Abstract
The human prolyl-4-hydroxylase domain (PHD) proteins 1–3 are known as cellular oxygen sensors, acting via the degradation of hypoxia-inducible factor (HIF) α-subunits. PHD2 and PHD3 genes are inducible by HIFs themselves, suggesting a negative feedback loop that involves PHD abundance. To identify novel regulators of the PHD2 gene, an expression array of 704 transcription factors was screened by a method that allows distinguishing between HIF-dependent and HIF-independent promoter regulation. Among others, the E-twenty six transcription factor ETS translocation variant 4 (ETV4) was found to contribute to PHD2 gene expression particularly under hypoxic conditions. Mechanistically, complex formation between ETV4 and HIF-1/2α was observed by mammalian two-hybrid and fluorescence resonance energy transfer analysis. HIF-1α domain mapping, CITED2 overexpression and factor inhibiting HIF depletion experiments provided evidence for cooperation between HIF-1α and p300/CBP in ETV4 binding. Chromatin immunoprecipitation confirmed ETV4 and HIF-1α corecruitment to the PHD2 promoter. Of 608 hypoxically induced transcripts found by genome-wide expression profiling, 7.7% required ETV4 for efficient hypoxic induction, suggesting a broad role of ETV4 in hypoxic gene regulation. Endogenous ETV4 highly correlated with PHD2, HIF-1/2α and several established markers of tissue hypoxia in 282 human breast cancer tissue samples, corroborating a functional interplay between the ETV4 and HIF pathways.
INTRODUCTION
Cellular adaptation to a shortage of oxygen is mainly governed by transcriptional regulation. Hypoxia-inducible factors (HIFs) are key players in the hypoxic cell and orchestrate the expression of hundreds of downstream target genes, adapting the cellular metabolism to a low oxygen environment (1). Heterodimeric HIFs consist of a tightly O2-regulated α-subunit (HIF-1α, HIF-2α or HIF-3α) and a constitutively expressed β-subunit (HIF-1β). At oxic conditions, HIF α-subunits are continuously marked for proteasomal degradation through hydroxylation of two key prolyl-residues by prolyl-4-hydroxylase domain (PHD) oxygen sensor proteins (2). PHD hydroxylation activity fades as a direct function of oxygen, thus reciprocally controlling the nuclear accumulation of HIFαs. Stabilized HIF-complexes bind to a cis-acting HIF binding site (HBS) conformed by a highly conserved core 5′-RCGTG-3′ motif present in all direct target genes (1,3,4). In analogy to destabilizing proline hydroxylation, the transcriptional activity of HIF is tuned by factor inhibiting HIF (FIH), which hydroxylates a distinct asparagine residue within the HIF-α carboxy terminus and consequently hinders its association with the transcriptional 300-kilodalton coactivator protein (p300) and cAMP response element-binding protein (CREB) binding protein (CBP) (5–7).
Among the three characterized HIF prolyl-4-hydroxylases, PHD2 is widely accepted as the most crucial isoform controlling basal activity of the HIF pathway in oxic cells (8). Underlining its dominant role, disruption of the Egln1 locus, encoding mouse PHD2, results in prenatal lethality, while PHD1 and PHD3 knock out mice are born normally (9). Broad-spectrum conditional deletions of all three PHDs in mice revealed a global hyperproliferative vascular phenotype uniquely when targeting PHD2, demonstrating an absolute requirement for PHD2, which is not confined to embryonic development (10). Accordingly, PHD2 abundance is considered as a critical factor in tumor angiogenesis, although divergent roles of stromal and tumor cell-derived PHD2 have been discussed (11–13). As PHD2 protein is strikingly stable and the de novo translated enzyme outlasts a period of transient hypoxia by more than 48 h, transcriptional regulation of the PHD2 locus must be considered as the main process defining cellular levels of PHD2 (14,15). Expression of PHD2 itself is delicately influenced by HIF transcriptional activity, forming a negative feedback loop which facilitates dynamic oxygen sensing (16–18).
To identify upstream regulatory pathways affecting PHD2 gene expression in an unbiased system, we developed a screening approach that allows the identification of transcriptional interactions with DNA-bound HIF complexes and HIF-independent promoter regulation at the same time. The herein described synthetic transactivation screening led to the identification of several members of the E-twenty six (ETS) and FBJ murine osteosarcoma viral oncogene homolog (FOS) families of transcription factors as novel activators of the human PHD2 promoter (P2P). Among those, ETS translocation variant 4 (ETV4; also known as E1A enhancer binding protein, E1AF, or polyoma-enhancing activator 3, PEA3), was found to be a potent coactivator of HIF-1-dependent transcription.
MATERIALS AND METHODS
Cell culture
Human HeLa cervix carcinoma and U2OS osteosarcoma cells were grown in Dulbecco's modified Eagle's medium (DMEM, Sigma). Human PC3 prostate cancer cells were cultured in Roswell Park Memorial Institute medium (RPMI-1640, Sigma). Media were supplemented with 10% fetal calf serum (FCS) and antibiotics (penicillin 50 IU/ml and streptomycin 100 µg/ml; Gibco-BRL). Hypoxic cell culture was carried out at 0.2% O2 (if not indicated differently) using a gas-controlled InvivO2 400 workstation (Ruskinn Technologies). Transfections were performed using polyethyleneimine (PEI; Polysciences) as described earlier (17).
P2P constructs
P2P constructs containing the wild-type and mutant HBS in the pGL3basic luciferase vector (Promega) were generated in earlier work (16). Serial 5′-truncations of P2P and a start codon fusion to the luciferase open reading frame (ORF) were used for both promoter versions using standard cloning techniques. Within the scope of the screening approach, the firefly reporter gene of pGL-P2P(−607/+3) variants was replaced with the renilla luciferase ORF cloned into NcoI and XbaI sites.
Transfection and synthetic transactivation screening
Reverse transfection (19) and screening were carried out using an arrayed expression library containing 704 transcriptionally relevant human full-length cDNAs from the Origene collection (FTCW 19603, GFC-transfection array in a 96-well format). An annotated list of all genes covered by this array is provided online by the manufacturer. Dried DNA (100 ng) of a distinct expression construct spotted per well was reconstituted at room temperature with 20 µl of serum-free medium containing a mixture of pGL-P2P(−607/+3) HBSwt firefly and pGL-P2P(−607/+3) HBSmut renilla reporter plasmids (100 ng DNA/each). Subsequently, 20 µl of diluted TransIT-LT1 transfection reagent (3:1, µg DNA/ µl TransIT-LT1; Mirus Bio LLC) was added and complex formation was allowed for 30 min at room temperature before 60 µl of a cell suspension containing 1 × 104 U2OS cells in DMEM supplemented with 10% FCS were plated in each well. Plates were incubated at 20% O2 for 24 h before being subjected to the screening conditions of 0.2% O2 for an additional 24 h. Cultures were lysed in 20 µl of passive lysis buffer (Promega) and luminescence was immediately analyzed with a microplate luminometer (Berthold) using a standard dual luciferase reporter assay system (Promega). Luciferase activities were normalized to the median calculated individually for each plate and luminescence source and expressed as induction factors (IF) according to the equation 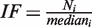 , with Ni indicating individual luciferase activity value of each well of plate i and mediani indicating median of luciferase activities of all 96-wells on plate i. To compare the distribution of replicate assays a standard z-score evaluation was performed following the equation
, with Ni indicating individual luciferase activity value of each well of plate i and mediani indicating median of luciferase activities of all 96-wells on plate i. To compare the distribution of replicate assays a standard z-score evaluation was performed following the equation 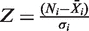 , with
, with 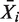 , plate mean of respective luminescence values and σi, standard deviation (SD) of plate mean of luminescence values.
, plate mean of respective luminescence values and σi, standard deviation (SD) of plate mean of luminescence values.
Reporter gene assays and mammalian two-hybrid analyses
Construction of pGLTfHRE wt and pGLTfHRE mut reporter plasmids carrying a hypoxia-responsive enhancer element derived from the human Transferrin gene was described previously (20). Transfections for standard reporter gene experiments were carried out on 100 mm culture plates essentially as described earlier (17). In brief, U2OS cells were cotransfected with 3 µg reporter plasmid or a mix of 1.5 µg reporter and 1.5 µg expression plasmids, respectively. Transfection efficiency was controlled by cotransfection of 20 ng pRLSV40 renilla luciferase reporter vector (Promega). RNA-interference (RNAi) mediated knock down of HIF-1α, FIH-1 or p300 was achieved by transiently transfecting once (HIF-1α, p300) or twice (FIH-1) U2OS cells with 100 nM stealth RNAi duplexes (HIF-1α, 5′-caggacaguacaggaugcuugccaa-3′; FIH-1, 5′-gaaacauugagaagaugcuuggaga-3′; p300, 5′-ggauucgucugugauggcuguuuaa-3′; control, 5′-gcuccggagaacuaccagaguauua-3′; sense strands) using Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen). Combined knock down and reporter gene analyses were performed by sequentially transfecting cells with stealth RNAi duplexes 24 h before subjecting them to polyethyleneimine (PEI)-mediated DNA transfection.
