Abstract
Malignant gliomas are the most frequent type of primary brain tumors. Patients' outcome has not improved despite new therapeutics, thus underscoring the need for a better understanding of their genetics and a fresh approach to treatment. The lack of reproducibility in the classification of many gliomas presents an opportunity where genomics may be paramount for accurate diagnosis and therefore best for therapeutic decisions. The aim of this work is to identify large and focal copy number abnormalities (CNA) and loss of heterozygosity (LOH) events in a malignant glioma population. We hypothesized that these explorations will allow discovery of genetic markers that may improve diagnosis and predict outcome. DNA from glioma specimens were subjected to CNA and LOH analyses. Our studies revealed more than 4000 CNA and several LOH loci. Losses of chromosomes 1p and/or 19q, 10, 13, 14, and 22 and gains of 7, 19, and 20 were found. Several of these alterations correlated significantly with histology and grade. Further, LOH was detected at numerous chromosomes. Interestingly, several of these loci harbor genes with potential or reported tumor suppressor properties. These novel genetic signatures may lead to critical insights into diagnosis, classification, prognosis, and design of individualized therapies.
Introduction
Diffuse gliomas are the most frequent type of primary tumors of the central nervous system. They affect more than 15,000 individuals in the United States each year and have a poor prognosis. In fact, more than 80% of patients die within a year after diagnosis (Deorah et al., 2006; Engelhard et al., 2002; Perry, 2001). Furthermore, in contrast to many other cancers, the mortality rate has not improved. These observations underscore the need for a better understanding of these neoplasms and novel approaches to treatment (Lee et al., 2008; Rich et al., 2005). Gliomas are classified into three histological subtypes: astrocytomas, oligodendrogliomas, and mixed oligoastrocytomas. Under the WHO classification of malignant gliomas, grade II includes tumors with low proliferative potential and have the best prognosis: oligodendroglioma (ODG), astrocytoma (ACG), and mixed oligoastrocytoma (MOA). Grade III encompasses anaplastic oligodendroglioma (AOD), anaplastic astrocytoma (AAC), and mixed anaplastic oligoastrocytoma (AMOA). Grade IV includes high proliferative tumors with the poorest prognosis: glioblastoma (GBM). ODGs and oligoastrocytomas represent nearly 33% of all gliomas with increasing numbers detected in recent years (Deorah et al., 2006). Patients with ODG have a relatively better prognosis whereas patients with glioblastoma have the worst outcome (Bello et al., 1995; Jeuken et al., 1999; Kraus et al., 1995; Reifenberger et al., 1994; Smith et al., 2000; Von Deimling et al., 1994). To this point, only histology, age, and Karnofsky performance status (KPS) are clinically used as valid prognostic factors (Lee et al., 2008; Rich et al., 2005). The correct diagnosis of these tumors is very important because some are very sensitive to chemotherapy, whereas the majority is drug resistant (Cairncross et al., 1998; Gupta et al., 2005; Kim et al., 1996). Moreover, there are serious challenges that result from variations in inter- and intraobserver diagnoses when using classic histology and limited molecular investigations (Gupta et al., 2005). Therefore, the potential to improve outcome is not promising (Smith et al., 2000). Compounding the problem is the inability to predict sensitivity or resistance to chemotherapy regimens (Gupta et al., 2005; Kim et al., 1996), which may result in early and unnecessary chemotherapy treatment with undesirable toxic effects and little therapeutic benefit.
Given that gliomas are very complex and genetically heterogeneous with multiple alterations in critical pathways (Gupta et al., 2005; Lee et al., 2008; Rich et al., 2005), genomic explorations may aid therapeutic decisions and constitute key diagnostic features. Defining glioma subtypes based on objective and unbiased molecular signatures will allow for a more rational patient specific approach to therapy (Smith et al., 2000).
The aim of this study is to identify major genetic alterations that occur in our glioma population in order to improve diagnosis and identify therapeutic outcome predictors. This study may lead to design of novel strategies to improve therapeutic outcome for gliomas by establishing a scientific basis for personalized therapies.
Materials and Methods
Tumor samples
Sixty tumor biopsies from patients with glioma (ODGs, ACGs, and glioblastomas) were harvested at the University of Iowa Department of Neurosurgery according to institutional review board (IRB) regulations and guidelines (IRB# 200707727). Biopsies were snap frozen within 1 to 2 h following surgery. Patients had not received chemotherapy or radiotherapy prior to surgery. Histology, diagnosis, fluorescence in situ hybridization (FISH), pathology, predictive clinical, and outcome data were recorded. For 24 recent patients, blood (5 mL) was also collected for DNA extraction (as control DNA) in a pairwise analysis for CNA confirmations.
Genomic analysis
Briefly, DNA was extracted from the biopsies using standard techniques (DNAeasy, Qiagen, Germany). Integrity and quality of extracted DNA was checked on a 1% agarose gel. Genotyping of DNA samples was performed with the Affymetrix GeneChipR Human SNP Mapping 6.0 array according to the GeneChip Mapping Assay Manual (Affymetrix, Santa Clara, CA). This microarray allows genotyping of approximately 1.8 million single nucleotide polymorphisms (SNPs) and CNAs permitting thus to detect abnormalities with a median intermarker distance of around 1 kb. Briefly, 250 ng of DNA was digested using either StyI or NspI (New England Biolabs, Ipswich, MA). StyI or NspI adaptors were subsequently ligated to the ends of all fragments using T4 DNA ligase (New England Biolabs). This was used as a template in PCR amplification using Titanium Taq (Clontech, Mountainview, CA) and a single primer complementary to the adaptor sequence. PCR products were purified from excess primers and salts by column filtration and the eluted products were fragmented using DNase I. An aliquot of the fragmented DNA was separated and visualized in a 4% agarose gel in 1× TBE buffer to ensure that the bulk of the product had been properly fragmented to a size less than 200 bp. The fragmented samples were end-labeled with biotin using terminal deoxynucleotidyl transferase before each sample was hybridized to the Human Genome wide 6.0 SNP arrays. The Human Genome wide 6.0 SNP arrays were hybridized for 16 h at 50°C. Following hybridization the arrays were washed and stained using a Affymetrix Fluidics Station 450. The most stringent wash was 0.6× SSPE, 0.01% Tween-20 at 45°C and the samples were stained with R-phycoerythrin (Life technologies, Carlsbad, CA). Imaging of the microarrays was performed using a GCS3000 high-resolution scanner (Affymetrix, Santa Clara, CA). The probe intensity data were collected using Affymetrix GCOS v1.4 software. PartekGS software (Partek, St. Louis, MO) and Nexus (BD Biosystems, El Segundo, CA) were used to evaluate copy number abnormalities and LOH status. Tumor data files (.Cel files) were compared to a baseline generated using 270 normal HapMap samples (www.HapMap.org) (for Partek) or using NCBI built 36.1 (for Nexus) and normalized using a Hidden Markov Model. Regions of increased or decreased copy number were determined by genomic segmentation. Chi-square test was used to determine statistically significant copy number changes by cancer type. Other comparisons were generated using blood samples (24 samples) for baseline and pairwise comparisons between paired blood and tumor samples.
