Abstract
DNA mutations are the inevitable consequences of errors that arise during replication and repair of DNA damage. Because of their random and infrequent occurrence, quantification and characterization of DNA mutations in the genome of somatic cells has been difficult. Random, low-abundance mutations are currently inaccessible by standard high-throughput sequencing approaches because they cannot be distinguished from sequencing errors. One way to circumvent this problem and simultaneously account for the mutational heterogeneity within tissues is whole genome sequencing of a representative number of single cells. Here, we show elevated mutation levels in single cells from Drosophila melanogaster S2 and mouse embryonic fibroblast populations after treatment with the powerful mutagen N-ethyl-N-nitrosourea. This method can be applied as a direct measure of exposure to mutagenic agents and for assessing genotypic heterogeneity within tissues or cell populations.
INTRODUCTION
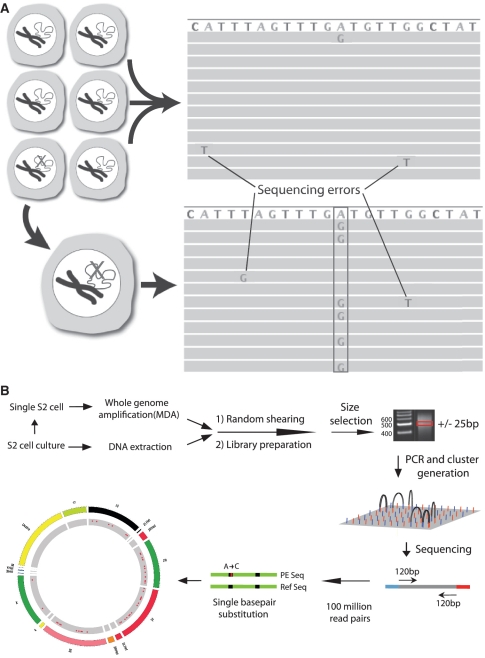
In spite of the enormous progress in genomics, the field is still very much focused on averages. Virtually all genome-wide analysis methods are geared towards clonally derived genomic DNA. For example, in sequencing cancer genomes, typically only a very small fraction of all mutations present in the tumor are detected (1). The far majority of mutations are low-abundance mutations present in a limited number of cells. In principle, such mutations could be detected by sequencing a large number of times across the same locus, i.e. at great sequencing depth. However, the high error rate of current high-throughput sequencing platforms (0.1–1%) effectively masks low-abundance mutations (2,3). To account for sequencing errors, current protocols for mutation detection are based on a consensus model, i.e. finding the same event in multiple, independent reads from the same locus. This allows only the detection of clonally amplified mutations present in most or all of the cells in a tissue sample and essentially constrains access to low-abundance mutations. One way to circumvent this problem is to sequence genomes of individual cells after whole genome amplification (WGA) (4). Every mutation in that cell at a particular locus now acts as the consensus sequence. This is schematically depicted in Figure 1A.
Figure 1.
Somatic mutation detection using single cell sequencing. (A) Somatic mutations in tissues are rare and therefore found only in single sequencing reads from which they are routinely filtered out as sequencing errors during post-alignment processing. Adopting a single cell approach overcomes this limitation by transforming each somatic event into a consensus variant call. (B) Schematic depiction of the single cell sequencing protocol used for Drosophila S2 cells.
To demonstrate that the accurate detection and quantification of somatic mutations in the genomes of single eukaryotic cells is feasible, we developed a protocol in which single cells are subjected to WGA followed by paired-end sequencing. After alignment to a reference sequence, mutations are detected as differences between the amplified single-cell genomic DNA and the reference sequence normalized to the non-amplified genomic DNA of the mother cell population. The efficacy of this protocol was validated by determining mutational spectra in single cells from Drosophila melanogaster (S2 cells) and mouse embryo's (mouse embryonic fibroblasts; MEF) exposed to the powerful point mutagen N-ethyl-N-nitrosourea (ENU).
