Abstract
Abstract
The amino terminal domain (ATD) of ionotropic glutamate receptor (iGluR) subunits resides at the extracellular region distal to the membrane. The ATD is structurally and functionally the most divergent region of the iGluR subunits. Structural studies on full-length GluA2 and the ATDs from three iGluR subfamilies have shed light on how the ATD facilitates subunit assembly, accommodates allosteric modulator compounds, and controls gating properties. Here recent developments in structural and functional studies on iGluR ATDs are reviewed.
Hiro Furukawa (Cold Spring Harbor Laboratory) earned his PhD from the University of Tokyo in 2001 after completing his chemistry degree at Tufts University. He then did his postdoctoral research at Columbia University and Vollum Institute where he worked with Eric Gouaux. He then moved to Cold Spring Harbor Laboratory as a faculty member in 2007 and is currently an Associate Professor. He has spent most of his career working on the structure and function of receptors and receptor ion channels.
 |
Ionotropic glutamate receptors
The majority of excitatory transmission in the mammalian brain is mediated by l-glutamate. This excitatory transmission is critical in brain development, as well as in basic functions including learning and memory formation (Kandel et al. 1995). The glutamate-mediated excitatory transmission is elicited by actions of metabotropic glutamate receptors (mGluRs) and ionotropic glutamate receptors (iGluRs), which are classified as G protein-coupled receptors and ligand-gated ion channels, respectively. There are four subfamilies of iGluRs, including α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA) receptors (GluA1–GluA4), kainate receptors (GluK1–GluK5), N-methyl-d-aspartate (NMDA) receptors (GluN1, GluN2A–D, GluN3A-B) and delta receptors (GluD1 and GluD2) (Traynelis et al. 2010). The combination of AMPA, kainate and NMDA receptors at the synaptic and extrasynaptic sites determines the amplitude and kinetics of excitatory postsynaptic currents (Lester et al. 1990), and thus the overall property of excitatory synaptic transmission. Therefore, understanding the regulatory mechanisms of these receptors is crucial in dissecting the rather complex pharmacology of the neuronal synapses.
All of the iGluR subunits are composed of four distinct domains: the ATD, ligand-binding domain (LBD), transmembrane domain (TMD), and C-terminal domain (CTD) (Traynelis et al. 2010) (Fig. 1A). Of all the domains, the ATD has the most divergent primary sequences among the iGluR subunits. While AMPA, kainate and delta receptor ATDs have approximately 20–25% sequence identity, there is little or no sequence identity between the non-NMDA receptors and NMDA receptors. The sequence identities are higher within the subfamilies: AMPA receptor subunits (∼55% among GluA1–4); kainate receptor subunits (∼75% among GluK1–3, 65% among GluK4–5 and ∼30% between GluK1–3 and GluK4–5); NMDA receptors (35–55% among GluN2A–D and ∼15% between GluN1 and GluN2A–D); and delta receptors (60% between GluD1 and GluD2). Compared to ATD, sequence identities are significantly higher within LBDs (80–90% within similar groups) or TMDs (80–95% within similar groups).
Figure 1. Organization of domains and subunits in iGluRs.
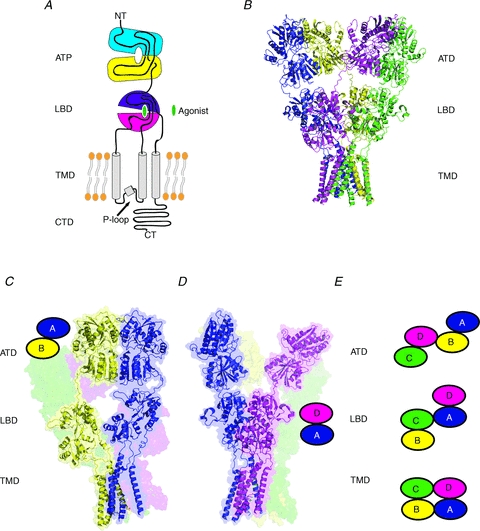
A, iGluR subunits are composed of distinct domains including the amino terminal domain (ATD), ligand-binding domain (LBD), transmembrane domain (TMD), and carboxyl terminal domain (CTD). B, crystal structure of the homotetrameric full-length GluA2 receptors (PDB code: 3KG2) showing the pattern of subunit arrangement and domain organization in the tetrameric assembly (Sobolevsky et al. 2009). The four subunits (A–D) are coloured as blue (A), yellow (B), green (C) and magenta (D). C–E, ATD dimers (panel C) and LBD dimers (panel D) are formed by an A–B (shown as a cartoon) or C–D pair and A–D (shown as a cartoon) or B–C pair, respectively. This results in a crossover of the dimer pairs in the ATD and LBD sections. Panels C, D, and E are modified from Hansen et al. (2010) with permission from the American Society for Pharmacology and Experimental Therapeutics.