Survival and statistical analyses
Overall survival plots between tumor subtype groups were performed according to Kaplan-Meier analysis and median survival were compared using the Log-Rank test. Variables that were significant in univariate analyses were also analyzed using multivariate Cox proportional hazard regression (KPS, age, and molecular events). Correlation analyses were performed using Pearson and Spearman correlation tests. The chi-square test was performed to segregate genomic data according to diagnoses and to study the association with molecular alterations. Bonferroni correction for multiple comparisons was performed. Statistical significance was considered as p<0.05. All statistical analyses were performed using PartekGS software (Partek, St. Louis, MO) and R-project.
RT-PCR and real-time Q-PCR
Significant results from genomic analysis were confirmed by semiquantitative RT-PCR and real-time Q-PCR. RNA was isolated from each biopsy using Trizol (Life Technologies, Carlsbad, CA). RNA integrity and quality were assessed on a 1% agarose gel and quantified by spectrometry at 260 nm/280 nm. RNA biopsies (5 μg) were reverse transcribed into cDNA using MuMLV retrotranscriptase and oligo dT (Life Technologies). Fifty nanograms of the cDNA mixture were used for further analyses. Selected cDNAs were amplified at the appropriate number of cycles with specific primers for human MGMT (CCT-GGC-TGA-ATG-CCT-ATT-TC and GAT-GAG-GAT-GGG-GAC-AGG-ATT), EGFR (CCA-CCA-AAT-TAG-CCT-GGA-CA and CGC-GAC-CCT-TAG-GTA-TTC-TG), and EGFRv3 (GAG-CTC-TTC-GGG-GAG-CAG and GTG-ATC-TGT-CAC-CAC-ATA-ATT-ACC-TTT-CT) with GAPDH (ACC-ACA-GTC-CAT-GCC-ATC-AC and TCC-ACC-ACC-CTG-TTG-CTG-TA) as a housekeeping gene. Real-time Q-PCR analysis of PTPRK transcripts levels was performed using RNA extracted from several biopsies along with RNA from nontumor control specimens from epilepsy patients using specific PTPRK primers (TGG-AGA-AAA-AGC-CAG-ACT-TCA and AGC-CAA-TCT-CTA-CCC-GTG-AAT). Chr.1p, chr.19q, and chr.7 genomic status were also confirmed using oligonucleotides that amplify specific microsatellites loci in genomic DNA as previously reported (Nigro et al., 2001). Quantitative real-time PCRs were performed on an ABI StepOne machine (Applied Biosystems, Foster City, CA).
Validation sets
To validate our results we used TCGA (http://tcga-portal.nci.nih.gov) and Rembrandt (http://rembrandt.nci.nih.gov) as independent data sets. TCGA is a huge data set that includes only GBM, whereas Rembrandt is a smaller bank which includes GBM and lower grade gliomas. Significant results were then first queried using the already analyzed data (online query) and next the raw data were reanalyzed with the same conditions as this study population using Partek and Nexus.
Results
Clinical data
This study population included 5 patients with ACG, 13 with anaplastic astrocytoma (AAC), 29 with GBM, 6 with ODG and 7 with AOD. The median age at diagnosis was 51 years and the median survival was 31 months. Histology, diagnosis, pathology, FISH, therapeutics, clinical and outcome data were recorded and the associations among clinical and biological elements were evaluated. Kaplan-Meier overall survival analyses indicated that outcome correlates significantly with age (cutoff age=51 years; p=0.0025, Log-Rank test) and with diagnosis (p=0.0001, Log-Rank test). Patients younger than 51 years have better prognosis, whereas patients with GBM have the worst outcome (18 months). Patients with low-grade ODG have the best outcome (56 months). Further, lineage showed significant correlation with survival (p=0.0010, Log-Rank test). The astrocytic lineage has the poorest prognosis compared to the oligodendrocytic lineage (22 months vs. 41 months, respectively). These results are in agreement with the literature (Aldape et al., 2007; Bello et al., 1995; Giannini et al., 2008; Jeuken et al., 1999; Kraus et al., 1995; Lo et al., 2007; Nigro et al., 2001; Reifenberger et al., 1994; Smith et al., 2000; Von Deimling et al., 1994).
Mapping arrays can be used as a diagnostic tool and can detect substantial genetic alterations occurring in diffuse gliomas
Chromosomal imbalances or instabilities can be identified by a variety of techniques such as karyotyping, FISH, microsatellites analysis, array comparative genomic hybridization (aCGH), and recently by microarray screening. Indeed, microarray investigations have been shown to surpass many classical techniques in cancer diagnosis (Acharya et al., 2008; Cancer Genome Atlas Research Network, 2008; Kleihues et al., 1994; Parsons et al., 2008; Rich et al., 2005), especially in identifying recurrent small regions of copy number alterations. Considering genetic heterogeneity of brain cancer, these genome-wide analyses appear to be ideal tools for profound explorations. A global exploration of these genetic abnormalities will establish molecular profiles of tumors that, in turn, may improve diagnosis. Additionally, it may allow prediction of sensitivity or resistance to chemotherapies based on genomic and transcriptomics differences.