Mammalian two-hybrid analyses were performed using the mammalian Matchmaker system (Clontech) as described previously (14). Expression vectors of HIF-1α NAD and carboxy-terminal activation domain (CAD) fused to Gal4 DBD (21) were a kind gift of Dr Sang (Drexel University, Philadelphia, PA, USA). U2OS cells were transiently cotransfected with 1.5 µg of Gal4 DBD and 1.5 μg of VP16 AD fusion protein vectors together with 500 ng of firefly luciferase reporter vector pGRE5xE1b and 20 ng of pRL-SV40. Total transfected DNA amounts were equalized in each experiment using the corresponding empty vector. Luciferase reporter gene activities were determined using the dual-luciferase reporter assay system (Promega).
Fluorescence resonance energy transfer
The full-length ORF of human ETV4 was cloned into pENTR4 and subsequently recombined with pECFP-C1-DEST (15) to obtain the expression vector for a cyan fluorescent ETV4 fusion protein. U2OS cells were transiently transfected with the pECFP-ETV4 and pEYFP-HIF1 or pEYFP-HIF2 plasmids as recently described (22). Fluorescence resonance energy transfer (FRET) was monitored under normoxic or hypoxic conditions (1% O2 for 4 h) 24–48 h post-transfection.
Protein extraction and immunoblot analysis
Cells were washed twice and scraped into ice-cold phosphate-buffered saline. Soluble cellular protein was extracted with a high salt extraction buffer containing 0.4 M NaCl, 0.1% Nonidet P-40, 10 mM Tris–HCl (pH 8.0), 1 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride and 1× protease inhibitor cocktail (Sigma). Protein concentrations were determined by the Bradford method and 50–80 µg of cellular protein were subjected to immunoblot analysis using the following antibodies: mouse monoclonal antibody (mAb), anti-human HIF-1α (clone 54/HIF-1α; BD Transduction Laboratories), mAb anti-ETV4 [PEA3 (16); Santa Cruz Biotechnology], rabbit anti-ETV4 (sdix20580002; Novus Biologicals), rabbit anti-human PHD2 (NB100-137; Novus Biologicals), mAb anti-FIH-1 (NBP1-30333; Novus Biologicals), mAb anti-p300 (554215; BD Pharmingen) and mAb anti-β-actin (clone AC-74; Sigma). Primary antibodies were detected with respective polyclonal anti-mouse or anti-rabbit sera conjugated to horseradish peroxidase (Pierce). Chemiluminescence signals were developed using Supersignal West Dura substrate (Pierce) and images were acquired with a digital light imaging system (LAS 4000; Fuji).
mRNA quantification
Complementary DNA was generated by reverse transcription (RT) of 1–5 µg of total RNA using AffinityScript reverse transcriptase (Agilent). Transcript levels were determined by real-time quantitative (q) polymerase chain reaction (PCR) using a SybrGreen qPCR reagent kit (Sigma) in combination with the MX3000P light cycler (Agilent). All RT-qPCR data are presented as ratios relative to ribosomal protein L28 mRNA values. Primer sets for human PHD2, CA9 and L28 have been described earlier (17,23).
TMA analysis
Clinicopathological characterization and immunohistochemical analyses of selected components of the HIF-pathway using a tissue microarray (TMA) consisting of 282 invasive breast cancer cases diagnosed at the Institute of Surgical Pathology (University Hospital, Zürich, Switzerland) have been described recently (13). Sections of the same TMA were stained with a rabbit polyclonal anti-ETV4 antibody (HPA005768, Sigma) in a 1:100 dilution using an automated immunohistochemistry platform (Ventana BenchMark, Roche). An immunoreactive score for ETV4 staining was calculated by multiplication of staining intensity (graded between 0 and 3) and the percentage of positive cells (graded between 0 and 4 with 0, nil; 1, <10%; 2, 10–50%; 3, 51–80%; 4, >80%) as quantified by a senior pathologist (G.K.). Nonparametric correlations between ETV4 expression and HIF-1/2α or HIF target genes were analyzed by calculating Spearman's rank correlation coefficient using predictive analysis software (IBM SPSS Statistics 18).
Short hairpin RNA constructs and lentiviral infections
Expression vectors encoding short hairpin RNA (shRNA) sequences targeting human ETV4 and a noncoding control driven by the U6 promoter in a pLKO.1-puro plasmid were purchased from Sigma. Viral particles were produced in HEK293T cells using the ViraPower lentiviral expression system according to the manufacturer's instructions (Invitrogen). Infected PC3 cells were cultured in RPMI-1640 supplemented with 0.5 μg/ml puromycin.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) assays from parental PC3 cells exposed to 20% or 0.2% O2 for 4 and 24 h were performed essentially as described previously (24). The following antibodies were used for immunoprecipitation: rabbit anti-HIF-1α (ab2185; Abcam) and rabbit anti-ETV4 (sdix20580002; Novus Biologicals). Rabbit serum (011-000-001; Jackson ImmunoResearch) served as unspecific control. Enrichment of P2P chromatin was determined by PCR using the following oligonucleotides: PHD2 forward 5′-gtatgccctgcgctcctc-3′, reverse 5′-gctgagagaatagggcctgtg-3′.
Gene array analysis
Total RNA was extracted from pools of shRNA expressing PC3 clones with RNeasy (Qiagen). RNA integrity was evaluated using the Agilent 2100 Bioanalyzer. Genome-wide RNA levels were quantified using the human gene expression ShurePrint GE3 (8 × 60 K) microarray according to the manufacturer's instructions (Agilent). All data were deposited in NCBI's Gene Expression Omnibus (GEO) and are publicly accessible through GEO accession number GSE32385 (http://www.ncbi.nlm.nih.gov/geo/).
Statistical analysis
If not otherwise indicated, results are presented as mean values ± standard error of the mean of at least three independent experiments. Column statistics applying paired Student's t-tests were calculated using GraphPad Prism version 4.0 (GraphPad Software).
RESULTS
A single HBS is sufficient for hypoxic induction of the human P2P but dispensable for basal promoter activity
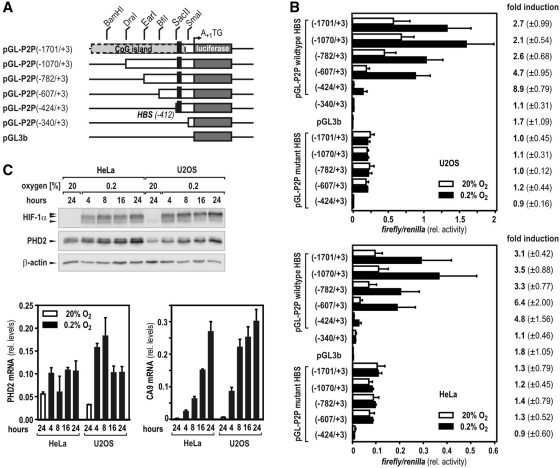
Previously, we have reported on a functional, single HIF-binding site located in the 5′-regulatory region of the human PHD2 gene (16). Using a series of 5′-truncated luciferase reporter genes (schematically depicted in Figure 1A), the minimal hypoxia-responsive region of the P2P was further mapped to an element spanning nucleotides −424 to +3 relative to the translational start site. While pGL-P2P(−424/+3) still revealed high hypoxic inducibility when transiently transfected into U2OS or HeLa cells, normoxic promoter activity of this region was largely lost when compared with the longer promoter variants, regardless of whether the HBS was wild-type or mutant (Figure 1B). Elongation of the promoter by at least 183 nts [construct pGL-P2P(−607/+3)] showed robust normoxic activity in both cell lines (Figure 1B). As basal induction of HBS-lacking constructs was observed between nucleotides −424 and −607 without any further effects upstream, the existence of transcriptionally active elements required for oxic expression of the PHD2 gene in this particular region is suggested (Figure 1B). Of note, the fold of hypoxic activation of pGL-P2P(−607/+3) resembled the oxygen-dependent IF of endogenous PHD2 mRNA and protein in both cell lines (Figure 1C).
Figure 1.
Identification of the minimal human P2P. (A) Schematic representation of P2P 5′-truncations and their cloning strategy as used in this study. The translational start site is designated ‘+1’. (B) Regulatory DNA regions of the human PHD2 gene were cloned into luciferase reporter vectors that were transiently transfected into human U2OS osteosarcoma cells. One day after transfection, cells were incubated for 24 h at 20 or 0.2% O2. Hypoxic IF (mean values ± SD) of relative luciferase activities were calculated from three independent experiments performed in triplicates. Mutation of a single HBS (black rectangles in A) completely abrogated hypoxic inducibility of all constructs. (C) HeLa and U2OS cells were incubated at 20 or 0.2% O2 for 4–24 h and protein levels of HIF-1α, PHD2 and β-actin were analyzed by immunoblotting. Total RNA was isolated from cultures treated as in (B) and mRNA levels of PHD2 and CA9 were determined by RT-qPCR. Transcript levels of CA9 served as positive control to confirm continuous hypoxic responses. Gene expression levels were expressed in relation to ribosomal L28 mRNA (rel. levels) calculated from three independent experiments (±SD).
