Abstract
The mechanism of DNA translocation by papillomavirus E1 and polyomavirus LTag hexameric helicases involves consecutive remodelling of subunit–subunit interactions around the hexameric ring. Our biochemical analysis of E1 helicase demonstrates that a 26-residue C-terminal segment is critical for maintaining the hexameric assembly. As this segment was not resolved in previous crystallographic analysis of E1 and LTag hexameric helicases, we determined the solution structure of the intact hexameric E1 helicase by Small Angle X-ray Scattering. We find that the C-terminal segment is flexible and occupies a cleft between adjacent subunits in the ring. Electrostatic potential calculations indicate that the negatively charged C-terminus can bridge the positive electrostatic potentials of adjacent subunits. Our observations support a model in which the C-terminal peptide serves as a flexible ‘brace’ maintaining the oligomeric state during conformational changes associated with ATP hydrolysis. We argue that these interactions impart processivity to DNA unwinding. Sequence and disorder analysis suggest that this mechanism of hexamer stabilization would be conserved among papillomavirus E1 and polyomavirus LTag hexameric helicases.
INTRODUCTION
The papillomavirus replication protein E1 is a member of helicase superfamily III (SFIII), a group of replication initiator proteins from small RNA and DNA viruses. These proteins are also members of the wider class of AAA+ proteins (ATPases associated with a variety of cellular activities) that normally function as oligomers (1). Oligomeric ATPase proteins belonging to the AAA+ superfamily commonly form hexameric rings that function as molecular motors powered by ATP binding and hydrolysis. AAA+ proteins are also P-loop NTPases, the defining sequence motifs of which are the Walker A and B boxes (2,3). The Walker A and B residues bind, respectively, the β and γ phosphates of ATP/Mg2+. Other motifs are also required for ATPase activity. These include an arginine finger and sensors 1–3 residues that interact with the bound cofactor and/or key residues of a neighbouring NTPase motif (2,4). An active site is generated at the interface between monomers. Some residues, such as those of the Walker A and B boxes, are said to act in cis since they coordinate the ATP/Mg2+ moiety within a subunit. Others, e.g. the arginine finger residue, are considered to act in trans since they contribute to the active site from an adjacent subunit (5,6). In hexameric helicases, the mechanism of coupling ATP hydrolysis to work has been linked to conformational changes and the mechanical repositioning of structural elements around the protein assembly. However, different ‘motor’ models have been proposed to describe the sequence of events. The sequential conformational change of subunits around the hexamer invites the proposition of a cyclical and sequential order of actions around the ring (7). Despite this, there is also evidence for random or ‘probabilistic’ hydrolysis (8,9).
The approximately 600 amino acid papillomavirus E1 protein (Figure 1A) is highly conserved between viral species. The SF III helicase homology resides in the C-terminal 300 residues and in bovine papillomavirus (BPV-1) this domain functions autonomously as a helicase with enzymatic properties similar to the full-length protein (10), hence the current study is limited to the helicase component of the E1 protein. The X-ray crystal structure of the BPV-1 helicase domain (E1HD) has been solved without and with ADP and single-stranded DNA (ssDNA) bound (9,11). Helicase domains in both structures are assembled into a hexameric ring composed of two tiers, the near-symmetrical N-terminal domain (residues ~300–378), and the highly asymmetric C-terminal AAA+ domain (residues 379–605). The ATP-binding sites adopt specific conformations linked to positional changes in two ssDNA binding elements (a β-hairpin and hydrophobic loop) that project into a central ssDNA binding tunnel. From these data, a model for ssDNA translocation has been proposed where the ssDNA binding elements capture and escort one base unit of DNA through the complex using the energy of ATP hydrolysis (11). Although the ‘coordinated escort’ model of ssDNA translocation is attractive, the current structural data do not sufficiently explain all the biochemical properties of the E1 helicase. For example, it remains unknown how DNA base pairs are melted or how the hexameric assembly is maintained during processive unwinding. Such complications arise because certain features of a bona fide replication fork or of the protein chain were missing or could not be modelled from the available structural data. In particular, our biochemical analysis indicates that a C-terminal segment missing in the crystal structures is crucial for maintaining the stability of the oligomeric assembly during processive DNA unwinding. Here, we describe the solution structure of the intact E1HD hexamer determined by small angle X-ray scattering (SAXS). SAXS is a low-resolution method employed to study macromolecules of different sizes under near native conditions (12). Importantly, the technique is able to provide structural information about the overall architecture of flexible particles and flexible fragments, not seen by high-resolution methods (13–16).
Figure 1.
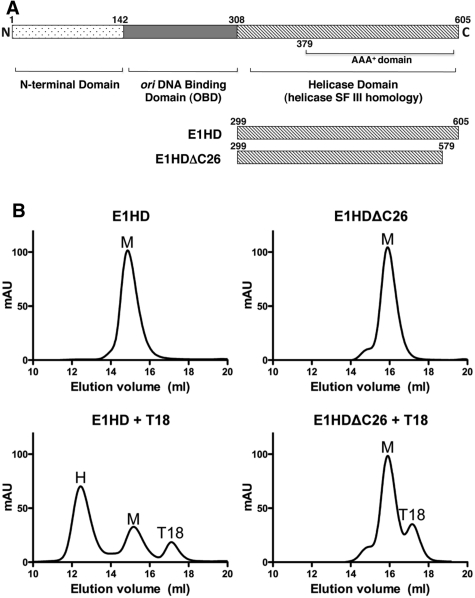
Oligomerization of E1HD and E1HDΔC26. (A) Domain organization of E1 helicase, with residue numbering corresponding to BPV E1. The C-terminal approximately 300 amino acid segment functions as a helicase with enzymatic properties similar to the full length E1. The N-terminal half contains a sequence-specific origin DNA binding domain (OBD) and an N-terminal domain with regulatory properties. The constructs and their amino acid coordinates used in this study are shown. (B) Protein (63.7 µM) was pre-incubated with or without a 1/6 molar equivalent of T18 oligonucleotide before resolution on a pre-calibrated gel-filtration column. The elution volume of each protein alone was consistent with monomer (top panels), although a small shoulder to the E1HDΔC26 peak indicated the presence of higher order species. T18 ssDNA induced significant hexamerization of E1HD but not E1HDΔC26 (lower panels; M = monomer, H = E1HD hexamer and T18 the elution peak of the oligonucleotide). (C) Oligomerization of HD proteins was induced by ATP/Mg2+ without ssDNA, but for E1HDΔC26, monomer dominated over the presence of oligomeric species (top panel). Similar results were obtained in the presence of T18 (bottom panel), although the equilibrium was driven further in favour of hexamer (E1HD) and higher order oligomeric species (E1HDΔC26). (D) Gel-shift analysis with 32P-labelled T30 ssDNA. 1 nM T30 was incubated with HD proteins (31.3, 62.5, 125 and 250 nM) without (left) and with (right) 5 mM ATP/Mg2+. A hexameric ssDNA complex formed with E1HD as described previously (10) but ssDNA binding was much reduced with E1HDΔC26. Two complexes formed and formation of the species of lowest mobility was enhanced in the presence of ATP. Also, the faster migrating species was unstable compared to the slower one, as determined by ssDNA competition assay (data not shown). Formation of this species could relate to the tendency of E1HDΔC26 to form low molecular weight oligomers as observed in gel filtration without cofactors (B, top right).
In the present study, SAXS was employed to determine the configuration of the intact E1HD helicase containing the previously unresolved C-terminal extension. The SAXS results provide strong corroborative evidence that the C-terminal 26 amino acids of E1 play a major role in monomer-to-monomer contacts permuting around the hexamer, thus facilitating processive DNA unwinding. The conserved nature of this structural element among the papillomaviruses E1 also extends to the large T antigen (LTag) hexameric helicases of polyomaviruses, suggesting that they employ a similar mechanism to maintain the hexameric assembly.
