Abstract
Previous studies have implicated SAGA (Spt-Ada-Gcn5-acetyltransferase) and TFIID (Transcription factor-IID)-dependent mechanisms of transcriptional activation in yeast. SAGA-dependent transcriptional activation is further regulated by the 19S proteasome subcomplex. However, the role of the 19S proteasome subcomplex in transcriptional activation of the TFIID-dependent genes has not been elucidated. Therefore, we have performed a series of chromatin immunoprecipitation, mutational and transcriptional analyses at the TFIID-dependent ribosomal protein genes such as RPS5, RPL2B and RPS11B. We find that the 19S proteasome subcomplex is recruited to the promoters of these ribosomal protein genes, and promotes the association of NuA4 (Nucleosome acetyltransferase of histone H4) co-activator, but not activator Rap1p (repressor-activator protein 1). These observations support that the 19S proteasome subcomplex enhances the targeting of co-activator at the TFIID-dependent promoter. Such an enhanced targeting of NuA4 HAT (histone acetyltransferase) promotes the recruitment of the TFIID complex for transcriptional initiation. Collectively, our data demonstrate that the 19S proteasome subcomplex enhances the targeting of NuA4 HAT to activator Rap1p at the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional stimulation, hence providing a new role of the 19S proteasome subcomplex in establishing a specific regulatory network at the TFIID-dependent promoter for productive transcriptional initiation in vivo.
INTRODUCTION
Transcriptional initiation is an important step of gene expression, and is promoted by gene-specific activators that bind to the specific DNA sequences upstream of the core promoter element (known as upstream activating sequence or UAS). Activators function by enhancing the assembly of the general transcription factors (GTFs) such as TFIIA, TFIIB, TFIID, TFIIE, TFIIF and TFIIH, as well as RNA polymerase II at the core promoter to form a pre-initiation complex (PIC) for transcriptional initiation. Such an enhanced PIC formation is mediated by the interaction of the activator with one or more transcription factors, termed as ‘target’ (1). Based on the target specificities of the activators, previous studies have revealed two distinct mechanisms of transcriptional activation that are mediated by the SAGA (Spt3-Ada-Gcn5-acetyltransferase) and TFIID complexes in Saccharomyces cerevisae (1). SAGA is a large multi-protein complex with two different enzymatic activities such as histone acetyltransferase (HAT) and histone deubiquitinase, while TFIID is composed of TBP (TATA-box binding protein) and 14 different TBP-associated factors (TAFs) (1). For SAGA-dependent transcriptional activation, the activator targets SAGA that subsequently promotes the PIC formation at the core promoter for transcriptional initiation (1–8). There are about 10% RNA polymerase II genes whose expression is regulated by SAGA (1,9–12). However, the expression of a vast majority of genes is regulated by the TFIID complex (1,9–12). At the TFIID-regulated genes, activator targets TFIID for transcriptional initiation (1,13–16). Importantly, TFIID has been implicated in regulating the transcription of ribosomal protein genes (1,13,15). Expression of ribosomal protein genes is crucial for ribosomal biogenesis and the subsequent translation of mRNA into proteins for normal cellular growth and functions (17). Thus, TFIID plays an important role in ribosome biogenesis, and hence cellular growth. Further, transcription of ribosomal protein genes is controlled by TOR (target of Rapamycin) signaling pathway that is highly conserved from yeast to humans (17,18). TOR inactivation by rapamycin (a macrocyclic lactone) through inhibition of a TOR-kinase containing protein complex impairs various anabolic as well as catabolic processes including ribosomal protein gene expression, thus regulating the growth and fate of eukaryotic cell.
In yeast, there are 137 ribosomal protein genes (~2% of the total genes), and ~50% of RNA polymerase II transcription is devoted to these genes in the TFIID and TOR-dependent fashions (17,19). Two TOR-dependent factors have been implicated to regulate the transcription of ribosomal protein genes in yeast in response to nutrient cues (17). These are Sfp1p and forkhead transcription factor Fhl1p. The co-activator and co-repressor of Fhl1p are Ifh1p and Crf1p, respectively (17,20–23). Sfp1p binds to the promoters of ribosomal protein genes to enhance transcription in a TOR-dependent manner. In the presence of rapamycin or nutrient starvation, Sfp1p is inactivated, leading to transcriptional downregulation of ribosomal protein genes. Likewise, Fhl1p binds to the promoters of ribosomal protein genes and activate them under nutrient-rich growth conditions in a TOR-dependent manner. Under such growth conditions, the co-repressor Crf1p stays in the cytoplasm via the action of TOR-dependent protein kinase A. Upon nutrient starvation, Crf1p moves into the nucleus and binds to Fhl1p, leading to the transcriptional repression of ribosomal protein genes. In addition to these regulations, TOR also regulates ribosomal protein gene expression by enhancing association of NuA4 (Nucleosome acetyltransferase of histone H4) HAT complex with the promoter and dissociation of Rpd3p histone deacetylase from the promoter, hence stimulating the transcription of the ribosomal protein genes (17,24). Following inhibition of the TOR signaling pathway, NuA4 HAT dissociates from the ribosomal protein genes and Rpd3p binds to the promoter, leading to the transcriptional repression of ribosomal protein genes (17,24,25). In addition to nutrient or TOR-dependent regulation, transcription of ribosomal protein genes is also controlled by other environmental insults such as heat and osmotic shocks. Thus, transcription of ribosomal protein genes is co-ordinately regulated in a complex manner, which has a major impact on overall capacity of protein synthesis and cellular growth.
Recently, DNA microarray analysis has implicated the proteasome complex in transcriptional regulation of ribosomal protein genes (1,26–29); further complicating ribosomal-protein gene expression. The 26S proteasome is a highly versatile protein degradation machine with a molecular chaperonin activity. It consists of 20S proteolytic core and 19S regulatory particles (CP and RP, respectively). The 19S RP is further composed of a ‘lid’ of eight non-ATPases, and a ‘base’ of six ATPases (Rpt1–Rpt6) and three non-ATPases. The 19S RP has the molecular chaperonin activity (30), and its ATPase activity is required for its association with 20S CP to form the 26S proteasome complex (31). The 19S ATPase activity is also crucial for the degradation of proteins marked by a chain of more than four lysine-48-linked ubiquitin molecules (32–34). The lid of the 19S RP binds to the polyubiquitin chain of the substrate protein, and the 19S ATPase activity subsequently unfolds the substrate protein and translocates it into the catalytic site of the 20S CP for proteolysis (34). Via such a degradation mechanism, the 26S proteasome complex regulates the functions and fates of many transcription factors, and hence transcription (34). In fact, ~70% of the genomic transcripts in yeast is altered in the temperature-sensitive (ts) inactivation of either the 19S RP or 20S CP (28).
The 19S RP has been previously shown to increase the interaction between activator Gal4p and co-activator SAGA at the SAGA-dependent GAL1 gene for stimulation of transcriptional initiation in a proteolysis-independent manner (35,36). However, it is not yet known whether transcriptional initiation of the TFIID-dependent ribosomal protein genes is also similarly regulated by the 19S RP in a proteolysis-independent manner. With this view, we performed a series of experiments at several ribosomal protein genes such as RPS5, RPL2B and RPS11B in S. cerevisae. We find that the 19S base is recruited to the promoters of the ribosomal protein genes, and enhances the recruitment of NuA4 HAT, but not activator Rap1p (repressor–activator protein1). Further, we show that NuA4 HAT promotes the recruitment of the TFIID complex, and hence transcription of ribosomal protein genes. Thus, the 19S base promotes the targeting of NuA4 HAT to enhance the recruitment of TFIID for stimulation of transcriptional initiation. These results provide a new regulatory mechanism of transcriptional activation of the TFIID-dependent ribosomal protein genes by the 19S proteasome subcomplex.
MATERIALS AND METHODS
Plasmids
The plasmid pFA6a-13Myc-KanMX6 (37) was used for genomic tagging of the proteins of interest by Myc epitope. The plasmid PRS406 was used for PCR-based disruption of PDR5.
Strains
The yeast (S. cerevisiae) strain bearing ts mutation in Rpt4p (rpt4-ts or sug2-13, Sc677) and its isogenic wild-type equivalent (Sc599) were obtained from the Kodadek and Johnston laboratories (38). The esa1-ts mutant (LPY3291) and wild-type (LPY3498) strains were obtained from the Pillus laboratory (39). Multiple Myc epitope tags were added at the original chromosomal loci of RPN9, PRS3, PRE6 and RPN12 in FY67 (40) to generate NSY5 (Rpn9p-Myc), NSY6 (Prs3p-Myc), NSY8 (Pre6p-Myc) and NSY7 (Rpn12p-Myc), respectively. Strains PSY17 (Rpt2p-Myc) and PSY18 (Rpt6p-Myc) were generated by adding multiple Myc epitope tags at the C-termini of Rpt2p and Rpt6p, respectively, in Sc599. Multiple Myc epitope tags were added at the original chromosomal locus of ESA1 in the rpt4-ts and wild-type strains to generate BUY13 (Esa1p-Myc in rpt4-ts) and BUY12 (Esa1p-Myc), respectively. The PDR5 gene was deleted from the wild-type strain by PCR-based gene disruption method to generate SLY16a (Δpdr5, Δura3).