A differential screening approach to identify site-specific transcription factor interplay
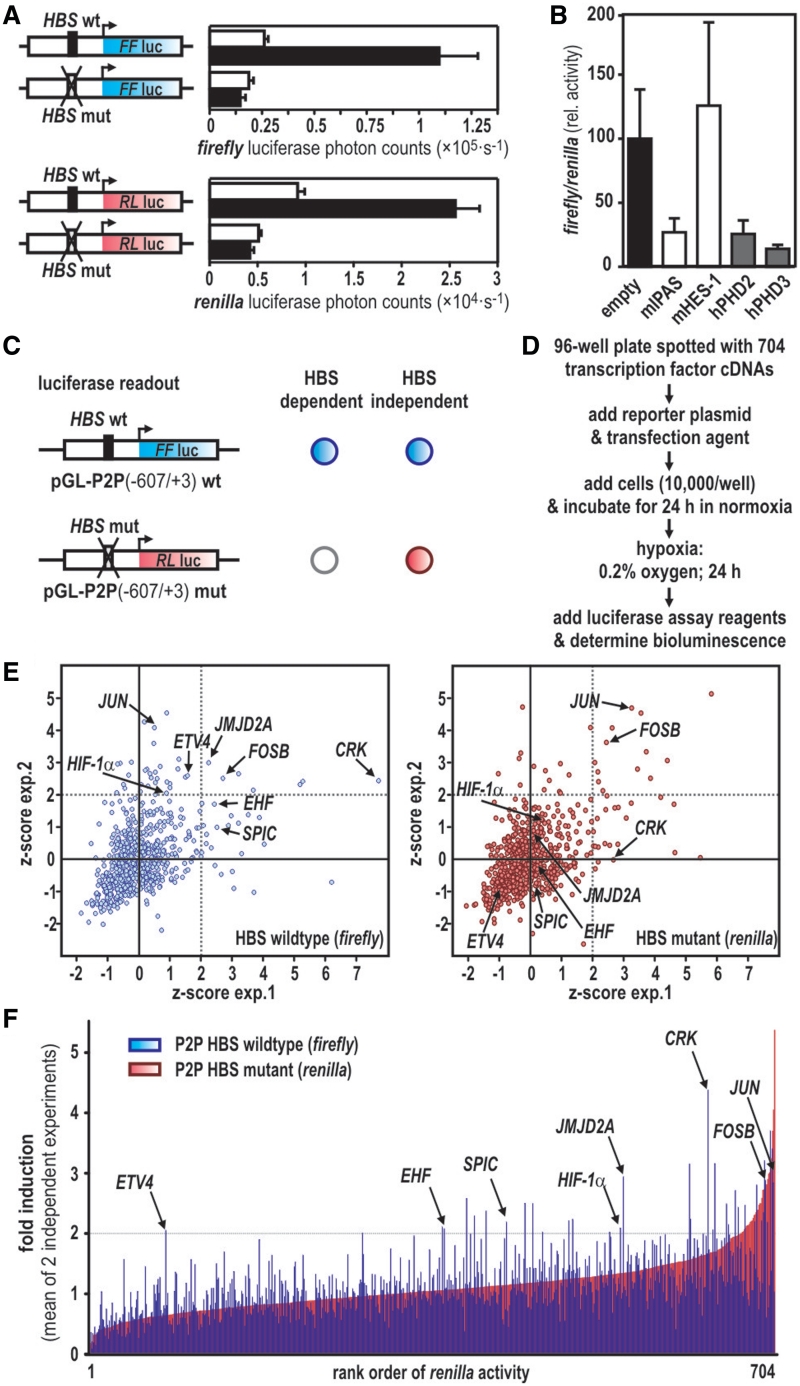
Multiplexed transfection of firefly and renilla reporter genes controlled by either wild-type or HBS-mutant P2Ps, respectively, allows to classify any reporter-modulating event as HIF-dependent or self-sufficient. Moreover, the dual reporter system provides a read-out that permits to screen under HIF-stabilizing conditions (e.g. hypoxia) while simultaneously assessing the noninduced background levels (simulated ‘normoxia’) in the very same cells, thereby reducing intra-assay variabilities and screening complexity. Nucleotides −607 to +3 of the P2P containing either the wild-type or mutant HBS were used to drive transcription of firefly or renilla luciferase cDNAs, respectively. The two luciferase reporter genes showed identical hypoxic responses in the respective reporter vectors (Figure 2A). As a proof of principle, this system was tested by coexpressing mIpas, encoding an inhibitory isoform of HIF-α (25), with the two reporter plasmids as described above. Indeed, a marked downregulation of hypoxic firefly versus renilla luciferase activity was observed when compared with cotransfection with the empty vector or an unrelated transcription factor (mHes-1). As expected (17), forced expression of PHD2 or PHD3 strongly attenuated the hypoxic activation of the reporter system, confirming that also post-translational mechanisms impairing the activation state of the HIF-pathway can be assessed by this method (Figure 2B).
Figure 2.
Synthetic transactivation screening allows to discriminate HBS-dependent and independent promoter activation. (A) P2P(−607/+3) driven firefly (blue) and renilla (red) luciferase reporter genes transfected into U2OS cells show similar hypoxic activation (0.2% O2 for 24 h, black bars) when compared with normoxic (20% O2, open bars) cultures. HBS mutation completely abrogated hypoxic induction of both constructs. (B) Coexpression of transcription factors (mIpas, mHes-1) or PHD enzymes with P2P(−607/+3)wt HBS driven firefly and P2P(−607/+3)mut HBS-driven renilla luciferase reporters, respectively. Dual luciferase activities were determined after 24 h of hypoxic exposure (0.2% O2) and expressed as relative luciferase activity normalized to controls receiving the empty expression vector. (C) Schematic overview of the P2P-luciferase constructs used for synthetic transactivation screening. Expected readouts for HBS-dependent (firefly reporter activation only, blue circle) and independent reporter gene activation (simultaneous activation of firefly and renilla reporter genes indicated by blue and red circles, respectively) are illustrated by a cross table. (D) Workflow of synthetic transactivation screening analyzing a commercial transcription factor expression library. (E) Z-scores of two independent screening experiments were plotted for both reporters. Thresholds for z > 2 are indicated by dotted lines. (F) Paired values for individual coexpressed cDNAs for firefly (blue) and renilla (red) read-outs are depicted. A fold of induction >2 was considered as lead for a re-screening. Positions of HIF-1α and seven new transcription factors positively re-evaluated at a secondary screening level are indicated. Data are given as the mean of two independent screening experiments. For calculations and full gene names refer to the text.
Members of the FOS and ETS transcription factor families transactivate the human P2P
An array of 704 cDNA expression vectors representing all commonly known transcription factor families was screened using the HBS-specific synthetic transactivation readout. Possible HIF coregulators alter firefly but not renilla luciferase expression, whereas HIF-independent factors interfere with both reporters (schematically depicted in Figure 2C). A work flow of the transfection and screening procedure is given in Figure 2D. Two independent screens were performed and reproducibility was visualized by plotting z-scores (Figure 2E). Solid correlations were observed for the vast majority of the coexpressed transcription factors, confirming that no significant deviation between the two experiments occurred. Reporter gene inductions by each coexpressed transcription factor were calculated as multiples of the respective 96-well plate median individually for firefly and renilla activities, averaged over the two experiments, and ranked according to the IF of the HBS-mutant renilla reporter (Figure 2F). The cut-off was defined as an increase in luciferase activity by a factor of at least 2, tolerating a deviation of 0.05. Factors that showed a reproducible increase in either firefly luminescence (Supplementary Table S1A) or firefly and renilla luminescence together (Supplementary Table S1B) were considered as leads. Underlining the validity of this approach, the only known transcriptional activator of PHD2 expression, namely HIF-1α, was among the cDNAs identified (Figure 2E and F). For reevaluation, the cDNAs of 43 leads were retrieved and partially sequenced to verify their identities. Seven of these cDNAs, namely ETS-variant 4 (ETV4), Spi-C transcription factor (SPIC), ETS homologous factor (EHF), the proto-oncogenes JUN and FOSB, v-crk sarcoma virus CT10 oncogene homolog (CRK) and Jumonji domain-containing protein 2A resulted in a reproducible HBS-dependent regulation of the P2P. Most of these factors can be attributed to two major groups: the ETS (ETV4, EHF and SPIC) and the FOS (JUN and FOSB) families.
ETV4 activates PHD2 and transferrin promoters synergistically with HIF-1
Expression vectors of the seven newly identified factors were cotransfected together with pGL-P2P(−607/+3) HBS wild-type or mutant reporter genes, cloned into identical backbones to exclude false positive effects, which may have resulted from the two different luciferase cDNAs in the original screen. In the presence of a functional HBS, coexpression of ETV4 resulted in a striking super-induction of the P2P under hypoxic conditions only (Figure 3A, left panel), which was fully lost with the mutant construct (Figure 3A, right panel). As ETV4 was the strongest hit identified, subsequent work focused on the role of this transcription factor as a putative transactivator of PHD2. Of note, the P2P region used in this screen lacks a consensus 5′-A/CGGAAGT-3′ ETV4 binding site (26). Thus, direct binding of ETV4 to P2P appears unlikely, although not fully excluded regarding the small residual stimulation of constitutive P2P activity following HBS mutation. However, the complete lack of hypoxic P2P stimulation by ETV4 suggests that HIF-1 might actually recruit ETV4 to enhance hypoxic induction. Accordingly, knock down of HIF-1α by RNA interference in U2OS cells abrogated hypoxia and ETV4-mediated induction of the wild-type P2P (Figure 3B, right panel), while an unrelated control siRNA had no effect (Figure 3B, left panel). A heterologous reporter construct driven by the minimal SV40 promoter in conjunction with either a wild-type (Figure 3C, left panel) or mutant (Figure 3C, right panel) hypoxia response element derived from the human Transferrin gene (20) recapitulated the strong hypoxic superinduction by exogenous ETV4, proposing a more general model of synergistic interaction between ETV4 and HIF-1. In line with our screening data, overexpressed ETV4 significantly upregulated endogenous PHD2 mRNA and protein levels in hypoxic U2OS cells, while PHD1 mRNA levels remained unaffected (Figure 3D and E).
