Abstract
RNA tetraloops can recognize receptors to mediate long-range interactions in stable natural RNAs. In vitro selected GNRA tetraloop/receptor interactions are usually more ‘G/C-rich’ than their ‘A/U-rich’ natural counterparts. They are not as widespread in nature despite comparable biophysical and chemical properties. Moreover, while AA, AC and GU dinucleotide platforms occur in natural GAAA/11 nt receptors, the AA platform is somewhat preferred to the others. The apparent preference for ‘A/U-rich’ GNRA/receptor interactions in nature might stem from an evolutionary adaptation to avoid folding traps at the level of the larger molecular context. To provide evidences in favor of this hypothesis, several riboswitches based on natural and artificial GNRA receptors were investigated in vitro for their ability to prevent inter-molecular GNRA/receptor interactions by trapping the receptor sequence into an alternative intra-molecular pseudoknot. Extent of attenuation determined by native gel-shift assays and co-transcriptional assembly is correlated to the G/C content of the GNRA receptor. Our results shed light on the structural evolution of natural long-range interactions and provide design principles for RNA-based attenuator devices to be used in synthetic biology and RNA nanobiotechnology.
INTRODUCTION
In nature, long-range RNA interactions involving sequence positions often located hundreds of nucleotides away from each other, contribute to the folding of stable RNAs into functional three-dimensional (3D) structures (1,2). The most abundant of all identified long-range interactions are A-minor packing interactions, which occur between stacked adenines and the shallow groove of small helical receptors composed of at least two Watson–Crick (WC) base pairs (bps) (3,4). In large ribozymes and riboswitches, A-minor interactions are often part of larger structural motifs involving GNRA tetraloops binding to helices or small receptors, with GYRA/helix and GAAA/11 nt receptor interactions being the most widespread (N stands for any base, Y stands for pyrimidine and R stands for purine) (5–16). This observation has raised the question whether GNRA tetraloops other than GYRA and GAAA tetraloops can form equivalent specific long-range interactions. This was addressed, at least partially, when several new receptors for GUAA, GUGA, GAAA and GGAA tetraloops were identified by in vitro selection experiments (8,15) and were shown to have thermodynamics and loop selectivity comparable to natural ones when tested in standard physiological conditions (15) (E. C., S. Baudrey, L. J., unpublished data). Most of these in vitro selected loop/receptor interactions, including the GAAA/C7.2 (8), GAAA/C7.10 (8), GUAA/B7.8 (8), GGAA/R1 (15) and GGAA/R2 (15) loop/receptors, are not observed in known group I (8,16,17) and group II introns (6,8), RNase P RNAs (9,18), molybdenum cofactor riboswitches (19), class I di-GMP riboswitches (20) and ribosomal RNAs. In order to explain the evolutionary bias toward natural GNRA/receptor interactions versus those obtained by in vitro selection, other selection pressures than those for particular biochemical and biophysical properties should be at work during the structural evolution of natural RNA molecules.
In vitro selected loop/receptor interactions are typically more ‘G/C-rich’ than their natural counterparts. Even naturally occurring GAAA/11 nt receptors, which can accommodate AA, AC and GU dinucleotide platforms, strongly favor the AA platform versus all the others (8,21). This suggests that natural RNA motif sequences might be selected for their robustness toward intra-molecular RNA misfolding rather than for their local thermodynamic stability or selectivity. Natural helical and ‘A/U-rich’ GNRA receptors are more likely to avoid kinetic and thermodynamic folding traps at the level of larger sequence contexts than their artificial counterparts. On the other side, ‘G/C-rich’ receptors might be more suited for designing artificial riboswitches able to attenuate formation of GNRA/receptor interactions.
Previously, we developed a self-assembling tectoRNA heterodimer system based on bimolecular GNRA/receptor interactions that was employed as building blocks for nano-constructions (3,15,22–27) and in vitro selection of novel GGAA receptors (15) (Figure 1). Inspired by working principles from natural transcription attenuators (28–31), we have engineered several tectoRNA riboswitches able to adopt mutually exclusive structures that promote or inhibit formation of GNRA/receptor interactions (Figure 1). These tectoRNA riboswitches are used to monitor the ability of several tetraloop/receptor motifs with different G/C content to be thermodynamically trapped by pseudoknot (PK) formation. The mechanism of attenuation of inter-molecular GNRA/receptor interactions by intra-molecular PK is investigated by gel-shift assays, lead cleavage probing, competition experiments and co-transcriptional assembly assays. While our data shed new light on the structural evolution of GNRA/receptor interactions, it also provides new design principles for RNA-based switching devices suitable for synthetic biology and nanobiotechnology (32–34).
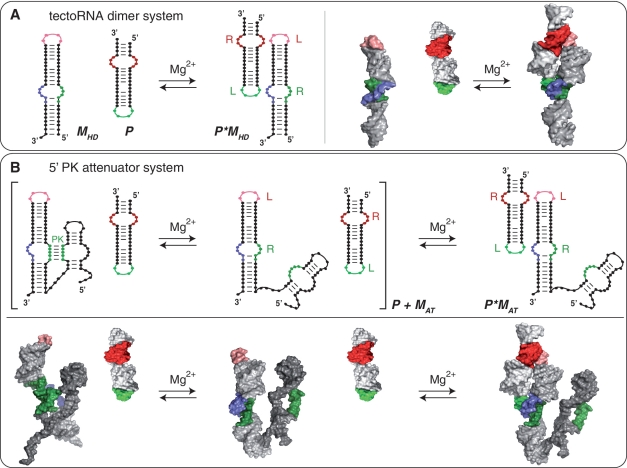
Figure 1.
Self-assembly equilibrium reactions for the tectoRNA systems reported. (A) TectoRNA heterodimer (HD) system: a HD-forming module (MHD) assembles with a probe (P) through GNRA/receptor interactions to form a heterodimer (P × MHD). This system is used as control. (B) 5′ PK attenuator system: the tectoRNA attenuator (MAT), consisting of a HD-forming module linked to a PK-forming module, can assemble with a probe (P) through its HD-forming module (equilibrium on the right) to form the heterodimer (P × MAT). Attenuation of inter-molecular self-assembly between the tectoRNA attenuator and the probe occurs when the PK-forming module interacts with the 5′ side (in green) of the receptor of the HD-forming module to form a 5′ PK (equilibrium reaction between brackets). Interacting receptor (R) and loop (L) motifs as well as pseudoknot (PK) are indicated. Equilibrium reactions and 3D stereo view for the 3′ PK attenuator system are provided Supplementary Figure S1.
MATERIALS AND METHODS
TectoRNA design and 3D modeling
3D atomic models were manually constructed using the program Swiss-Pdb Viewer (35) following the RNA architectonics guidelines (24). All tectoRNA attenuators contain a heterodimer-forming module that assembles with a probe through two inter-molecular receptor/GNRA interactions (Figures 1 and 2). This module was modeled after the tectoRNA heterodimer (HD) (15,22,23) for which atomic model structures are presently available [PDB_ID: 2adt] (36,37). The 5′ and 3′ PK forming modules leading to the formation of 5′ and 3′ intra-molecular pseudoknots (PK), respectively, were modeled after the NMR structure of the Box H/ACA snoRNA bound to its rRNA target [PDB code: 2p89, 2pcv] (38,39) (Figure 1B and C and Supplementary Figure S1). In order to form the PK, the PK-forming module with a 10-bp apical stem, is linked through 4 nt to the HD-forming module, which includes a 3-bp stem (Figures 1 and 2). TectoRNA sequences (listed Supplementary Table S1) were checked for proper folding with the program Mfold (40,41) to maximize the stability of their secondary structure while minimizing the occurrence of alternative secondary structure folds. All tectoRNA attenuators are predicted to fold into a unique secondary structure prone to assemble with the probe. PKs with one or more GC bp are accurately predicted with Kinefold (42) (Supplementary Table S2).
