Abstract
Saccharomyces cerevisiae Mph1 is a 3–5′ DNA helicase, required for the maintenance of genome integrity. In order to understand the ATPase/helicase role of Mph1 in genome stability, we characterized its helicase activity with a variety of DNA substrates, focusing on its action on junction structures containing three or four DNA strands. Consistent with its 3′ to 5′ directionality, Mph1 displaced 3′-flap substrates in double-fixed or equilibrating flap substrates. Surprisingly, Mph1 displaced the 5′-flap strand more efficiently than the 3′ flap strand from double-flap substrates, which is not expected for a 3–5′ DNA helicase. For this to occur, Mph1 required a threshold size (>5 nt) of 5′ single-stranded DNA flap. Based on the unique substrate requirements of Mph1 defined in this study, we propose that the helicase/ATPase activity of Mph1 play roles in converting multiple-stranded DNA structures into structures cleavable by processing enzymes such as Fen1. We also found that the helicase activity of Mph1 was used to cause structural alterations required for restoration of replication forks stalled due to damaged template. The helicase properties of Mph1 reported here could explain how it resolves D-loop structure, and are in keeping with a model proposed for the error-free damage avoidance pathway.
INTRODUCTION
Mph1 is a DNA-dependent ATPase and DNA helicase with seven specific motifs characteristic of the subfamily of helicase superfamily 2 (SF2) called DEAH/DExH (1–3). The MPH1 (mutator phenotype 1) gene was first identified as a mutator gene that increased mutation rates when deleted (4). It showed an epistatic relationship with homologous recombination proteins. Genetic interactions between MPH1 and other genes involved in recombination and repair indicated that MPH1 is involved in an error-free DNA damage avoidance pathway involving stalled replication forks (1,5). In vivo, it was shown that MPH1 regulates crossover frequency during studies of genes synthetic lethal with srs2Δ. In vitro, purified Mph1 was shown to displace D-loop structure in support of its in vivo role promoting non-crossover in mitotic recombination (6). Recently, we proposed that Mph1 is involved in Okazaki fragment processing by stimulating Fen1 nuclease to promote formation of ligatable nicks (7).
Three eukaryotic orthologs of Mph1 have been identified; Fml1 and Fml2 in fission yeast and FANCM in humans (8,9). FANCM is a component of the Fanconi anemia (FA) core complex that confers DNA damage tolerance in cells with stalled DNA replication forks (11). FANCM catalyzes D-loop displacement, branch migration and replication fork regression. It has been proposed that FANCM counteracts the movement of forks when they approach damaged sites and remodels the fork (10,11). Fml1 promotes recombination at a stalled replication fork and controls crossover in this particular recombination event (9). Like Mph1, Fml1 possesses D-loop displacement activity and unwinds a variety of replication fork-like substrates. It also catalyzes branch migration and replication fork regression, which are thought to support template switching when leading strand synthesis is blocked by DNA damage.
In lagging strand DNA synthesis, the polymerase (pol)-α primase complex synthesizes short RNA–DNA primers that are extended by pol δ to full-length Okazaki fragments (12). Because of its ability to carry out displacement DNA synthesis, pol δ generates single-stranded 5′-flaps by displacing the 5′-end regions of downstream Okazaki fragments that are subsequently processed by the combined action of Fen1 and Dna2. The action of Dna2 is important especially when flaps are long enough to bind replication-protein A (RPA) since RPA-bound flaps are not cleaved by Fen1. The nicks generated by Fen1 are finally sealed by DNA ligase I. It has been suggested that the correct and timely processing of Okazaki fragments is critical to ensure genome stability because faulty processing inevitably leaves unsealed nicks or unprocessed flaps that can lead to DNA double-strand breaks. We have isolated a number of additional factors involved in Okazaki fragment processing by screenings for genetic suppressors; In vitro studies revealed that some of these suppressors stimulate the nuclease activity of Dna2 or Fen1. Others are likely to work in parallel with these two processing enzymes (12). We showed that Mph1 stimulated the structure-specific endonuclease activity of Fen1, but its ATPase/helicase activity was not required for this effect (7). During the course of these studies, we found that Mph1 catalyzed the displacement of 5′-flaps from 5′-flap substrates. This finding is unexpected since Mph1 translocates in the 3–5′ direction along single-stranded DNA to which it binds (2,7). This unexpected finding prompted us to investigate the substrate specificity of Mph1 using synthetic DNA substrates containing three- or four-way junctions. We found that Mph1 is a versatile helicase that utilizes many different substrates including 5′-flap, 3′-flap and fixed-double flap substrates. Moreover, like Fml1, Mph1 reversed replication forks, reminiscent of the action of RecQ-type helicases that maintain genome stability in eukaryotes and prokaryotes (9,13). We showed that Mph1 promotes flap equilibration towards formation of 5′-flap, the in vivo substrate of Fen1 nuclease. This property of Mph1 is consistent with our previous finding that it plays a role in flap processing. Since equilibrating flaps can arise from the resolution of recombination intermediate, it appears that one important role of helicase activity of Mph1 is to produce DNA structure readily processed by Fen1 or Dna2 endonuclease. These and other properties of Mph1 described in this study are in keeping with its role in recombination and DNA damage avoidance, which are critical for the maintenance of genome stability.
MATERIALS AND METHODS
Proteins and nucleotides
Polynucleotide kinase was purchased from Enzynomics (Daejeon, Korea). The oligonucleotides used in this study were commercially synthesized from Genotech (Daejeon, Korea). ATP was obtained from Sigma (St Louis, MO, USA). Ribonucleotides (NTPs), deoxyribonucleotides (dNTPs) and adenosine-5′-(γ-thio)-triphosphate (ATPγS) were from Roche (Basel, Switzerland). [γ-32P] and [α-32P] ATP (3000 Ci/mmol) were purchased from Izotop (Budapest, Hungary). Budding yeast Mph1 protein was purified as described before (7).
Preparation of DNA substrates
The sequences of oligonucleotides used for substrate preparations are listed in Table 1. Partial-duplex substrates were prepared as follows; one of the two oligonucleotides (10 pmol) in a partial-duplex DNA substrate was 5′-labeled with 3.3 pmol of [γ-32P] ATP and polynucleotide kinase according to the manufacturer's protocol. The labeled oligonucleotides were then annealed to the second oligonucleotide in 50 mM HEPES–NaOH/pH 7.5 and 200 mM NaCl, using a PCR machine (95°C: 5 min, 65°C: 30 min and −0.5°C/min to 25°C). For the preparation of flap-structured or nicked-duplex DNA substrates, one oligonucleotide was 5′-end labeled as described above and then annealed to the template and upstream oligonucleotides using a molar ratio of 1:2:4, respectively. To prepare DNA substrates containing two 5′-flaps (as shown in Figure 3), four-way junction (Figure 7) and replication fork-like substrates (fixed or partially movable double-strand three-way junction and regressed replication fork; shown in Figures 8A and 9), one of the oligonucleotides was labeled as described above and annealed with other three oligonucleotides at a molar ratio of 1:1:1:1. For the preparation of movable double-stranded three-way junction substrate (Figure 8B, C and D), two oligonucleotides 11 and 35 were first labeled and then annealed (1:1 ratio) separately to oligonucleotides 14 and 34, respectively. Subsequently, both partial-duplex DNAs were mixed together (1:1) and allowed to anneal for 12 h at 25°C. All annealed substrates were purified by 10% PAGE and the amount of recovered substrate was determined as previously described (14).
Table 1.
