Abstract
Twenty-first century life sciences have transformed into data-enabled (also called data-intensive, data-driven, or big data) sciences. They principally depend on data-, computation-, and instrumentation-intensive approaches to seek comprehensive understanding of complex biological processes and systems (e.g., ecosystems, complex diseases, environmental, and health challenges). Federal agencies including the National Science Foundation (NSF) have played and continue to play an exceptional leadership role by innovatively addressing the challenges of data-enabled life sciences. Yet even more is required not only to keep up with the current developments, but also to pro-actively enable future research needs. Straightforward access to data, computing, and analysis resources will enable true democratization of research competitions; thus investigators will compete based on the merits and broader impact of their ideas and approaches rather than on the scale of their institutional resources. This is the Final Report for Data-Intensive Science Workshops DISW1 and DISW2. The first NSF-funded Data Intensive Science Workshop (DISW1, Seattle, WA, September 19–20, 2010) overviewed the status of the data-enabled life sciences and identified their challenges and opportunities. This served as a baseline for the second NSF-funded DIS workshop (DISW2, Washington, DC, May 16–17, 2011). Based on the findings of DISW2 the following overarching recommendation to the NSF was proposed: establish a community alliance to be the voice and framework of the data-enabled life sciences. After this Final Report was finished, Data-Enabled Life Sciences Alliance (DELSA, www.delsall.org) was formed to become a Digital Commons for the life sciences community.
Contributors
Ruben Abagyan, University California San Diego, ruben@ucsd.edu
George Adams, Purdue University, gba@purdue.edu
Patrick Allen, Annai Systems, patricka@annaisystems.com
Ilkay Altintas, University California San Diego, altintas@sdsc.edu
Gordon Anderson, Pacific Northwest National Laboratory, gordon@pnl.gov
Ioannis Androulakis, Rutgers University, yannis@rci.rutgers.edu
Ron Arnold, Geospiza, roba@3rdmountain.com
Peter Arzberger, University California San Diego, parzberg@sdsc.edu
Dan Atkins, University of Michigan, atkins@umich.edu
Magdalena Balazinska, University of Washington, magda@cs.washington.edu
Roger Barga, Microsoft Research, barga@microsoft.com
Bill Barnett, Indiana University, barnettw@iu.edu
Chaitan Baru, University California San Diego, baru@sdsc.edu
Andrew Bauman, VLST Corporation, baumanab@gmail.com
Reed Beaman, University of Florida, rbeaman@flmnh.ufl.edu
David Beck, University of Washington, dacb@u.washington.edu
Jacek Becla, SLAC National Accelerator Laboratory, becla@slac.stanford.edu
Philip Bernstein, Microsoft Research, philbe@microsoft.com
Kathleen Bongiovanni, Seattle Children's Research Institute (SCRI), kathleen.bongiovanni@seattlechildrens.org
Phil Bourne, University California San Diego, bourne@sdsc.edu
Stuart Bowers, Microsoft Research, sbowers@microsoft.com
H Leon Bradlow, Hackensack University Medical Center, hlbradlow@gmail.com
Olga Brazhnik, National Center for Research Resources, NIH, brazhnik@nih.gov
Bill Broomall, Quantum Linux, billb@quantumlinux.com
Olga Castaner, Institut Municipal d'Investigacio Medica, Barcelona, Spain, olgacastaner@hotmail.com
Tim Clark, Harvard University, tim_clark@harvard.edu
Guy Coates, Wellcome Trust Sanger Institute, U.K., gmpc@sanger.ac.uk
Vicky Cohn, Mary Ann Liebert, Publishers, vcohn@liebertpub.com
Milton Corn, National Library of Medicine, NIN, cornm@mail.nih.gov
Robert Cottingham, Oak Ridge National Laboratory, cottinghamrw@ornl.gov
Maribel Covas, Institut Municipal d'Investigacio Medica, Barcelona, Spain, mcovas@imim.es
David Cullen, SCRI, david.cullen@seattlechildrens.org
Chinh Dang, Allen Insitute, chinhda@alleninstitute.org
Neil Davies, University of California Berkeley, ndavies@moorea.berkeley.edu
Chinh Dang, Allen Institute for Brain Science, chinhda@alleninstitute.org
Ewa Deelman, University of Southern California, deelman@isi.edu
Christiana DelloRusso, Washington Biotechnology and Biomedical Association, christiana@washbio.org
Fransisco DeMayo, Baylor College of Medicine, fdemaoy@bcm.tmc.edu
William Dobyns, SCRI, william.dobyns@seattlechildrens.org
Jack Faris, North West Diabetes Research Institute, jfaris@pndri.org
Wu Feng, Virginia Tech, feng@cs.vt.edu
Jose Fortes, University of Florida, fortes@ufl.edu