To identify major CNA that occur during gliomagenesis, we profiled DNA from histologically confirmed glioma biopsies. We used multiple algorithms (Partek and Nexus) to reduce false positive results and increase the statistical power of our analyses. We performed multiple analyses: (1) pair-wise analysis for 24 samples for which corresponding blood DNAs were available, and (2) using HapMap samples and NCBI built 36.1 as controls. The HapMap and NCBI built 36.1 comparisons had similar statistical power and provided similar sensitivity and relevant data under study conditions for the pairwise comparisons. Therefore, we used HapMap (Partek) and NCBI built 36.1 data (Nexus) as a baseline because we did not have control blood DNA from majority of the patients.
Our studies revealed more than 4000 CNA spanning the entire genome (p<0.0001, ANOVA test; Supplementary Fig. 1 and Supplementary Table 1). Some of these genetic alterations were concordant with previously published studies using FISH and aCGH analyses (Aldape et al., 2007; Cancer Genome Atlas Research Network, 2008; Kleihues et al., 1994; Parsons et al., 2008). However, the majority of them are novel. The patterns of these chromosomal alterations were variable in frequency as well as size across our population of glioma patients. Their size extended from a few hundred base pairs to full chromosome gains or losses. Patients with higher grade tumors (AAC, AOD, and GBM) had more CNA and the average molecular event was much larger. Such alterations include chromosomes 1p and/or 19q deletions, and whole chromosome 7 gains. Chr.1p and 19q events correspond to a complete loss of the p-arm (1p13–1pter) and q-arm (19q12–19qter). Also, 9p losses (9p11–9pter) and 9q gains (9q21–9qter) were identified. Partial or complete losses of chromosomes 10, 13, 14, and 22 and whole gains of 19 and 20 were additionally found in a significant number of patients. Specifically, we estimate that many tumors have gained up to 11 copies of chr.7 (15 tumors), 4 copies of chr.19 (6 tumors), and 5 copies of chr.20 (5 tumors), whereas other tumors have lost one or both copies of chr.10 (18 tumors), chr.13 (2 tumors), chr.14 (2 tumors), and chr.22 (3 tumors). Moreover, the majority of CNAs were focal abnormalities that affect relatively small chromosomal regions that harbor many genes particularly relevant to gliomagenesis and to oncogenesis in general (Supplementary Table 1).
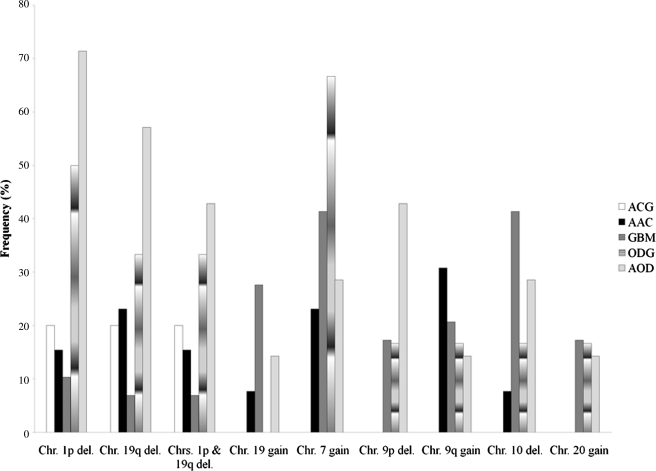
Using a chi-square test, we segregated these data according to the clinical diagnoses (Fig. 1). As expected, Chr.1p/19q losses were seen more frequently in the oligodendrocytic lineage (∼61%, p=0.0260; Pearson test) than in the astrocytic lineage (∼17%, p=0.0048; Pearson test). Gains of Chr. 7 were detected in both astrocytic (∼47%) and oligodendrocytic lineages (∼30%). This observation disagrees with previously published works that show gains of chr.7 mostly in astrocytic lineages (Aldape et al., 2007; Giannini et al., 2008; Lo et al., 2007; Nigro et al., 2001). We found that classification of grade II and III gliomas based on 1p/19q status is a better outcome predictor than histology in our population (Figs. 2 and 3). Indeed, it is widely believed that 1p and 19q assessment must be added to the diagnostic evaluation (Aldape et al., 2007; Giannini et al., 2008; Gupta et al., 2005; Smith et al., 2000). Specifically, it is advocated that anaplastic gliomas should be divided into anaplastic 1p/19q deleted gliomas and anaplastic non-1p/19q deleted glioma independently of lineage (Giannini et al., 2008). Our analysis shows that mapping arrays are ideal tools to cluster patients in these two categories.
FIG. 1.
Frequency of major chromosome alterations in the studied glioma population. Chi-square test was used to determine statistically significant copy number changes by glioma subtype. ACG, astrocytoma grade II; AAC, anaplastic astrocytoma grade III; GBM, glioblastoma grade IV; ODG, oligodendroglioma grade II; AOD, anaplastic oligodendroglioma grade III.
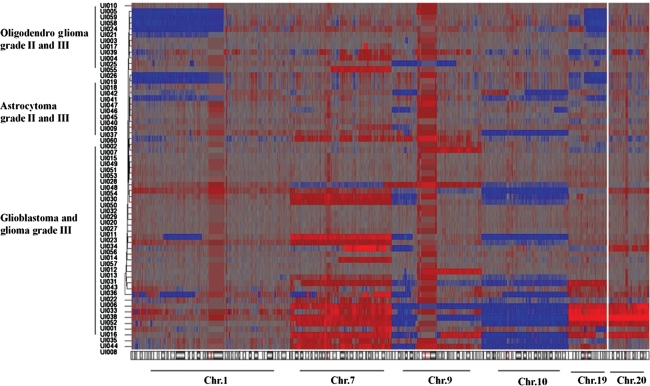
FIG. 2.
Summary of gain (red) and loss (blue) events observed in selected chromosomes that showed highly statistically significance associations with diagnosis (p<0.0001, ANOVA); chromosome 1, chromosome 7, chromosome 9, chromosome 10, chromosome 19, and chromosome 20 are shown. Specific genetic losses or gains have been detected and shown to correlate significantly with histology and subtype.