MATERIALS AND METHODS
Protein expression and purification
The BPV-1 E1 helicase domain (E1HD) was expressed and purified to homogeneity as previously described (10). E1HDΔC26, lacking 26 amino acids at the C-terminus, was purified similarly. After ammonium sulphate precipitation, the protein was purified by size exclusion chromatography (Sephacryl S100 HR; 20 mM sodium phosphate pH 7.2, 500 mM NaCl, 2 mM DTT, 10% glycerol, 0.1 mM PMSF), IMAC chromatography (His-Trap, GE Healthcare; 50 mM Tris–Cl pH 7.5, 500 mM NaCl, 2 mM DTT, 10% glycerol, 1 mM PMSF, 10–270 mM imidazole gradient) and ion exchange chromatography (Source S, GE Healthcare; 20 mM phosphate pH 6.8, 2.5 mM DTT, 0.5 mM EDTA, 10% glycerol, 1 mM PMSF; 50–350 mM NaCl gradient). For biochemical characterization, both proteins were dialysed against 20 mM sodium phosphate pH 7.2, 300 mM NaCl, 1 mM DTT, 10% glycerol, 0.1 mM PMSF and stored at −80°C. Details of the buffers used to stabilize high concentration samples of E1HD and E1HDΔC26 for SAXS experiments are described in the Supplementary Materials and Methods. Protein concentrations were determined by Abs 280 nm using the calculated molar extinction coefficients.
Oligomerization, DNA binding and enzymatic assays
Gel filtration (20 mM Na phosphate pH 7.2, 200 mM NaCl, 10% glycerol, 1 mM DTT, 1 mM PMSF) was performed as previously described using a Superdex 200 column calibrated against protein standards (10), with a protein concentration of 63.7 µM and T18 oligonucleotide concentration of 10.6 µM. Where appropriate, 5 mM ATP/5 mM MgCl2 was added to the protein pre-incubation (10 min, 4°C) and 1 mM ATP/3 mM MgCl2 to the column buffer. Assays were performed in triplicate at least.
DNA binding reactions (20 mM Na phosphate pH 7.2, 135 mM NaCl, 10% glycerol, 0.1% NP40, 0.1 mg/ml BSA, 1 mM DTT, 1 mM PMSF) employed a T30 oligonucleotide (1 nM) end-labelled with 32P using standard procedures. Products were resolved on 5% 80:1 acrylamide:bis-acrylamide gel, 0.25× TBE buffer, as described previously (10). Dried gels were exposed to phosphorimager plates for image capture and quantification (Fuji FLA 3000, image gauge V3.3 software). Data shown are for assays performed at least in triplicate and expressed as the mean and standard error of the mean.
ATPase assays (4 µM HD protein) were performed at 22°C in 20 mM HEPES-NaOH pH 7.5, 135 mM NaCl, 1 mM DTT, 0.01% NP40, 7.5 mM ATP, 8.5 mM MgCl2 containing 35 nmols/µl [γ-32P]ATP (7000 Ci/mmol). Pi release was determined by the charcoal binding assay of Iggo and Lane (17). Data shown are for assays performed at least in triplicate and expressed as the mean and standard error of the mean.
The substrates for the helicase assay were partially single- and double-stranded DNA. The single-stranded component was a 3′ T55 overhang and the dsDNA components either 25, 76 or 153 bp. To make the substrate with 25 bp, the following oligonucleotides were annealed and ligated: 5′-CGCGCTGAGGTGCGGTGTGAAATAC (O1), 5′-GTATTTCACACCGCACC (O2) and 5′-TCAGCGCG(T)55 (O3). O1 was 32P end-labelled and O3 synthesized 5′ phosphorylated. The substrates with 76 and 152 bp dsDNA were generated from PCR products amplified from pUC19 DNA. One primer (5′-CGCGCTGAGGTGCGGTGTGAAATACCG, 32P end-labelled), contained a cleavage site for the nicking endonuclease Nt.BbvCI (underlined, complementary strand). PCR products were cleaved with Nt.BbvCI, purified by phenol/chloroform extraction and ethanol precipitation and ligated with an excess of oligonucleotide O3. All substrates were gel-purified before use. Helicase assays were performed in 25 mM HEPES-NaOH pH 7.5, 20 mM NaCl, 1 mM DTT, 1 mM ATP, 3 mM MgCl2 as described previously (10). Imaging and quantification was done as described above. Data shown are for assays performed at least in triplicate and expressed as the mean and standard error of the mean.
SAXS data collection and analysis
SAXS data were collected at the EMBL X33 beamline at the DESY storage ring DORIS III, Hamburg (18,19). Detailed description of E1HD, E1HDΔC and E1HD/ssDNA samples is provided in Supplementary Materials and Methods. Notably, the buffer used for gel-filtration analysis was not appropriate for SAXS analysis due to the high glycerol concentration. Hence, a number of buffer conditions were tested to prevent or reduce polydispersity. Samples were measured at 10°C at a minimum of three solute concentrations. E1HD/ATP was measured in the range 1.8–6.7 mg/ml. E1HD and E1HDΔC concentrations are detailed in Supplementary Table S1. Data were processed using PRIMUS (20). The buffer subtracted data were extrapolated to infinite dilution where appropriate and further used for analysis and modelling (Supplementary Figure S1). The radii of gyration (Rg) were determined from the Guinier approximation (21), where slight sample polydispersity was ameliorated by truncation of scattering data at the lowest scattering angles. Maximum complex dimensions Dmax and the interatomic distance distribution functions P(r) were calculated using GNOM (22). The excluded (Porod) particle volumes were calculated using PRIMUS. The molecular weight (MW) of the particle was estimated using the SAXSMoW applet (www.if.sc.usp.br/~saxs/) (23). Evaluation of the theoretical scattering curves using crystal structures of E1HD (PDB codes 2V9P and 2GXA, chains A–F) (9,11) and fitting to the experimental scattering data was performed using CRYSOL (24). The coordinates of DNA and small molecule cofactors were removed from the structures prior to the calculations.
Ab initio modelling
Ten ab initio bead models were calculated using DAMMIN (25), with and without P6 symmetry, where data was fitted to s = 2.5 nm−1. For the hexameric E1HD/ATP, the models were generated with P6 symmetry and oblate particle anisometry. Ten higher resolution ab initio models were constructed using GASBOR (26) with P6 symmetry imposed, where the asymmetric part comprised 307 ‘dummy’ residues. Different ab initio models were aligned using DAMAVER (27), which provides a value of Normalized Spatial Discrepancy (NSD). NSD values close to one indicate that the models are similar. The GASBOR model with the best fit to the data was aligned with the crystal structure of E1 helicase using Supcomb13 (28).
Combined ab initio and rigid body modelling
The E1HD protein used for SAXS included extensions at the N-terminus (six amino acids) and C-terminus (26 amino acids) that have previously not been structurally defined. These regions, and a loop at position 551–553, were modelled using the program BUNCH (29) with P6 symmetry imposed. BUNCH models the missing parts with interatomic distances and angles between ‘dummy’ residues to mimic a Cα chain. Ten BUNCH models were generated and CRYSOL was used to determine the model with lowest discrepancy to scattering data in the range 0.02–2.5 nm−1. This model was further refined using normal mode analysis (43), and subsequently aligned with the crystallized hexamer (PDB code: 2V9P hexamer 1) using CCP4MG (30).