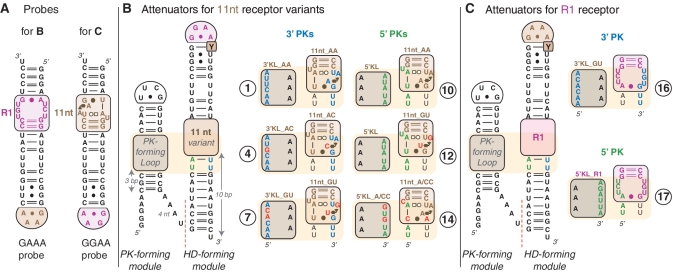
Figure 2.
Secondary structure diagrams and nomenclature of tectoRNA attenuators reported. (A) GAAA and GGAA molecular probes. (B) TectoRNA attenuators based on 11 nt receptor variants: their HD-forming module can assemble with the GAAA probe. (C) TectoRNA attenuators based on the R1 receptor motif and assembling with the GGAA probe. Indicated RNA constructs (labeled 1–17) are those with combinations of PK-forming modules and HD-forming modules that can form 3′PK (between nucleotides in blue) or 5′PK (between nucleotides in green). Red nucleotides are positions that vary from molecules 1 and 10 (with the classic 11 nt receptor and corresponding 3′ and 5′ PK-forming loops). Most constructs tested have a U at the pyrimidine position, which is labeled ‘Y’ within the HD-forming module. Additional constructs with combinations of PK-forming modules and HD-forming modules leading to mismatched PKs or with Y = C have been tested (see Figures 3 and 5 and Supplementary Table S1). Base-pairings are indicated according to Leontis and Westhof (43) annotation. The 11nt_GU receptor has been reported in previous studies as receptor C7.10 (8,15). For a description of the modularity of 11 nt receptor variants, see Supplementary Figure S2.
TectoRNA synthesis and assembly
TectoRNAs were synthesized by in vitro T7 run-off transcription from PCR generated templates, purified by denaturing polyacrylamide gel electrophoresis (PAGE) and labeled at their 3′-end using 3′-[32P]pCp as previously described (15,23). For the determination of equilibrium constants of dissociation (Kd) by titration experiments, RNA samples were typically prepared by mixing equimolar amounts of each tectoRNA at various concentrations (1 nM to 50 µM) in water. After denaturation (2 min, 95°C; 2 min, 4°C; 2 min, 30°C), samples were renatured by addition of magnesium buffer [89 mM Tris-borate pH 8.3 (TB), 50 mM KCl, 2 or 15 mM Mg(OAc)2 final concentration] at 30°C for 20 min before incubation at 10°C. For tri-molecular competition experiments, attenuator tectoRNA molecules were first assembled with linear switching (SW) RNA molecules at 15 mM Mg(OAc)2 for 15 min, before further incubation with the RNA probe for 30 min. One of the tectoRNAs used in the self-assembly mix (usually the probe) contained a fixed amount of 3′-end [32P]pCp-labeled RNA (1–10 nM final) for visual monitoring on native 10% (29:1) PAGE gels. Samples were cooled on ice before addition of blue loading buffer (magnesium buffer, 0.01% bromophenol blue, 0.01% xylene cyanol, 50% glycerol) and migration at a maximum temperature of 10°C for 3 h on PAGE gels with 2 or 15 mM Mg(OAc)2 and running buffer [89 mM Tris–borate, pH 8.3, 2 or 15 mM Mg(OAc)2].
Dissociation constants (Kd) and free energy (ΔG) calculations
Kd's were experimentally derived from titration experiments at 10°C performed as described above. Monomers [Probe (P), HD-forming module (MHD) or attenuator tectoRNA (MAT)] and heterodimers (P × MHD or P × MAT) were quantified using ImageQuant software (15,22,23,44). Kd's for the equilibrium reaction P + M → P × M (with M = MHD or MAT), were determined from a non-linear fit of the experimental data to equation: f = [2βM0 + Kd − (4M0βKd + Kd2)0.5]/2M0, where f is the fraction of RNA heterodimer, defined as the weight-in-weight (w/w) ratio of the dimer (P × M) to the total RNA species (P + M + P × M) (45). M0 is the total concentration of the probe (or attenuator tectoRNA). β is the maximum fraction of RNA able to dimerize. With β typically equal to 1 for most molecules tested, the Kd's equation is: Kd = [(M0)(1 − f)2]/f. Therefore, Kd's correspond to M0/2 when 50% of bi-molecular assemblies are formed (15,23). For each set of molecules, Kd values correspond to the average calculated from three independent experiments. The corresponding free energy variations of dimerization (ΔGHD) between tectoRNA attenuators and cognate RNA probes are determined from the equation, ΔGHD = RTlnKd, where R is the gas constant (1.985 cal/K/mol) and T is the temperature (283°K). The apparent free energy variation of attenuation at 10°C (ΔΔGAT) can be derived from the equation, ΔΔGAT = ΔGHD(MAT + P) − ΔGHD(MHD + P), where, ΔGHD(MAT + P) is the free energy of dimerization between the tectoRNA attenuator (comprising attenuator PK-forming and HD-forming modules) and its cognate RNA probe, and ΔGHD(MHD + P) is the free energy variation of dimerization between the corresponding HD-forming module alone and its cognate probe. All Kd's and associated ΔGHD and ΔΔGAT are reported in Supplementary Tables S2–S4.
Co-transcriptional assembly
PCR-generated DNA templates coding for a tectoRNA attenuator and its cognate RNA probe (GAAA2 or GGAA2; Supplementary Table S1) were mixed at equimolar concentrations in presence of the transcription mixture [50 mM Tris pH 7.5, 10 mM MgCl2, 2 mM spermidine, 2.5 mM NTPs, 10 mM DTT, α[32P]-ATP (10 mCi/ml)]. Transcription was initiated by addition of home-made T7 RNA polymerase (10 U/µl final) at 37°C. Small aliquots of the transcription mix were taken at 15-, 30-, 45- and 60-min time intervals and quenched by incubation with RQ1 RNase-free DNase (0.3 U/µl final) for 15 min at 37°C, just before native PAGE analysis at 10°C in presence of 10 mM Mg(OAc)2 as described above.
Lead Pb(II)-induced cleavage
RNA samples (4 µM final with 10 nM of 3′-end labeled RNA) assembled as described above, were incubated in presence of Pb(OAc)2 (8 mM final) for 2 min before addition of 50 mM EDTA and ethanol precipitation (15,23). Lead-induced cleavage patterns were visualized on 8 M Urea/20% PAGE (see also Supplementary Data). Cleaved positions were identified using RNA samples treated by RNase T1 digestion and alkaline hydrolysis.
