Abstract
The influenza A virus genome consists of eight viral RNAs (vRNAs) that form viral ribonucleoproteins (vRNPs). Even though evidence supporting segment-specific packaging of vRNAs is accumulating, the mechanism ensuring selective packaging of one copy of each vRNA into the viral particles remains largely unknown. We used electron tomography to show that the eight vRNPs emerge from a common ‘transition zone’ located underneath the matrix layer at the budding tip of the virions, where they appear to be interconnected and often form a star-like structure. This zone appears as a platform in 3D surface rendering and is thick enough to contain all known packaging signals. In vitro, all vRNA segments are involved in a single network of intermolecular interactions. The regions involved in the strongest interactions were identified and correspond to known packaging signals. A limited set of nucleotides in the 5′ region of vRNA 7 was shown to interact with vRNA 6 and to be crucial for packaging of the former vRNA. Collectively, our findings support a model in which the eight genomic RNA segments are selected and packaged as an organized supramolecular complex held together by direct base pairing of the packaging signals.
INTRODUCTION
The packaging machinery of RNA viruses must discriminate the genomic viral RNA (vRNA) against a variety of cellular RNAs, and, most often, other viral RNA species. Usually, selective packaging of the vRNA is achieved by the specific binding of a viral structural protein to a packaging signal solely present in this RNA. This general problem reaches an additional level of complexity in viruses whose genome is segmented, as in the Orthomyxoviridae family. Among these viruses, Influenza A viruses are an important threat for public health (1). The influenza A virus genome consists of eight single-stranded negative-sense RNA segments ranging from 890 to 2341 nt that are associated with nucleoprotein (NP) and a heterotrimeric polymerase complex (2), and appear as individual ribonucleoprotein complexes (vRNPs) inside an enveloped filamentous or spherical structure (3–7). The eight vRNAs have a common organization: a long central coding region (in antisense orientation) is flanked by relatively short segment-specific untranslated regions (UTRs) and by two partially complementary terminal promoters (12 nt at the 3′-end and 13 nt at the 5′-end). These are conserved between segments and impose a panhandle structure to the vRNPs (8,9). As the natural transmission of influenza A viruses is initiated at low multiplicity of infection (m.o.i.), viral particles that do not incorporate at least one copy of each genomic segment will fail to replicate (10).
Two models have been proposed for the packaging of the vRNAs into influenza A viral particles: a random model and a segment-specific model, but growing evidence supports the existence of a specific packaging mechanism (10). First, genetic evidence indicated that influenza A virions normally incorporate exactly eight vRNAs (11–13). Second, electron microscopy of a variety of influenza A virus strains repeatedly showed that seven vRNPs are organized around a central one (3–5,7). In elongated and filamentous viral particles, the vRNPs contact the matrix layer at one of the hemispherical cap of the virions, which has been identified as their budding tips (3,7). Third, defective interfering RNAs generated from the vRNA segments revealed the presence of segment-specific packaging signals within ~100 to ~300 nt from each end of the vRNAs [for a review, see Ref. (10)]. Forth, reverse genetics experiments using segments in which a reporter gene was substituted for most of the central coding region pointed to the existence of segment-specific packaging signals located in the 3′ and 5′ terminal regions of every influenza A genomic vRNA (14–21). Finally, sequence analysis showed that the most conserved codons tend to accumulate in the terminal packaging regions, and point mutations identified single nucleotides or short nucleotide clusters required for optimal packaging of several vRNAs (5,22,23).
Despite these studies, the molecular basis underlying segment-specific packaging of influenza A vRNAs remains elusive. Here, we used electron tomography, native gel electrophoresis and capture of RNA complexes on magnetic beads to visualize vRNPs inside virions and analyse the interactions between vRNAs. Collectively, our findings support a model in which the eight genomic RNA segments are selected and packaged as an organized supramolecular complex held together by direct base pairing of the packaging signals.
MATERIALS AND METHODS
Electron tomography
Madin–Darby canine kidney (MDCK) cells were infected with A/Moscow/10/99 (H3N2) virus at low m.o.i. After 24 h, cells and viruses were fixed, stained and embedded in epon as described (6). Tilt series of uranyl-stained 150–200 nm thick sections were acquired on a Tecnai G2 Sphera electron microscope (FEI Company, Eindhoven, The Netherlands) operating at 200 kV. Digital images were automatically recorded on a CCD camera (Gatan Ultrascan 1000) at ×25 000 nominal magnification (pixel size 0.46 nm) over a tilt range of −60 to +60 with 1° increments. The images in the tilt series were pre-processed and aligned with or without gold fiducial markers. The eTomo graphical user interface of the IMOD tomography package was used to calculate the 3D reconstruction (24). The final tomograms were calculated by weighted backprojection. Individual virions were extracted from the calculated tomograms and denoised using an iterative median filter (25). The 3D volumes were imported into the 3D visualization software AMIRA (Mercury Computer Systems, Mérignac, France) for visualization, manual segmentation and surface rendering.
Plasmids and cloning
A/Moscow/10/99 (H3N2) cDNAs were cloned between the BsmbI sites of pUC2000 vectors starting from the reverse genetic plasmids (6). These vectors were derived from pUC19 by introducing a PstI restriction site, removing the multiple cloning site cassettes and inserting a T7 promoter, the cloning cassette of pHW2000 containing two BsmbI sites, and a unique restriction site (Eco47III, Bsh1236I or Ecl136II) between the PstI and EcoRI sites.
In vitro transcription and electrophoretic mobility shift assay
DNA templates digested with Eco47III, Bsh1236I or Ecl136II were co-transcribed in vitro for 3 h and RNA complexes were analysed by native agarose gel electrophoresis as described (26), except that the samples were treated with 5 U of RNase-free DNaseI (Roche) and that phenol–chloroform extraction was omitted. Briefly, linerarized plasmids were incubated in 10 µl reactionnal volume containing 40 mM Tris–HCl pH 7.5, 50 mM NaCl, 15 mM MgCl2, 1 mM spermidine, supplemented with 5 mM DTT, 4 mM of each NTP, 1 U RNasine (Promega), 0.05 mg/ml BSA, 0.05% Triton X-100 and 0.5 µl of home-made T7 RNA polymerase. After 3 h of incubation, samples were treated with 5 U of RNAse-free DNase I (Roche) for 1 h at 37°C, supplemented with 2 µl loading buffer and loaded on a 0.8% agarose gel containing 0.01% ethidium bromide. Electrophoresis was performed for 4 h at 150 V at 4°C in a buffer containing Tris 100 mM, borate 85 mM, MgCl2 0.1 mM. Gels were analysed using a Gel Doc (Bio-Rad) imager and the Quantity One software. The weight fraction (%) of the RNA mass corresponding to each band in a lane was determined, and the percentage of intermolecular complex was determined by dividing the weight fraction of the corresponding band by the sum of the weight fractions of all bands in the lane. Integrity of the RNAs was routinely checked by denaturing gel electrophoresis.