Geoffrey Fox, Indiana University, gcf@indiana.edu
Peter Fox, Rensselaer Polytechnic Institute, pfox@cs.rpi.edu
Robert Franza, Myoonet, bfranza@myoonet.com
Dmitrij Frishman, Technical University of Munich, Germany, d.frishman@wzw.tum.de
Michael Galperin, National Center for Biotechnology Information, NIH, mygalperin@gmail.com
Lawrence Ganeshalingham, Annai Systems, lawrenceg@annaisystems.com
Jeffrey Gardner, University of Washington, gardnerj@phys.washington.edu
Christy Geraci, American Association for the Advancement of Science, hydropsychid@gmail.com
Damian Gessler, University of Arizona, dgessler@iplantcollaborative.org
Jack Gilbert, Argonne National Laboratory, gilbertjack@anl.gov
Paul Gilna, Oak Ridge National Laboratory, gilnap@ornl.gov
Stephen Goff, iPlant Collaborative, University of Arizona, sgoff@iplantcollaborative.org
Anthony Goldbloom, Kaggle, anthony.goldbloom@kaggle.com
Mark Gomelsky, University of Wyoming, gomelsky@uwyo.edu
Corinna Gries, University of Wisconsin-Madison, cgries@wisc.edu
Matt Griffin, Pine Street Group, matt@pinest.com
Stinger Guala, U.S. Geological Survey, gguala@usgs.gov
Winston Haynes, SCRI, winston.haynes@seattlechildrens.org
Matthias Hebrok, University California San Francisco, mhebrok@diabetes.ucsf.edu
Joseph Hellerstein, Google, jlh@google.com
Gerd Herber, The HDF Group, gherber@hdfgroup.org
Tony Hey, Microsoft Research, tony.hey@microsoft.com
Roger Higdon, SCRI, roger.higdon@seattlechildrens.org
Jason Hogan, Bristol Myers Squibb, jason.hogan@bms.com
Russ Hobby, Internet2, rdhobby@internet2.edu
Chris Howard, SCRI, chrisrahoward@gmail.com
Bill Howe, University of Washington, billhowe@cs.washington.edu
Michael Huerta, National Library of Medicine, NIH, mhuert1@mail.nih.gov
Lee Huntsman, Life Sciences Discovery Fund, huntsman@lsdfa.org
John Hutton, Cincinnati Children's, University of Cincinnati, john.hutton@cchmc.org
Jared Jackson, Microsoft Research, jaredj@microsoft.com
Jeremy Jaech, jeremy@jaech.com
Shantenu Jha, Rutgers University, shantenu.jha@rutgers.edu
Tatiana Karpinets, Oak Ridge National Laboratory, karpinetstv@ornl.gov
Kerstin Kleese van Dam, Pacific Northwest National Laboratory, kerstin.kleesevandam@pnl.gov
Rob Knight, University of Colorado Boulder, rob.knight@colorado.edu
Bartha M. Knoppers, McGill University, Canada, bartha.knoppers@mcgill.ca
Evelyne Kolker, University of Washington, evelynek@u.washington.edu
Natali Kolker, SCRI, natali.kolker@seattlechildrens.org
Ashok Krishnamurthy, Ohio Supercomputer Center, ashok@osc.edu
Julia Krushkal, University of Tennessee Memphis, jkrushka@uthsc.edu
Maode Lai, Zhenjian University, Hangzhou, China, lmd@zju.edu.cn
Doron Lancet, Weizmann Institute of Science, Rehovot, Israel, doron.lancet@weizmann.ac.il
Trudie Lang, Oxford University, U.K., trudie@globalhealthtrials.org
Hilmar Lapp, Nescent, Duke University, hlapp@nescent.org
Ed Lazowska, University of Washington, Lazowska@cs.washington.edu
Michael Lesk, Rutgers University, lesk@acm.org
Biaoyang Lin, Zhejiant Univerisity, Hangzhou, China, biaoylin@gmail.com
Andrew Lowe, SCRI, andrew.lowe@seattlechildrens.org
Julio Lopez, Carnegie Mellon University, jclopez@cs.cmu.edu
Sonya Lowry, iPlant Collaborative, University of Arizona, sonya@iplantcollaborative.org
Peter Lyster, National Institute of General Medical Sciences, NIH, lysterpe@nigms.nih.gov
Sean MacLeod, Stratos, macleod@stratos.com
Courtney MacNealy-Koch, SCRI, courtney.macnealykoch@seattlechildrens.org
Philip Maechling, University of Southern California, maechlin@usc.edu
Susannah Malarkey, Technology Alliance, susannahm@technology-alliance.com
Dan Maltbie, Annai Systems, danm@annaisystems.com
Dinesh Manocha, Indian Institute of Technology, Delhi, India, dm@cs.unc.edu
Stephen Marcus, National Institute of General Medical Sciences, NIH, marcusst@mail.nih.gov
Ronald Margolis, National Institute of Diabetes and Digestive Kidney Diseases, NIH, margolisr@mail.nih.gov
Neo Martinez, Computational Ecology Lab, neo@PEaCElab.net
Andrea Matsunaga, University of Florida, ammatsun@ufl.edu
Suzanne Matthews, Texas A&M, sjm@cse.tamu.edu
Daniel McDonald, University of Colorado, mcdonadt@colorado.edu
Michael McLennan, Purdue University, mmclennan@purdue.edu
Craig McLuckie, Google, craigmcl@google.com
Chris Mentzel, Gordon and Betty Moore Foundation, chris.mentzel@moore.org
Simon Mercer, Microsoft Research, simon.mercer@microsoft.com
Carol Beaton Meyer, Foundation for Earth Science, carolbmeyer@esipfed.org
Folker Meyer, Argonne National Laboratory, folker@mcs.anl.gov