Oligonucleotide sequences used in this study
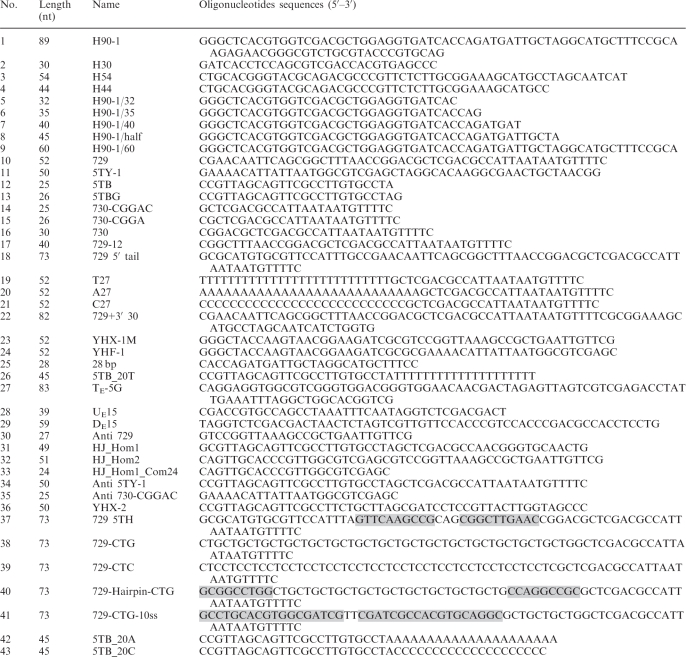 |
Sequences in shaded boxes form hairpin structures.
Competing sequences for annealing in equilibrating flap substrate are underlined.
Oligonucleotide sequences for preparing equilibrating flap substrate (27, 28 and 29) are as previously described (24).
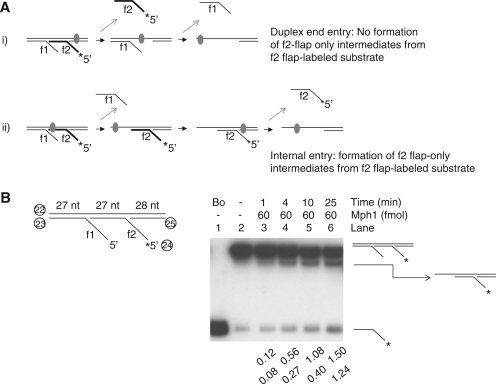
Figure 3.
Mph1 DNA helicase activity on substrates containing two 5′-flaps. (A) Schematic models of Mph1 unwinding mechanism. (B) Helicase activity of Mph1 on substrates containing two 5′-flaps. Assays were performed for various incubation periods (1, 4, 10 and 25 min). f1, flap1; f2, flap2.
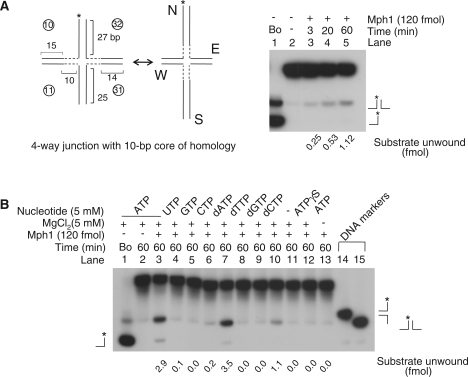
Figure 7.
Mph1 DNA helicase activity on four-way junction substrates. (A) Left panel: the structure of the partially movable four-way junction substrate (containing 10-bp core of homology). Dotted lines indicate homology core and each four arm is named as N (North), S (South), E (East) and W (West) as indicated. Right panel: Branch migration activity of Mph1 as a function of time (3, 20 and 60 min). Reactions were performed with 120 fmol of Mph1 in 10 μl reaction volume. The amount of product formed was measured and indicated at bottom of the panel. (B) Branch migration activity of Mph1 in the presence of various ribo- and deoxyribonucleotides.
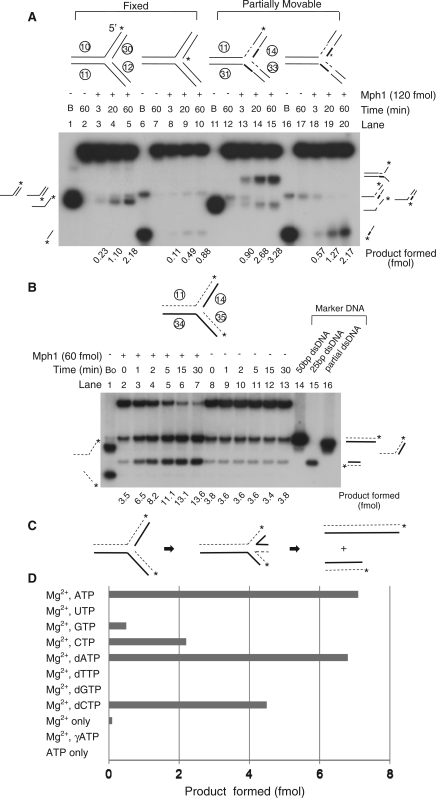
Figure 8.
Mph1 DNA helicase activity on replication fork substrates. (A) Helicase activity of Mph1 on fixed or partially movable double-stranded three-way junction substrates. Mph1 (120 fmol) was incubated using reaction conditions in Figure 6. (B) Replication fork regression activity of Mph1 on completely movable double-stranded three-way junction substrate. Assays were performed with 60 fmol of Mph1 using standard reaction conditions (20 μl) for various incubation periods (0, 1, 2, 5, 15 and 30 min). The amount of total product (slower and faster migrating band) formed by regression is indicated below each lane. In this case, the level of product formed in the absence of Mph1 was not subtracted. (C) Illustration how two dsDNA products are formed by regression of replication forks via chicken-foot structure. (D) Replication fork regression activity of Mph1 with various ribo- and deoxyribonucleotides as cofactors. Mph1 (50 fmol) was incubated using standard reaction condition for 15 min and the amount of product formed is shown as a bar graph. The level of product formed in the absence of Mph1 was subtracted from the values presented.
Figure 9.
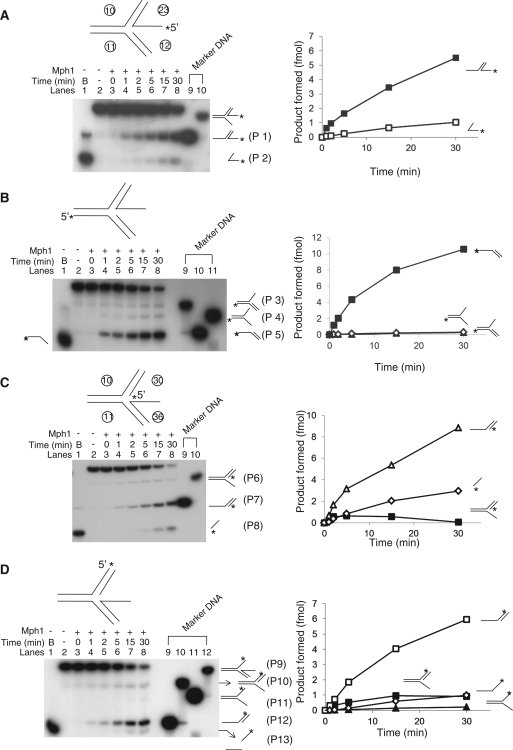
Mph1 helicase activity on regressed lagging or leading strand replication fork substrate. (A) Mph1 helicase activity on regressed lagging strand replication fork substrate. Assays were performed with 60 fmol of Mph1 using standard reaction condition (20 μl reaction volume) for various incubation periods (0, 1, 2, 5, 15 and 30 min). The amount of product formed was plotted against the time of incubation. (B) Assays were performed with same substrate as in (A), but labeled differently as illustrated. (C) Mph1 helicase activity on regressed leading strand replication fork substrate. Assays were performed as in (A) and the amount of product formed was plotted against incubation time. (D) Assays were performed with same substrate as in (C), but labeled differently as illustrated. (E) The mechanism of Mph1 unwinding of regressed replication fork substrates are shown based on the results from (A–D). Arrows with solid line indicate the preferred pathway while dotted lines denote the minor pathways.
Helicase assay
Unless indicated, standard helicase assays of Mph1 were performed in reactions (20 µl) containing 25 mM Tris–HCl/pH 7.8, 25 mM NaCl, 5 mM ATP, 5 mM MgCl2, 2 mM DTT, 0.25 mg/ml BSA and 15 fmol of DNA substrate (7). Mph1 was diluted in buffer (25 mM HEPES–NaOH/pH 7.6, 500 mM NaCl, 1 mM DTT, 0.25 mg/ml BSA, 0.01% NP-40, 10% glycerol) prior to addition. Reactions were incubated at 30°C, as indicated and 6× stop solution [60 mM EDTA/pH 8.0, 40% (w/v) sucrose, 0.6% SDS, 0.25% xylene cyanol, 0.25% bromophenol blue] was added to terminate reactions. Products were subjected to electrophoresis at 150 V through 10% polyacrylamide gel in 1× TBE (89 mM Tris–base, 89 mM Boric acid and 2 mM EDTA), after which gels were dried on DEAE-cellulose paper and autoradiographed. The resolved DNA products were quantified using a PhosphorImager (Molecular Dynamics) and their levels (as femtomoles) indicated below each lane of the gel in figures. Background level of unwound substrate was subtracted from quantified value of product formed except for Figure 2B. When two products were formed, the amount of the slower migrating (larger) product formed in the gel is indicated above the level of the faster migrating (smaller) product.