Electrostatic potential analysis and subunit–subunit interface characterization
A hybrid E1HD hexamer structure was constructed using the 2V9P (chains A–F; residues 301–305, 310–546, 559–579) hexamer, where the disordered side chain of K425 was modelled for subunits A–D; and six copies of 2GXA, chain F, residues 547–558. This hybrid model was used to calculate the electrostatic potential of the E1HD hexamer using the CHARMM PBEQ electrostatics server (31) with default settings, except the grid spacing, which was increased to 50 Å. The continuum electrostatic isopotential was visualized using PyMOL, contoured at ±0.25 kT/e. Interface contact areas, hydrogen bonding and salt bridge formation in the E1HD hexamer were analysed using the Protein Interfaces, Surfaces and Assemblies service (PISA), European Bioinformatics Institute (http://www.ebi.ac.uk/pdbe/prot_int/pistart.html) (32). The hydrogen bond cutoff distance was 3.5 Å.
Sequence alignments and disorder prediction
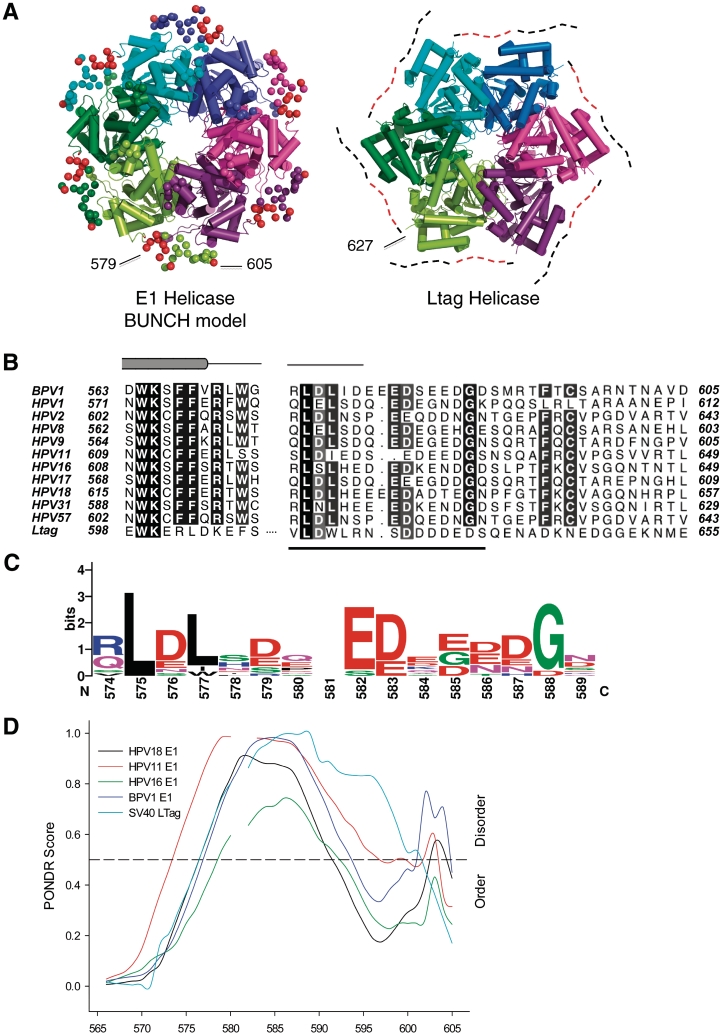
A search for homologous helicases was performed using PSI-BLAST against the BPV1 E1 helicase (378–605). The SV40 LTag helicase aligned with 27% sequence identity through E1 residues 416–545. A global alignment of E1 helicase sequences with the LTag using Kalign (33) identified a conserved acidic patch of residues in the C-terminal segment of these hexameric helicases. Protein disorder was predicted using the RONN server (34), DisEMBL (35), Disopred (36) and PONDR VL-XT (37–39).
RESULTS
Oligomerization of E1HD
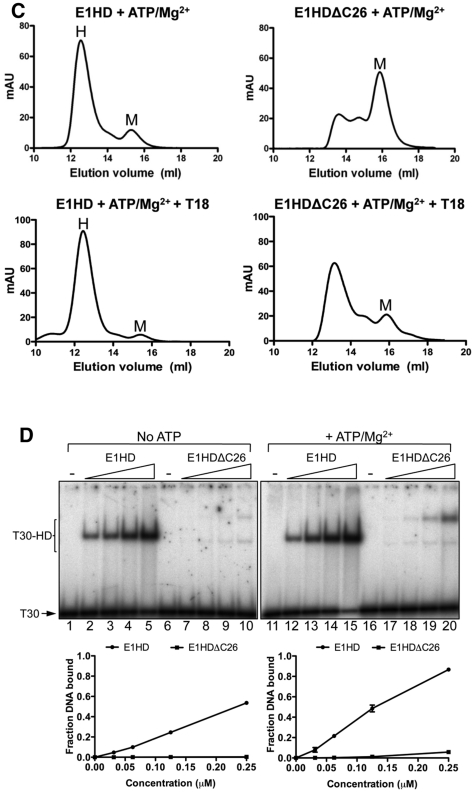
In our previous X-ray structure of E1HD, the final refined model included residues 300–579 of the crystallized segment 299–605 (9). There was no electron density for the 26 C-terminal residues, indicating that they are disordered or have a flexible nature. Overall, this structure could be readily superimposed with the structure of the C-terminally truncated E1HD (residues 308–577) (11) determined in complex with ADP and ssDNA, demonstrating that the integrity of the protein fold is maintained regardless of the presence of the C-terminal amino acids. This observation is substantiated by the fact that both purified E1HD (residues 299–605) and its C-terminally truncated version E1HDΔC26 (residues 299–579) have virtually identical circular dichroism (CD) spectra (data not shown). However, our analysis of oligomerization induced by nucleotide cofactor or ssDNA in solution revealed significant differences between E1HD and E1HDΔC26. Consistent with our previous analysis (10), hexamerization of E1HD (63.7 μM) was readily induced in the presence of ssDNA (T18; 10.6 μM) as determined by gel filtration (Figure 1B, left). In contrast, little oligomerization was detected with E1HDΔC26 (Figure 1B, right). However, we note that even in the absence of any cofactors, E1HDΔC26 (Figure 1B, top right) exhibited a tendency to form some low molecular weight oligomer, evident as the small shoulder to the monomeric protein peak. Similarly, hexamerization of E1HD was induced in the presence of ATP/Mg2+, but with E1HDΔC26 oligomerization was reduced and characterized by the presence of multiple species (Figure 1C). For both proteins, the combination of ssDNA and ATP/Mg2+ is most efficient for inducing oligomerization compared to each ligand alone. Furthermore, the hexameric E1HD complex formed with ATP/Mg2+ remained stable after reapplication to the gel-filtration column. In contrast, the oligomeric complex formed with E1HDΔC26 under the same conditions and also reapplied to the column in the same time frame almost completely dissociated to monomer (Supplementary Figure S2). Thus, the 26 C-terminal residues of E1 influence both assembly and stability of the oligomeric state.
Similar results to the above were obtained by gel-shift analysis using a 32P end-labelled T30 oligonucleotide and protein titrated in the nanomolar range (Figure 1D). E1HD formed a single discrete protein–DNA complex without and with ATP/Mg2+ (lanes 2–5 and 12–15 and the graphed data below). With E1HDΔC26 (lanes 7–10), DNA binding in the absence of ATP/Mg2+ was minimal compared to E1HD and two protein DNA complexes formed. However, when the binding reactions were challenged with excess unlabelled ssDNA competitor after complex formation, only the species with the slowest mobility was stable (data not shown). With ATP/Mg2+, the DNA binding activity of E1HDΔC26 increased significantly. Two protein–DNA complexes were also observed but the complex with the slowest mobility dominated (lanes 17–20). These data therefore demonstrate that oligomerization of E1HD induced by either nucleotide or ssDNA cofactors are significantly impaired when the C-terminal ‘tail’ of 26 amino acids are deleted.