Figure 3.
Hypoxic transactivation of the P2P by ETV4 requires HIF-1α activity. (A) Standard dual luciferase reporter gene assays of seven reevaluated hits from the transcription factor overexpression array. Wild-type (left panel) or HBS mutant (right panel) P2P regions controlling firefly luciferase reporter plasmids were cotransfected into U2OS cells together with expression constructs of the aforementioned factors. Transfection of an empty expression vector (empty) served as negative control and differences in transfection efficiency were controlled by cotransfecting SV40 promoter driven renilla luciferase. Cells were cultured at 20% or 0.2% oxygen for 24 h before dual luciferase activities were determined. (B) Transient RNAi mediated knock down of HIF-1α fully abrogated hypoxic activation of the P2P by ETV4. U2OS cells were transiently transfected with siRNA oligonucleotides targeting HIF-1α (siHIF1α, right panel) or a control sequence having no human target (siControl, left panel). Reporter gene experiments using the P2P reporter construct with only wild-type HBS were performed as described in (A). The inset shows an immunoblot confirming the robust knock down of HIF-1α in U2OS cells. (C) ETV4 and HIF-1 synergism in hypoxic gene activation is not restricted to the P2P. A heterologous hypoxia responsive reporter gene containing two functional HBS from the human Transferrin hypoxia response element (pGL-TfHRE wt) was tested in luciferase reporter assays as described in (A). Mutation of both HBS (pGL-TfHRE mut) caused an abrogation of the signal as seen in (A). (D and E) Forced expression of ETV4 in U2OS cells upregulates endogenous PHD2 protein and transcript levels. (D) Whole cell lysates were prepared from cells exposed for 16 h to 20 or 0.2% oxygen and analyzed for HIF-1α, ETV4, PHD2 and β-actin levels by immunoblotting. (E) Total RNA was extracted of similarly treated cells and mRNA levels of PHD1, PHD2 and L28 were quantified by RT-qPCR. Data are shown in relation to ribosomal L28 mRNA (rel. levels) calculated from three independent experiments (**P < 0.01, paired Student's t-test).
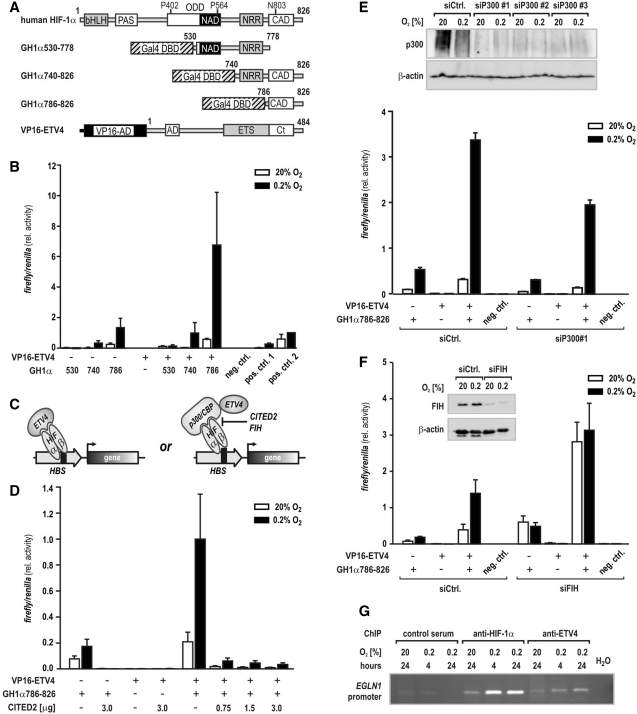
ETV4 interaction with the carboxy-terminal transactivation domain of HIF-1α depends on corecruitment of p300
Intrigued by the HIF-dependent ETV4 effects, we aimed for the characterization of a putative physical interaction between ETV4 and HIF-1α. Using a mammalian two-hybrid system, expression plasmids encoding for ETV4 fused to the activation domain (AD) of viral protein 16 (VP16-ETV4) were cotransfected with HIF-1α oxygen regulatory domains (21,27) fused to a Gal4-DNA binding domain (DBD; Figure 4A). Due to its intrinsic transactivation activity, constructs containing the CAD of HIF-1α [amino acids 775-826 (27)] were sufficient to activate the Gal4-responsive promoter (Figure 4B). Coexpression of ETV4 strikingly superinduced GH1α740-826 and GH1α786-826, particularly under hypoxic conditions, suggesting that HIF-1α CAD and ETV4 cooperate to transactivate target genes (Figure 4B). ETV4 effects on the aminoterminal activation domain (27) (NAD; HIF-1α amino acids 549-582) were negligible. Both, HIF-1α and ETV4 have been demonstrated to interact with the ubiquitous transcriptional coactivators p300/CBP (28,29). To address the question whether the two factors directly interact or whether a ternary complex between HIF-1, p300/CBP and ETV4 is formed (schematically depicted in Figure 4C), binding of HIF-1α CAD to p300 was disrupted by forced overexpression of CBP/p300-interacting transactivator 2 (CITED2), known to negatively regulate HIF function (30). Structural analyses revealed that CITED2 and HIF-1α share an overlapping binding interface in the p300 cysteine–histidine-rich 1 (CH1) domain and competition assays showed a 33-fold higher affinity of CITED2 for binding to p300 CH1 than a corresponding HIF-1α CAD peptide, indicating that CITED2 is a dominant inhibitor of HIF-1α:p300/CBP complex formation (31). HIF-1α CAD:ETV4 interplay was totally abrogated by CITED2 in mammalian two-hybrid experiments (Figure 4D), underscoring the assumption that ETV4 coactivation of HIF-1 requires functional interaction of the latter with p300/CBP.
Figure 4.
Transcriptional cooperation between ETV4 and HIF-1 is disrupted by CITED2. (A) Schematic representation of HIF-1α and ETV4 domain structure and fusion constructs used in mammalian two-hybrid assays. PAS, PER-ARNT-SIM; bHLH, basic helix–loop–helix domain; ODD, oxygen-dependent degradation domain; NRR, negative regulatory region; NAD and CAD, amino-carboxy-terminal activation domain and CADs, respectively. A GAL4-DBD was fused to regions encompassing the HIF-1α NAD and CAD. Full-length ETV4 bearing two activation domains (AD, acidic domain; Ct, carboxy-terminal tail) flanking a central ETS domain was fused to a VP16 activation domain (VP16-AD). Numbers indicate the amino acids present in the respective constructs. (B) U2OS cells were cotransfected with a Gal4-responsive reporter plasmid and Gal4-HIF-1α (GH1α) constructs alone or in combination with VP16-ETV4. The GH1α fusion constructs are specified by the aminoterminal starting amino acid of the truncated HIF-1α regions (530, 740 and 786, respectively). Following transfection, cells were evenly split and incubated at 20 or 0.2% O2 before luciferase activities were determined 24 h later. Noninteracting Gal4 DBD-p53 and VP16-AD-CP1 served as negative control (neg. ctrl.), while the interactions between Gal4 DBD-PHD2 and VP16-AD-HIF-2α(ODD) or VP16-AD-FKBP38 were used as positive controls (pos. ctrl. 1 and pos. ctrl. 2, respectively). (C) Scheme of the potential interactions between HIF-1, p300/CBP and ETV4 as assessed by mammalian two-hybrid assays. Both CITED2 and FIH can block the interaction between HIF-1α and p300/CBP. (D) Cotransfection of the indicated amounts of a CITED2 expression construct together with the mammalian two-hybrid expression vectors followed by hypoxic exposure and luciferase activity determination as described for (B). (E) Cotransfection of siRNA directed against p300 together with the mammalian two-hybrid expression vectors followed by hypoxic exposure and luciferase activity determination as described for (B). The p300 knock down efficiency of different siP300 oligonucleotides was analyzed by immunoblotting (upper panel) and siP300#1 was chosen for further experiments. (F) Cotransfection of siRNA directed against FIH together with the mammalian two-hybrid expression vectors followed by hypoxic exposure and luciferase activity determination as described for (B). The efficiency of the siFIH mediated FIH knock down was confirmed by immunoblotting as shown in the inset. (G) ChIP of normoxic or hypoxic PC3 cells using antibodies directed against HIF-1α or ETV4, or control serum. The amount of coprecipitated chromatin derived from the human P2P region (encoded by EGLN1) containing the HBS was determined by PCR followed by agarose gel electrophoresis.
