Abstract
Background
Chromosomal translocations are usually analyzed as a single entity, and are associated with a poor outcome in chronic lymphocytic leukemia. Translocations involving immunoglobulin genes are recurrent, but uncommon (<5%), and their individual prognosis is not clear. The two most frequent partners are BCL2 (18q21) and BCL3 (19q13).
Designs and methods
Herein, 75 cases are reported of chronic lymphocytic leukemia and t(14;18) (BCL2-CLLs). Our series benefits from morphological, immunological and cytogenetical reviews. The IGHV status analyses were performed by referring laboratories. Comparison was made with our previously published series of chronic lymphocytic leukemia patients with t(14;19) (BCL3-CLLs, n=29).
Results
Compared with BCL3-CLLs, lymphocytosis was lower in BCL2-CLLs (p<0.008), and splenomegaly was less frequent (p<0.0001). There were more “typical” morphologies (p<0.005) and Matutes scores >4 (p<0.001) in the BCL2-CLLs group, and less CD38 expression (p<0.04). More variant BCL2-translocations were observed (t(18;22), n=11; 2t(2;18), n=2; p<0.02), and BCL2-translocation was frequently single (p<0.002). Complex karyotypes (p<0.02), trisomy 12 (p<0.03), 6q deletion (p<0.002) and TP53 deletion (p<0.02) were less frequent in BCL2-CLLs, whereas 13q deletion was more frequent (p<0.005). The IGHV gene was frequently mutated in BCL2-CLLs (p<0.0001). Treatment-free survival was longer in BCL2-CLLs (p<0.0001).
Conclusions
BCL2-CLL.S express CD5 and lack expression of CD38, and have a Matutes score ≥4, frequent trisomy 12, no ATM and 6q deletions, and a mutated IGHV status. Compared to BCL3-CLLs, BCL2-CLLs are much less aggressive; indicating that identifying individual translocations and cytogenetic partners would allow improved patient stratification.
Keywords: Chronic lymphocytic leukemia, IGH, BCL2, chromosomal translocation
Introduction
Chromosomal translocations involving the immunoglobulin (IG) loci are found in some, but not all, forms of B-cell malignancies. Some IG translocations are seen in almost all cases of a specific disease subgroup and, therefore, may be useful as a diagnostic marker. In contrast, cytogenetically identical translocations may be found in several types of disease. Chromosomal translocations are usually associated with poor prognosis in chronic lymphocytic leukemia (CLL) [1-3]. Translocations involving IG genes are recurrent, but uncommon (<5%) in CLL. The result of these translocations is deregulated expression of the partner genes. Recurrent known partner genes in CLL include BCL2 (18q21), BCL3 (19q13), BCL11A (2p11), and MYC (8q24), with BCL2 and BCL3 being the two most frequent [4]. In most studies, CLL cases with IG translocations are analyzed as one group; consequently, the significance of these translocations remains poorly understood.[5, 6] The target gene that becomes overexpressed may be relevant to the outcome. We have recently observed that the t(14;19) translocation, which involves the BCL3 gene, is associated with an aggressive subgroup of atypical CLLs [7], in accordance with previous publications [8-10].
Herein, we report a clinical and biological study of 75 cases where CLL harbors a t(14;18), or its variants (BCL2-CLLs), the t(2;18)(p11;q21) and t(18;22)(q21;q11). This is the largest reported series to date, and is compared with our previously published series of 29 cases with CLL and t(14;19)(BCL3-CLLs).