Christopher Moss, SCRI, christopher.moss@seattlechildrens.org
Alexey Nesvizhskii, University of Michigan, nesvi@med.umich.edu
Sean Nolan, Microsoft, sean.nolan@microsoft.com
Gene Oates, Intellectual Ventures, goates@intven.com
Bert O'Malley, Baylor college of Medicine, berto@bcm.edu
Peter O'Neil, National LambdaRail, poneil@nlr.net
Douglas Orr, Google, orr@google.com
Cynthia Parr, Smithsonian Institution, parrc@si.edu
Valerio Pascucci, University of Utah, pascucci@acm.org
Mette Peters, University of Washington, mpeters1@u.washington.edu
Vladimir Poroikov, Institute of Biomedical Chemistry, Moscow, Russian Federation, vladimir.poroikov@ibmc.msk.ru
Judy Qiu, Indiana University, xqiu@indiana.edu
Sean Rapson, SCRI, sean.rapson@seattlechildrens.org
Deborah Penque, Instituto Nacional de Saúde Dr Ricardo Jorge, Lisbon, Portugal, deborah.penque@gmail.com
Chance Reschke, University of Washington, reschke@u.washington.edu
Chris Rivera, Washington Biotechnology and Biomedical Association, chris@washbio.org
Jean-Baptiste Riviere, SCRI, jbriviere01@yahoo.fr
Dan Rigden, University of Liverpool, U.K., drigden@liverpool.ac.uk
Galina Riznichenko, Moscow State University, riznich46@mail.ru
Robert Robbins, Electronics Scholarly Publishing Project, rjr8222@gmail.com
Allen Rodrigo, Nescent, Duke University, a.rodrigo@nescent.org
Lynn Rose, SCRI, lynn.rose@seattlechildrens.org
Christian Roth, SCRI, christian.roth@seattlechildrens.org
Evelyne Rozner, Pine Street Group, evelyne@pinest.com
Donna Russell, SCRI, donna.russell@seattlechildrens.org
Isabel Sa Correia, Institute for Biotechnology and BioEngineering, Lisbon, Portugal, isacorreia@ist.utl.pt
Marilyn Safran, Weizmann Institute of Science, Rehovot, Israel, marilyn.safran@weizmann.ac.il
Charles Schmitt, Renaissance Computing Institute, cschmitt@renci.org
Arnold Smith, SCRI, arnold.smith@seattlechildrens.org
Charles Smith, SCRI, charles.smith@seattlechildrens.org
Burton Smith, Microsoft Research, burtons@microsoft.com
Michael Snyder, Stanford University, mpsnyder@stanford.edu
Sanjeeva Srivastava, Indian Institute of Technology, Bombay, sanjeeva@iitb.ac.in
Larissa Stanberry, SCRI, larissa.stanberry@gmail.com
Al Stephan, Stratos, als@stratos.com
John Sterling, Genetic Engineering & Biotechnology News, jsterling@genengnews.com
Jesse Stombaugh, University of Colorado Boulder, jesse.stombaugh@colorado.edu
Alex Szalay, John Hopkins University, szalay@jhu.edu
Peter Tarczy-Hornoch, University of Washington, pth@u.washington.edu
Kaitlin Thaney, Digital Science, k.thaney@digital-science.com
Anne Thessen, Data Conservancy, athessen@eol.org
Mauricio Tsugawa, University of Florida, tsugawa@ufl.edu
Pamela Vagata, Microsoft Research, pavaga@microsoft.com
Dave Vieglais, University of Kansas, vieglais@ku.edu
Daniel Wang, SLAC National Accelerator Laboratory, danielw@slac.standford.edu
Dave Wecker, Microsoft Research, wecker@microsoft.com
Dean Welch, SCRI, deanwelch77@gmail.com
Fredric Wolf, University of Washington, wolf@u.washington.edu
John Wooley, University California San Diego, jwooley@ucsd.edu
Gene Yee, Fenwick and West, genehyee1@yahoo.com
Bülent Yener, Rensselaer Polytechnic Institute, yener@cs.rpi.edu
Yi-Kuo Yu, National Center for Biotechnology Information, NIH, yyu@ncbi.nlm.nih.gov
Kenneth Zaret, University of Pennsylvania, zaret@upenn.edu
Jiang Zheng, SCRI, jiang.zheng@seattlechildrens.org
Introduction
The transition of life sciences to the cloud paradigm involves aspects of science, computation, and even the cultural mindset within the scientific community. The first NSF-funded Data-Intensive Science Workshop (DISW1, Seattle, WA, September 19–20, 2010) had six working groups (Policy, Communication, Biology, Education, Technology, and Bioinformatics) that identified the challenges and opportunities within the topic and summarized findings in order to build a platform for the second workshop (Barga et al., 2011; Bernstein et al., 2011; Faris et al., 2011; Kolker, 2011a; Ozdemir et al., 2011a; Smith et al., 2011; Wolf et al., 2011).
Challenges and opportunities identified included:
1. The research necessity of the life sciences community to work across diverse domains and with computer, cyberinfrastructure, and data experts to leverage opportunities in data-enabled science (DES).
2. Scientific progress and accelerated rate of data production in life sciences result in a pressing need for validation and reproducibility of results through new standards and data sharing capabilities.