Figure 2.
Mph1 DNA helicase activity on 5′-flap-structured DNA. (A) helicase activity of Mph1 on 5′-flap-structured substrates of various lengths (1, 5 and 15-nt) in the flap region. The amount of substrate unwound plotted against the incubation period (1, 4, 10 and 25 min). (B) Helicase assays were performed with fork-structured substrates of various lengths (0-, 27- and 48-nt) in their 5′-extension region. The amount of substrate unwound was measured and plotted against the incubation period (1, 4, 10 and 25 min). (C) Unwinding rate of 5′-flap-structured substrates of different nucleotides sequences in the 27-nt 5′-flap region (random, dT, dA and dC). The amount of substrate unwound was measured and plotted against the incubation period (2, 8 and 25 min). (D) Unwinding rate of fork-structured substrates with sequence variations in the 27-nt 5′ tail region (random, dT, dA and dC). The amount of substrate unwound was measured and plotted against incubation period (2, 8 and 25 min). In all assays (A–D), 60 fmol of Mph1 was used. Note that one representative result was shown from several experiments carried out similarly.
Gel mobility shift assay
Reaction mixtures (20 μl) containing 25 mM Tris–HCl/pH 7.8, 25 mM NaCl, 5 mM MgCl2, 2 mM DTT, 0.25 mg/ml BSA, 15 fmol of DNA substrate and indicated levels of protein were incubated at 30°C for 15 min, followed by the addition of glycerol and bromphenol blue to 10% (v/v) and 0.05% (w/v), respectively (7). Reaction products were subjected to electrophoresis through a 6% polyacrylamide gel for 1.5 h in 0.5× TBE at 4°C. Gels were dried on DEAE-cellulose paper and autoradiographed. Levels of nucleoprotein complexes formed were quantified using a PhosphorImager.
ATPase assay
Reaction mixtures (20 μl) containing 25 mM Tris–HCl/pH 7.8, 25 mM NaCl, 5 mM MgCl2, 2 mM DTT, 0.25 mg/ml BSA, 5 mM cold ATP, 16.5 nM [α-32P] ATP (3000 Ci/mmol), 5 pmol of unlabeled DNA substrate and 100 fmol of Mph1 were incubated at 30°C for indicated time periods. At each time point, reaction mixture (5 μl) was removed and mixed with an equal volume of 0.5 M EDTA to stop the reaction. One microliter of each sample was spotted on a polyethyleneimine-cellulose plate (Baker, J.T. Inc.) and products were resolved in 0.5 M LiCl/1.0 M formic acid. The amounts of products were analyzed using a PhosphorImager.
RESULTS
The requirement of single-stranded region for Mph1 DNA helicase activity
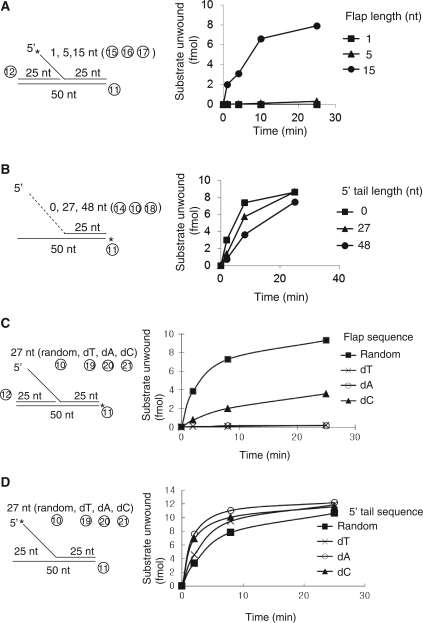
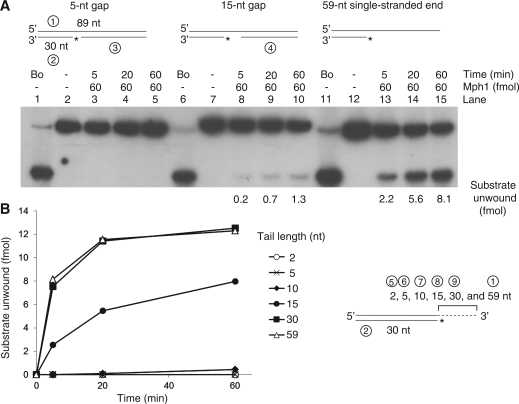
We previously noticed that the 3–5′ Mph1 helicase displaced the 5′- flap strands from flap-structured substrates (7). Unwinding of a 5′-flap strand was unexpected since this required Mph1 to translocate in the 5′ to 3′ direction along ssDNA. Though most helicases load onto ssDNAs using either free end or ssDNA flanked by duplex DNA, some helicases, like Hjm of Sulfolobus tokodaii or UvrD and RecBCD of E. coli unwind blunt ended-duplex DNA substrates lacking ssDNA regions by invading blunt ends of duplex DNA (15–17). In addition, it was shown that UvrD catalyzed the unwinding of nicked circular DNA. In order to exclude the possibility that Mph1 unwinds blunt-ended duplex DNA leading to dissociation of the 5′-flap strand from the substrate, helicase assays were carried out using nicked-duplex DNA with blunt ends prepared with synthetic oligonucleotides. We first examined whether Mph1 could utilize gapped-duplex DNA as helicase substrates (Figure 1). The substrates tested contained 5- and 15-nt-ssDNA regions flanked by duplex. Another substrate containing 3′-overhang (59-nt) ssDNA was used as positive control (Figure 1A, lanes 11–15). Following 60 min of incubation with 60 fmol Mph1, displacement of the labeled 5-nt gapped substrate was not detected (Figure 1A, lanes 1–5), while low levels of labeled oligonucleotide were released (1.3 fmol after 60 min of incubation) from the 15-nt gapped substrate (Figure 1A, lanes 6–10). These results indicate that Mph1 can unwind duplex DNA after being loaded onto a ssDNA gap >5 nt. As expected, the partial-duplex DNA substrate with a 3′ 59-nt ssDNA overhang was efficiently utilized by Mph1 with more than half of the substrate (8.1 fmol) unwound after 60 min of incubation (Figure 1A, lanes 11–15). We also analyzed Mph1 helicase activity as a function of 3′ ssDNA tail length, and found that lengthening of the 3′ tail up to 30 nt promoted unwinding (Figure 1B). Further increase in the 3′ tail size, however, did not increase the unwinding efficiency (Figure 1B). From these results, we concluded that Mph1 can unwind duplex DNA containing >15-nt ssDNA regions. Most likely, this ssDNA region acts as an entry site for Mph1 helicase.
Figure 1.
Single-stranded regions are required for Mph1 DNA helicase activity. (A) Helicase assays were carried out with partial-duplex DNA substrates containing 5-, 15-nt gap and 59-nt 3′ single-stranded overhang under standard reaction conditions for various incubation periods (5, 20 and 60 min). The circled numbers in substrates denote the oligonucleotide described in Table 1. Asterisk in substrate indicate the 32P-labeled end. The amount of substrate unwound is indicated below the panel. Bo: boiled substrate. (B) Duplex DNA substrates containing various lengths of 3′-tail (as described at the right of the panel) were incubated with Mph1 under reaction conditions used above for various incubation periods (0, 5, 20 and 60 min) and the results plotted.