History of structural studies on iGluRs
The structural study of iGluRs started in the late 1990s with the isolated LBD of GluA2 AMPA receptor (Armstrong et al. 1998). A series of GluA2 LBD structures in complex with different ligands and allosteric modulators or structures of mutant GluA2 LBDs has provided insights into receptor activation, deactivation and desensitization (Armstrong & Gouaux, 2000; Sun et al. 2002; Jin et al. 2005; Armstrong et al. 2006). Extensive studies have also been conducted on kainate receptor and NMDA receptor LBDs (Furukawa & Gouaux, 2003; Furukawa et al. 2005; Inanobe et al. 2005; Mayer, 2005; Yao et al. 2008; Vance et al. 2011). The common findings in those studies are as follows: (1) iGluR LBDs have bi-lobed clamshell-like architecture; (2) opening and closing of the LBD clamshell structures are coupled to gating activities; (3) non-NMDA receptor and NMDA receptor LBDs form homodimers in crystals while a GluN1–GluN2 heterodimer has also been observed; and (4) the dimer interface regulates speed of deactivation and the extent of desensitization. It is worth mentioning that some non-NMDA receptors can function as homotetramers in heterologous expression systems; however, they exist mostly as heterotetramers in the mammalian brain. Although there have not been any reports of heterodimeric structures of non-NMDA receptor LBDs to date, such studies may provide some important insights. It has been difficult to conduct studies of heterodimeric assembly in LBDs due to weak association between subunits. Assessment of oligomerization has relied on crystal packing since the wild-type LBD proteins for all of the iGluR families exist as monomers in solution, which is in contrast to ATD proteins that form dimers in solution. Nevertheless, the dimeric arrangement of LBDs observed in those crystallographic studies has proven to be physiological as the recent full-length GluA2 structure contains the same LBD dimers.
Meanwhile, an image of an intact AMPA receptor was revealed by a single particle electron microscopy analysis, providing the insight into how native AMPA receptors adopt various conformations and exist with membrane proteins belonging to the Stargazin/TARP family (Nakagawa et al. 2005). After an enormous number of crystallographic studies on iGluR LBDs, the structures of ATDs from all of the subfamilies became available in 2009 and later, as discussed in the following section (Hansen et al. 2010). Finally, completion of the full-length GluA2 AMPA receptor crystal structure in late 2009 marked a historical end to the mystery involving the subunit stoichiometry and the domain organization, and started a new era in the structural and functional studies of iGluRs (Sobolevsky et al. 2009). Perhaps the most surprising aspect of the structure is the presence of two conformers (A/C and B/D types) of four subunits with a crossover at the ATD and LBD sections, which results in staggering of ATD and LBD dimers (A–B and C–D dimers at ATD and A–D and B–C dimers at LBD; Fig. 1B–D). Importantly, with this full-length structure, one can now predict how the conformational movement of each modular domain in the iGluR subunits may couple to function in a much more precise manner than before. AMPA, kainate and NMDA receptors have similar architectures in the LBD and probably TMD and thus, have a similar LBD–TMD inter-domain orientation. However, the recent crystallographic studies showed that NMDA receptor ATDs are clearly different from non-NMDA receptor ATDs, not only in basic architecture (Karakas et al. 2009, 2011; Farina et al. 2011), but also in the pattern of subunit arrangement in the ATD dimers (Karakas et al. 2011). Thus, ATD is structurally the most diverse region among the iGluR subunits and the tetrameric arrangement in ATD is also expected to differ significantly between non-NMDA receptors and NMDA receptors. This is understandable considering the low sequence identity between non-NMDA receptor ATDs and NMDA receptor ATDs (<10%).
Comparison of amino terminal domain structures from different subfamilies
A series of iGluR ATD structures have emerged in the last two years including the ones for GluA2 and 3 AMPA receptors (Clayton et al. 2009; Jin et al. 2009; Sukumaran et al. 2011), GluK2, 3 and 5 kainate receptors (Kumar et al. 2009; Kumar & Mayer, 2010; Kumar et al. 2011) and GluN1 and GluN2B NMDA receptors (Karakas et al. 2009, 2011; Farina et al. 2011). Both AMPA and kainate receptor ATDs have bi-lobed clamshell-like architectures that are composed of R1 (upper lobe) and R2 (lower lobe) domains and are similar to leucine/isoleucine/valine-binding protein (LIVBP) and mGluR LBDs (Fig. 2A). While there are robust conformational changes in LIVBP (Quiocho & Ledvina, 1996) and mGluR LBDs (Kunishima et al. 2000; Tsuchiya et al. 2002) featuring opening and closing of clamshell-like structures upon ligand binding and unbinding, there appears to be no such conformational variability in non-NMDA receptor ATDs. That is, all of the non-NMDA receptor ATD structures obtained to date adopt similar intermediate conformations that reside between the open-cleft and closed-cleft of LIVBP or mGluR LBDs. The non-NMDA receptor ATDs are organized as homodimers in crystals as well as in solution, indicating that dimers are basic units in ATDs (Fig. 2C) (Clayton et al. 2009; Jin et al. 2009; Kumar et al. 2009; Kumar & Mayer, 2010; Sukumaran et al. 2011) except for the recently reported heterodimeric structure of GluK2 and GluK5 (Kumar et al. 2011). In general, the non-NMDA receptor subunits are symmetrically arranged within the ATD dimers in a side-by-side orientation mediated by strong R1–R1 and R2–R2 interactions with the inter-R1–R2 clefts facing the front and back side of the dimers (Fig. 2C). The recent structure of the GluK2–GluK5 ATD heterodimer shows a pattern of dimeric subunit arrangement similar to that observed in GluK2 or GluK3 ATD homodimers with strong R1–R1 and R1–R2 interactions (Kumar et al. 2011). In contrast, substantially weaker R1–R1 interactions are observed for GluK5 ATD homodimers due to a 16 deg tilt in the R1 (Kumar & Mayer, 2010; Kumar et al. 2011). GluK5 subunits are obligate heteromers that form functional ion channels when combined with GluK1–3. Thus, it is understandable that heteromeric assembly of GluK2–GluK5 ATDs is considerably favoured over homomeric assembly of GluK5 ATDs. Furthermore, the tetrameric assembly of GluK2 kainate receptors or GluK2–GluK5 at the ATD in the crystals is shown to be similar to the one observed in the crystal structure of the full-length GluA2 AMPA receptor by extensive disulfide based cross-linking experiments (Das et al. 2010; Kumar et al. 2011).