RESULTS
Modular design of tectoRNA attenuators
Each tectoRNA attenuator contains a ‘pseudoknot (PK)-forming’ module linked in 5′ to a ‘heterodimer (HD)-forming’ module (Figure 1 and Supplementary Figure S1). The ‘HD-forming’ module is based on a previous self-dimerizing tectoRNA construct (15,22,23) that consists of a GNRA receptor tectoRNA unit assembling through bimolecular GNRA/receptor interactions with a tectoRNA probe (Figures 1 and 2). Because of the high recognition specificity of GAAA and GGAA tetraloops by their cognate receptors, HD-forming modules cannot self-assemble in the absence of probe. The ‘PK-forming’ module contains an internal loop called the PK-forming loop (or PKL) and promotes formation of an intra-molecular PK with the 5′ or 3′ side of the receptor from the ‘HD-forming’ module. This PK competitively inhibits assembly with the GRAA probe (Figure 1). In other words, pseudoknot formation attenuates tectoRNA dimer formation. While tectoRNA attenuators are based on the same structural scaffold, they essentially differ from one another at the level of their receptor and PKL sequences (Figures 2 and 3C, Supplementary Table S1). We designed a total of 17 different attenuators (numbered 1–17) by combining five HD-forming modules based on the 11 nt and R1 receptor sequences (11nt_AA, 11nt_AC, 11nt_GU and 11nt_A/CC) (8,15), with various PK-forming modules containing different PKL loop sequences (3′KL_AA, 3′KL_AC, 3′KL_GU, 5′KL, 5′KL_A/CC and 5′KL_R1), with up to 5 to 6 nt complementary to the 3′ or 5′ sides of the receptor. The resulting intra-molecular PKs are structurally and conceptually similar to the binding modality of stable inter-molecular paranemic RNA molecules previously shown to require a minimum of 5 bps for self-assembly (46). The effects of additional nucleotide variations were investigated within the context of some of these constructs (Supplementary Table S1). To check the influence of PKL size on PK formation, two additional adenines were introduced in the PKL, on the strand opposite to the PK forming strand (Molecules of the 1a–17a series). To modulate the binding affinity of the HD-forming module for its cognate probe, the gGRAAu terminal loop of the HD-forming module was changed into a gGRAAc loop (Molecules of the 1″–17″ and 1a″–17a″ series).
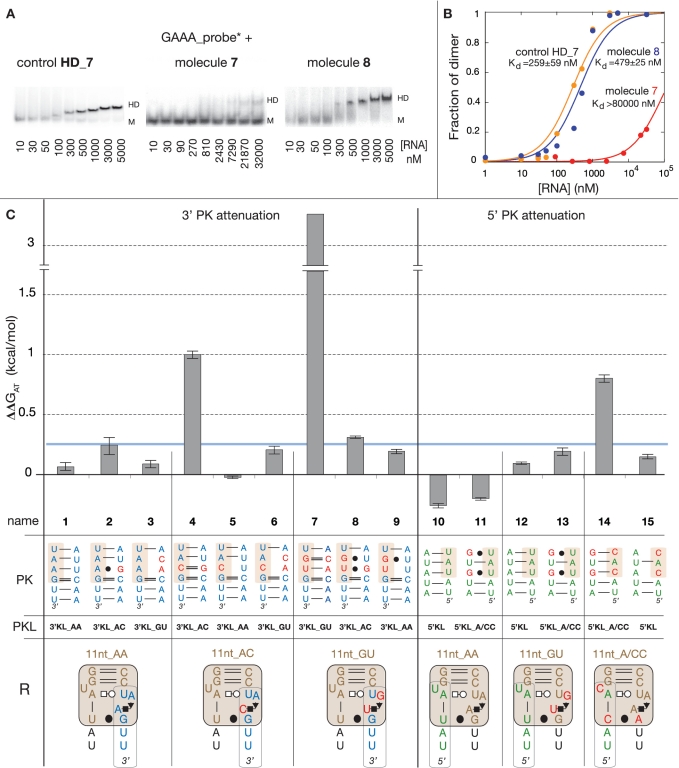
Figure 3.
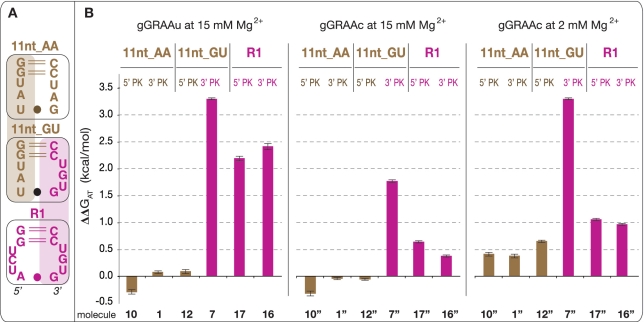
Thermodynamic analysis of tectoRNA attenuators based on 11 nt receptor variants. (A) Typical examples of native PAGE titration experiments at 15 mM Mg(OAc)2 and 10°C, for the control DF_7 (11nt_GU HD-forming module alone), molecule 7 (11nt_GU HD-forming and 3′KL_GU PK-forming modules) and molecule 8 (11nt_GU HD-forming and 3′KL_AC PK-forming modules) in presence of equimolar concentrations of radiolabeled GAAA probe. M and HD indicate the position of monomers and heterodimers, respectively. (B) Titration curves with calculated equilibrium constants of dissociation (Kd's) corresponding to the tectoRNA assemblies in (A). (C) Free energies of attenuation of heterodimer formation for all attenuator constructs based on the 11 nt receptor variants (see also Supplementary Table S3). The free energies of attenuation (ΔΔGAT) were estimated at 10°C and 15 mM Mg(OAc)2 as described in the Materials and Methods section. The sequence of the intra-molecular 3′ or 5′ PK, which competes with heterodimer formation by sequestering either the receptor 3′ or 5′ side, the name of the PK-forming module (PKL) and the 2D structure and name of the receptor (R) from the HD-forming module are indicated for each tectoRNA attenuator tested (numbered 1–15). All constructs with ΔΔG values below the threshold of 0.25 kcal/mol (indicated by the blue line) are considered to have no significant attenuation. This threshold was estimated based on the range of standard error deviations observed through the study (Supplementary Table S1).
Characterization of tectoRNA attenuators based on GAAA/11 nt receptor interactions
Free energies of dimerization (ΔGHD) between HD-forming modules and their cognate probe can be derived from equilibrium constants of dissociation estimated by native PAGE gel-shift assays as indicated in the Materials and Methods section (Figure 3A and B, Supplementary Table S2). By comparing ΔGHD of a particular HD-forming module in the tectoRNA attenuator context with the one in absence of linked PK-forming module, we can estimate for each tectoRNA attenuator the variation of free energy (ΔΔGAT) that corresponds to attenuation by PK formation (see Materials and Methods section).
At 15 mM Mg(OAc)2 and 10°C, most HD-forming modules based on the 11 nt receptor variants (Supplementary Figure S2) assemble to the GAAA probe with very similar ΔGHD. ΔGHD's for the 11nt_AA, 11nt_AC and 11nt_GU modules are almost undistinguishable (see HD_1, HD_4 and HD_7 in Supplementary Table S2) while the 11nt_A/CC receptor, which differs by three point mutations from the classic 11nt_AA receptor, leads to a minor decrease of 0.43 kcal/mol in binding affinity when compared to the 11nt_AA receptor (HD_14 in Supplementary Table S2). Overall, this result corroborates the isosteric nature of the AA, GU and AC dinucleotides platforms, which structurally contribute in a similar way to the stabilization of the GAAA tetraloop/11 nt receptor interaction within the HD-forming module context.