Growth media
For the ChIP studies at the ribosomal protein genes in the wild-type strain, yeast cells were grown in YPD (yeast extract–peptone plus 2% dextrose) at 30°C up to an OD600 of 1.0 prior to formaldehyde-based in vivo cross-linking. However, the rpt4-ts and esa1-ts mutants and their isogenic wild-type equivalents were grown in YPD at 23°C up to an OD600 of 0.85 and then transferred to 37°C for 1- or 2-h before cross-linking. For experiments at INO1, yeast cells were initially grown in synthetic complete medium (yeast nitrogen base and complete amino acid mixture plus 2% dextrose) containing 100 µM inositol at 30°C up to an OD600 of 0.45, and then switched to the same medium without inositol for 2 h prior to MG132 (75 µM) treatment for 2 h.
Chromatin Immunoprecipitation assay
The Chromatin Immunoprecipitation (ChIP) assay was performed as described previously (2,4,41,42). Briefly, yeast cells were treated with 1% formaldehyde for 15 min, collected and resuspended in lysis buffer. Following sonication, cell lysate (400 µl lysate from 50 ml of yeast culture) was pre-cleared by centrifugation and then 100 µl lysate was used for each immunoprecipitation. Immunoprecipitated protein–DNA complexes were treated with proteinase K, the cross-links were reversed and DNA was purified. Immunoprecipitated DNA was dissolved in 10 µl TE 8.0 (10 mM Tris-HCl pH 8.0 and 1 mM EDTA), and 1 µl of immunoprecipitated DNA was analyzed by PCR. PCR reactions contained (α-32P)dATP (2.5 µCi for each 25 µl reaction), and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, ‘input’ DNA was isolated from 5 µl lysate without going through the immunoprecipitation step and dissolved in 100 µl TE 8.0. To compare PCR signal arising from the immunoprecipitated DNA with the input DNA, 1 µl of input DNA was used in the PCR analysis.
The association of Esa1p with ribosomal protein genes was analyzed by modified-ChIP assay as described in our previous publication (42). For ChIP analysis of the proteasome components, we modified the above ChIP protocol as follows. 1600 μl of lysate was prepared from 200 ml of yeast culture following formaldehyde-based in vivo cross-linking for 25 min. 600 μl of lysate was used for each immunoprecipitation (using 10 μl of anti-Myc antibody and 100 μl of protein A/G plus agarose beads from Santa Cruz Biotechnology, Inc.), and immunoprecipitated DNA sample was dissolved in 5 μl of TE 8.0 of which 1 μl was used in PCR analysis. In parallel, PCR for input DNA was performed using 1 μl of DNA that was prepared by dissolving purified DNA from 5 μl of lysate in 100 μl of TE 8.0. Autoradiograms were scanned and quantitated by the National Institutes of Health image 1.62 program. Immunoprecipitated DNAs were quantitated as the ratio of IP to input.
The primer pairs used for PCR analysis were as follows:
| RPS5 (UAS): | 5′-AGAAACAATGAACAGCCTTGAGTTCTC-3′ |
| 5′-GCAGGGCCATTCTCATCTGA-3′ | |
| RPS5(Core): | 5′-GGCCAACTTCTACGCTCACGTTAG-3′ |
| 5′-CGGTGTCAGACATCTTTGGAATGGTC-3′ | |
| RPS5 (ORF): | 5′-AGGCTCAATGTCCAATCATTGAAAG-3′ |
| 5′-CAACAACTTGGATTGGGTTTTGGTC-3′ | |
| RPL2B (UAS): | 5′-TACCGATTACCAAGTTTTCAGACTA-3′ |
| 5′-AATTCCTTCTTTTTCTCCCTAGCGG-3′ | |
| RPL2B (Core): | 5′-TGGTGGATTCTGCTCTGGAAACTAT-3′ |
| 5′-CTTTGTGGTTTCTTGGTGAGTTTAT-3′ | |
| RPL2B (ORF): | 5′-GTGCTTTCCACAAGTACAGATTGAA-3′ |
| 5′-TTTGACCAGAAACGGCACCTCTAGA-3′ | |
| RPS11B (UAS): | 5′-GATATACACAAGAATTTCTGGAAGA-3′ |
| 5′-CACTTCCTCATTTCACAAAGACACT-3′ | |
| RPS11B (Core): | 5′-AAGTCCAATAGCTTTACGTTTCCCT-3′ |
| 5′-CTTTTTCCCTGGCTTGATACGTTTC-3′ | |
| RPS11B (ORF): | 5′-GCACCGTACCATTGTCATCAGAAGA-3′ |
| 5′-GGTCTACATTGACCAACGGTAACAA-3′ | |
| ACT1 (Core): | 5′-AACCGTTTTGAAACCAAACTCGCCT-3′ |
| 5′-TTCTTGGTTTGAGTAGAAAGGGGAA-3′ | |
| INO1 (Core): | 5′-TTCACATGGAGCAGAGAAAGCGCA-3′ |
| 5′-GGATAAAACTAACATTAGGAAC CCGAC-3′ | |
| Chr-V: | 5′-GGCTGTCAGAATATGGGGCCGTAGTA-3′ |
| 5′-CACCCCGAAGCTGCTTTCACAATAC-3′. |
UAS, upstream activating sequence; Core, core promoter; ORF, open reading frame; and Chr-V, Chromosome-V.
Total mRNA preparation
The total mRNA was prepared from yeast cell culture as described by Peterson et al. (43). Briefly, 10 ml yeast culture was harvested and suspended in 100 µl RNA preparation buffer (500 mM NaCl, 200 mM Tris–HCl, 100 mM Na2EDTA and 1% SDS) along with 100 µl phenol/chloroform/isoamyl alcohol and 100 µl volume equivalent of glass beads (acid washed; Sigma). Subsequently, yeast cell suspension was vortexed with a maximum speed (10 in VWR mini-vortexer; Cat. No. 58816-121) for five times (30 s each). Cells suspension was put in ice for 30 s between pulses. After vortexing, 150 µl RNA preparation buffer and 150 µl phenol/chloroform/isoamyl alcohol were added to yeast cell suspension followed by vortexing for 15 s with a maximum speed on VWR mini-vortexer. The aqueous phase was collected following 5 min centrifugation at a maximum speed in microcentrifuge machine. The total mRNA was isolated from aqueous phase by ethanol precipitation.