Mph1 unwinds 5′-flap-structured DNA
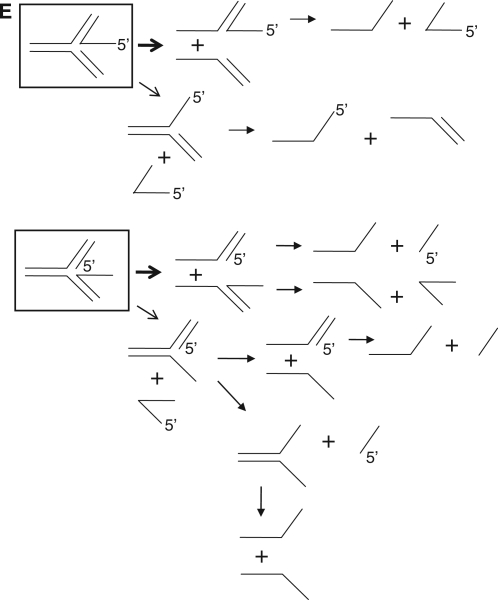
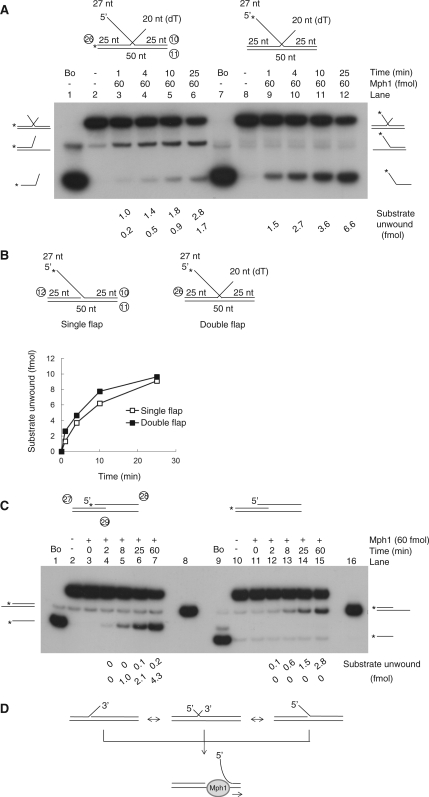
We showed previously that Mph1 displaced 5′-flap strands from a 5′-flap-structured substrate (7). This unusual unwinding activity has been also reported for other 3–5′ helicases including the S. pombe Fml1, E. coli RecQ and Bloom and Werner proteins (9,18–20). In order to understand the unwinding properties of Mph1, we first examined the unwinding of 5′-flap and simple Y fork-DNA structures that differed only by the presence or absence of a 25-nt oligonucleotide upstream of the labeled 5′-flap strand. We also examined substrates devoid of flaps that are equivalent to nicked duplex DNAs. All of these substrates except for the nicked duplex DNA were unwound identically as a function of time or level of Mph1 added (data not shown). When we examined the influence of the length of 5′-flap on unwinding, the substrate containing the 1- or 5-nt flaps was virtually inactive like the nicked duplex, whereas the 15-nt flap substrate was efficiently unwound (Figure 2A). In contrast to the marked effects noted with the length of the 5′-flap structures, the 5′ tail length of forked substrates did not influence the efficiency of the Mph1 unwinding reaction as much as the length of the 5′-flap (compare Figure 2A and B; Supplementary Figures S1A and B). Rather, longer 5′ tailed substrates were slightly less effectively unwound compared to the substrate without a 5′ tail. Thus, Mph1 was not stimulated by fork-structured DNA, suggesting that the unwinding mechanism of Mph1 differs from replicative or other helicases such as the S. pombe Pfh1 and Mcm4-6-7 (21–24).
The results above suggest that Mph1 translocates along ssDNA and then unwinds duplex DNA regions that it subsequently encounters or that it recognizes junction structure directly and unwinds it; the presence of 5′-branched structures is essential for unwinding when there is no ssDNA for translocation. Once Mph1 binds to the junction in 5′-flap substrate, it invades into the junction to reach the other ssDNA strand and then translocates along the ssDNA in the 3–5′ direction. If this were the case, interactions between the 5′-flap structure and Mph1 could be affected by the nucleotide sequences of the flap. We tested this notion with identically structured substrates that contained different nucleotide sequences either within the 5′-flap (flap substrates) or the 5′-tail (fork-structured substrates) regions (Figure 2C and D; gel pictures in Supplementary Figures S2A and D). For this purpose, four substrates containing a 27-nt random, homopolymeric dT, dA and dC sequence (random, dT27, dA27 and dC27 in Figure 2C) were prepared. A 5′ tail dG27 was not used in these experiments because such oligonucleotide sequences were difficult to synthesize and could form high-ordered, inter- or intramolecular structures such as G-quadruplex (data not shown). As shown in Figure 2C, the sequences of the 5′-flap structure markedly affected the helicase activity of Mph1; flap substrates containing a random sequence were most efficiently unwound, followed by the dC27 flap structure. In contrast, the dA27 and dT27 flap substrates were hardly unwound; the Mph1 helicase activity observed with fork-structured DNAs was hardly affected by the 5′-tail sequence (Figure 2D). The differences in unwinding efficiency were not due to differences in substrate binding efficiency, since the random tail, dT27 and dA27 flap substrates bound Mph1 with similar efficiencies (Supplementary Figure S2B). Moreover, it was not due to differences in ATPase activities because ATP hydrolysis rates was comparable when unlabeled 5′ random, dT, dA and dC flap substrates were used as cofactors (Supplementary Figure S2C). Therefore, our results support the notion that the mechanisms by which these two substrates are unwound are not the same.
In vivo, 5′-flap structures most likely exist temporarily in various DNA intermediates arising from Okazaki fragment synthesis as well as base excision repair and homologous recombination process. Furthermore, flap structures are likely to be present in the middle of linear duplex DNA and not near free duplex DNA ends like the in vitro oligonucleotide-based substrates tested in this study. Therefore, we investigated whether Mph1 can be loaded directly onto a 5′-flap structure present in the middle of a duplex DNA. To this end, we prepared a substrate containing two 5′-flaps as illustrated in Figure 3A; one of the flaps (f2) was labeled at its 5′-end. If the unwinding activity of Mph1 were absolutely dependent on free duplex DNA ends for entry, we would not expect formation of the f2-flap only intermediate (with the f1 flap displaced) from the f2-flap-labeled substrates [Figure 3A, i). The formation of the f2 flap only intermediate would require the displacement of the f1 flap prior to the displacement of the f2 flap (Figure 3A, ii). We found that incubation of Mph1 with f2-labeled substrate resulted in the formation of two different products; the labeled f2 flap itself and the f2 flap only intermediate (Figure 3B). For the formation of latter, Mph1 should be loaded directly onto dsDNA between the two flaps or be able to recognize the f1 flap directly (Figure 3A, ii). Ultimately, however, the results above suggest that the ability of Mph1 to displace flap structure is independent of free duplex DNA ends.
Mph1 DNA helicase activity on double flap-structured DNA
We next examined the action of Mph1 helicase on double-flap substrates that contained both 5′ and 3′-flaps (Figure 4). Theoretically, either or both flaps could be displaced by Mph1 in view of its 3–5′ translocation along ssDNA (2,7) and its ability to displace the 5′-flap in flap substrates. In order to determine which flap was displaced more efficiently when present in one substrate, a double flap-structured DNA substrate was prepared with a 3′ dT20 tail (see Figure 4A for structure). The rate of displacement of two identical but differentially radiolabeled substrates (one at the 3′-flap strand, and the other at the 5′-flap strand) was determined (Figure 4A). The labeled 3′-flap substrate yielded distinct products; a fork-structured DNA generated by displacement of 5′-flap, and the displaced 3′-flap itself (Figure 4A, lanes 1–6). When the rate of displacement of the 5′- and 3′-flap strands was compared, we found that the 5′-flap was released more rapidly (2- to 5-fold) than the 3′-flap (Figure 4A, lanes 1–6). When the 5′-flap-labeled substrate was used, the only product detected was the labeled 5′-flap strand (Figure 4A, lanes 7–12). This finding, in keeping with our previous data, indicates that 5′-flaps are readily dissociated from template by Mph1. This also suggests that the release of the free 3′-flap strand (observed with the 3′-flap labeled substrate) most likely arises from fork-structures generated by the initial dissociation of the 5′-flap. We excluded the possibility that release of the 5′-flap by Mph1 could be inhibited by the presence of 3′-flap since the unwinding rates of the 5′-flap- and double-flap substrates were comparable (Figure 4B) and the sequence of 3′-flap did not alter meaningfully the unwinding rate of the 5′-flap when we repeated similar experiments with homopolymeric 3′ dT, dA or dC flap substrates (Supplementary Figure S3).