While transient knock down of p300 slightly reduced the intrinsic transactivation activity of GH1α786-826 in mammalian two-hybrid experiments, robust superinduction of this construct occurred when VP16-ETV4 was cotransfected, irrespective of the presence of p300 (Figure 4E). Thus, we assume that both p300 and CBP can function as bridging molecules for HIF-1α CAD:ETV4 interplay. Underlining the essential requirement of p300/CBP for HIF-1α:ETV4 interaction, normoxic transactivation activity of cotransfected GH1α786-826 and VP16-ETV4 was similar to hypoxic activation levels in U2OS cells transiently depleted of FIH (Figure 4F). FIH has been described previously as oxygen-dependent negative regulator of HIF-α:p300/CBP interaction (7). ChIP experiments using either anti-HIF-1α or anti-ETV4 antibodies revealed oxygen-dependent enrichment of the HRE-containing PHD2 (EGLN1) promoter region in both precipitations (Figure 4G), providing further evidence for corecruitment of the two transcription factors to the endogenous PHD2 locus.
Both HIF-α isoforms are capable of forming a complex with ETV4
Fluorescence resonance energy transfer (FRET) analyses of coexpressed ETV4 and HIF-1α marked with cyan or yellow fluorescent protein tags (CFP and YFP, respectively) resulted in a robust energy transfer between both factors. Similar FRET efficiencies were observed when YFP-labeled HIF-2α was used together with CFP-ETV4 (Figure 5A and B). The intracellular distance of the two nuclear proteins was calculated to be 5.6–5.7 nm and did not differ in oxygenated or hypoxic cells, which might be explained by saturation of the HIF-α degradation pathways by exogenous overexpression of the transcription factors. Notably, efficient energy transfer between HIF-1α and p300 at ambient oxygen tensions has been reported previously (32).
Figure 5.
Both HIF-1α and HIF-2α colocalize with ETV4 to the nucleus within molecular proximity. U2OS cells were transiently transfected with the indicated CFP or YFP plasmids, and FRET analysis was performed at 20% O2 or 1% O2 24–48 h post-transfection. (A) Microscopic images showing the subcellular localization of the exogenous proteins. Fluorescence intensity of FRET signals is visualized by false colors on a color bar from low (blue) to high (white) intensity. (B) FRET efficiencies for CFP-ETV4 and YFP-HIF-1α (upper panel) or YFP-HIF-2α (lower panel) fusion protein pairs were calculated from 20 to 40 randomly selected cells which displayed various fluorescent acceptor/donor ratios. Scatter plots were fit to a single-site binding model. FRET efficiencies are given as the percentage of transferred energy relative to the energy absorbed by the donor.
A broad role for ETV4 in hypoxic gene expression
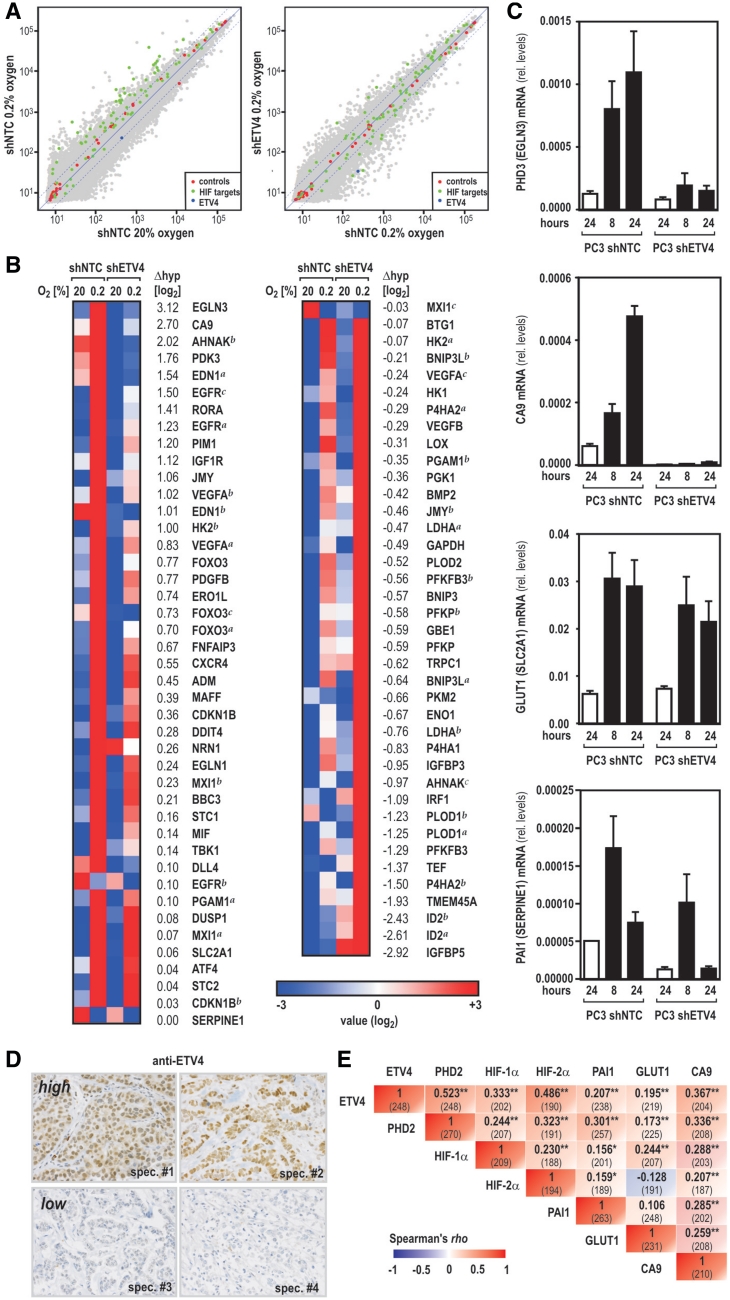
Recent work reported high ETV4 expression levels in the human prostate cancer cell line PC3 that lacks the constitutive photomorphogenic protein COP1 acting as E3 ubiquitin ligase for a variety of ETS proteins (33,34). As endogenous ETV4 expression levels in U2OS cells were close to the detection limit, the PC3 cell model was chosen to study the involvement of ETV4 in the hypoxic response by applying a genome-wide expression array screening. PC3 cells lentivirally infected with shRNA expression constructs targeting ETV4 (shETV4) revealed a robust knock down of mRNA and protein levels, while a nontarget control shRNA (shNTC) did not affect ETV4 expression (Figure 6A and B). ETV4 depleted PC3 cells showed robustly reduced mRNA levels of the known ETV4 target gene cyclooxygenase-2 (COX2) (29,35), confirming loss of ETV4 function in these cells (Figure 6B). Total RNA was isolated from PC3 shETV4 and shNTC control cells exposed to 20 or 0.2% oxygen for 24 h and samples from three independent experiments were labelled for microarray analysis. When compared with normoxic control cells, 977 mRNAs and large intergenic noncoding RNAs (lincRNAs) were more than twofold downregulated (P < 0.05) in normoxic PC3 cells lacking ETV4 (Figure 6C; green). Hypoxia alone upregulated 608 mRNAs/lincRNAs by more than twofold (P < 0.05; Figure 6C; red). Interestingly, 450 mRNAs/lincRNAs showed a more than twofold reduction (P < 0.05) of the hypoxic expression levels in cells lacking ETV4 when compared with hypoxic control cells (Figure 6C; blue), out of which a group of 47 mRNAs/lincRNAs was found to be simultaneously hypoxia-inducible. Individual expression levels of these 47 transcripts centered on the mean of the three normoxic control samples (PC3 shNTC) are depicted in a heatmap in Figure 6D. Array data were validated by RT-qPCR of four randomly chosen transcripts (Figure 6E).
Figure 6.
Genome-wide microarray expression analysis reveals a broad role for ETV4 in HIF mediated hypoxic gene regulation. (A) Efficient knock down of ETV4 in human PC3 prostate cancer cells. PC3 cells were stably transduced with lentiviral shRNA expression vectors encoding either a nontarget control (shNTC) or shETV4. Following 24 h of exposure to 20% O2 or 0.2% O2, ETV4, HIF-1α, PHD2 and β-actin protein levels were analyzed by immunoblotting. (B) Total RNA was isolated from cultures treated as in (A) and mRNA levels of ETV4 and its target gene COX2 were determined by RT-qPCR. Gene expression levels were expressed in relation to ribosomal L28 mRNA (rel. levels) calculated from three independent experiments. (C) Venn diagram showing the number of transcripts regulated by either an at least twofold induction by hypoxia alone (red), an at least twofold reduction in normoxic cells by the knock down of ETV4 (green), or an at least twofold reduction in hypoxic cells by the knock down of ETV4 (blue), respectively. (D) Heatmap of the individual expression levels of the 47 transcripts that required ETV4 for efficient hypoxic induction. (E) Expression levels of four randomly chosen transcripts shown in (D) were confirmed by RT-qPCR as described for (B).
