Abstract
Pyrrole–imidazole (Py–Im) hairpin polyamides are a class of programmable, sequence-specific DNA binding oligomers capable of disrupting protein–DNA interactions and modulating gene expression in living cells. Methods to control the cellular uptake and nuclear localization of these compounds are essential to their application as molecular probes or therapeutic agents. Here, we explore modifications of the hairpin γ-aminobutyric acid turn unit as a means to enhance cellular uptake and biological activity. Remarkably, introduction of a simple aryl group at the turn potentiates the biological effects of a polyamide targeting the sequence 5′-WGWWCW-3′ (W = A/T) by up to two orders of magnitude. Confocal microscopy and quantitative flow cytometry analysis suggest this enhanced potency is due to increased nuclear uptake. Finally, we explore the generality of this approach and find that aryl-turn modifications enhance the uptake of all polyamides tested, while having a variable effect on the upper limit of polyamide nuclear accumulation. Overall this provides a step forward for controlling the intracellular concentration of Py–Im polyamides that will prove valuable for future applications in which biological potency is essential.
INTRODUCTION
Hairpin pyrrole–imidazole (Py–Im) polyamides are a class of heterocyclic amino acid oligomers that can be programmed to bind a wide repertoire of DNA sequences with high affinity and specificity (1,2). Sequence-selective recognition of the minor groove of DNA is achieved through side-by-side stacked ring pairings: Im/Py distinguishes G·C from C·G, while Py/Py is degenerate for T·A and A·T. In recent years, our group has focused on the biological evaluation of eight-ring polyamides arranged in a hairpin configuration through a γ-aminobutyric acid linker (Figure 1) (3,4). These compounds are of modest (~1300 Da) molecular weight and recognize 6 bp of DNA, similar to the size of many eukaryotic transcription factor binding motifs (5). When applied to living cells, hairpin polyamides can disrupt protein–DNA interactions and modulate the expression of genes induced by many transcription factors, including the ligand-activated nuclear receptors glucocorticoid receptor (GR) and androgen receptor (AR) (6–10). However, one persistent challenge encountered when applying Py–Im polyamides to new biological systems is cellular uptake. Previous studies have shown that the nuclear localization of fluorescently labeled polyamides can be influenced by several variables including molecular weight, modifications to the C-terminal moiety, and composition of Py/Im content (11–13). In particular, Py–Im polyamides incorporating multiple (>2–3) N-methylimidazole subunits show reduced nuclear localization, limiting the ability to target GC rich sequences in vivo (14). Therefore, new solutions for enhanced uptake are important for advancing Py–Im polyamides as probes of transcription factor binding and, potentially, as therapeutic agents.
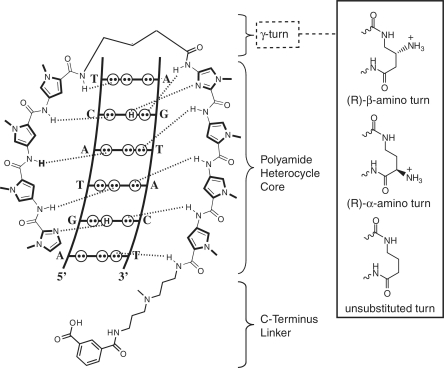
Figure 1.
Schematic diagram of eight-ring Py–Im polyamides targeting the sequence 5′-WGWWCW-3′ (W = A/T). Dashed lines indicate hydrogen bonds between the polyamide and DNA base pairs. The γ-aminobutyric acid turn unit enforces an antiparallel hairpin configuration, and codes for W (16). Substitution of the chiral turn functionality can have substantial effects on DNA-binding and biological activity.
While a large amount of work has studied the effect of C-terminal modifications on polyamide nuclear localization (13,15), relatively few studies have explored the impact of modifications to the γ-aminobutyric acid (GABA) turn moiety on biological activity. In addition to imposing the pairing of Py/Im heterocycles and increasing DNA-binding affinity, the GABA turn demonstrates sequence selectivity for A,T over G,C base pairs, presumably due to a steric clash between the aliphatic turn and the exocyclic amine of guanine (16). The incorporation of a chiral (R)-α-amino substituent on the GABA turn enforces polyamide binding in a N-terminus → C-terminus orientation with respect to the 5′ → 3′ direction of the adjacent DNA strand (17). More recently, we introduced hairpin polyamides bearing (R)-β-amino GABA turns which show increased DNA-binding affinity and, importantly for the purposes of this study, a negligible decrease in DNA-binding affinity upon acetylation (Figure 1) (17). Structural studies suggest this is due to the unique stereochemical presentation of the chiral β-amino moiety, which is directed up and out of the minor groove floor, thereby providing a chemical handle for introduction of functionality at the turn position (18). Furthermore, biological evaluation of a β-acetylated polyamide targeted to the consensus androgen response element (ARE) half site 5′-WGWWCW-3′ (W = A/T) showed inhibition of prostate specific antigen (PSA) gene expression at 10-fold lower concentrations relative to polyamides bearing unsubstituted β-amino turns, illustrating the ability of turn substitution to potentiate gene regulatory effects (19).
Guided by these results, we explored modifications of the Py–Im polyamide hairpin γ-aminobutyric acid turn unit as a means to enhance uptake and biological activity. Our strategy applied cytotoxicity analysis at an extended time point to identify biologically active polyamides from small focused libraries. This led to the discovery that for a hairpin polyamide targeting the DNA sequence 5′-WGWWCW-3′ (W = A/T), simple conversion of the β-amino turn to a β-benzamide turn lead to a 100-fold increase in gene regulatory activity. To better understand the mechanism of this phenomenon, fluorescent analogues of β-amino and β-aryl polyamides were synthesized and analyzed by confocal microscopy and quantitative flow cytometry. Our results suggest the enhanced potency of β-aryl polyamides is due to increased nuclear uptake of these compounds. Finally, we explore the generality of this modification and find that aryl-turn substitution capably enhances the uptake of other polyamides cores, but has a variable effect on the upper limits of polyamide nuclear accumulation. Overall, this provides a new direction for controlling the intracellular concentration of Py–Im polyamides that will prove essential for future applications in which biological potency is required.
MATERIALS AND METHODS
Synthesis of polyamides
All polyamide cores were synthesized by manual solid-phase synthesis on Kaiser oxime resin (Nova Biochem) according to the previously published protocol (20). Polyamides were cleaved from resin by aminolysis with 3,3′-diamino-N-methyldipropylamine for 3 h at 55°C. Repeated cycles of precipitation and washing with diethyl ether were used to remove excess 3,3′-diamino-N-methyldipropylamine from polyamides prior to purification by reverse phase HPLC (21). The purified polyamide cores were modified at the C-terminal tail position by isophthalic acid (IPA) or fluorescein-5-isothiocyanate (FITC isomer I; Invitrogen) as previously described (15). Polyamides incorporating the β-Cbz-γ-aminobutyric acid turns were subjected to repeated cycles of precipitation and washing with diethyl ether to remove excess reagents, resuspended in 9:1 CF3COOH/TFMSA (0.9 ml, 5 min) to remove the benzyl carbamate group (22), and purified by HPLC to afford polyamides bearing a β-amino group at the turn. These compounds were coupled to the designated acids by PyBOP and subjected to a final step of HPLC purification to yield β-aryl turn conjugates. Purity of all compounds was verified by analytical HPLC and matrix-assisted, LASER desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry. Detailed synthetic schemes, yields, procedures can be found in the Supplementary Data.
