Abstract
The phosphoesterase (PE) domain of the bacterial DNA repair enzyme LigD possesses distinctive manganese-dependent 3′-phosphomonoesterase and 3′-phosphodiesterase activities. PE exemplifies a new family of DNA end-healing enzymes found in all phylogenetic domains. Here, we determined the structure of the PE domain of Pseudomonas aeruginosa LigD (PaePE) using solution NMR methodology. PaePE has a disordered N-terminus and a well-folded core that differs in instructive ways from the crystal structure of a PaePE•Mn2+• sulfate complex, especially at the active site that is found to be conformationally dynamic. Chemical shift perturbations in the presence of primer-template duplexes with 3′-deoxynucleotide, 3′-deoxynucleotide 3′-phosphate, or 3′ ribonucleotide termini reveal the surface used by PaePE to bind substrate DNA and suggest a more efficient engagement in the presence of a 3′-ribonucleotide. Spectral perturbations measured in the presence of weakly catalytic (Cd2+) and inhibitory (Zn2+) metals provide evidence for significant conformational changes at and near the active site, compared to the relatively modest changes elicited by Mn2+.
INTRODUCTION
Many bacteria have two independent mechanisms to repair DNA double-strand breaks (DSBs)—homologous recombination and non-homologous end joining (NHEJ) (1,2). DNA ligase D (LigD) is the central catalyst of bacterial NHEJ in vivo (3). LigD is composed of three autonomous enzymatic modules: an ATP-dependent ligase (LIG) domain that joins 3′-OH and 5′-phosphate ends (4,5); a polymerase (POL) domain that adds ribonucleotides or deoxyribonucleotides to DSB ends and gaps (6–11); and a 3′-phosphoesterase (PE) domain (12) that hydrolyzes 3′-monophosphate termini to 3′-OH ends that can either be extended by POL or sealed by LIG. The PE domain also has a unique 3′-phosphodiesterase activity that resects short 3′ ribonucleotide tracts created at DSB ends by the POL domain, to leave a 3′-OH monoribonucleotide end that is the preferred substrate for sealing by bacterial NHEJ ligases (12–16). Mutational studies of the PE domain of Pseudomonas aeruginosa LigD (PaePE) have identified residues essential for activity (13). Whereas His42, His48, Asp50, Arg52, His84 and Tyr88 are important for both the monoesterase and diesterase activities, Arg14, Asp15, Glu21 and Glu82 are implicated specifically in the monoesterase activity.
In order to understand the structural basis for catalysis by the PE domain, we pursued crystallographic and solution NMR studies of PaePE in parallel. The 1.9-Å crystal structure of the active phosphodiesterase core of PaePE revealed a novel fold in which an 8-strand β-barrel with a hydrophobic interior supports a distinctive crescent-shaped hydrophilic active site on its outer surface, wherein six of the essential side chains coordinate manganese and a sulfate anion (17). Subsequent crystal structures of two archaeal PE proteins at high resolution—1.1 Å for Candidatus Korarchaeum cryptofilm PE (CkoPE) and 2.1 Å for Methanosarcina barkeri PE (MbaPE)—higlighted conservation of the PaePE β-barrel fold, active site constituents and metal-binding site (18). Thus, PaePE exemplifies a novel family of 3′-end healing enzymes found in diverse taxa of the bacterial and archaeal domains of life. Putative PE homologs were also detected in several eukaryal proteomes (17).
Missing from the PaePE crystal structure was an N-terminal peptide segment, deletion of which selectively diminished the 3′-phosphomonoesterase activity of PaePE without affecting the 3′-phosphodiesterase activity (16). The MbaPE protein has the homologous N-terminal peptide, but electron density corresponding to this segment was missing in the MbaPE crystal structure, suggesting that it is disordered (18). CkoPE naturally lacks an equivalent of the N-peptide, and the associated 3′-phosphomonoesterase activity. It was notable that the crystallization of PaePE, CkoPE and MbaPE could only be achieved in the presence of Mn2+ and an oxyanion (either sulfate or phosphate), both of which were present as a Mn2+•oxyanion complex in the respective PE active sites (18). The phosphate and sulfate anions in the crystal structures were taken to be mimetics of the scissile 3′-phosphate of the PE nucleic acid substrate.
Our aim in the present study was to obtain a solution structure of PaePE in order to determine its conformation in the absence of metals or substrate mimetics and, if possible, to visualize the N-terminal peptide, in the absence of any perturbations including external non-physiological additives and crystal-packing forces. Initial NMR studies were performed with PaePE-(1–187), which produced spectra that were suitable for obtaining backbone assignments, but were not of high enough quality to yield a sufficient number of sidechain 1H assignments to allow structure determination. Serial C-terminal truncations identified PaePE-(1–177) as a catalytically active 3′-monoesterase/3′-diesterase that produced spectra of considerably higher quality and yielded sufficient number of 1H assignments (19) suitable for calculating an NMR structure of this highly dynamic enzymatic module. Here, we present the solution structure of PaePE-(1–177) determined using standard NOE-based approaches. By investigating spectral changes in the presence of each of three different oligonucleotide primer–template duplexes and a series of bivalent metal ions, we provide insight to the DNA binding and metal ion recognition properties of PaePE.
MATERIALS AND METHODS
Protein expression and purification
PaePE-(1–177) (referred to hereafter as PaePE) was produced in E. coli as an N-terminal His10-Smt3 fusion; isotope labeling and purification of the tag-free PaePE protein are described in detail elsewhere (19). PaePE samples for NMR experiments were prepared in a buffer containing 50 mM Bis–Tris (pH 6.5), 200 mM NaCl and 5 mM DTT (referred to as the NMR buffer from hereon forward) and typically contained ~300 µM protein. The lack of chemical shift changes in the 15N,1H TROSY spectra acquired with and without a 10-fold excess of EDTA argued against the retention or acquisition of enzyme-bound bivalent metal ions during the purification process. Thus, all subsequent spectra were acquired in the absence of EDTA.
Resonance assignment
Backbone 1H, 15N, 13Cα, 13C′ and sidechain 13Cβ assignments were obtained via TROSY-based HNCO, HN(CA)CO, HNCACB and HN(CO)CACB experiments (20) utilizing uniformly 2H,13C,15N-labeled PaePE. Sidechain 13C and 1H assignments were obtained via (H)C(CO)NH and H(CCO)NH experiments utilizing uniformly 13C,15N-labeled PaePE (21). Experimental details have been described at length elsewhere (19).
Distance and angular constraints
Backbone 13Cα, 13Cβ, 13C′ and 15N chemical shifts were converted into dihedral ϕ and ψ angular restraints using the TALOS+ software suite (22). NOE-derived distance constraints to the backbone amide 1H nuclei were obtained using a 15N-edited NOESY-HSQC (utilizing a uniformly 15N-labeled sample) with a 150-ms mixing time at a field strength of 800 MHz. Distance restraints involving sidechain protons were obtained from 3-dimensional 13C-edited NOESY experiments in D2O (150 ms mixing time, 800 MHz), separately optimized for aliphatic and aromatic regions of the spectrum. Uniformly 15N,13C-labeled samples were utilized in both cases. NOESY crosspeak intensities were obtained using NMRView (23) and converted into distance restraints using the ambiguous distance restraints (ADR) protocol as implemented in the ARIA (24) software suite. Manual NOE assignments were supplemented with those assigned automatically using the ARIA scheme as described previously (24).