HIF target genes divide into two groups, either ETV4 coactivated or independent
Because hypoxically upregulated gene sets are highly variable between different cellular models, we next focused on a predefined gene set of 61 hypoxia-inducible transcripts. This gene set has previously been reported based on established HIF target genes (36). In line with this publication, the majority of these genes was found to be hypoxically upregulated in PC3 shNTC control cells (green dots in Figure 7A, left panel). Comparing hypoxic expression levels of the same genes in PC3 shETV4 knock down with shNTC control cells, the group of established HIF target genes roughly clustered into two halves, representing transcripts which either remained unaffected or which did not respond to hypoxia anymore in the absence of ETV4 (Figure 7A, right panel). Interestingly, following ranking of the HIF target genes according to their requirement for ETV4, the HIF-dependent PHD3 oxygen sensor (encoded by EGLN3) showed the highest ETV4 sensitivity for hypoxic induction in PC3 cells (Figure 7B and C), demonstrating that ETV4 plays a major role in the feedback control of mammalian oxygen sensing. However, a number of established HIF-responsive genes was only slightly affected (e.g. PAI1, encoded by SERPINE1) or completely resistant (e.g. GLUT1, encoded by SLC2A1) to ETV4 depletion (see Figure 7B and C), suggesting that the HIF pathway can be divided into two branches according to the requirement for ETV4.
Figure 7.
Role of ETV4 in the regulation of established HIF target genes in vitro and in vivo. (A) Dot plots showing the correlation between transcripts in normoxic versus hypoxic control cells (left panel) or in hypoxic control versus hypoxic ETV4 knock down cells (right panel) as derived from the gene array data (grey dots). Red dots refer to internal controls and the blue dot shows ETV4 which is downregulated in shETV4 cells. Green dots indicate the positions of a predefined set of 61 well-established HIF target genes. (B) Heat map of the 61 HIF target genes ranked by the magnitude of ETV4 requirement for hypoxic induction according to differences in hypoxic expression levels with Δhyp = log2(shNTC_hypoxia) − log2(shETV4_hypoxia) and mean hypoxic expression levels centered to the mean of normoxic control cells. (C) Exemplary mRNA levels of HIF target genes which either require ETV4 for efficient hypoxic induction (PHD3 and CA9) or which remain unaffected by the ETV4 knock down (GLUT1 and PAI1). mRNA was quantified as described for Figure 6B. (D and E) Correlation between ETV4 and established markers for tissue hypoxia in human breast cancer. (D) Independent specimens (spec.) of immunohistochemical evaluation of ETV4 expression in primary mammary carcinoma with high (upper panel) or low (lower panel) ETV4 expression levels. (E) Rank-order correlations (Spearman's rho) for ETV4 and PHD2 as well as known markers reflecting tissue hypoxia (HIF-1α, HIF-2α, PAI1, GLUT1 and CA9) are summarized in a cross table. The number of cases where both of the correlated markers could be assessed is displayed in parentheses. Asterisks indicate statistical significance with *P < 0.05 and **P < 0.01.
ETV4 expression levels correlate with HIF-α accumulation in human breast cancer
Elevated ETV4 transcript levels have been reported in a variety of human neoplastic diseases including breast cancer (37). Moreover, the onset of spontaneous mammary tumor development has been shown to be profoundly delayed in MMTV-neu transgenic mice which express a dominant negative variant of the mouse homologue of ETV4, suggesting that ETV4 may possess tumor promoting effects (38). We recently characterized protein levels of a variety of hypoxic marker genes, including HIF-1α, in tumor samples of 282 patients diagnosed with primary breast carcinoma (13,24). When the same specimens were immunostained for ETV4 protein levels, a strong and highly significant correlation between ETV4 and PHD2 was observed in samples where both factors were assessed (P < 0.01, Spearman's rho, N = 243). Consistent with our model of synthetic action of ETV4 and HIF-α, a solid association was also observed between these transcription factors and three well described target genes (PAI1; GLUT1; CA9) of the HIF pathway. Correlation coefficients between ETV4, PHD2 and HIF-1α and HIF-2α were considerably higher than those observed among both HIF-αs and the three markers for tissue hypoxia (Figure 7D and E), demonstrating a putative role for ETV4 in the regulation of PHD2 expression also in vivo.
DISCUSSION
Unique responsiveness to altered oxygen environments and broad conservation of all components of the PHD/HIF oxygen sensing pathway in multicellular life clearly indicate the central role of HIFs as literally hypoxia-inducible transcription factors (39). However, eukaryotic gene expression is a multistep process requiring the complex transcription machinery to interact with promoter DNA and initiate transcription (40). Not surprisingly, numerous studies have identified other nuclear regulators that contribute to the full spectrum of transcriptional changes in response to hypoxia. Yet, general patterns of direct interplay among HIFs and other transcriptional regulators are largely unknown and interactions were often found to rely on specific cell models (41). Here, we report on a novel screening approach that in combination with overexpression of arrayed transcription factors aimed for the systematic analysis of HBS-specific transcription factor interplay. The core P2P was used as a paradigm for HIF-dependent gene regulation, as it embeds a single HBS conferring hypoxic activation and because the endogenous locus is ubiquitously expressed.
We identified various members of the activating protein-1 (AP-1) family as novel activators of the PHD2 gene, although JUN/FOS has been found to enhance hypoxic gene expression previously (41). However, to the best of our knowledge, our study is the first to link ETV4 with HIF-dependent transcription. As shown by the use of different reporter genes, ETV4 function as facilitator of HIF-1 transactivation activity is not restricted to the P2P. While nuclear distances between ETV4 and HIF-1/2α, as calculated by FRET experiments, support a close interaction, our data favor a model where p300/CBP serves as essential bridging molecule between the two factors. This conclusion is based on the following features of the interaction between ETV4 and HIF-1α: (i) oxygen sensitivity in the absence of the oxygen-dependent degradation domain; (ii) mapping to the C-terminal activation domain; (iii) competition by CITED2; and (iv) requirement of FIH for oxygen sensitivity. Such a ternary complex is still in line with the FRET data, as previous findings suggest binding of HIF-1α to both CH1 and CH3 domains of p300 (42). ETV4 is known to interact with p300 at its CH3 domain and, thus, might well get into close or even physical contact to HIF-1α (43). Interestingly, out of 371 known interactors of human p300/CBP (44), 101 (27.2%) were present in the synthetic transactivation screen, but only 10 (9.9%) of them met the criteria to be considered as reproducible activators of the P2P. Apparently, there is no simple redundancy among the p300/CBP interactors to serve as transcriptional coactivators, and the target gene context is thought to play an important role in p300/CBP complex formation (44). The latter is of particular importance, as it provides some reliability regarding the specificity of our screening approach, underlining its general applicability.
Based on literature searches, ETV4 and HIF pathways share several common target genes, including matrix metalloproteases (MMP) 1, MMP-3, MMP-7, MMP-9, iNOS and COX-2 (45–48). Hence, one might speculate that the two factors directly cooperate at regulatory elements of these genes, jointly boosting invasive properties of malignant cells. Notably, ETV4 and its close relatives ETV1 and ETV5 are frequently overexpressed in prostate cancer due to gene fusion with androgen-responsive gene loci (49). Similarly, loss of the tumor suppressor PTEN causes high normoxic expression levels of HIF-1α, a key feature of invasive prostate cancers (50). Recent studies using animal models propose a mutational sequence, where early loss of PTEN and overactivation of ETS target genes collectively promote prostatic cancer progression (34,51). Thus, it will be highly interesting to explore a putative synthetic HIF-1/ETV4 role in these pathologies.
Due to its high endogenous expression levels in the PC3 prostate cancer cell line, gene array analyses in ETV4 wild-type and knock down PC3 cells were undertaken to explore the general role of ETV4 in hypoxic gene regulation. Remarkably, 47 of 608 hypoxically induced transcripts depend on ETV4 for efficient upregulation. Further analysis concentrating on a set of 61 established HIF target genes revealed 14 genes whose hypoxic induction was at least twofold higher in the presence of ETV4 than in its absence. For example, hypoxic induction of carbonic anhydrase 9 (CA9) was strongly dependent on the presence of ETV4, as it was largely absent in ETV4 knock down cells. This finding further explains the unusually strong hypoxic inducibility of CA9 which has previously been attributed to the cooperation between HIF and ATF-4 (52), another transcription factor that we found to be involved in oxygen signaling (53). Somewhat unexpected, PHD2 did not fulfil, at least in PC3 cells, our stringency criteria for ETV4-dependent hypoxically induced genes. Individual inspection revealed a 2.7-fold hypoxic induction that was only reduced by 16% in the absence of ETV4. Low hypoxic inducibility in this cell type was also seen on the protein level (see also Figure 6A) and might explain the rather weak response of this gene to ETV4 depletion, despite the fact that forced expression of ETV4 strongly induced the P2P in the U2OS cell model. However, out of 61 established HIF target genes, PHD3 was most sensitive to ETV4 depletion, suggesting that in PC3 cells PHD3 rather than PHD2 might represent the primary oxygen sensor targeted by ETV4.
In vivo, ETV4 has been implicated in kidney branching morphogenesis, differentiation of spinal motor neurons and mammary gland development (54–57). Importantly, ETV1 (ER81), ETV4 (PEA3/E1AF) and ETV5 (ERM) are highly similar and constitute the PEA3 subfamily among the ETS-domain family of transcription factors (58). ETV4 and ETV5 are functionally highly redundant and a double knock out was required to reveal the role of ETV4 in kidney development (54,55). Thus, we tested the ability of these additional subfamily members to induce the P2P. While ETV1 did not have any effect, ETV4 and ETV5 super-induced the hypoxic PHD2 and transferrin promoters in a HBS-dependent manner, suggesting functional redundancy of these two subfamily members in hypoxic gene regulation (Supplementary Figure S1). As the DNA-binding/ETS domain is highly conserved between all three PEA3 subfamily members, we further conclude that the interaction with HIFα takes place outside of the ETS domain of ETV4.