Cell culture
All cell lines were purchased from ATCC (Manassas, VA, USA) and maintained in the following media: A549 cells (F-12K); LNCaP (RPMI 1640); HCT-116 (McCoy's 5a Medium Modified); MCF-7 (Eagle's Minimum Essential Medium). All media were supplemented with 10% FBS and cultured at 37°C under 5% CO2.
Sulforhodamine B assay of polyamide cytotoxicity
IC50 values for cytotoxicity were determined in 96-well microplates using the sulforhodamine B colorimetric assay for cellular protein content as previously described (23). All polyamide stock solutions were prepared in neat DMSO and dosed to give a final concentration of ≤0.3% DMSO. Briefly, cell lines were plated in 100 µl of the defined media at the following densities: A549 (1000 cells/well); LNCaP (5000 cells/well), HCT-116 (750 cells/well), MCF-7 (3000 cells/well). After 24 h, polyamides were added to adhered cells in 100 µl of media by serial dilution. Quadruplicate wells were used for each polyamide concentration. After 72 h, the medium was replaced with 100 µl fresh medium, and cells were allowed to recover for 24 h. Following recovery, cells were fixed with 100 µl 10% trichloroacetic acid solution, washed, stained and dried as described. For 24 and 48 polyamide treatments, the procedure was followed as above with A549 cells plated at 3000 and 2000 cells per well, respectively. After solubilization of the bound dye in 10 mM Tris (pH 10.5), the absorbance was measured at 490 nm on a Victor microplate reader (PerkinElmer). The data are charted as a percentage of untreated controls, corrected for background absorbance. IC50 is defined as the concentration that inhibits 50% of control cell growth. These values were determined by non-linear least-squares regression fit to Y = A + (B − A)/(1+10⁁((Log EC50 − X) × H, where A = max., B = min. and H = Hill Slope. All calculations were performed using Prism 4 (GraphPad) software. Three independent biological replicates were averaged; stated IC50 values represent the mean and standard deviation.
Thermal melting temperature analysis
Melting temperature analysis was performed on a Varian Cary 100 spectrophotometer equipped with a thermo-controlled cell holder possessing a cell path length of 1 cm. An aqueous solution of 10 mM sodium cacodylate, 10 mM KCl, 10 mM MgCl2 and 5 mM CaCl2 at pH 7.0 was used as analysis buffer. Oligonucleotides (0.1 mM stock solutions dissolved in 10 mM Tris–Cl, 0.1 mM EDTA, pH 8.0) were purchased from Integrated DNA Technologies. DNA duplexes and hairpin polyamides were mixed to a final concentration of 2 and 2.4 µM, respectively, for each experiment. Prior to analysis, samples were heated to 90°C and cooled to a starting temperature of 25°C with a heating rate of 5°C/min for each ramp. Denaturation profiles were recorded at λ = 260 nm from 25°C to 90°C with a heating rate of 0.5°C/min. The reported melting temperatures were defined as the maximum of the first derivative of the denaturation profile, and represent the average of four measurements.
Quantitative real-time polymerase chain reaction analysis of nuclear receptor-mediated gene expression
To analyze the effects of polyamide-treatment on dexamethasone (DEX)-induced gene expression, A549 cells were plated in 24-well plates at a density of 15–25 × 103 cells per well (30–50 × 103 cells/ml) (8). After 24 h, the medium was replaced by F-12K containing 10% FBS, and polyamides were added to the specified concentrations (1–10 000 nM). Cells were incubated with polyamides for 12, 24 or 48 h, followed by induction with DEX (100 nM). After 6 h, cells were harvested and mRNA was isolated (RNEasy 96 kit—Qiagen) and reverse transcribed (Transcriptor First Strand cDNA Synthesis Kit—Roche). Quantitative real-time PCR (qRT-PCR) was performed with FastStart Universal SYBR Green Master Mix (Roche) on an ABI 7300 qPCR instrument (Applied Biosystems) following the manufacturer's protocol. A similar protocol was utilized to measure 4,5α-dihydrotestosterone (DHT)-induced gene expression in LNCaP cells, with the following modifications: (i) the initial plating density was 20–30 × 103 cells per well (40–60 × 103 cells/ml), (ii) cells were incubated with polyamides for 48 h and (iii) cells were induced with DHT (1 nM) for 16 h (7). In both cases, cDNA corresponding to the genes of interest was measured relative to β-glucuronidase as an endogenous control. Primer sequences are provided in the Supplementary Data.
Confocal microscopy analysis
For microscopy experiments, A549 cells in F-12K medium were plated into culture dishes equipped with glass bottoms for direct imaging (MatTek product no. P35G-0-14-C) at a density of 20 × 103 cells per dish (100 × 103 cells/ml). Cells were grown in the glass-bottom dishes for 24 h. Medium was then removed and replaced with 200 µl of fresh medium supplemented with FITC-labeled polyamides (1 µM) in DMSO (final concentration 0.1%). Cells were incubated at 37°C for 16 h, followed by removal of media, gentle washing with 100 µl PBS and addition of fresh medium immediately prior to imaging. For colocalization experiments, 15 µM Hoechst 33342 (0.5 mM stock in ddH2O) and 1 µM Mitotracker Red CM-H2XRos (1 mM stock in DMSO) were added 2 h prior to imaging. For time-course experiments, cells were imaged directly after polyamide addition using an environmentally controlled microscopy chamber (37°C, 5% CO2). Imaging was performed at the Caltech Beckman Imaging Center using a Zeiss LSM 510 Meta NLO 2-photon inverted laser scanning microscope equipped with a 40× oil-immersion objective lens. Polyamide–fluorescein conjugates and Mitotracker were imaged in multi-track mode using 488 and 543 nm laser excitation with a pinhole of 375 µm and standard filter sets for fluorescein and rhodamine, respectively. Hoechst was imaged using 800 nm two-photon excitation with an HFT KP680 dichroic and a 390- to 465-nm bandpass filter with a fully open pinhole. All images were analyzed using Zeiss LSM software.