Reverse transcriptase–PCR analysis
Reverse transcriptase (RT)–PCR analysis was performed following the standard protocols (44,45). Briefly, total mRNA was prepared from 10 ml yeast culture, and was used in the reverse transcription assay. mRNA was treated with RNase-free DNase (M610A, Promega) and then reverse-transcribed into cDNA using oligo(dT) as described in the protocol supplied by Promega (A3800, Promega). PCR was performed using synthesized first strand as template and the primer pairs targeted to the RPS5, RPL2B, RPS11B and ACT1 ORFs. RT–PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. The primer pairs used in the PCR analysis were as follows:
| RPS5: | 5′-AGGCTCAATGTCCAATCATTGAAAG-3′ |
| 5′-CAACAACTTGGATTGGGTTTTGGTC-3′ | |
| RPL2B: | 5′-GTGCTTTCCACAAGTACAGATTGAA-3′ |
| 5′-TTTGACCAGAAACGGCACCTCTAGA-3′ | |
| RPS11B: | 5′-GCACCGTACCATTGTCATCAGAAGA-3′ |
| 5′-GGTCTACATTGACCAACGGTAACAA-3′ | |
| ACT1: | 5′-TCCACCACTGCTGAAAGAGAAATTG-3′ |
| 5′-AATAGTGATGACTTGACCATCTGGA-3′ |
RESULTS
The 19S base, but not lid or 20S CP, is recruited to the RPS5 promoter
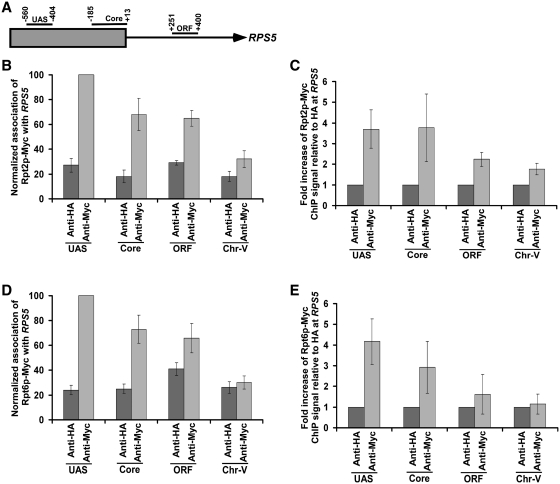
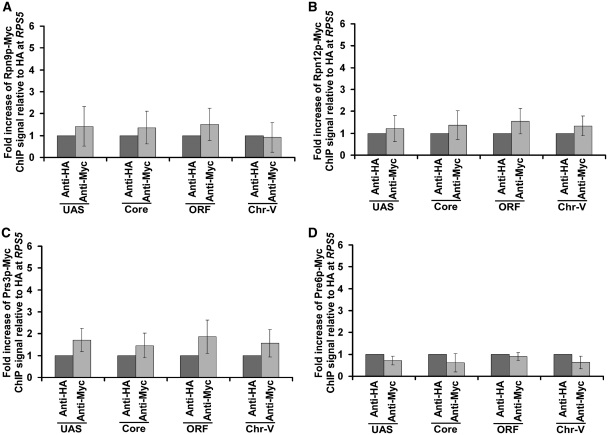
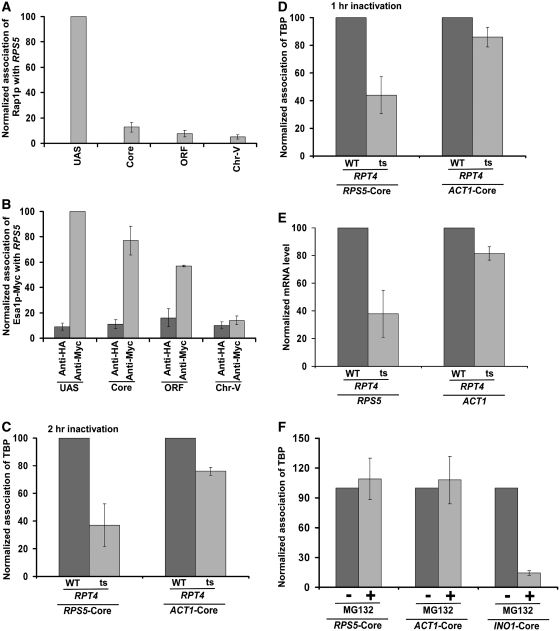
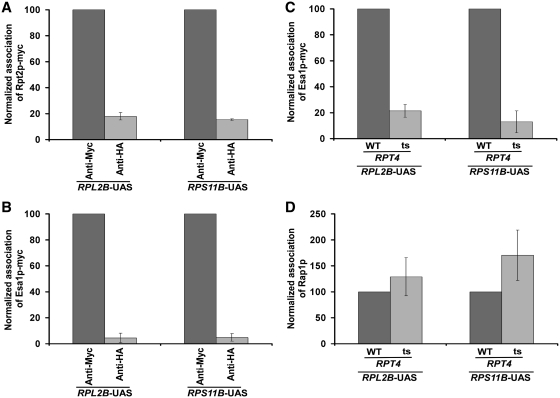
To determine the role of the 26S proteasome in regulation of transcriptional initiation of the TFIID-dependent ribosomal protein genes, we first analyzed its association with the promoter of a well-characterized ribosomal protein gene, RPS5. In view of this, we tagged the Rpt6p (19S base), Rpt2p (19S base), Rpn9p (19S lid), Rpn12p (19S lid), Prs3p (20S CP) and Pre6p (20S CP) components of the 26S proteasome complex by Myc epitope in their endogenous chromosomal loci. Using these epitope-tagged strains, we performed the ChIP assay at the RPS5 UAS, core promoter and coding sequence (ORF) (Figure 1A). The inactive region of chromosome V (Chr-V) was used as a non-specific DNA control. An anti-HA served as a non-specific antibody in the ChIP assay. We find that Rpt2p and Rpt6p components of the 19S base were recruited to the RPS5 promoter (Figure 1B–E and Supplementary Figure S1). However, a relatively higher association of Rpt2p and Rpt6p was observed at the UAS (Figure 1B and D). The 19S base was also found at the coding sequence (Figure 1B and D), consistent with its known role in transcriptional elongation (46). The 19S lid (Rpn9p and Rpn12p) or 20S CP (Prs3p and Pre6p) was not recruited to the RPS5 promoter (Figure 2A–D). Together, these results demonstrate that the 19S base is recruited to the RPS5 promoter independently of the 19S lid or 20S CP. Likewise, the 19S base has also been previously shown to be associated with the GAL1 promoter (36,47). These results support the existence of the 19S base independently of lid or 20S CP in vivo. Consistent with these in vivo observations, the 19S base without lid or 20S CP has been biochemically characterized (48). Moreover, previous studies have also demonstrated the existence of the 19S base independently of the lid or 20S CP in the nucleus (49).
Figure 1.
The 19S base is recruited to the RPS5 promoter. (A) The schematic diagram of the RPS5 promoter with the PCR amplification regions (UAS, Core and open reading frame or ORF) in the ChIP assay. (B) Analysis of recruitment of the Rpt2p component of the 19S base to RPS5. The yeast strain expressing myc-tagged Rpt2p was grown at 30°C in YPD (yeast extract, peptone plus 2% dextrose) up to an OD600 of 1.0 prior to formaldehyde-based in vivo crosslinking. The ChIP assay was performed as described in the ‘Materials and Methods’ section. Primer-pairs (‘Materials and Methods’ section) located at the UAS, core promoter and ORF regions of RPS5 were used for PCR analysis of the immunoprecipitated DNA samples. Immunoprecipitation was performed using a mouse monoclonal antibody against the c-myc epitope-tag (9E10; Santa Cruz Biotechnology, Inc.). The anti-HA (Santa Cruz Biotechnology, Inc.) was used as a non-specific antibody. A specific primer pair spanning an inactive region in the chromosome V (Chr-V) was used as a non-specific DNA control. The maximum ChIP signal was set to 100, and other signals were normalized with respect to the maximum ChIP signal. (C) The results of the (B) were presented as a fold increase of the ChIP signal of Rpt2p-myc relative to non-specific anti-HA antibody. (D) Analysis of recruitment of the Rpt6p component of the 19S base to RPS5. The yeast strain expressing myc-tagged Rpt6p was grown, cross-linked and immunoprecipitated as in (B). (E) The results of the (C) were presented as a fold increase of the ChIP signal of Rpt6p-myc relative to non-specific anti-HA antibody.
Figure 2.
Analysis of recruitment of the 19S Lid or 20S CP to RPS5. (A and B) The 19S Lid is not recruited to RPS5. The yeast strains expressing myc-tagged Rpn9p and Rpn12p were grown, cross-linked and immunoprecipitated as in Figure 1B. (C and D) The 20S CP is not recruited to RPS5. The yeast strains expressing myc-tagged Prs3p and Pre6p were grown, cross-linked, and immunoprecipitated as in Figure 1B.