In contrast, some of the tectoRNA attenuators based on these receptors display markedly different behaviors (Figure 3). Attenuators 1, 4 and 7 have PK-forming modules designed to form a PK of 6 WC bps with the 3′ sides of receptors 11nt_AA, 11nt_AC and 11nt_GU, respectively. While no significant change in binding affinity is observed for molecule 1 with respect of HD_1, molecules 4 and 7 attenuate heterodimer formation by 1 and 3.3 kcal/mol, respectively. Other 3′PK attenuators with combinations of HD-forming and PK-forming modules that introduce G:U bp and/or WC mismatches in the PK do not present significant attenuation (molecules 2, 3, 5, 6, 8 and 9). This data suggests that attenuation is correlated to the stability of the PK that requires at least two G:C bps to efficiently compete with heterodimer formation. Similar results are provided by 5′PK attenuators 10, 12 and 14, designed to form pairings of 5 WC bps with the 5′ sides of receptors 11nt_AA, 11nt_AC and 11nt_GU, respectively. Molecules 10 and 12 able to form a PK of 5 A:U bps are unable to trap the receptors but molecule 14, which can form a PK with 2 G:C bps, attenuates heterodimer formation by 0.80 kcal/mol (Figure 3). Not surprisingly, none of the attenuator combinations with PKs with G:U or A:C bps are able to compete with heterodimer formation.
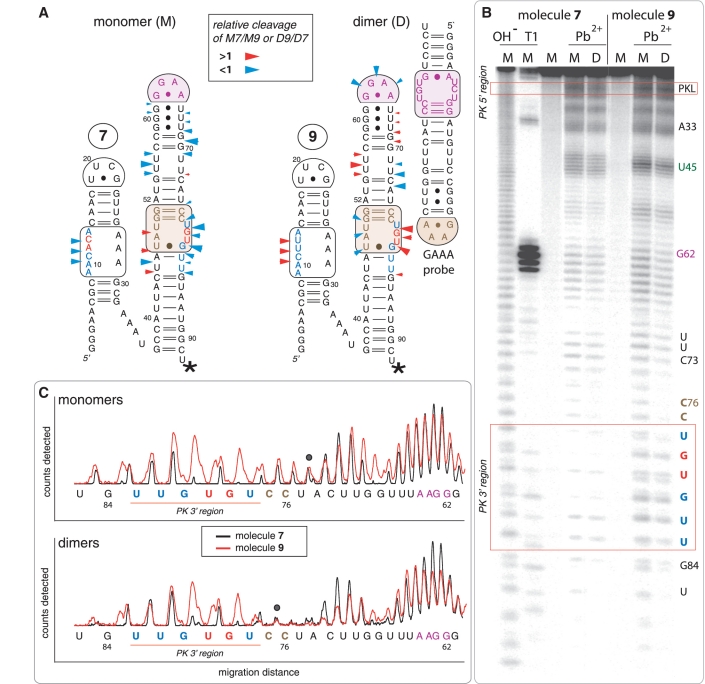
Additional structural evidences for intra-molecular PK formation in attenuator 7 are provided by lead cleavage experiments (Figure 4 and Supplementary Figure S3). Lead is widely used as a conformational probe for RNA because it preferentially cleaves the phosphodiester backbone in flexible regions or non-canonically paired motifs of RNA molecules (15,22,23,25,46). Irrespective from absence or presence of the GAAA probe, the 3′ strand of the 11nt_GU receptor of attenuator 7 is strongly protected toward cleavage in comparison to the one of molecules 8 (or 9). Additionally, the 5′ PK strand within the PK-forming module of 7 also shows enhanced protection toward lead cleavage relative to 8 (or 9). This strongly suggests that the PK is formed in attenuator 7 but not in 8 (or 9). In contrast, in presence of the GAAA probe, molecules 8 and 9 display partial protection of the receptor and tetraloop regions from the HD-forming module, corroborating their assembly with the probe.
Figure 4.
Lead(II)-induced cleavage patterns for tectoRNA attenuators 7 and 9 in their monomeric and heterodimeric states. (A) 2D diagrams of tectoRNA attenuators with reported differential Pb(II) cleavage patterns in the monomeric (M) and heterodimeric (D) states. Phosphate positions in monomer 7 (M7) that show enhanced or reduced Pb(II) cleavage with respect to monomer 9 (M9) are indicated by red or blue arrows on the 2D diagram of 7, respectively. Phosphate positions in heterodimer 9 (D9) that show enhanced or reduced Pb(II) cleavage with respect to heterodimer 7 (D7) are indicated by red or blue arrows on the 2D diagram of 9, respectively. The size of the arrows is roughly proportionate to the difference in cleavage for M7 versus M9 or D9 versus D7. A star indicates the radiolabeled RNA 3′-end. (B) Pb(II) cleavage patterns of 32P radiolabeled molecules 7 and 9 either alone or bound to their non-radioactive cognate GAAA probe [as shown in (A)]. M and D correspond to monomer and dimer lanes, respectively. Cleavage experiments (indicated by Pb2+) were carried out as described in the Materials and Methods section; OH− indicates alkaline hydrolysis ladder; T1 indicates RNase T1 digestion. (C) Superposed lead cleavage profiles for monomers 7 and 9 (top) and for the corresponding heterodimers in presence of GAAA probe (bottom). Black dots indicate positions used for normalization. Similar results are obtained by comparing attenuators 7 and 8 (Supplementary Figure S3).
In summary, the 11 nt receptor can easily accommodate sequence variations that are all able to efficiently promote self-assembly with the cognate GAAA tetraloop. However, the ability to trap its sequence in an alternative conformation like a PK, is highly dependent of the presence of Gs or Cs, G:C bps being much more effective than U:A bps for stabilizing alternative WC pairings.
Modulating tectoRNA attenuation with an artificial receptor, point mutations and magnesium
The GGAA/R1 receptor interaction was isolated by in vitro selection (15). It is highly selective for the GGAA tetraloop and its affinity is comparable to GAAA/11 nt receptor interactions (15). Interestingly, R1 is four mutations away from the 11nt_AA receptor but only two mutations away from the 11nt_GU with which it shares an identical 3′-side (Figure 5A). R1 is, however, more ‘G/C-rich’ than any of the 11 nt variants. Consequently, the resulting attenuators 16 and 17 are expected to form stable intra-molecular 3′ and 5′ PKs, respectively: according to Freier's (47) table, the calculated stability of the 3′PK of 16 and 5′PK of 17 is −5.3 and −3.5 kcal/mol, respectively. As shown in Figure 5B, molecules 16 and 17 attenuate heterodimer formation with their cognate GGAA probe by 2.16 and 1.91 kcal/mol, respectively. While both molecules 7 and 16 form the same 3′PK, attenuation with 16 is less dramatic than with 7 in presence of their respective probes; this could be explained by the fact that the HD_16 heterodimer complex involving the GGAA/R1 interaction, is 0.5 kcal/mol more stable than the HD_7 heterodimer complex involving the GAAA/11nt_GU interaction (Supplementary Table S2). Therefore, for 16, heterodimer formation is advantaged with respect of PK formation.