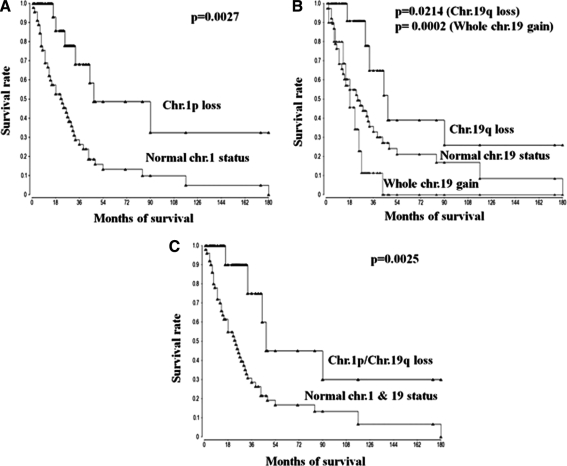
FIG. 3.
Kaplan-Meier estimates of probability of survival stratified according to chr.1p deletion signature (A), to chr.19 gain or chr.19q loss status signature (B), and to chr.1p/chr.19q losses signature (C).
Furthermore, hierarchical clustering of the 60 tumor specimen resulted in the formation of distinct groups (Fig. 2). These genetic signatures cluster and correlate significantly with diagnosis, histology, and grade. However, few patients clustered away from the clinically assigned diagnosis. These cases may constitute misdiagnosed patients or may belong to the classical mixed oligoastrocytoma type that harbors characteristics of both lineages. Therefore, our data underscore additional evidence of the superiority of microarray analyses over classic histology diagnostic tests (Fritz et al., 2002). Moreover, we observed that many genetic abnormalities have a specific pattern of distribution. Indeed, tumors with amplifications of chr.7 also show losses of chr.10 and chr.9p, whereas tumors with chr.1p and or chr.19q deletions have no alteration in chr.7 or chr.9p. Further, tumors with whole chr.19 gains also show chr.7 gains along with losses of chr.10 and gains of chr.20 (Fig. 2). These latter tumors are all high grade tumors (grade III or IV).
The prognostic significance of this clustering was then evaluated using Kaplan-Meier survival analysis. Deletion of 1p, 19q, and loss of 1p in association with 19q correlate better with outcome than intact chromosomes 1 and 19 in both oligodendrocytic and astrocytic lineages (Fig. 3A–C) (p=0.0027 (33 vs. 18 months), p=0.0214 (31 vs. 23 months), and p=0.0025 (20 vs. 83 months), respectively, Log-Rank test). Surprisingly, whole chr.19 gain signature was correlated with a poorer outcome (Fig. 3B) (p=0.0002, 16 vs. 31 months, Log-Rank test). Correlation between amplification of chr.7 and outcome did not reach significance (p=0.0825, 29 vs. 37 months, Log-Rank test; not shown) but showed trends to be associated with a poor outcome in both lineages.
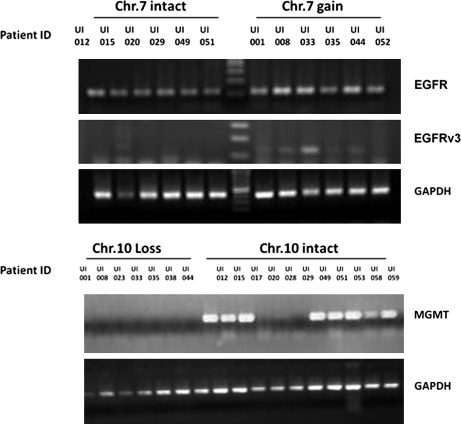
It was therefore decided to confirm several data alterations using real-time Q-PCR or using FISH results that were previously performed as a diagnostic exploration for some patients. As expected, high correlation between FISH, confirmatory real-time PCR (microsatellite analysis using real-time Q-PCR on 6 markers located at chr.1p, chr.19q, and chr.7 as described) (Nigro et al., 2001) and mapping array results for 1p and 19q detection was observed (data not shown). RT-PCR analyses were performed to further validate the data by showing that these CNA are accompanied by a change also at the transcript levels of major significant genes at the particular loci. It was then decided to analyze EGFR, EGFGRv3, and MGMT status because their loci were significantly altered as shown by the analysis. In a significant number of glioma biopsies gains of chromosome 7 and losses of chromosome 10 and the consequent amplification of EGFR and deletion of MGMT loci were observed. The analyses confirmed that EGFR was expressed at higher levels in the astrocytic lineage and that some biopsies also expressed the splice variant EGFRv3 (Fig. 4). EGFRv3 was shown in patients that have highest copy number of chr.7. These data positively correlate with amplification of chr.7. Moreover, MGMT was silenced (Fig. 4) in many astrocytic specimens, which seems to be closely associated with loss of chr.10.
FIG. 4.
EGFR, EGFRv3, and MGMT gene expressions comparisons in representative specimens using semiquantitative RT-PCR showing variable levels of expression depending on chr.7 and chr.10 status. GAPDH serves as a housekeeping gene for normalization.
Last, segmentation analysis revealed deletions or amplifications of many important genes in oncogenesis at significant frequency (Supplementary Table 1). Genes playing a critical role in several important oncogenic pathways were found to be altered (angiogenesis, NOTCH, Hedgehog, MAPK, cell cycle, ubiquitination, cell–cell adhesion, and inflammation). Some of these alterations were also independently identified in the TCGA and Rembrandt data sets. Specifically, our analyses revealed deregulations of many critical pathways implicating p53, Rb, and RTK/PI3K and allowed individualizing six major pathways that are frequently deregulated: (1) receptor tyrosine kinases (RTK) pathway, (2) focal adhesion pathway, (3) DNA repair/MGMT status, (4) mTOR pathway, (5) angiogenesis pathway, and (6) proteasome/ubiquitination pathway. Individualizing therapies for patients based on these alterations may help improve outcomes. Some of these findings are in agreement with recently published works (Freije et al., 2004; Cancer Genome Atlas Research Network, 2008; Parsons et al., 2008; Phillips et al., 2006) and with the data obtained from TCGA and Rembrandt data sets.