Flow cytometry analysis
For flow cytometry experiments A549 cells were plated in 6-well plates at a density of 500 × 103 cells per well (133 × 103 cells/ml), and allowed to adhere for 16–24 h before treatment with polyamide-FITC conjugates (100–10 000 nM). Cells were grown for 6, 12, 24 or 48 h, the medium removed, washed with cold PBS and trypsinized for 5 min at 37°C. The trypsinized cells were combined with the cell culture supernatant and wash solution, and centrifuged for 5 min at 300g. This pellet was resuspended, washed with cold PBS, pelleted for 5 min at 300g and resuspended in 800 µl Hank's Balanced Salt Solution (2.5 mg/ml BSA, 10 mM HEPES, pH 7.0, no Mg2+, no Ca2+, no phenol red). Cell viability was checked with trypan blue stain and found to be ≥90–95% in all cases. Live cells were then diluted to a concentration of 5 × 105 cells/ml and pipetted through a 40 -µm cell strainer (BD Falcon) into 5 ml polystyrene round-bottom tubes (BD Falcon). Just prior to analysis, cells were stained for viability using 7-amino-actinomycin D (7-AAD—eBioscience). Analyses were performed on a BD Bioscience FACSCalibur instrument at the Caltech Flow Cytometry Cell Sorting Facility using standard filter sets for fluorescein and 7-AAD. SPHERO Rainbow Calibration Particles (6 peaks, 3.0–3.4 µm—Spherotech) were used as calibration standards. For each condition 10 000 cells were analyzed. Fluorescence values are representative of the relative median fluorescence (RMF) intensity of the main population, gated for viability based on 7-AAD dye exclusion. Comparison of RMFs of polyamide-labeled cells with SPHERO Rainbow Calibration Particles was used to calculate molecules of equivalent fluorescein per cell, which was converted to a nuclear concentration based on modeling the A549 nucleus as a cylinder with radius 10 µm and height 5 µm to give a calculated nuclear volume of 1 × 10−12 l (estimates based on confocal microscopy). All data were analyzed using FlowJo v8.8.2 (TreeStar) and indicate the average and standard deviation of two trials.
RESULTS
Synthesis, DNA-binding and cytotoxicity of β-aryl polyamides
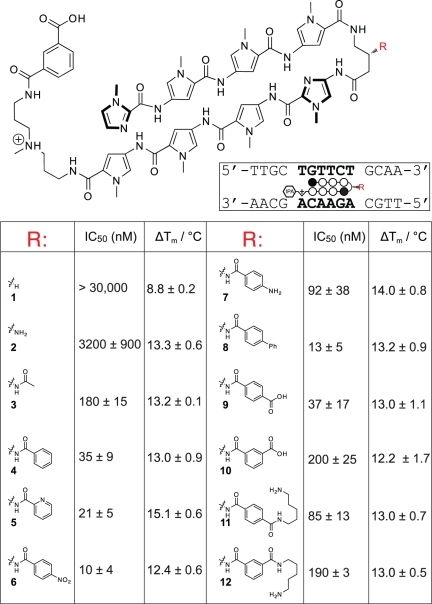
Following an unanticipated observation that a hairpin polyamide conjugated to an aryl group at the β-amino position showed greater activity in cell culture, we synthesized a small panel of β-aryl substituted polyamides (4–12) targeting the sequence 5′-WGWWCW-3′ and benchmarked their DNA-binding affinities and biological activity against unsubstituted parent (1), β-amino (2) and β-acetylated (3) GABA turns in the A549 human lung carcinoma cell line (Figure 2). We used cytotoxicity at 96 h as a proxy for uptake following the observation that polyamide uptake, gene regulatory activity and cytotoxicity are often highly correlated (13). Thermal denaturation analysis of a DNA duplex of the sequence 5′-TTGCTGTTCTGCAA-3′ (polyamide match site in bold) shows all polyamides containing a β-amino GABA group (2–14) similarly increase the melting temperature by ~13–15°C, suggesting no substantial energetic penalty for appendage of the bulky β-aryl groups (Figure 2). Cytotoxicity analyses demonstrate that β-amino GABA incorporating polyamide 2 (IC50 = 3.2 µM) is considerably more cytotoxic than its unsubstituted counterpart 1 (IC50 > 30 µM). While this trend mirrors the relative duplex stabilization of these molecules (ΔTm 1 = 8.8°C; ΔTm 2 = 13.3°C), simple N-acetylation of the β-amino turn (3) results in another order of magnitude increase in growth inhibition while not greatly affecting binding affinity (ΔTm 3 = 13.2°C). Replacement of the acetyl unit of 3 with a benzoyl functionality (4) results in approximately another order of magnitude increase in cytotoxicity (IC50 = 35 nM), again without concomitant change in the duplex stabilizing ability of this minor groove binder. Within the aryl series (4–12) several trends are seen, including increased cytotoxicity of p-substituted benzoic acids (compare 9 and 10) and a preference for electron-withdrawing groups at the p-position (compare 6 and 7). Remarkably, significantly increasing the steric bulk of the β-aryl turn, as in polyamides 8 and 11, does not greatly affect either DNA-binding or cytotoxicity, arguing against interaction of the β-aryl turn with a small pocket of a specific protein partner as a mechanism of cytotoxicity. Since previous studies have noted that polyamide activity can be strongly influenced by cell type (12), we tested the generality of the increased cytotoxicity of 4 and 9 in LNCaP prostate cancer, MCF-7 breast cancer and HCT-116 colon cancer cell lines. All three cell lines showed a similar increase in potency for β-aryl compared to β-amino polyamides as was observed in A549 cells (Table 1).
Figure 2.
Biological activity and DNA-binding of β-substituted hairpin polyamides. Cytotoxicity analyses were conducted 96 h following polyamide treatment in the A549 lung carcinoma cell line. IC50 values representing the mean of three biological replicates. ΔTm denotes the shift in melting temperature following polyamide treatment for the 5′-WGWWCW-3′ duplex DNA sequence shown.
Table 1.
Cytotoxicity of β-amino (2) and β-aryl (4, 9) polyamides targeting the sequence 5′-WGWWCW-3′ toward alternative cancer cell lines
| Polyamide | Cell line |
|||
|---|---|---|---|---|
| A549 | LNCaP | HCT-116 | MCF-7 | |
| 2 | 3155 ± 1895 | 2550 ± 91 | 4680 ± 954 | 24400 ± 9530 |
| 4 | 35 ± 9 | 28 ± 12 | 106 ± 18 | 666 ± 168 |
| 9 | 37 ± 17 | 109 ± 19 | 136 ± 13 | 470 ± 77 |
Growth inhibition values represent the mean and standard deviation of three biological replicates.