MATERIALS AND METHODS
Cells and treatment
The Drosophila S2 cell line was acquired from Invitrogen. S2 cells were cultured in Schneider's insect medium (Sigma) supplemented with 10% FBS and penicillin/streptomycin (GIBCO) at 29°C. Primary MEFs were collected from LacZ transgenic C57BL/6 mice and cultured in DMEM (GIBCO), supplemented with 10% FBS, penicillin and streptomycin. S2 cells and MEFs (passage 5) were treated with 500 mg/ml (4.3 mM) ENU or solvent control in serum-free medium for 30 min. Following treatment, cells were washed three times in PBS and cultured in serum-supplemented medium for an additional 72 h before proceeding with mutation analysis.
Single cell isolation and WGA
Single S2 cells or MEFs were collected under an inverted microscope by hand-held capillaries, deposited in PCR tubes along with 1 µl PBS, and immediately frozen on dry ice. The remaining cells were frozen on dry ice and stored at −80°C. Multiple displacement amplification was performed on single cells using the REPLI-g UltraFast Mini kit (Qiagen, Santa Clara, CA, USA) according to the manufacturer's instructions with some modification. After lysis, an initial 30-min amplification at 30°C was used, followed by a 23.5-h amplification at 37°C. The DNA was purified using AMPure XP magnetic beads (Agencourt, Beverly, MA, USA) and the yield measured using the NanoDrop 1000 spectrophotometer (Nanodrop Technologies LLC, Wilmington, DE, USA). Single cell reactions with yields >1 µg were tested for locus dropout at 10 (S2) or 8 (MEF) loci using real-time PCR. Aliquots from the single cell reactions and from an unamplified control sample were diluted to 1 ng/µl. Real-time PCR (StepOne Plus, Applied Biosystems, Foster City, CA, USA) was performed with Fast SYBR® Green Master Mix (Applied Biosystems, Foster City, CA, USA) using 2 ng of input from each of the diluted samples and a final primer concentration of 187.5 nM. The relative abundance of each locus was estimated by comparing the Ct values from the unamplified control and each individual single cell. Cells with the highest number of loci present at a sufficient level (relative abundance > 0.25) were chosen for Illumina sequencing.
Sequencing library construction
Up to 5 µg of DNA was used as input to make Illumina libraries (5). Unamplified control DNA was extracted from the frozen cell culture mixtures isolated after the Drosophila S2 and MEF treatments. Drosophila S2 unamplified control DNA or single cell DNA was randomly fragmented using the Covaris S220 instrument (Covaris, Woburn, MA, USA). The fragmented DNA was end-repaired and size-selected to 475–525 bp using 1.5% agarose (Certified Molecular Biology Agarose, Bio-Rad, Hercules, CA, USA). The size-selected material was A-tailed and ligated to Illumina paired-end adapters (IDT, Coralville, IA, USA). A second size-selection was performed under the same conditions to reduce the frequency of chimeras. Approximately 10 ng of the product was used as input for 10 cycles of enrichment PCR with 2 U of Platinum Pfx DNA polymerase (Invitrogen, Carlsbad, CA, USA), 1× Pfx buffer, 2 mM MgSO4, 400 µM dNTPs and 1 µM Illumina enrichment PCR primers (IDT, Coralville, IA, USA). MEF unamplified control DNA or single cell DNA was digested with 50 U of MseI (NEB, Ipswich, MA, USA), end-repaired using 5 U of Mung Bean Nuclease (NEB, Ipswich, MA, USA) and size-selected using 1.5% agarose to 250–350 bp. The size-selected material was A-tailed and ligated to Illumina paired-end adapters. Approximately 10 ng of the product was used as input for 10 cycles of enrichment PCR.