Loss of heterozygosity analysis revealed multiple tumor suppressor candidates
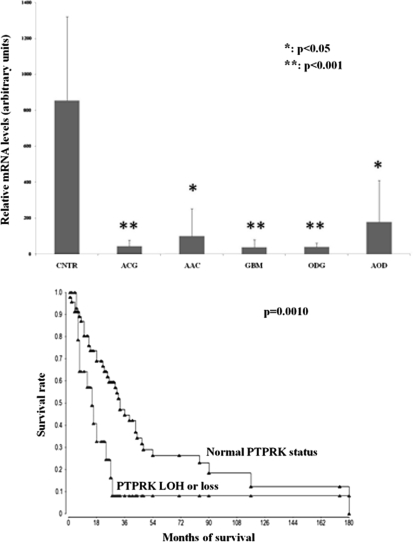
SNP arrays, in opposition to aCGH explorations, can also assess loss of heterozygosity (LOH), which could allow identification of genome-wide patterns of allelic imbalance with potential prognostic and diagnostic value. They can detect copy neutral LOH along with LOH resulting from amplifications or deletions. These programs use a hidden Markov model-based method to identify LOH from unpaired tumor samples by calculating and comparing SNP intermarker distances, documented SNP-specific heterozygosity rates, and the haplotype structure of the human genome (Beroukhim et al., 2006). Moreover, these calculations allow identification of LOH even in patients for whom no corresponding blood DNA was available. Using Partek and Nexus algorithms, we were able to highlight many chromosomal regions showing significant LOH (copy neutral or with deletion), thus revealing multiple gene candidates as potential tumor suppressors (Supplementary Table 1). The most significant is PTPRK [protein tyrosine kinase receptor kappa (6q22.33)]. Gene expression analysis using real-time Q-PCR of this selected candidate (lowest p-value and highest frequency) revealed significant lower PTPRK mRNA levels in tumor than nontumor brain specimens (Fig. 5A; p<0.05 and p<0.001; ANOVA). Corresponding healthy brain tissues from patients with glioma could not be harvested. As a consequence, biopsies from patients with epilepsy were used as nontumor control specimens. Moreover, the prognostic significance of these altered genes were evaluated using Kaplan-Meier survival analysis, which revealed strong associations between PTPRK molecular status and median survival (p=0.0010, 30 vs. 14 months, Log-Rank test) (Fig. 5B). The data indicate that patients with inactivated or deleted PTPRK (23%) have poorer outcomes compared to patients with normal locus. PTPRK status at both gene and transcriptomic levels was also shown to predict outcome in Rembrandt glioma data (not shown). However, it did not reach significance in the TCGA data mainly because of the absence of lower grade tumor specimens in this databank.
FIG. 5.
(A) PTPRK gene expression comparison in representative specimens using real-time Q-PCR showing lower levels of PTPRK expression in various tumor specimen compared to nontumor brain samples (epilepsy biopsies; CNTR). ACG, astrocytoma grade II (n=5); AAC, anaplastic astrocytoma grade III (n=13); GBM, glioblastoma grade IV (n=29); ODG: oligodendroglioma grade II (n=6); AOD, anaplastic oligodendroglioma grade III (n=7). (B) Kaplan-Meier estimates of probability of survival stratified according to PTPRK genomic status.
Discussion
Although limited by a relatively small sample size, this study reveals major genomic alterations that significantly correlate with diagnosis and clinical outcome. Furthermore, it underscores the great value of unbiased genomic analyses in the molecular characterization of adult brain cancers. The data show that the SNP 6.0 mapping array (and equivalent arrays) can be used as a diagnostic tool to detect deletions or gains of chromosomes 1, 7, 10, 19, and 20, which are the most constant genetic alterations in diffuse gliomas. Based on the results of this analysis, patients with glioma could be assigned to lineage and genetic subtypes on the basis of genome-wide signatures with high accuracy. Moreover, many of these genetic alterations also predict therapeutic response. Loss of 1p has been reported as a significant predictor of tumor sensitivity to chemotherapy (Bauman et al., 2000; McDonald et al., 2005). Combined allelic losses of 1p and 19q have been statistically associated with better survival after chemotherapy. Further, the loss of 1p has been shown to predict longer survival after radiotherapy, with or without chemotherapy (Bauman et al., 2000), thus constituting possible therapy-independent prognostic factors.
LOH analysis and overall survival analyses have identified PTPRK as a potential tumor suppressor candidate, as a prognostic marker, and perhaps a potential predictive marker. PTPRK, a cell surface receptor protein tyrosine phosphatase, is a homophilic cell adhesion molecule expressed in glial cells that is known to regulate β-Catenin/Cadherin-dependent adhesion and to dephosphorylate EGFR (Novellino et al., 2008; Ostman et al., 2006; Xu et al., 2005). Although it seems that PTRPK plays an essential role in controlling kinase signaling networks, the role of this protein in oncogenesis is not well understood. Nevertheless, the ability of PTPRK to mediate interactions among cells, together with the observation that its expression is upregulated by cell density strongly suggest a crucial role of PTPRK in modulating cell–cell interactions and adhesion, which are a hallmark of diffuse gliomas (Novellino et al., 2008). Accordingly some studies have now shown that PTPRK maps to a putative tumor suppressor gene locus (Nakamura et al., 2003; Novellino et al., 2008; Starr et al., 2009; Xu et al., 2005). Consequently, additional studies are needed to explore PTPRK's roles in gliomagenesis and especially the diffusion properties of gliomas in the surrounding brain tissue.