Figure 2. Structures of iGluR ATDs.
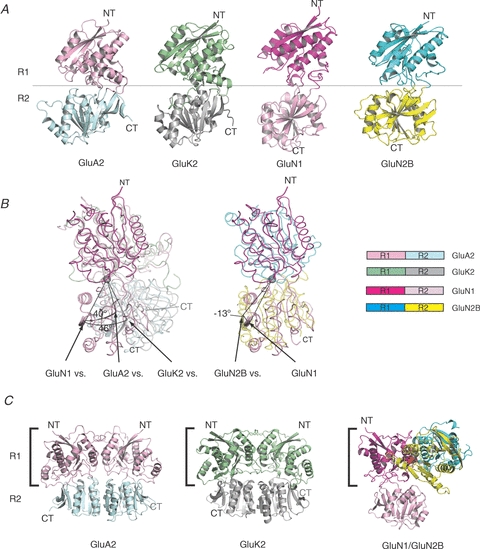
A, structures of ATD monomers from the AMPA, kainate and NMDA receptor subfamilies. The overall architecture of iGluR ATDs is shaped like a bi-lobed clamshell composed of the upper lobe (R1) and the lower lobe (R2), coloured differently. The structures (PDB codes are 3H5V, 3H6G, 3QEK and 3JPYB for GluA2, GluK2, GluN1 and GluN2B, respectively) are aligned with the similar R1 orientation. B, distinct R1–R2 orientation in NMDA receptors. Superposition of the R1 domains shows that the ATD clamshells from NMDA receptor subunits (both GluN1 and GluN2B) are substantially ‘twisted’ compared to those from non-NMDA receptors. The pivotal points for the R1–R2 twist are displayed as grey spheres. C, comparison of GluA2 ATD homodimer, GluK2 ATD homodimer, and GluN1–GluN2B ATD heterodimer. The R1s of ATDs on the left (square bracket; GluN1 R1 in GluN1–GluN2B ATD heterodimer) are similarly oriented. Note a substantial difference in the subunit orientation of GluN1–GluN2B ATDs compared to those of GluA2 or GluK2 ATDs. Panels B and C are modified from Karakas et al. (2011).
In contrast to non-NMDA receptor ATDs, the GluN2B NMDA receptor ATD exists as monomers in crystals and in solution (Karakas et al. 2009). The GluN2B ATD also has an overall clamshell-like structure, but with a strikingly different R1–R2 orientation that involves twisting by ∼50 deg (Fig. 2A and B) (Karakas et al. 2011). A similar twist is also observed in recent structures of GluN1 ATD (Farina et al. 2011; Karakas et al. 2011) and is suggested to exist in the GluN2A ATD based on functional experiments (Stroebel et al. 2011) indicating that the twist in R1–R2 orientation is a specific structural feature of NMDA receptor ATDs.
It is now known that GluN1 and GluN2 ATDs form heterodimers in mature NMDA receptors (Fig. 2C). This has been concluded based on the following observations: (1) GluN1 and GluN2B ATD proteins form dimers in solution only when they are mixed together (Karakas et al. 2011); (2) GluN1 and GluN2B can be cross-linked by disulfide bonds (Karakas et al. 2011; Lee & Gouaux, 2011); and (3) GluN1 and GluN2B ATDs form heterodimers in crystals (Karakas et al. 2011). Furthermore, an allosteric modulator, ifenprodil, significantly strengthens the heteromeric interaction in solution (Karakas et al. 2011). This observation is consonant with the crystallographic study that identified an ifenprodil binding site at the GluN1–GluN2B heterodimer interface (Karakas et al. 2011). The GluN1–GluN2B ATD heterodimer has a subunit arrangement that is highly distinct from those observed in non-NMDA receptors. While, non-NMDA receptor ATDs associate with each other symmetrically through R1–R1 and R2–R2 interactions, GluN1 and GluN2B ATDs do so through asymmetrical interactions involving R1–R1 and R1(GluN1)–R2(GluN2B) (Karakas et al. 2011) (Fig. 2C). This mode of subunit association results in the complete lack of R2–R2 interactions in NMDA receptors in contrast to non-NMDA receptors with strong R2–R2 interactions. The lack of R2–R2 interaction in NMDA receptor ATDs and the resulting freedom in the motion of R2 is perhaps an important structural feature that facilitates ATD-mediated allosteric regulation. Indeed, the recent study of Karakas et al. (2011) shows that trapping the movement of GluN2B R2 by disulfide cross linking prohibits allosteric inhibition by ifenprodil. Thus, although multiple conformations of clamshells have not yet been captured by crystallography, it is plausible that the ATD clamshells undergo conformational changes.
