahler AM. The conformation of microRNA seed regions in native microRNPs is prearranged for presentation to mRNA targets. Nucleic Acids Res. 2011;39:4827–4835. doi: 10.1093/nar/gkr077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulkarni M, Ozgur S, Stoecklin G. On track with P-bodies. Biochem. Soc. Trans. 2010;38:242–251. doi: 10.1042/BST0380242. [DOI] [PubMed] [Google Scholar]
- 14.Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat. Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 15.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 16.Hwang H-W, Wentzel EA, Mendell JT. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 17.Kren BT, Wong PY-P, Sarver A, Zhang X, Zeng Y, Steer CJ. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009;6:65–72. doi: 10.4161/rna.6.1.7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krützfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 19.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Vermeulen A, Robertson B, Dalby AB, Marshall WS, Karpilow J, Leake D, Khvorova A, Baskerville S. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjärn M, Hansen HF, Berger U, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 22.Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KGC, Enright AJ, Gait MJ, Vigorito E. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–4475. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Ørum H. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hutvágner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 27.Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res. 2007;35:2885–2892. doi: 10.1093/nar/gkm024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis S, Lollo B, Freier S, Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davis S, Propp S, Freier SM, Jones LE, Serra MJ, Kinberger G, Bhat B, Swayze EE, Bennett CF, Esau C. Potent inhibition of microRNA in vivo without degradation. Nucleic Acids Res. 2009;37:70–77. doi: 10.1093/nar/gkn904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabani MM, Gait MJ. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oh SY, Ju Y, Park H. A highly effective and long-lasting inhibition of miRNAs with PNA-based antisense oligonucleotides. Mol. Cells. 2009;28:341–345. doi: 10.1007/s10059-009-0134-8. [DOI] [PubMed] [Google Scholar]
- 32.Torres AG, Threlfall RN, Gait MJ. Potent and sustained cellular inhibition of miR-122 by Lysine-derivatised peptide nucleic acids (PNA) and phosphorothioate Locked Nucleic Acid (LNA)/2′-O-methyl (OMe) mixmer anti-miRs in the absence of transfection agents. Artificial DNA:PNA & XNA. 2011;2 doi: 10.4161/adna.17731. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elmén J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjärn M, Hansen JB, Hansen HF, Straarup EM, et al. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 2008;36:1153–1162. doi: 10.1093/nar/gkm1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lennox KA, Behlke MA. A direct comparison of anti-microRNA oligonucleotide potency. Pharm. Res. 2010;27:1788–1799. doi: 10.1007/s11095-010-0156-0. [DOI] [PubMed] [Google Scholar]
- 35.Jarikote DV, Köhler O, Socher E, Seitz O. Divergent and linear solid-phase synthesis of PNA containing thiazole orange as artificial base. European J. Org. Chem. 2005;2005:3187–3195. [Google Scholar]
- 36.Wojciechowski F, Hudson RHE. Fluorescence and hybridization properties of peptide nucleic acid containing a substituted phenylpyrrolocytosine designed to engage Guanine with an additional H-bond. J. Am. Chem. Soc. 2008;130:12574–12575. doi: 10.1021/ja804233g. [DOI] [PubMed] [Google Scholar]
- 37.Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- 38.Steuble M, Gerrits B, Ludwig A, Mateos JM, Diep T-M, Tagaya M, Stephan A, Schätzle P, Kunz B, Streit P, et al. Molecular characterization of a trafficking organelle: dissecting the axonal paths of calsyntenin-1 transport vesicles. Proteomics. 2010;10:3775–3788. doi: 10.1002/pmic.201000384. [DOI] [PubMed] [Google Scholar]
- 39.Torres AG, Fabani MM, Vigorito E, Gait MJ. MicroRNA fate upon targeting with anti-miRNA oligonucleotides as revealed by an improved Northern-blot-based method for miRNA detection. RNA. 2011;17:933–943. doi: 10.1261/rna.2533811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Köhler O, Seitz O. Thiazole orange as fluorescent universal base in peptide nucleic acids. Chem. Commun. 2003;2003:2938–2939. doi: 10.1039/b308299g. [DOI] [PubMed] [Google Scholar]