3. A perceived gap between the needs of data-enabled life sciences and current funding initiatives.
4. A specific need to integrate data-enabled life sciences with major international and national initiatives.
As the second NSF-funded Data-Intensive Science Workshop (DISW2, Washington, DC, May 16–17, 2011) progressed, animated discussions of the transitional issues highlighted a need for a pivotal infrastructure that organizes, supports, and provides resources and services to the scientific community. Indeed, this need for infrastructure has not gone unnoticed. In March 2011, a multipart report was published by the NSF Advisory Committee for Cyberinfrastructure (ACCI; http://www.nsf.gov/od/oci/taskforces/) on the needs of 21st century science and education given the present era of the 4th paradigm of scientific inquiry (NSF_CIF21, www.nsf.gov/about/budget/fy2012/pdf/40_fy2012.pdf).
Challenges and Opportunities
The 4th Paradigm data intensive scientific discovery was originally proposed by Jim Gray and colleagues as a 4th paradigm of scientific research, following and interacting with the three other paradigms—theory, experimentation, and simulation (modeling) (Hey et al., 2009). DES, defined by NSF as science that depends on data, is firmly part of the 4th paradigm era. The NSF report detailed the issues and challenges of the current situation and potential solutions. In addition to these reports, the NSF developed the Cyberinfrastructure Framework for 21st Century Science & Engineering (CIF21), an NSF-wide vision crafted to address these issues (NSF_CIF21, 2011). Other Federal agencies such as the National Institutes of Health, the Department of Defense, and the Department of Energy are also contributing their experience, expertise, and efforts to addressing these issues.
Notably, the rate of data generation in the life sciences has now exceeded the growth of computational power predicted by Moore's law (Moore, 1965). Furthermore, existing data storage resources and tools for analysis and visualization lack integration and can be difficult to disseminate and maintain because the resources (both people and cyberinfrastructure) are not organized to sustain them. Many analysis tools are not adapted to handle large data sets, and are not implemented on platforms that can support such big data sets. Many tools are built with a single purpose in mind (i.e., disposable software), but it has become imperative to consider the level of effort put into such tools. Further, those tools that are built to handle large data sets were not always done so with the specific needs of the life sciences community in mind, and as such, are either intractable or unavailable. Thus, the return on investments made in generating data and tools has yet to realize its full potential. In a recent analysis of U.S. science with a comparison to the EU and China, the United States has, by most metrics, maintained its position of relative preeminence in the sciences (Hather et al., 2010). However, this inability to realize full potential must be addressed if the United States wishes to stay at the top and continue enabling infrastructure science, sustainable knowledge-based advancement, and innovative collaboration (Hather et al., 2010; Kolker, 2010; Kolker, 2011; Ozdemir et al., 2011a).
Cloud computing could help realize this potential as it can integrate networks, servers, storage, applications, and services, thereby enabling convenient, on-demand access to a shared pool of configurable computing resources. More importantly, the cloud components can be rapidly provisioned and released in a centralized manner with minimal management effort and service provider interaction. Cloud resources could also provide access to data repositories and advanced technology and tools, as well as the ability to scale and augment existing compute resources. The cloud computing paradigm shifts the costs of high-performance computing and large data storage away from individual organizations to distributed compute centers with skilled support personnel. Currently, cloud computing services are being provided by commercial vendors, academic centers, and government agencies. Several publications have presented promising results for life science computations on the cloud (e.g., Kolker et al., 2011a; Qiu et al., 2010; Taylor, 2010).
Modern life sciences are DES that seek to understand biological processes through data-intensive techniques. Our goal was to identify challenges and opportunities for new avenues of growth as DES begins to utilize clouds to transition to a new level of collaborative science and collective innovation. The issues identified will serve to inform the scientific community and other DES stakeholders for short-term action that will contribute to a strong foundation for long-term scientific progress. Already, a number of groups have been exploring the potential of cloud-based computing, discussing issues such as tool transition, data transfer, computing power, and economics (e.g., Dudley et al., 2010; Schadt et al., 2010; Schatz et al., 2010; Stein, 2010).
As acknowledged in the newly released National Science Board report on Digital Research Data Sharing and Management, “A core expectation of the scientific method is the documentation and sharing of results, underlying data, and methodologies” (NSB, 2011). Truly, in the era of immense data generation, we find ourselves seemingly without the capacity to take full advantage of the data potential. Yet the challenges we are facing are the ones we can meet, with appropriate organization and innovation.
New technologies generate terabytes of data and are expected to reach petabyte scale in the next several years. For life scientists, future success already depends upon the ability to leverage and utilize large-scale data. Data analysis is the final, most complex and compute-intensive step for the translation of large-scale data into knowledge-based innovations. The cost of computational analyses is projected to far exceed that of data generation, threatening current data mining infrastructures. Currently, research progress is severely impeded by heterogeneity of acquisition formats, lack of integration among commonly used tools and, most importantly, by the scale and computational challenges related to mining and analysis of these vast data sources. Hence, there is a pressing need for adequate cyberinfrastructure that could consolidate computing and analytic resources, provide tools for exploration and analysis of large, heterogeneous data and, ultimately, allow the building of complex models of biological systems. For the research community in general and bioinformatics in particular, the cloud computing paradigm can be the quantum leap to meet this crucial need thereby improving research efficiency and enabling breakthroughs in data analysis and modeling.
The transition from local computing environments to clouds or other technologies is a multifaceted technological and organizational challenge and as such, demands thorough planning and oversight as well as long-term investments. The establishment and maintenance of the cloud-accessible resources requires a centralized effort by the community. Dedicated partnerships and coordinated leadership need to be established to determine access protocols, cloud content, and structure, to specify the appropriate incentives and to provide a long-term funding solution. Budgeting for the compute centers (clouds) and the maintenance costs can be shared by all stakeholders and realized via subscription services for academic institutions, governed access rights for industry, and designated budgets in biomedical grants issued by federal and private funding agencies.