It is also suggested that the GluN1 subunit forms homodimers through ATD in the initial stage of NMDA receptor assembly before being replaced by GluN2 subunits to form mature tetrameric NMDA receptors (Atlason et al. 2007; Farina et al. 2011). In the recent study, a small portion (∼5%) of the purified GluN1a ATD protein sample was shown to form dimers by negative staining and single particle electron microscope analyses (Farina et al. 2011). Crystallographic study of GluN1a ATD identified a dimer in an asymmetric unit, which may represent the premature form of NMDA receptors (Farina et al. 2011).
The strikingly different subunit arrangement in the GluN1–GluN2B ATD dimers places the C-terminal ends rather far apart (by ∼79 Å) compared to the equivalent distance in non-NMDA receptors (Fig. 3A). With an assumption that the GluN1 and GluN2 subunits are arranged in the GluN1–GluN2–GluN1–GluN2 orientation, and that the domain swap between ATD and LBD observed in the GluA2 AMPA receptor structure occurs in NMDA receptors, the distance at the N-terminal ends of non-dimer forming LBDs (A/B or C/D in Fig. 1D) is ∼68 Å on the average (Fig. 3B). While there are 11 and 9 linker residues between the end of ATD structures and the beginning of LBD structures of GluN1 and GluN2B, respectively, how they fill this ∼11 Å distance gap is an unanswered question. One possible explanation may be that the recent crystal structure of GluN1–GluN2B ATDs is in the phenylethanolamine-bound form, and thus may represent a state similar to a desensitized state (Kew et al. 1996). It is plausible that the tetrameric arrangement of LBDs in the ifenprodil-bound GluN1–GluN2B NMDA receptors may be different from the antagonist-bound state that is represented by the recent full-length GluA2 AMPA receptor structure. Nevertheless, the highly distinct heterodimeric arrangement of the GluN1–GluN2B ATDs implies that the inter-domain organization between ATD and LBD may be significantly different between non-NMDA receptors and NMDA receptors, and that the pattern of the inter-subunit and inter-domain organization in NMDA receptors may not be precisely predicted by simply extrapolating the structural information obtained from the crystallographic study of the full-length GluA2 homotetramer. More structural or equivalent work capturing different functional states may be necessary to further understand the mode of inter-domain arrangement and cross talk between ATD and LBD in NMDA receptors.
Figure 3. Possible subunit arrangement at the extracellular region of NMDA receptors.
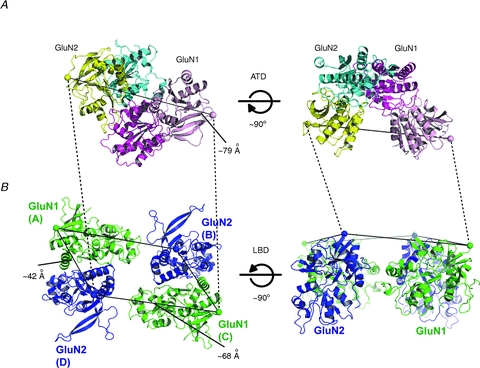
A, GluN1–GluN2B ATD dimer viewed from the C-terminal ends (spheres; left panel) or the side of the C-termini. The colour code of GluN1 and GluN2B is as in Fig. 2. B, tetrameric model of NMDA receptor LBDs assuming the GluN1–GluN2–GluN1–GluN2 orientation viewed from the sides of the N-terminal ends (spheres; left panel) and the side of N-termini. This model is built by superposing the top portion (domain 1) of the GluN1 and GluN2A LBD bi-lobed structures (PDB code: 2A5T) (Furukawa et al. 2005) onto the equivalent portion of the full length GluA2 receptor structure. Specifically, two GluN1 LBDs are superposed to the A and C subunits of GluA2 AMPA receptor structure in Fig. 1 whereas two GluN2 LBDs are superposed to the B and D subunits. The distance between the C-terminal ends of GluN1 and GluN2B ATDs is ∼79 Å whereas the GluN1–GluN2 distance between the N-termini of non-dimer forming LBDs is ∼68 Å.