Nucleoprotein overexpression and purification
The expression plasmid coding for NP was a king gift of Dr Yizhi Jane Tao (Department of Biochemistry and Cell Biology, Rice University, Houston, USA). Overexpression and purification were performed according to their published protocols (27). Briefly, NP was expressed in BL21-DE3 Escherichia coli cells. After centrifugation, the cell pellet was resuspended in 50 mM Tris–HCl pH 7.5, 300 mM NaCl, 5 mM β-mercaptoethanol, treated with lysozyme (1 µg/ml) for 30 min at 4°C and sonicated. After treatment with DNAse I (2.5 µg/ml) and RNase A (5 µg/ml), and centrifugation (30 min at 17 000 g at 4°C), NP was recovered from the supernatant by chromatography using a Ni-NTA affinity column (Qiagen) followed by a HiTrap Heparin HP column (Pharmacia) as described (27). The purified protein was concentrated on a centricon 30 K (Amicon) device and stored at 4°C in 50 mM Tris–HCl pH 7.5, 100 mM NaCl.
Analysis of vRNA complexes on magnetic beads
Wild-type vRNA 7 and vRNA 7Δ100 5′ with a 3′ extension were transcribed in vitro from plasmids linearized with AatII. After transcription, 20 pmol of a 16-mer biotinylated oligonucleotide complementary to the 3′ extension, NP (1 protein/24 nt) and NaCl (100 mM) were added to the reaction mixture (20 µl final volume) and incubated at 37°C. vRNA 6 was incubated with NP in parallel and after 30 min, the two vRNAs were mixed and incubated together for a further 30 min at 37°C. M-280 magnetic beads (Invitrogen, 20 µl) were added and the complexes were allowed to bind for 30 min at room temperature. The beads were washed twice, then treated with 100 µl formaldehyde 95%, EDTA 10 mM. After ethanol precipitation, vRNA aliquots were reverse transcribed with AMV RT (MP Biomedicals) and amplified by PCR using Dream-Taq polymerase (Fermentas) and segment-specific primers.
Oligonucleotide mapping experiments
Oligodeoxyribonucleotides complementary to vRNA 7 were added to the in vitro transcription reaction (10 µM final concentration, including 10 000 cpm of purified 5′-[32P]-labelled oligonucleotide). After electrophoresis and visualization of the RNA species under UV light, gels were fixed in 10% (v/v) trichloroacetic acid and dried at room temperature.
Cell culture, reverse genetics, virus growth kinetics and competition experiments
MDCK cells were purchased from Cambrex Bioscience (ATCC, CCL34), Walkersville, MD, USA. Cells were passaged twice weekly in serum free Ultra-MDCK medium (Cambrex Bioscience) supplemented with 2 mM l-glutamine (Sigma Aldrich, St Louis, MO, USA), penicillin (225 U/ml) and streptomycin (225 µg/ml) (Cambrex Bioscience, Walkersville, MD, USA). 293T cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% foetal calf serum and supplemented with 2 mM l-glutamine (Sigma Aldrich, St Louis, MO, USA), penicillin (225 U/ml) and streptomycin (225 µg/ml) (Cambrex Bioscience, Walkersville, MD, USA). All cells were maintained at 37°C with 5% CO2.
Wild-type and mutant A/Moscow/10/99 (H3N2) recombinant viruses were produced by reverse genetics as described earlier (6,28). Briefly, the cDNA corresponding to the different genes of the wild-type and mutant viruses were cloned into the pHW2000 vector (28) allowing expression of both corresponding vRNAs and viral proteins. Plasmids were mixed with Superfect reagent (Qiagen) in Opti-MEM (GIBCO-BRL), according to the manufacturer's instructions and added to 293T cells in six-well tissue culture plates. At 48-h post-transfection, viruses in the culture supernatant were harvested and used to infect MDCK cells. After two passages on MDCK cells, titres were measured using standard methods (6).
In competition experiments, 1 µg of wild-type plasmid and 1 µg of the plasmid coding for segment 7 with the S71–R73 mutation were mixed with Superfect reagent (Qiagen) in Opti-MEM (GIBCO-BRL) and added to 293T cells. To define the rate of incorporation of the mutated vRNP into infectious viral particles, plaque purification in MDCK cells was carried out from the supernatant of the transfected cells. Plaque-purified viruses from two independent experiments were analysed by RT-PCR specific for the M segment and sequencing.
RESULTS
The 8 vRNPs are interconnected in a transition zone located underneath the matrix layer at the budding tip of the influenza A viral particles
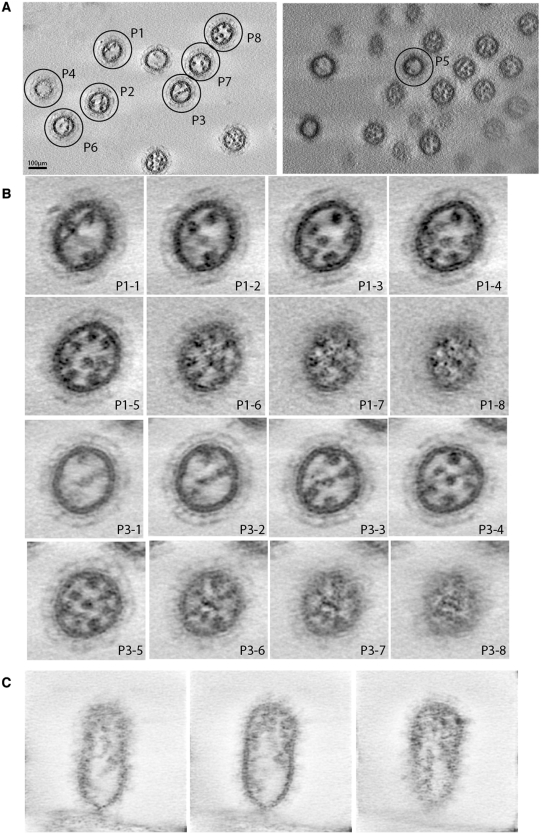
Electron microscopy and tomography previously revealed the typical ‘7 + 1’ disposition of the influenza A vRNPs within viral particles (3–5,7) and showed that the vRNPs contact the matrix layer at the budding tip of the virions (3,7). In some images, pairs of vRNPs appeared to be locally in close proximity, but whether such contacts represented specific interactions with functional significance could not be determined (7). Here, we used electron tomography to visualize the internal organization of virions of the contemporary human influenza A/Moscow/10/99 (H3N2) virus budding from MDCK cells 24-h post-infection. Two large fields of view revealed 10 and 12 viral particles in which individual vRNPs were visible as dots (Figure 1A). Eight particles, named P1–P8, with a low background and distinguishable vRNPs were selected for detailed analysis (Figure 1B; Supplementary Figure S1 and Supplementary Movies S1A and S1B). As previously observed by electron microscopy (7), the number of dots gradually increases from the bottom to the top of the viral particles, up to a point where the eight individual vRNPs can be distinguished (Figure 1B, panels P1-1–P1-5 and P3-1–P3-5, and Supplementary Movies S1A and S1B). We also recorded a tomogram providing longitudinal views of a viral particle still attached to the cell from which it is budding (Figure 1C). These views show that the vRNPs contact the matrix layer at the budding tip of the virion, in agreement with previous electron microscopy data (7). In the cross sections, between the zone where the eight vRNPs are individually visible as round dots and the matrix layer, there is a ‘transition zone’ where the dots progressively become smaller and elongated. Density progressively becomes visible between these dots, which appear to be interconnected, often giving rise to a star-like structure (Figure 1B, panels P1-6–P1-8 and P3-6–P3-8, and Supplementary Figure S1). These data strongly suggest that vRNP/vRNP interactions take place in this transition zone.