Figure 4.
Mph1 DNA helicase activity on double flap-structured and equilibrating flap DNA. (A) Helicase assays were performed on double flap-structured substrates for various incubation periods (1, 4, 10 and 25 min). Two identical substrates but labeled on different oligonucleotides were used in these assays; one was labeled on the upstream oligonucleotide (lanes 1–6) and the other on the downstream oligonucleotide (lanes 7–12). (B) Helicase assays were performed on 5′-flap and double flap-structured substrates as a function of time. The amount of substrate unwound was plotted against the incubation period (0, 2, 5, 10 and 25 min). (C) Mph1 helicase assays were performed with equilibrating flap substrates using standard reaction conditions with increasing time of incubation (0, 2, 8, 25 and 60 min). Substrates were radio-labeled at the 5′-end of one of the two oligonucleotides annealed to the template as indicated with asterisks. (D) The mode of displacement of equilibrating flaps by Mph1 is illustrated.
Mph1 DNA helicase activity on equilibrating flap-structured DNA
We next examined how Mph1 unwound equilibrating flaps. The displaced 5′-flaps formed from the 5′-end region of a downstream Okazaki fragment by pol δ-catalyzed displacement reaction can compete for annealing to the template strand with the newly synthesized 3′-end of upstream Okazaki fragments, generating both 5′ and 3′-flaps that vary in length. The numerous intermediates arising in this manner are called ‘equilibrating flaps’, and can be efficiently removed by the combined action of Fen1 and Dna2 (25,26). In order to investigate whether Mph1 utilized this structure, we prepared equilibrating-flap substrates labeled at the 5′-end of one of the two annealed oligonucleotides. When we carried out a time-course experiment with an equilibrating flap substrate containing a labeled downstream oligonucleotide, we found that Mph1 displaced the labeled oligonucleotide preferentially, while the upstream oligonucleotide was hardly displaced (Figure 4C, lanes 1–7), since the 3′-overhang partial-duplex intermediates hardly accumulated. We obtained the same result when the experiment was carried out with an equilibrating flap substrate labeled on the upstream oligonucleotide (Figure 4C, lanes 9–15). These observations are consistent with our findings above that Mph1 displaced the 5′-flap strand in fixed double flap substrates preferentially (Figure 4A and B). It is likely that this property of the Mph1 helicase would generate 5′-flaps from equilibrating flaps that can be preferentially cleaved by Fen1 or Dna2 as shown in Figure 4D.
Mph1 DNA helicase activity on 3′-flap-structured DNA
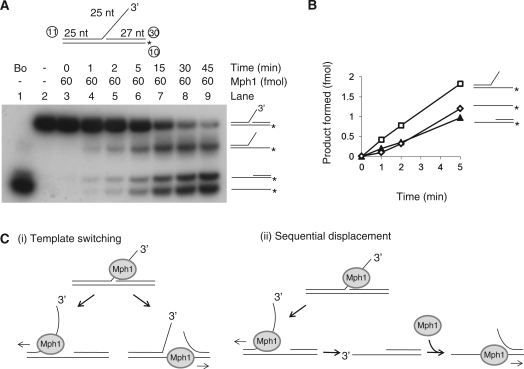
The action of Mph1 on 3′-flap substrates was examined in a similar way as described above with the 5′-flap substrate. As shown in Figure 5A, the substrate was labeled on the template strand. When a time-course experiment was done, three different unwound products were formed (Figure 5A); a Y-structured DNA lacking the downstream 27-nt oligonucleotide (=oligonucleotide 30 in the substrate shown above the autoradiography) and a simple partial-duplex DNA lacking the upstream 3′-flap strand (oligonucleotide 11) and the labeled template strand. Although it appeared that they all formed with a similar kinetics (Figure 5A), analysis of early time points revealed that Mph1 displaced the downstream oligonucleotide (products, Y-structured DNA) more rapidly than the 3′-flap strand (products, the simple partial duplex) (Figure 5B). The presence of the Y-structured DNA and the simple partial duplex at early time points suggests two possible ways for Mph1 to utilize the substrate DNA tested. The formation of products lacking the downstream oligonucleotide is consistent with the mechanism involving template switching as shown (Figure 5C, i); by this mode of action, the translocation of Mph1 helicase bound at the 3′-flap has two alternative fates; it can translocate along the 3′-flap strand and displace the annealed template, generating a 3′-overhang partial duplex. In the template-switching mechanism, it can switch from the 3′-flap to the template strand and displace the downstream oligonucleotide, generating forked-structured products. Thus, the template-switching mechanism allows Mph1 to displace the downstream oligonucleotide without unwinding the 3′-flap strand. The higher levels of the Y-structured DNA than the simple partial-duplex products suggest that template switching of Mph1 could occur rather efficiently. The alternative mechanism involves sequential displacement (Figure 5C, ii) in which Mph1 would dissociate the 3′-flap strand first, producing the 3′-overhang partial duplex that can be unwound further by Mph1. Thus, the accumulation of the 3′-overhang would be expected to precede formation of the downstream 27-nt oligonucleotide. Taken together, the results derived from these experiments further indicate that Mph1 can unwind flap structure regardless of their polarity.
Figure 5.
Mph1 DNA helicase activity on 3′-flap-structured DNA. (A) Helicase assays were carried out on 3′-flap-structured substrates as a function of incubation time (0, 1, 2, 5, 15, 30 and 45 min). (B) The results obtained in (A) were plotted against incubation period; note that results from the early time period (0–5 min) are shown. (C) predicted modes of unwinding of 3′-flap-structured DNA by Mph1 are illustrated.
Mph1 DNA helicase activity on secondary-structured flap DNA
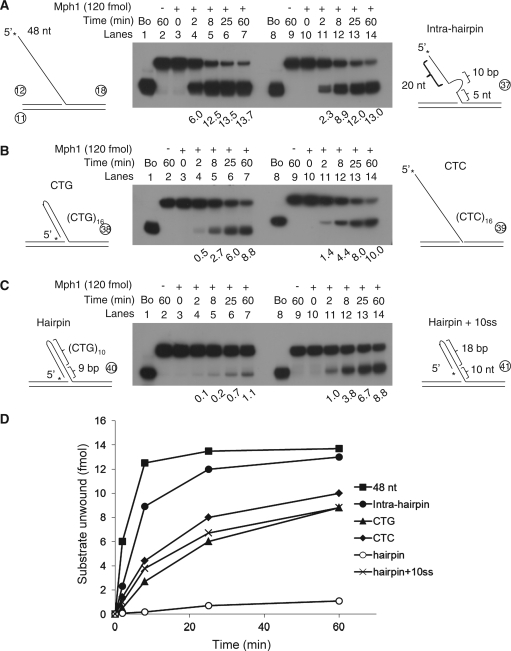
Recently, we have reported that Mph1 was involved in Okazaki fragment processing by virtue of its ability to stimulate Fen1 activity in an ATPase-independent manner (9). Although the catalytic activity was not directly required for Fen1 stimulation, it is likely that the unique property of Mph1 helicase activity might still contribute to processing of Okazaki fragments, for example, by removing any secondary-structure flap from the template that is resistant to cleavage by either Dna2 or Fen1. In order to test this idea, we prepared several flap substrates in which flaps were identical in length, but differed in their structures (Figure 6) in a similar manner as reported (24). We found that Mph1 was able to displace, downstream flap-containing oligonucleotides in all substrates, but with varying efficiencies. Non-structured flaps were displaced most efficiently as expected (Figure 6A, lanes 1–7). Insertion of a 10-bp hairpin in the middle of flap hardly interfered with unwinding (Figure 6A, lanes 8–14). A (CTG)16 flap containing 16 repeats of CTG that forms a hairpin in solution (27–30) reduced the unwinding rate greatly (12-fold) especially in early time points; for example, 6 and 0.5 fmol in 2 min with random and (CTG)16 flap, respectively (compare lanes 1–7 in Figure 6A and B). However, Mph1 showed 2- to 3-fold increase in the unwinding rate in early time points with non-structured (CTC)16 flap with respect to the (CTG)16 flap (Figure 6B, compare lanes 1–7 and 8–14). We believe that the slow rate of unwinding is inherent to the non-structured (CTC)16 repeats, because flaps composed of dC or dT only were the poor substrates for Mph1 as shown in Figure 2C. When we tested a 5′-duplex flap consisting of (CTG)10 and 9-bp duplex (which is identical to a nicked three-way junction), Mph1 unwound, but very poorly (Figure 6C, lanes 1–7). In contrast, the 5′-duplex flap containing 10-nt ssDNA near the junction was efficiently unwound by Mph1 (Figure 6C, lanes 8–14). This result emphasizes the importance of ssDNA for Mph1 helicase activity in this substrate. As shown in Figure 6D, the 5′ duplex flap substrate (indicated as ‘hairpin’) was least efficiently unwound by Mph1. In addition, this finding suggests that unwinding of the (CTG)16 flap substrate (Figure 6B, left) could be due to ssDNA near the junction, most likely because self-annealed hairpins failed to form a perfect duplex, thus leaving a short stretches of ssDNA region available for binding of Mph1.