Design and methods
The Groupe Francophone de Cytogénétique Hématologique (GFCH) collected data from 80 B chronic lymphoproliferative disorders with circulating blood cells, excluding follicular lymphoma. The cases had t(14;18) or variant BCL2-translocations, and were diagnosed between 1985 and 2009. All patients gave their informed consent in agreement with the Helsinki declaration, and the Institutional Ethics Committee at Pitie-Salpetriere Hospital approved this study. Morphological review was performed for 62 cases by two cytologists (KM and CS), an immunological review was performed for all cases by HM-B. Cytogenetic analyses were performed on peripheral blood or bone-marrow lymphocytes cultured for 72h with 12-O-tetradecanoylphorbol-13-acetate (TPA) or CpG-oligonucleotides + interleukin 2 (IL-2). All karyo-types were reviewed by members of the GFCH. Chromosomes were classified according to the International System for Human Cytogenetic Nomenclature (ISCN 2009). Fluorescence in situ hybridization (FISH) was performed on interphase nuclei and metaphases, following standard procedures and using specific probes: IGH, BCL2, IGL, IGK (Dako, Trappes, France), CEP12, 13q14 (D13S319), ATM, p53, 6q21, MALT1, (Abbott, Rungis, France). Analyses of the mutational status of the variable region of the immunoglobulin gene (IGHV) were performed by the referring laboratories. Statistical analyses were carried out using Fisher’s exact test, and continuous data were analyzed using the Mann-Whitney test. The chi2-test was used to compare our data with that from the literature. An effect was considered statistically significant if the p value was 0.05 or less. Overall survival (OS) and treatment-free survival (TFS) calculated from diagnoses were estimated using Kaplan-Meier methodology, and statistical significance was determined using a log-rank test. All tests were two-sided.
Results
The 80 patients had a gender ratio of 61 males and 19 females. Their median age at diagnosis was 66 years (range 32-83). Seventeen out of 66 (26%) patients had lymphnodes, and four (6%) had splenomegaly. The median level of lymphocytosis was 13.9×109/l (range 5.4-106.2×109/l). Of the 62 cases that had a cytological review, 29 (47%) had “typical” CLL, 28 (45%) had “atypical” CLL, with more than 10% of lymphoplasmacytoid cells and/or large cells, 4 (6%) had marginal zone lymphoma (MZL), and 1 (2%) had unclassified low-grade lymphoma. The 18 cases that could not be reviewed were classified as CLLs by referring laboratories. In total, there were 75 CLLs and 5 lymphomas. Of the 68 CLLs with available data at diagnosis, 63 (93%) were classified as Binet stage A, 4 (6%) as Binet B, and 1 (1%) as Binet C (Supplementary Table 1).
Regarding CLLs, all tested cases (58/58) were CD10 negative, 69/73 (94%) were CD5 positive and 61/70 (87%) were CD23 positive. Of the 68 CLL cases with an available Matutes score, 57 (84%) had a score ≥4, seven (10%) a score of 3, and four (6%) a score <3. Of 64 analyzed cases, 45 (70%) were CD38 negative. We observed 62 t(14;18) translocations, and 13 variant translocations 11 t(18;22), 2 t(2;18) (Table 1 and Supplementary Table 1). The involvement of IG was confirmed by FISH in all tested cases (54 IGH, 6 IGL), and the involvement of BCL2 was confirmed by FISH in 69/70 (98%) of tested cases. Of note, the case without a proven BCL2-rearrangement displayed a t(18;22), with a rearrangement of IGL in 6% of the interphase nuclei, and did not involve the MALT1 gene. The median percentage of interphase nuclei carrying the BCL2-rearrangement was 81% (range 15-100%). The t(14;18) or variant-t was observed as the sole cytogenetic aberration in 25/74 (34%) cases. When associated with other chromosomal abnormalities in the karyotype, t(14;18) (or variant-t) was the primary change in 15/49 (31%) cases, in the same clone in 29/49 (59%) of cases, and as the subclone change in 5/49 (10%) cases. Trisomy 12 was the primary change in these five latter cases. The karyotype was complex (≥3 abnormalities) in 15/74 (20%) cases. There were 33/75 (44%) cases with trisomy 12, 32/68 (47%) with 13q14 deletion (9 observed by karyotype and FISH, 23 detected by FISH only), 1 (out of 72) (1%) TP53 deletion, no (0/72) ATM deletion, and no (0/59) 6q21 deletion. Of note, the majority of cytogenetic analyses was obtained before any treatment (63/71, 89%).
Table 1.