Figure 5.
Thermodynamic analysis of tectoRNA attenuators based on the 11 nt and R1 receptors. (A) Sequence relationships between the R1 (15), 11nt_GU [or C7.10 (8,15)] and 11 nt receptors. (B) Free energies of attenuation of heterodimer formation for all attenuator constructs based on the 11 nt and R1 receptors (see also Supplementary Table S4). The free energies of attenuation (ΔΔG) were estimated at 10°C and 2 or 15 mM Mg(OAc)2 as described in the Materials and Methods section. Attenuator molecules 1″, 7″, 10″, 12″, 16″ and 17″ differ from molecules 1, 7, 10, 12, 16 and 17 by the presence of gGRAAc terminal loops (instead gGRAAu). This single nucleotide variation increases heterodimer stability. A similar series of attenuator constructs with PK-forming loops involving five As (instead of three) show similar attenuation results (Supplementary Figure S4).
TectoRNA heterodimer assembly, which occurs through two GNRA/receptor interactions, is favored by a point mutation that changes the gGRAAu terminal loop of the HD-forming module into a gGRAAc loop. The thermodynamic stability of the resulting HD heterodimers is increased by 0.5–1.2 kcal/mol at 10°C and 15 mM Mg(OAc)2 (Supplementary Table S3). This is likely due to small structural variations that favor the local stabilization of gGRAAc/receptor interactions versus gGRAAu/receptor interactions. When incorporated within attenuators 7, 16 and 17 (to give 7″, 16″ and 17″), this mutation leads to a reduction of attenuation by 2- to 5-fold (Figure 5B).
TectoRNA assembly is particularly sensitive to small variations in magnesium concentration (3,22,23). By reducing magnesium concentration from 15 to 2 mM, the affinity between heterodimer modules and corresponding probes decreases by 1.5–2 kcal/mol (Supplementary Table S4). Intra-molecular formation of 3′ or 5′ PK, which relies on the formation of canonical WC bps, should not be as sensitive to magnesium as inter-molecular formation of GNRA/receptor interactions. As expected, molecule 7″ and to a lesser extent, molecules 17″ and 16″, attenuate HD formation more effectively at 2 mM than at 15 mM magnesium (Figure 5B).
TectoRNA attenuators of the 1a–17a series, with two additional adenines in their PKL, were also tested in order to determine whether the size of the PKL could affect PK formation in conjunction with the nucleotide composition of the PK. The behavior of the 1a–17a attenuator series is overall comparable to the one of the 1–17 attenuator series (Figure 5 and Supplementary Figure S4). At 15 mM magnesium, a small enhancement of attenuation is noticeable for molecules 1a, 10a and 12a versus 1, 10 and 12. This indicates that ‘A/U-rich’ PKs form better when the size of PKL is increased, probably because of less steric hindrance.
In summary, these results indicate that the most effective tectoRNA attenuators are those based on ‘G/C-rich’ receptors such as the R1 and 11 nt-GU receptors. The extent of tectoRNA attenuation can be modulated as a function of magnesium concentration as well as peripheral single point mutations in a predictable manner. From a rational design point of view, our results suggest that artificial ‘G/C-rich’ receptors are better suited than ‘A/U-rich’ receptors for designing riboswitches that require folding into alternative RNA structures. However, from an evolutionary point of view, ‘G/C-rich’ receptors might be disadvantageous because they are more prone to trap native RNA sequences into alternative undesirable structures.
TectoRNA attenuation during in vitro transcription
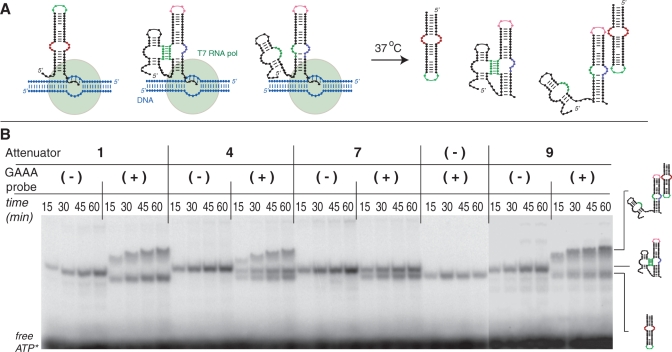
We have also investigated how tectoRNA attenuation could occur during in vitro RNA transcription in isothermal conditions (37°C) (see ‘Materials and Methods’ section). While all the experiments described above were performed in conditions usually favoring thermodynamic control versus kinetic control, co-transcriptional assembly experiments should be more representative of folding and assembly processes taking place within the cell (48,49). During the linear phase of RNA transcription, three different types of products are observed on native PAGE: the RNA probe, the tectoRNA attenuator and the complex resulting from the inter-molecular assembly between the probe and attenuator molecules (Figure 6A and Supplementary Figure S5). Because of its smaller size, the probe product is transcribed in larger quantity than the attenuator product, explaining why a portion of it always remains unassembled. In presence of GAAA probe, the totality of attenuator 1 (and 10) products assembles to the probe (Figure 6B). In contrast, only ~60% of the attenuator 4 product forms a stable complex with the probe, suggesting that the remaining 40% is blocked into the PK conformation state (Figure 6B). In perfect agreement with previous data, attenuator 7 (and 7″) demonstrates full attenuation of tectoRNA assembly, while attenuator 9, which differs from molecule 7 by only two point mutations within its intra-molecular PK, assembles with the probe to its full extent (Figure 6B and Supplementary Figure S5). Interestingly, attenuators 14, 16 and 17 assemble with their cognate probe to form complexes with faster gel mobility than those obtained with attenuator 1, 9 and 10 (Supplementary Figure S5). We have observed that tectoRNA complexes with higher Kd's (or lower affinities) typically migrate faster at lower RNA concentrations than those with lower Kd's (or higher affinities) (15,23). This behavior has been described as resulting from monomers and heterodimers being in dynamic equilibrium (15,23). Our observation corroborates the fact that attenuators 14, 16 and 17 bind less efficiently their cognate probe than their corresponding HD_forming modules. In these attenuators, formation of a transient intra-molecular PK likely displaces the inter-molecular assembly equilibrium toward the monomers.
Figure 6.
Co-transcriptional assemblies of tectoRNA attenuators 1, 4, 7 and 9 in presence (or absence) of cognate GAAA probe. (A) Schematic illustrating the possible molecular states adopted by the tectoRNA attenuator system during its transcription from DNA templates (in blue) by T7 RNA polymerase (in green) at 37°C in presence of 10 mM Mg2+. (B) Native PAGE analysis of different tectoRNA attenuator transcription mixtures at various times in presence (+) or absence (−) of GAAA probe: co-transcriptional assembly is monitored by RNA body-labeling with α[P32]ATP and native PAGE is performed at 10°C and 10 mM Mg(OAc)2 after quenching the transcription with DNase as described in the ‘Materials and Methods’ section. See also Supplementary Figure S5.