Enzymatic activities of E1HD and E1HDΔC26
The ATPase activity of E1HD and E1HDΔC26 was determined by measuring the release of 32Pi from [γ-32P]ATP, in the absence and presence of T30 ssDNA, over time (Figure 2A). Without the T30 oligonucleotide, ATP hydrolysis was reduced by ~50% for E1HDΔC26 compared to E1HD (the turnover numbers determined from initial rates are 0.29 and 0.14/s for E1HD and E1HDΔC26 respectively). In the presence of excess T30 ssDNA (1:1.5 T30:HD monomer), the rate of ATP hydrolysis increased by ~1.6-fold for E1HD. However, for E1HDΔC26 the increase was 2.9-fold (E1HD 0.47/s and E1HDΔC26 0.4/s determined from the initial rates) and therefore the difference in enzymatic activity between the two forms of the enzyme is small in the presence of ssDNA. These data reflect the data in Figure 1B and C above, where formation of the hexameric E1 assembly, the active form of the enzyme, is most efficient with ssDNA and ATP/Mg2+ combined. They also argue that the C-terminal 26 amino acids of E1HD have a significant indirect effect on ATP turnover by influencing the hexameric assembly state of the enzyme rather than the active site directly.
Figure 2.
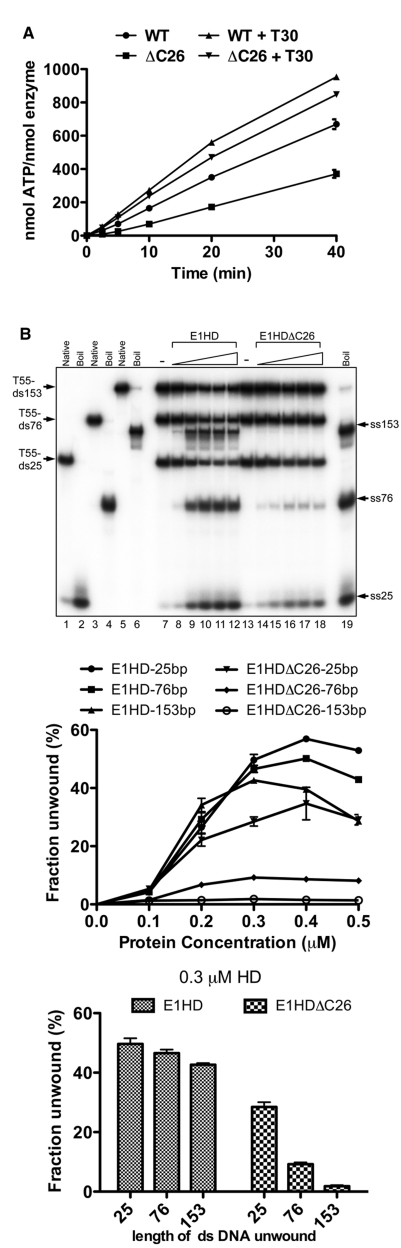
Helicase domain enzymatic assays. (A) ATPase assays (4 µM HD protein) with or without T30 ssDNA oligonucleotide (1:1.5 T30:HD monomer) were sampled over time (2.5, 5, 10, 20 and 40 min) and phosphate release determined. Without T30 ssDNA, the ATPase activity of E1HDΔC26 was reduced ~50% relative to E1HD. With ssDNA, the activity of E1HDΔC26 was ~85% that of E1HD. Turnover numbers (per second) were determined from the slope of the graph using the values up to 20 min where ATP hydrolysis is in the linear range. (B) Helicase assays (0.1, 0.2, 0.3, 0.4 and 0.5 µM HD protein) were performed with substrates with a 55 base 3′ poly T tail and duplex portions of 25, 76 and 153 base (T55-ds25, T55-ds76 and T55-153; 0.5 nM each) combined in the same reaction. The short strand of each was end-labelled with 32P and reaction products were resolved by polyacrylamide gel electrophoresis. The line graph shows unwinding as a function of protein concentration. The bar graph shows the data for the intermediate concentration (0.3 µM). Compared to E1HD, unwinding of dsDNA by E1HDΔC26 was significantly and progressively impaired with increasing DNA length.
We next asked if the C-terminal 26 amino acids of E1 influence helicase activity. The substrates used consisted of a 55 poly T 3′ ssDNA component required for helicase loading and either 25, 76 or 153 bases of dsDNA to be unwound (T55-ds25, -ds76 and -ds153). The three substrates, each with one strand 32P radiolabelled, were combined in a single reaction and strand displacement measured by gel electrophoresis of reaction products. In Figure 2B, lanes 1–6 show the electrophoretic mobility of each substrate in the native and denatured (boiled) state. Lanes 7–12 demonstrate DNA unwinding by the intact helicase domain, E1HD. There was little difference in the ability of E1HD to unwind the substrates with 25, 76 and 153 base pairs of dsDNA (see the line graph below for all protein concentration and the bar graph for the intermediate protein concentration, 0.3 µM HD). In contrast, for E1HDΔC26 unwinding of T55-ds25 was reduced to ~0.6 times that of E1HD, T55-ds76 was reduced further (~5-fold) and unwinding of the substrate with 153 bases of dsDNA was minimal. Therefore, relative to E1HD, E1HDΔC26 demonstrates a progressive defect in its ability to unwind dsDNA substrates of increasing length. Considering the data above, the most likely explanation of this outcome is that the C-terminal 26 amino acids influence the assembly and stability of the HD hexamer and hence its residence time once engaged with a substrate. E1HDΔC26 therefore has defective processivity. Consistent with this result, E1HDΔC26 completely fails to support replication of BPV ori plasmids in in vivo replication assays (data not shown).
Solution structures of E1HDΔC26 and E1HD
An analysis of E1HDΔC26 and E1HD was undertaken in the absence of ATP, to investigate the solution structure of the monomeric species. We first studied E1HDΔC26, which is not expected to oligomerize significantly in solution. To maintain monodispersity at the high protein concentrations used for SAXS, numerous buffer conditions were investigated (see ‘Materials and Methods’ section and Supplementary Table S2). The overall parameters calculated from the scattering patterns, summarized in Supplementary Table S1, indicated significant concentration dependence, pointing to formation of larger oligomeric species at higher concentrations in the range 72–215 μM, particularly evident at higher salt concentrations. Notably, these effects were not significant at the lower protein concentrations (63.7 μM) used for gel-filtration analysis of oligomerization. We first checked if the data could be fitted with available crystallographic models. The PDB structure (2V9P) of hexamer 1 was split into six monomers and their scattering patterns were calculated by CRYSOL (24). The individual chains from 2V9P showed a reasonable agreement with the experimental scattering from E1HDΔC26 in solutions below 3 mg/ml (94 μM), but displayed significant deviations with increasing protein concentration. Second, we tested the possible presence of dimeric species by creating six dimers and validating their scattering data using CRYSOL (24). The fits for the dimeric species were still far from perfect, in the entire concentration range (data not shown), clearly indicating that yet higher oligomers are present in solution. To determine the oligomeric composition of E1HDΔC26 in solution, different higher order oligomers were generated (trimers, tetramers, hexamers, double hexamers) and their scattering patterns were employed together with those of monomers and dimers to fit the experimental data using OLIGOMER (20). The fits indicated that only monomers, dimers, hexamers and double hexamers were present in significant amounts (see Supplementary Table S1). The OLIGOMER results allowed us to conclude that (i) E1HDΔC26 is mainly present as a monomer at low concentrations, in agreement with gel-filtration data; whereas (ii) high concentration data could only be well fitted with a mixture of monomers and a small fraction of hexamers or double hexamers (see Supplementary Table S1).