Role of amino terminal domain in iGluRs
Despite numerous studies, the exact role of non-NMDA receptor ATDs on ion channel activities remains unknown to date. The lack of allosteric regulation in non-NMDA receptors is suggested to stem from inflexibility of the lower lobe of the ATD clamshell (R2) through tight R2–R2 interaction within ATD dimers of both GluA2 and GluK2 subunits (Clayton et al. 2009; Jin et al. 2009; Kumar et al. 2009). In contrast, three different dimeric arrangements with significantly weaker R2–R2 interaction are observed within the GluA3 ATD crystal structures (Sukumaran et al. 2011). This, along with results of dynamic studies suggests that opening and closing as well as the rearrangement of the subunit interface similar to that observed in mGluR1 LBD can occur in the AMPA receptor ATD (Sukumaran et al. 2011). Additionally, unidentified electron density is observed at the clamshell cleft of the GluA2 ATD structure in a crystallization condition indicating the possible existence of an allosteric modulator that may bind AMPA receptor ATDs (Sukumaran et al. 2011). The deposited coordinate of the crystal structure accounts for this electron density by a sulfate ion. Furthermore, a phosphate ion in the crystallization condition along with water molecules can perhaps explain this density. Thus, whether or not an authentic ligand for non-NMDA receptors exists is an issue that remains to be resolved. Further work needs to be carefully conducted to assess a regulatory role of the non-NMDA receptor ATD on the ion channel activities.
In contrast to non-NMDA receptors, the functional roles of NMDA receptor ATDs are widely known (Hansen et al. 2010; Paoletti, 2011). There are two basic roles of NMDA receptor ATDs: (1) regulation of open probability and deactivation; and (2) allosteric regulation of the ion channel activity by binding to modulator compounds. An important feature of NMDA receptors is their functional diversity, including different open probability and deactivation kinetics, which relies upon which of the four GluN2 subunits (A–D) is present in the tetrameric receptors (Paoletti, 2011). Recent studies using chimeric receptors revealed that the ATD and the short linker between the ATD and LBD are at least in part responsible for controlling both open probability and deactivation rates (Gielen et al. 2009; Yuan et al. 2009). For example, substitution of ATDs between GluN2A and GluN2B or GluN2D shifts the open probability and deactivation rates toward those of the subunit providing the ATD (Gielen et al. 2009; Yuan et al. 2009). Perhaps the most distinct feature of NMDA receptor ATDs is that they bind allosteric modulators and regulate the ion channel activities. One of the allosteric modulators, zinc, binds both GluN2A (Paoletti et al. 1997) and GluN2B (Rachline et al. 2005) ATDs and allosterically inhibits the ion channel activity in a voltage-independent manner with IC50 values at low nanomolar and micromolar, respectively. Within the GluN2B ATD, zinc was shown to bind to the clamshell cleft and stabilize a ‘closed’ conformation in a recent crystallographic study (Karakas et al. 2009). Based on numerous mutagenesis studies, it is known that the high-affinity zinc binding site is also located at the similar cleft of the GluN2A ATD (Fayyazuddin et al. 2000; Stroebel et al. 2011). Binding of zinc to the ATD increases sensitivity to protons that inhibit NMDA receptor activities (Choi & Lipton, 1999; Low et al. 2000). Although several regions of the receptors, including the LBD dimer interface (Gielen et al. 2008) and the region adjacent to the gate (Low et al. 2003), have been proposed to serve as proton sensors, there is currently no clear view on how zinc binding at the ATD can affect those proton sensors.
Phenylethanolamines are di-aryl compounds that show neuroprotective effects by specifically targeting and allosterically inhibiting GluN2B containing NMDA receptors (>100-folds over GluN2A), and thus there has been substantial enthusiasm for applying them for treatment of a number of neurological diseases (Mony et al. 2009; Koller & Urwyler, 2010). The recent crystallographic study showed that the binding site for phenylethanolamine compounds, ifenprodil and Ro 25-6981, is located at the GluN1–GluN2B ATD subunit interface rather than the previously predicted site within the clamshell cleft (Karakas et al. 2011). The phenylethanolamine binding site has no positional overlap with the zinc binding site, which is located within the clamshell cleft of GluN2B ATD (Fig. 4). While, this crystallographic study revealed the precise architecture of the correct phenylethanolamine binding site, the mechanism underlying specific binding to GluN2B over GluN2A remains unresolved. Surprisingly, GluN2A and GluN2B differ only by one residue at the phenylethanolamine binding site (Ile111 in GluN2B is Met112 in GluN2A) and interchanging this residue between GluN2A and GluN2B does not abolish or confer ifenprodil sensitivity (Karakas et al. 2011). One possible explanation is that the patterns of subunit interaction between the GluN1–GluN2A heterodimer and the GluN1–GluN2B ATD heterodimer may be different from each other. The correlation between the subtype specific patterns of GluN1–GluN2 subunit interactions and binding of allosteric modulators needs to be clarified by further crystallographic studies on other NMDA receptor subtypes. Nevertheless, the recent structural identification of the phenylethanolamine binding site should facilitate the development of ATD-targeting compounds in the right direction. A further challenge in the development of phenylethanolamine-based compounds for therapeutic usage includes minimizing off-target effects towards the human ether-a-go-go channel and α1-adrenergic receptors.