Figure 6.
Mph1 DNA helicase activity on secondary-structured flap substrates. Mph1 helicase assays were performed with six different secondary- or non-structured substrates in standard reaction condition with increasing the time of incubation (0, 2, 8, 25 and 60 min). The standard helicase assays were carried out with simple and intra-hairpin flap substrates (A), with (CTG)16- and (CTC)16-flap substrates (B) and with substrates containing perfect hairpin or hairpin plus 10-nt ssDNA in the flap (C). The substrates used are illustrated in each side of the figure and see the text for the detailed description. The amount of substrate unwound is indicated below each lane (fmol). (D) the amount of substrate unwound in the assay A was plotted against the incubation periods. One representative result was shown from several experiments carried out similarly.
Mph1 activity on four-way junction DNA
Both Fml1 and FANCM were shown to catalyze branch migration with a four-way junction substrate that mimics Holliday junctions (9,10). The branch migration activities of Fml1 and FANCM require homologous ‘movable’ sequences in a four-way junction substrate like human Rad54 and E. coli RuvA-RuvB proteins which are either inactive or poorly active with four heterologous armed substrates (9,10,31,32). In the course of this study, we noted that Mph1 supported branch migration of non-movable four-way junction substrate although the activity was relatively weak compared to other known branch-migrating helicases (Fml1, RecQ1, Bloom and Werner) (6,33–35). When we performed assays using a non-movable four-way junction substrate which had 24- to 27-bp arms, we detected formation of low levels of branch-migrated products with 12 nM Mph1 (data not shown), similarly to the substrate containing the complete duplex flap (three-way junction substrate) in Figure 6C and D. However, when the DNA sequences in the junction region were replaced with a 10-bp homologous movable core, the level of branch-migrated products formed increased with incubation time (Figure 7A). In order to confirm that this activity was intrinsic to Mph1 helicase, we determined the nucleotide(s) requirement for this reaction (Figure 7B). Products formed by branch migration activity were detected mainly in the presence of ATP and dATP (Figure 7B; lanes 3 and 7, respectively), while much less activity was observed in the presence of dCTP (lane 10). This result is identical to those obtained with partial-duplex DNA substrates (data not shown). In the presence of a non-hydolyzable ATP (ATPγS) or in the absence of Mg2+, no unwound product was detected, indicating that ATP hydrolysis is essential for this activity (Figure 7B, lanes 12 and 13). Thus, it appears that the branch migration activity is intrinsic to Mph1 helicase.
Depending on the direction of junction migration, the branch-migration activity of Mph1 could generate two different fork-structured DNAs from the 10-bp homologous movable core substrate; one formed by horizontal junction migration that results in the shortening of the two horizontal arms (E, east or W, west), and the other by migration of the two vertical junction arms (N, north or S, south) as illustrated in Figure 7A. The two products derived from horizontal or vertical junction migration can be distinguished by gel analysis (Figure 7B, lanes 14 and 15 for vertical and horizontal junction migration control, respectively) since the duplex length of each arm is different; 15, 14, 27 and 25 bp for E, W, N and S arm, respectively (Figure 7A). Experimentally, we detected only one kind of fork-structured DNA produced by horizontal junction migration based on its size (Figure 7B). This most likely resulted from the difference in size of each arm; the longer the double-stranded DNA arm, the more difficult it was for Mph1 to migrate branch. Thus, Mph1 appears to have a limited ability to catalyze branch migration, which will be discussed in detail below.
Mph1 activity on replication fork DNA structure
Although both 5′- and 3′-flaps were efficiently unwound in the presence of 3 nM Mph1 (60 fmol) under standard assay conditions, double-stranded three-way junction substrates that mimicked replication forks required higher concentrations of Mph1 for detectable unwinding (data not shown; see also Figure 6C). When we increased the concentration of Mph1 to 12 nM (~120 fmol in 10 μl), unwound products were easily detected (Figure 8A, lanes 1–10). The products formed were largely ssDNA, indicating that the fork structures were first dissociated into the two partial-duplex products (5′- and 3′-overhang partial duplexes). The 3′-overhang partial-duplex DNA underwent further unwinding to form ssDNA products. When we used a substrate with a partially movable branch point (by introducing homologous sequences indicated in thick and dotted lines; Figure 8A, lanes 11–20), Mph1 generated unwound products that were markedly different from those formed from the fixed three-way junction substrate (Figure 8A, compare lanes 1–10 and 11–20). The most prominent products formed from the template-labeled substrate were forked structures (the slowest migrating products in lanes 13–15) that had lost the two annealed oligonucleotides corresponding to nascent DNA. This was confirmed by the increased release of the labeled nascent strand (Figure 8A, lanes 16–20). We also tested a three-way junction substrate that resembled replication forks in vivo, in which nascent and parental DNA strands were completely complementary. The branch point in such substrate can move or regress, creating four-way junction structures, until two double-stranded DNAs are formed (see Figure 8C for illustration). When Mph1 was incubated with this substrate, it promoted ‘fork regression’ as shown in Figure 8B (compare lanes 2–7 and 8–13). This activity required the same nucleotides that supported helicase and branch migration activities as summarized in Figure 8D. These data suggest that Mph1, like Rad5, can remodel replication fork structures by using a template-switching mechanism for DNA damage avoidance (36).
We also examined how Mph1 acted on regressed leading or lagging strand replication forks (Figure 9). Since regressed replication forks are equivalent to the presence of a ssDNA overhang at the branch point in a fixed three-way junction substrate, we speculated that such a DNA substrate would be efficiently utilized by Mph1. For this purpose, two identical time-course experiments were performed using each type of substrate labeled differently and the unwound products formed were analyzed. With regressed lagging strand replication forks (Figure 9A and B), Mph1 dissociated the parental duplex regions more rapidly than other duplex regions, producing mostly the products labeled P1 (Figure 9A) and P5 (Figure 9B). Mph1 dissociated the 5′-overhang strand, forming P2 (Figure 9A) and P3 (Figure 9B). In addition, Mph1 also displaced both strands corresponding to newly synthesized DNA at replication forks, producing P4 (Figure 9B). The 5′-overhang ssDNA product (P2), formed by regression of the newly synthesized lagging strand was more prominent than the other minor products (P3 and P4). We repeated the same experiment with regressed leading strand replication fork substrates (Figure 9C and D) and found that Mph1 still efficiently displaced the parental duplex regions, producing P7 and P12 (Figure 9C and D, respectively). Like regressed lagging strand replication fork substrates, Mph1 displaced one or both of the newly synthesized strands at replication forks, producing P6 and P8 (Figure 9C) and P9, P10 and P11 (Figure 9D). We noted the slow release of P13, which required the unwinding of two different duplex regions (Figure 9D). As illustrated in Figure 9E, Mph1 supported strand separation in a variety of ways on regressed leading or lagging replication forks. Among them, the unwinding of the parental duplex DNA region (that had not as yet been replicated) at replication forks was most efficiently unwound by Mph1. These results suggest that one potential function of Mph1 is to remodel replication forks, permitting cells to cope with various types of stalled or damaged DNA replication forks.