Overall Recommendation
Based on the findings of DISW1 and DISW2, we have developed the following overarching recommendation to NSF:
Establish a community alliance to be the voice and framework of the community. The immediate goals of the alliance would be to: (1) synergize research and educational efforts across the life sciences using contemporary compute approaches to comprehend large and diverse data; (2) make the alliance an integral part of the international and national projects to address the challenges of data-enabled life sciences; (3) cohesively address the development, research, and educational needs of the community through creation of the supporting ecosystem of federal agencies, foundations, academic institutions, and industrial partners; and (4) implement topic recommendations found in the following pages of this report (Tables 1–3). This recommendation is in line with the CIF21 Community Research Networks recommendations to develop new, multidisciplinary research communities to address challenges that require diverse inputs (NSF_CIF21, 2011).
Table 1.
Data Accessibility: Challenges, Opportunities, and Recommendations
| Challenges | Opportunities | Recommendations |
|---|---|---|
| Variations in acquisition standards Differences in data formats Lack of access to existing data Lack of metadata Lack of incentives to share and disseminate data High curation and archiving costs |
Establishing unified data format(s) Providing straightforward access to repositories Increasing analytical abilities and breadth of approaches Increasing use and integration of data Creating a truly global platform, for example, through the emerging cloud-computing technologies and a new DES alliance, for data access and sharing including in rural communities in resource-limited settings, to help catapult the United States as a global leader in data-enabled sciences |
Survey scientists/develop multiple distributed data and meta-data repositories based on the determined needs Develop a community-wide effort to catalog and monitor core data resources/wiki-style may be effective Develop/adapt an open source reusable identity management system linked to access control. Security will be increasingly important as data are moved to shared resources |
Table 3.
Development of Education and Funding Policies to Enable DES: Challenges, Opportunities, and Recommendations
| Challenges | Opportunities | Recommendations |
|---|---|---|
| Implementation requires advanced computing skills that are not readily available Slow sharing of technology with lack of incentives |
Enabling community evaluation to ensure quality |
Use the community's collective strength to craft solutions by recommending challenges to approach: prizes, data journals, competitions Initiate ecosystem of funding agencies, academia, and industry and outside expertise groups to address the needs of the community Adjust funding consideration and merit evaluations to include key components of DES infrastructure and management resources: IT, data, meta-data, software, personnel. Reward data-oriented scientists Update scientist training to include expanded instruction in computer science, statistics, and collaborative research |
In the remainder of the report we outline three major discussion topics central to the transition of the life sciences to fully data-enabled life sciences, including providing highly accessible data, the establishment of tool repositories, the development of enabling funding strategies, and training scientists to develop and utilize these resources. We identify existing challenges and outline opportunities and recommendations to improve data accessibility, enable the transition of analysis tools to high performance computing (HPC)/Cloud resources, and to develop policies for education and funding that are in step with the DES community needs. More money cannot be expected from funding sources; we must look to innovative, collaborative, and transformative solutions to our current and future challenges (Kolker, 2010). We have to more effectively utilize the reduced funding support, while at the same time being able to achieve better sustainable outcomes (Hather et al., 2010; Kolker, 2010; Kolker, 2011; Ozdemir et al., 2011b).
Three Specific Recommendations
1. Data accessibility: the goal of bioinformatics is the understanding of biological processes through models and algorithms of mathematics, statistics, and computer science. Bioinformatics leverages the increasingly vast volumes of data generated by new technologies to increase knowledge. The challenges of data sharing and dissemination can be addressed using clouds or similar technologies. Highly accessible data will be an invaluable resource for bioinformatics researchers, enabling algorithmic and analytical developments. High accessibility of data will also increase collaborative and crossdisciplinary efforts. We emphasize that a potential cloud paradigm for data sharing does not imply archiving in a dedicated repository, but rather it requires establishing high-capacity, distributed access from locally hosted services, for example, university clusters, existing archives, and even from rural community settings from developing countries in an increasingly interconnected and globalized world. The challenge is to organize and catalog the data, information, and knowledge and to establish fast and reliable access to data repositories to best enable opportunities for sustainable collective innovation (Hather et al., 2010; Kolker, 2010; Kolker, 2011; Ozdemir et al., 2011b).
Currently, comprehensive data sharing practices are virtually nonexistent. Locally hosted data are rarely distributed amongst the global community of DES researchers due to differences in acquisition protocols, varying formatting standards, absence of sharing incentives, and inadequate cyberinfrastructure to stably host and disseminate the data. Lack of access to these diverse resources hinders research progress and stalls the scientific progress within and across the national borders. To alleviate this problem, the NSF established requirements for data deposition both prior to publication and in association with NSF funding [NSF, General Grant Conditions (GC-1), 2001). The compliance, however, is impeded by lack of adequate guidance for, and deposition of, metadata and a reliable infrastructure.
The shift to a cloud paradigm for distributed data faces a number of hurdles. The successful transition would require standardized data formats, unified acquisition protocols, and appropriate incentives for resource sharing. Current existing resources need to be prioritized, cataloged, and curated, while newly collected data must be acquired in compliance with predetermined standards and made available in a timely manner.
Table 1 summarizes the challenges, opportunities, and recommendations for this topic.