We then studied the intact E1HD containing the C-terminal segment. This species required higher salt to maintain monodispersity for SAXS analysis (see Supplementary Table S2 and Supplementary Materials and Methods). Interestingly, even at the lowest protein concentration, the data could not be fitted by the scattering from a monomeric protein alone, irrespective of whether the missing peptide was added using BUNCH (see analysis below). OLIGOMER was employed to fit the SAXS data with oligomeric mixtures as described above; possible oligomers were generated using the hexamer BUNCH model (see below). The analysis of the full-length E1HD revealed that the fractions of higher order oligomeric species are covariant with protein concentration (see Supplementary Table S1). Compared to the deletion mutant, the full-length E1HD displayed lower amounts of monomers and systematically higher fractions of dimers and hexamers. However, neither trimers nor tetramers were present in the samples we studied. This was not surprising as both these species are thought to form only on a double-stranded DNA scaffold, with the interaction mediated by the E1 origin DNA binding domain with its binding site (40,41). Differences in oligomerization states observed by gel filtration (Figure 1B) are likely due to the comparatively lower protein and salt concentrations used during gel-filtration analysis.
Overall, our SAXS analysis indicates that E1HDΔC26 has a much lower ability to form higher order oligomers compared to E1HD. The significantly greater propensity for higher order oligomerization of the full-length E1HD indicates that the C-terminus plays a major role in the oligomerization.
The C-terminal segments of E1HD form a flexible monomer–monomer bridge
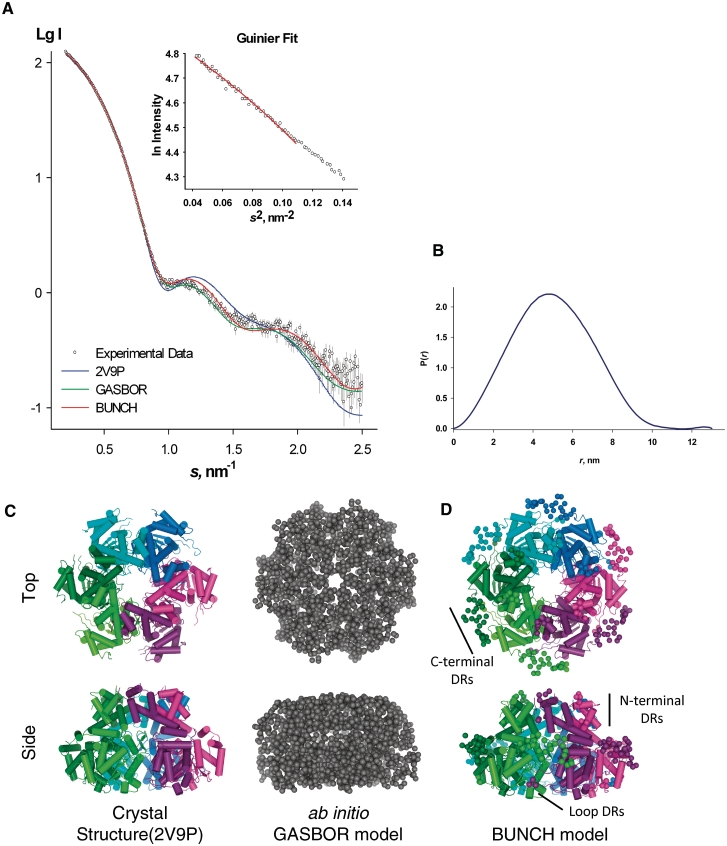
Next, we induced hexamerization of E1HD with ATP and collected the SAXS data (Figure 3A, black circles). The Guinier plot of the data extrapolated to infinite dilution indicated monodisperse particles (Figure 3A, inset) with a radius of gyration (Rg) of 3.9 ± 0.1 nm. Molecular weight (MW) estimation using Porod analysis indicated that the MW equals 230 ± 20 kDa, in agreement with the calculated 209 kDa mass of the hexamer. Further, as described below, ab initio modelling using DAMMIN identified an excluded volume, Vp = 380 ± 40 nm3, which is consistent with a hexameric assembly. Finally, given Dmax = 13 nm (Figure 3B), the SAXS MoW applet (23) was used to approximate the MW of the E1HD complex, yielding 210 ± 20 kDa, the equivalent of six monomers per assembly. X-ray scattering intensity predicted on the basis of the crystal structure (residues 301–579; PDB code 2V9P), gave Rg = 3.5 nm, Dmax = 11.4 nm and Vp = 300 nm3, all values being smaller than those observed by SAXS. Computed scattering for the E1HD hexamer (residues 308–577, PDB code 2GXA) yielded similar results (Rg = 3.5 nm, Dmax = 11.3 nm and Vp = 300 nm3). This size difference was likely due to the absence of the C-terminal extensions present in the protein used for SAXS analysis. Further, scattering calculated for the crystal structures of E1HD showed a discrepancy in fit to the experimental data (s range from 0.2 to 2.5 nm−1), where χ = 2.79 and χ = 2.77 for 2V9P (Figure 3A, blue line) and 2GXA, respectively. Scattering of a hybrid model of 2V9P and 2GXA (see ‘Materials and Methods’ section) was also computed, resulting in a similar size particle with a slightly improved fit to the experimental data (data not shown). All scattering parameters and χ fits are summarized in Table 1.
Figure 3.
SAXS ab initio and rigid body modelling of the E1HD/ATP hexamer (A) SAXS profile at infinite dilution for the E1HD hexamer (Experimental data, black circles), overlaid with the crystal structure, ab initio and combined ab initio/rigid body model fits. The linear Guinier plot shown inset confirmed the sample was monodisperse. Predicted scattering for the crystal structure of the E1HD hexamer 2V9P chains A–F (blue), GASBOR ab initio model (green), ab initio/rigid body BUNCH model (red) are shown as solid lines. (B) Distance distribution P(r) function calculated using GNOM for E1HD/ATP hexamer, Dmax = 13 nm. (C) The GASBOR model (χ = 1.34) (grey) was aligned with 2V9P chains A–F (cartoon representation, (left) and translated along the x-axis, and both viewed from the N-terminal oligomerization domain (top), and rotated about the x-axis by 90° (bottom). The hexamer is coloured lime, green, cyan, blue, pink and purple from chain A–F, respectively. (D) BUNCH symmetrical hexameric model constructed using 2V9P chain A, refined using Normal Mode Analysis (χ = 1.57) (top view) shows ‘dummy’ residue (DRs) C-terminal tails (spheres) are located between monomers. N-terminal and loop DRs are also indicated (side view). Figures were prepared using PyMOL (42).
Table 1.
Summary of SAXS data and modelling of E1HD/ATP hexamer
| Guinier | P(r) | Porod | SAXS MoW | CRYSOL |
Ab initio |
BUNCH | |||
|---|---|---|---|---|---|---|---|---|---|
| 2V9P | 2GXA | DAMMIN | GASBOR | ||||||
| Rg (nm) | 3.9 ± 0.1 | 3.8 | – | 3.8 | 3.5 | 3.5 | 3.8 | 3.8 | 3.6 |
| Dmax (nm) | – | 13 | – | – | 11.4 | 11.3 | 10.9 | 13 | 11.4 |
| Vp (nm3) | – | – | 370 ± 40 | 300 | 300 | 380 ± 40 | 310 | 325 | |
| MW (kDa) | – | – | 230 ± 20 | 210 ± 20 | 180 | 180 | 220 ± 20a | – | 209 |
| Chi | – | – | – | – | 2.79 | 2.77 | 1.19b | 1.34 | 1.57 |
Chi discrepancy was calculated for experimental data in the range s = 0.02–2.5 nm−1.
aExcluded volume and MW estimate from model with P1 symmetry, no imposed anisometry.
bChi discrepancy to the experimental data for model calculated with P6 symmetry and oblate anisometry imposed.