Suppression of nuclear receptor-mediated gene expression by β-aryl polyamides
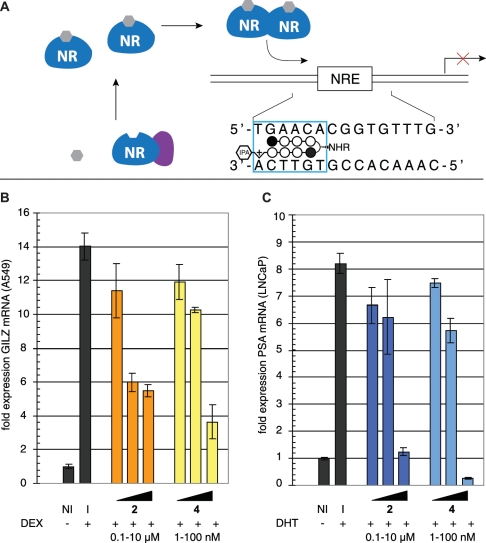
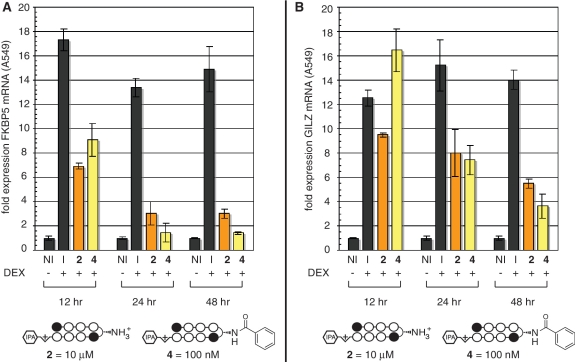
A549 lung epithelial cells have been widely applied as a model for inflammatory gene expression mediated by the transcription factor glucocorticoid receptor (GR) (24–26). GR is a member of larger family of nuclear receptors that utilize activation by small molecule ligands in order to effect release from cytoplasmic inhibitory complexes, after which they traffic to the nucleus, multerimize with their cognate protein partners, and activate (or repress) transcription (Figure 3A). Hairpin polyamides have been previously shown to inhibit nuclear receptor–DNA interactions in cell culture, making them promising agents for mechanistic studies of nuclear receptor–DNA binding and therapeutic modulation of nuclear receptor activity in diseases such as prostate cancer. Polyamides 1–12 target the 5′-WGWWCW-3′ sequence found in the consensus glucoroticoid response element (GRE). Therefore, as an initial test of whether the increased activity of β-aryl polyamides might be extended to gene regulatory studies, we analyzed the effects of β-aryl polyamide 4 and parent polyamide 2 on endogenous GR-mediated transcription in A549 cells. Following 48-h treatment with 2, 4 or vehicle DMSO, A549 cells were induced with DEX before isolation of total RNA and analysis by qRT-PCR. As seen in Figure 3B, β-aryl polyamide 4 blunts GR-driven transcription of the canonical GR-regulated gene GILZ (24,27) in a dose-dependent fashion. Remarkably, inhibition of gene expression by 4 occurs at polyamide concentrations 100× lower than parent compound 2. Similar results are seen for expression of FKBP5, another prototypical GR target (Supplementary Figure S1) (28). Notably, these effects are not expected to be due to cytotoxicity, as these experiments utilize a fivefold higher cell plating density than cytotoxicity analyses and are normalized to a housekeeping gene (GUSB) that remains stable to polyamide treatment (Supplementary Figure S2). Time course experiments show substantial inhibition of DEX-induced transcription as early as 12 h after polyamide treatment, far prior to the onset of cytotoxicity (Figure 4). Since the sequence targeted by 4, 5′-WGWWCW-3′, is also found in the androgen response element (ARE), we next tested whether 4 showed similarly enhanced inhibition of androgen receptor (AR) regulated gene expression (7). LNCaP prostate cancer cells were exposed to polyamides 2 or 4 for 48 h prior to induction with the AR-activating ligand 4,5α-dihydrotestosterone (DHT). Quantitative PCR analysis of reverse-transcribed mRNA shows a drastic decrease in transcription of the known AR target gene prostate specific antigen (PSA, also known as KLK3) following exposure to 4 (Figure 3C) (29,30). Once again, this inhibition is greater than that observed when parent compound 2 is applied at 100× greater concentrations, reducing PSA mRNA to below basal (non-induced) levels. These results highlight the activity of β-aryl polyamides as potent antagonists of nuclear receptor-mediated gene expression in living cells.
Figure 3.
Inhibition of nuclear receptor-mediated gene expression by β-aryl polyamides. (A) General schematic of nuclear receptor-mediated gene expression and polyamide inhibition of the protein–DNA interface. (B) Effect of β-turn polyamides 2 and 4 on DEX-induced GILZ gene expression as measured by qRT-PCR analysis. I = DEX-induced. NI = non-induced. Polyamide 2 concentrations: 100, 1000, 10 000 nM, polyamide 4 concentrations: 1, 10, 100 nM. (C) Effect of β-substituted polyamides 2 and 4 on dihydrotestosterone (DHT)-induced PSA gene expression as measured by qRT-PCR analysis. Polyamide 2 concentrations: 100, 1000, 10 000 nM, polyamide 4 concentrations: 1, 10, 100 nM. I = induced. NI = not induced.
Figure 4.
Time course analysis of polyamide-mediated inhibition of DEX-induced gene expression. (A) Effect of β-turn polyamides 2 and 4 on DEX-induced GILZ gene expression as measured by qRT-PCR analysis. I = DEX induced. NI = non-induced. Polyamide 2 concentration: 10 000 nM, polyamide 4 concentration: 100 nM. These concentrations of 2 and 4 were chosen because they show approximately equivalent inhibition of GR-mediated gene expression at 48 h. (B) Identical time course analysis of polyamide inhibition of FKBP5 gene expression over time.