Structure calculations
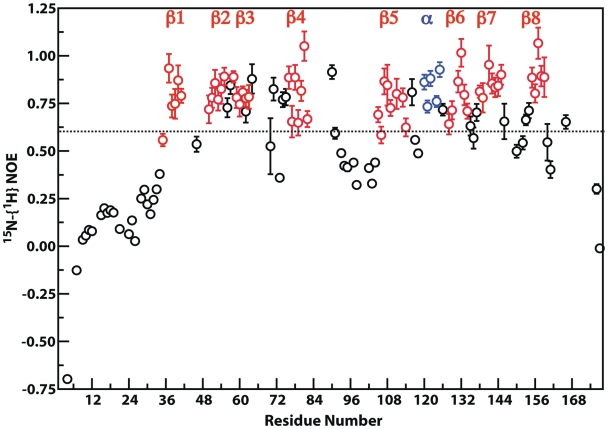
To facilitate structure calculation, the ordered regions of PaePE were determined using an improved pulse sequence to measure the 15N-{1H} steady-state NOE at 600 MHz (25). Spectra were acquired with several repetitions of a τ-π-τ (τ = 11 ms) symmetric 1H saturation scheme for a total of 3 s. An additional delay of 2 s was used to allow lock stabilization. The reference experiment included a 5 s pre-delay. Spectra were acquired using sweep-widths of 14 ppm (512 complex points) and 32 ppm (128 complex points) for the 1H and 15N dimensions respectively, using 64 transients per t1 point. Based on the 15N-{1H} steady-state NOE values, structure calculations were initially performed excluding the flexible N- and C-termini of PaePE and including only residues Thr32-Gly163. After the ordered ‘core’ had been suitably refined, the N-terminal 31 residues were included in the structure calculations to obtain the final structural ensemble. Few resonance assignments were available for the disordered C-terminal region (164–177) that is dispensable for catalytic activity. Consequently, this polypeptide fragment was not included in the structure calculations.
Manually assigned NOEs were combined with dihedral and hydrogen-bonding constraints into a standard assignment/structure-calculation protocol using the ARIA 2.3/CNS 1.1 (24) software suite. Pro65 and Pro72 were constrained to be in the cis conformation using the cis-proline patch during the structure calculation. Additional assignments that were obtained automatically during a run were inspected for correctness and incorporated into subsequent runs. Structure calculations were performed using a Cartesian dynamics simulated annealing protocol that consisted of: (i) high temperature dynamics at 2000 K (10 000 steps); (ii) 4000 steps of refinement; (iii) a Cartesian dynamics cooling stage from 2000 to 1000 K (6000 steps); and (iv) a second Cartesian dynamics cooling stage from 1000 to 50 K (5000 steps). Force constants of 5, 25 and 200 kCal mol−1 Å−2 for the dihedral constraints and 10, 10 and 50 kCal mol−1 Å−2 for the distance constraints (ambiguous, unambiguous and hydrogen bond) were used during the three temperature stages of the Cartesian dynamics protocol. Prochiral Hβ and Hγ from long-chain amino acids and methyl groups from Val and Leu, were treated using the floating chirality method (24). For the final refinement step, a subset of the lowest energy structures were solvated in a 7.5 -Å box of explicit solvent and refined using the full Lenard-Jones non-bonded potential and electrostatic interactions from the OPLS force-field using a protocol described in detail previously (26). In the final run, 2048 structures were calculated and the resultant 128 lowest energy structures were used in the water-refinement protocol. Fifteen of the lowest energy water-refined structures with the least number of violations of experimental constraints were chosen to represent the final structural ensemble of PaePE (note that only residues 1–163 were included in the final round of structure calculations). The quality of the resulting NMR structures was evaluated with PROCHECK (27). Regions of secondary structure were defined using Promotif 3.0 (28,29).
Preparation of oligonucleotides
HPLC purified DNA oligos D16F1R1 (5′-TTTTTTGGGACCGGTC(C2′F)(rC); wherein the penultimate nucleoside is 2′-fluorocytidine and the 3′ terminal nucleoside is ribocytidine), D18 (5′-TTTTTTGGGACCGGTCCC) and D18p (5′-TTTTTTGGGACCGGTCCCp: where p is a 3′-monophosphate) were purchased from Integrated DNA Technologies Incorporated. Stock solutions containing 100 µl of aqueous D16F1R1 (9.2 mM) or 50 µl of aqueous D18 (11.6 mM) or 50 µl of aqueous D18p (6 mM) were annealed in an Eppendorf thermocycler at 95°C for 3 min, at 60°C for 2 min, at 40°C for 10 min and finally at 25°C for 10 min, to generate duplex primer–templates each containing a 6-nt single-stranded tail. The concentrations of the annealed primer–template duplexes were estimated from absorption measurements at wavelengths of 260 and 280 nm (see Figure 4a for details of the annealed duplexes).
Figure 4.
(A) The three primer-template constructs used to measure spectral perturbations in PaePE: D18p contains a 3′-phosphate; D16F1R1, contains a 2′-fluorocytidine (to closely mimic a ribose) in the penultimate position and a 3′-terminal ribocytidine. (B) Results of the NMR titration of PaePE with D18. 15N,1H TROSY spectra acquired at 600 MHz in the absence (red) and the presence of equimolar amounts of D18 (blue). The inset shows the trajectory of the resonance corresponding to Leu157 that lies on β8 during the course of the titration.
NMR measurements of protein–oligonucleotide interactions
Samples of uniformly 2H, 15N-labeled PaePE (100 µM) in NMR buffer were used for NMR titrations. For all the primer-templates, 15N,1H TROSY spectra were acquired for PaePE:oligo ratios of 1:0.05, 1:0.12, 1:0.2, 1:0.3, 1:0.5, 1:1, 1:2 and 1:3 for D18; 1:0.05, 1:0.1, 1:0.2, 1:0.3, 1:0.5, 1:1, 1:2 and 1:3 for D18p. Data were acquired at 600 MHz at 25°C using sweep-widths of 15 ppm (512 complex points) and 36.2 ppm (128 complex points) in the direct and indirect dimensions, respectively, using a recycle delay of 1.5 s and 48 transients per t1 point. For the D16F1R1 primer-template, the following PaePE:oligo ratios were used 1:0.05, 1:0.1, 1:0.2, 1:0.3, 1:0.5, 1:1, 1:2 and 1:3 and 15N,1H TROSY spectra were acquired at 800 MHz at 25°C using sweep-widths of 14 ppm (512 complex points) and 32 ppm (128 complex points) in the direct and indirect dimensions respectively using a recycle delay of 1.5 s and 64 transients per t1 point.
Scaled chemical shift differences were calculated using the following equation:
| (1) |
where δo.H and δo.N are the 1H and 15N chemical shifts for a given residue in apo PaePE, respectively, while δi.H and δo.N are the corresponding shifts at the ith titration point.
NMR measurements of divalent cation binding
Samples of uniformly 15N,2H labeled PaePE (100 µM) in the NMR buffer (excluding DTT; removal of DTT produced no spectral perturbations) were used for the metal ion titrations (Mn2+, Cd2+ and Zn2+). Metal ions were added as dilutions from 7.5 mM stock solutions of MnCl2, CdCl2 or ZnCl2. A series of 15N, 1H HSQC datasets were obtained at 700 MHz (25°C) with sweep-widths of 14 ppm (512 complex points) and 32 ppm (128 complex points) in the direct and indirect dimensions respectively, using recycle delays of 1.5 s (5.5 s for Mn2+; to allow a more accurate comparison between the peak intensities in the reference state and those in the presence of the paramagnetic metal ion resulting in increased R1 rates and enhanced recovery toward equilibrium between scans) and 32 transients per t1 point. The following protein-to-metal ratios were used: Cd2+—1:0.25, 1:0.5, 1:1 and 1:2; Zn2+—1:0.5, 1:1; Mn2+—1:0.25, 1:0.5 and 1:1. In addition, the effects of phosphate were analyzed using a 10-fold excess of phosphate in buffer containing equimolar ratios of PaePE and each of the three divalent cations. An additional dataset was collected at 600 MHz (25°C) for Zn2+ in the presence of DTT using the following ratios of PaePE and Zn2+—1:0.2, 1:0.4, 1:0.6, 1:0.8, 1:1, 1:1.5 and 1:2. These data were fully consistent with those collected in the absence of DTT. The reversibility of the spectral perturbations induced by metal ion binding in all cases was tested by addition of an excess of EDTA.