Figure 1.
Electron tomography of budding H3N2 influenza A virions. (A) Two single fields of view indicating eight virions, named P1–P8, that have been selected for further analysis. (B) Transversal sections of viral particles P1 (Panels P1-1–P1-8) and P3 (Panels P3-1–P3-8). (C) Longitudinal sections of a viral particle attached to the cell from which it is budding.
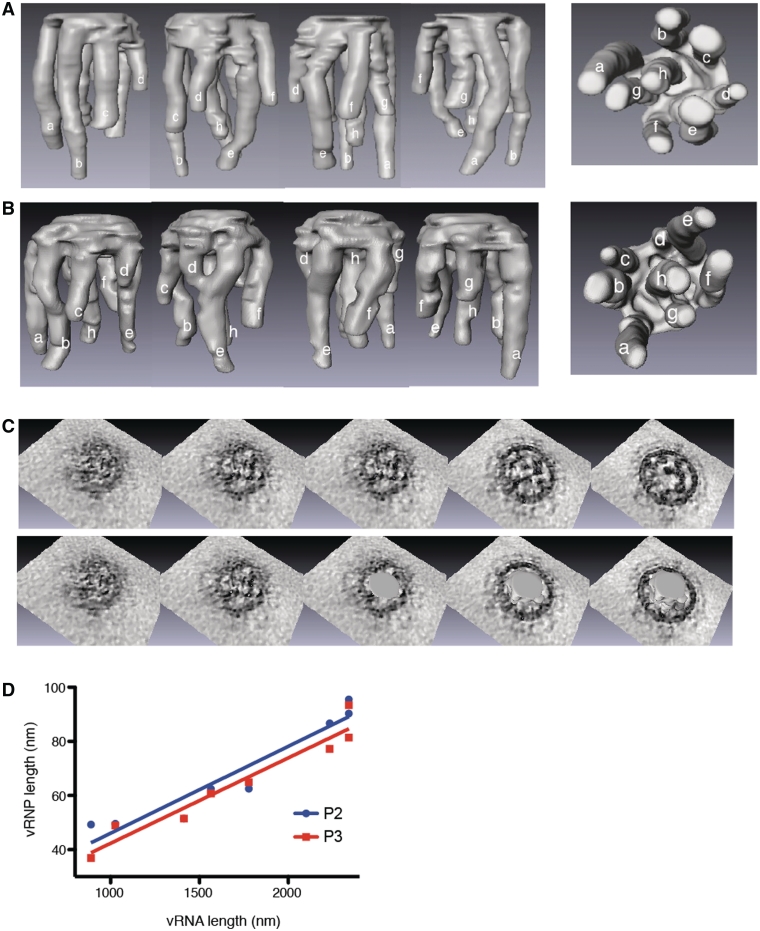
We were able to obtain 3D rendering of the vRNP surface of particles P1–P4 (Figure 2; Supplementary Figure S2 and Supplementary Movies S2A and S2B). In the four particles, eight vRNPs of different length hang from a common platform that includes the ‘transition zone’ of the tomograms close to the top of the viral particles, below the matrix layer, where the vRNPs appear to be interconnected (Figure 2C). The density of the tomograms corresponding to the matrix layer, which is visible all around the viral particles has been omitted from the 3D surface rendering, but the density between the ‘transition zone’ and the matrix layer is continuous (Figure 2C). We systematically measured the thickness of the platform between adjacent outer vRNPs in P1, P2 and P3. The platform thickness varied from 5 nm between segments a and b of P2 to 23.1 nm between segments d and e in P1. The mean thickness of the platform was 13.3 ± 2 nm in P1, 13.8 ± 2.1 nm in P2 and 17.5 ± 0.8 nm in P3. The platform of P4 was thicker (up to 30 nm), and this was likely the consequence of a lower signal over noise ratio for this particle. We thus omitted this particle when comparing the thickness (averaged over P1, P2 and P3) of the platform between successive vRNPs: it ranged from 9.9 ± 1.9 nm between segments f and g to 20.0 ± 1.6 nm between segments d and e. Importantly, in general, no contact between vRNPs was observed outside the platform. The only exception was observed in particle P3, in which the tip of vRNP d and the middle of vRNP e are in close proximity (Figure 1C and Supplementary Figure S2B). As this contact was not reproducibly observed, its significance, if any, is unclear.
Figure 2.
3D surface rendering of the vRNPs in budding H3N2 influenza A virions. (A and B). Side and bottom views of the 3D surfaces of the interior of particles P2 (A) and P3 (B). vRNPs are labelled anticlockwise from a to g, starting with the two longest adjacent vRNPs, and h is the central vRNP. (C) Views of the top of P2 without (upper panel) or with superimposition of the 3D surfaces. (D) Correlation between the length of the vRNPs and the length of the vRNAs. The length of the vRNPs measured in the 3D-reconstructions of P2 (blue dots) and P3 (red squares) is plotted against the length of the vRNAs. The compaction factor of the vRNAs in the vRNP is calculated from the slope of the lines (slope = 0.032 ± 0.004 for P2 and 0.032 ± 0.003 for P3).
Within the virions, vRNPs adopt the hairpin conformation previously observed at high resolution for an artificial mini-vRNP (8,9), and the ‘transition zone’ most likely contains the polymerase complex associated with the ends of each vRNA, as well as the terminal regions of these RNAs. To determine the compaction factor of the vRNPs in particles P2 and P3, we plotted the length of the vRNPs determined from the 3D-reconstructions as a function of the known length of the vRNAs (Figure 2D). The mean compaction value derived from the slope of the lines is 31–32 nt/nm. Taking this compaction factor into account, our data indicate that the platform of virions P1, P2 and P3 can incorporate a mean value of 206–271 nt, ranging from 153 nt between segments f and g to 310 nt between segments d and e.