Figure 4. Binding sites for allosteric modulators in GluN1–GluN2B ATDs.
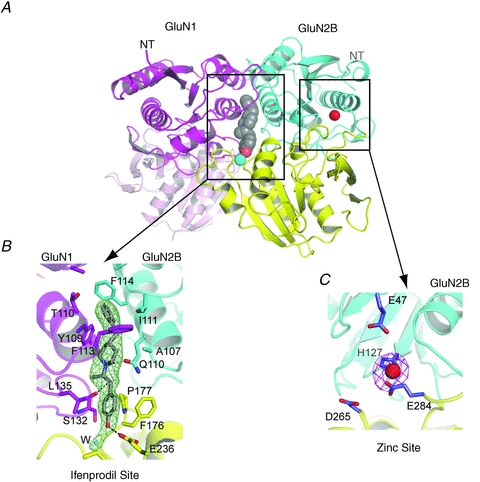
A, GluN1–GluN2B ATDs with the binding site for ifenprodil (grey sphere) at the subunit interface and for zinc (red sphere) within the GluN2B cleft. The cartoon represents the composite structure of GluN1–GluN2B ATDs in complex with ifenprodil (PDB code: 3QEL) and GluN2B ATD in complex with zinc (PDB code: 3JPY). Cyan sphere represents a water molecule at the ifenprodil binding site. B, blow-up view of the ifenprodil binding site. Binding of ifenprodil involves residues from both GluN1 and GluN2B subunits, which form hydrophobic and polar interactions. Shown in mesh is the Fo-Fc omit electron density map contoured at 3σ. A cyan sphere represents a water molecule. C, zinc binding site at the clamshell cleft of GluN2B ATD. His127 and Glu284 coordinate directly to zinc. Glu47 and Asp265 are proximal to the zinc binding site and have been previously shown to affect zinc sensitivity (Rachline et al. 2005). Water molecules that are likely to be present but not visible in this crystal structure due to limited resolution of the crystallographic data may play an important role in zinc coordination along with Glu47 and Asp265. Shown in magenta mesh is the anomalous difference Fourier map at 6σ.
Another role of the iGluR ATD is to serve as sites for interaction with extracellular proteins and cis- or trans-synaptic proteins. For example, the AMPA receptor ATD interacts with N-cadherin either cis- or trans-synaptically and promotes formation of dendritic spines (Saglietti et al. 2007). In addition, binding of EphrinB to EphB receptor tyrosine kinase facilitates interaction with NMDA receptors through the extracellular domain of the EphB receptor and ATD (Dalva et al. 2000; Takasu et al. 2002). This interaction stabilizes the surface expression of NMDA receptors (Nolt et al. 2011). Recent work reports that binding of β-amyloid to the EphB receptor reduces the number of NMDA receptors on the cell surface, perhaps by interfering with the EphB receptor–NMDA receptor interaction thereby reducing the synaptic function (Cisse et al. 2011). Thus, these findings suggest a potentially important role of EphB receptor–NMDA receptor interaction in Alzheimer's disease (Cisse et al. 2011).
Conclusions
Recent structural studies on iGluR ATDs provide important insights into how this domain assembles and functions. However, several fundamental questions remain unanswered: (1) can non-NMDA receptors be allosterically regulated by binding of small compounds or proteins to the ATD? (2) How does the ATD mediate allosteric regulation in GluN2A and GluN2B containing NMDA receptor ion channels? (3) Are there small compounds that allosterically regulate GluN2A, GluN2C and GluN2D NMDA receptors in a subtype-specific manner? Finding answer to these questions will not only deepen our understanding of iGluR functions, but will also pave the way to developing iGluR-based compounds with therapeutic potentials for neurological disorders and diseases.
Acknowledgments
This work was supported by NIH (MH085926) and Alzheimer's Association to H.F.