We used ab initio modelling to reconstruct the hexamer model of E1HD. Ten GASBOR models were generated, with normalized spatial discrepancy (NSD) values in the range 1.04–1.09. Further, 10 DAMMIN models were calculated, with NSD values in the range 0.64–0.74. Both GASBOR and DAMMIN modelling converged to a similar shape as judged by low NSD values (Supplementary Table S3). The best fit to the experimental scattering showed an improved fit with χ = 1.19 and χ = 1.34 for DAMMIN and GASBOR, respectively (Figure 3A, green line). The GASBOR model (Figure 3C, grey) correlated well with the crystal structure, having NSD = 1.05 (PDB code 2V9P; Figure 3C, left).
Given the particle in solution contained C-terminal extensions not visible in the crystal structure, a combined ab initio and rigid body modelling approach was also employed. An analysis of intersubunit contacts (see below) suggested that the weakest interface within the hexamer occurs between chains A and F. We therefore focused on chain A as the rigid body for symmetrical BUNCH modelling. Generated BUNCH models showed reasonable agreement to the scattering data (s range from 0.020 to 2.5 nm−1) with χ in the range 1.74–1.93, NSD in the range 0.43–0.71, illustrating that all models were similar, with the best fitting model having a Cα RMSD of 2.22 Å to the crystallized hexamer (PDB code: 2V9P) (Supplementary Table S3 and Supplementary Figure S3). As some movements are expected in the hexamer, we further refined the BUNCH model using normal mode analysis (43). The resulting model had a discrepancy of 1.57 and RMSD of 2.4 Å to the BUNCH model; and an RMSD of 2.72 Å when compared with the crystallized hexamer structure (PDB code: 2V9P hexamer 1). Notably, the ab initio/rigid body model showed an improvement in the fit in the range s = 1–2 nm−1 relative to the 2V9P, 2GXA and hybrid fit (Figure 3A, red line). The ab initio modelled ‘dummy’ residues at the C-terminus were found proximal to the inter-monomer interface, where the terminus extends to contact the neighbouring subunit (Figure 3D). We note that the C-terminus is very rich in charged amino acids, containing 11 negatively charged and four positively charged residues per C-terminal segment (see below).
To test whether the E1 helicase forms similar assemblies in the presence of DNA, ADP and Mg2+, we performed SAXS experiments on E1HD complexes formed with two ssDNA fragments (14 and 16 bp), in the presence and absence of ADP/Mg2+. For all samples, clear formation of hexamers was observed even at the lowest protein concentration. Similarity of scattering patterns (Supplementary Figure S4) indicates that all complexes have the same architecture as E1HD/ATP.
Consecutive remodelling of subunit–subunit interactions around the hexameric ring
An analysis of the inter-monomer interface contact areas of the E1HD/ADP/ssDNA (2GXA) hexamer using PISA (Table 2) showed that the most extensive area is buried between subunits B and C (1525 Å2, 17% of a total subunit surface) with most of the contacts (~1000 Å2) made between AAA+ domains. This interface (Supplementary Figure S5A, chain B green, chain C cyan), has been previously characterized as an ‘ATP-bound’ interface. In contrast, the interface between subunits F and A, described as ‘empty’ (11), is significantly smaller (889 Å2, 11% of a total subunit surface), with most of the contacts (550 Å2) generated by the oligomerization domain and with only minimal contacts contributed by the AAA+ domain (Supplementary Figure S5A, chain A lime, chain F purple). A structural alignment of AAA+ domains of subunits B and F illustrates the difference in interface contacts with partner subunits C and A, where the arrow indicates the relative direction of movement of subunit A away from the interface (Supplementary Figure S5A). The significant difference in subunit–subunit contacts is also reflected in the number of direct hydrogen bonding interactions. Subunits B and C are bridged by 14 hydrogen bonds, 11 of which are within the AAA+ domain (378–605). In comparison, subunits A and F of the ‘open’ interface make only three hydrogen bonding interactions, all formed between oligomerization domains and notably no hydrogen bonding interactions form between the C-terminal ATPase domains. Thus, the additional bridging of subunits in the case of the ‘open’ A/F interface, as indicated by BUNCH modelling (Supplementary Figure S5B, chain A lime, chain F purple), would be particularly important for oligomerization.
Table 2.
Intersubunit surface area calculated for E1 hexamer (PDB code 2GXA)
| Monomer pair | Interface area (Å2) | Hydrogen bonds | Salt bridges |
|---|---|---|---|
| ABa | 1409 | 9 | 6 |
| BCa | 1525 | 14 | 5 |
| CDa | 1499 | 12 | 8 |
| DEb | 1392 | 12 | 6 |
| EFb | 1252 | 6 | 2 |
| FAc | 889 | 3 | 4 |
Interface pairs were previously characterized as: aATP bound, bADP bound and cempty (11).
Sequence and disorder conservation
The E1 helicase is structurally similar to the SV40 LTag helicase (Figure 4A). Notably, the C-terminus of neither helicase has been characterized structurally by X-ray crystallography, but both feature a conserved acidic C-terminal region immediately following the last α-helix defined in both structures (Figure 4B and C). Sequence-based analysis identified potential disorder in BPV1 E1 encompassing residues 577–603 (RONN), 573–605 (DisEMBL), 579–593 and 598–605 (Disopred), 577–593 and 602–605 (PONDR). In particular, a sequence motif comprising amino acids 572–589 associated with the intrinsic disorder (44) appears to be conserved, as illustrated in Figure 4C. The position of the predicted disorder is conserved in other HPV E1 proteins and also in SV40 LTag helicase (Figure 4D) and other polyomavirus LTag helicases, aligning with the previously identified sequence motif (BPV1 E1 572–589, Figure 4C). In agreement with this analysis, in the crystal structure of E1 helicase (2V9P), there was no electron density observed for residues 580–605 and there was no difference between E1HD and E1HDΔC26 CD spectra. Taken together, the data indicate that the negatively charged, conserved C-terminus has a flexible character.
Figure 4.
Sequence and disorder conservation. (A) E1 and LTag helicase structures are shown side by side. The acidic residues of the C-terminal segment of E1 (BUNCH model) are shown as red spheres (left). The corresponding C-terminal extension of LTag is represented as a dotted line (right), with the acidic region represented as a red dotted line. The structurally characterized C-terminal residues are shown as spheres. (B) Alignment of E1 helicase sequences (BPV1 and HPVs 1, 2, 8, 9, 11, 16, 17, 18, 31, 57) and SV40 LTag shows C-terminal sequence conservation following the last α-helix. The secondary structure in this region of BPV1 E1 is illustrated above the alignment. A 16 residue insertion in this region of LTag, not present in the E1 sequences, is represented by a dotted line. The underlined section indicates the position of the sequence logo. (C) A sequence logo covering the region 572–589 (BPV 1 E1) of the MSA produced using WebLogo (45). This representation of the alignment in (B) shows maintenance of residues typically associated with disorder. (D) Disorder prediction (PONDR) based on aligned C-terminal sequences of HPV 11, 16, 18; BPV1 E1 and LTag illustrates homologous disorder potential. The sequence numbering corresponds to BPV1 E1.
Electrostatic potential calculations
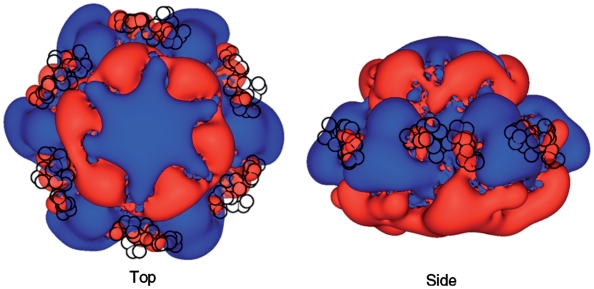
As the C-terminus contains a conserved segment of negatively charged residues, we reasoned that its role could be to stabilize the hexamer through electrostatic interactions. We investigated this possibility using electrostatic potential calculations, focusing on continuum electrostatic effects (46) (Figure 5). This analysis has highlighted the positive potential of the central tunnel, previously characterized as the region of the hexamer that binds ssDNA. Surrounding the central tunnel, at the top and the base of the hexamer, are areas of negative potential framing large extended areas of positive potential emerging at the surface of each subunit (Figure 5, right). This positive potential is a dominant feature around the outside of the hexamer. Notably, the electrostatic potential has asymmetric features at each subunit interface, possibly conferring a variable effect on the position of the C-terminal peptide. We note that the BUNCH modelling positioned the conserved acidic C-termini between extended basic potentials of adjacent subunits (Figure 5, black circles).