Sequencing and data analysis
Libraries were sequenced using 108-bp paired-end sequencing (S2 cells) or 121-bp single-end sequencing (MEFs) on the HiSeq 2000 (Illumina, San Diego, CA, USA). Raw sequencing data was aligned to the dm3 (S2 cells) and mm9 (MEFs) reference sequences using BWA (6) with default parameters. Reads that did not align uniquely to the genome were discarded to eliminate false positives due to mapping artifacts. The aligned sequence data was processed using GATK (7) to realign reads containing indels or a high entropy of mismatches, recalibrate the base quality scores and to compute coverage data and statistics. Somatic point mutations and germline variation were scored using a pipeline composed of SAMtools (8) and the VarScan (9) somatic command. A minimum base quality score of 20 and a minimum mapping quality score of 20 were set in the VarScan command. For the Drosophila S2 cells, a minimum read depth of 20 was required for both the unamplified sample and the single cell along with a minimum mutant allele frequency of 20% for point mutations found in the single cell. Additionally, a strand bias script was also used that filtered out events where the variant allele was biased towards reads aligning to one strand. For the MEFs, a minimum read depth of 10 was required for both the unamplified sample and the single cell, along with a minimum mutant allele frequency of 40% for point mutations found in the single cell. The VarScan parameters used for somatic mutation discovery in the S2 cells and MEFs were found to achieve the appropriate balance between reducing false positives and genotyping a high fraction of the genome/target region. More stringent parameters did not lead to a considerable change in the measured mutation frequency. Somatic events found in multiple single cells were discarded, as were events found in at least one read in the unamplified control sample. Filtered somatic point mutations were visually validated using a custom IGV (10) batch script that recorded images of aligned reads at each locus containing a somatic point mutation. Select point mutations were chosen for further validation using Sanger sequencing. Primers were designed to flank either side of the mutant of interest. DNA from the single cell containing the somatic mutation and the cell line were tested and the trace images were inspected to confirm that the wild type and mutant alleles (trace peaks) were found at the expected ratio.
Mutation spectra and localization
Analysis of the localization and spectra of point mutations was performed using GATK. Functional classification of point mutations was performed using GATK's GenomicAnnotator tool and ANNOVAR (11). Strand specificity was measured for those mutations falling within genic regions based on the assumption that ENU-induced mutagenic lesions only occur at guanine and thymine bases (12). The expression levels (RPKM values) of Drosophila genes containing detected point mutations were obtained from an RNA-Seq experiment (13) with the S2 cell line, and the distribution of these values was compared to the genome-wide distribution to test for a correlation. The analysis was repeated for detected point mutations in MEFs but a microarray data set (14) was used due to lack of an appropriate RNA-Seq data set. The proximity of detected point mutations to replication origins was tested by annotating the single cell point mutation tracks with high-throughput replication timing data from the S2 cell line (15) and MEFs (16). A genome accessibility data set (17) was used to test for a correlation between point mutation localization and chromatin state in the S2 cells.
RESULTS
Estimation of the mutation load in ENU-treated and untreated Drosophila S2 cells
To assess somatic mutation spectra in single cells in a genome-wide manner we first used S2 cells derived from D. melanogaster, an organism with a genome size of 160 MB. The strategy followed is outlined in Figure 1B. Individual single cells were picked from an S2 cell culture 72-h after treatment with 4.2 mM of the powerful point mutagen ENU or mock-treated with solvent (control). At 72-h post-treatment virtually no lesions remain (18) and cell survival is >90% (not shown). Each single cell was lysed and subjected to WGA using an optimized multiple displacement amplification (MDA) protocol (19). Using a qPCR protocol, specifically designed for the purpose, amplified products for each cell were first screened to verify successful amplification by using primer pairs distributed over the different chromosomes (Supplementary Figure S1). Only samples showing amplification for all or most tested chromosomes were further processed to generate sequencing libraries for the Illumina HiSeq platform. In this way we prepared libraries of three untreated and three ENU-treated cells. For comparison, a similar library was made from unamplified total genomic DNA from the untreated S2 cell population.
To identify all possible mutations, either spontaneously formed in the unexposed, control cells or induced by ENU in the treated cells, the libraries were sequenced using a paired-end module. Variant analysis revealed point mutations, small indels and genome rearrangements. Since ENU is a point mutagen our subsequent analysis was based on this type of mutation only. Aligned sequence data from the unamplified sample and the MDA-amplified single cells were compared and all differences with the reference sequence were recorded as variants. Variants with sufficient coverage (20×) in both the unamplified and the single cell sample were classified as ‘germline’ or ‘somatic’ based on whether the variant was shared between the unamplified sample and the single cell. Somatic variants were further processed using a strand-bias filter and visually validated using the Integrative Genomics Viewer (IGV).