The 19S base promotes the recruitment of TFIID to the RPS5 promoter for transcriptional initiation
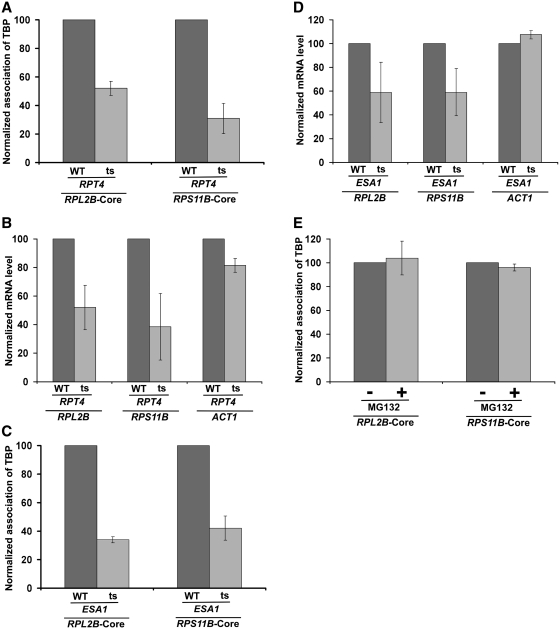
Rap1p recognizes the RPG box upstream of the RPS5 core promoter, and thus, is recruited to the UAS, but not core promoter, of RPS5 (Figure 3A). Subsequently, it activates transcription (13,15,16). Rap1p also functions as a transcriptional activator of other ribosomal protein genes (19,50). In addition to its role in transcriptional activation, it further plays an important role in silencing at telomeres and mating-type loci (51,52). Previous studies (13,15,16) have demonstrated that Rap1p targets TAFs to recruit the TFIID complex at the core promoter for transcriptional initiation, indicating that TAFs are the essential targets of the transcriptional activator Rap1p in vivo. Consistent with these in vivo results, Weil and colleagues (53,54) have also demonstrated biochemically the interaction of Rap1p with TAFs. Collectively, these studies have implicated TAFs as the target of transcriptional activator at the ribosomal protein genes. However, previous studies (55,56) have also demonstrated the requirement of NuA4 HAT for transcription of ribosomal protein genes. NuA4 is highly conserved among eukaryotes, and is required for acetylation of histones H4 and H2A (55,57,58). The catalytic subunit of NuA4 is Esa1p which is essential for cellular viability. We find that like the 19S base, NuA4 HAT (Esa1p-Myc) is recruited to the RPS5 promoter (Figure 3B), consistent with previous studies (55). However, it is also recruited to the coding sequence (Figure 3B). Likewise, Ginsburg et al. (59) have demonstrated the association of NuA4 HAT with the coding sequence for transcriptional elongation. Further, Reid et al. (55) have implicated the role of Rap1p in targeting NuA4 HAT. Consistently, co-immunoprecipitation analysis revealed the interaction of Rap1p with NuA4 (Supplementary Figure S2). Likewise, previous biochemical studies have also demonstrated the interaction of NuA4 with acidic activators (6,56,60). Taken together, Rap1p, NuA4 HAT and the 19S base are recruited to RPS5, and Rap1p and NuA4 HAT are essential for transcription of RPS5. However, the role of the 19S base in regulation of the RPS5 transcription is not known. Like Rap1p and NuA4 HAT, the 19S base might be playing a crucial role in regulating the transcriptional initiation of RPS5. To test this possibility, we analyzed the role of the 19S base in recruitment of the TFIID complex at the RPS5 core promoter, since our previous studies (13) have correlated the recruitment of the TFIID complex with transcriptional initiation. We previously demonstrated that TBP and TAFs components of the TFIID complex are recruited to the RPS5 core promoter (13), and these components are essential for RPS5 transcription (13). Further, we have shown previously that TAFs are essential for recruitment of TBP to the RPS5 core promoter for transcriptional initiation (10). Thus, we have used TBP as a representative core component of the TFIID complex for ChIP analysis at the RPS5 promoter. We find that the recruitment of TBP to the RPS5 core promoter was significantly decreased in the ts mutant strain of Rpt4p ATPase subunit of the 19S base (Figure 3C and D). The rpt4-ts mutant encodes point mutation (L231R) in the ATPase module of Rpt4p (38). Rpt4p is degraded at the non-permissive temperature in the rpt4-ts strain (data not shown), and is essential for the structural integrity of the 19S base (61). As a control, we analyzed the recruitment of TBP to the ACT1 core promoter as its transcription is independent of the proteasome complex (62). As expected, the recruitment of TBP to the ACT1 core promoter was not significantly altered in the rpt4-ts mutant strain (Figure 3C and D). These results strongly support the role of the 19S base in recruitment of TFIID to RPS5, and thus transcription of RPS5 was significantly impaired in the rpt4-ts mutant strain (Figure 3E). Consistently, the recruitment of RNA polymerase II to the RPS5 core promoter was also decreased in the rpt4-ts mutant strain (Supplementary Figure S3). Further, we demonstrate that the inhibition of the proteolytic function of the proteasome complex by MG132 did not alter the recruitment of TBP to the RPS5 core promoter (Figure 3F). Likewise, the treatment of MG132 did not alter the recruitment of TBP to the proteasome-independent ACT1 gene (Figure 3F). As a positive control, we show that the recruitment of TBP to the proteasome-dependent INO1 gene (62) was significantly decreased following MG132 treatment (Figure 3F). Thus, our results revealed the non-proteolytic role of the proteasome in transcriptional initiation of the RPS5 gene. Consistently, we find that 20S CP was not recruited to the RPS5 promoter (Figure 2C and D).
Figure 3.
The 19S base stimulates the recruitment of TBP (and hence transcription) at the RPS5 promoter. (A) Analysis of recruitment of Rap1p to RPS5. Immunoprecipitation was performed using an antibody against Rap1p (SC-6663; Santa Cruz Biotechnology, Inc.). (B) Analysis of recruitment of Esa1p to RPS5. The Esa1p component of NuA4 was tagged by myc-epitope at the C-terminal of its chromosomal locus for immunoprecipitation. (C) Analysis of the role of 19S base in recruitment of TBP to the RPS5 core promoter. The wild-type and rpt4-ts mutant strains were grown in YPD at 23°C up to an OD600 of 0.85, and then switched to 37°C for 2 h prior to cross-linking. Immunoprecipitation was performed using an anti-TBP antibody against TBP (obtained from the Green laboratory; 13). (D) Similar to the (C). But, Rpt4p was inactivated for 1 h. (E) RT–PCR analysis of RPS5 and ACT1 transcripts in the rpt4-ts mutant and its isogenic wild-type equivalent following 1 h ts inactivation at the non-permissive temperature. (F) Treatment of yeast cells carrying null mutation of PDR5 with MG132 (75 µM) for 2 h does not alter the recruitment of TBP to the RPS5 core promoter. Yeast cells were grown in YPD at 30°C up to an OD600 of 0.7, and then treated with MG132 for 2 h prior to cross-linking.
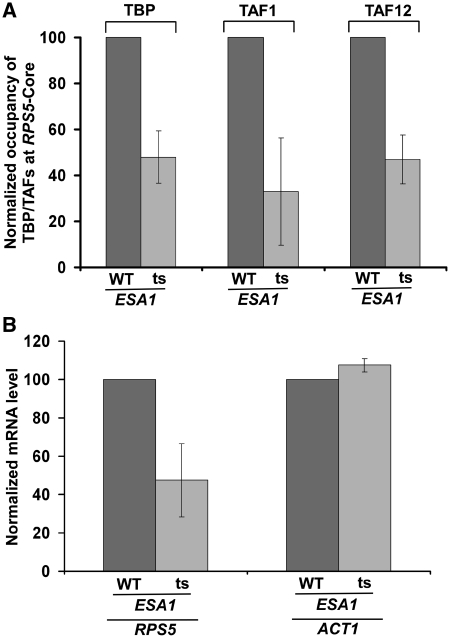
Recruitment of TFIID to the RPS5 promoter is dependent on NuA4 HAT
To determine whether, like the 19S base, NuA4 HAT also facilitates the recruitment of TFIID, we analyzed the association of TBP and TAFs components (TAF1 and TAF12) of the TFIID complex with the RPS5 core promoter in the wild-type and ts mutant strains of the Esa1p HAT component of NuA4. We find that the recruitment of the TFIID complex to the RPS5 core promoter was significantly impaired in the esa1-ts mutant strain as compared with the wild-type equivalent (Figure 4A). Such an impaired recruitment of TFIID in the esa1-ts mutant significantly lowered the transcription of RPS5 (Figure 4B). As a control, we have used ACT1, since its transcription is not dependent on NuA4 HAT (55). As expected, we find that transcription of ACT1 was not altered in the esa1-ts mutant strain (Figure 4B). As a whole, our data demonstrate that NuA4 HAT enhances the recruitment of TFIID (and hence transcription) at the core promoter of the ribosomal protein gene, RPS5. Consistently, previous studies (55,56) have also implicated the role of NuA4 HAT in stimulation of transcription of the ribosomal protein genes. However, the mechanism of such transcriptional stimulation was not known. Here, we show that NuA4 HAT promotes transcription of the ribosomal protein gene, RPS5, by enhancing the recruitment of the TFIID complex. Further, NuA4 HAT has been shown to be targeted to the promoters of ribosomal protein genes by the activator Rap1p (55). Therefore, the recruitment of Rap1p is essential for association of TFIID with promoter. Indeed, previous studies (13,15,16) have demonstrated the role of Rap1p in recruitment of TFIID. Here, we demonstrate that like Rap1p, NuA4 HAT and 19S base are both required to facilitate the recruitment of TFIID for transcriptional initiation of RPS5.
Figure 4.
NuA4 HAT is required for recruitment of the TFIID complex to the RPS5 promoter. (A) Analysis of the recruitment of TBP and TAFs components of the TFIID complex to the RPS5 core promoter in the esa1-ts mutant and its isogenic wild-type equivalent. Yeast cells were grown as in Figure 3C, but Esa1p was inactivated for 1 h at the non-permissive temperature. Immunoprecipitation was performed using anti-TBP, anti-TAF1 and anti-TAF12 antibodies against TBP, TAF1 and TAF12 (obtained from the Green laboratory; 13). (B) RT–PCR analysis of RPS5 and ACT1 transcripts in the esa1-ts mutant and its isogenic wild-type equivalent following 1 h ts inactivation at the non-permissive temperature.