In vitro, the packaging signals mediate interactions between the viral RNA segments
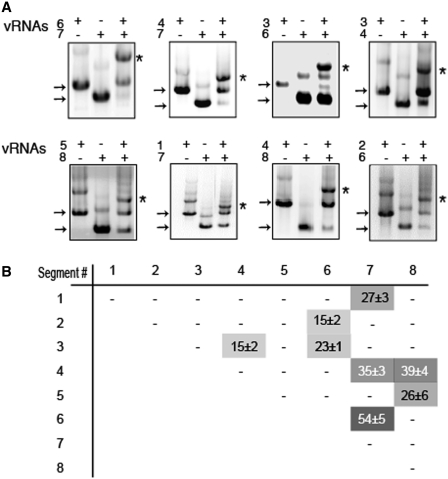
It has been proposed that the packaging signals might be involved in intermolecular base pairing (16). As identification of intermolecular interactions between vRNAs using virology approaches has not been successful yet, we tried to identify such interactions using an in vitro approach. We cloned the segments of the human influenza A/Moscow/10/99 (H3N2) virus under the control of a T7 promoter, and we co-transcribed all possible RNA pairs in vitro and analysed the RNA/RNA interactions by native gel electrophoresis (Figure 3 and Supplementary Figure S3). Even though some vRNAs formed low amounts of homodimers when transcribed alone, these complexes almost completely disappeared to the expense of intermolecular complexes in the co-transcription experiments. Each vRNA formed clearly visible complexes with at least one other RNA segment. In most cases, the band corresponding to the complex was easily identified owing to its migration relative to the monomeric and homodimeric bands and to the fact that it was only present when two vRNAs were co-transcribed (Figure 3A). Only in the cases of vRNAs with similar lengths, were we unable to discriminate between the heterodimeric complex and homodimers (Supplementary Figure S3). However, in these cases, when both vRNAs were co-transcribed, the percentage of homo/heterodimer was lower that the added percentages of homodimers when each vRNAs were transcribed individually, suggesting these vRNA pairs did not interact (Supplementary Figure S3). Out of 28 possible intermolecular interactions, 8 were reproducibly detected, while the remaining ones were weak or undetectable (Figure 3 and Supplementary Figure S3). Not all interactions have the same stability, as the RNA mass engaged in intermolecular complexes ranged from 15% to 54% (Figure 3). Noticeably, the eight strongest interactions are sufficient to interconnect all vRNA segments in a single interaction network.
Figure 3.
In vitro interactions between vRNAs. (A) Analysis of the RNA/RNA interactions by native agarose gel electrophoresis. Individual vRNAs are indicated by arrows and intermolecular complexes are marked by asterisks. (B) Quantification of the complexes. The weight fraction (%) of the RNA mass migrating as an intermolecular complex is expressed as mean ± SEM (n = 3−19). −, <10% complex. The grey levels (on a 0–100 scale) directly correspond to the percentage of complex formed.
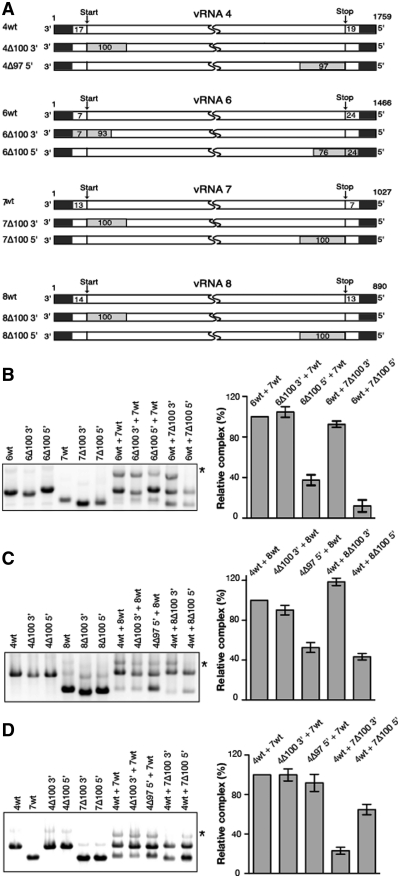
To test whether this network might be relevant to the selective packaging of the vRNPs, we analysed the effect of deletions in the terminal regions of the vRNAs, where the segment-specific packaging signals are located (5,15,16,19,22,23), on the three strongest interactions detected in our assay, namely the interactions between vRNAs 6 and 7, 4 and 8, and 4 and 7 (Figure 3B). Deleting 100 nt in the 5′ region of vRNA 6 reduced the interaction with vRNA 7 by 2.5-fold (Figure 4A and B). Similarly, this interaction was reduced 10-fold by a 100 nt deletion in the 5′ region of vRNA 7 (Figure 4B). On the other hand deletions in the 3′ region of either vRNA 6 or vRNA 7 had no significant effect on the interaction between these vRNAs. Concerning the interaction between vRNAs 4 and 8, deleting 97 or 100 nt at the 5′-end of the coding region of vRNA 4 or 8 reduced complex formation by 2- and 2.5-fold, respectively, whereas deletions at the 3′-end of the coding regions of vRNA 4 or 8 did not impair this interaction (Figure 4A and C). A somewhat different picture was observed with the interaction between vRNAs 4 and 7 (Figure 4A and D). Deletions in the 3′ and 5′ regions of vRNA 7 both impacted this interaction, even though the deletion in the 3′ region had a more dramatic effect (4-fold, versus a 35% reduction). On the contrary, deletions at the 3′ or the 5′-end of the coding region of vRNA 4 had no significant impact on this interaction. It is possible that the region(s) of vRNA 4 interacting with vRNA 7 is (are) located either in the UTRs or in a more central part of the coding region. However, we cannot exclude the existence of stable redundant interactions involving both terminal domains of the coding region of vRNA 4.
Figure 4.
Effect of terminal deletions on the three strongest in vitro vRNA/vRNA interactions. (A) Schematic representation and nomenclature of the wild-type and mutant vRNAs. Deletions are represented by grey rectangles. Numbering of the genomic vRNAs is from 3′- to 5′-end. (B–D) Representative gels and quantifications are shown for interactions between wild-type or mutant vRNAs 6 and 7 (B), 4 and 8 (C) and 4 and 7 (D). Intermolecular complexes are marked by asterisks. The weight fraction (%) of the RNA mass migrating as an intermolecular complex was determined for each vRNA pair, and for each panel the intermolecular complex obtained with mutant vRNAs was normalized relative to the complex formed by the two wild-type vRNAs. Quantifications are expressed as mean ± SEM (n = 3−10).
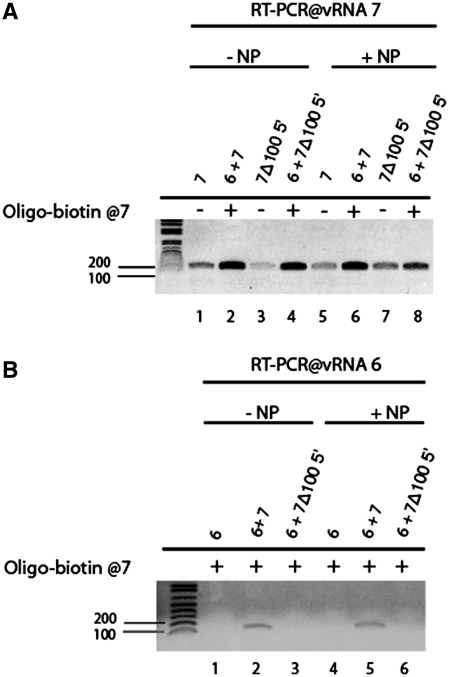
To corroborate the idea that packaging signals mediate interactions between vRNPs via direct vRNA-vRNA interaction, we next focused on vRNA 7 and its interaction with vRNA 6. The analysis of the RNA complexes formed in vitro described above did not allow the study of the intermolecular interactions taking place in the presence of NP: this protein inhibited in vitro transcription, and if using purified vRNAs, the vRNA/NP complexes could not be resolved on agarose gels due to aggregation (data not shown). We therefore developed an alternative protocol in which a modified vRNA 7 without or with a 100 nt deletion at the 5′-end of the coding sequence was modified to include a 3′ overhang complementary to a biotinylated DNA oligonucleotide. Modified vRNA 7 and vRNA 6 were first transcribed and incubated separately with saturating amounts of NP. They were then incubated together, and the NP-covered vRNA 7 and vRNA 7/vRNA 6 complexes were retained on streptavidin-coated beads. Finally, vRNAs retained on the beads were detected by segment-specific RT-PCR. Even though some background binding of vRNA 7 and vRNA 7Δ100 5′ to the beads was detected when the biotinylated DNA oligonucleotide was omitted, the PCR signal was stronger when it was included in the reaction mixture (Figure 5A). Binding of vRNA 6 to the beads was detected in the presence of vRNA 7, but not in its absence or in the presence vRNA 7Δ100 5′ (Figure 5B) in agreement with the analysis by native gel electrophoresis (Figure 4B). Importantly, similar results were observed with ‘naked’ vRNAs and NP-covered RNAs (Figure 5), indicating that NP does not prevent the interaction between vRNPs 6 and 7 and that it does not affect its specificity.