The results indicated sufficient coverage (20×) for genotyping at between 40% and 80% of target bases (Table 1). The incomplete coverage is due to amplification bias, which can be pronounced especially with small template amounts (19). While we optimized the MDA protocol, locus dropout was still observed, as was a significant level of allele dropout. The latter was measured using both heterozygous SNPs present in our S2 cell line population and the mutant read frequency distribution, which produced similar results. A comparison of the mutation load in the control and ENU-treated cells showed a 7.5-fold induction of point mutations by ENU on average in the exposed cells (Figure 2A, Table 1). We chose multiple somatic mutations for validation with Sanger sequencing using the remaining amplified material from our single cells and found no evidence of false positives (for an example, see Supplementary Figure S2).
Table 1.
Single cell sequencing data
| Single cell | Point mutations | Bases in genome with sufficient coverage (MB) | Fraction of target region (%) | Alleles represented (%) | Mutations per MB |
|---|---|---|---|---|---|
| S2 Cont. 1 | 45 | 58.97 | 50.56 | 56.68 | 0.34 |
| S2 Cont. 2 | 43 | 53.00 | 45.44 | 55.95 | 0.36 |
| S2 Cont. 3 | 40 | 37.17 | 31.87 | 54.33 | 0.50 |
| S2 ENU 1 | 938 | 97.74 | 83.80 | 73.36 | 3.27 |
| S2 ENU 2 | 482 | 82.58 | 70.80 | 57.44 | 2.54 |
| S2 ENU 3 | 690 | 90.05 | 77.16 | 60.27 | 3.18 |
| MEF Cont. 1 | 9 | 85.17 | 38.71 | ~60 | 0.09 |
| MEF Cont. 2 | 14 | 89.42 | 40.65 | ~60 | 0.13 |
| MEF ENU 1 | 426 | 89.69 | 40.77 | 59.89 | 3.97 |
| MEF ENU 2 | 446 | 92.98 | 42.27 | 61.34 | 3.91 |
Figure 2.
Observed somatic point mutations. (A) Genome-wide sequence coverage and mutation localization in an untreated cell and an ENU-treated cell. The outer track represents binned coverage with an upper cutoff of 50×. The inner track shows the location of detected point mutations (represented as dark points). (B) Histograms of mutant read frequencies for point mutations on chr2L and chrX. The dotted lines indicate a normal distribution with mean of 25 for chr2L and 50 for chrX with standard deviations of 22.
While spontaneous mutations present in the untreated cells could be expected to occasionally be homozygous, all ENU-induced mutations are likely to involve one allele only. The S2 cell line is known to be tetraploid (13) (Supplementary Figure S3) and assuming an equal representation of each allele in the whole genome-amplified material from our single cells, we would expect a quarter of the reads aligning across a spontaneous or induced mutation to contain the mutant base. Figure 2B shows the mutant read frequencies across chr2L for the ENU-induced mutations. While we did find the expected peak read frequency of 25% for chr2L, the significant tail to higher frequencies indicates the unequal amplification across the four alleles. Since the S2 cell line is male, there are two X chromosomes in addition to the four sets of autosomes. Hence, we would expect a read frequency peak at 50% rather than 25% for chrX and this is indeed what was found (Figure 2B).
Estimation of the mutation load in ENU-treated and untreated MEFs
To apply the same strategy to mammalian cells is significantly more expensive because of the larger genome size. Therefore, we slightly modified the procedure shown in Figure 1B, applying a reduced representation approach. For this purpose we used mouse embryonic fibroblast (MEF) populations, either treated with 4.2 mM ENU or mock-treated with solvent only. Instead of preparing sequencing libraries directly using randomly fragmented DNA, we digested whole genome-amplified DNA from two treated and two control MEFs as well as unamplified genomic DNA from the MEF population with MseI, a four-base cutter with a TTAA cleavage site.
Following digestion, a fragment size range (250–350 bp) corresponding to a target region of ~300 MB was isolated from agarose gels and sequenced using 121-bp single-end reads. The processed data revealed a significant number of point mutations induced by ENU in the two cells from the exposed population, similar to what was observed for the S2 cells (Table 1). Due to the nature of the reduced representation library, the strand bias filter could not be used and therefore a more stringent mutant read frequency cutoff (>40%) was adopted. The results indicate a 35-fold induction of point mutations in the ENU-treated MEFs. Previously, using a lacZ reporter in the same cell population, a significantly smaller number of ENU-induced mutations was observed (20), underscoring the reduced sensitivity of reporter systems, which can only detect mutations that alter the phenotype to a considerable extent (21).