Analyzing the effect of β-aryl substitution on polyamide uptake by quantitative fluorescence analysis
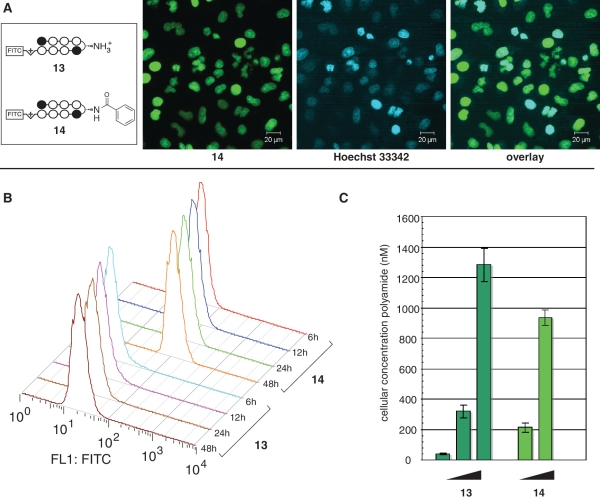
Hypothetically, the increased biological activity of β-aryl turn polyamides could be attributed to either (i) an improved ability to impede protein–DNA interactions, (ii) increased uptake or (iiii) reduced efflux. The former seemed unlikely given that our initial structure–activity analysis showed no correlation between steric bulk of the β-aryl turn, which would be expected to affect the interaction of groove-binding proteins, and cytotoxicity (Figure 2). Therefore, to examine cellular uptake in a systematic fashion we synthesized fluorescent analogues of polyamides 2 and 4 and analyzed their accumulation by confocal microscopy and flow cytometry (11,31). Polyamide-FITC conjugates 13 and 14 show similar trends in terms of biological activity compared to parent compounds, although the observed gap in cytotoxicities is decreased from ~100× to ~10× (Supplementary Figure S3). Following addition to growth media, polyamides 13 and 14 penetrate the membrane and localize to the nucleus of A549 cells, as verified by colocalization with the well-known DNA stain Hoechst (Figure 5A). However, flow cytometry analysis reveals quantitative differences in the kinetics and degree of uptake. Cells treated with 100 nM β-aryl polyamide 14 demonstrate a rapid increase in fluorescence intensity between 0–12 h, compared to much slower accumulation of β-amino polyamide 13 dosed under identical conditions (Figure 5B). Analyzing the overall percentage of fluorescently labeled cells as compared to a DMSO-treated control shows that treatment with 100 nM 14 results in fluorescent labeling of ~88% of A549 cells after 6 h, while cells treated with 100 nM 13 show labeling of only ~5% of cells over the same time period (Supplementary Figure S4). In order to gain a more quantitative view of the fluorescence increase, we calculated the nuclear concentrations of fluorescent polyamides 13 and 14 through comparison to fluorescent beads functionalized with known amounts of the FITC fluorophore (14,31). Using this methodology, at 48 h we observe a >4× greater accumulation of 14 than 13 in A549 nuclei when dosed at identical concentrations (100 nM). However, these concentrations can be shifted by increasing polyamide concentration, as a 10× greater dosage concentrations results in a ~3× increase in polyamide concentration values over 48 h (Figure 5C). Notably, this is not due merely to decreased cell growth, as analysis of cell count and viability prior to flow cytometry revealed no differences between treated samples. Finally, to differentiate uptake and efflux, we compared the effect of verapamil on uptake of β-aryl and β-amino polyamides. Verapamil is an inhibitor of the p-glycoprotein transporter that has previously been implicated in cellular efflux of polyamides (32). If β-aryl polyamide 4 is attaining higher concentrations through reduced efflux, verapamil treatment should have little or no effect on intracellular polyamide concentration, whereas if β-aryl polyamide 4 is attaining higher concentrations through enhanced uptake, verapamil will have an additive effect on nuclear accumulation. Our results are consistent with the latter mechanism, as we observed similarly higher fluorescent labeling by both 2 and 4 in A549 cells co-treated with a non-toxic (10 µM) dose of verapamil (Supplementary Figure S5). Overall these findings have two implications: (i) β-aryl turns can significantly increase the rate of polyamide uptake at submicromolar concentrations and (ii) polyamide cytotoxicity and cellular uptake are well correlated, as the 10× increase in cytotoxicity of 14 relative to 13 is mirrored by its accumulation in cells at 10× lower concentrations.
Figure 5.
Quantitative fluorescence analysis of β-turn substitution on Py–Im polyamide nuclear uptake. (A) Nuclear localization of β-aryl polyamide 13, as verified by colocalization with Hoechst 33342. (B) Influence of incubation time on fluorescence for A549 cells treated with 100 nM polyamide 13 or 14. X-axis: relative median fluorescence (FL1: FITC channel); Y-axis: hours of polyamide treatment. (C) Influence of dosage concentration on nuclear accumulation of polyamides. Polyamide 13 concentration: 100, 1000, 10 000 nM. Polyamide 14 concentration: 100, 1000 nM.
Exploring the utility of β-aryl substitution in promoting uptake of alternative polyamide cores
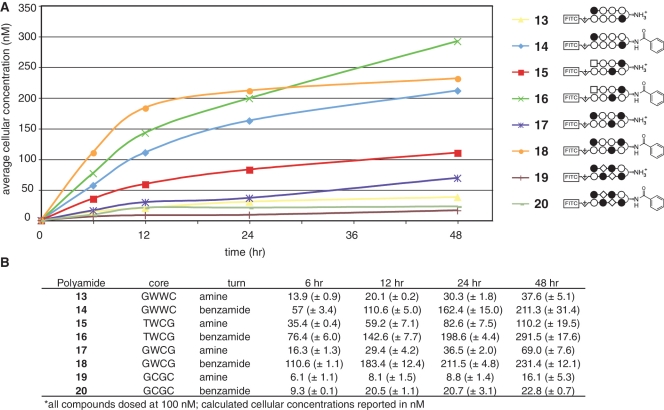
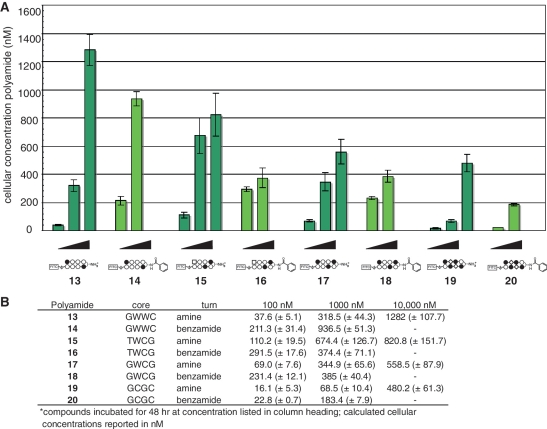
Finally, we examined the ability of β-aryl turns to influence the uptake of polyamide cores targeting alternative sequence motifs. We synthesized fluorescent β-amino and β-benzamide polyamides targeting the sequences 5′-WTWCGW-3′ (15–16), 5′-WGWCGW-3′ (17–18) and 5′-WGCGCW-3′ (19–20). Compound 15 is a high-affinity binder of the 5′-ATACGT-3′ sequence found within the hypoxia response element (HRE) of the VEGF enhancer (6), while compounds 17–20 probe the ability of β-aryl turns to facilitate the uptake of polyamides with increased N-methylimidazole content, a known negative determinant of polyamide nuclear localization (12,14). After verifying their nuclear localization by confocal microscopy (Supplementary Figure S6), each compound was analyzed for time and concentration-dependent uptake by quantitative flow cytometry (Figures 6 and 7). When added to media at 100 nM, time-course experiments demonstrate that polyamide-FITC conjugates 15–18 rapidly accumulate in A549 cells (Figure 6). Analysis of Im-rich polyamides 19–20 at 100 nM was less informative, as these compounds required dosing at 1000 nM to label a significant percentage of treated cells (Supplementary Figure S4). Under these treatment conditions (100 nM 15–18; 1000 nM 19–20) each β-aryl polyamide shows increased cellular uptake relative to its β-amino counterpart, demonstrating the general utility of this modification in promoting increased uptake (Figure 7). This enhanced uptake is accompanied by a large increase in the cytotoxicity of non-fluorescent analogues of Im-rich polyamides 18 and 20, but surprisingly not 5′-WTWCGW-3′ targeting polyamide 16 (Supplementary Figure S7). Once again flow cytometric analysis proved informative in explaining this unexpected observation, as A549 cells dosed at increasing concentrations (0.1–10 µM) of 15–16 show concentration-dependent accumulation of parent polyamide 15, while β-aryl polyamide 16 shows relatively equivalent uptake at both 100 and 1000 nM (Figure 7). This is consistent with the small, but constant, inhibitory effect polyamide 15 has on HIF1-α-mediated transcription, as judged by qRT-PCR analysis of VEGF gene expression (Supplementary Figure S8). Therefore, while our studies show β-aryl turns provide a generally applicable approach to increase the uptake of polyamides at reduced concentrations, this modification can have a variable consequence on the upper limits of polyamide nuclear accumulation which may be required for biological effects.