Fluorescence measurements of protein–DNA interactions
A 1 µM sample of PaePE in the NMR buffer was used for the fluorescence measurements using a 3-ml cuvette that allowed continuous mixing. All measurements were performed on a Spex Fluorolog-3 (Horiba Jobin Yvon) at 25°C. Tryptophan emission was monitored following excitation at 280 nm. Emission spectra were recorded from 300 to 450 nm in 1 nm increments using a 0.5-s integration time and averaged over four scans. For the D16F1R1 primer-template, the following PaePE:oligo ratios were used—1:0, 1:0.05, 1:0.1, 1:0.2, 1:0.3, 1:0.5, 1:0.75, 1:1, 1:1.5, 1:2, 1:3, 1:5, 1:8, 1:12, 1:18, 1:25 and 1:40. Experiments were performed first in the absence of divalent metal ions and then in the presence of 100 µM Mn2+. Apparent Kd values were obtained by fitting the change in fluorescence intensity at the emission maximum in the absence of oligos (331 nm) with increasing D16F1R1 concentration to a quadratic binding isotherm [Equation (2)] using in-house software that utilized the ODRPACK libraries (30).
 |
(2) |
where I is the intensity at 331 nm at a dsDNA concentration of [D], I0 is the corresponding intensity in the absence of DNA, I∞ is the intensity difference at saturation, [P]0 is the protein concentration (held constant at 1 µM). Additionally, fluorescence titrations of 1 µM PaePE with Mn2+ were performed in the absence of oligonucleotide using the following PaePE:metal ion ratios—1:0, 1:0.05, 1:0.1, 1:0.2, 1:0.5, 1:1, 1:2, 1:5, 1:10, 1:20, 1:40 and 1:80.
RESULTS AND DISCUSSION
Solution structure of PaePE
Inspection of the final NMR ensemble of 15 structures (see Table 1 for statistics) reveals a disordered N-terminus (1–31). This fact is also borne out by the extremely low 15N-{1H} NOE values (0.08 ± 0.21; see Figure 1) for this region. However, some degree of local ordering is seen encompassing residues Asp15–Thr19 (15N-{1H} NOE: 0.18 ± 0.01). This region contains Asp15 and is flanked by Arg14 and Glu21; these three sidechains are important for the 3′-monoesterase activity of PaePE (16). The distribution of ϕ/ψ angles for the Asp15–Thr19 segment indicates an overall extended structure (Supplementary Figure S1). A hydrogen bond between the carbonyl oxygen of Thr19 and the amide of Glu21 is found in all 15 structures of the final ensemble.
Table 1.
NMR restraints and structure statistics
| Restraints and statistics | |
|---|---|
| Total number of restraints | 2380 |
| NOE restraints | 2080 |
| Unambiguous | 1676 |
| Intraresidue | 563 |
| Sequential (|i - j| = 1) | 488 |
| Short-range (|i - j| = 2,3) | 153 |
| Medium-range (|i - j| = 4,5) | 53 |
| Long-range (|i - j| > 5) | 419 |
| Ambiguous | 404 |
| Dihedral angle restraints | 212 |
| Hydrogen bond restraintsa | 88 |
| Constraint violationsb | |
| NOE violations > 0.5 Å | 0 |
| Dihedral violations > 5° | 4 |
| Deviation from idealized geometry | |
| Bond lengths (Å) | 5.55 × 10−3 ± 0.12 × 10−3 |
| Bond angles (°) | 0.74 ± 0.02 |
| Impropers (°) | 2.05 ± 0.10 |
| Energies (kcal mol−1) | |
| Total | −4334.8 ± 106.7 |
| Van der Waals | −303.8 ± 23.9 |
| Distance restraints | 321.5 ± 17.3 |
| RMSD from average structurec | |
| All residues (236–298) | |
| Backbone (N, Cα, C) (Å) | 0.46 ± 0.07 |
| Heavy atoms (Å) | 1.29 ± 0.19 |
| Ramachandran statisticsd | |
| Most favored region (%) | 65.0 (82.1) |
| Additionally allowed (%) | 28.0 (17.2) |
| Generously allowed (%) | 5.5 (0.6) |
| Disallowed (%) | 1.5e (0.1) |
aHydrogen bond restraints were HN-O distance of 1.8–2.3 Å and an N–O distance of 2.8–3.3 Å.
bTotal number of violated constraints for the final ensemble of 15 water-refined structures including residues 1–163.
cRMSD (from the mean) for residues in regions of definite secondary structure 35–42, 48–55, 58–63, 76–84, 105–115, 119–125, 127–134, 138–144, 155–159.
dRamachandran statistics for residues 33–163 for the 15 structures in the final water refined ensemble. The numbers in the parenthesis indicate the statistics for residues in regions of definite secondary structure: 35–42, 48–55, 58–63, 76–84, 105–115, 119–125, 127–134, 138–144, 155–159.
eA majority of the residues found in the disallowed regions of the Ramachandran plot belong to Asp71 that flanks the cis Pro72. The remainder of the disallowed residues preceded or followed those for which no mainchain 1H,15N assignments were available.
Figure 1.
15N-{1H} Steady-state NOE for PaePE. Data was acquired at 600 MHz. NOE values for the β-strands and the α-helix are shown in red and blue respectively.
The core region (Gly33–Gly163) of PaePE forms a β-barrel consisting of eight antiparallel β-stands (Figure 2) as follows: Leu35–His42 (β1), His48–Leu55 (β2), Thr58–Ala63 (β3), Arg76–His84 (β4), Val105–Pro115 (β5), His127–Gly134 (β6), Ser138–Arg145 (β7) and Trp155–Lys159 (β8). A single α-helix (Pro119–Lys125) bridges the β5 and β6 strands. The locations of the secondary structural elements are consistent with those predicted from chemical shift data (19) and those seen in the crystal structure (in complex with Mn2+ and sulfate) (17). A ninth β-strand, which packs against the N-terminus of β5 and includes residues Gly93 and Ser94, is seen in the majority of the structures in the solution ensemble; this strand extends to residues Phe91 and Ile95 in several of the structures in the ensemble. The crystal structure also shows a strand between residues Gly93 and Ile95. However, the low 15N-{1H} NOE values for this region (0.38 ± 0.15 for residues 91–95; Figure 1) and the elevated local displacement in the NMR ensemble (Supplementary Figure S2) suggest its disorder in solution. This disordered strand (β9, Gly93–Ser94) is colored purple in Figure 2b. In addition, there are three instances of 310 helices, comprising residues Tyr88–Asp90 (in 40% of the structures of the final ensemble), Gln97–His99 (20% of structures) and Thr146–Leu148 (20% of the structures). Only the first of these 310 helices is seen in the crystal structure (17), albeit between residues Leu86–Asp90. There is no electron density in the crystal structure corresponding to the second 310 helix, indicating significant disorder of this segment, as confirmed by the elevated spin–spin relaxation rates (R2 at 900 MHz; data not shown) suggesting µs–ms timescale dynamics.