The much higher fold induction of mutations in the ENU-treated MEFs than in the S2 cells is almost entirely due to a lower baseline mutation frequency in the two untreated MEFs. This is not surprising since the S2 cell line used has a long history of passaging during which mutations are likely to have accumulated. Baseline levels of somatic mutation frequencies are obviously very difficult to determine with high accuracy and in this case also depend on the cut-offs used to filter out potential artifacts. Here, we were more interested in comparing the absolute number of mutations per MB induced by ENU in cells from the two species, which proved remarkably similar. Indeed, the ENU-induced mutation frequency in the MEFs was only 30% higher than that found in the S2 cells (Table 1). Somatic mutation frequencies are a measure for the efficiency of an organism to cope with DNA damage and it is somewhat surprising that cells from such disparate species are very similar in this respect. More extensive studies with different, freshly isolated cell types from these species should provide a definite answer.
Cross species comparison of the spectra and localization of ENU-induced and spontaneous mutations
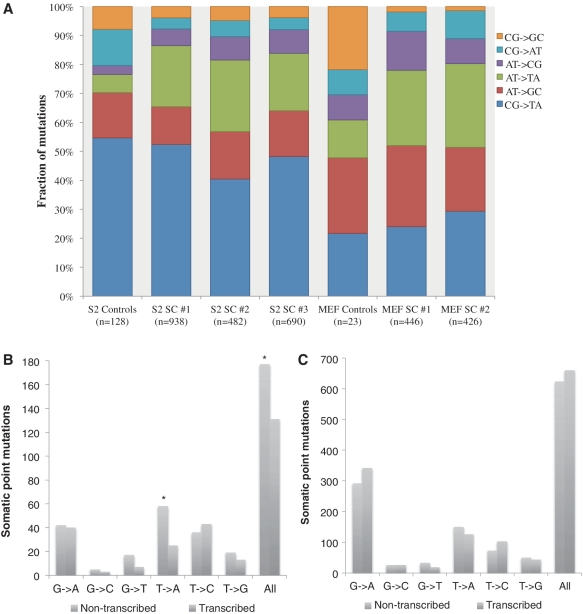
A major advantage of direct sequencing is that the mutation spectra can immediately be visualized across the genome. The ENU-induced spectrum was highly consistent across individual cells from the same population, but a larger fraction of C:G →T:A mutations was found in the S2 cells (Figure 3A). This may be due to a more efficient repair of O6-ethyl-guanine adducts by the mouse O6-alkylguanine-DNA alkyltransferase gene (Mgmt) compared with its Drosophila homologue. In spite of this difference, the similarity between the two species also at this level is striking. The spontaneous mutation spectra observed in the untreated cells showed considerably lower levels of A:T →T:A mutations, which are known to be highly enriched following treatment with alkylating agents (12,22).
Figure 3.
Somatic point mutation spectra and localization. (A) Mutation spectra for the control and ENU-treated S2 and MEF single cells. Due to a limited number of point mutations, the control cells were combined for the analysis. (B) Strand of origin for ENU-induced mutations found in genic regions of MEFs. (C) Strand of origin for ENU-induced mutations found in genic regions of S2 cells.
Since ENU is a small, direct-acting agent, we did not expect to see a large bias for mutations localized in accessible regions of the genome. By comparing data on the accessibility of the S2 cell line with the coordinates of our point mutations, we determined that there was no correlation between mutation localization and genome accessibility. There was also no correlation in either the MEFs or S2 cells between the mutation frequency and proximity to a replication origin or localization in a functional category (exon, intron or intergenic region). Analysis of the ENU-induced mutations falling within genic regions in the two MEF cells showed evidence of transcription-coupled repair, with a lower fraction of ENU-induced mutations occurring at T and G bases, the predominant adduct bases, on the transcribed strand than the non-transcribed strand (P < 0.01) (Figure 3B). This bias appeared strongest for TA transversions (P < 0.0002), supporting previous results at the endogenous HPRT gene locus (22). No evidence for any transcription-coupled repair process (Figure 3C) was seen in the S2 cells, in keeping with results obtained with Drosophila Kc cells (23) and the absence of homologues of either CSA or CSB, the main TCR genes, in Drosophila (24).