Figure 6.
Time course analysis of uptake of polyamide-FITC conjugates incorporating diverse DNA sequence-recognition elements. (A) Cellular concentration of polyamide-FITC conjugates 13–20 as a function of time incubated with A549 cells. Polyamide structures are represented as ball and stick models according the shorthand code: closed circle, Im monomer; open circle, Py monomer; diamond, β-alanine; square, 3-chlorothiophene 2-carboxylic acid. Complete structures can be found in Supplementary Data. Cellular concentration calculated from flow cytometry data as described in materials and methods. (B) Data displayed in tabular form with standard deviations. Core = DNA sequence targeted by the hairpin polyamide core heterocylic ring pairs. Turn = identity of β-turn modification.
Figure 7.
Influence of polyamide dosage on cellular concentration at 48 h. (A) Graphical depiction of relative cellular concentrations of polyamides analyzed in this study. β-Amino polyamides 13, 15, 17 and 19 were dosed at 100, 1000 and 10 000 nM, respectively (left to right). β-Aryl polyamides 14, 16, 18 and 20 were dosed at 100 and 1000 nM, respectively (left to right). Attempts to dose β-aryl compounds at higher concentrations were hindered by insolubility. Cellular concentration calculated from flow cytometry data as described in materials and methods. (B) Data displayed in tabular form with standard deviations. Core = DNA sequence targeted by the hairpin polyamide core heterocylic ring pairs. Turn = identity of β-turn modification.
DISCUSSION
Chemical approaches for controlling gene expression at the protein–DNA interface require efficient nuclear delivery of gene regulatory agents. Thus far, comprehensive efforts to define structure-uptake relationships for hairpin polyamides in cell culture have largely focused on the optimization of the C-terminus. Insights from these studies are reflected in our use of polyamides modified with a C-terminal isophthalic acid modification for gene regulation and cytotoxicity studies (1–12, 21–26), with the higher molecular weight FITC reserved for direct analysis of polyamide uptake (14–20) (12,15). Here, we explore integration of an additional uptake determinant at the hairpin polyamide β-aminobutyric acid turn. Introduction of functionality at the polyamide turn position has previously been most well-explored in the design of covalent sequence-selective DNA alkylating agents (33,34). Our findings here represent an initial inquiry into the effect of turn modification on non-covalent sequence-selective DNA-binding agents, and have lead to the discovery of a polyamide (4) that exhibits excellent affinity for DNA and nanomolar inhibition of DEX and DHT-induced gene expression in human cancer cells. This molecule represents one of the most biologically potent members of this compound-class (eight-ring hairpin polyamides) identified to date.
The second phase of this study examined the mechanism and generality of β-aryl turn modification as a vehicle for increasing polyamide potency using fluorescent polyamide conjugates and quantitative flow cytometry analysis. The power of this approach lies in its relatively simple calibration and ability to sample a large number of cells for any given condition. Our findings indicate the β-aryl turn of 14 aids polyamide uptake, allowing rapid permeation and nuclear accumulation as compared to β-amino-modified 13. When extended to hairpin polyamides with alternative heterocycle composition and DNA-binding preferences (15–20), uptake of polyamide-FITC conjugates was well-correlated with cytotoxicity and gene regulatory effects, implicating membrane permeability as a primary determinant of the biological activity for this class of molecules. However, while informative, these methods are not without caveats. First, the calculated nuclear concentrations cannot be taken as absolute values, as they do not take into account the known fluorescence enhancements exhibited by polyamide-FITC conjugates upon binding to DNA (35), subcellular localization, fluorescein photostabilty, or differences in the optical properties of cells as compared to fluorochrome-coated beads. Second, fluorescein modification can significantly alter the biological properties of hairpin polyamides (compare the IC50 of 4 with 14; Supplementary Figure S3). Integration of a low molecular weight reporter into the hairpin polyamide scaffold is therefore attractive from the standpoint of streamlining activity and uptake assays. This approach may benefit from recent incorporation of click chemistry methods into many flow cytometry workflows (36,37).
It is interesting to speculate as to the mechanism by which β-aryl turns expedite polyamide uptake. Studies of oligonucleotide-based therapeutics have shown that modification of these agents with highly lipophilic moieties, such as cholesterol, can facilitate association with the cell membrane and endocytosis (38,39). It is possible a similar effect mediates the delivery of β-aryl polyamides. This is consistent with the increased cytotoxicity observed as we progress from β-amine 2, to β-acetamide 3, to β-aryl benzamide 4 (Figure 2). Also interesting is the lack of nuclear accumulation of β-aryl polyamide 16 at higher concentrations. The finding that β-aryl modification promotes increased uptake at 100 nM but not 1000 nM suggests this may result from a physical phenomenon such as insolubility (due to aggregation) at the higher concentration. In general, β-aryl polyamides show decreased solubility relative to β-amino compounds owing to their reduced charge at physiological pH. Combined with a better understanding of such physical properties, this approach may be useful in promoting the uptake of polyamides targeting extended (>8 bp) sequence motifs (10,40,41). Separate from delivery, perhaps the most important future challenge lies in developing new methods to define the concentration-dependent effects of Py–Im polyamides on gene expression in living cells. Analytical techniques such as MPE footprinting and affinity cleavage have proven essential to the design of sequence-selective DNA-binding agents (42,43), and reveal dose-dependent binding patterns that can be used to directly guide applications in vitro (44,45). Quantitative fluorescence analysis of polyamide uptake, as performed here, combined with recently developed high-throughput sequencing strategies for analysis of protein–DNA binding (46,47) and gene expression (46) represent promising approaches to similarly footprint polyamide-induced perturbations and binding events in vivo, and thereby define the relationship between nuclear concentration and gene regulatory effects.
Overall, these findings highlight hairpin turn modification as a promising new strategy for intracellular delivery of Py–Im polyamides. In terms of applications, the increased potency of these analogues should prove immediately useful for testing in animal models, where the ability to work at lower concentrations will help overcome technical challenges of polyamide solubility and formulation. The amenability of the β-aryl turn to substitution also raises the possibility of using it as a selective handle to optimize polyamide pharmacokinetic properties, such as plasma protein binding, through attachment of pendant chemical functionalities. Finally, β-aryl turns may prove useful for the delivery of molecular probes using polyamides as tethered DNA-binding domains into living cells. The relevance of such strategies is highlighted by the numerous studies which have used polyamides to target the activity of alkylating agents (33), chromatin remodeling enzymes (48) and transcriptional activation domains (49,50) to subsets of genomic loci. Future work will focus on characterizing the mechanism of β-aryl polyamide uptake and applying this technology for the manipulation of protein–DNA interactions in living systems.