Because these developmental processes often occur in tissues with low oxygenation, our data point to a role of tissue hypoxia in physiological ETV4/ETV5 function. Similarly, high expression levels of ETV4 have been linked to metastasis or bad prognosis in a variety of human cancers (59). These clinical features are well-known for hypoxic tumors expressing high levels of HIF-1 (60). Supporting our screening results, we found a strikingly good correlation between ETV4 and PHD2 protein levels in breast cancer tissues, in line with a potentially relevant function of ETV4 in hypoxic tissues in vivo.
In summary, synthetic transactivation screening as exemplarily demonstrated for HIF-dependent gene expression proved to be a powerful tool to unravel novel interactions among common signaling pathways. The general setup of this method may be easily adapted to study other transcriptional pathways. A multiplexed single-well readout system predestines this approach for extensive screening projects, including small molecule library analyses and genome-wide gene silencing approaches, where inter-well variances are technically difficult to control.
ACCESSION NUMBER
NCBI's Gene Expression Omnibus (GEO) accession number GSE32385.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1, Supplementary Figure 1.
FUNDING
Deutsche Forschungsgemeinschaft Grants FA225/22 (to U.B.-P. and J.F.) and GRK1431/2 (to J.F.); Hartmann Müller-Stiftung (to D.P.S.); Forschungskredit of the University of Zürich (to D.P.S.); Swiss National Science Foundation Grant 31003A_129962/1 (to R.H.W. and D.P.S.). Funding for open access charge: Regular budget of the Wenger group at the Institute of Physiology of the University of Zurich.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank N. Sang, L. Poellinger and J.-L. Baert for providing expression plasmids; S. Behnke and M. Storz for assistance with TMA analyses; S. Hafen-Wirth for the labeling of the microarray samples and P. Spielmann for general technical assistance.
REFERENCES
- 1.Wenger RH, Stiehl DP, Camenisch G. Integration of oxygen signaling at the consensus HRE. Sci. STKE. 2005;2005:re12. doi: 10.1126/stke.3062005re12. [DOI] [PubMed] [Google Scholar]
- 2.Kaelin WG, Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- 4.Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ. Genome-wide association of hypoxia-inducible factor (HIF)-1α and HIF-2α DNA binding with expression profiling of hypoxia-inducible transcripts. J. Biol. Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1α and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hewitson KS, McNeill LA, Riordan MV, Tian YM, Bullock AN, Welford RW, Elkins JM, Oldham NJ, Bhattacharya S, Gleadle JM, et al. Hypoxia-inducible factor (HIF) asparagine hydroxylase is identical to factor inhibiting HIF (FIH) and is related to the cupin structural family. J. Biol. Chem. 2002;277:26351–26355. doi: 10.1074/jbc.C200273200. [DOI] [PubMed] [Google Scholar]
- 7.Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouysségur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeda K, Ho VC, Takeda H, Duan LJ, Nagy A, Fong GH. Placental but not heart defects are associated with elevated hypoxia-inducible factor α levels in mice lacking prolyl hydroxylase domain protein 2. Mol. Cell. Biol. 2006;26:8336–8346. doi: 10.1128/MCB.00425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeda K, Cowan A, Fong GH. Essential role for prolyl hydroxylase domain protein 2 in oxygen homeostasis of the adult vascular system. Circulation. 2007;116:774–781. doi: 10.1161/CIRCULATIONAHA.107.701516. [DOI] [PubMed] [Google Scholar]
- 11.Mazzone M, Dettori D, Leite de Oliveira R, Loges S, Schmidt T, Jonckx B, Tian YM, Lanahan AA, Pollard P, Ruiz de Almodovar C, et al. Heterozygous deficiency of PHD2 restores tumor oxygenation and inhibits metastasis via endothelial normalization. Cell. 2009;136:839–851. doi: 10.1016/j.cell.2009.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan DA, Kawahara TL, Sutphin PD, Chang HY, Chi JT, Giaccia AJ. Tumor vasculature is regulated by PHD2-mediated angiogenesis and bone marrow-derived cell recruitment. Cancer Cell. 2009;15:527–538. doi: 10.1016/j.ccr.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bordoli MR, Stiehl DP, Borsig L, Kristiansen G, Hausladen S, Schraml P, Wenger RH, Camenisch G. Prolyl-4-hydroxylase PHD2- and hypoxia-inducible factor 2-dependent regulation of amphiregulin contributes to breast tumorigenesis. Oncogene. 2011;30:548–560. doi: 10.1038/onc.2010.433. [DOI] [PubMed] [Google Scholar]
- 14.Barth S, Nesper J, Hasgall PA, Wirthner R, Nytko KJ, Edlich F, Katschinski DM, Stiehl DP, Wenger RH, Camenisch G. The peptidyl prolyl cis/trans isomerase FKBP38 determines hypoxia-inducible transcription factor prolyl-4-hydroxylase PHD2 protein stability. Mol. Cell. Biol. 2007;27:3758–3768. doi: 10.1128/MCB.01324-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barth S, Edlich F, Berchner-Pfannschmidt U, Gneuss S, Jahreis G, Hasgall PA, Fandrey J, Wenger RH, Camenisch G. Hypoxia-inducible factor prolyl-4-hydroxylase PHD2 protein abundance depends on iIntegral membrane anchoring of FKBP38. J. Biol. Chem. 2009;284:23046–23058. doi: 10.1074/jbc.M109.032631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metzen E, Stiehl DP, Doege K, Marxsen JH, Hellwig-Bürgel T, Jelkmann W. Regulation of the prolyl hydroxylase domain protein 2 (phd2/egln-1) gene: identification of a functional hypoxia-responsive element. Biochem. J. 2005;387:711–717. doi: 10.1042/BJ20041736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stiehl DP, Wirthner R, Köditz J, Spielmann P, Camenisch G, Wenger RH. Increased prolyl 4-hydroxylase domain proteins compensate for decreased oxygen levels. Evidence for an autoregulatory oxygen-sensing system. J. Biol. Chem. 2006;281:23482–23491. doi: 10.1074/jbc.M601719200. [DOI] [PubMed] [Google Scholar]
- 18.Ginouvès A, Ilc K, Macìas N, Pouysségur J, Berra E. PHDs overactivation during chronic hypoxia “desensitizes” HIFα and protects cells from necrosis. Proc. Natl Acad. Sci. USA. 2008;105:4745–4750. doi: 10.1073/pnas.0705680105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziauddin J, Sabatini DM. Microarrays of cells expressing defined cDNAs. Nature. 2001;411:107–110. doi: 10.1038/35075114. [DOI] [PubMed] [Google Scholar]
- 20.Rolfs A, Kvietikova I, Gassmann M, Wenger RH. Oxygen-regulated transferrin expression is mediated by hypoxia-inducible factor-1. J. Biol. Chem. 1997;272:20055–20062. doi: 10.1074/jbc.272.32.20055. [DOI] [PubMed] [Google Scholar]
- 21.Sang N, Fang J, Srinivas V, Leshchinsky I, Caro J. Carboxyl-terminal transactivation activity of hypoxia-inducible factor 1α is governed by a von Hippel-Lindau protein-independent, hydroxylation-regulated association with p300/CBP. Mol. Cell. Biol. 2002;22:2984–2992. doi: 10.1128/MCB.22.9.2984-2992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wotzlaw C, Otto T, Berchner-Pfannschmidt U, Metzen E, Acker H, Fandrey J. Optical analysis of the HIF-1 complex in living cells by FRET and FRAP. FASEB J. 2007;21:700–707. doi: 10.1096/fj.06-6280com. [DOI] [PubMed] [Google Scholar]
- 23.Nytko KJ, Maeda N, Schläfli P, Spielmann P, Wenger RH, Stiehl DP. Vitamin C is dispensable for oxygen sensing in vivo. Blood. 2011;117:5485–5493. doi: 10.1182/blood-2010-09-307637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stiehl DP, Bordoli MR, Abreu-Rodríguez I, Wollenick K, Schraml P, Gradin K, Poellinger L, Kristiansen G, Wenger RH. Non-canonical HIF-2α function drives autonomous breast cancer cell growth via an AREG-EGFR/ErbB4 autocrine loop. Oncogene. 2011 doi: 10.1038/onc.2011.417. doi: 10.1038/onc.2011.1417. [DOI] [PubMed] [Google Scholar]
- 25.Makino Y, Cao R, Svensson K, Bertilsson G, Asman M, Tanaka H, Cao Y, Berkenstam A, Poellinger L. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 26.Xin JH, Cowie A, Lachance P, Hassell JA. Molecular cloning and characterization of PEA3, a new member of the Ets oncogene family that is differentially expressed in mouse embryonic cells. Genes Dev. 1992;6:481–496. doi: 10.1101/gad.6.3.481. [DOI] [PubMed] [Google Scholar]