Figure 5.
Interaction of wild-type vRNA 7 and vRNA 7Δ100 5′ with vRNA 6 in the presence of NP. vRNA 7 without or with a 100 nt deletion at the 5′-end of the coding sequence was modified to include a 3′ overhang complementary to a biotinylated DNA oligonucleotide. Modified vRNA 7 and vRNA 6 were first transcribed and incubated separately with saturating amounts of NP, then incubated together. The biotinylated DNA oligonucleotide was used to retain the complexes on magnetic beads, and segment-specific PCR was used to detect vRNA 7 (A) and vRNA 6 (B) retained on the beads in the absence (−NP) or in the presence of NP (+NP).
A short sequence of vRNA 7 interacts with vRNA 6 and modulates packaging of vRNA 7
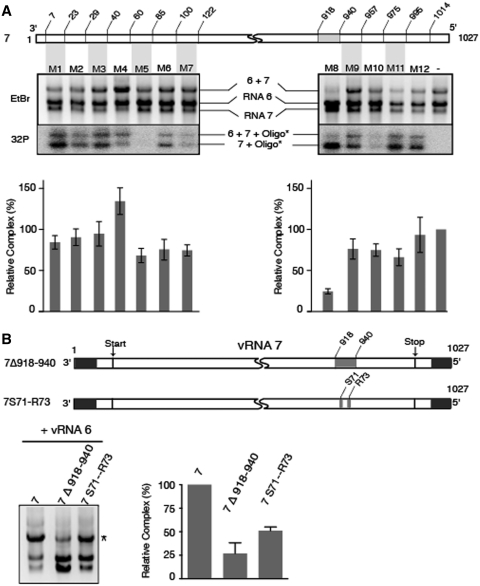
Next, we used the native gel electrophoresis assay to map the region of vRNA 7 interacting with vRNA 6 more precisely by including a series of DNA oligonucleotides complementary to the former vRNA during in vitro transcription (Figure 6A). All oligonucleotides except M5 bound vRNA 7 to some degree under the conditions of in vitro transcription, suggesting that the M5 target sequence is involved in a stable intramolecular structure. Oligonucleotides M1–M7, which bind to the 3′ region of vRNA 7, did not significantly inhibit the interaction with vRNA 6. One of them even increased complex formation (Figure 6A), suggesting that interactions might take place between the 5′ and 3′ regions of vRNA 7, and that binding of an oligonucleotide to the 3′ region might facilitate intermolecular interactions involving the 5′ region. Most oligonucleotides binding to the 5′ region of vRNA 7 also had limited effects, except M8, which reduced formation of the vRNA 6/vRNA 7 complex by 4-fold (Figure 6A). M8 was hardly detected in the complex, further demonstrating that nucleotides 918–940 of vRNA 7 played a central role in the interaction with vRNA 6. Accordingly, deleting the corresponding sequence in vRNA 7 decreased complex formation by 4-fold (Figure 6B, mutant 7Δ918–940). Noticeably, this region encompasses the highly conserved codons S71 and R73 of protein M2 that affect influenza A/PR/8/34 virus growth, virion assembly and genome packaging (5). Importantly, when introduced in our in vitro system, mutations S71–R73 (i.e. substitution of nucleotides 924, 925, 926, 930 and 932) produced a 2-fold reduction of the complex formed between vRNAs 6 and 7 (Figure 6B). Thus, our results show that both codons S71 and R73 and the flanking nucleotides of segment 7 are important for the interaction with vRNA 6.
Figure 6.
Precise mapping of a region of vRNA 7 interacting with vRNA 6. (A) A region interacting with vRNA 6 was identified using oligonucleotides complementary to the 3′ and 5′ regions of vRNA 7. A representative gel is shown. Ethidium bromide (EtBr) staining allowed quantification of the complex formed between vRNAs 6 and 7, while autoradiography of the 32P radiolabelled oligonucleotides allowed to monitor binding of the DNA oligonucleotides to vRNA 7 during in vitro transcription. (B) A deletion or substitutions in the 918–940 region of vRNA 7 affect interaction with vRNA 6. Mutation S71–R73 correspond to substitution of nucleotides 924, 925 926, 930 and 932. In A and B, intermolecular complexes are marked by asterisks. Relative variations of the amount of complex were determined as in Figure 4 and are given as mean ± SEM (n = 3−7).
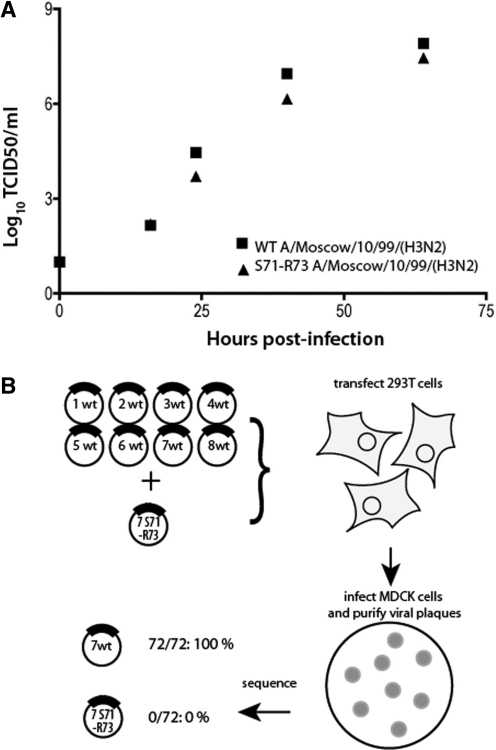
As the importance of codons S71 and R73 in genome packaging has only been demonstrated for a laboratory adapted strain of H1N1 influenza A virus (5), we introduced the same silent mutations in recombinant A/Moscow/10/99 (H3N2), which is a strain that recently circulated in the human population. As compared to the wild-type virus, mutant H3N2 virus produced 5- to 10-fold less infectious virions (Figure 7A). We checked that S71–R73 mutations have no effect on vRNA synthesis or viral gene expression 8-h post-infection (data not shown). Expression of proteins M1 and M2 was analysed by Western blot using mouse monoclonal antibodies after infection at a m.o.i. of 4. To prove that they affect packaging of vRNA 7, we produced recombinant viruses from 293T cells transfected with equimolar amounts of the eight wild-type reverse genetics cDNA clones plus the reverse genetic cDNA clone of segment 7 containing the S71–R73 mutation (Figure 7B). This set-up allows competition between wild-type and mutant vRNA 7 for incorporation into viral particles. To determine the incorporation rate of the mutated segment 7 vRNPs into infectious viral particles, plaque purification was carried out from the supernatant of the transfected cells. A total of 72 randomly chosen plaque-purified viruses were analysed by sequencing: all incorporated wild-type segment 7, demonstrating that codons S71 and R73 have a major impact on the selective incorporation of segment 7 (Figure 7B).