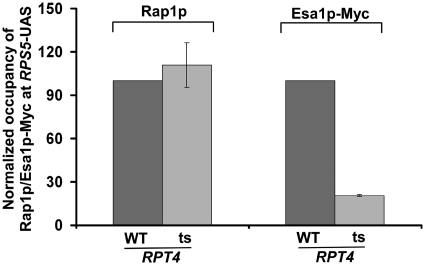
The 19S base enhances the targeting of NuA4 HAT, but not Rap1p, to the RPS5 promoter
So far, it is clear that Rap1p, NuA4 HAT and 19S base play crucial roles to recruit TFIID for transcriptional initiation of RPS5. However, the specific regulatory network of these factors at the RPS5 promoter is not known. It has been demonstrated previously that the 19S base enhances the targeting of SAGA HAT co-activator to the activator Gal4p at the SAGA-dependent GAL1 gene for transcriptional initiation of GAL1 (35,36). Based on these results, we hypothesize that the 19S base might be similarly enhancing the targeting of NuA4 HAT to the activator Rap1p at the RPS5 UAS to facilitate the recruitment of TFIID for transcriptional initiation. To test this possibility, we analyzed the recruitment of Rap1p and NuA4 HAT to the RPS5 UAS in the wild-type and ts mutant strains of the Rpt4p ATPase component of the 19S base. If the 19S base increases the targeting of NuA4 HAT to Rap1p at the RPS5 UAS, we would observe a significant decrease in the recruitment of NuA4 HAT, but not Rap1p, at the RPS5 UAS in the rpt4-ts mutant strain as compared with the wild-type equivalent. Indeed, we find that Rap1p recruitment to the RPS5 UAS was not altered in the rpt4-ts mutant strain (Figure 5). However, the recruitment of NuA4 HAT (Esa1p-Myc) to the RPS5 UAS was significantly impaired in the rpt4-ts mutant strain (Figure 5). These results strongly support that the 19S base facilitates the targeting of NuA4 HAT to Rap1p at the RPS5 UAS. This observation is remarkably consistent with previous studies (35,36) that demonstrated the role of the 19S base in enhancing the targeting of SAGA HAT to the activator Gal4p at the SAGA-dependent GAL1 gene. Although previous studies (35,36) have shown the role of the 19S base in targeting SAGA to the SAGA-dependent gene, the role of the 19S base at the promoter of the TFIID-dependent gene remained unknown. This study demonstrates for the first time the role of the 19S base in enhanced targeting of NuA4 HAT to the activator of a TFIID-dependent ribosomal protein gene, RPS5 (Figure 5), and subsequently facilitates the recruitment of the TFIID complex for transcriptional initiation (Figure 4A and B).
Figure 5.
Analysis of recruitment of Rap1p and NuA4 HAT (Esa1p-myc) to the RPS5 promoter in the wild-type and rpt4-ts mutant strains following ts inactivation of Rpt4p at 37°C for 1 h. Immunoprecipitations were performed as described in Figures 3A and B.
Since the 19S base enhances the targeting of NuA4, it would promote histone H4 acetylation at the RPS5 promoter via NuA4 HAT. To test this, we analyzed histone H4 acetylation at the RPS5 promoter in the rpt4-ts mutant and its isogenic wild-type equivalent. We find that histone H4 acetylation at the RPS5 core promoter was decreased in the rpt4-ts mutant strain as compared to the wild-type equivalent (Supplementary Figure S4A and S4B). Similarly, histone H4 acetylation was impaired in the esa1-ts mutant strain (Supplementary Figure S4B), consistent with previous studies (55). Thus, the 19S base regulates histone H4 acetylation by modulating the targeting of NuA4 HAT to the promoter.
Previous studies have demonstrated that NuA4 HAT is recruited to the promoters of the ribosomal protein genes in a TOR-dependent manner (24). We thus asked whether the recruitment of the 19S base to the RPS5 promoter is also regulated by the TOR pathway. To address this question, we analyzed the recruitment of the 19S base and Rap1p to the RPS5 UAS in the presence and absence of rapamycin. Rapamycin inhibits TOR-kinase activity, and thus impairs TOR pathway. We find that the recruitment of the 19S base (Rpt2p and Rpt6p), but not Rap1p, was significantly decreased in the presence of rapamycin (Supplementary Figure S5A and Supplementary S5B). Thus, the 19S base is recruited to the RPS5 UAS in a TOR-dependent manner. Similarly, the recruitment of NuA4 HAT to the RPS5 UAS was also decreased in the presence of rapamycin (Supplementary Figure S5B), consistent with previous studies (24).
The 19S base enhances the targeting of NuA4 HAT, but not Rap1p, to the RPL2B and RPS11B promoters to facilitate the recruitment of TFIID for stimulated transcription
Although the above results implicated an important role of the 19S base in transcriptional stimulation of RPS5, it is not known whether other ribosomal protein genes are also similarly regulated by the 19S base. In view of this, we next analyzed the role of the 19S base in recruitment of TBP to the core promoters of two other ribosomal protein genes such as RPL2B and RPS11B in the rpt4-ts mutant strain and its isogenic wild-type equivalent. Interestingly, we find that the recruitment of TBP to the core promoters of these genes was significantly impaired in the rpt4-ts mutant strain (Figure 6A). Consistently, we find that the transcription of these genes was also significantly decreased in the rpt4-ts mutant strain (Figure 6B). Thus, similar to the results obtained at RPS5, we find that the 19S base facilitates the recruitment of TBP (and hence TFIID) to the core promoters of RPL2B and RPS11B to promote transcriptional initiation.
Figure 6.
Both 19S base and NuA4 HAT promote the recruitment of TBP (and hence transcription) at the RPL2B and RPS11B core promoters. (A) Analysis of recruitment of TBP to the RPL2B and RPS11B core promoters in the rpt4-ts mutant and its isogenic wild-type equivalent following 1 h ts inactivation at the non-permissive temperature. (B) RT–PCR analysis of RPL2B, RPS11B and ACT1 transcripts in the rpt4-ts mutant and its isogenic wild-type equivalent following 1 h ts inactivation at the non-permissive temperature. (C) Analysis of recruitment of TBP to the RPL2B and RPS11B core promoters in the esa1-ts mutant and its isogenic wild-type equivalent following ts inactivation for 1 h. (D) RT–PCR analysis of RPL2B, RPS11B and ACT1 transcripts in the esa1-ts mutant and its isogenic wild-type equivalent following 1 h ts inactivation at the non-permissive temperature. (E) Treatment of yeast cells carrying null mutation of PDR5 with MG132 (75 µM) for 2 h does not alter recruitment of TBP to the RPL2B and RPS11B core promoters. Yeast cells were grown and cross-linked as in Figure 3F.
Next, we asked whether recruitment of TBP to the core promoters of RPL2B and RPS11B is also facilitated by NuA4 HAT. In this direction, we analyzed the recruitment of TBP to the core promoters of these genes in the esa1-ts mutant strain and its isogenic wild-type equivalent. We find that like the results at RPS5, the recruitment of TBP to the core promoters of RPL2B and RPS11B was also significantly decreased in the esa1-ts mutant strain as compared with the wild-type equivalent (Figure 6C). Since the recruitment of TBP is directly correlated with transcription, the RPL2B and RPS11B transcription would be decreased in the esa1-ts mutant strain. Indeed, we find that the transcription of these genes was also reduced in the esa1-ts mutant strain (Figure 6D). Thus, similar to the results obtained at RPS5, we find that NuA4 HAT facilitates the recruitment of TBP (and hence TFIID) to the core promoters of the RPL2B and RPS11B genes to promote transcription. Further, we demonstrate that the recruitment of TBP to the core promoters of these genes was not altered in the presence of MG132 (Figure 6E). Therefore, the proteolytic function of the proteasome is dispensable for recruitment of TBP to these ribosomal protein genes.
We find that both NuA4 HAT and 19S base enhance the recruitment of TFIID to the core promoters of RPL2B and RPS11B. Thus, similar to the results at RPS5, it is likely that the 19S base is recruited to the RPL2B and RPS11B promoters, and promotes the association of NuA4 HAT for facilitating the recruitment of TFIID to stimulate transcriptional initiation. To test this possibility we next analyzed the recruitment of the 19S base at the RPL2B and RPS11B promoters, and subsequently determined the role of the 19S base in recruitment of NuA4 HAT. We find that the 19S base was recruited to the RPL2B and RPS11B promoters (Figure 7A), and facilitated the association of NuA4 HAT (Esa1p-Myc) (Figure 7B and C). However, the recruitment of Rap1p to these genes was not altered in the rpt4-ts mutant strain (Figure 7D). Thus, the 19 S base specifically promotes the recruitment of NuA4 HAT, but not activator Rap1p, to the RPL2B and RPS11B promoters. Subsequently, NuA4 HAT stimulates the recruitment of TFIID to the core promoter (Figure 6C), hence facilitating transcription (Figure 6D). Taken together, our data strongly support that the 19 S base enhances the targeting of NuA4 HAT to the activator Rap1p at RPL2B and RPS11B to promote the recruitment of TFIID for stimulated transcription.
Figure 7.
The 19S base enhances the targeting of NuA4 HAT, but not activator Rap1p, to the promoters of RPL2B and RPS11B. (A) Analysis of recruitment of the 19S base (Rpt2p-myc) to the promoters of RPL2B and RPS11B. Yeast cells were grown, cross-linked and immunoprecipitated as in Figure 1B. (B) Analysis of recruitment of NuA4 HAT (Esa1p-myc) to the promoters of RPL2B and RPS11B. Yeast cells were grown, cross-linked and immunoprecipitated as in Figure 3B. (C) Analysis of recruitment of NuA4 HAT (Esa1p-myc) to the promoters of RPL2B and RPS11B in the rpt4-ts mutant and its wild-type equivalent following 1 h ts inactivation at the non-permissive temperature. (D) Analysis of recruitment of Rap1p to the promoters of RPL2B and RPS11B in the rpt4-ts mutant and its wild-type equivalent following 1 h ts inactivation at the non-permissive temperature.