Figure 5.
Continuum electrostatics of the BPV1 E1 hexamer. Calculations were made for a hybrid E1 hexamer generated using PDB structures 2V9P and 2GXA. Electrostatic potentials were contoured at +0.25 kT/e (blue) and −0.25 kT/e (red). E1 is shown along the oligomer axis (left) and rotated by 90°about the X-axis (right). The C-terminal ‘dummy’ residues are shown as black circles, overlaid on the electrostatic continuum images.
DISCUSSION
Our biochemical data reveal defective oligomerization for E1HDΔC26 compared to a full-length E1HD in the presence of ATP/Mg2+ and ssDNA (Figure 1). This defect appears also to manifest itself at the level of hexamer stability, as the deletion mutant E1HDΔC26 poorly unwinds long, but not short, dsDNA substrates in comparison to the intact helicase domain (Figure 2). These data indicated a direct involvement of the C-terminal residues in maintaining a processive DNA unwinding complex. To understand the structure–function relationship of the C-terminal 26 residues of the E1HD hexamer, we determined the solution structure of the intact BPV E1 helicase domain using SAXS. This structure includes a C-terminal peptide which was not resolved in the previous crystal structure (9). The hexamer model shows the position of the C-terminal 26 amino acids (580–605) in the assembly (Figure 3). The acidic portion (residues 580–590) of the tail is proximal to the C-terminal lobe of the same protein subunit, projecting towards the neighbouring subunit, where residues 591–605 extend and make interface contacts. Structural observations are in agreement with sequence-based disorder prediction and consistent with the absence of electron density corresponding to these residues in crystals (9). Furthermore, sequence alignments of other E1 helicases have revealed the conservation of a predicted region of intrinsic disorder within the C-terminal peptide. This conservation appears to extend to polyomavirus LTag helicases (Figure 4). We note that the C-terminal residues are positioned between adjacent subunits indicating their potential involvement in bridging interactions. This is particularly important for the A to F ‘open’ interface, where the inter-subunit contacts are most loose. Our analysis suggests an electrostatic nature of interactions with the C-terminus due to the presence of intensely positive electrostatic potentials extending from adjacent subunits.
Previous structural data showed that each subunit of the E1HD hexamer is in a distinct conformational state (9,11). A mechanism of DNA translocation was proposed (11) in which each subunit undergoes a conformational change to sample each NTP hydrolysis state, with changes permuting around the hexameric ring in a wave-like motion. Transitions between different states around the ring of E1 subunits are coupled with ATP hydrolysis events. These transitions result in substantial remodelling of subunit–subunit interactions with the surface area of inter-subunit contacts varying between 1525 and 890 Å2 (Table 2). Most of this difference is due to changes in the contact area between AAA+ domains, which varies between 960 and 198 Å2. Notably, the decrease in the inter-subunit contact area within this region of the interface is characterized by a complete loss of direct inter-subunit hydrogen bonds. Given the dramatic differences in interface contacts, the question arises as to how the hexameric assembly can be maintained over many catalytic cycles in such a dynamic system? The same considerations would also apply for the SV40 LTag helicase. In contrast to the E1 helicase, LTag has been proposed to unwind DNA using a concerted mechanism, where ATP is hydrolysed simultaneously around the hexameric ring (47). Post-hydrolysis, the apo LTag hexamer undergoes an ‘iris-like’ conformational change, resulting in a dramatic decrease in inter-subunit contact area, transitioning from 4344 Å2 in the ATP bound state to 2474 Å2 in the apo state. Therefore, the percentage decrease in inter-subunit contacts for E1 helicase (42%) and LTag (44%) are similar. Our data on E1 indicate a requirement for a flexible ‘brace’ at the C-terminus of the protein that appears to have a functional counterpart in LTag, which acts as a dynamic bridge between subunits to maintain hexamer stability. Supporting this mechanism is the observation that LTag C-terminal truncation mutants missing residues in the sequence range 591–669 fail to form higher MW oligomers in vivo (48). The conserved C-terminal intrinsic flexibility motif identified by alignment with E1 helicases lies within this region (633–644), with predicted flexibility (PONDR) extending to residue 678. Further, cryoelectron microscopy difference maps of wild type versus truncated LTag (108–627) show strong peaks of density between subunits (49). Interestingly, the involvement of N-terminal segments in contacting adjacent subunits has previously been reported for bacteriophage T7 gp4 protein, bacteriophage phi12 p4 protein and repA helicase (7,50,51). We note, however, that the N-terminal bridging motif in all these cases is fixed on the adjacent subunit, making several specific short-range interactions. Further, these proteins form obligate hexamers in the absence of DNA/RNA and nucleotide cofactors, which has not been observed for E1 and LTag helicases. The ‘fixed bridge’ mechanism of the oligomer stabilization in these proteins thus contrasts the proposed ‘flexible brace’ mechanism of E1 and LTag, where subunit assembly is maintained by a long range electrostatic interaction between the negatively charged C-terminus and the positive electrostatic potentials of two adjacent subunits.
The ability to maintain a stable assembly on DNA for many rounds of catalysis is a distinguishing feature of replicative helicases. The ring-like assembly of some hexameric helicases around ssDNA suggests that subunit interactions contribute significantly to processivity. Although protein–protein interactions between protein domains with distinct three-dimensional architecture have been more widely considered, the contribution of unstructured domains is becoming increasingly recognized. Intrinsically unstructured proteins, protein domains and short linear motifs are common in the proteomes of most organisms (44,52,53). Of this class of protein/protein domain, many function in molecular recognition in key cellular processes such as transcription (54), serving to assemble, stabilize and regulate multi-protein complexes (52). In such cases, the absence of secondary structure elements and a stable, defined fold may confer certain advantages such as increased speed and freedom in orientational search for the binding target. Interactions through short linear motifs are often of low affinity and mediated by only a few key residues, allowing them to act as transient molecular switches (55–57). These short linear motifs are of a mixed sequence composition but with underlying flexibility, often enriched with hydrophobic and charged residues and also potential phosphorylation sites that could provide an opportunity for regulation (53). The features of the C-terminal tail of E1 that we describe physically and functionally bare a clear resemblance to this class of peptide. On the basis of our data, we propose that the C-terminal tails of the E1 helicase dynamically bridge interactions between monomers, adjusting their conformation in accord with positional and angular movements between adjacent subunits. We note that the C-terminal segment is located between adjacent subunits. It is rich in acidic residues and is flexible (disordered), while the adjacent subunits have an intensely positive electrostatic potential, thus indicating the electrostatic nature of interactions that stabilize the oligomeric state. By this mechanism, the hexamer is maintained in the processive phase, reducing the likelihood of disassembling the active DNA unwinding complex. Additionally, we propose that polyomavirus LTag helicases utilize a similar C-terminal dynamic brace mechanism to E1 to perpetuate their hexameric assembly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–3, Supplementary Figures 1–5 and Supplementary Materials and Methods.
FUNDING
Yorkshire Cancer Research (grant number S302 to C.S. and A.A.A.); Wellcome Trust (fellowship number 081916 to A.A.A.); Bundesministerium für Bildung und Forschung (grant SYNC-LIFE contract number 05K10YEA to A.V.S. and D.I.S.); European Union FP7 e-Infrastructure (grant WeNMR, contract number 261572). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Clement Blanchet for technical support at the X33 beamline and Peter Konarev for help during the SAXS data analysis. We acknowledge the EMBL-Hamburg for synchrotron beamtime allocation at DORIS storage ring, EMBL/DESY Hamburg. Access to the synchrotron facility is supported by the European Community's Seventh Framework Programme (FP7/2007-2013) under grant agreement Number 226716.