Glossary
- ATD
amino terminal domain
- CTD
carboxyl terminal domain
- LBD
ligand-binding domain
- TMD
transmembrane domain
References
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. doi: 10.1038/27692. [DOI] [PubMed] [Google Scholar]
- Atlason PT, Garside ML, Meddows E, Whiting P, McIlhinney RA. N-Methyl-D-aspartate (NMDA) receptor subunit NR1 forms the substrate for oligomeric assembly of the NMDA receptor. J Biol Chem. 2007;282:25299–25307. doi: 10.1074/jbc.M702778200. [DOI] [PubMed] [Google Scholar]
- Carron C, Jullien A, Bucher B. Synthesis and pharmacological properties of a series of 2-piperidino alkanol derivatives. Arzneimittelforschung. 1971;21:1992–1998. [PubMed] [Google Scholar]
- Choi YB, Lipton SA. Identification and mechanism of action of two histidine residues underlying high-affinity Zn2+ inhibition of the NMDA receptor. Neuron. 1999;23:171–180. doi: 10.1016/s0896-6273(00)80763-1. [DOI] [PubMed] [Google Scholar]
- Cisse M, Halabisky B, Harris J, Devidze N, Dubal DB, Sun B, Orr A, Lotz G, Kim DH, Hamto P, et al. Reversing EphB2 depletion rescues cognitive functions in Alzheimer model. Nature. 2011;469:47–52. doi: 10.1038/nature09635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton A, Siebold C, Gilbert RJ, Sutton GC, Harlos K, McIlhinney RA, Jones EY, Aricescu AR. Crystal structure of the GluR2 amino-terminal domain provides insights into the architecture and assembly of ionotropic glutamate receptors. J Mol Biol. 2009;392:1125–1132. doi: 10.1016/j.jmb.2009.07.082. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Das U, Kumar J, Mayer ML, Plested AJ. Domain organization and function in GluK2 subtype kainate receptors. Proc Natl Acad Sci U S A. 2010;107:8463–8468. doi: 10.1073/pnas.1000838107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farina AN, Blain KY, Maruo T, Kwiatkowski W, Choe S, Nakagawa T. Separation of domain contacts is required for heterotetrameric assembly of functional NMDA receptors. J Neurosci. 2011;31:3565–3579. doi: 10.1523/JNEUROSCI.6041-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fayyazuddin A, Villarroel A, Le Goff A, Lerma J, Neyton J. Four residues of the extracellular N-terminal domain of the NR2A subunit control high-affinity Zn2+ binding to NMDA receptors. Neuron. 2000;25:683–694. doi: 10.1016/s0896-6273(00)81070-3. [DOI] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. Mechanisms of activation, inhibition and specificity: crystal structures of the NMDA receptor NR1 ligand-binding core. EMBO J. 2003;22:2873–2885. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Singh SK, Mancusso R, Gouaux E. Subunit arrangement and function in NMDA receptors. Nature. 2005;438:185–192. doi: 10.1038/nature04089. [DOI] [PubMed] [Google Scholar]
- Gielen M, Le Goff A, Stroebel D, Johnson JW, Neyton J, Paoletti P. Structural rearrangements of NR1/NR2A NMDA receptors during allosteric inhibition. Neuron. 2008;57:80–93. doi: 10.1016/j.neuron.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gielen M, Siegler Retchless B, Mony L, Johnson JW, Paoletti P. Mechanism of differential control of NMDA receptor activity by NR2 subunits. Nature. 2009;459:703–707. doi: 10.1038/nature07993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen KB, Furukawa H, Traynelis SF. Control of assembly and function of glutamate receptors by the amino-terminal domain. Mol Pharmacol. 2010;78:535–549. doi: 10.1124/mol.110.067157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inanobe A, Furukawa H, Gouaux E. Mechanism of partial agonist action at the NR1 subunit of NMDA receptors. Neuron. 2005;47:71–84. doi: 10.1016/j.neuron.2005.05.022. [DOI] [PubMed] [Google Scholar]
- Jin R, Clark S, Weeks AM, Dudman JT, Gouaux E, Partin KM. Mechanism of positive allosteric modulators acting on AMPA receptors. J Neurosci. 2005;25:9027–9036. doi: 10.1523/JNEUROSCI.2567-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Singh SK, Gu S, Furukawa H, Sobolevsky AI, Zhou J, Jin Y, Gouaux E. Crystal structure and association behaviour of the GluR2 amino-terminal domain. EMBO J. 2009;28:1812–1823. doi: 10.1038/emboj.2009.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessel TM. Essentials of Neural Science and Behavior. East Norwalk: Appelton & Lange; 1995. [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Structure of the zinc-bound amino-terminal domain of the NMDA receptor NR2B subunit. EMBO J. 2009;28:3910–3920. doi: 10.1038/emboj.2009.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakas E, Simorowski N, Furukawa H. Subunit arrangement and phenylethanolamine binding in GluN1/GluN2B NMDA receptors. Nature. 2011;475:249–253. doi: 10.1038/nature10180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kew JN, Trube G, Kemp JA. A novel mechanism of activity-dependent NMDA receptor antagonism describes the effect of ifenprodil in rat cultured cortical neurones. J Physiol. 1996;497:761–772. doi: 10.1113/jphysiol.1996.sp021807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller M, Urwyler S. Novel N-methyl-D-aspartate receptor antagonists: a review of compounds patented since 2006. Expert Opin Ther Pat. 2010;20:1683–1702. doi: 10.1517/13543776.2010.533656. [DOI] [PubMed] [Google Scholar]