Figure 7.
Effect of silent mutations S71 and R73 on viral replication and on incorporation of vRNA 7. (A) MDCK cells were infected at a m.o.i. of 10−4 with wild-type recombinant A/Moscow/10/99 (H3N2) virus or a virus bearing silent mutations at M2 codons 71 and 73. The release of viral progeny into the supernatant was monitored by determining the tissue culture infective dose (TCID50). Points correspond to the mean of two experiments; the data ranges are smaller than the symbol size. (B) Strategy and output of the competition experiment.
A model for the internal organization of human H3N2 influenza A virions
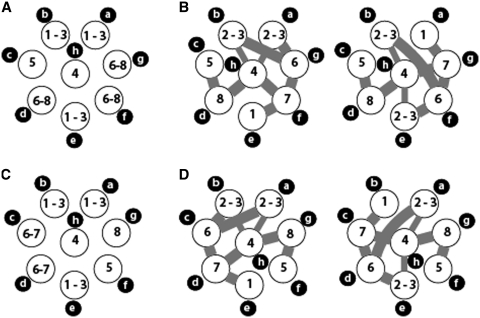
As our results suggest that the intermolecular vRNAs interactions we detected in vitro are relevant to vRNP packaging, we combined these data with our tomography data to analyse the relative disposition of vRNPs within human H3N2 influenza virions (Figure 8). According to Figure 2D, it is possible to discriminate vRNAs 1, 2 and 3 from the other vRNAs, based on the length of the vRNPs, but not between them. Similarly, it is possible to discriminate vRNAs 4 and 5, on one hand, and vRNAs 6–8 (for P2) or 6 and 7 (for P3), on the other hand, from the other vRNAs. In all four 3D reconstructions, two of the three longest vRNPs are contiguous and were used as starting points for anticlockwise labelling of the vRNPs, while the third one is opposite to them (Figure 2; Supplementary Figure S2 and Supplementary Movies S1A, S1B, S2A and S2B). Even though there is little difference between the length of the fourth and fifth vRNPs, the fourth one was always the central one, strongly suggesting that vRNP h contains vRNA 4 (Figure 1B, panels P1-1, P1-2 and P3-2; Figure 2A–D; Supplementary Figure S2 and Supplementary Movies S1A, S1B, S2A and S2B). The central location of vRNA 4 fits with the observation that this vRNA interacts with three different partners in vitro, whereas vRNA 5 has only one partner (Figures 3, 8B and D). This in turn allows identification of vRNA 5 in the 3D reconstructions of P2 and P3. In P3, vRNA 8 can be identified as the shortest vRNP (Figures 2B, C and 8C), and it is located next to vRNA 5 in agreement with the in vitro interaction data (Figures 3 and 8D). Noticeably, the tomography data indicate that all influenza A H3N2 virions do not share the same internal organization, as vRNA 5 has different locations in P2 and P3 (Figure 8A and C). Note that the interaction network defines a block of four external vRNPs comprising vRNAs 1, 7, 6 and 2 or 3, but that the orientation of this block can be deduced neither from tomography nor from the in vitro interaction network. Nevertheless, by combining the tomography data with the in vitro interaction network, and assuming that, in virions, interactions preferentially take place between close neighbours, the number of possible arrangements of the vRNPs in P2 and P3 is reduced from 5 760 to 4 (Figure 8C and D). As there are two vRNAs that are each involved in three intermolecular interactions while not interacting together (vRNAs 4 and 6), there will always be at least one interaction linking non-adjacent vRNAs, and we selected arrangements with exactly one such interaction (Figure 8C and D). Measuring the length of the vRNPs in P4 was difficult, as their upper end could not always be identified precisely. However, our data suggest that the internal organization of the vRNAs in P1 and P4 is similar to P3.
Figure 8.
Possible arrangements of the vRNPs within budding H3N2 influenza A virions. Top views of the possible arrangements of the vRNPs in P2 (A and B) and P3 (C and D) based on tomography data alone (a and c) or incorporating the in vitro interaction data (B and D). The intermolecular RNA interactions identified in vitro are indicated by thick grey line. The two thin lines correspond to the interaction between vRNAs 3 and 4 and are mutually exclusive, depending on the actual location of vRNAs 2 and 3.
DISCUSSION
Increasing evidence supports the notion that the eight vRNAs constituting the influenza A genome are specifically packaged into virions, but despite numerous efforts, the molecular mechanism underlying this process remains largely unknown (10). Some virology data (5,19,23,29) suggested the existence of a ‘viral genome complex’ that might involve direct or indirect interactions between vRNAs, but no direct evidence was provided.
Our electron tomography data (Figures 1 and 2; Supplementary Figures S1 and S2 and Supplementary Movies S1A, S1B, S2A and S2B) revealed that in budding viral particles, there is a transition zone between the matrix layer at the budding tip of the virions and the region where the eight vRNPs adopt the classical ‘7 + 1’ arrangement. In this zone, which appears as a platform in the 3D surface rendering, the dots corresponding to the vRNPs progressively become elongated and interconnected in a star-like structure. The common platform from which the 8 vRNPs emerge can accommodate a mean value of 206–271 nt, ranging from 153 nt between segments f and g to 310 nt between segments d and e. Thus, the platform is thick enough to accommodate all previously defined packaging signals, which are usually located within 100 nt or less of the vRNA ends (14–23) (Figure 2). In most particles, no close contact between vRNPs was observed outside this platform, suggesting that specific interactions between vRNPs, which are a prerequisite for specific packaging, take place within the platform. This idea is supported by the connections observed between the vRNPs in the transition zone of the tomograms. We suggest that the supramolecular complex depicted in Figure 2, Supplementary Figure S1 and Supplementary Movies S2A and S2B is a key intermediate in the selective packaging of influenza A vRNPs. Several groups reported that the typical ‘7 + 1’ arrangement of the vRNPs is more frequently observed in budding virions than in purified viral particles (4,7). In keeping with these observations, we have been unable to obtain a clear view of the interior of purified viruses, suggesting that the vRNP complex might disorganize or dissociate after budding.
Comparison of four individual viral particles showed that while distribution of the four longest vRNPs seems to be conserved amongst virions, the disposition of the shorter vRNPs is not absolutely conserved (Figures 2 and 8). The limited variability existing in the relative disposition of vRNPs between virions does not contradict the existence of segment-specific intermolecular interactions, but implies some flexibility of the vRNPs.