- 42.Köhler O, Jarikote DV, Seitz O. Forced intercalation probes (FIT Probes): thiazole orange as a fluorescent base in peptide nucleic acids for homogeneous single-nucleotide-polymorphism detection. ChemBioChem. 2005;6:69–77. doi: 10.1002/cbic.200400260. [DOI] [PubMed] [Google Scholar]
- 43.Kummer S, Knoll A, Socher E, Bethge L, Herrmann A, Seitz O. Fluorescence imaging of influenza H1N1 mRNA in living infected cells using single-chromophore FIT-PNA. Angew. Chem. 2011;50:1931–1934. doi: 10.1002/anie.201005902. [DOI] [PubMed] [Google Scholar]
- 44.Heuser JE, Anderson RG. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J. Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuchler-Bopp S, Dietrich JB, Zaepfel M, Delaunoy JP. Receptor-mediated endocytosis of transthyretin by ependymoma cells. Brain Res. 2000;870:185–194. doi: 10.1016/s0006-8993(00)02413-6. [DOI] [PubMed] [Google Scholar]
- 46.Abes S, Williams D, Prevot P, Thierry A, Gait MJ, Lebleu B. Endosome trapping limits the efficiency of splicing correction by PNA-oligolysine conjugates. J. Controlled Release. 2006;110:595–604. doi: 10.1016/j.jconrel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 47.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated TAT peptide involves clathrin-dependent endocytosis and heparan sulfate receptors. J. Biol. Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 48.Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu. Rev. Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- 49.Granseth B, Odermatt B, Royle SJ, Lagnado L. Clathrin-mediated endocytosis is the dominant mechanism of vesicle retrieval at hippocampal synapses. Neuron. 2006;51:773–786. doi: 10.1016/j.neuron.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 50.Ivanova GD, Arzumanov A, Abes R, Yin H, Wood MJa, Lebleu B, Gait MJ. Improved cell-penetrating peptide-PNA conjugates for splicing redirection in HeLa cells and exon skipping in mdx mouse muscle. Nucleic Acids Res. 2008;36:6418–6428. doi: 10.1093/nar/gkn671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saleh AF, Arzumanov A, Abes R, Owen D, Lebleu B, Gait MJ. Synthesis and splice-redirecting activity of branched, arginine-rich peptide dendrimer conjugates of peptide nucleic acid oligonucleotides. Bioconj. Chem. 2010;21:1902–1911. doi: 10.1021/bc100275r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shiraishi T, Pankratova S, Nielsen PE. Calcium ions effectively enhance the effect of antisense peptide nucleic acids conjugated to cationic tat and oligoarginine peptides. Chem. Biol. 2005;12:923–929. doi: 10.1016/j.chembiol.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 53.He Z, Fan J, Kang L, Lu J, Xue Y, Xu P, Xu T, Chen L. Ca2+ triggers a novel clathrin-independent but actin-dependent fast endocytosis in pancreatic beta cells. Traffic. 2008;9:910–923. doi: 10.1111/j.1600-0854.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- 54.Sankaranarayanan S, Ryan TA. Calcium accelerates endocytosis of vSNAREs at hippocampal synapses. Nat. Neurosci. 2001;4:129–136. doi: 10.1038/83949. [DOI] [PubMed] [Google Scholar]
- 55.Gibbings D, Voinnet O. Control of RNA silencing and localization by endolysosomes. Trends Cell Biol. 2010;20:491–501. doi: 10.1016/j.tcb.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 56.Hu J, Dodd DW, Hudson RHE, Corey DR. Cellular localization and allele-selective inhibition of mutant huntingtin protein by peptide nucleic acid oligomers containing the fluorescent nucleobase [bis-o-(aminoethoxy)phenyl]pyrrolocytosine. Bioorg. Med. Chem. Lett. 2009;19:6181–6184. doi: 10.1016/j.bmcl.2009.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Franks TM, Lykke-Andersen J. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 2007;21:719–735. doi: 10.1101/gad.1494707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eystathioy T, Chan EKL, Tenenbaum SA, Keene JD, Griffith K, Fritzler MJ. A phosphorylated cytoplasmic autoantigen, GW182, associates with a unique population of human mRNAs within novel cytoplasmic speckles. Mol. Biol. Cell. 2002;13:1338–1351. doi: 10.1091/mbc.01-11-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shin D, Nam M, Yoon Y, Kim M. Membrane-based hybridization capture of intracellular peptide nucleic acid. Anal. Biochem. 2010;399:135–137. doi: 10.1016/j.ab.2009.11.031. [DOI] [PubMed] [Google Scholar]