Morphologic, immunophenotypic, cytogenetic and molecular data
| No | Morphology | Matutes score | CD38 | Treated before K | Time Diagnosis-K (months)* | Sample | Karyotype (according to ISCN2009) | IGHV mutation % identity | V rearrangement |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Atypical CLL | 3 | 95% | No | 98 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[13]/47,idem,?del(3)(p14p22),add(9)(q34),del(10)(q23),add(17)(q23)[6] | nd | nd |
| 2 | CLL | 4 | 1% | No | 4 | Pb | 46,XY,t(14;18)(q32;q21)[15]/46,XY[16] | nd | nd |
| 3 | CLL | 2 | 1% | No | 0 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[2]/46,XX[13] | nd | nd |
| 4 | CLL | nd | 2% | No | 80 | Pb | 46,XY,del(13)(q13q22),t(14;18)(q32;q21)[20] | nd | nd |
| 5 | CLL | 5 | 2% | No | 30 | Pb | 46,XY,t(14;18)(q32;q21)[9]/46,XY[5] | nd | nd |
| 6 | CLL¤ | 4 | 0% | No | 18 | Pb | 46,XY,t(14;18)(q32;q21)[16]/46,XY[3] | M | V3-23 |
| 7 | CLL | 4 | 5% | No | 5 | Pb | 46,XX,t(14;18)(q32;q21)[7]/46,idem,del(10)(q23q24)[7]/46,idem,der(1)t(1;1)(p12;q34),add(5)(p15),del(20)(q12)[7]/46,XX[4] | M | V3-23 |
| 8 | CLL | 4 | 0% | No | 0 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[10]/46,XY[13] | M | V4-4 |
| 9 | CLL | 5 | 26% | No | 39 | Pb | 46,XX,t(14;18)(q32;q21)[15]/47,idem, + 12[5] | nd | nd |
| 10 | CLL | 4 | 57% | No | 102 | Pb | 47,XY,?add(1)(p36), + 12,t(14;18)(q32;q21)[19]/46,XY[2] | nd | nd |
| 11 | CLL | 4 | nd | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[2]/46,XY[20] | nd | nd |
| 12 | Atypical CLL | 5 | 0% | No | 0 | Pb | 47,XY, + 12,?del(13)(q14q21)[2]/47,idem,t(14;18)(q32;q21)[7]/46,XY[10]** | nd | nd |
| 13 | Atypical CLL | 3 | Pos^ | No | 0 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[16]/46,XY[4] | M | V2-5 |
| 14 | CLL | 4 | Neg^ | No | 0 | Pb | 47,XY,+X,t(14;18)(q32;q21)[18]/47,idem,der(5)del(5)(p13p14)del(5)(q12q2?1),der(12)(12p13->p12::12q13->p11::12q14->q24::?)[4] | M | V3-9 |
| 15 | Atypical CLL | 4 | 10% | No | 0 | Bm | 47,XY,+X,t(14;18)(q32;q21)[16]/46,XY[4] | nd | nd |
| 16 | Atypical CLL | 5 | Neg^ | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[16]/46,idem,?del(13)(q14q24)[2]/46,XY[14]** | nd | nd |
| 17 | Atypical CLL | 5 | nd | Yes | 165 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[11]/46,XY[9] | nd | nd |
| 18 | Atypical CLL | 4 | Neg^ | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[2]/46,XY[18] | nd | nd |
| 19 | Atypical CLL | 4 | Neg^ | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[3]/46,XY[18] | nd | nd |
| 20 | CLL¤ | nd | nd | Un | 40 | Pb | 47,XY, + 12,t(18;22)(q21;q11)[19]/46,XY[1] | nd | nd |
| 21 | CLL¤ | nd | nd | No | 36 | Pb | 46,XY,t(18;22)(q21;q11)[17]/46,XY[3] | nd | nd |
| 22 | Atypical CLL | 5 | 2% | No | 90 | Pb | 46,XY,t(14;18)(q32;q21)[17]/46,idem,del(13)(q14q21)[3]/46,XY[2] | M | V3-33 |
| 23 | CLL¤ | 4 | 11% | No | 78 | Pb | 46,XY,t(14;18)(q32;q21)[4]/46,idem,t(13;22)(q14;p11)[9]/46,XY[9] | nd | nd |
| 24 | CLL¤ | 4 | 65% | Yes | 28 | Pb | 46,XX,t(1;14)(q31;q32),t(2;14)(p12;q32),t(18;22)(q21;q11)[20] | NM | V3-30 |