Data access and management have been an afterthought for too long. A flexible approach to proactive management is a federated network of partnerships that pulls together expertise and resources regardless of physical location. A successful example is the Library of Congress, which has built a distributed network of partnerships to overcome challenges and take advantage of new opportunities and emerging technology [The National Digital Information Infrastructure and Preservation Program (NDIIPP), 2010). The NSF-funded Data Conservancy group also investigated data management challenges and partnerships for solving these challenges (Thessen and Patterson, 2011), while DataOne for environmental science and ICPSR for the social sciences are leading data management in those fields (DataOne, www.dataone.org; ICPSR, www.ispsr.umich.edu).
In these times of severe budget cuts, a data access solution would provide added value for every funding dollar as data collected in one lab can be used by many others (Hather et al., 2010; Kolker, 2010). The access to quality data resources will be a notable educational asset as well. Highly accessible data will necessarily lead to scientific advances and collaborative research efforts. For example, the data from the Sloan Digital Sky Survey are used throughout the globe, and the project's new methods of data management have led the way for similar efforts (National Virtual Observatory, http://www.us-vo.org/).
2. Tools and Cyberinfrastructure Utilization: Bioinformatics uses vast arrays of computational tools and databases to analyze and interpret biological data. Cloud-based implementation of these tools can alleviate many issues facing bioinformatics researchers. Currently, the available resources are decentralized and dispersed across multiple sites. Investigators must consistently rely upon expert installation and continuous maintenance of databases and software packages. Furthermore, the discord between the data and software formats makes it difficult to integrate the two.
The in-lab software development typically focuses on relatively specialized problems that make it difficult to scale-up the analyses or to transport them to different environments (Baxter et al., 2006). In-lab solutions are rarely shared across the community due to differences in data formats, lack of incentives, high development costs, and an inability to provide for adequate support. Furthermore, in bioinformatics, the analysis typically requires the establishment of a pipeline consisting of multiple software applications intertwined with custom code.
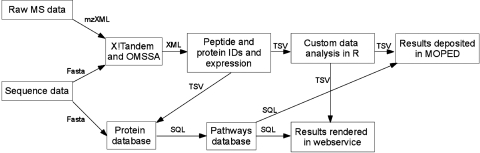
Figure 1 shows an example of a proteomics data analysis through SPIRE (Systematic Protein Investigative Research Environment) (Kolker et al., 2011b) along with the deposition of results in MOPED (Model Organism Protein Expression Database) (Kolker et al., in press). SPIRE has integrated software from many different sources and required the development of numerous scripts, tools, packages, and algorithms to make these components compatible. Development of the software to join the components has required extensive time and effort. Similar analysis pipelines are being generated across disciplines and are maintained with great, and often duplicated, effort by researchers.
FIG. 1.
Illustration of data flow for SPIRE, Systematic Protein Investigative Research Environment (Kolker et al., 2011b). Boxes indicate the processing environment being utilized and arrows indicate the file formats being transferred between the steps.
Table 2 summarizes the challenges, opportunities, and recommendations for this topic.
Table 2.
Tools and Cyberinfrastructure Utilization: Challenges, Opportunities, and Recommendations
| Challenges | Opportunities | Recommendations |
|---|---|---|
| Data aggregation and formatting is time-consuming Tools are format-specific Simple analyses are not automated Slow tool sharing with lack of incentives Duplicated installation and maintenance costs Inability to utilize most advanced technologies and analysis methods Difficulty to generate analysis pipelines |
Enabling readily available analysis tools Supplying readily available pipelines Allowing prompt tool sharing and technology proliferation Centralizing maintenance costs Disseminating advanced analysis methods developed by community Mining and analysis of data repositories |
Develop an Analysis Tool Shop for simplified, standardized, and documented access to analysis tools (starting with Alignment, clustering and R tools). Leverage and curate existing collections. Deploy tools on a cloud-like resource. Provide a support team to maintain and troubleshoot these tools. An active community-driven Shop will be the best approach. Funding could come from community pool/government grants/private research support/use fees |
For the scientific community, HPC/Cloud-enabled tools will make standard analysis and pipelines immediately available. The tools will be prioritized by the community and the list will vary by discipline. For bioinformatics, a repository will include such tools as BLAST, R, XTandem, Python, etc. Another valuable asset for the researchers is a FlexPipe (flexible pipeline), an arbitrary chain of applications interlaced with user code (e.g., R or Python scripts) that complies with input/output data structures. In addition, these enabled tools should have rapid access to data repositories, for analysis or mining or data mining.
The maintenance and support costs for the analytic component of the research will be shifted from the lab to the tool repository. Standardized data formats will simplify the development of HPC and cloud-based analysis pipelines. It is crucial that both the data accessibility and the tool accessibility challenges be addressed in concert. An example of a standardized and widely used Bioinformatics resource is the Taverna workbench (Taverna, www.taverna.org.uk). Taverna is open-source software for designing and executing work flows that addresses the tool accessibility challenge. Developed under the e-Science program, the software is used by more than 350 organizations throughout the world. It tightly integrates with myExperiment, a social Web site that enables reuse of work flows while also facilitating scientific collaborations and sharing of research expertise. Finally, the BioCatalogue site provides a curated catalog of Life Science Web Services (BioCatalogue, www.biocatalogue.org). All three of these Web sites serve as a unifying resource for collaborative bioinformatics for both researchers and developers to enable collective innovation (Hather et al., 2010; Kolker, 2010; Ozdemir et al., 2011b).