SUPPLEMENTARY DATA
Supplementary Data is available at NAR Online: Supplementary Figures S1–S8, Supplementary Methods, Supplementary Schemes S1–S4 and Supplementary References (51,52).
FUNDING
National Institutes of Health (grant numbers GM51747 and GM27681); American Cancer Society (grant number PF-10-015-01-CDD to J.L.M, postdoctoral fellowship) and National Institutes of Health Cellular, Biochemical, and Molecular Sciences Predoctoral Research training grant (grant number 5T32GM007616 to D.C.M.). Funding for open access charge: National Institutes of Health (grant numbers GM51747).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
Mass spectrometry analyses were performed in the Mass Spectrometry Laboratory of the Division of Chemistry and Chemical Engineering at the California Institute of Technology. Flow cytometry analyses were performed at the Caltech Flow Cytometry Cell Sorting Facility. We thank Rochelle Diamond for support and technical expertise in performing flow cytometry experiments, and William Dempsey for helpful discussions regarding confocal microscopy.
REFERENCES
- 1.Dervan PB. Molecular recognition of DNA by small molecules. Bioorg. Med. Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 2.Dervan PB, Edelson BS. Recognition of the DNA minor groove by pyrrole-imidazole polyamides. Curr. Opin. Struct. Biol. 2003;13:284–299. doi: 10.1016/s0959-440x(03)00081-2. [DOI] [PubMed] [Google Scholar]
- 3.Mrksich M, Parks ME, Dervan PB. Hairpin peptide Motf. A new class of oligopeptides for sequence-specific recognition in the minor groove of double-helical DNA. J. Am. Chem. Soc. 1994;116:7983–7988. [Google Scholar]
- 4.Hsu CF, Phillips JW, Trauger JW, Farkas ME, Belitsky JM, Heckel A, Olenyuk BZ, Puckett JW, Wang CC, Dervan PB. Completion of a programmable DNA-binding small molecule library. Tetrahedron. 2007;63:6146–6151. doi: 10.1016/j.tet.2007.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 6.Olenyuk BZ, Zhang GJ, Klco JM, Nickols NG, Kaelin WG, Jr, Dervan PB. Inhibition of vascular endothelial growth factor with a sequence-specific hypoxia response element antagonist. Proc. Natl Acad. Sci. USA. 2004;101:16768–16773. doi: 10.1073/pnas.0407617101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nickols NG, Dervan PB. Suppression of androgen receptor-mediated gene expression by a sequence-specific DNA-binding polyamide. Proc. Natl Acad. Sci. USA. 2007;104:10418–10423. doi: 10.1073/pnas.0704217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muzikar KA, Nickols NG, Dervan PB. Repression of DNA-binding dependent glucocorticoid receptor-mediated gene expression. Proc. Natl Acad. Sci. USA. 2009;106:16598–16603. doi: 10.1073/pnas.0909192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Modulating hypoxia-inducible transcription by disrupting the HIF-1-DNA interface. ACS Chem. Biol. 2007;2:561–571. doi: 10.1021/cb700110z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matsuda H, Fukuda N, Ueno T, Tahira Y, Ayame H, Zhang W, Bando T, Sugiyama H, Saito S, Matsumoto K, et al. Development of gene silencing pyrrole-imidazole polyamide targeting the TGF-beta1 promoter for treatment of progressive renal diseases. J. Am. Soc. Nephrol. 2006;17:422–432. doi: 10.1681/ASN.2005060650. [DOI] [PubMed] [Google Scholar]
- 11.Best TP, Edelson BS, Nickols NG, Dervan PB. Nuclear localization of pyrrole-imidazole polyamide-fluorescein conjugates in cell culture. Proc. Natl Acad. Sci. USA. 2003;100:12063–12068. doi: 10.1073/pnas.2035074100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelson BS, Best TP, Olenyuk B, Nickols NG, Doss RM, Foister S, Heckel A, Dervan PB. Influence of structural variation on nuclear localization of DNA-binding polyamide-fluorophore conjugates. Nucleic Acids Res. 2004;32:2802–2818. doi: 10.1093/nar/gkh609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacobs CS, Dervan PB. Modifications at the C-terminus to improve pyrrole-imidazole polyamide activity in cell culture. J. Med. Chem. 2009;52:7380–7388. doi: 10.1021/jm900256f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nishijima S, Shinohara K, Bando T, Minoshima M, Kashiwazaki G, Sugiyama H. Cell permeability of Py-Im-polyamide-fluorescein conjugates: Influence of molecular size and Py/Im content. Bioorg. Med. Chem. 18:978–983. doi: 10.1016/j.bmc.2009.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Nickols NG, Jacobs CS, Farkas ME, Dervan PB. Improved nuclear localization of DNA-binding polyamides. Nucleic Acids Res. 2007;35:363–370. doi: 10.1093/nar/gkl1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swalley SE, Baird EE, Dervan PB. Effects of γ-turn and β-tail amino acids on sequence-specific recognition of DNA by hairpin polyamides. J. Am. Chem. Soc. 1999;121:1113–11120. [Google Scholar]
- 17.Herman DM, Baird EE, Dervan PB. Stereochemical control of the dna binding affinity, sequence specificity, and orientation preference of chiral hairpin polyamides in the minor groove. J. Am. Chem. Soc. 1998;120:1382–1391. [Google Scholar]
- 18.Chenoweth DM, Dervan PB. Allosteric modulation of DNA by small molecules. Proc. Natl Acad. Sci. USA. 2009;106:13175–13179. doi: 10.1073/pnas.0906532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chenoweth DM, Harki DA, Phillips JW, Dose C, Dervan PB. Cyclic pyrrole-imidazole polyamides targeted to the androgen response element. J. Am. Chem. Soc. 2009;131:7182–7188. doi: 10.1021/ja901309z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Belitsky JM, Nguyen DH, Wurtz NR, Dervan PB. Solid-phase synthesis of DNA binding polyamides on oxime resin. Bioorg. Med. Chem. 2002;10:2767–2774. doi: 10.1016/s0968-0896(02)00133-5. [DOI] [PubMed] [Google Scholar]
- 21.Krutzik PO, Chamberlin AR. Synthesis of DNA-binding polyamides. Robust solid-phase methods for coupling heterocyclic aromatic amino acids. Methods Mol. Biol. 2002;201:77–92. doi: 10.1385/1-59259-285-6:77. [DOI] [PubMed] [Google Scholar]