- 60.Rabinovich E, Kerem A, Frohlich K-U, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parker JS, Roe SM, Barford D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature. 2005;434:663–666. doi: 10.1038/nature03462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma J-b, Yuan Y-r, Meister G, Pei Y, Tuschl T, Patel DJ. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, Patel DJ. Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature. 2009;461:754–761. doi: 10.1038/nature08434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Aubry S, Burlina F, Dupont E, Delaroche D, Joliot A, Lavielle S, Chassaing G, Sagan S. Cell-surface thiols affect cell entry of disulfide-conjugated peptides. FASEB J. 2009;23:2956–2967. doi: 10.1096/fj.08-127563. [DOI] [PubMed] [Google Scholar]
- 65.Console S, Marty C, García-Echeverría C, Schwendener R, Ballmer-Hofer K. Antennapedia and HIV transactivator of transcription (TAT) “protein transduction domains” promote endocytosis of high molecular weight cargo upon binding to cell surface glycosaminoglycans. J. Biol. Chem. 2003;278:35109–35114. doi: 10.1074/jbc.M301726200. [DOI] [PubMed] [Google Scholar]
- 66.Simeoni F, Morris MC, Heitz F, Divita G. Insight into the mechanism of the peptide-based gene delivery system MPG: implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crombez L, Morris MC, Dufort S, Aldrian-Herrada G, Nguyen Q, Mc Master G, Coll J-L, Heitz F, Divita G. Targeting cyclin B1 through peptide-based delivery of siRNA prevents tumour growth. Nucleic Acids Res. 2009;37:4559–4569. doi: 10.1093/nar/gkp451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weller K, Lauber S, Lerch M, Renaud A, Merkle HP, Zerbe O. Biophysical and biological studies of end-group-modified derivatives of Pep-1. Biochemistry. 2005;44:15799–15811. doi: 10.1021/bi051535d. [DOI] [PubMed] [Google Scholar]
- 69.Dikmen ZG, Wright WE, Shay JW, Gryaznov SM. Telomerase targeted oligonucleotide thio-phosphoramidates in T24-luc bladder cancer cells. J. Cell. Biochem. 2008;104:444–452. doi: 10.1002/jcb.21635. [DOI] [PubMed] [Google Scholar]
- 70.Zhao Q, Matson S, Herrera CJ, Fisher E, Yu H, Krieg AM. Comparison of cellular binding and uptake of antisense phosphodiester, phosphorothioate, and mixed phosphorothioate and methylphosphonate oligonucleotides. Antisense Res. Dev. 1993;3:53–66. doi: 10.1089/ard.1993.3.53. [DOI] [PubMed] [Google Scholar]
- 71.Middleton RJ, Kellam B. Fluorophore-tagged GPCR ligands. Curr. Op. Chem. Biol. 2005;9:517–525. doi: 10.1016/j.cbpa.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 72.Yoo H, Juliano RL. Enhanced delivery of antisense oligonucleotides with fluorophore-conjugated PAMAM dendrimers. Nucleic Acids Res. 2000;28:4225–4231. doi: 10.1093/nar/28.21.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Puckett CA, Barton JK. Fluorescein redirects a ruthenium-octaarginine conjugate to the nucleus. J. Am. Chem. Soc. 2009;131:8738–8739. doi: 10.1021/ja9025165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Szeto HH, Schiller PW, Zhao K, Luo G. Fluorescent dyes alter intracellular targeting and function of cell-penetrating tetrapeptides. FASEB J. 2005;19:118–120. doi: 10.1096/fj.04-1982fje. [DOI] [PubMed] [Google Scholar]
- 75.Barnett EM, Elangovan B, Bullok KE, Piwnica-Worms D. Selective cell uptake of modified Tat peptide-fluorophore conjugates in rat retina in ex vivo and in vivo models. Invest. Ophthalmol. Vis. Sci. 2006;47:2589–2595. doi: 10.1167/iovs.05-1470. [DOI] [PubMed] [Google Scholar]
- 76.Koller E, Vincent TM, Chappell A, De S, Manoharan M, Bennett CF. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hassane FS, Ivanova GD, Bolewska-Pedyczak E, Abes R, Arzumanov AA, Gait MJ, Lebleu B, Gariépy J. A peptide-based dendrimer that enhances the splice-redirecting activity of PNA conjugates in cells. Bioconj. Chem. 2009;20:1523–1530. doi: 10.1021/bc900075p. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.