Also available is Meandre, a semantic Web-driven data-intensive flow execution environment that provides basic infrastructure for data-intensive science (Meandre, www.seasr.org.meandre). Built at the National Center for Supercomputing Applications, University of Illinois at Urbana–Champaign, Meandre was developed to take advantage of HPC resources.
3. Development of Education and Funding Policies: As the need for multidisciplinary teams grows it has become obvious that the education, funding, and career development environment of science must adapt in order to attract and retain the best researchers in the Data Intensive Approaches. Young researchers need more training in the possibilities and potential of open source collaboration and collective innovation approaches (Ozdemir et al., 2011a, b). These new approaches hold great promise in enabling scientists to work together but require a shift in mind set from the one-scientist, one-project approach so frequently taught. In addition, they must be shown that there are strong career trajectories that can involve large-scale data projects and collaborative teams. Credit toward tenure or funding must be given for development of tools and data sets that have value to the community, and resources must be in place to support sharing of those data sets and tools. Developing an infrastructure that embraces sharing will enable new discovery through collective innovation (Hather et al., 2010; Kolker, 2010; Kolker, 2011; Ozdemir et al., 2011b) (for details, see Table 3).
As discussed in the workshop, life sciences research produces vast resources of diverse data, yet the tools and cyber infrastructure to handle these data are largely inadequate. What is needed is a community of life scientists, computer scientists, data and cyber infrastructure experts, and others. The alliance would be established to be a voice and framework to address the current 4th paradigm changes in life sciences. The goals of the alliance would be to synergize research efforts across the life sciences, explore scalable compute approaches enabling interpretation of multifaceted data, and transform them to knowledge-based innovations addressing the pressing needs of global society.
The key challenges to be addressed include: (1) improved community-wide data sharing and dissemination, (2) establishment of appropriate HPC- and cloud-based cyberinfrastructure, (3) development and use of scalable informatics tools, (4) adoption of new standards and practices in data and tools sharing and evaluation, (5) establishment of funding and merit evaluation policies adapted to the needs and opportunities of data-enabled sciences, and (6) development of data-enabled life sciences educational, training, and collaborative research practices. A community alliance will engage federal agencies, research foundations, and industrial partners to enable and accelerate crossdisciplinary collaborations in life sciences.
Table 3 summarizes the challenges, opportunities and recommendations for this topic.
Conclusions
Twenty-first century life sciences have undergone a transformation that brings new challenges and opportunities to the forefront. Data-enabled sciences now use data-, computation-, and instrumentation-intensive approaches to seek meaningful knowledge and deeper understanding of wide ranging problems from the environment to global health. The NSF leadership in this transformation has been a crucial part of addressing the challenges and opportunities that have arisen. Looking into the future it has become obvious that research needs will require even more extensive efforts.
These efforts should be coordinated and relevant to the community. Based on the findings of DISW1 and DISW2, an overarching recommendation to the NSF has been proposed: establish a community alliance to be the voice and framework of the data-enabled life sciences. To fulfill such a mission, three immediate goals of this community alliance are:
1. synergize research and educational efforts across the life sciences using contemporary compute approaches to comprehend large and diverse data;
2. make the alliance an integral part of the international and national developments to address challenges and explore opportunities of data-enabled life sciences; and
3. cohesively address the development, research, and educational needs of the community through creation of the supporting ecosystem of federal agencies, foundations, academic institutions, and industrial partners.
Research success largely depends upon the reliable and speedy access to the best existing practices, methods, and data resources. Currently, there is an urgent need to both better utilize existing tools and develop new scalable approaches capable of handling current and future volumes of data. The comprehensive, crossdisciplinary, community resources will inspire collective innovation, advance scientific developments, and improve research outcomes in the life sciences (Hather et al., 2010; Kolker, 2010; Kolker, 2011; Ozdemir et al., 2011b). Straightforward, equal, and sustainable access to data, computing, and analysis resources will enable true democratization of research competitions; thus investigators will compete based on merits and broader impact of their ideas and approaches rather than on the scale of their institutional resources. The progression of data to knowledge to action will be accelerated in all parts of the community, from premier universities to government centers to school classrooms and citizen scientists' laptops. It is our timely response to the challenges of DES that will ultimately determine whether we would ride this wave of new information or are overpowered by it.