Figure 2.
(A) NMR ensemble of 15 water-refined structures of PaePE (shown in cross-eyed stereo view) showing only residues Ser31-Gly163. The β-strands and the α-helix are shown in red and blue, respectively. The disordered segment that lies between Phe91 and Asp104 is shown in yellow (this includes a disordered 9th β-strand between Gly93 and Ser94). The β3–β4 loop containing the two proline residues in cis conformation (Pro65 and Pro72) is shown in green. A 310 helix (indicated in cyan) forms in ~40% of structures in the final NMR ensemble. (B) Ribbon diagram for a representative structure (closest to the mean) of the NMR ensemble. A ninth β-strand that forms in majority of the structures between Gly93 and Ser94 is colored purple.
Analysis of the final NMR ensemble reveals high local displacement (Supplementary Figure S2) in the β-hairpin between β1 and β2 (Asp43–Tyr49, 0.69 ± 0.18 Å, including the N-terminus of β2) and in the loop between β4 and β5 (Phe91–Asp104, 0.40 ± 0.13 Å) that includes a dynamic 310 helix as discussed above. Some disorder is also seen in the loop between β7 and β8 (0.30 ± 0.09 Å). Regions of definite secondary structure are generally well defined (0.10 ± 0.08 Å) except the extreme N-terminus of β2. The regions of high disorder, i.e. elevated mean local displacement values in the loop regions, also display low 15N-{1H} NOE values (Figure 1) indicating significant subnanosecond timescale dynamics.
Differences between the NMR and crystal structures of PaePE
Whereas the overall fold in solution is very similar to the crystal structure (17), significant differences can be seen, with the largest deviations being near the active site. The crystal structure (colored red in Figure 3) adopts a more closed conformation with a concave active site, compared to a more open conformation seen in the NMR ensemble in solution (the member of the NMR ensemble closest to the mean is colored cyan in Figure 3). The open–closed conformational switch is indicated by the dashed line with arrowheads in Figure 3. The largest distortions occur at the face of the β-barrel comprising strands β1, β2 and β5. Significantly smaller deviations are seen on the opposite face comprising strands β6, β7 and β8. In the crystal structure, the catalytic Mn2+ ion is coordinated with octahedral geometry by the sidechains of His42 (β1), His48 (β2) and Asp50 (β2), the sulfate anion and two waters. This network might favor a relatively rigid closed structure, whereas a more open structure, on average, forms in the absence of the metal ion and crystal packing forces. The C-terminal tail (177–187) in the crystal structure, missing from our PaePE NMR construct, tucks in under the β1–β2–β5 sheet and thus might also stabilize the closed conformation seen in crystallo. Note that the NMR ensemble (Figure 2a) includes both open as well as closed structures, though the former constitute a larger fraction of the ensemble. The archaeal PE crystal structures (Supplementary Figure S3a) illustrate varying degrees of closure around the active site. In the present study, mainchain 1H,15N assignments could not be made for PaePE residues His42, Ala44, Ser45, Leu47, His48, Tyr49 and Tyr88 (though mainchain 13Cα, 1Hα assignments could be obtained for all of these residues except His48). 15N-{1H} NOE data for residues in this region where 1H, 15N resonance assignments are available, suggest overall rigidity on the ps–ns timescale. However, substantial line broadening (confirmed by elevated R2 values, data collected at 900 MHz; data not shown) associated with assigned resonances in this region indicate significant conformational dynamics on the µs–ms timescale. The conformational flexibility seen at the active site in the NMR ensemble is not evident in the crystal structure of the PaePE•Mn2+•sulfate complex. It therefore appears that the catalytic metal ion and the phosphomimetic sulfate are necessary to induce order at the active site and orient the important sidechains into their catalytically competent conformations. The extensive dynamics is also the likely reason that PaePE (or any other PE domain) could not be crystallized in the absence of metal ions.
Figure 3.
Differences between the NMR and X-Ray structures of PaePE. The X-Ray structure (red) was solved in the presence of a catalytic Mn2+ ion and a phosphomimetic sulfate anion. The key residues that lie in the co-ordination sphere of the cation and anion are shown in green and blue, respectively. The regions where the differences are the largest are indicated—the β1–β2–β5 face near the catalytic site (dashed arrow), the β3–β4 loop (blue dashed oval) that contains the two cis prolines (trans in the X-Ray structure) and the β7–β8 loop (red dashed oval). Only the core region is shown in both cases.
Notable differences between the crystal and NMR structures are also seen in the long loop between β3 and β4 (circled in blue in Figure 3, right panel). Two proline residues (Pro65 and Pro72) in this segment were predicted to be in the cis conformation (albeit in a trans configuration in the crystal structure), based on the differences between their 13Cβ and 13Cγ chemical shifts (19). Whereas the cis conformation for Pro65 could be confirmed by the presence of Hαi-1-Hαi) NOE cross-peaks (Supplementary Figure S4), extensive spectral overlap prevented the unambiguous assignment of either Hαi-1-Hαi) (characteristic of cis proline) or Hαi-1-Hδi (characteristic of trans proline) crosspeaks corresponding to Pro72. Nevertheless, the two prolines residues were locked in cis conformation for the structure calculations. The presence of cis prolines in the β3-β4 loop does not lead to significant deviations between the crystal and NMR structures for the strands β3 and β4 themselves, except for some minor differences at the extreme C- and N-terminal ends of β3 and β4, respectively. The biological role, if any, of the cis prolines is not immediately obvious though this loop contains two to four prolines in bacterial and archaeal PE domains (Supplementary Figure S5).
The loop between β7 and β8 also displays differences between the crystal structure and the solution structure (circled in red in Figure 3). In the NMR ensemble, this loop folds back at an angle of ~90° about residues Asn147 and Ser153. This region is also quite dynamic on the ps–ns timescale with moderately low 15N-{1H} NOE values (0.44 ± 0.09) compared with the flanking regions, a fact that is reflected by the relatively high mean local displacement values in the NMR ensemble (Supplementary Figure S2). Different degrees of closure for this loop are also seen in the case of MbaPE and CkoPE (Supplementary Figure S3a).
Comparison of the observed 13Cα chemical shifts and those calculated from the crystal structures of PaePE using the program SHIFTX2 (31) averaged over three residues (Supplementary Figure S3b) reveals that largest differences occur around the regions that also display the largest structural deviations between the crystal and NMR structures. Most notable among these include the β1–β2 hairpin and the β4–β5 loop. The largest residue-specific deviations occur for Leu86 (calculated to be shifted 3.4 ppm downfield from that observed in solution) and Asp87 (3.13 ppm downfield) suggesting a higher degree of helicity in crystallo compared to that in solution, in agreement with that seen comparing the NMR and crystal structures (Figure 3).
The sidechains of the catalytic residues in the NMR ensemble sample orientations similar to those seen in the crystal structure (Supplementary Figure S6). It is likely that the catalytically optimal orientations are locked in by the presence of the metal ion as discussed earlier. However, Glu82 (implicated in the monoesterase activity) samples sidechain orientations that are quite different from those seen in the crystal structure. The reasons for this are not immediately apparent. Orientations for His48, for which no resonance assignments are available, were not calculated.
In order to further probe the differences between the NMR and crystal structures of PaePE, we tested several alignment media (32) in an attempt to obtain a set of residual dipolar couplings. However, in spite of our best efforts, all these attempts failed because PaePE was unstable under all conditions tested.