| 25 | Atypical CLL | 4 | 7% | Yes | 188 | Pb | 46,XY,t(14;18)(q32;q21)[2]/46,idem,del(5)(q13q31),inv(12)(q13q24)[11]/46,idem,add(4)(p11),add(10)(q22),-11,+mar[8]/46,XY[1] | M | V2-5 |
| 26 | CLL¤ | nd | 95% | Yes | 211 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[2]/47,XX,+X[2]/46,XX[39] | M | V5-51 |
| 27 | Atypical CLL | nd | 92% | No | 6 | Pb | 46,XX,t(14;18)(q32;q21)[1]/47,idem, + 12[1]/46,XX[40] | M | V3-74 |
| 28 | CLL | 3 | 0% | No | 15 | Pb | 46,XY,t(14;18)(q32;q21)[3]/46,XY[14]/47,idem,+X[5]/46,XY[1] | M | V3-23 |
| 29 | CLL¤ | 2 | 9% | No | 0 | Bm | 46,XX,t(14;18)(q32;q21)[5]/6,XX[16] | M | V1-69 |
| 30 | CLL¤ | 4 | 84% | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[10]/46,idem,r(7)(?)[4]/47,idem, + 12[2]/46,XY[1] | NM | V1-69 |
| 31 | Atypical CLL | 4 | 95% | No | 0 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[19]/46,XY[6] | nd | nd |
| 32 | Atypical CLL | 5 | 6% | No | 0 | Bm | 46,XY,t(14;18)(q32;q21)[11]/46,XY[9] | nd | nd |
| 33 | CLL | 4 | 3% | No | 19 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[20] | nd | nd |
| 34 | CLL¤ | 4 | 8% | Yes | 12 | Pb | 46,XY,t(14;18)(q32;q21)[20] | nd | nd |
| 35 | CLL¤ | 5 | 0% | Yes | 69 | Pb | 46,XY,t(6;17)(p21;p13),t(14;18)(q32;q21)[20] | nd | nd |
| 36 | CLL¤ | 4 | nd | No | 17 | Pb | 46,XY,del(13)(q13q22),t(14;18)(q32;q21)[17]/46,XY[3] | nd | nd |
| 37 | CLL | 4 | 7% | No | 7 | Pb | 45,XY,der(1)t(1;8)(q41;q13),del(7)(p15p22),-8,t(18;22)(q21;q11)[20] | nd | nd |
| 38 | CLL | 5 | 7% | No | 40 | Pb | 46,XX,t(14;18)(q32;q21)[11]/46,XX[16] | M | V5-51 |
| 39 | CLL | 4 | nd | No | 46 | Pb | 46,XY,t(18;22)(q21;q11)[19]/46,XY[2] | M | V4-39 |
| 40 | CLL | 4 | 3% | No | 41 | Pb | 46,XY,t(14;18)(q32;q21)[4]/46,XY[13] | M | V4-34 |
| 41 | CLL | 4 | 5% | No | 4 | Pb | 46,XY,t(2;18)(p11;q21)[9]/46,XY[2] | M | V3-9 |
| 42 | CLL | 5 | nd | No | 56 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[13]/46,XX[1] | M | V3-30 |
| 43 | CLL | 2 | 90% | No | 26 | Pb | 46,XY,t(14;18)(q32;q21)[6]/46,XY[11]£ | nd | nd |
| 44 | Atypical CLL | 5 | nd | No | 47 | Pb | 47,XY, + 12[21]/47,idem,t(14;18)(q32;q21)[4] | NM | V3-7 |
| 45 | Atypical CLL | 4 | 3% | No | 5 | Pb | 47,XY, + 12,t(18;22)(q21;q11)[16]/46,XY[1] | NM | V2-70 |
| 46 | CLL | 5 | 0% | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[17]/46,idem,del(13)(q14q24)[2] | nd | nd |
| 47 | CLL | 4 | 7% | No | 0 | Bm | 46,XY,t(14;18)(q32;q21)[9]/46,XY[1] | M | V3-48 |
| 48 | CLL | 5 | 13% | No | 88 | Pb | 46,XY,t(14;18)(q32;q21)[7]/46,XY[3] | M | V5-51 |
| 49 | CLL | 4 | 0% | No | 81 | Pb | 46,XX,-4,add(8p)(p21),der(14)del(14)(q21q24)t(14;18)(q32;q21),der(18)t(14;18),+mar[10] | M | V3-48 |
| 50 | Atypical CLL | 5 | 1% | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[17]/46,XY[3] | M | V3-23 |
| 51 | CLL | 5 | 1% | Un | 27 | Pb | 46,XY,del(13)(q14q2?1),t(18;22)(q21;q11)[12]/46,XY[3] | M | V4-34 |
| 52 | Atypical CLL | 5 | 4% | Yes | 145 | Pb | 46,XY,t(14;18)(q32;q21)[11]/46,idem,del(13)(q14q3?1)[6]/46,XY[2] | M | V3-07 |