This and other recent genomic profiling studies have demonstrated that at least three major pathways are frequently altered in gliomas (Cancer Genome Atlas Research Network, 2008; Parsons et al., 2008; Pelloski et al., 2006; Sarkaria et al., 2007). These include: (1) amplification or mutation of receptor tyrosine kinases (EGFR, VEGFR, PDGFR, cKIT) and consequent activation of PI3K and RAS pathways (RTK/PI3K/Ras); (2) mutation and inactivation of P53 pathway (p53/MDM2/p14); and (3) mutation and inactivation of retinoblastoma (Rb/p16) pathway. Furthermore, other key pathways such as mTOR, PTEN, SRC, ubiquitination, NOTCH, hedgehog, and angiogenesis are also altered. These pathways play crucial roles in gliomagenesis as they are involved in cell cycle, transcription, translation, angiogenesis, cell proliferation, cell growth, and drug resistance (Zhu et al., 2009). Characterization may help to individualize therapies by directing selected patients to treatment targeted for their specific genetic makeup. Indeed, patients with alterations in the receptor tyrosine kinase (RTK) pathway, related to amplification of chromosome 7 and the subsequent EGFR locus amplification, are expected to respond better to RTK inhibitors, AKT inhibitors, SRC, or RAS modulators. Another altered pathway is DNA repair that is frequently targeted in gliomas via induction of DNA breaks. Glioma patients are treated mainly using alkylating agents and it is widely believed that numerous patients receiving these agents may not benefit from them, but may experience unwanted toxic effects (Huang et al., 2009). Indeed, non- or low MGMT expressers, that are associated frequently with Chr.10 deletion signature in this study population, were shown to respond well to alkylating agents and specifically to temozolomide, whereas patients with higher levels of MGMT are resistant or refractory to temozolomide (Hegi et al., 2005).
Microvascular proliferation and necrosis are the characteristic features of high-grade glioma tumors that result from proneural to a mesenchymal gene expression signature transition occurring at the stem cell niche level (Sathornsumetee and Reardon, 2009). This signature is closely associated in this study to the chr.19 amplification signature and the subsequent NOTCH3 (19p13.12) overactivation. NOTCH seems to play a major role in the proneural to a mesenchymal transition and therefore in angiogenesis. Targeting NOTCH and VEGF with specific inhibitors such as gamma secretases inhibitors and bevacizumab, respectively, may improve therapeutic outcomes specifically in patients with chr.19 genetic signature.
Transcriptomic analysis has shown several limitations and poor reproducibility (Colman et al., 2010; Freije et al., 2004; Lee et al., 2008; Li et al., 2009; Lo et al., 2007; Phillips et al., 2006; Rich et al., 2005; Zhu et al., 2009). Gene expression analyses provide a snapshot at a certain time, whereas genomics (CNA and SNP arrays) provide data that are specific, constant, and highly reproducible (Aldape et al., 2007; Cancer Genome Atlas Research Network, 2008; Parsons et al., 2008). This is demonstrated by the high data reproducibility published in the literature that were obtained using aCGH and older versions of CNA microarray, compared to gene expression results, which are characterized by very low and deceptive reproducibility and standardization. Although gene expression profiling has proven useful in diagnosis and subtyping tumors, it seems that SNP arrays are more powerful than expression arrays for many obvious reasons; DNA stability, quantity, quality, availability, control (blood), inability to harvest healthy surrounding tissues from the same patient, circadian rhythm, localization, time between surgery and RNA extractions, and degree of infiltration by inflammatory cells. These factors can adversely influence gene expression analysis and interpretation. However, based on these and other results, genomic exploration of CNA and SNP appear to be highly reproducible, independent of laboratory or technology (aCGH, Affymetrix, Illumina, next-generation sequencing) (Cowell et al., 2010; Cancer Genome Atlas Research Network, 2008; Parsons et al., 2008). Moreover, the recent progress in cancer treatment has been realized mainly based on genomic data (Her2/trastuzumab/breast cancer) (Pegram et al., 1998), EGFR/erlotinib/cetuximab (Perez-Soler et al., 2004), gefitinib/lung cancer in nonsmoking Asian females (Pao et al., 2004; Tamura and Fukuoka, 2005), and imatinib/t(9-22)-positive leukemia (Kantarjian et al., 2002). Indeed, transcriptomic signatures have been successful only using retrospective studies that have not been independently validated to date. Specifically, there are no distinct studies that have identified the same set of transcriptomic signatures that allow reproducible diagnosis, prognosis, and/or therapeutic prediction. Although several studies tried to identify specific and reproducible transcriptomic signatures that predict survival in different population data sets using different normalization and statistical analyses (Colman et al., 2010; Gravendeel et al., 2009; Li et al., 2009) great discordances are still observed.
Conclusion
This study not only provides data that are in agreement with recently published works but also reveals significant novel genetic alterations that may help improve diagnosis and establish prognosis. Moreover, this work is more complete because the analysis is not limited to glioblastomas or to lower grades gliomas and shows analyses of genomic aberrations in all glioma subgroups. The results reveal significant genetic alterations that are lineage specific, grade specific, and prognostic accurate. Furthermore, these identified genetic signatures could be potentially used as predictors of therapeutic outcome that reveal sensitivity or resistance to anticancer drugs prior to administration and may provide new insight into molecular mechanisms underlying the severity and heterogeneity of gliomas. Superiority of SNP arrays and thereby cytogenetic arrays, over standard techniques and gene expression profiling, highlight a need for further validation prior to introduction into clinical practice and utilization to realize the tremendous potential for personalized medicine.
Supplementary Material
Acknowledgments
We thank Dr. Jeffrey C. Murray and Dr. Aline Lourenco Petrin from the University of Iowa for the help during this study by providing us access to their bioinformatics tools. We are grateful to the DNA core facility of the University of Iowa for the technical help with microarrays hybridizations and analyses. This work was supported by the Institute of Clinical and Translational Science at the University of Iowa award. This publication was made possible by Grant Number UL1RR024979 from the National Center for Research Resources (NCRR), a part of the National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the CTSA or NIH. This research was supported through the Institutional Research Grant Number IRG-77-004-34 from the American Cancer Society, administered through The Holden Comprehensive Cancer Center at the University of Iowa.
Author Disclosure Statement
The authors declare that no conflicting financial interests exist.