NMR analysis of the interactions of PaePE with DNA
Positively charged residues distributed over the surface of PaePE (Supplementary Figure S7) could potentially act as loci for DNA binding. In order to determine the DNA-binding surface of PaePE, we analyzed spectral perturbations induced in 15N,1H TROSY spectra of uniformly 15N,2H-labeled protein by increasing concentrations of the 18-mer primer–template duplex, D18 (Figure 4a). Many spectral perturbations were seen in the presence of D18 (Figure 4b) reflecting the alteration of the chemical environment of the main chain nuclei due the combined effects of direct binding and induced conformational changes. Many residues were broadened beyond the threshold of detection early in the titration course (Supplementary Table S1). This extensive line broadening indicates exchange on the intermediate timescale (33). The broadened resonances correspond to residues situated near the active site, on the β3–β4 loop, on strands β4, β5 and β7, and on the β7–β8 loop. The positively charged residues His127, Lys136 and Arg140 were also completely broadened out early during the titration. From this group of perturbed residues, those on β5 (Asp109 and Gly111 flank the positively charged Arg110), and the set of positively charged residues that show localized perturbations (Lys136, Arg140) are quite distant from the catalytic site, all lying on the opposite face of the protein (Figure 5c). If one or more of these residues were to take part in a productive binding event, a substantial distortion would be required of the duplex bound to this patch to access the catalytic site. A conformational change in this region in response to DNA binding at the opposite face, while also possible, is difficult to rationalize without further evidence. It is therefore likely that the spectral perturbations seen in the presence of the D18 primer–template result from both productive and non-productive binding events. In addition to these extensive line-broadening effects, significant chemical shift changes (Supplementary Figure S8) were also seen for several residues in and around strands β4 and β8.
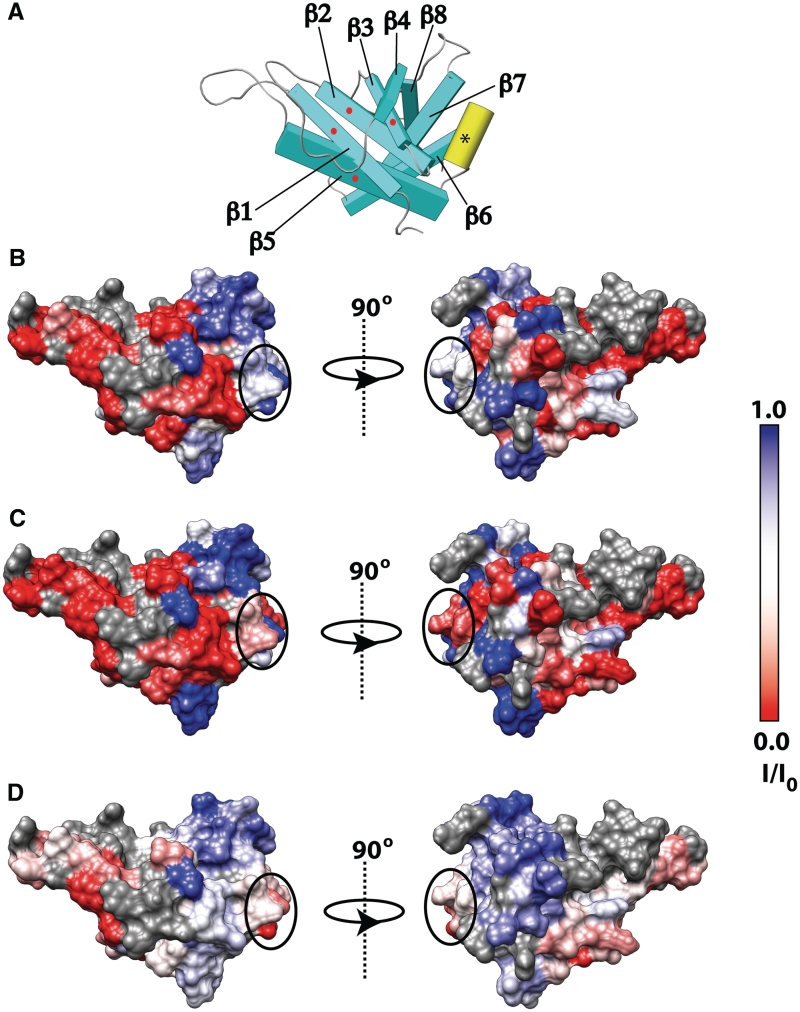
Figure 5.
(A) Residues important for the catalytic activity of PaePE. Gln40 and Glu82 (along with Arg14 and Glu15 on the disordered N-terminus, not shown) important for the 3′-phosphomonoesterase activity are colored magenta; Lys66 and Arg76 that influence the 3′-phosphodiesterase activity are colored blue; His42, His48, Asp50, His84 and Trp88 influence both activities, are colored cyan. (B) Schematic representation of PaePE showing the secondary structural elements in the same orientation (looking down into the catalytic cavity) as in (a) and the left panels of (C–E). The location of the catalytic residues as in (a) is indicated by the dotted oval. The strands comprising the β3–β4–β7–β8 face that shows the largest perturbations in the presence of oligonucleotide is indicated by the red dots. (c) Influence of D18 on PaePE. Residues that are broadened out beyond detection during the titration course are colored blue and residues that display chemical shift changes [Δδ, Equation (1)] >0.11 ppm at an equimolar protein:oligonucleotide ratio, are colored red. The circled patches represent perturbations involving residues near pockets of positive charges on the face opposite to that bearing the catalytic residues. These perturbations are likely due to non-productive binding events. Residues for which data were not analyzed due to missing assignments, spectral overlap or weak peaks in the reference state, are colored green. Residues that show no significant spectral perturbations in the presence of oligonucleotide are shown in gray. The effects of D18p are quite similar to those of D18. (d) While the overall spectral perturbations on the catalytic face, in the presence of D16F1R1, are similar as in D18 and D18p, the perturbations on the face opposite to that bearing the catalytic residues, are not seen. (e) Sequence conservation in bacterial and archaeal PE domains are depicted on the PaePE surface using a cyan (least conserved) to maroon (most conserved) gradient. The catalytic residues and a large part of the putative oligonucleotide binding surface are well conserved.
Given the ability of PaePE to catalyze the hydrolysis of the 3′-PO4 of an all-DNA primer–template, we tested whether the presence of a phosphate group at the 3′-end (primer–template D18p) resulted in any alteration in the spectral perturbations. The overall distribution of the perturbations (Supplementary Figure S8 and Supplementary Table S1) was quite similar to those seen for D18. Thus, the presence of a 3′-PO4 group did not seem to have a substantial effect on the nature of spectral perturbations, at least in the absence of the catalytic Mn2+ ions.