| 53 | Atypical CLL | 4 | 61% | No | 5 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[7]/46,XX[13] | NM | V6-1 |
| 54 | CLL¤ | 5 | 61% | Un | 32 | Pb | 47,XY, + 12[18]/47,idem,t(18;22)(q21;q11)[2] | NM | V1-69 |
| 55 | Atypical CLL | 5 | 4% | No | 1 | Pb | 47,XY, + 12[2]/47,idem,t(14;18)(q32;q21)[14]/46,XY[5] | nd | nd |
| 56 | Atypical CLL | 4 | 47% | No | 31 | Pb | 46,XX,t(18;22)(q21;q11)[8]/46,idem,t(4;20)(q13;q11)[16]/47,idem, + 12[2] | M | V3-7 |
| 57 | Atypical CLL | 3 | nd | No | 6 | Pb | 46,XY,t(14;18)(q32;q21)[10]/46,XY[10] | M | V3-30 |
| 58 | CLL | 5 | 1% | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[2]/46,XY[18] | M | V3-74 |
| 59 | Atypical CLL | 5 | 70% | No | 6 | Pb | 46,XY,t(14;18)(q32;q21)[2]/46,XY[18] | M | V3-23 |
| 60 | Atypical CLL | 3 | nd | No | 16 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[2]/46,XX[18] | M | V4-34 |
| 61 | Atypical CLL | 4 | 20% | Yes | 22 | Pb | 47,XX, + 12[15]/47,idem,t(14;18)(q32;q21)[6]/46,XX[1] | NM | V1-69 |
| 62 | CLL¤ | 5 | 59% | No | 34 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[2]/46,XY[18] | NM | V3-11 |
| 63 | Atypical | nd | nd | No | 26 | Pb | 47,XY,add(4)(q?34), + 12,t(14;18)(q32;q21)[16]/46,XY[4] | nd | nd |
| 64 | Atypical CLL | nd | 3% | Un | Un | Pb | 46,XY,t(2;18)(p11;q21)[8]/49,idem,+6, + 12,+21[3]/46,XY[9] | nd | nd |
| 65 | Atypical CLL | 5 | 0% | No | 0 | Pb | 46,XY,t(14;18)(q32;q21)[6]/46,XY[24] | M | V3-53 |
| 66 | CLL¤ | 4 | 2% | No | 84 | Pb | 46,XY,t(18;22)(q21;q11)[27]/46,XY[3] | nd | nd |
| 67 | CLL | 4 | 0% | No | 98 | Pb | 46,XY,t(14;18)(q32;q21)[20] | M | V3-23 |
| 68 | CLL | 5 | 0% | No | 26 | Pb | 46,XY,t(14;18)(q32;q21)[18]/46,XY[2] | nd | nd |
| 69 | CLL | 4 | 1% | No | 0 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[11]/47,idem,del(13)(q11)[2]/46,XX[7] | nd | nd |
| 70 | CLL | 3 | 79% | No | 12 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[8] | M | V3-7 |
| 71 | CLL¤ | 4 | 3% | No | 1 | Pb | 46,XY,t(14;18)(q32;q21)[10] | nd | nd |
| 72 | CLL¤ | 5 | 4% | No | 29 | Pb | 46,XY,t(14;18)(q32;q21)[5]/47,idem, + 12[7] | nd | nd |
| 73 | Atypical CLL | 2 | 85% | No | 0 | Pb | Failure££ | M | V3-48 |
| 74 | CLL¤ | 3 | 59% | No | 188 | Pb | 47,XY,+12,del(13)(q13q21),t(18;22)(q21;q11)[5]/47,idem,del(10)(q24)[6]/47,idem,t(3;12)(p21;q24)[5]/47,idem,t(2;11)(q22;q22)[4] | M | V4-34 |
| 75 | CLL¤ | 4 | 8% | No | 10 | Pb | 46,XX,t(14;18)(q32;q21)[5]/46,idem,del(13)(q13q22)[3]/46,XX[10] | M | V1-69 |
| 76 | MZL | 1 | 99% | No | 0 | Pb | 47,XX, + 12,t(14;18)(q32;q21)[12]/46,XX[8] | M | V3-23 |
| 77 | MZL | 3 | 38% | No | 4 | Pb | 47,XY,t(2;18)(p11;q21), + 12[11]/46,XY[9] | nd | nd |
| 78 | MZL | 1 | 1% | No | 0 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[5]/46,XY[15] | nd | nd |
| 79 | MZL | 2 | 95% | No | 0 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[12]/46,XY[7] | M | V3-23 |
| 80 | MZL | 0 | 83% | No | 33 | Pb | 47,XY, + 12,t(14;18)(q32;q21)[18]/46,XY[1] | nd | nd |
CLL: chronic lymphocytic leukemia. MZL: marginal zone lymphoma.