DISCUSSION
In the present work, we addressed the problem as to how low-abundant DNA mutations, for example, as typically induced by environmental mutagens, can be detected directly using ultra-high-throughput sequencing. In principle, such low abundant mutations can be detected by single-molecule sequencing (25). However, this necessarily involves the sequencing of fragments derived from different cells and precludes insight into the mosaic basis of somatic mutations in tissues or cell populations. Indeed, the absence of robust methods for analyzing genomes at the level of the single cell essentially constrains access to the complexity of biological tissues (26). For example, cancerous tissue is notoriously heterogeneous with each cell carrying its own unique capabilities for growing into a full-blown tumor (1). The ability to analyze subclonal genetic diversity will greatly expand the accessible clinical information about a particular cancer in a particular patient (27).
Another area of application of single-cell genome sequencing is in monitoring stem cells. The genomic integrity of human embryonic stem cells and induced pluripotent stem cells has been questioned due to a high observed rate of point mutations (28), copy number variations (29) and changes in genome-wide CpG methylation (30). These studies identified potentially hazardous somatic mutations/epi-mutations that had clonally expanded through the population. However, there may also be low-abundance mutations present in the stem cell populations missed by the aforementioned analyses that need to be addressed before clinical use can be considered.
Finally, during development, maturation and aging the genome of somatic cells is subject to the continuous occurrence of random genome sequence alterations, which gradually diminish intra-organ homogeneity and may lead to loss of coordination of expression among multiple genes in functional pathways or networks (31). Such emerging cell-to-cell variability is only amenable through single-cell genomics approaches.
In this study, we have taken the first step towards comprehensively analyzing mammalian single-cell genomes using next-generation sequencing. We first showed evidence of an induced mutation load in single ENU-treated cells from a Drosophila cell line. The high level of induction and the consistency of the results across the three ENU-treated cells provide strong evidence that the experimental results for the ENU-induced cells are accurate. Next, we developed a reduced-representation assay to repeat the experiment using MEFs. We observed a consistent mutation frequency across the two single ENU-treated MEFs, similar to the levels found in the treated S2 cells. The mutation frequencies observed in the treated MEFs were two-fold higher than those previously estimated using a lacZ transgenic reporter gene (20). The discrepancy can be explained, at least in part, by the fact that the reporter gene cannot detect mutations that do not inactivate the β-galactosidase enzymatic activity (21).
While our results conclusively indicate elevated mutation frequencies in both S2 cells and MEFs after exposure to ENU, our data does not allow for accurate estimates of the frequency of spontaneous mutations in control cells. For the control MEF cells, it is possible to make a comparison with spontaneous mutation frequencies observed with our lacZ reporter system in these same cells (32). While we do find higher mutation frequencies by direct sequencing than with the reporter system (in keeping with the inability of the lacZ positive selection system to detect silent mutations), they are in the same range.
Background mutation frequencies are equal to the sum of the spontaneous mutation frequencies within the individual cells and the background error rate of the assay due to mutations introduced during the WGA step. An error introduced early in MDA would be found in 12.5% of the sequences in a diploid cell and 6.25% of the sequences in a tetraploid cell on average (25). The kinetics of the amplification process, i.e. the fact that multiple polymerase molecules may be operating on the original template strand when the initial error is introduced, may further reduce the probability of significant errors occurring. The percentages listed above are averages however, and it is possible that an error produced early in MDA could be randomly selected for, leading to a false positive call present in the majority of reads aligning at a locus. Additionally, the high degree of allele dropout observed in many of our samples increases the probability that some artifacts produced early in MDA could be found in a significant proportion of reads aligning at a locus. In order to obtain a more quantitative estimate of the spontaneous mutation frequency, more extensive studies are needed, using maximum-likelihood methods for estimating mutation frequencies from the high-throughput sequencing data, for example, as described by Lynch (33).