DISCUSSION
Here, we demonstrate that the 19S base is recruited to the promoter of the ribosomal protein gene independently of the lid and 20S CP. Such a recruitment of the 19S base enhances the targeting of NuA4 HAT to the promoters of ribosomal protein genes. Subsequently, NuA4 HAT facilitates the recruitment of the TFIID complex for stimulation of the transcriptional initiation of the ribosomal protein genes. Collectively, these results demonstrate an important role of the 19S base in establishing a specific regulatory network at the promoters of ribosomal protein genes for promoting transcriptional initiation, hence significantly advancing our current understanding of the regulation of ribosomal protein gene activation in vivo.
NuA4 is a multi-subunit protein complex in yeast. The essential HAT subunit (Esa1p) of NuA4 and its human and Drosophila homologues [Tip60 (Tat-interacting protein, 60 kDa) and MOF (males absent on the first), respectively] belong to a conserved family of acetylases that are involved in acetylation of histone H4. Tip60 is a catalytic subunit of human TIP60 complex that augments Tat-mediated transcription (63), and is also involved in DNA repair and apoptosis (64). MOF, a catalytic subunit of the MSL complex, is required for histone H4 hyperacetylation that is associated with an increase in X-linked transcription in Drosophila males (65,66). Further, MOF has also been shown to facilitate transcription in yeast when artificially targeted to a promoter (66). Human ortholog of Drosophila MOF is involved in histone H4 acetylation at lysine 16 (67–70). In humans, MOF is associated with transcriptional activation in coordination with histone H3 lysine 4 methyltransferase (67). Further, Pandita and colleagues (68,70) have implicated human MOF in DNA repair. They have demonstrated that MOF interacts with the ATM (ataxia-telangiectasia-mutated) protein, and acetylates histone H4 following ionizing radiation (68,70). Such modification plays an important role in cellular DNA damage response and repair (70). MOF has also been implicated in mammalian embryogenesis (69). Like Tip60 and MOF, NuA4 HAT has been shown to promote transcription (55, 56). Further, previous studies have demonstrated the recruitment of NuA4 HAT to the promoters of ribosomal protein genes in an activator-dependent manner (55). Consistently, NuA4 HAT has been shown to be the target of transcriptional activators (6,56,60). However, it is not known how NuA4 HAT promotes transcription of the ribosomal protein genes. Here, we show that NuA4 HAT is recruited to the promoters of ribosomal protein genes (Figures 3B and 7B). Subsequently, it facilitates the recruitment of the TFIID complex to the promoter (Figures 4A and 6C). TFIID nucleates the assembly of the GTFs at the promoter to form the PIC for transcriptional initiation (1, 34). Thus, an increased association of TFIID by NuA4 HAT enhances transcriptional initiation of the ribosomal protein genes (Figures 4B and 6D). Therefore, our results demonstrate that NuA4 HAT promotes transcriptional initiation by facilitating the recruitment of the TFIID complex at the promoters of ribosomal protein genes.
Although previous studies (13,15,16) have implicated TFIID as the target of the activator Rap1p, our current results demonstrate that Rap1p cannot efficiently target or recruit TFIID to the ribosomal protein genes when the HAT activity of NuA4 is impaired in the esa1-ts mutant strain (Figures 4A and 6C). Further, previous studies have demonstrated a high level of histone H4 acetylation at the promoters of the ribosomal protein genes, RPS5, RPL2B and RPS11B, in the presence of NuA4 HAT (55). Thus, it is likely that the HAT activity of NuA4 or histone H4 acetylation is essential to recruit TFIID for transcriptional initiation. Such a HAT activity is not essential for recruitment of the activator Rap1p (data not shown). These results support a model where NuA4 HAT is targeted by the activator Rap1p, which then acetylates histone H4 at the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for stimulation of transcriptional initiation.
Intriguingly, we also demonstrate here that like NuA4 HAT, the 19S base promotes the association of TFIID with the promoters of the ribosomal protein genes (Figures 3C, D and 6A). Consistently, we find that the transcription of ribosomal protein genes is significantly impaired in the ts mutant of the Rpt4p ATPase subunit of the 19S base (Figures 3E and 6B). Thus, we show here for the first time that 19S proteasome subcomplex promotes the recruitment of TFIID to the promoters of the ribosomal protein genes to initiate transcription. Such function of the 19S proteasome subcomplex is mediated via an enhanced targeting of NuA4 HAT as discussed below. Further, we demonstrate that the proteolytic function of the proteasome is dispensable for recruitment of TFIID to the promoters of these genes (Figures 3F and 6E). Similarly, previous studies have demonstrated the role of 19S proteasome subcomplex in mammalian transcriptional regulation in a proteolysis-independent manner (71,72). Several other studies in yeast have also implicated the non-proteolytic role of the proteasome in transcriptional regulation (1,34).
We show here that both NuA4 and 19S base facilitate the recruitment of TFIID to the promoters of the ribosomal protein genes for transcriptional stimulation. NuA4 HAT promotes the recruitment of TFIID, presumably via histone H4 acetylation as mentioned above and discussed below. However, it is not clear how the 19S base enhances the recruitment of TFIID. We find here that the 19S base increases the recruitment of NuA4 HAT, but not the activator Rap1p, to the promoters of the ribosomal protein genes (Figures 5, 7C and D). These observations support that the 19S base promotes the targeting of NuA4 HAT to the activator Rap1p in vivo. Analogous to this observation, previous studies have demonstrated an increased targeting of SAGA HAT to the activator Gal4p at the SAGA-dependent GAL1 gene (35,36). Thus, like previous studies at the SAGA-dependent gene (35,36), we demonstrate here a new role of the 19S base in targeting NuA4 HAT to the promoters of the TFIID-dependent ribosomal protein genes for stimulation of transcriptional initiation. How does the 19S base promote the targeting of NuA4 HAT to the activator Rap1p? As mentioned above, the 19S base has a molecular chaperonin activity (30). Presumably, the molecular chaperonin activity of the 19S base is playing an important role to increase the targeting of NuA4 HAT to the activator Rap1p, possibly by enhancing proper folding or assembly of NuA4 components. However, such a model remains to be further elucidated biochemically. Nonetheless, this study uncovered an important role of the 19S base in facilitating the targeting of NuA4 HAT to the ribosomal protein genes in promoting transcription in vivo. Since NuA4 HAT is involved in histone H4 acetylation at the RPS5, RPL2B and RPS11B genes (55), our study thus implicates the role of the 19S base in regulation of histone H4 acetylation via NuA4 HAT. Indeed, we find that the 19S base promotes histone H4 acetylation (Supplementary Figure S4A and S4B). Similarly, previous studies have demonstrated the function of the 19S proteasome subcomplex in promoting histone H3 acetylation by facilitating the recruitment of SAGA HAT in yeast (35). Likewise, acetylation of histones H3 and H4 in human has also been shown to be regulated by the 19S proteasome subcomplex (71,72).
Importantly, our data implicate the role of NuA4 HAT or histone H4 acetylation in promoting recruitment of TFIID. However, NuA4 HAT is not solely required for association of TFIID, since a dramatically impaired recruitment of NuA4 HAT to the ribosomal protein genes (~6-fold) in the rpt4-ts strain (Figures 5 and 7C) decreased the occupancy of TFIID only by ~2.5-fold (Figures 3D and 6A). Further, even though the recruitment of NuA4 HAT to the ribosomal protein genes was decreased by ~6-fold in the rpt4-ts mutant strain (Figures 5 and 7C), transcription was impaired only by ~2.5-fold (Figures 3E and 6B), consistent with ~2.5-fold reduction in the recruitment of TFIID in the rpt4-ts mutant strain (Figures 3D and 6A). Together, these results implicate that NuA4 HAT or histone H4 acetylation is not solely required for recruitment of TFIID, but rather facilitates TFIID recruitment (and hence transcription). Such stimulation of TFIID recruitment or transcription is likely to have a great effect in cellular differentiation and development in higher eukaryotes, since temporal and spatial regulation of gene expression is tightly correlated with cell fate.