References
- Acharya C.R. Hsu D.S. Anders C.K. Anguiano A. Salter K.H. Walters K.S., et al. Gene expression signatures, clinicopathological features, and individualized therapy in breast cancer. JAMA. 2008;299:1574–1587. doi: 10.1001/jama.299.13.1574. [DOI] [PubMed] [Google Scholar]
- Aldape K. Burger P.C. Perry A. Clinicopathologic aspects of 1p/19q loss and the diagnosis of oligodendroglioma. Arch Pathol Lab Med. 2007;131:242–251. doi: 10.5858/2007-131-242-CAOQLA. [DOI] [PubMed] [Google Scholar]
- Bauman G.S. Ino Y. Ueki K. Zlatescu M.C. Fisher B.J. Macdonald D.R., et al. Allelic loss of chromosome 1p and radiotherapy plus chemotherapy in patients with oligodendrogliomas. Int J Radiat Oncol Biol Phys. 2000;48:825–830. doi: 10.1016/s0360-3016(00)00703-3. [DOI] [PubMed] [Google Scholar]
- Bello M.J. Leone P.E. Vaquero J. De Campos J.M. Kusak M.E. Sarasa J.L., et al. Allelic loss at 1p and 19q frequently occurs in association and may represent early oncogenic events in oligodendroglial tumors. Int J Cancer. 1995;64:207–210. doi: 10.1002/ijc.2910640311. [DOI] [PubMed] [Google Scholar]
- Beroukhim R. Lin M. Park Y. Hao K. Zhao X. Garraway L.A., et al. Inferring loss-of-heterozygosity from unpaired tumors using high-density oligonucleotide SNP arrays. PLoS Comput Biol. 2006;2:e41. doi: 10.1371/journal.pcbi.0020041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairncross J.G. Ueki K. Zlatescu M.C. Lisle D.K. Finkelstein D.M. Hammond R.R., et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90:1473–1479. doi: 10.1093/jnci/90.19.1473. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman H. Zhang L. Sulman E.P. McDonald J.M. Shooshtari N.L. Rivera A., et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell J.K. Lo K.C. Luce J. Hawthorn L. Interpreting aCGH-defined karyotypic changes in gliomas using copy number status, loss of heterozygosity and allelic ratios. Exp Mol Pathol. 2010;88:82–89. doi: 10.1016/j.yexmp.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deorah S. Lynch C.F. Sibenaller Z.A. Ryken T.C. Trends in brain cancer incidence and survival in the United States: Surveillance, Epidemiology, and End Results Program, 1973 to 2001. Neurosurg Focus. 2006;20:E1. doi: 10.3171/foc.2006.20.4.E1. [DOI] [PubMed] [Google Scholar]
- Engelhard H.H. Stelea A. Cochran E.J. Oligodendroglioma: pathology and molecular biology. Surg Neurol. 2002;58:111–117. doi: 10.1016/s0090-3019(02)00751-6. [DOI] [PubMed] [Google Scholar]
- Freije W.A. Castro-Vargas F.E. Fang Z. Horvath S. Cloughesy T. Liau L.M., et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- Fritz B. Schubert F. Wrobel G. Schwaenen C. Wessendorf S. Nessling M., et al. Microarray-based copy number and expression profiling in dedifferentiated and pleomorphic liposarcoma. Cancer Res. 2002;62:2993–2998. [PubMed] [Google Scholar]
- Giannini C. Burger P.C. Berkey B.A. Cairncross J.G. Jenkins R.B. Mehta M., et al. Anaplastic oligodendroglial tumors: refining the correlation among histopathology, 1p 19q deletion and clinical outcome in Intergroup Radiation Therapy Oncology Group Trial 9402. Brain Pathol. 2008;18:360–369. doi: 10.1111/j.1750-3639.2008.00129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravendeel L.A.M. Kouwenhoven M.C. Gevaert O. de Rooi J.J. Stubbs A.P. Duijm J.E., et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69:9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- Gupta M. Djalilvand A. Brat D.J. Clarifying the diffuse gliomas: an update on the morphologic features and markers that discriminate oligodendroglioma from astrocytoma. Anat Pathol. 2005;124:755–768. doi: 10.1309/6JNX-4PA6-0TQ5-U5VG. [DOI] [PubMed] [Google Scholar]
- Hegi M.E. Diserens A.C. Gorlia T. Hamou M.F. de Tribolet N. Weller M., et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- Huang T.T. Sarkaria S.M. Cloughesy T.F. Mischel P.S. Targeted therapy for malignant glioma patients: lessons learned and the road ahead. Neurotherapeutics. 2009;6:500–512. doi: 10.1016/j.nurt.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken J.W. Sprenger S.H. Wesseling P. Macville M.V. Von Deimling A. Teepen H.L., et al. Identification of subgroups of high-grade oligodendroglial tumors by comparative genomic hybridization. J Neuropathol Exp Neurol. 1999;58:606–12. doi: 10.1097/00005072-199906000-00005. [DOI] [PubMed] [Google Scholar]
- Kantarjian H. Sawyers C. Hochhaus A. Guilhot F. Schiffer C. Gambacorti-Passerini C., et al. International STI571 CML Study Group. Hematologic and cytogenetic responses to imatinib mesylate in chronic myelogenous leukemia. N Engl J Med. 2002;346:645–652. doi: 10.1056/NEJMoa011573. [DOI] [PubMed] [Google Scholar]
- Kim L. Hochberg F.H. Thornton A.F. Harsh G.R., 4th Patel H. Finkelstein D., et al. Procarbazine, lomustine and vincristine (PCV) chemotherapy for grade III and grade IV oligoastrocytomas. J Neurosurg. 1996;85:602–607. doi: 10.3171/jns.1996.85.4.0602. [DOI] [PubMed] [Google Scholar]
- Kleihues P. Lubbe J. Watanabe K. von Ammon K. Ohgaki H. Genetic alterations associated with glioma progression. Verh Dtsch Ges Pathol. 1994;78:43–47. [PubMed] [Google Scholar]
- Kraus J.A. Koopmann J. Kaskel P. Maintz D. Brandner S. Schramm J., et al. Shared allelic losses on chrom. 1p and 19q suggest a common origin of oligodendroglioma and oligoastrocytoma. J Neuropathol Exp Neurol. 1995;54:91–95. doi: 10.1097/00005072-199501000-00011. [DOI] [PubMed] [Google Scholar]