Selective packaging of the eight vRNAs in influenza A virions require a minimum of seven intersegment interactions. In principle, these interactions could be mediated by direct base pairing between the vRNAs, or by RNA binding proteins. As not a single cellular or viral protein able to selectively recognize a packaging signal of influenza A virus vRNAs has been identified to date (10), direct RNA interactions are our favourite hypothesis. A similar, albeit more simple, example is found in retroviruses, which package their genome as RNA homodimers (30). Here, we tested the existence of intermolecular vRNA interactions in vitro, following an approach that allowed the identification of the dimer linkage sequences of many retroviral RNAs (30,31).
In vitro, each vRNA establishes at least one intermolecular interaction and the eight vRNAs are involved in a single interaction network (Figure 3). Currently, we cannot ascertain that all interactions observed in vitro take place in vivo, and that, conversely, all inter-vRNA interactions potentially taking place in vivo can be observed in our system. We analysed the interactions at 37°C in a buffer containing 90 mM monovalent cations, 15 mM Mg2+ and 1 mM spermidine. Under the in vitro transcription conditions, ~6 mM Mg2+ ions are bound to the NTPs. Hence, the ionic and temperature conditions of our assay do not dramatically differ from those supposed to prevail in vivo. Moreover, the fact that the regions involved in the three strongest inter-vRNAs interactions in vitro correspond to known packaging regions (Figure 4), even when they could be narrowed down to a few nucleotides (Figure 6), strongly argues in favour of the relevance of our findings for the segment-specific mechanism of incorporation of the vRNPs in influenza A virions.
Surprisingly, two of the interactions we analysed involve vRNA pairs that have a globally parallel orientation (they involve the 5′ region of both vRNAs). This does not preclude that locally the two vRNAs might adopt an anti-parallel orientation and interact via classical base pairing (Supplementary Figure S4). An analogous situation exists in retroviruses. EM pictures revealed a globally parallel orientation of the two copies of genomic RNA molecules, and such an orientation likely exists in virions to allow efficiency recombination. However, it is now known that this globally parallel orientation is maintained by locally anti-parallel interactions (30,31).
In the case of vRNA 7, we showed that a limited set of nucleotides (mutations S71–R73) is important for interaction with vRNA 6 (Figure 6). Mutations S71–R73 reduced replication of our contemporary H3N2 circulating strain by 5- to 10-fold (Figure 7A), in agreement with previous results obtained with a H1N1 laboratory strain (5). In addition, our competition experiment indicated that this mutation has a strong negative impact on the selective incorporation of vRNA7 (Figure 7B).
Globally, terminal deletions revealed that regions containing the previously identified packaging signals play a crucial role in the vRNA/vRNA interactions we detected in vitro. Interestingly, the 5′ and 3′ regions of vRNA 7 are involved in interactions with different vRNA partners, an observation that fits with the fact that both terminal regions of the coding sequences are required for optimal packaging of the influenza A vRNAs (14,16,17,19–23). Note that none of the mutations that reduced intermolecular RNA interactions abolished them completely, suggesting a certain level of redundancy. Similarly, most deletions have rather limited effects on vRNA packaging (14–21).
A technical limitation of the in vitro analysis resides in the analysis of the RNA interactions in the presence of NP. Our data obtained using this protein shows that it does not affect the specificity of the interaction between vRNAs 6 and 7 (Figure 5). Unfortunately, unspecific binding of some vRNAs to the magnetic beads hindered generalization of this assay (Figure 5 and data not shown). Crystal structures of NP suggested that RNA binds at the surface of the protein oligomers (27,32), and chemical probing indicated that NP increases the accessibility of the vRNA bases (33). Thus, NP might facilitate intermolecular interactions between vRNAs, rather than prevent them. The presence of stable intramolecular RNA structures within vRNPs is unclear. A previous study with purified vRNP 8 indicated that the 5′ region of this vRNA is accessible to single-strand specific probes, except for the terminal promoter (34). However, vRNPs are susceptible to RNase V1, a clear indication of the existence of base paired regions (35).
Our in vitro approach opens up the possibility to identify the intermolecular interactions between influenza A vRNAs at the nucleotide level. This would in turn allow testing of the functional role of these interactions by introducing point mutations in the interacting sequences. This work is in progress in our laboratory but is complicated by several factors. First, the bioinformatics approach is of limited use as the number of possible interactions is huge, even when deletion mutants have identified the region of the vRNAs that are involved. Second, the oligonucleotide mapping strategy we used to identify a short region of vRNA 7 interacting with vRNA 6 does not always work because some oligonucleotides do not anneal to their target in the condition of the in vitro assay. Third, as discussed above, some interactions seem to be at least partially redundant, masking the effects of deletions.
Our combined structural and biochemical approaches strongly favour a model in which the eight genomic vRNAs are selected and packaged as an organized supramolecular complex held together by base pairing between previously identified packaging regions. As packaging of orthologous vRNAs is a prerequisite for genetic reassortment of influenza viruses, our findings also suggest that interactions between heterologous vRNAs is crucial for this process (36). Conservation of the sequences involved in the inter-vRNA interactions is likely to be a key factor in the emergence of reassortant viruses, which can generate pandemic viruses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figure 1–4; Supplementary Movies 1A, 1B, 2A and 2B.
FUNDING
Centre National de la Recherche Scientifique (CNRS) (fellowships to E.F.); Fondation de la Recherche Médicale (FRM) (fellowships to E.F.) and SANOFI-PASTEUR France (to E.F., partial). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Redmond Smyth for critical reading of the manuscript.