The mutant spectra observed in the treated S2 and MEF cells agrees with data obtained using reporter genes (34–36), providing additional evidence for the accuracy of our measured ENU-induced mutation loads. A common argument against the use of reporter genes is that they may not be representative of genome-wide events, due to both sequence specificity and their dependence on a phenotypic change. ENU is a small direct-acting agent with a lack of sequence specificity and, therefore, it was not surprising that no major differences were found between reporter gene data and our genome-wide unbiased approach.
While this analysis was limited to point mutations, the same methodology can be applied to investigate small insertions and deletions (InDels) and structural variation. Indeed, the paired-end sequencing approach allows us to detect structural alterations as invalid alignments to the reference genome sequence (37). If the mapped locations of the ends of a paired-read have abnormal distances, orientation or chromosomal localization, then a genomic rearrangement is suggested. While we were clearly able to detect such events, ENU is a point mutagen and we did not find an increase of genome rearrangements in the treated cells (not shown).
In summary, these results show for the first time how massively parallel sequencing can be used effectively for measuring random, low-abundance mutations in somatic cells. This opens up the possibility to analyze intra-tissue heterogeneity of cellular genotypes. Importantly, our methodology provides a direct, comprehensive approach for estimating an individual's risk from exposure to mutagenic agents, such as radiation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures S1–S3.
FUNDING
National Institutes of Health (NIH) (grant numbers R01AG034421, P01AG17242, R21AG030567, R21ES019520, U54RR024346). Funding for open access charge: NIH, National Institute of Aging (R01AG034421).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Donald Conrad, Alex Maslov, Brent Calder and Silvia Gravina for many helpful discussions of the data and Francesca Faggioli for karyotyping the S2 cell line.
REFERENCES
- 1.Salk JJ, Fox EJ, Loeb LA. Mutational heterogeneity in human cancers: origin and consequences. Annu. Rev. Pathol. 2010;5:51–75. doi: 10.1146/annurev-pathol-121808-102113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center's improvements to the Illumina sequencing system. Nat. Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Druley TE, Vallania FL, Wegner DJ, Varley KE, Knowles OL, Bonds JA, Robison SW, Doniger SW, Hamvas A, Cole FS, et al. Quantification of rare allelic variants from pooled genomic DNA. Nat. Methods. 2009;6:263–265. doi: 10.1038/nmeth.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geigl JB, Speicher MR. Single-cell isolation from cell suspensions and whole genome amplification from single cells to provide templates for CGH analysis. Nat. Protoc. 2007;2:3173–3184. doi: 10.1038/nprot.2007.476. [DOI] [PubMed] [Google Scholar]
- 5.Quail MA, Swerdlow H, Turner DJ. Improved protocols for the illumina genome analyzer sequencing system. Curr. Protoc. Hum. Genet. 2009 doi: 10.1002/0471142905.hg1802s62. Chapter 18, Unit 18 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–2285. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res.. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosal L, Comendador MA, Sierra LM. In vivo repair of ENU-induced oxygen alkylation damage by the nucleotide excision repair mechanism in Drosophila melanogaster. Mol. Genet. Genomics. 2001;265:327–335. doi: 10.1007/s004380000419. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Y, Malone JH, Powell SK, Periwal V, Spana E, Macalpine DM, Oliver B. Expression in aneuploid Drosophila S2 cells. PLoS Biol. 2010;8:e1000320. doi: 10.1371/journal.pbio.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen J, Liu J, Yang J, Chen Y, Ni S, Song H, Zeng L, Ding K, Pei D. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 2011;21:205–212. doi: 10.1038/cr.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eaton ML, Prinz JA, MacAlpine HK, Tretyakov G, Kharchenko PV, MacAlpine DM. Chromatin signatures of the Drosophila replication program. Genome Res. 2011;21:164–174. doi: 10.1101/gr.116038.