Overall, co-transcriptional assembly data corroborate those obtained previously. To effectively attenuate tectoRNA assembly, the 3′ and 5′ PK base pairings need to have a calculated thermodynamic stability lower than −4 and −3 kcal/mol, respectively. Co-transcriptional data also suggest that attenuation can occur through two distinct mechanisms. In the mechanism shared by attenuators 4, 7 and 7″, the attenuator RNA product folds into stable PK_forming and HD_forming conformers, which are unable to interchange into one another. This is probably due to the PK_forming conformer acting as a folding trap, unable to switch into the HD_conformer. In the second mechanism shared by attenuator 14, 16 and 17, the PK_forming and HD_forming conformers are in dynamical equilibrium with one another, allowing the PK_forming conformer to switch into the HD_forming conformer. In the future, further work will be needed to unravel the dynamical and structural constraints favoring one mechanism versus the other.
Controlling tectoRNA attenuation with small RNA switches
Moving toward more complex tectoRNA attenuator devices, we designed a tri-molecular system aiming at controlling tectoRNA attenuation with small RNA inhibitors acting as molecular switches (Figure 7A). TectoRNA attenuator 18 is derived from attenuator 7, from which it differs by four point mutations in the PKL, on the strand opposite to the PK forming sequence (Figure 7B). Two small RNAs, SW(I) and SW(II), are designed to favor heterodimer formation between 18 and the GAAA probe by preventing intra-molecular PK formation (Figure 7A): they assemble to the PK-forming loop according to two equivalent structural modalities expected to perfectly mimic the assembly of the U65 ψ pocket (U65hp) of human U65 H/ACA snoRNA with its rRNA substrate (38,39) (Figure 7B). In presence of the GAAA probe, attenuator 18 has a more modest attenuation potential than attenuator 7 (−0.84 kcal/mol versus −3.24 kcal/mol): this possibly results from the formation of additional non-canonical bps, which might stabilize the internal structure of the PKL of 18 and disfavor PK formation. Nevertheless, when either SW(I) or SW(II) are added to the mix, the affinity of 18 for the GAAA probe significantly increases to be similar to the one observed for the HD_7:GAAA probe complex (~0.2 kcal/mol). Molecules SW(I) and SW(II) bind to the PK_forming module of 18 with Kd's of 7.3 and 16.8 nM, respectively. While these values are consistent with previously published results for similar binding interactions (46), they indicate that SW(I) and SW(II) can completely switch off the PK_forming module of 18 and prevent the formation of the intra-molecular 3′PK with the 11nt_GU receptor of the HD_forming module. Therefore, these results provide further evidence in support of the mechanism of attenuation of heterodimer formation through intra-molecular PK formation.
Figure 7.
Switching off a tectoRNA attenuator with small RNA oligonucleotides. (A) Schematic of the self-assembly equilibrium reaction of tectoRNA attenuator 18 in presence of two small RNAs [SW(I) and SW(II)] that favor heterodimer formation by preventing internal PK formation. The two small RNAs switch off the PK attenuator module by assembling to the PK-forming loop in a way expected to perfectly mimic the NMR structures of the pseudouridylation pocket of the Box H/ACA snoRNA bound to its rRNA substrate [PDB codes: 2p89 (38) and 2pcv (39)]. (B) 2D structure diagrams of the PK-forming module of attenuator 18, bound to SW(I) and SW(II) small switching RNAs. Two possible equivalent binding modalities (boxed in grey) are shown. (C) Titration curves with calculated equilibrium constants of dissociation (Kd's) corresponding to the assembly of attenuator 18 with the GAAA probe in absence (black circles) or presence of switching RNAs [magenta circles for SW(I) and blue circles for SW(II)]. Experiments were carried out at 15 mM Mg(OAc)2 and 10°C as described in the ‘Materials and Methods’ section.
DISCUSSION
Implications for the rational design of RNA 3D structures
Using a rationally designed molecular system based on tectoRNA self-assembly, we have demonstrated that the formation of GNRA/receptor tertiary interactions can be attenuated by formation of alternative PK structures: the higher the G/C content of the sequence signature of the GNRA receptor motif, the more easily this sequence can be trapped into an alternative pseudoknot structure that attenuates its ability to recognize a GNRA loop target. From a rational design point of view, these data make perfect sense because ‘G/C-rich’ base pairings are thermodynamically more stable than ‘U/A-rich’ base pairings. However, the data also highlight that it is a rather narrow thermodynamic threshold that determines whether the PK can effectively attenuate the formation of GNRA/receptor interactions in vitro. For instance, only one additional C or G in the nucleotide platform of the 11 nt motif (11nt_GU and 11nt_AC versus 11nt_AA) is sufficient for stabilizing by 1.5–2 kcal/mol the resulting PKs and lead to attenuation. Nevertheless, if most PKs (with at least one G:C bp) were accurately predicted with the Kinefold program (42), the overall quantitative extent of attenuation cannot yet be predicted from purely theoretical thermodynamic analysis. Indeed, our tectoRNA attenuation system depends on the thermodynamic stability of RNA tertiary interactions and structure motifs that essentially involve non-canonical bp interactions and that are also very sensitive to divalent ion concentration. In vitro cotranscriptional self-assembly revealed that attenuation could proceed in isothermal conditions according to two mechanisms that could be distinguished based on whether the PK_forming and the HD_forming conformers are in dynamical equilibrium or not. Previous studies (48,49) have shown that kinetics and thermodynamics make different contributions to RNA folding in vitro and in vivo: it is therefore possible that the exchange between stable alternative tertiary structures might be more rapid in vivo than in vitro (48,49). Clearly, further work will be necessary to understand tectoRNA attenuation mechanisms in more detail, especially within the context of cells. In any case, our data already provide RNA modules and design principles that can be used for developing controllable artificial RNA nano-switches with tunable binding properties for nanobiotechnology and synthetic biology applications. For instance, we have demonstrated that RNA tertiary interactions can be specific target locations for designing RNA switches that allow precise modulation of the folding and assembly of these RNA molecules. Because of the large number of topologically equivalent GNRA/receptor interactions presently available (8,14,15), these interactions offer high structure designability for the rational design of RNA nanodevices (27). The choice of a particular RNA self-assembling motif can vary depending on the intended design goal. A/U-rich RNA motifs can maximize a unique folding pathway by minimizing undesirable folding traps resulting from the formation of alternative bps. Alternatively, G/C-rich tertiary motifs with one or two Gs (or Cs) localized on the same strand, can be used to design artificial RNAs with distinct alternative conformational states. Pseudoknots involving 5–6 bps with two G:C bps are sufficient for competing with the formation of GNRA/receptor interactions. However, as a difference of a few kcals can displace the equilibrium toward a unique molecular state, a good empirical understanding of the energetic balance between the thermodynamic strength of competing tertiary interactions is necessary for designing truly tunable devices.