DISCUSSION
We previously found that Mph1 contributed to the processing of Okazaki fragments and noted that the 3–5′ Mph1 helicase dissociated the 5′-flap strand from 5′-flap substrate in an ATP-dependent reaction, an unexpected property for a 3–5′ DNA helicase (7). These observations prompted us to further analyze the mechanism of its helicase action. To this end, we prepared various DNA structures that could arise during DNA replication, repair and recombination. We first determined the nucleoside triphosphate required to support the Mph1 DNA helicase activity. With 3′-overhang ssDNA substrates, Mph1 was most and almost equally active with ATP and dATP (data not shown) and poorly active with dCTP and CTP (18 and 7% compared to ATP, respectively; data not shown). With other substrates, including equilibrating flap and four-way junction substrates, Mph1 displayed the same nucleotide requirements (data not shown; Figures 7B and 8D), suggesting that the intrinsic helicase activity of Mph1 was responsible for unwinding activity observed with all these substrates.
We showed that Mph1 unwound fixed double-flap DNA (an intermediate form of equilibrating flaps) that could arise in vivo through an equilibrium process from unprocessed long 5′-flaps, in such a way that the displacement of 5′-flap among the two flaps occurs first (Figure 4C). This property indicates that Mph1 converts double-flap structures into a single 5′-flap and displaces the 5′-flap-containing strand. Based on this property and the fact that Fen1 can generate ligatable nicks only from 5′-flaps, we propose that the Mph1 helicase activity contributes to Okazaki fragment processing by directing the flap equilibration reaction towards 5′-flap formation illustrated in Figure 4D. Even with the 3′-flap-only substrate, Mph1 displaced the downstream duplex (Figure 5). These observations support our proposal that Mph1 helicase generates 5′-flap structures for subsequent Fen1 cleavage. This model is in keeping with our previous findings that Mph1 stimulates Fen1 activity (7). We believe that the unique properties of Mph1 helicase allow cells to cope with different flap structures that could arise following many DNA transactions. The helicase converts these into products that can be processed by Fen1, an essential step required to form a continuous duplex DNA.
Although the catalytic activity of Mph1 was not required to stimulate the Fen1 activity, the unique properties of the Mph1 helicase activity might still contribute to Okazaki fragment processing in other way; for example, by displacing any secondary-structure flap from the template that is resistant to cleavage by either Dna2 or Fen1. We tested this possibility with several flap substrates in which flaps were identical in length, but differed in their structures as shown in Figure 6. We found that Mph1 displaced flap-containing oligonucleotides in all substrates efficiently, regardless of its secondary structures, as long as short (~5-nt) ssDNA regions were present at the ssDNA–dsDNA junction. This displacement activity could be particularly useful in the processing of hairpin structures 5′-flaps that are resistant to nuclease cleavage or unwinding by other DNA helicases including Dna2. The ability of Mph1 to displace 5′ secondary-structure flap may allow cells to displace the chronically defective Okazaki fragments from the template; this would lead to the generation of a new flap different in nucleotide sequence and/or structure that could be more readily processed by Dna2 and Fen1.
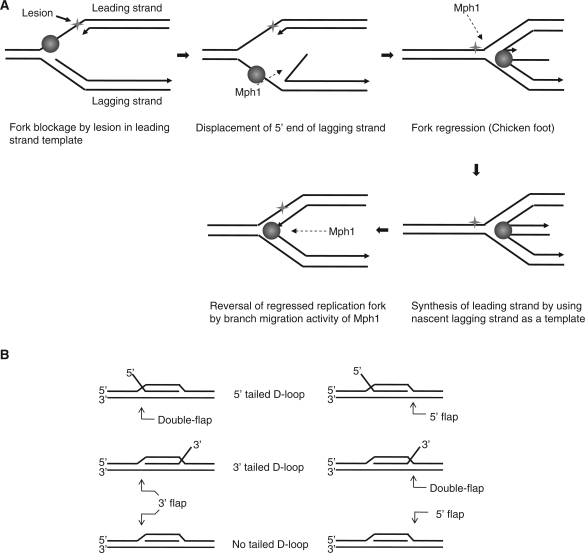
Mph1 was shown to play a role in damage avoidance when DNA replication forks are stalled due to damage lesions in the template as (5). A similar role for the fission yeast homolog Fml1 in this regard was recently proposed (9); it was suggested that stalled forks lead to the formation of a four-way junction by the regression of stalled replication forks when leading strand DNA synthesis is blocked by a damage on template DNA. Our analyses of Mph1 helicase activity suggest that the action of Mph1 leads to fork regression as illustrated in Figure 10A. (i) Mph1 first recognizes stalled replication forks with ssDNA regions exposed. (ii) Mph1 binds to the exposed ssDNA region and displaces the 5′-end of the nascent lagging strand. (iii) This then permits both template strands to re-anneal, resulting in the regression of replication forks for error-free repair of the lesion. This scenario is compatible with a proposed model based on studies with Fml1 in which DNA lesions were shown to be bypassed by using the nascent lagging strand as template (9). (iv) The final step in our model leads to the restoration of regressed replication forks. This could be achieved by Mph1 catalyzing the reversal of the regressed replication fork. We showed that Mph1 could branch-migrate a four-way junction substrate in vitro in the direction leading to the preferential removal of the shortest arm (Figure 7B and C). These are also discussed in recent publications (37–39).
Figure 10.
Model of Mph1 function on stalled replication forks arising from leading strand damage template and mode of Mph1 action in D-loop displacement. (A) A model describing the role of Mph1 in restarting stalled replication forks (see the text for detailed description). (B) Illustration of three representative D-loop substrates (5′-, 3′- or no-tailed) used previously (6). The arrows indicate the structure equivalent to flap structure that can be used for displacement of invaded strand (3′ or double flap as described).
Both Mph1 and Fml1 have also been implicated in the regulation of DNA recombination process by suppressing crossover events (6,9). This has been attributed to Mph1's ability to displace the invaded strand from a D-loop structure. The helicase properties of Mph1, based on our results, further support this hypothesis. Prakash and his colleagues (6) used three different D-loop substrates that differed only in the invaded strand; a D-loop substrate with the invaded strand lacking any tail, while the other two substrates containing only 5′-tail or 3′-tail. They found that the rate of displacement of these invaded strands by Mph1 was almost identical with all three substrates. All three D-loop substrates, regardless of the presence or polarity of the ssDNA tail, contained two junctions where three strands converged. The upstream (or left-side) junction in each substrate (Figure 10B) is structurally equivalent to a double flap in the 5′ tailed D-loop and to a 3′-flap in the 3′ and no tailed D-loop substrates. In these structures, Mph1 would be expected to unwind invaded strands preferentially as shown in Figures 4 and 5. The downstream (right-side) junction is structurally identical to a 5′-flap in the 5′ and no tailed D-loop and to a double flap in the 3′ tailed substrate, but these junctions would not support displacement of invaded strands based on our data (Figures 4 and 5). Collectively, the properties of Mph1 helicase activity described here are well suited to displace the invaded strand preferentially, preventing crossover events during recombination.
During the preparation of this manuscript, several genetic studies on the biochemical role of MPH1 in conjunction with other repair/recombination genes were published. In these studies, it was shown that Mph1 may support formation of template switching intermediates through Rad51-dependent strand invasion, fork regression and the resolution of such intermediates (37,40,41). Our findings in this study are consistent with these observations and provide biochemical explanations for these events. In light of the complex and multiple functions of Mph1 in DNA replication, repair and recombination, it is conceivable that Mph1, like its homologous helicase present in other organisms, could be regulated by or jointly works together with a number of other protein factors. For example, it was shown that branch migration activity of Rad54 and WRN helicase was stimulated by Rad51 and RPA, respectively (42,43). Therefore, further studies are necessary to understand how the helicase activity of Mph1 is influenced and/or regulated by interacting proteins involved in the damage avoidance pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–3.
FUNDING
Funding for open access charge: National Research Foundation of Korea (Grant No. 20100000009) funded by the Ministry of Education, Science and Technology.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Jerard Hurwitz for critical reading of the manuscript.