REFERENCES
- 1.Horimoto T, Kawaoka Y. Influenza: lessons from past pandemics, warnings from current incidents. Nat. Rev. Microbiol. 2005;3:591–600. doi: 10.1038/nrmicro1208. [DOI] [PubMed] [Google Scholar]
- 2.Palese P, Shaw M. Orthomyxoviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia: Lippincott, Williams and Wilkins; 2006. pp. 1647–1689. [Google Scholar]
- 3.Calder LJ, Wasilewski S, Berriman JA, Rosenthal PB. Structural organization of a filamentous influenza A virus. Proc. Natl Acad. Sci. USA. 2010;107:10685–10690. doi: 10.1073/pnas.1002123107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc. Natl Acad. Sci. USA. 2006;103:19123–19127. doi: 10.1073/pnas.0607614103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hutchinson EC, Curran MD, Read EK, Gog JR, Digard P. Mutational analysis of cis-acting RNA signals in segment 7 of influenza A virus. J. Virol. 2008;82:11869–11879. doi: 10.1128/JVI.01634-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moules V, Ferraris O, Terrier O, Giudice E, Yver M, Rolland J, Bouscambert-Duchamp M, Bergeron C, Ottmann M, Fournier E, et al. In vitro characterization of naturally occurring influenza H3NA-viruses lacking the NA gene segment: toward a new mechanism of resistance. Virology. 2010;404:215–224. doi: 10.1016/j.virol.2010.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Noda T, Sagara H, Yen A, Takada A, Kida H, Cheng RH, Kawaoka Y. Architecture of ribonucleoprotein complexes in influenza A virus particles. Nature. 2006;439:490–492. doi: 10.1038/nature04378. [DOI] [PubMed] [Google Scholar]
- 8.Coloma R, Valpuesta JM, Arranz R, Carrascosa JL, Ortin J, Martin-Benito J. The structure of a biologically active influenza virus ribonucleoprotein complex. PLoS Pathog. 2009;5:e1000491. doi: 10.1371/journal.ppat.1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu MT, Parvin JD, Gupta S, Krystal M, Palese P. Genomic RNAs of influenza viruses are held in a circular conformation in virions and in infected cells by a terminal panhandle. Proc. Natl Acad.Sci. USA. 1987;84:8140–8144. doi: 10.1073/pnas.84.22.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutchinson EC, von Kirchbach JC, Gog JR, Digard P. Genome packaging in influenza A virus. J. Gen. Virol. 2010;91:313–328. doi: 10.1099/vir.0.017608-0. [DOI] [PubMed] [Google Scholar]
- 11.Laver WG, Downie JC. Influenza virus recombination: I. Matrix protein markers and segregation during mixed infections. Virology. 1976;70:105–117. doi: 10.1016/0042-6822(76)90240-3. [DOI] [PubMed] [Google Scholar]
- 12.Lubeck M, Palese P, Schulman J. Nonrandom association of parental genes in influenza A virus recombinants. Virology. 1979;95:269–274. doi: 10.1016/0042-6822(79)90430-6. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima K, Sugiura A. Three-factor cross of influenza virus. Virology. 1977;81:486–489. doi: 10.1016/0042-6822(77)90165-9. [DOI] [PubMed] [Google Scholar]
- 14.Fujii K, Fujii Y, Noda T, Muramoto Y, Watanabe T, Takada A, Goto H, Horimoto T, Kawaoka Y. Importance of both the coding and the segment-specific noncoding regions of the influenza A virus NS segment for its efficient incorporation into virions. J. Virol. 2005;79:3766–3774. doi: 10.1128/JVI.79.6.3766-3774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujii K, Ozawa M, Iwatsuki-Horimoto K, Horimoto T, Kawaoka Y. Incorporation of influenza A virus genome segments does not absolutely require wild-type sequences. J. Gen. Virol. 2009;90:1734–1740. doi: 10.1099/vir.0.010355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujii Y, Goto H, Watanabe T, Yoshida T, Kawaoka Y. Selective incorporation of influenza virus RNA segments into virions. Proc. Natl Acad. Sci. USA. 2003;100:2002–2007. doi: 10.1073/pnas.0437772100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang Y, Hong Y, Parslow TG. cis-Acting packaging signals in the influenza virus PB1, PB2, and PA genomic RNA segments. J. Virol. 2005;79:10348–10355. doi: 10.1128/JVI.79.16.10348-10355.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang Y, Huang T, Ly H, Parslow TG, Liang Y. Mutational analyses of packaging signals in influenza virus PA, PB1, and PB2 genomic RNA segments. J. Virol. 2008;82:229–236. doi: 10.1128/JVI.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marsh GA, Hatami R, Palese P. Specific residues of the influenza A virus hemagglutinin viral RNA are important for efficient packaging into budding virions. J. Virol. 2007;81:9727–9736. doi: 10.1128/JVI.01144-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muramoto Y, Takada A, Fujii K, Noda T, Iwatsuki-Horimoto K, Watanabe S, Horimoto T, Kida H, Kawaoka Y. Hierarchy among viral RNA (vRNA) segments in their role in vRNA incorporation into influenza A virions. J. Virol. 2006;80:2318–2325. doi: 10.1128/JVI.80.5.2318-2325.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozawa M, Maeda J, Iwatsuki-Horimoto K, Watanabe S, Goto H, Horimoto T, Kawaoka Y. Nucleotide sequence requirements at the 5′ end of the influenza A virus M RNA segment for efficient virus replication. J. Virol. 2009;83:3384–3388. doi: 10.1128/JVI.02513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gog JR, Afonso EDS, Dalton RM, Leclercq I, Tiley L, Elton D, von Kirchbach JC, Naffakh N, Escriou N, Digard P. Codon conservation in the influenza A virus genome defines RNA packaging signals. Nucleic Acids Res. 2007;35:1897–1907. doi: 10.1093/nar/gkm087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marsh GA, Rabadán R, Levine AJ, Palese P. Highly conserved regions of influenza a virus polymerase gene segments are critical for efficient viral RNA packaging. J. Virol. 2008;82:2295–2304. doi: 10.1128/JVI.02267-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 25.van der Heide P, Xu XP, Marsh BJ, Hanein D, Volkmann N. Efficient automatic noise reduction of electron tomographic reconstructions based on iterative median filtering. J. Struct. Biol. 2007;158:196–204. doi: 10.1016/j.jsb.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 26.Sinck L, Richer D, Howard J, Alexander M, Purcell DF, Marquet R, Paillart JC. In vitro dimerization of human immunodeficiency virus type 1 (HIV-1) spliced RNAs. RNA. 2007;13:2141–2150. doi: 10.1261/rna.678307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Q, Krug RM, Tao YJ. The mechanism by which influenza A virus nucleoprotein forms oligomers and binds RNA. Nature. 2006;444:1078–1082. doi: 10.1038/nature05379. [DOI] [PubMed] [Google Scholar]
- 28.Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl Acad. Sci. USA. 2000;97:6108–6113. doi: 10.1073/pnas.100133697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutchinson EC, Wise HM, Kudryavtseva K, Curran MD, Digard P. Characterisation of influenza A viruses with mutations in segment 5 packaging signals. Vaccine. 2009;27:6270–6275. doi: 10.1016/j.vaccine.2009.05.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paillart J-C, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 31.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng AK-L, Zhang H, Tan K, Li Z, Liu J-h, Chan PK-S, Li S-M, Chan W-Y, Au SW-N, Joachimiak A, et al. Structure of the influenza virus A H5N1 nucleoprotein: implications for RNA binding, oligomerization, and vaccine design. FASEB J. 2008;22:3638–3647. doi: 10.1096/fj.08-112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baudin F, Bach C, Cusack S, Ruigrok RW. Structure of influenza virus RNP. I. Influenza virus nucleoprotein melts secondary structure in panhandle RNA and exposes the bases to the solvent. EMBO J. 1994;13:3158–3165. doi: 10.1002/j.1460-2075.1994.tb06614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klumpp K, Ruigrok RW, Baudin F. Roles of the influenza virus polymerase and nucleoprotein in forming a functional RNP structure. EMBO J. 1997;16:1248–1257. doi: 10.1093/emboj/16.6.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yamanaka K, Ishihama A, Nagata K. Reconstitution of influenza virus RNA-nucleoprotein complexes structurally resembling native viral ribonucleoprotein cores. J. Biol. Chem. 1990;265:11151–11155. [PubMed] [Google Scholar]
- 36.Neumann G, Noda T, Kawaoka Y. Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature. 2009;459:931–939. doi: 10.1038/nature08157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.