Acknowledgments
This policy report and DISW workshops were supported by the NSF Grant DBI-0969929 and SCRI internal funding to E. Kolker (Principal Investigator). Special thanks go to Anne Maglia, David Lipman, Drex DeFord, James Hendrix, Judith Verbeke, Peter McCartney, and Thomas Hanson for numerous discussions, encouragement, and support. Special thanks also go to Courtney MacNealy-Koch and Andrew Lowe for organizational support. The views expressed in this article are entirely personal opinions of the authors and do not necessarily represent positions of their affiliated institutions or the National Science Foundation.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Barga R. Howe B. Beck D. Bowers S. Dobyns W. Haynes W., et al. Bioinformatics and data-intensive scientific discovery in the beginning of the 21st century. OMICS. 2011;15:199–201. doi: 10.1089/omi.2011.0024. [DOI] [PubMed] [Google Scholar]
- Baxter S.M. Day S.W. Fetrow J.S. Reisinger S.J. Scientific software development is not an oxymoron. PLoS Comput Biol. 2006;2:e87. doi: 10.1371/journal.pcbi.0020087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P.A. Wecker D. Krishnamurthy A. Manocha D. Gardner J. Kolker N., et al. Technology and data-intensive science in the beginning of the 21st century. OMICS. 2011;15:203–207. doi: 10.1089/omi.2011.0013. [DOI] [PubMed] [Google Scholar]
- Dudley J.T. Pouliot Y. Chen R. Morgan A.A. Butte A.J. Translational bioinformatics in the cloud: an affordable alternative. Genome Med. 2010;2:51. doi: 10.1186/gm172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris J. Kolker E. Szalay A. Bradlow L. Deelman E. Feng W., et al. Communication and data-intensive science in the beginning of the 21st century. OMICS. 2011;15:213–215. doi: 10.1089/omi.2011.0008. [DOI] [PubMed] [Google Scholar]
- Hather G. Haynes W. Higdon R. Kolker N. Stewart E.A. Arzberger P., et al. The United States of America and scientific research. PLoS One. 2010;5:e12203. doi: 10.1371/journal.pone.0012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey T., editor; Tansley S., editor; Tolle K., editor. The Fourth Paradigm. Data-Intensive Scientific Discovery. Redmond, WA: Microsoft Research; 2009. [Google Scholar]
- Kolker E. A vision for 21st century U.S. Policy to support sustainable advancement of scientific discovery and technological innovation. OMICS. 2010;14:333–335. doi: 10.1089/omi.2010.0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolker E. Special issue on data-intensive science. OMICS. 2011;15:197–1988. doi: 10.1089/omi.2011.02ed. [DOI] [PubMed] [Google Scholar]
- Kolker N. Higdon R. Broomall W. Stanberry L. Welch D. Lu W., et al. Classifying proteins into functional groups based on all-versus-all BLAST of 10 million proteins. OMICS. 2011a;15:513–521. doi: 10.1089/omi.2011.0101. [DOI] [PubMed] [Google Scholar]
- Kolker E. Higdon R. Welch D. Bauman A. Stewart E.A. Haynes W., et al. SPIRE: Systematic Protein Investigative Research Environment. www.proteinspire.org. J. Proteomics. 2011b;75:122–126. doi: 10.1016/j.jprot.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Kolker E. Higdon R. Haynes W. Welch D. Broomall W. Lancet D., et al. MOPED: Model Organism Protein Expression Database. Nucleic Acids Res. moped.proteinspire.org. moped.proteinspire.org [DOI] [PMC free article] [PubMed]
- Moore G. Cramming more components onto integrated circuits. Electronics. 1965;38:114–117. [Google Scholar]
- Ozdemir V. Smith C. Bongiovanni K. Cullen D. Knoppers B.M. Lowe A., et al. Policy and data-intensive scientific discovery in the beginning of the 21st century. OMICS. 2011a;15:221–225. doi: 10.1089/omi.2011.0007. [DOI] [PubMed] [Google Scholar]
- Ozdemir V. Rosenblatt D.S. Warnich L. Srivastava S. Tadmouri G. Aziz R., et al. Towards an ecology of collective innovation: human variome project (HVP), rare disease consortium for autosomal loci (RaDiCAL) and data-enabled life sciences alliance (DELSA) Curr Pharmacogenomics Person Med. 2011b;9:243–251. doi: 10.2174/187569211798377153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J. Ekanayake J. Gunarathne T. Choi J. Seung-Hee B. Hui L., et al. Hybrid cloud and cluster computing paradigms for life science applications. BMC Bioinf. 2010;11(Suppl 12):S3. doi: 10.1186/1471-2105-11-S12-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt E.E. Linderman M.D. Sorenson J. Lee L. Nolan G. Computational solutions to large-scale data management and analysis. Nat Rev Genet. 2010;11:647. doi: 10.1038/nrg2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz M.C. Langmead B. Salzberg S.L. Cloud computing and the DNA data race. Nat Biotechnol. 2010;28:691. doi: 10.1038/nbt0710-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. Balazinska M. Baru C. Gomelsky M. McLennan M. Rose L., et al. Biology and data-intensive scientific discovery in the beginning of the 21st century. OMICS. 2011;15:209–212. doi: 10.1089/omi.2011.0006. [DOI] [PubMed] [Google Scholar]
- Stein L.D. The case for cloud computing in genome informatics. Genome Biol. 2010;11:207. doi: 10.1186/gb-2010-11-5-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R.C. An overview of the Hadoop/MapReduce/HBase framework and its current applications in bioinformatics. BMC Bioinformatics. 2010;11(Suppl 12):S1. doi: 10.1186/1471-2105-11-S12-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The National Digital Information Infrastructure and Preservation Program [NDIIPP] 2010. Report: Preserving Our Digital Heritage.
- Digital Research Data Sharing and Management. Report from the Task Force on Data Policies. National Science Board, National Science Foundation. 2011. www.nsf.gov/nsb/publications/2011/nsb1124.pdf www.nsf.gov/nsb/publications/2011/nsb1124.pdf
- Thessen A. Patterson D. Data issues in the life sciences, a White Paper. 2011. [DOI] [PMC free article] [PubMed]
- Wolf F. Hobby R. Lowry S. Bauman A. Franza B. Lin B., et al. Education and data-intensive science in the beginning of the 21st century. OMICS. 2011;15:217–219. doi: 10.1089/omi.2011.0009. [DOI] [PubMed] [Google Scholar]