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiratani I, Ryba T, Itoh M, Rathjen J, Kulik M, Papp B, Fussner E, Bazett-Jones DP, Plath K, Dalton S, et al. Genome-wide dynamics of replication timing revealed by in vitro models of mouse embryogenesis. Genome Res. 2010;20:155–169. doi: 10.1101/gr.099796.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat. Struct. Mol. Biol. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- 18.Bielas JH, Heddle JA. Proliferation is necessary for both repair and mutation in transgenic mouse cells. Proc. Natl Acad. Sci. USA. 2000;97:11391–11396. doi: 10.1073/pnas.190330997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lasken RS. Genomic DNA amplification by the multiple displacement amplification (MDA) method. Biochem. Soc. Trans. 2009;37:450–453. doi: 10.1042/BST0370450. [DOI] [PubMed] [Google Scholar]
- 20.Mahabir AG, Zwart E, Schaap M, van Benthem J, de Vries A, Hernandez LG, Hendriksen CF, van Steeg H. lacZ mouse embryonic fibroblasts detect both clastogens and mutagens. Mutat. Res. 2009;666:50–56. doi: 10.1016/j.mrfmmm.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 21.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Op het Veld CW, van Hees-Stuivenberg S, van Zeeland AA, Jansen JG. Effect of nucleotide excision repair on hprt gene mutations in rodent cells exposed to DNA ethylating agents. Mutagenesis. 1997;12:417–424. doi: 10.1093/mutage/12.6.417. [DOI] [PubMed] [Google Scholar]
- 23.de Cock JG, van Hoffen A, Wijnands J, Molenaar G, Lohman PH, Eeken JC. Repair of UV-induced (6-4)photoproducts measured in individual genes in the Drosophila embryonic Kc cell line. Nucleic Acids Res. 1992;20:4789–4793. doi: 10.1093/nar/20.18.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sekelsky JJ, Brodsky MH, Burtis KC. DNA repair in Drosophila: insights from the Drosophila genome sequence. J Cell Biol. 2000;150:F31–F36. doi: 10.1083/jcb.150.2.f31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dear PH. One by one: single molecule tools for genomics. Brief Funct. Genomic Proteomic. 2003;1:397–416. doi: 10.1093/bfgp/1.4.397. [DOI] [PubMed] [Google Scholar]
- 26.Kalisky T, Quake SR. Single-cell genomics. Nat. Methods. 2011;8:311–314. doi: 10.1038/nmeth0411-311. [DOI] [PubMed] [Google Scholar]
- 27.Fox EJ, Salk JJ, Loeb LA. Cancer genome sequencing–an interim analysis. Cancer Res. 2009;69:4948–4950. doi: 10.1158/0008-5472.CAN-09-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gore A, Li Z, Fung HL, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, et al. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, et al. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohi Y, Qin H, Hong C, Blouin L, Polo JM, Guo T, Qi Z, Downey SL, Manos PD, Rossi DJ, et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell. Biol. 2011;13:541–549. doi: 10.1038/ncb2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijg J. Aging of the Genome: The Dual Role of the DNA in Life and Death. Oxford; New York: Oxford University Press; 2007. [Google Scholar]
- 32.Busuttil RA, Rubio M, Dolle ME, Campisi J, Vijg J. Oxygen accelerates the accumulation of mutations during the senescence and immortalization of murine cells in culture. Aging Cell. 2003;2:287–294. doi: 10.1046/j.1474-9728.2003.00066.x. [DOI] [PubMed] [Google Scholar]
- 33.Lynch M. Estimation of nucleotide diversity, disequilibrium coefficients, and mutation rates from high-coverage genome-sequencing projects. Mol. Biol. Evol. 2008;25:2409–2419. doi: 10.1093/molbev/msn185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pastink A, Vreeken C, Nivard MJ, Searles LL, Vogel EW. Sequence analysis of N-ethyl-N-nitrosourea-induced vermilion mutations in Drosophila melanogaster. Genetics. 1989;123:123–129. doi: 10.1093/genetics/123.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vivian JL, Chen Y, Yee D, Schneider E, Magnuson T. An allelic series of mutations in Smad2 and Smad4 identified in a genotype-based screen of N-ethyl-N-nitrosourea-mutagenized mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2002;99:15542–15547. doi: 10.1073/pnas.242474199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takahasi KR, Sakuraba Y, Gondo Y. Mutational pattern and frequency of induced nucleotide changes in mouse ENU mutagenesis. BMC Mol. Biol. 2007;8:52. doi: 10.1186/1471-2199-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.