Implications for RNA structural evolution
More importantly, our data provide possible clues for RNA structural evolution as they can explain the existence of particular sequence patterns coding for RNA tertiary interactions. GNRA mediated interactions in stable RNAs are essentially dominated by two families: GYRA/helix and GAAA/receptor interactions. For example, in the class I di-GMP riboswitches (20,50,51), ~35% are GYRA/helix motifs and 65% are GAAA/11 nt-like receptor motifs (Supplementary Table S5A). Within the GAAA/11 nt-like receptors, 64% have no more than one G or C, 24% have two G or C and only 12.3% have three G or C or more (Supplementary Table S5B). Therefore, in the 11 nt receptor family, ‘U/A-rich’ rather than ‘G/C-rich’ nucleotide compositions are favored at the level of the 11 nt internal loop, with AA platforms (70.4%) being more abundant than AC (8.6%) and GU (11.3%) platforms. Interestingly, the thermodynamic stability and GNRA selectivity of the ‘G/C-rich’ 11 nt receptor variants are not significantly different from those of the more ‘A/U-rich’ 11 nt receptors (Supplementary Table S2 and Supplementary Figure S6). Moreover, in addition to class I di-GMP riboswitches, several other natural RNA contexts like RNase P RNAs (9,18), group I (16,17) and group II introns (6,8,52), contain GYRA/helix interactions, which are thermodynamically less stable than most 11 nt receptors, in place of GAAA/11 nt motifs. Considering the range of observed riboswitch behaviors in response to the evolutionary need for precise genetic regulation, the sequence of the GNRA/receptor interaction from the di-GMP riboswitch may be tuned so that the riboswitch functions more as a dimmer or rheostat than a binary on/off switch (53).
In natural RNAs, the GAAA tetraloop is universally more abundant than any of the other GNRA loops (54,55) (Supplementary Table S5A). However, the predominance of the GUAA tetraloop over other GYRA and GRRA tetraloops is not consistent and might depend on the molecular and genomic context (54,55). As exemplified for the di-GMP riboswitch of class I, it is particularly striking that <1.5% of the GNRA/receptor interactions take advantage of other GRRA tetraloops (Supplementary Table S5A), while it has been recently demonstrated that artificial GGRA receptors, such as the highly stable and selective R1 and R2 receptors, could be isolated by SELEX (15). In fact, using the RNAmotif software (56), we searched for the R1, R2 and 11nt_GU (C7.10) sequence signatures and did not identify any of them in known natural RNA sequences such as class I di-GMP riboswitch (20), molybdenum cofactor riboswitch (19), group I introns, group II introns and RNAse P RNA sequences from the rfam database (57). While we cannot rule out the possibility that these receptors exist in some genomes, our search already suggests that they are much less common than the 11 nt receptor.
Based on the mere consideration of thermodynamics, kinetics and loop selectivity, it is not obvious to explain the strong bias toward ‘A/U-rich’ GAAA/receptors or GYRA/helix receptors in natural stable RNAs. Clearly, higher order selection pressures imposed by the larger structural context of natural RNA molecules, the kinetics constraints on the global folding of RNA inside the cell or the possible involvement of additional cellular components are likely at play (58). Based on our present data, the most straightforward explanation is that the preferred occurrence of natural RNA motif sequences stems from an evolutionary adaptation that make them less prone to misfolding and therefore less likely to interfere with the folding of a large RNA sequence (through formation of alternative pairings or interactions with other regions of the RNA sequence). As we have clearly demonstrated that ‘G/C-rich’ receptors are more likely to be trapped into alternative PK structures than ‘A/U-rich’ receptors, we propose that, in cells, the natural GYRA/helix and ‘A/U-rich’ GAAA/11 nt receptor interactions result from two evolutionary strategies that minimize kinetic and thermodynamic folding traps in large RNA structural contexts. The first strategy, best exemplified by the ‘classic’ GYRA/helix interaction, takes advantage of receptors that use Cs and Gs to maximize the formation of stable local WC helical regions, preventing them to form long-range alternative pairings. The other strategy takes advantage of AU-rich internal loop motifs, like the 11 nt receptor motif, that minimize the formation of stable alternative base pairings. Interestingly, the IC3 receptor motif, a natural GNRA receptor identified in IC3 group I introns (14), can be seen as a mix of both strategies, with Gs and Cs involved in local bps and U and As involved in a small asymmetrical internal loop (14,15).
Recently, Mitra et al. (58) proposed that, in group I introns, the greater thermodynamic stability of a native conformation over non-native structures, achieved through selection of strong tertiary interactions, comes at the expense of slower folding to the catalytic conformation due to formation of long lived intermediates. Efficient folding is therefore achieved by balancing the gain in structural stability due to tertiary contact formation with the probability of misfolding due to loss of conformational freedom. As such, in contrast to other strong but G/C-rich GNRA/receptor interactions selected in vitro (8,15), the GAAA/11 nt motif offers a unique sequence pattern with great thermodynamic strength and lower probability to create stable alternative structures.
In conclusion, when a structural motif is part of a large structural network, the avoidance of alternative folding traps could be a significant selective advantage during evolution. We have therefore an example of how the sequence information of the whole RNA molecule could affect from the top-down the sequence information of the small local structural part (59). Similar to the usage of codons in cells, many synonymous RNA structural motifs exist but they are not identical after all (60). A good understanding of the structural designability of RNA motifs is therefore key for RNA architectonics (3,24,44,61,62) and the future development of RNA synthetic biology and nanobiotechnology, especially when artificial RNA molecules need to operate in vivo.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–6 and Supplementary References (63,64).
FUNDING
This work was funded by the National Institutes of Health (R01-GM079604 to L.J.). Funding for open access charge: The open access publication charge for this paper has been waived by Oxford University Press—NAR Editorial Board members are entitled to one free paper per year in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
L.J. wishes to dedicate this article to Professor Jérôme Lejeune. We wish to thank Dr Namhee Kim for her technical help in the RNA motif search. K.A. and L.J. designed research; K.A. and Y.-P.L performed biochemical and biophysical experiments; E.C. performed sequence comparative analysis and search: K.A., Y.-P.L. and L.J. analyzed data; K.A. and L.J. wrote the article.