- Lee Y. Scheck A.C. Cloughesy T.F. Lai A. Dong J. Farooqi H.K., et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A. Walling J. Ahn S. Kotliarov Y. Su Q. Quezado M., et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69:2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo K.C. Rossi M.R. LaDuca J. Hicks D.G. Turpaz Y. Hawthorn L., et al. Candidate glioblastoma development gene identification using concordance between copy number abnormalities and gene expression level changes. Genes Chromosomes Cancer. 2007;46:875–894. doi: 10.1002/gcc.20474. [DOI] [PubMed] [Google Scholar]
- McDonald J.M. See S.J. Tremont I.W. Colman H. Gilbert M.R. Groves M., et al. The prognostic impact of histology and 1p/19q status in anaplastic oligodendroglial tumors. Cancer. 2005;104:1468–1477. doi: 10.1002/cncr.21338. [DOI] [PubMed] [Google Scholar]
- Nakamura M. Kishi M. Sakaki T. Hashimoto H. Nakase H. Shimada K., et al. Novel tumor suppressor loci on 6q22–23 in primary central nervous system lymphomas. Cancer Res. 2003;63:737–741. [PubMed] [Google Scholar]
- Nigro J.M. Takahashi M.A. Ginzinger D.G. Law M. Passe S. Jenkins R.B., et al. Detection of 1p and 19q loss in oligodendroglioma by quantitative microsatellite analysis, a real-time quantitative polymerase chain reaction assay. Am J Pathol. 2001;158:1253–1262. doi: 10.1016/S0002-9440(10)64076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novellino L. De Filippo A. Deho P. Perrone F. Pilotti S. Parmiani G., et al. PTPRK negatively regulates transcriptional activity of wild type and mutated oncogenic β-catenin and affects membrane distribution of β-catenin/E-cadherin complexes. Cell Signal. 2008;20:872–883. doi: 10.1016/j.cellsig.2007.12.024. [DOI] [PubMed] [Google Scholar]
- Ostman A. Hellberg C. Bohmer F.D. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- Pao W. Miller V. Zakowski M. Doherty J. Politi K. Sarkaria I., et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proc Natl Acad Sci USA. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D.W. Jones S. Zhang X. Lin J.C. Leary R.J. Angenendt P., et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pegram M.D. Lipton A. Hayes D.F. Weber B.L. Baselga J.M. Tripathy D., et al. Phase II study of receptor-enhanced chemosensitivity using recombinant humanized anti-p185HER2/neu monoclonal antibody plus cisplatin in patients with HER2/neu-overexpressing metastatic breast cancer refractory to chemotherapy treatment. J Clin Oncol. 1998;16:2659–2671. doi: 10.1200/JCO.1998.16.8.2659. [DOI] [PubMed] [Google Scholar]
- Pelloski C.E. Lin E. Zhang L. Yung W.K. Colman H. Liu J.L., et al. Prognostic associations of activated mitogen-activated protein kinase and Akt pathways in glioblastoma. Clin Cancer Res. 2006;12:3935–3941. doi: 10.1158/1078-0432.CCR-05-2202. [DOI] [PubMed] [Google Scholar]
- Pérez-Soler R. Chachoua A. Hammond L.A. Rowinsky E.K. Huberman M. Karp D., et al. Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J Clin Oncol. 2004;22:3238–3247. doi: 10.1200/JCO.2004.11.057. [DOI] [PubMed] [Google Scholar]
- Perry A. Oligodendroglial neoplasms: current concepts, misconceptions, and folklore. Adv Anat Pathol. 2001;8:183–199. doi: 10.1097/00125480-200107000-00001. [DOI] [PubMed] [Google Scholar]
- Phillips H.S. Kharbanda S. Chen R. Forrest W.F. Soriano R.H. Wu T.D., et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Reifenberger J. Reifenberger G. Liu L. James C.D. Wechsler W. Collins V.P. Molecular genetic analysis of oligodendroglial tumors shows preferential allelic deletions on 19q and 1p. Am J Pathol. 1994;145:1175–1190. [PMC free article] [PubMed] [Google Scholar]
- Rich J.N. Hans C. Jones B. Iversen E.S. McLendon R.E. Rasheed B.K., et al. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65:4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- Sarkaria J.N. Yang L. Grogan P.T. Kitange G.J. Carlson B.L. Schroeder M.A., et al. Identification of molecular characteristics correlated with glioblastoma sensitivity to EGFR kinase inhibition through use of an intracranial xenograft test panel. Mol Cancer Ther. 2007;6:1167–1174. doi: 10.1158/1535-7163.MCT-06-0691. [DOI] [PubMed] [Google Scholar]
- Sathornsumetee S. Reardon D.A. Targeting multiple kinases in glioblastoma multiforme. Expert Opin Invest Drugs. 2009;18:277–292. doi: 10.1517/13543780802692603. [DOI] [PubMed] [Google Scholar]
- Smith J.S. Perry A. Borell T.J. Lee H.K. O'Fallon J. Hosek S.M., et al. Alterations of chromosome arms 1p and 19q as predictors of survival in oligodendrogliomas, astrocytomas, and mixed oligoastrocytomas. J Clin Oncol. 2000;18:636–645. doi: 10.1200/JCO.2000.18.3.636. [DOI] [PubMed] [Google Scholar]
- Starr T.K. Allaei R. Silverstein K.A. Staggs R.A. Sarver A.L. Bergemann T.L., et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323:1747–1750. doi: 10.1126/science.1163040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K. Fukuoka M. Gefitinib in non-small cell lung cancer. Expert Opin Pharmacother. 2005;6:985–993. doi: 10.1517/14656566.6.6.985. [DOI] [PubMed] [Google Scholar]
- Von Deimling A. Nagel J. Bender B. Lenartz D. Schramm J. Louis D.N., et al. Deletion mapping of chromosome 19 in human gliomas. Int J Cancer. 1994;57:676–680. doi: 10.1002/ijc.2910570511. [DOI] [PubMed] [Google Scholar]
- Xu Y. Tan L. Grachtchouk V. Voorhees J.J. Fisher G.J. Receptor-type protein-tyrosine phosphatase-κ regulates epidermal growth factor receptor function. J Biol Chem. 2005;280:42694–42700. doi: 10.1074/jbc.M507722200. [DOI] [PubMed] [Google Scholar]
- Zhu H. Acquaviva J. Ramachandran P. Boskovitz A. Woolfenden S. Pfannl R., et al. Oncogenic EGFR signaling cooperates with loss of tumor suppressor gene functions in gliomagenesis. Proc Natl Acad Sci USA. 2009;106:2712–2716. doi: 10.1073/pnas.0813314106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.