REFERENCES
- 1.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 2.Saraste M, Sibbald PR, Wittinghofer A. The P-loop–a common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 1990;15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Vetter IR, Wittinghofer A. Nucleoside triphosphate-binding proteins: different scaffolds to achieve phosphoryl transfer. Q. Rev. Biophys. 1999;32:1–56. doi: 10.1017/s0033583599003480. [DOI] [PubMed] [Google Scholar]
- 5.Ogura T, Whiteheart SW, Wilkinson AJ. Conserved arginine residues implicated in ATP hydrolysis, nucleotide-sensing, and inter-subunit interactions in AAA and AAA+ ATPases. J. Struct. Biol. 2004;146:106–112. doi: 10.1016/j.jsb.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu. Rev. Biophys. Biomol. Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 7.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 8.Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 9.Sanders CM, Kovalevskiy OV, Sizov D, Lebedev AA, Isupov MN, Antson AA. Papillomavirus E1 helicase assembly maintains an asymmetric state in the absence of DNA and nucleotide cofactors. Nucleic Acids Res. 2007;35:6451–6457. doi: 10.1093/nar/gkm705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castella S, Burgin D, Sanders CM. Role of ATP hydrolysis in the DNA translocase activity of the bovine papillomavirus (BPV-1) E1 helicase. Nucleic Acids Res. 2006;34:3731–3741. doi: 10.1093/nar/gkl554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 12.Mertens HD, Svergun DI. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Bielnicki JA, Shkumatov AV, Derewenda U, Somlyo AV, Svergun DI, Derewenda ZS. Insights into the molecular activation mechanism of the RhoA-specific guanine nucleotide exchange factor, PDZRhoGEF. J. Biol. Chem. in press;286:35163–35175. doi: 10.1074/jbc.M111.270918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verstraete K, Vandriessche G, Januar M, Elegheert J, Shkumatov AV, Desfosses A, Van Craenenbroeck K, Svergun DI, Gutsche I, Vergauwen B, et al. Structural insights into the extracellular assembly of the hematopoietic Flt3 signaling complex. Blood. 2011;118:60–68. doi: 10.1182/blood-2011-01-329532. [DOI] [PubMed] [Google Scholar]
- 15.Mylonas E, Hascher A, Bernado P, Blackledge M, Mandelkow E, Svergun DI. Domain conformation of tau protein studied by solution small-angle X-ray scattering. Biochemistry. 2008;47:10345–10353. doi: 10.1021/bi800900d. [DOI] [PubMed] [Google Scholar]
- 16.Shkumatov AV, Chinnathambi S, Mandelkow E, Svergun DI. Structural memory of natively unfolded tau protein detected by small-angle X-ray scattering. Proteins. 2011;79:2122–2131. doi: 10.1002/prot.23033. [DOI] [PubMed] [Google Scholar]
- 17.Iggo RD, Lane DP. Nuclear protein p68 is an RNA-dependent ATPase. EMBO J. 1989;8:1827–1831. doi: 10.1002/j.1460-2075.1989.tb03577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roessle MW, Klaering R, Ristau U, Robrahn B, Jahn D, Gehrmann T, Konarev P, Round A, Fiedler S, Hermes C, et al. Upgrade of the small-angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J. Appl. Crystallogr. 2007;40:s190–s194. [Google Scholar]
- 19.Round AR, Franke D, Moritz S, Huchler R, Fritsche M, Malthan D, Klaering R, Svergun DI, Roessle M. Automated sample-changing robot for solution scattering experiments at the EMBL Hamburg SAXS station X33. J. Appl. Crystallogr. 2008;41:913–917. doi: 10.1107/S0021889808021018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: a Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 21.Guinier A. La diffraction des rayons X aux tres petits angles: application a l'etude de phenomenes ultramicroscopiques. Ann. Phys. 1939;12:161–237. [Google Scholar]
- 22.Svergun D. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 23.Fischer F, de Oliveira Neto M, Napolitano HB, Craievich AF, Polikarpov I. The molecular weight of proteins in solution can be determined from a single SAXS measurement on a relative scale. J. Appl. Cryst. 2010;43:101–109. [Google Scholar]
- 24.Svergun D, Barberato C, Koch MHJ. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 25.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- 29.Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McNicholas S, Potterton E, Wilson KS, Noble MEM. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jo S, Vargyas M, Vasko-Szedlar J, Roux B, Im W. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 2008;36:W270–W275. doi: 10.1093/nar/gkn314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Lassmann T, Sonnhammer EL. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005;6:298. doi: 10.1186/1471-2105-6-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 35.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 36.Ward JJ, McGuffin LJ, Bryson K, Buxton BF, Jones DT. The DISOPRED server for the prediction of protein disorder. Bioinformatics. 2004;20:2138–2139. doi: 10.1093/bioinformatics/bth195. [DOI] [PubMed] [Google Scholar]
- 37.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 38.Romero P, Obradovic Z, Dunker K. Sequence data analysis for long disordered regions prediction in the Calcineurin Family. Genome Inform. Ser. Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 39.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 40.Enemark EJ, Stenlund A, Joshua-Tor L. Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex. EMBO J. 2002;21:1487–1496. doi: 10.1093/emboj/21.6.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuck S, Stenlund A. ATP-dependent minor groove recognition of TA base pairs is required for template melting by the E1 initiator protein. J. Virol. 2007;81:3293–3302. doi: 10.1128/JVI.02432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DeLano WLT. San Carlos, CA. USA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System. [Google Scholar]
- 43.Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34:W52–W56. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 45.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Im W, Beglov D, Roux B. Continuum solvation model: computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comp. Phys. Commun. 1998;111:59–75. [Google Scholar]
- 47.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 48.Montenarh M, Vesco C, Kemmerling G, Müller D, Henning R. Regions of SV40 large T antigen necessary for oligomerization and complex formation with the cellular oncoprotein p53. FEBS Lett. 1986;204:51–55. doi: 10.1016/0014-5793(86)81386-2. [DOI] [PubMed] [Google Scholar]
- 49.Cuesta I, Nunez-Ramirez R, Scheres SH, Gai D, Chen XS, Fanning E, Carazo JM. Conformational rearrangements of SV40 large T antigen during early replication events. J. Mol. Biol. 2010;397:1276–1286. doi: 10.1016/j.jmb.2010.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mancini EJ, Kainov DE, Grimes JM, Tuma R, Bamford DH, Stuart DI. Atomic snapshots of an RNA packaging motor reveal conformational changes linking ATP hydrolysis to RNA translocation. Cell. 2004;118:743–755. doi: 10.1016/j.cell.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 51.Niedenzu T, Roleke D, Bains G, Scherzinger E, Saenger W. Crystal structure of the hexameric replicative helicase RepA of plasmid RSF1010. J. Mol. Biol. 2001;306:479–487. doi: 10.1006/jmbi.2000.4398. [DOI] [PubMed] [Google Scholar]
- 52.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 53.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 54.Triezenberg SJ. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 1995;5:190–196. doi: 10.1016/0959-437x(95)80007-7. [DOI] [PubMed] [Google Scholar]
- 55.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem. Sci. 2005;30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 56.Demchenko AP. Recognition between flexible protein molecules: induced and assisted folding. J. Mol. Recognit. 2001;14:42–61. doi: 10.1002/1099-1352(200101/02)14:1<42::AID-JMR518>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 57.Hegyi H, Schad E, Tompa P. Structural disorder promotes assembly of protein complexes. BMC Struct. Biol. 2007;7:65. doi: 10.1186/1472-6807-7-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.