The phosphodiesterase activity of PaePE requires a 2′-OH in the penultimate nucleobase (12). Thus, successive base removal occurs until a single ribonucleotide remains at the 3′-end. Given this scenario, we tested the influence of a primer–template containing a 3′-terminal ribonucleotide on the spectral perturbations in PaePE. A primer–template construct (D16F1R1, Figure 4a) that was also 2′-fluoro modified at the penultimate base (to mimic closely an RNA base) was used in the NMR titrations. In the presence of D16F1R1, substantial changes in the spectral perturbations were seen compared to D18 and D18p. The perturbations for residues comprising β5 and the isolated perturbations (seen in the cases of the D18 and D18p constructs) near the positively charged residues His127, Lys136 and Arg140 were no longer seen (highlighted in Supplementary Table S1). While Lys151 on the catalytic face was no longer significantly perturbed, the perturbation in Gly150 persisted, albeit at a much higher concentration of DNA (Supplementary Table S1). Line broadening effects were limited to residues near the active site (Tyr37, Lys41 and Val105), the β3–β4 loop (Val64, Leu70), β4 (Arg76, Leu77, Ala78, Val79, Val81, Glu82 and His84) and on the β7–β8 loop (Thr146, Asn147 and Gly150). In addition, large chemical shift changes (>0.11 ppm at an equimolar ratio with D16F1R1, Figure 5d and Supplementary Figure S8) were seen near the catalytic site (Glu40: 0.12 ppm), the β3–β4 loop (Val74: 0.15 ppm; Lys75: 0.25 ppm), β4 (Gln80: 0.11 ppm), β7 (Ile144: 0.16 ppm), the β7–β8 loop (Gln152: 0.22 ppm; Ser153: 0.12 ppm and Gln154: 0.33 ppm) and β8 (Trp155: 0.37 ppm; Phe156: 0.13 ppm; Leu157: 0.2 ppm and Lys159: 0.15 ppm). Based on these observations, it appears that the binding site for DNA that is productive for catalysis involves a surface composed of portions of strands β3, β4, β7, β8 and intervening loops (Figure 5b and d). This region shows a high degree of conservation in PE domains found across the bacterial and archaeal taxa (Figure 5e). In the absence of a preferred ribose base at the 3′-end, detectable oligonucleotide binding seems to occur at additional putatively non-productive sites at the opposite face of the protein. The inability to fit the chemical shift or intensity changes with increasing oligonucleotide concentration to simple quadratic binding also suggests a more complex interaction dynamics than a single-site binding event (also suggested by the fluorescence-based assays described below). It is tempting to speculate that the extensive line broadening effects seen could in part be due to exchange between the productive and non-productive sites. The presence of the ribose enables a higher degree of selectivity in the occupation of the productive sites.
No significant changes in the chemical shifts were seen for the N-terminal segment with any of the primer-templates used in this study. No spectral perturbations were seen in the Gly98-Gly101 segment (missing from the crystal structure); it was conjectured that this segment might grip the primer-template and position it at the catalytic site (18).
No perturbations were detected for the N-terminal tail in a complex consisting of PaePE, D16F1R1 and Mn2+ in a 1:0.5:0.5 ratio in a buffer containing 2 mM phosphate (to mimic a state immediately following the 3′-monoesterase activity in the presence of the product inorganic phosphate). Sub-stoichiometric amounts of the paramagnetic Mn2+ and D16F1R1 were used to minimize the line-broadening effects due to the former and allow the visualization of a maximal number of resonances (Supplementary Table S1). Based on the lack of observed perturbations at the N-terminus, it is conceivable that an intact phosphorylated 3′-end is necessary to observe the conformational changes, if any, required for the monoesterase activity. It is also possible that the conformational changes are quite fast or transient on the chemical shift timescale and thus do not give rise to any observable chemical shift changes or line broadening effects.
Fluorescence studies of the interactions of PaePE with DNA
In addition to the NMR titration experiments discussed above, we measured native tryptophan fluorescence for PaePE to probe its interactions with D16F1R1. The protein contains five tryptophan residues (Trp62, Trp108, Trp113, Trp141 and Trp155), all of which are buried in the structure, as confirmed by a fluorescence emission maximum around 331 nm (34) following excitation at 280 nm. The change in fluorescence emission seen in the presence of D16F1R1 (Figure 6) was fit to a simple one-site binding model [Equation (2)] yielding an apparent Kd value of 5.6 ± 0.6 µM. However, the quality of the fit to the single-site binding model is quite poor (Figure 6; black symbols), as evidenced by the nature of the residuals, clearly suggesting that this simple model is not suitable for PaePE/DNA interactions. Note, it is unlikely that this change in fluorescence emission is due to direct quenching by DNA, but rather due to a conformational change in the protein. Surface exposure of the tryptophan residues to allow contact with DNA would result in a red shift of the fluorescence maximum to around 340 nm (34). Based on the NMR titrations, the most perturbed tryptophan residues are likely to be Trp62, Trp155 and Trp158.
Figure 6.
Change in PaePE tryptophan fluorescence (measured at 331 nm) in the presence of D16F1R1 with (red) and without (black) 100 µM Mn2+. Experimental data are shown as filled circles and fits to a quadratic isotherm [Equation (2)] by solid lines. Almost no change in tryptophan fluorescence is seen with an excess of Mn2+ ions alone (blue circles). The inset depicts the residuals for the fits in the absence (black dashed line) or the presence (red dashed line) of Mn2+. Note the reduced deviation from a simple quadratic binding isotherm and the improvement in the quality of the fits seen in the presence of Mn2+.
Better fits to a quadratic isotherm were obtained in the presence of an excess of Mn2+ (see Figure 6, red symbols) with greatly improved residuals and a concomitant decrease in the χ2 value per degree of freedom from 2509 in the absence of Mn2+ to 995 in its presence, albeit with a similar apparent Kd value of 3.0 ± 0.1 µM. This suggests that the presence of the catalytic Mn2+ ions in addition to a ribose base at the 3′-end further reduces binding at the non-productive site and helps localize the oligonucleotide substrate more efficiently near the active site at the productive binding surface. No significant change in fluorescence emission was seen with Mn2+ ions in the absence of DNA (see Figure 6; blue symbols). As stated earlier, PaePE requires Mn2+ for catalytic activity but our results indicate that Mn2+ alone does not induce conformational changes that are detectable by tryptophan fluorescence. A comparison of the NMR ensemble with the crystal structure of PaePE•Mn2+•sulfate complex indicates that the orientations of the tryptophan residues are quite similar. Therefore it is not surprising that the metal induced conformational changes are not reflected in our fluorescence data. However, as will be demonstrated in the following section, the overall conformational changes induced by the catalytic Mn2+ are far more modest compared to those induced by Cd2+ that partially supports catalysis and Zn2+ that is highly inhibitory (12).
Interactions with metal ions
As mentioned previously, PaePE depends on Mn2+ for its phosphomonoesterase and phosphodiesterase activities. Cd2+ weakly supports the phosphodiesterase activity but not the subsequent hydrolysis of the 3′-PO4 (12). In order to test whether Cd2+ also engaged the residues that constitute the Mn2+ binding site revealed by the crystal structure, we analyzed spectral perturbations in PaePE in the presence of increasing amounts of Cd2+ (Supplementary Figure S9). Resonances corresponding to almost all residues (for which data could be analyzed) that lie on sheets β1, β2, β3 and β5 together with the intervening loops (see Figure 7b and Supplementary Figure S10) were progressively attenuated during the course of the titration and finally broadened beyond the threshold of detection at an equimolar ratio of protein to Cd2+ ions. Minimal further changes were seen upon increasing the metal ion concentration, suggesting a single ion-binding site on PaePE. The Cd2+-induced spectral perturbations could be reversed by addition of EDTA. Based on the nature of the spectral perturbations seen (no significant chemical shift changes or the appearance of new resonances), it appears that the PaePE-Cd2+ interaction occurs on the intermediate exchange regime on the chemical shift timescale.
Figure 7.