REFERENCES
- 1.Scheller J, Schürer A, Rudolph C, Hettwer S, Kramer W. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics. 2000;155:1069–1081. doi: 10.1093/genetics/155.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prakash R, Krejci L, Van Komen S, Anke Schürer K, Kramer W, Sung P. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J. Biol. Chem. 2005;280:7854–7860. doi: 10.1074/jbc.M413898200. [DOI] [PubMed] [Google Scholar]
- 3.Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu. Rev. Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- 4.Entian KD, Schuster T, Hegemann JH, Becher D, Feldmann H, Güldener U, Götz R, Hansen M, Hollenberg CP, Jansen G, et al. Functional analysis of 150 deletion mutants in Saccharomyces cerevisiae by a systematic approach. Mol. Gen. Genet. 1999;262:683–702. doi: 10.1007/pl00013817. [DOI] [PubMed] [Google Scholar]
- 5.Schürer KA, Rudolph C, Ulrich HD, Kramer W. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from homologous recombination, but not from postreplicative repair. Genetics. 2004;166:1673–1686. doi: 10.1534/genetics.166.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. Yeast Mph1 helicase dissociates Rad51-made D-loops: implication for crossover control in mitotic recombination. Genes Dev. 2009;23:67–79. doi: 10.1101/gad.1737809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang YH, Kang MJ, Kim JH, Lee CH, Cho IT, Hurwitz J, Seo YS. The MPH1 gene of Saccharomyces cerevisiae functions in Okazaki fragment processing. J. Biol. Chem. 2009;284:10376–10386. doi: 10.1074/jbc.M808894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 2005;37:921–922. doi: 10.1038/ng1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun W, Nandi S, Osman F, Ahn JS, Jakoveleska J, Lorenz A, Whitby MC. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-stranded break repair. Mol. Cell. 2008;32:118–128. doi: 10.1016/j.molcel.2008.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gari K, Décaillet C, Stasiak AZ, Stasiak A, Constantinou A. The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell. 2008a;29:141–148. doi: 10.1016/j.molcel.2007.11.032. [DOI] [PubMed] [Google Scholar]
- 11.Gari K, Décaillet C, Delannoy M, Wu L, Constantinou A. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl Acad. Sci. USA. 2008b;105:16107–16112. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kang YH, Lee CH, Seo YS. Dna2 on the road to Okazaki fragment processing and genome stability in eukaryotes. Crit. Rev. Biochem. Mol. Biol. 2010;45:71–96. doi: 10.3109/10409230903578593. [DOI] [PubMed] [Google Scholar]
- 13.Opresko PL, Cheng WH, Bohr VA. Junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 2004;30:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 14.Kim JH, Seo YS. In vitro assays for studying helicase activities. Methods. Mol. Biol. 2009;521:361–379. doi: 10.1007/978-1-60327-815-7_20. [DOI] [PubMed] [Google Scholar]
- 15.Runyoun GT, Bear DG, Lohman TM. Escherichia coli helicase II (UvrD) protein initiates DNA unwinding at nicks and blunt ends. Proc. Natl Acad. Sci. USA. 1990;87:6383–6387. doi: 10.1073/pnas.87.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Z, Lu S, Hou G, Ma X, Sheng D, Ni J, Shen Y. Hjm/Hel308A DNA helicase from Sulfolobus tokodaii promotes replication fork regression and interacts with Hjc endonuclease in vitro. J. Bacteriol. 2008;190:3006–3017. doi: 10.1128/JB.01662-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor AF, Smith GR. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J. Mol. Biol. 1985;185:431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- 18.Brosh RM, Jr, Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 19.Hishida T, Han YW, Shibata T, Kubota Y, Ishino Y, Iwasaki H, Shinagawa H. Role of the Escherichia coli RecQ DNA helicase in SOS signaling and genome stabilization at stalled replicaton forks. Genes Dev. 2004;18:1886–1897. doi: 10.1101/gad.1223804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matson SW, Kaiser-Rogers KA. DNA Helicases. Annu. Rev. Biochem. 1990;59:289–329. doi: 10.1146/annurev.bi.59.070190.001445. [DOI] [PubMed] [Google Scholar]
- 22.Lee JK, Hurwitz J. Processive DNA helicase activity of the minichromosome maintenance proteins 4, 6 and 7 complex requires forked DNA structures. Proc. Natl Acad. Sci. USA. 2001;98:54–59. doi: 10.1073/pnas.98.1.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.You Z, Ishimi Y, Mizuno T, Sugasawa K, Hanaoka F, Masai H. Thymine-rich single-stranded DNA activates Mcm4/6/7 helicase on Y-forked and bubble like substrate. EMBO J. 2003;17:6148–6160. doi: 10.1093/emboj/cdg576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu GH, Tanaka H, Kim DH, Kim JH, Bae SH, Kwon YN, Rhee JS, MacNeill SA, Seo YS. Genetic and biochemical analysis of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res. 2004;32:4205–4216. doi: 10.1093/nar/gkh720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu Y, Zhang H, Veeraraghavan J, Bambara RA, Freudenreich CH. Saccharomyces cerevisiae flap endonuclease 1 uses flap equilibration to maintain triplet repeat stability. Mol. Cell. Biol. 2004;24:4049–4064. doi: 10.1128/MCB.24.9.4049-4064.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JH, Kim HD, Ryu GH, Kim DH, Hurwitz J, Seo YS. Isolation of human Dna2 endonuclease and characterization of its enzymatic properties. Nucleic Acids Res. 2006;34:1854–1864. doi: 10.1093/nar/gkl102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gacy MN, Goellner G, Juranić N, Macura S, Mcmurray CT. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81:533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 28.Pearson CE, Tam M, Wang Y-H, Montgomery SE, Dar AC, Cleary JD, Nichol K. Slipped-strand DNAs formed by long (CAG)•(CTG) repeats: slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petruska J, Arnheim N, Goodman MF. Stability of intrastrand hairpin structures formed by the CAG/CTG class of DNA triplet repeats associated with neurological diseases. Nucleic Acids Res. 1996;24:1992–1998. doi: 10.1093/nar/24.11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith GK, Jie J, Fox GE, Gao X. DNA CTG triplet repeats involved in dynamic mutations of neurologically related gene sequences form stable duplexes. Nucleic Acids Res. 1995;23:4303–4311. doi: 10.1093/nar/23.21.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 32.Iwasaki H, Takahagi M, Nakata A, Shinagawa H. Escherichia coli RuvA and RuvB proteins specifically interact with Holliday junctions and promote branch migration. Genes Dev. 1992;6:2214–2220. doi: 10.1101/gad.6.11.2214. [DOI] [PubMed] [Google Scholar]
- 33.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. USA. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bugreev DV, Brosh RM, Jr, Mazin AV. RECQ1 possesses DNA branch migration activity. J. Biol. Chem. 2008;283:20231–20242. doi: 10.1074/jbc.M801582200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blastyák A, Pintér L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication for regression. Mol. Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt KH, Viebranz EB, Harris LB, Mirzaei-Souderjani H, Syed S, Medicus R. Defects in DNA lesion bypass lead to spontaneous chromosomal rearrangements and increased cell death. Eukaryot. Cell. 2009;9:315–324. doi: 10.1128/EC.00260-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ede C, Rudolph CJ, Lehmann S, Schürer KA, Kramer W. Budding yeast Mph1 promotes sister chromatid interactions by a mechanism involving strand invasion. DNA Repair. 2011;10:45–55. doi: 10.1016/j.dnarep.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 39.Whitby MC. The FANCM family of DNA helicases/traslocases. DNA Repair. 2010;9:224–236. doi: 10.1016/j.dnarep.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 40.Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl Acad. Sci. USA. 2009;106:21252–21257. doi: 10.1073/pnas.0908258106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panico ER, Ede C, Schildmann M, Schürer KA, Kramer W. Genetic evidence for a role of Saccharomyces cerevisiae Mph1 in recombinational DNA repair under replicative stress. Yeast. 2009;27:11–27. doi: 10.1002/yea.1727. [DOI] [PubMed] [Google Scholar]
- 42.Rossi MJ, Mazin AV. Rad51 protein stimulates the branch migration activity of Rad54 protein. Nucleic Acids Res. 2008;283:24698–24706. doi: 10.1074/jbc.M800839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sowd G, Wang H, Pretto D, Chazin WJ, Opresko PL. Replication protein A stimulates the Werner syndrome protein branch migration activity. J. Biol. Chem. 2009;284:34682–34691. doi: 10.1074/jbc.M109.049031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.