REFERENCES
- 1.Tinoco I, Jr, Bustamante C. How RNA folds. J. Mol. Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 2.Noller HF. RNA structure: reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 3.Geary C, Chworos A, Jaeger L. Promoting RNA helical stacking via A-minor junctions. Nucleic Acids Res. 2011;39:1066–1080. doi: 10.1093/nar/gkq748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl. Acad. Sci. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J. Mol. Biol. 1994;236:1271–1276. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 6.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 8.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massire C, Jaeger L, Westhof E. Phylogenetic evidence for a new tertiary interaction in bacterial RNase P RNAs. RNA. 1997;3:553–556. [PMC free article] [PubMed] [Google Scholar]
- 10.Abramovitz DL, Pyle AM. Remarkable morphological variability of a common RNA folding motif: the GNRA tetraloop-receptor interaction. J. Mol. Biol. 1997;266:493–506. doi: 10.1006/jmbi.1996.0810. [DOI] [PubMed] [Google Scholar]
- 11.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 12.Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of ribonuclease P–a universal ribozyme. Curr. Opin. Struct. Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Ikawa Y, Naito D, Aono N, Shiraishi H, Inoue T. A conserved motif in group IC3 introns is a new class of GNRA receptor. Nucleic Acids Res. 1999;27:1859–1865. doi: 10.1093/nar/27.8.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geary C, Baudrey S, Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36:1138–-1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J. Mol. Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 17.Lehnert V, Jaeger L, Michel F, Westhof E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem. Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 18.Massire C, Jaeger L, Westhof E. Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol. 1998;279:773–793. doi: 10.1006/jmbi.1998.1797. [DOI] [PubMed] [Google Scholar]
- 19.Regulski EE, Moy RH, Weinberg Z, Barrick JE, Yao Z, Ruzzo WL, Breaker RR. A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol. 2008;68:918–932. doi: 10.1111/j.1365-2958.2008.06208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- 22.Jaeger L, Leontis NB. TectoRNA: one-dimensional self-assembly through tertiary interactions. Angew. Chem. Int. Ed. Engl. 2000;39:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 23.Jaeger L, Westhof E, Leontis NB. TectoRNA: modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Afonin KA, Leontis NB. Generating new specific RNA interaction interfaces using C-loops. J. Am. Chem. Soc. 2006;128:16131–16137. doi: 10.1021/ja064289h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res. 2006;34:1381–1392. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishikawa J, Fujita Y, Maeda Y, Furuta H, Ikawa Y. GNRA/receptor interacting modules: versatile modular units for natural and artificial RNA architectures. Methods. 2011;54:226–238. doi: 10.1016/j.ymeth.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 28.Dawid A, Cayrol B, Isambert H. RNA synthetic biology inspired from bacteria: construction of transcription attenuators under antisense regulation. Phys. Biol. 2009;6:025007. doi: 10.1088/1478-3975/6/2/025007. [DOI] [PubMed] [Google Scholar]
- 29.Gutierrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E. Biochemical features and functional implications of the RNA-based T-box regulatory mechanism. Microbiol. Mol. Biol. Rev. 2009;73:36–61. doi: 10.1128/MMBR.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wachter A. Riboswitch-mediated control of gene expression in eukaryotes. RNA Biol. 2011;7:67–76. doi: 10.4161/rna.7.1.10489. [DOI] [PubMed] [Google Scholar]
- 31.Breaker RR. Riboswitches and the RNA World. Cold Spring Harb. Perspect. Biol. 2010 doi: 10.1101/cshperspect.a003566. November 24 (doi:10.1101/cshperspect.a003566; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hess H, Jaeger L. Nanobiotechnology. Curr. Opin. Biotechnol. 2010;21:373–375. doi: 10.1016/j.copbio.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Topp S, Gallivan JP. Emerging applications of riboswitches in chemical biology. ACS Chem. Biol. 2011;5:139–148. doi: 10.1021/cb900278x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Las Heras A, Carreno CA, Martinez-Garcia E, de Lorenzo V. Engineering input/output nodes in prokaryotic regulatory circuits. FEMS Microbiol. Rev. 2011;34:842–865. doi: 10.1111/j.1574-6976.2010.00238.x. [DOI] [PubMed] [Google Scholar]
- 35.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 36.Davis JH, Foster TR, Tonelli M, Butcher SE. Role of metal ions in the tetraloop-receptor complex as analyzed by NMR. RNA. 2007;13:76–86. doi: 10.1261/rna.268307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Davis JH, Tonelli M, Scott LG, Jaeger L, Williamson JR, Butcher SE. RNA helical packing in solution: NMR structure of a 30 kDa GAAA tetraloop-receptor complex. J. Mol. Biol. 2005;351:371–382. doi: 10.1016/j.jmb.2005.05.069. [DOI] [PubMed] [Google Scholar]
- 38.Wu H, Feigon J. H/ACA small nucleolar RNA pseudouridylation pockets bind substrate RNA to form three-way junctions that position the target U for modification. Proc. Natl Acad. Sci. USA. 2007;104:6655–6660. doi: 10.1073/pnas.0701534104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jin H, Loria JP, Moore PB. Solution Structure of an rRNA Substrate Bound to the Pseudouridylation Pocket of a Box H/ACA snoRNA. Mol. Cell. 2007;26:205–215. doi: 10.1016/j.molcel.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 40.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 41.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xayaphoummine A, Bucher T, Isambert H. Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots. Nucleic Acids Res. 2005;33:W605–W610. doi: 10.1093/nar/gki447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop ‘kissing’ complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl Acad. Sci. USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J. Am. Chem. Soc. 2008;130:93–102. doi: 10.1021/ja071516m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freier SM, Kierzek R, Jaeger JA, Sugimoto N, Caruthers MH, Neilson T, Turner DH. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl Acad. Sci. USA. 1986;83:9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahen EM, Harger JW, Calderon EM, Fedor MJ. Kinetics and thermodynamics make different contributions to RNA folding in vitro and in yeast. Mol. Cell. 2005;19:27–37. doi: 10.1016/j.molcel.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 49.Mahen EM, Watson PY, Cottrell JW, Fedor MJ. mRNA secondary structures fold sequentially but exchange rapidly in vivo. PLoS Biol. 2010;8:e1000307. doi: 10.1371/journal.pbio.1000307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smith KD, Lipchock SV, Ames TD, Wang J, Breaker RR, Strobel SA. Structural basis of ligand binding by a c-di-GMP riboswitch. Nat. Struct. Mol. Biol. 2009;16:1218–1223. doi: 10.1038/nsmb.1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulshina N, Baird NJ, Ferre-D'Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nat. Struct. Mol. Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toor N, Keating KS, Fedorova O, Rajashankar K, Wang J, Pyle AM. Tertiary architecture of the Oceanobacillus iheyensis group II intron. RNA. 2010;16:57–69. doi: 10.1261/rna.1844010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baird NJ, Kulshina N, Ferre-D'Amare AR. Riboswitch function: flipping the switch or tuning the dimmer? RNA Biol. 2010;7:328–332. doi: 10.4161/rna.7.3.11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prathiba J, Malathi R. Group I introns and GNRA tetraloops: remnants of ‘The RNA world'? Mol. Biol. Rep. 2008;35:239–249. doi: 10.1007/s11033-007-9076-4. [DOI] [PubMed] [Google Scholar]
- 55.Sheehy JP, Davis AR, Znosko BM. Thermodynamic characterization of naturally occurring RNA tetraloops. RNA. 2010;16:417–429. doi: 10.1261/rna.1773110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Macke TJ, Ecker DJ, Gutell RR, Gautheret D, Case DA, Sampath R. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 2001;29:4724–4735. doi: 10.1093/nar/29.22.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, Griffiths-Jones S, Finn RD, Nawrocki EP, Kolbe DL, Eddy SR, et al. Rfam: wikipedia, clans and the ‘decimal’ release. Nucleic Acids Res. 2011;39:D141–D145. doi: 10.1093/nar/gkq1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitra S, Laederach A, Golden BL, Altman RB, Brenowitz M. RNA molecules with conserved catalytic cores but variable peripheries fold along unique energetically optimized pathways. RNA. 2011;17:1589–1603. doi: 10.1261/rna.2694811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jaeger L, Calkins ER. Downward causation by information control in micro-organisms. Interface Focus. 2011 doi: 10.1098/rsfs.2011.0045. September 29 (doi:10.1098/rsfs.2011.0045; epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plotkin JB, Kudla G. Synonymous but not the same: the causes and consequences of codon bias. Nat. Rev. Genet. 2011;12:32–42. doi: 10.1038/nrg2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. A polyhedron made of tRNAs. Nat. Chem. 2010;2:772–779. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 63.Bindewald E, Kluth T, Shapiro BA. CyloFold: secondary structure prediction including pseudoknots. Nucleic Acids Res. 2010;38:W368–W372. doi: 10.1093/nar/gkq432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaeger L, Verzemnieks EJ, Geary C. The UA_handle: a versatile submotif in stable RNA architectures. Nucleic Acids Res. 2009;37:215–230. doi: 10.1093/nar/gkn911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.