- Kumar J, Mayer ML. Crystal structures of the glutamate receptor ion channel GluK3 and GluK5 amino-terminal domains. J Mol Biol. 2010;404:680–696. doi: 10.1016/j.jmb.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Jin R, Mayer ML. The N-terminal domain of GluR6-subtype glutamate receptor ion channels. Nat Struct Mol Biol. 2009;16:631–638. doi: 10.1038/nsmb.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Schuck P, Mayer ML. Structure and assembly mechanism for heteromeric kainate receptors. Neuron. 2011;71:319–331. doi: 10.1016/j.neuron.2011.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- Lee CH, Gouaux E. Amino terminal domains of the NMDA receptor are organized as local heterodimers. PloS One. 2011;6:e19180. doi: 10.1371/journal.pone.0019180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester RA, Clements JD, Westbrook GL, Jahr CE. Channel kinetics determine the time course of NMDA receptor-mediated synaptic currents. Nature. 1990;346:565–567. doi: 10.1038/346565a0. [DOI] [PubMed] [Google Scholar]
- Low CM, Lyuboslavsky P, French A, Le P, Wyatte K, Thiel WH, Marchan EM, Igarashi K, Kashiwagi K, Gernert K, et al. Molecular determinants of proton-sensitive N-methyl-D-aspartate receptor gating. Mol Pharmacol. 2003;63:1212–1222. doi: 10.1124/mol.63.6.1212. [DOI] [PubMed] [Google Scholar]
- Low CM, Zheng F, Lyuboslavsky P, Traynelis SF. Molecular determinants of coordinated proton and zinc inhibition of N-methyl-D-aspartate NR1/NR2A receptors. Proc Natl Acad Sci U S A. 2000;97:11062–11067. doi: 10.1073/pnas.180307497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML. Crystal structures of the GluR5 and GluR6 ligand binding cores: molecular mechanisms underlying kainate receptor selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Mony L, Kew JN, Gunthorpe MJ, Paoletti P. Allosteric modulators of NR2B-containing NMDA receptors: molecular mechanisms and therapeutic potential. Br J Pharmacol. 2009;157:1301–1317. doi: 10.1111/j.1476-5381.2009.00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Cheng Y, Ramm E, Sheng M, Walz T. Structure and different conformational states of native AMPA receptor complexes. Nature. 2005;433:545–549. doi: 10.1038/nature03328. [DOI] [PubMed] [Google Scholar]
- Nolt MJ, Lin Y, Hruska M, Murphy J, Sheffler-Colins SI, Kayser MS, Passer J, Bennett MV, Zukin RS, Dalva MB. EphB controls NMDA receptor function and synaptic targeting in a subunit-specific manner. J Neurosci. 2011;31:5353–5364. doi: 10.1523/JNEUROSCI.0282-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti P. Molecular basis of NMDA receptor functional diversity. Eur J Neurosci. 2011;33:1351–1365. doi: 10.1111/j.1460-9568.2011.07628.x. [DOI] [PubMed] [Google Scholar]
- Paoletti P, Ascher P, Neyton J. High-affinity zinc inhibition of NMDA NR1-NR2A receptors. J Neurosci. 1997;17:5711–5725. doi: 10.1523/JNEUROSCI.17-15-05711.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiocho FA, Ledvina PS. Atomic structure and specificity of bacterial periplasmic receptors for active transport and chemotaxis: variation of common themes. Mol Microbiol. 1996;20:17–25. doi: 10.1111/j.1365-2958.1996.tb02484.x. [DOI] [PubMed] [Google Scholar]
- Rachline J, Perin-Dureau F, Le Goff A, Neyton J, Paoletti P. The micromolar zinc-binding domain on the NMDA receptor subunit NR2B. J Neurosci. 2005;25:308–317. doi: 10.1523/JNEUROSCI.3967-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglietti L, Dequidt C, Kamieniarz K, Rousset MC, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, et al. Extracellular interactions between GluR2 and N-cadherin in spine regulation. Neuron. 2007;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Sobolevsky AI, Rosconi MP, Gouaux E. X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature. 2009;462:745–756. doi: 10.1038/nature08624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroebel D, Carvalho S, Paoletti P. Functional evidence for a twisted conformation of the NMDA receptor GluN2A subunit N-terminal domain. Neuropharmacology. 2011;60:151–158. doi: 10.1016/j.neuropharm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Sukumaran M, Rossmann M, Shrivastava I, Dutta A, Bahar I, Greger IH. Dynamics and allosteric potential of the AMPA receptor N-terminal domain. EMBO J. 2011;30:972–982. doi: 10.1038/emboj.2011.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Takasu MA, Dalva MB, Zigmond RE, Greenberg ME. Modulation of NMDA receptor-dependent calcium influx and gene expression through EphB receptors. Science. 2002;295:491–495. doi: 10.1126/science.1065983. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R, et al. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya D, Kunishima N, Kamiya N, Jingami H, Morikawa K. Structural views of the ligand-binding cores of a metabotropic glutamate receptor complexed with an antagonist and both glutamate and Gd3+ Proc Natl Acad Sci U S A. 2002;99:2660–2665. doi: 10.1073/pnas.052708599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Simorowski N, Traynelis SF, Furukawa H. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nature Commun. 2011;2:294. doi: 10.1038/ncomms1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Harrison CB, Freddolino PL, Schulten K, Mayer ML. Molecular mechanism of ligand recognition by NR3 subtype glutamate receptors. EMBO J. 2008;27:2158–2170. doi: 10.1038/emboj.2008.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Hansen KB, Vance KM, Ogden KK, Traynelis SF. Control of NMDA receptor function by the NR2 subunit amino-terminal domain. J Neurosci. 2009;29:12045–12058. doi: 10.1523/JNEUROSCI.1365-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]