The fact that significant amount of TFIID is recruited to the promoters of ribosomal protein genes in the absence of NuA4 HAT or histone H4 acetylation (Figures 4A and 6C), supports that Rap1p targets TAFs to recruit TFIID for transcriptional initiation of the ribosomal protein genes, consistent with previous studies (13,15,16,53,54). However, the presence of NuA4 HAT enhances the recruitment of TFIID. How does NuA4 HAT or histone H4 acetylation enhance TFIID recruitment to the promoters of the ribosomal protein genes? Previous studies have implicated the role of histone H4 acetylation in recruitment of bromodomain protein, Bdf1p (Bromodomain factor 1) (73–78). Bdf1p interacts with acetylated-histone H4, and thus, it is functionally connected to NuA4 HAT in vivo (73–78). Moreover, Bdf1p interacts with TFIID (74,79). Therefore, NuA4 HAT or histone H4 acetylation appears to enhance the recruitment of TFIID to the ribosomal protein genes via Bdf1p. Further, Bdf1p interacts with SWR1, an ATP-dependent chromatin remodeling complex that is involved in exchanging histone H2A–H2B dimer by histone H2A.Z–H2B dimer during transcriptional activation (80–84). Using the energy of ATP hydrolysis through the Swr1p subunit, the SWR1 complex replaces histone H2A–H2B dimer by histone H2A.Z–H2B dimer during transcriptional induction when nucleosomes are disassembled to allow the passage of RNA polymerase II followed by reassembly (74). Some evidence suggests that such incorporation of H2A.Z might promote disassembly of nucleosome during transcriptional initiation (85,86), and thus, they are largely promoter specific (80,85–90). Further, several studies suggest that H2A.Z-containing nucleosomes form a special chromatin structure, and this structure pre-sets nucleosomes for dismantling upon transcriptional activation (80,86,91,92). Therefore, deposition of histone H2A.Z plays an important role in transcriptional initiation. Bdf1p-mediated recruitment of SWR1 complex is essential for histone H2A.Z deposition. Consistently, histone H2A.Z occupancy has been shown to be dependent on Bdf1p and SWR1 complex in global genome-wide analysis (86). Further, previous studies have linked histone H4 acetylation with the occupancy of Bdf1 and H2A.Z at specific loci (88,93,94). Thus, NuA4 HAT or histone H4 acetylation plays a crucial role in H2A.Z deposition. Moreover, previous studies have also demonstrated the role of NuA4 HAT in acetylation of histone H2A.Z of deposited histone H2A.Z–H2B dimer (93,95,96). This acetylation of histone H2A.Z is believed to favor the specific destabilization of H2A.Z-containing nucleosomes on promoters during gene activation (93,95–97). Collectively, these studies implicate that NuA4 HAT or histone H4 acetylation might be facilitating the recruitment of TFIID (and hence transcription) at the ribosomal protein genes in similar manner, which remain to be elucidated to better understand the role of the NuA4 HAT or histone H4 acetylation in ribosomal-protein gene activation.
In summary, our results define for the first time the role of the 19S proteasome subcomplex in promoting the recruitment of NuA4 HAT that subsequently facilitates the association of TFIID for stimulation of transcriptional initiation of the ribosomal protein genes, thus shedding much light on the regulation of ribosomal-protein gene expression in vivo. Since ribosomal protein genes, NuA4 HAT, 19S base and TFIID are highly conserved from yeast to humans, similar regulatory mechanism of ribosomal-protein gene expression is likely to exist in humans. Further, as mentioned above, ribosomal-protein gene expression is strongly correlated with protein biosynthesis, and hence cellular growth, differentiation and development. Therefore, our study points to the role of the 19S proteasome subcomplex in controlling cellular growth, muscle and cardiac development through the regulation of ribosomal-protein gene expression in higher eukaryotes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures 1–5 and Supplementary Reference [98].
FUNDING
National Institutes of Health grant (1R15GM088798-01 to Bhaumik laboratory); American Heart Association (Greater Midwest Affiliate) grant-in-aid (10GRNT4300059); and Excellence in Academic Medicine grant from Southern Illinois University School of Medicine. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Michael R. Green and Thomas Kodadek for antibodies; Thomas Kodadek, Stephen Johnston, and Loraine Pillus for yeast strains; and Sarah Frankland-Searby for editorial assistance.
REFERENCES
- 1.Bhaumik SR. Distinct regulatory mechanisms of eukaryotic transcriptional activation by SAGA and TFIID. Biochim. Biophys. Acta, Gene Regul. Mech. 2011;1809:97–108. doi: 10.1016/j.bbagrm.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaumik SR, Green MR. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhaumik SR, Green MR. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Gene Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larschan E, Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown CE, Howe L, Sousa K, Alley SC, Carrozza MJ, Tan S, Workman JL. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science. 2001;292:2333–2337. doi: 10.1126/science.1060214. [DOI] [PubMed] [Google Scholar]
- 7.Reeves WM, Hahn S. Targets of the Gal4 transcription activator in functional transcription complexes. Mol. Cell. Biol. 2005;25:9092–9102. doi: 10.1128/MCB.25.20.9092-9102.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Swanson MJ, Qiu H, Sumibcay L, Krueger A, Kim SJ, Natarajan K, Yoon S, Hinnebusch AG. A multiplicity of coactivators is required by Gcn4p at individual promoters in vivo. Mol. Cell. Biol. 2003;23:2800–2820. doi: 10.1128/MCB.23.8.2800-2820.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TI, Causton HC, Holstege FC, Shen WC, Hannett N, Jennings EG, Winston F, Green MR, Young RA. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- 10.Shen WC, Bhaumik SR, Causton HC, Simon I, Zhu X, Jennings EG, Wang TH, Young RA, Green MR. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003;22:3395–3402. doi: 10.1093/emboj/cdg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 12.Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 13.Li XY, Bhaumik SR, Green MR. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science. 2000;288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 14.Kuras L, Kosa P, Mencia M, Struhl K. TAF-containing and TAF-independent forms of transcriptionally active TBP in vivo. Science. 2000;288:1244–1248. doi: 10.1126/science.288.5469.1244. [DOI] [PubMed] [Google Scholar]
- 15.Mencía M, Moqtaderi Z, Geisberg JV, Kuras L, Struhl K. Activator-specific recruitment of TFIID and regulation of ribosomal protein genes in yeast, Mol. Cell. 2002;9:823–833. doi: 10.1016/s1097-2765(02)00490-2. [DOI] [PubMed] [Google Scholar]
- 16.Li XY, Bhaumik SR, Zhu X, Li L, Shen WC, Dixit B, Green MR. Selective recruitment of TAFs by yeast upstream activating sequences: implications for eukaryotic promoter structure. Curr. Biol. 2002;12:1240–1244. doi: 10.1016/s0960-9822(02)00932-6. [DOI] [PubMed] [Google Scholar]
- 17.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 18.Schmelzle T, Hall MN. TOR, a central controller of cell growth. Cell. 2000;103:253–262. doi: 10.1016/s0092-8674(00)00117-3. [DOI] [PubMed] [Google Scholar]
- 19.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 20.Martin DE, Soulard A, Hall MN. TOR regulates ribosomal protein gene expression via PKA and the Forkhead transcription factor FHL. Cell. 2004;119:969–979. doi: 10.1016/j.cell.2004.11.047. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen P, Rupes I, Sharom JR, Schneper L, Brouch JR, Tyers M. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 2004;18:2491–2505. doi: 10.1101/gad.1228804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schawalder SB, Kabani M, Howald I, Choudhury U, Werner M, Shore D. Growth-regulated recruitment of the essential yeast ribosomal protein gene activator Ifh1. Nature. 2004;432:1058–1061. doi: 10.1038/nature03200. [DOI] [PubMed] [Google Scholar]
- 23.Rudra D, Zhao Y, Warner JR. Central role of Ifh1p-Fhl1p interaction in the synthesis of yeast ribosomal proteins. EMBO J. 2005;24:533–542. doi: 10.1038/sj.emboj.7600553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rohde J, Cardenas ME. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol. Cell. Biol. 2003;23:629–635. doi: 10.1128/MCB.23.2.629-635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Humphrey EL, Shamji AF, Bernstein BE, Schreiber SL. Rpd3p relocation mediates a transcriptional response to rapamycin in yeast. Chem. Biol. 2004;11:295–299. doi: 10.1016/j.chembiol.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 26.Dembla-Rajpal N, Seipelt R, Wang Q, Rymond BC. Proteasome inhibition alters the transcription of multiple yeast genes. Biochim. Biophys. Acta. 2004;1680:34–45. doi: 10.1016/j.bbaexp.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 27.Fleming JA, Lightcap ES, Sadis S, Thoroddsen V, Bulawa CE, Blackman RK. Complementary whole-genome technologies reveal the cellular response to proteasome inhibition by PS-341. Proc. Natl Acad. Sci. USA. 2002;99:1461–1466. doi: 10.1073/pnas.032516399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikder D, Johnston SA, Kodadek T. Widespread, but nonidentical, association of proteasomal 19 and 20S proteins with yeast chromatin. J. Biol. Chem. 2006;281:27346–27355. doi: 10.1074/jbc.M604706200. [DOI] [PubMed] [Google Scholar]
- 29.Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Mol. Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 30.Braun BC, Glickman M, Kraft R, Dahlmann B, Kloetzel PM, Finley D, Schmidt M. The base of the proteasome regulatory particle exhibits chaperone-like activity. Nat. Cell Biol. 1999;1:221–226. doi: 10.1038/12043. [DOI] [PubMed] [Google Scholar]
- 31.Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol. Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Finley D, Sadis S, Monia BP, Boucher P, Ecker DJ, Crooke ST, Chau V. Inhibition of proteolysis and cell cycle progression in a multiubiquitination-deficient yeast mutant. Mol. Cell. Biol. 1994;14:5501–5509. doi: 10.1128/mcb.14.8.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang M, Cheng D, Peng J, Pickart CM. Molecular determinants of polyubiquitin linkage selection by an HECT ubiquitin ligase. EMBO J. 2006;25:1710–1719. doi: 10.1038/sj.emboj.7601061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhaumik SR, Malik S. Diverse regulatory mechanisms of eukaryotic transcriptional activation by the proteasome complex. Crit. Rev. Biochem. Mol. Biol. 2008;43:419–433. doi: 10.1080/10409230802605914. [DOI] [PubMed] [Google Scholar]