- 22.Dose C, Farkas ME, Chenoweth DM, Dervan PB. Next generation hairpin polyamides with (R)-3,4-diaminobutyric acid turn unit. J. Am. Chem. Soc. 2008;130:6859–6866. doi: 10.1021/ja800888d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat. Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 24.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Adamio F, Zollo O, Moraca R, Ayroldi E, Bruscoli S, Bartoli A, Cannarile L, Migliorati G, Riccardi C. A new dexamethasone-induced gene of the leucine zipper family protects T lymphocytes from TCR/CD3-activated cell death. Immunity. 1997;7:803–812. doi: 10.1016/s1074-7613(00)80398-2. [DOI] [PubMed] [Google Scholar]
- 28.Magee JA, Chang LW, Stormo GD, Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- 29.Cleutjens KB, van der Korput HA, van Eekelen CC, van Rooij HC, Faber PW, Trapman J. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol. Endocrinol. 1997;11:148–161. doi: 10.1210/mend.11.2.9883. [DOI] [PubMed] [Google Scholar]
- 30.Riegman PH, Vlietstra RJ, van der Korput JA, Brinkmann AO, Trapman J. The promoter of the prostate-specific antigen gene contains a functional androgen responsive element. Mol. Endocrinol. 1991;5:1921–1930. doi: 10.1210/mend-5-12-1921. [DOI] [PubMed] [Google Scholar]
- 31.Hsu CF, Dervan PB. Quantitating the concentration of Py-Im polyamide-fluorescein conjugates in live cells. Bioorg. Med. Chem. Lett. 2008;18:5851–5855. doi: 10.1016/j.bmcl.2008.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crowley KS, Phillion DP, Woodard SS, Schweitzer BA, Singh M, Shabany H, Burnette B, Hippenmeyer P, Heitmeier M, Bashkin JK. Controlling the intracellular localization of fluorescent polyamide analogues in cultured cells. Bioorg. Med. Chem. Lett. 2003;13:1565–1570. doi: 10.1016/s0960-894x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- 33.Dickinson LA, Burnett R, Melander C, Edelson BS, Arora PS, Dervan PB, Gottesfeld JM. Arresting cancer proliferation by small-molecule gene regulation. Chem. Biol. 2004;11:1583–1594. doi: 10.1016/j.chembiol.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 34.Tsai SM, Farkas ME, Chou CJ, Gottesfeld JM, Dervan PB. Unanticipated differences between alpha- and gamma-diaminobutyric acid-linked hairpin polyamide-alkylator conjugates. Nucleic Acids Res. 2007;35:307–316. doi: 10.1093/nar/gkl1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rucker VC, Foister S, Melander C, Dervan PB. Sequence specific fluorescence detection of double strand DNA. J. Am. Chem. Soc. 2003;125:1195–1202. doi: 10.1021/ja021011q. [DOI] [PubMed] [Google Scholar]
- 36.Bradford JA, Clarke ST. Dual-pulse labeling using 5-ethynyl-2’-deoxyuridine (EdU) and 5-bromo-2’-deoxyuridine (BrdU) in flow cytometry. Curr. Protoc. Cytom. Chapter 7 doi: 10.1002/0471142956.cy0738s55. Unit 7 38. [DOI] [PubMed] [Google Scholar]
- 37.Poulin-Kerstien AT, Dervan PB. DNA-templated dimerization of hairpin polyamides. J. Am. Chem. Soc. 2003;125:15811–15821. doi: 10.1021/ja030494a. [DOI] [PubMed] [Google Scholar]
- 38.Gissot A, Camplo M, Grinstaff MW, Barthelemy P. Nucleoside, nucleotide and oligonucleotide based amphiphiles: a successful marriage of nucleic acids with lipids. Org. Biomol. Chem. 2008;6:1324–1333. doi: 10.1039/b719280k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bijsterbosch MK, Rump ET, De Vrueh RL, Dorland R, van Veghel R, Tivel KL, Biessen EA, van Berkel TJ, Manoharan M. Modulation of plasma protein binding and in vivo liver cell uptake of phosphorothioate oligodeoxynucleotides by cholesterol conjugation. Nucleic Acids Res. 2000;28:2717–2725. doi: 10.1093/nar/28.14.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Edwards TG, Koeller KJ, Slomczynska U, Fok K, Helmus M, Bashkin JK, Fisher C. HPV episome levels are potently decreased by pyrrole-imidazole polyamides. Antiviral Res. 91:177–186. doi: 10.1016/j.antiviral.2011.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yao EH, Fukuda N, Ueno T, Matsuda H, Matsumoto K, Nagase H, Matsumoto Y, Takasaka A, Serie K, Sugiyama H, et al. Novel gene silencer pyrrole-imidazole polyamide targeting lectin-like oxidized low-density lipoprotein receptor-1 attenuates restenosis of the artery after injury. Hypertension. 2008;52:86–92. doi: 10.1161/HYPERTENSIONAHA.108.112797. [DOI] [PubMed] [Google Scholar]
- 42.Van Dyke MW, Hertzberg RP, Dervan PB. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II) Proc. Natl Acad. Sci. USA. 1982;79:5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultz PG, Taylor JS, Dervan PB. Design and synthesis of a sequence-specific DNA cleaving molecule. (Distamycin-EDTA)iron(II) J. Am. Chem. Soc. 1982;104:6861–6863. [Google Scholar]
- 44.Schmidt TL, Heckel A. Pyrrole/imidazole-polyamide anchors for DNA tertiary interactions. Small. 2009;5:1517–1520. doi: 10.1002/smll.200900147. [DOI] [PubMed] [Google Scholar]
- 45.Cohen JD, Sadowski JP, Dervan PB. Programming multiple protein patterns on a single DNA nanostructure. J. Am. Chem. Soc. 2008;130:402–403. doi: 10.1021/ja0772400. [DOI] [PubMed] [Google Scholar]
- 46.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 47.Hesselberth JR, Chen X, Zhang Z, Sabo PJ, Sandstrom R, Reynolds AP, Thurman RE, Neph S, Kuehn MS, Noble WS, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods. 2009;6:283–289. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohtsukia A, Kimurab MT, Minoshimaa M, Suzukib T, Ikedab M, Bandoa T, Nagase H, Shinohara K, Sugiyama H. Synthesis and properties of PI polyamide–SAHA conjugate. Tetrahedron Lett. 2009;50:7288–7292. [Google Scholar]
- 49.Kwon Y, Arndt HD, Mao Q, Choi Y, Kawazoe Y, Dervan PB, Uesugi M. Small molecule transcription factor mimic. J. Am. Chem. Soc. 2004;126:15940–15941. doi: 10.1021/ja0445140. [DOI] [PubMed] [Google Scholar]
- 50.Mapp AK, Ansari AZ, Ptashne M, Dervan PB. Activation of gene expression by small molecule transcription factors. Proc. Natl Acad. Sci. USA. 2000;97:3930–3935. doi: 10.1073/pnas.97.8.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baird EE, Dervan PB. Solid phase synthesis of polyamides containing imidazole and pyrrole amino acids. J. Am. Chem. Soc. 1996;118:6141–6146. [Google Scholar]
- 52.Chenoweth DM, Harki DA, Dervan PB. Solution-phase synthesis of pyrrole–imidazole polyamides. J. Am. Chem. Soc. 2009;131:7175–7181. doi: 10.1021/ja901307m. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.