(A) Schematic representation of PaePE showing the secondary structural elements in the same orientation as in the left panels of (B–D). The largest conformational changes in the presence of metal ions involve the face formed by the strands β1, β2, β3 and β5 (indicated by the red dots). Some weak binding is also seen in the α-helical segment [indicated by an asterisk, also circled in panels (b–d)]. Spectral perturbations in the presence of (b) Cd2+, (c) Zn2+ and (d) Mn2+ ions are mapped onto the PaePE surface. Reduction in peak intensity (I/I0) in 15N,1H HSQC spectra of PaePE in the presence of an equimolar ratio of metal ions is depicted using a red (maximum attenuation in the presence of metal with respect to the reference state) to blue (minimum attenuation) scale (also see Supplementary Figure S10). Residues for which perturbations could not be analyzed due to missing assignments, spectral overlap or weak peaks in the reference state are shown in gray. Note that the signal attenuation in the presence of Mn2+ is due to a combination of conformational changes and paramagnetic relaxation enhancement effects.
Next, we tested the ability of PaePE to bind Zn2+ that has been shown to be inhibitory in the presence of Mn2+ (12). As in the case of Cd2+, strong spectral perturbations (Supplementary Figure S9), that reached their maximal value around a 1:1 stoichiometry, reversible by EDTA, were noted on the β1–β2–β3–β5 face of the protein (Figure 7c). Additionally, some smaller but significant perturbations, not present in the case of Cd2+ ions (indicated by the black oval in Figure 7), were seen on the α-helix. More importantly, unlike in the case of Cd2+, a new set of resonances appeared during the course of the titration (reaching their maximum intensity around an equimolar protein-to-metal ratio) suggesting an interaction in the slow exchange regime on the chemical shift timescale (see Supplementary Figure S11 for a representative example). Given the extensive nature of the spectral perturbations seen in the case of both Cd2+ and Zn2+, it appears they are the likely result of a conformational change involving the β1–β2–β3–β5 face of the protein. This is also the region where the largest differences were seen between the NMR and crystal structures (Figure 3).
Finally, we analyzed the spectral changes for PaePE in the presence of Mn2+ (Supplementary Figure S9). Modest perturbations (Supplementary Figure S10), a combination of the effects of paramagnetic relaxation enhancement and conformational changes, were seen. The location of the line-broadening effects (see Figure 7d), though smaller in magnitude, were consistent with those seen in the cases of Cd2+ and Zn2+, suggesting, as expected, a common binding site and similar conformational changes in the presence of Mn2+. As in the case of Zn2+, additional line-broadening effects were also seen (Figure 7d, indicated by the black oval) localized on the α-helical segment. It is therefore likely that the presence of several charged residues on this helix facilitate a weaker, non-specific interaction with bivalent metal ions. This binding is observable with Zn2+ that appears to have the highest overall affinity toward PaePE and with Mn2+ where the spectral perturbations due to binding events, even weak or transient ones, are greatly exaggerated due to relaxation enhancement induced by the paramagnetic nature of Mn2+. No significant differences in spectral perturbations were seen at an equimolar protein-to-metal ratio in the presence or absence of a 10-fold excess of phosphate for any of the metal ions except perhaps in the case of Mn2+ where some additional attenuation involving Lys41 and Asp43 (that flank the catalytic His42) was noted at the active site (Supplementary Figure S10). It is plausible that a 3′-PO4 facilitates the proper orientation of the catalytic Mn2+ at the active site. Overall, our data suggest that whereas PaePE relies on Mn2+ for catalysis, Mn2+ induces only modest conformational changes compared to Cd2+ or Zn2+.
A crystal structure of CkoPE bound to Zn2+ and phosphate has recently been determined (35). The authors found that the structure of CkoPE•Zn2+•phosphate complex is essentially identical to that of the previously determined CkoPE•Mn2+•sulfate complex. However, the main difference lies in the coordination geometry of the metal ions: Zn2+ and Mn2+ were found to have tetrahedral and octahedral coordination geometries, respectively. This is in contrast to the large-scale spectral perturbations seen in our NMR data for PaePE in the presence of Zn2+ (and to a certain extent Cd2+) but not for Mn2+ when compared to the metal-free enzyme. These differences cannot be explained simply by an alteration of the co-ordination sphere of the metal, rather points to a significant conformational change involving a large part of the catalytic face. While it is possible that the conformational changes induced by Zn2+ are different for the PaePE and CkoPE domains [Zn2+ acts as inhibitor for both proteins (12,35)], it is more likely that the conformers that display these changes are unstable under crystal packing forces.
CONCLUSIONS
We have obtained the solution structure of the PE domain of Pseudomonas aeruginosa LigD using solution NMR techniques. PE in solution displays the 8-stranded β-barrel fold observed in the crystal structures of PaePE and two archaeal PEs. However significant differences between the solution and crystal structures are seen near the catalytic site. This involves strands β1, β2 and β5 that form a dynamic structure that samples multiple conformations on a µs–ms timescale leading to an open conformation, on average, compared to a more closed conformation seen in the crystal structure in the presence of Mn2+ and sulfate. Additional differences are seen in the proline-rich β3-β4 loop where the two flanking proline residues are found to be in the cis conformation while trans conformations are seen in the crystal structure.
Measurement of the spectral perturbations in the presence of three different primer–template duplexes reveal the location of the DNA binding surface of PaePE to be at a large positively charged patch on the face that hosts the catalytic residues. In addition what appears to be non-productive binding of DNA is also seen around positively charged residues in two of the all-DNA primer–template duplexes. However this mode of binding is greatly reduced in the presence of a terminal ribonucleotide base that is the preferred substrate for LigD, as evident from the NMR spectral perturbations, and further decreased in the presence of Mn2+ ions, as revealed by fluorescence measurements. It therefore appears that a ribose 3′-end and the catalytic metal ion help localize the oligonucleotide efficiently near the catalytic site. In contrast, no DNA-induced perturbations are seen for the disordered N-terminus that has been implicated in the mononoesterase activity of LigD, whether in the absence or the presence of substoichiometric amounts of Mn2+. Thus, the nature of the contribution of this segment to the 3′-monoesterase activity of LigD remains a mystery.
Whereas Mn2+ is essential for the catalytic activity of PaePE, we have found evidence for significant conformational changes in the presence of both Cd2+ and Zn2+ ions, with spectral features in the latter confirming the existence of an alternate conformation(s) in the presence of metal ions, this is in contrast to the minimal conformational changes seen by comparing the crystal structures of the CkoPE•Mn2+•sulfate and CkoPE•Zn2+•phosphate complexes, beyond the co-ordination sphere of the metal cation. Our spectral perturbation data opens up an intriguing possibility—it is plausible that within the conformational ensemble sampled by PaePE in solution, there exists a dynamic equilibrium between conformers that are catalytically competent and others that are not. Zn2+ ions help stabilize states compromised in their catalytic ability and greatly reduce the rate of exchange with the catalytically competent states (or equivalently, shifts the equilibrium toward the catalytically incompetent states) to an extent that this process is far slower than the catalytic timescale leading to inhibition even in the presence of Mn2+ ions. This would imply that the new resonances that appear in the presence of Zn2+ correspond to an ensemble that is dominated by conformers unsuitable to induce catalysis. This shift in equilibrium is not as drastic in the case of Cd2+ ions where the exchange between the conformers appears to be in the intermediate regime on the chemical shift timescale and is still fast enough to support some level of catalytic activity (12).
ACCESSION NUMBERS
The coordinates for the core residues for the 15 structures of the final ensemble have been deposited in the PDB with accession code 2LJ6.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–11 and Supplementary Table 1.
FUNDING
National Science Foundation (MCB 083141 to R.G.); National Institutes of Health (GM63611 to S.S. and 5G12 RR03060, partial support of the core facilities at CCNY). S.S. is an American Cancer Society Research Professor. R.G. is a member of the New York Structural Biology Center, a STAR center supported by the New York State Office for Science, Technology and Academic Research. Funding for open access charge: National Science Foundation (MCB 083141).