- 27.Pugh CW, O'Rourke JF, Nagao M, Gleadle JM, Ratcliffe PJ. Activation of hypoxia-inducible factor-1; definition of regulatory domains within the α subunit. J. Biol. Chem. 1997;272:11205–11214. doi: 10.1074/jbc.272.17.11205. [DOI] [PubMed] [Google Scholar]
- 28.Arany Z, Huang LE, Eckner R, Bhattacharya S, Jiang C, Goldberg MA, Bunn HF, Livingston DM. An essential role for p300/CBP in the cellular response to hypoxia. Proc. Natl Acad. Sci. USA. 1996;93:12969–12973. doi: 10.1073/pnas.93.23.12969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Borchert GL, Phang JM. Polyoma enhancer activator 3, an ets transcription factor, mediates the induction of cyclooxygenase-2 by nitric oxide in colorectal cancer cells. J. Biol. Chem. 2004;279:18694–18700. doi: 10.1074/jbc.M308136200. [DOI] [PubMed] [Google Scholar]
- 30.Bhattacharya S, Michels CL, Leung MK, Arany ZP, Kung AL, Livingston DM. Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev. 1999;13:64–75. doi: 10.1101/gad.13.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freedman SJ, Sun ZY, Kung AL, France DS, Wagner G, Eck MJ. Structural basis for negative regulation of hypoxia-inducible factor-1α by CITED2. Nat. Struct. Biol. 2003;10:504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- 32.Ruas JL, Berchner-Pfannschmidt U, Malik S, Gradin K, Fandrey J, Roeder RG, Pereira T, Poellinger L. Complex regulation of the transactivation function of hypoxia-inducible factor-1α by direct interaction with two distinct domains of the CREB-binding protein/p300. J. Biol. Chem. 2010;285:2601–2609. doi: 10.1074/jbc.M109.021824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baert JL, Monte D, Verreman K, Degerny C, Coutte L, de Launoit Y. The E3 ubiquitin ligase complex component COP1 regulates PEA3 group member stability and transcriptional activity. Oncogene. 2010;29:1810–1820. doi: 10.1038/onc.2009.471. [DOI] [PubMed] [Google Scholar]
- 34.Vitari AC, Leong KG, Newton K, Yee C, O'Rourke K, Liu J, Phu L, Vij R, Ferrando R, Couto SS, et al. COP1 is a tumour suppressor that causes degradation of ETS transcription factors. Nature. 2011;474:403–406. doi: 10.1038/nature10005. [DOI] [PubMed] [Google Scholar]
- 35.Howe LR, Crawford HC, Subbaramaiah K, Hassell JA, Dannenberg AJ, Brown AM. PEA3 is up-regulated in response to Wnt1 and activates the expression of cyclooxygenase-2. J. Biol. Chem. 2001;276:20108–20115. doi: 10.1074/jbc.M010692200. [DOI] [PubMed] [Google Scholar]
- 36.Ortiz-Barahona A, Villar D, Pescador N, Amigo J, del Peso L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010;38:2332–2345. doi: 10.1093/nar/gkp1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benz CC, O'Hagan RC, Richter B, Scott GK, Chang CH, Xiong X, Chew K, Ljung BM, Edgerton S, Thor A, et al. HER2/Neu and the Ets transcription activator PEA3 are coordinately upregulated in human breast cancer. Oncogene. 1997;15:1513–1525. doi: 10.1038/sj.onc.1201331. [DOI] [PubMed] [Google Scholar]
- 38.Shepherd TG, Kockeritz L, Szrajber MR, Muller WJ, Hassell JA. The pea3 subfamily ets genes are required for HER2/Neu-mediated mammary oncogenesis. Curr. Biol. 2001;11:1739–1748. doi: 10.1016/s0960-9822(01)00536-x. [DOI] [PubMed] [Google Scholar]
- 39.Loenarz C, Coleman ML, Boleininger A, Schierwater B, Holland PW, Ratcliffe PJ, Schofield CJ. The hypoxia-inducible transcription factor pathway regulates oxygen sensing in the simplest animal, Trichoplax adhaerens. EMBO Rep. 2011;12:63–70. doi: 10.1038/embor.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kornberg RD. The molecular basis of eukaryotic transcription. Proc. Natl Acad. Sci. USA. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Eur. J. Physiol. 2005;450:363–371. doi: 10.1007/s00424-005-1413-7. [DOI] [PubMed] [Google Scholar]
- 42.Ruas JL, Poellinger L, Pereira T. Role of CBP in regulating HIF-1-mediated activation of transcription. J. Cell Sci. 2005;118:301–311. doi: 10.1242/jcs.01617. [DOI] [PubMed] [Google Scholar]
- 43.Matsui K, Sugimori K, Motomura H, Ejiri N, Tsukada K, Kitajima I. PEA3 cooperates with c-Jun in regulation of HER2/neu transcription. Oncol Rep. 2006;16:153–158. [PubMed] [Google Scholar]
- 44.Bedford DC, Kasper LH, Fukuyama T, Brindle PK. Target gene context influences the transcriptional requirement for the KAT3 family of CBP and p300 histone acetyltransferases. Epigenetics. 2010;5:9–15. doi: 10.4161/epi.5.1.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Firlej V, Bocquet B, Desbiens X, de Launoit Y, Chotteau-Lelievre A. Pea3 transcription factor cooperates with USF-1 in regulation of the murine bax transcription without binding to an Ets-binding site. J. Biol. Chem. 2005;280:887–898. doi: 10.1074/jbc.M408017200. [DOI] [PubMed] [Google Scholar]
- 46.Yang S, Kim J, Ryu JH, Oh H, Chun CH, Kim BJ, Min BH, Chun JS. Hypoxia-inducible factor-2α is a catabolic regulator of osteoarthritic cartilage destruction. Nat. Med. 2010;16:687–693. doi: 10.1038/nm.2153. [DOI] [PubMed] [Google Scholar]
- 47.Palmer LA, Semenza GL, Stoler MH, Johns RA. Hypoxia induces type II NOS gene expression in pulmonary artery endothelial cells via HIF-1. Am. J. Physiol. 1998;274:L212–L219. doi: 10.1152/ajplung.1998.274.2.L212. [DOI] [PubMed] [Google Scholar]
- 48.Kaidi A, Qualtrough D, Williams AC, Paraskeva C. Direct transcriptional up-regulation of cyclooxygenase-2 by hypoxia-inducible factor (HIF)-1 promotes colorectal tumor cell survival and enhances HIF-1 transcriptional activity during hypoxia. Cancer Res. 2006;66:6683–6691. doi: 10.1158/0008-5472.CAN-06-0425. [DOI] [PubMed] [Google Scholar]
- 49.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat. Rev. Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang BH, Jiang G, Zheng JZ, Lu Z, Hunter T, Vogt PK. Phosphatidylinositol 3-kinase signaling controls levels of hypoxia-inducible factor 1. Cell Growth Differ. 2001;12:363–369. [PubMed] [Google Scholar]
- 51.Squire JA. TMPRSS2-ERG and PTEN loss in prostate cancer. Nat. Genet. 2009;41:509–510. doi: 10.1038/ng0509-509. [DOI] [PubMed] [Google Scholar]
- 52.van den Beucken T, Koritzinsky M, Niessen H, Dubois L, Savelkouls K, Mujcic H, Jutten B, Kopacek J, Pastorekova S, van der Kogel AJ, et al. Hypoxia-induced expression of carbonic anhydrase 9 is dependent on the unfolded protein response. J. Biol. Chem. 2009;284:24204–24212. doi: 10.1074/jbc.M109.006510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Köditz J, Nesper J, Wottawa M, Stiehl DP, Camenisch G, Franke C, Myllyharju J, Wenger RH, Katschinski DM. Oxygen-dependent ATF-4 stability is mediated by the PHD3 oxygen sensor. Blood. 2007;110:3610–3617. doi: 10.1182/blood-2007-06-094441. [DOI] [PubMed] [Google Scholar]
- 54.Lu BC, Cebrian C, Chi X, Kuure S, Kuo R, Bates CM, Arber S, Hassell J, MacNeil L, Hoshi M, et al. Etv4 and Etv5 are required downstream of GDNF and Ret for kidney branching morphogenesis. Nat. Genet. 2009;41:1295–1302. doi: 10.1038/ng.476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuure S, Chi X, Lu B, Costantini F. The transcription factors Etv4 and Etv5 mediate formation of the ureteric bud tip domain during kidney development. Development. 2010;137:1975–1979. doi: 10.1242/dev.051656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Livet J, Sigrist M, Stroebel S, De Paola V, Price SR, Henderson CE, Jessell TM, Arber S. ETS gene Pea3 controls the central position and terminal arborization of specific motor neuron pools. Neuron. 2002;35:877–892. doi: 10.1016/s0896-6273(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 57.Kurpios NA, MacNeil L, Shepherd TG, Gludish DW, Giacomelli AO, Hassell JA. The Pea3 Ets transcription factor regulates differentiation of multipotent progenitor cells during mammary gland development. Dev. Biol. 2009;325:106–121. doi: 10.1016/j.ydbio.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 58.Sharrocks AD. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell. Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 59.de Launoit Y, Baert JL, Chotteau-Lelievre A, Monte D, Coutte L, Mauen S, Firlej V, Degerny C, Verreman K. The Ets transcription factors of the PEA3 group: transcriptional regulators in metastasis. Biochim. Biophys. Acta. 2006;1766:79–87. doi: 10.1016/j.bbcan.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29:625–634. doi: 10.1038/onc.2009.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.