- 35.Lee D, Ezhkova E, Li B, Pattenden SG, Tansey WP, Workman JL. The proteasome regulatory particle alters the SAGA coactivator to enhance its interactions with transcriptional activators. Cell. 2005;123:423–436. doi: 10.1016/j.cell.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 36.Malik S, Shukla A, Sen P, Bhaumik SR. The 19S proteasome subcomplex establishes a specific protein interaction network at the promoter for stimulated transcriptional initiation in vivo. J. Biol. Chem. 2009;284:35714–35724. doi: 10.1074/jbc.M109.035709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pingle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 38.Russell SJ, Johnston SA. Evidence that proteolysis of Gal4 cannot explain the transcriptional effects of proteasome ATPase mutations. J. Biol. Chem. 2001;276:9825–9831. doi: 10.1074/jbc.M010889200. [DOI] [PubMed] [Google Scholar]
- 39.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol. Cell. Biol. 1999;19:2515–2526. doi: 10.1128/mcb.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts SM, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhaumik SR, Green MR. Interaction of Gal4p with components of transcription machinery in vivo. Methods Enzymol. 2003;370:445–454. doi: 10.1016/S0076-6879(03)70038-X. [DOI] [PubMed] [Google Scholar]
- 42.Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson CL, Kruger W, Herskowitz I. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell. 1991;64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 44.Shukla A, Durairaj G, Schneider J, Duan Z, Shadle T, Bhaumik SR. Stimulation of mRNA export by an F-box protein, Mdm30p, in vivo. J. Mol. Biol. 2009;389:238–247. doi: 10.1016/j.jmb.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 45.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 2001. [Google Scholar]
- 46.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston SA. The 19S regulatory particle of the proteasome is required for efficient transcription elongation by RNA polymerase II. Mol. Cell. 2001;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2001;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 48.Sun L, Johnston SA, Kodadek T. Physical association of the APIS complex and general transcription factors. Biochem. Biophys. Res. Commun. 2002;296:991–999. doi: 10.1016/s0006-291x(02)02026-0. [DOI] [PubMed] [Google Scholar]
- 49.Isono E, Nishihara K, Saeki Y, Yashiroda H, Kamata N, Ge L, Ueda T, Kikuchi Y, Tanaka K, Nakano A, et al. The assembly pathway of the 19S regulatory particle of the yeast 26S proteasome. Mol. Biol. Cell. 2007;18:569–580. doi: 10.1091/mbc.E06-07-0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lascaris RF, Mager WH, Planta RJ. DNA-binding requirements of the yeast protein Rap1p as selected in silico from ribosomal protein gene promoter sequences. Bioinformatics. 1999;15:267–277. doi: 10.1093/bioinformatics/15.4.267. [DOI] [PubMed] [Google Scholar]
- 51.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 52.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr. Opin. Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 53.Garbett KA, Tripathi MK, Cencki B, Layer JH, Weil PA. Yeast TFIID serves as a coactivator for Rap1p by direct protein–protein interaction. Mol. Cell. Biol. 2007;27:297–311. doi: 10.1128/MCB.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Layer JH, Miller SG, Weil PA. Direct transactivator-transcription factor IID (TFIID) contacts drive yeast ribosomal protein gene transcription. J. Biol. Chem. 2010;285:15489–15499. doi: 10.1074/jbc.M110.104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reid JL, Iyer VR, Brown PO, Struhl K. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol. Cell. 2000;6:1297–1307. doi: 10.1016/s1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 56.Knutson BA, Hahn S. Domains of Tra1 important for activator recruitment and transcription coactivator functions of SAGA and NuA4 complexes. Mol. Cell. Biol. 2011;31:818–31. doi: 10.1128/MCB.00687-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grant PA, Duggan L, Cote J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 58.Allard S, Utley RT, Savard J, Clarke A, Grant P, Brandl CJ, Pillus L, Workman JL, Cote J. NuA4, an essential transcription adaptor/histone H4 acetyltransferase complex con-taining Esa1p and the ATF-related cofactor Tra1p. EMBO J. 1999;18:5108–5119. doi: 10.1093/emboj/18.18.5108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ginsburg DS, Govind CK, Hinnebusch AG. NuA4 lysine acetyltransferase Esa1 is targeted to coding regions and stimulates transcription elongation with Gcn5. Mol. Cell. Biol. 2009;29:6473–6487. doi: 10.1128/MCB.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Utley RT, Ikeda K, Grant PA, Côté J, Steger DJ, Eberharter A, John S, Workman JL. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 61.Park S, Roelofs J, Kim W, Robert J, Schmidt M, Gygi SP, Finley D. Hexameric assembly of the proteasomal ATPases is templated through their C termini. Nature. 2009;459:866–870. doi: 10.1038/nature08065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lipford JR, Smith GT, Chi Y, Deshaies RJ. A putative stimulatory role for activator turnover in gene expression. Nature. 2005;438:113–116. doi: 10.1038/nature04098. [DOI] [PubMed] [Google Scholar]
- 63.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Identification of a cellular protein that specifically interacts with the essential cysteine region of the HIV-1 Tat transactivator. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 64.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 65.Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC. The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol. Cell. Biol. 2000;20:312–318. doi: 10.1128/mcb.20.1.312-318.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophilia. Mol. Cel. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 67.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 68.Gupta A, Sharma GG, Young CS, Agarwal M, Smith ER, Paull TT, Lucchesi JC, Khanna KK, Ludwig T, Pandita TK. Involvement of human MOF in ATM function. Mol. Cell. Biol. 2005;25:5292–5305. doi: 10.1128/MCB.25.12.5292-5305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gupta A, Guerin–Peyrou TG, Sharma GG, Park C, Agarwal M, Ganju RK, Pandita S, Choi K, Sukumar S, Pandita RK, et al. The mammalian ortholog of Drosophila MOF that acetylates histone H4 lysine 16 is essential for embryogenesis and oncogenesis. Mol. Cell. Biol. 2008;28:397–409. doi: 10.1128/MCB.01045-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharma GG, So S, Gupta A, Kumar R, Cayrou C, Avvakumov N, Bhadra U, Pandita RK, Porteus MH, Chen DJ, et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell. Biol. 2010;30:3582–3595. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koues OI, Dudley RK, Truax AD, Gerhardt D, Bhat KP, McNeal S, Greer SF. Regulation of acetylation at the major histocompatibility complex class II proximal promoter by the 19S proteasomal ATPase Sug1. Mol. Cell. Biol. 2008;28:5837–5850. doi: 10.1128/MCB.00535-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Truax AD, Koues OI, Mentel MK, Greer SF. The 19S ATPase S6a (S6'/TBP1) regulates the transcription initiation of class II transactivator. J. Mol. Biol. 2010;395:254–269. doi: 10.1016/j.jmb.2009.10.035. [DOI] [PubMed] [Google Scholar]
- 73.Altaf M, Auger A, Monnet-Saksouk J, Brodeur J, Piquet S, Cramet M, Bouchard N, Lacoste N, Utley RT, Gaudreau L, et al. NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 2010;285:15966–15977. doi: 10.1074/jbc.M110.117069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Durant M, Pugh BF. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell. Biol. 2007;27:5327–5335. doi: 10.1128/MCB.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAF(II)250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 76.Ladurner AG, Inouye C, Jain R, Tjian R. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell. 2003;11:365–376. doi: 10.1016/s1097-2765(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 77.Matangkasombut O, Buratowski S. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell. 2003;11:353–363. doi: 10.1016/s1097-2765(03)00033-9. [DOI] [PubMed] [Google Scholar]
- 78.Pamblanco M, Poveda A, Sendra R, Rodriguez-Navarro S, Perez-Ortin JE, Tordera V. Bromodomain factor 1 (Bdf1) protein interacts with histones. FEBS Lett. 2001;496:31–35. doi: 10.1016/s0014-5793(01)02397-3. [DOI] [PubMed] [Google Scholar]
- 79.Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- 80.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 82.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 83.Altaf M, Auger A, Covic M, Côté J. Connection between histone H2A variants and chromatin remodeling complexes. Biochem. Cell. Biol. 2009;87:35–50. doi: 10.1139/O08-140. [DOI] [PubMed] [Google Scholar]
- 84.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 86.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li B, Pattenden SG, Lee D, Gutiérrez J, Chen J, Seidel C, Gerton J, Workman JL. Preferential occupancy of histone variant H2AZ at inactive promoters influences local histone modifications and chromatin remodeling. Proc. Natl Acad. Sci. USA. 2005;102:18385–18390. doi: 10.1073/pnas.0507975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 90.Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gévry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–733. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 95.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ishibashi T, Dryhurst D, Rose KL, Shabanowitz J, Hunt DF, Ausió J. Acetylation of vertebrate H2A.Z and its effect on the structure of the nucleosome. Biochemistry. 2009;48:5007–5017. doi: 10.1021/bi900196c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hall DB, Struhl K. The VP16 activation domain interacts with multiple transcriptional components as determined by protein-protein crosslinking in vivo. J. Biol. Chem. 2002;277:46043–46050. doi: 10.1074/jbc.M208911200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.