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Gupta R, Barkan D, Redelman-Sidi G, Shuman S, Glickman MS. Mycobacteria exploit three genetically distinct DNA double-strand break repair pathways. Mol. Microbiol. 2011;79:316–330. doi: 10.1111/j.1365-2958.2010.07463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shuman S, Glickman MS. Bacterial DNA repair by non-homologous end joining. Nat. Rev. Microbiol. 2007;5:852–861. doi: 10.1038/nrmicro1768. [DOI] [PubMed] [Google Scholar]
- 3.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–527. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akey D, Martins A, Aniukwu J, Glickman MS, Shuman S, Berger JM. Crystal structure and nonhomologous end-joining function of the ligase component of Mycobacterium DNA ligase D. J. Biol. Chem. 2006;281:13412–13423. doi: 10.1074/jbc.M513550200. [DOI] [PubMed] [Google Scholar]
- 5.Gong C, Martins A, Bongiorno P, Glickman M, Shuman S. Biochemical and genetic analysis of the four DNA ligases of mycobacteria. J. Biol. Chem. 2004;279:20594–20606. doi: 10.1074/jbc.M401841200. [DOI] [PubMed] [Google Scholar]
- 6.Zhu H, Nandakumar J, Aniukwu J, Wang LK, Glickman MS, Lima CD, Shuman S. Atomic structure and nonhomologous end-joining function of the polymerase component of bacterial DNA ligase D. Proc. Natl Acad. Sci. USA. 2006;103:1711–1716. doi: 10.1073/pnas.0509083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu H, Shuman S. A primer-dependent polymerase function of pseudomonas aeruginosa ATP-dependent DNA ligase (LigD) J. Biol. Chem. 2005;280:418–427. doi: 10.1074/jbc.M410110200. [DOI] [PubMed] [Google Scholar]
- 8.Zhu H, Shuman S. Gap filling activities of Pseudomonas DNA ligase D (LigD) polymerase and functional interactions of LigD with the DNA end-binding Ku protein. J. Biol. Chem. 2010;285:4815–4825. doi: 10.1074/jbc.M109.073874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brissett NC, Pitcher RS, Juarez R, Picher AJ, Green AJ, Dafforn TR, Fox GC, Blanco L, Doherty AJ. Structure of a NHEJ polymerase-mediated DNA synaptic complex. Science. 2007;318:456–459. doi: 10.1126/science.1145112. [DOI] [PubMed] [Google Scholar]
- 10.Pitcher RS, Brissett NC, Picher AJ, Andrade P, Juarez R, Thompson D, Fox GC, Blanco L, Doherty AJ. Structure and function of a mycobacterial NHEJ DNA repair polymerase. J. Mol. Biol. 2007;366:391–405. doi: 10.1016/j.jmb.2006.10.046. [DOI] [PubMed] [Google Scholar]
- 11.Brissett NC, Martin MJ, Pitcher RS, Bianchi J, Juarez R, Green AJ, Fox GC, Blanco L, Doherty AJ. Structure of a preternary complex involving a prokaryotic NHEJ DNA polymerase. Mol. Cell. 2011;41:221–231. doi: 10.1016/j.molcel.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 12.Zhu H, Shuman S. Novel 3′-ribonuclease and 3′-phosphatase activities of the bacterial non-homologous end-joining protein, DNA ligase D. J. Biol. Chem. 2005;280:25973–25981. doi: 10.1074/jbc.M504002200. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Shuman S. Substrate specificity and structure-function analysis of the 3′-phosphoesterase component of the bacterial NHEJ protein, DNA ligase D. J. Biol. Chem. 2006;281:13873–13881. doi: 10.1074/jbc.M600055200. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Shuman S. Characterization of Agrobacterium tumefaciens DNA ligases C and D. Nucleic Acids Res. 2007;35:3631–3645. doi: 10.1093/nar/gkm145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu H, Shuman S. Bacterial nonhomologous end joining ligases preferentially seal breaks with a 3′-OH monoribonucleotide. J. Biol. Chem. 2008;283:8331–8339. doi: 10.1074/jbc.M705476200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu H, Wang LK, Shuman S. Essential constituents of the 3′-phosphoesterase domain of bacterial DNA ligase D, a nonhomologous end-joining enzyme. J. Biol. Chem. 2005;280:33707–33715. doi: 10.1074/jbc.M506838200. [DOI] [PubMed] [Google Scholar]
- 17.Nair PA, Smith P, Shuman S. Structure of bacterial LigD 3′-phosphoesterase unveils a DNA repair superfamily. Proc. Natl Acad. Sci. USA. 2010;107:12822–12827. doi: 10.1073/pnas.1005830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith P, Nair PA, Das U, Zhu H, Shuman S. Structures and activities of archaeal members of the LigD 3′-phosphoesterase DNA repair enzyme superfamily. Nucleic Acids Res. 2011;39:3310–3320. doi: 10.1093/nar/gkq1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dutta K, Natarajan A, Nair PA, Shuman S, Ghose R. Sequence-specific 1H, 13C and 15N assignments of the phosphoesterase (PE) domain of Pseudomonas aeruginosa DNA ligase D (LigD) Biomol. NMR Assign. 2011;5:151–155. doi: 10.1007/s12104-010-9289-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc. Natl Acad. Sci. USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cavanagh J, Fairbrother WJ, Palmer AG, III, Rance M, Skelton NJ. Protein NMR Spectroscopy. 2nd edn. San Diego: Academic Press; 2007. [Google Scholar]
- 22.Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol. Biol. 2004;278:313–352. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 24.Habeck M, Rieping W, Linge JP, Nilges M. NOE assignment with ARIA 2.0: the nuts and bolts. Methods Mol. Biol. 2004;278:379–402. doi: 10.1385/1-59259-809-9:379. [DOI] [PubMed] [Google Scholar]
- 25.Ferrage F, Cowburn D, Ghose R. Accurate sampling of high-frequency motions in proteins by steady-state 15N-{1H} nuclear Overhauser effect measurements in the presence of cross-correlated relaxation. J. Am. Chem. Soc. 2009;131:6048–6049. doi: 10.1021/ja809526q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 28.Hutchinson EG, Thornton JM. PROMOTIF - a program to identify and analyze structural motifs in proteins. Protein Sci. 1996;5:212–220. doi: 10.1002/pro.5560050204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laskowski RA. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boggs PT, Donaldson JR, Byrd RH, Schnabel RB. ODRPACK software for weighted orthogonal distance regression. ACM Trans. Math. Software. 1989;15:348–364. [Google Scholar]
- 31.Han B, Liu Y, Ginzinger SW, Wishart DS. SHIFTX2: significantly improved protein chemical shift prediction. J. Biomol. NMR. 2011;50:43–57. doi: 10.1007/s10858-011-9478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prestegard JH, Bougault CM, Kishore AI. Residual dipolar couplings in structure determination of biomolecules. Chem. Rev. 2004;104:3519–3540. doi: 10.1021/cr030419i. [DOI] [PubMed] [Google Scholar]
- 33.Zuiderweg ERP. Mapping Protein-Protein Interactions in Solution by NMR Spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 34.Burstein EA, Vedenkina NS, Ivkova MN. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973;18:263–279. doi: 10.1111/j.1751-1097.1973.tb06422.x. [DOI] [PubMed] [Google Scholar]
- 35.Das U, Smith P, Shuman S. Structural insights to the metal specificity of an archaeal member of the LigD 3′-phosphoesterase DNA repair enzyme family. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr767. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.