not rewieved. nd: not done. Un: unknown.
% not communicated, the cut off being 20%.of cells. Bm: bone marrow. Pb: peripheral blood. K:karyotype. NM : unmutated, M: mutated.
time between Diagnosis and Karyotype.
: Using FISH, trisomy 12 was detected in a sub-clone.
Using FISH, trisomy 12 was present.
No 13q14 deletion was detected using FISH.
IGHV mutation analysis was performed in 41 patients. Based on a conventional 98% identity cutoff level, 33/41 (80%) cases were mutated. None showed use of the VH3-21 gene. After a median follow up of 48 months (range 4-264 months), 2 of the 62 patients who had available data had died from disease (at 48 and 204 months after diagnosis) (Supplementary Table 1). The median TFS interval was 48 months (+/-12 months), and the median OS was not reached. Finally, there was no clinical or biological difference between t(14;18)-CLLs and variant-CLLs (data not shown).
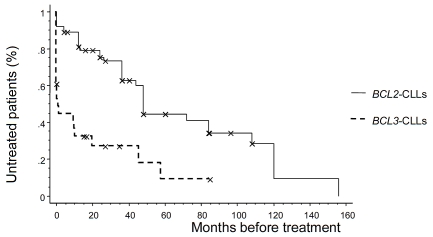
Comparisons between the 75 BCL2-CLLs and the 29 previously published BCL3-CLLs showed no difference in gender ratio, age or Binet stages (Table 2). Lymphocytosis was lower in BCL2-CLLs (p<0.008), and splenomegaly was less frequent (p<0.0001). There were more “typical” morphologies in the BCL2-CLL group (p<0.005), more Matutes score ≥4 (p<0.001), and more CD38-negative cases (p<0.04). There were more variant BCL2-r: tanslocations compared with variant BCL3-translocations (p<0.02). BCL2-translocations were more frequently observed as single aberrations (p<0.002). There were less complex karyotypes (p<0.02), more 13q14 deletions (p<0.005), and less occurrences of trisomy 12 (p<0.03), 6q deletion (p<0.002), and TP53 deletions (p<0.02) in BCL2-CLLs. The IGHV status of BCL2-CLLs was more frequently mutated (p<0.0001). BCL2-CLLs less frequently expressed IGHV4-39 genes (p<0.02). The TFS interval was longer in BCL2-CLLs (p<0.0001, median 48 vs 1.2 months) (Figure 1).
Table 2.
Comparison between BCL2-CLLS and BCL3-CLLS
| Patients | BCL2-CLLS n=75 | BCL3-CLLS n=29 | p |
|---|---|---|---|
| Morphology CLL | 47 (63%) | 9 (31%) | <0.005 |
| Atypical CLL | 28 (37%) | 20 (69%) | |
| Gender ratio: M/F | 57/18 | 18/11 | 0.22 |
| Age at diagnosis | 66 [32-83] | 62 [39-79] | 0.14 |
| Binet stage | 63A/4B/1C | 16A/5B/4C | 0.37 |
| Splenomegaly | 3/61 (5%) | 13/28 (46%) | <0.0001 |
| Lymphocytes count x109/l (median, range) | 14.6 [5.4-106.2] | 24.4 [5.5-514] | <0.008 |
| Matutes score | |||
| ≥4 | 57/68(84%) | 5/20 (25%) | <0.001 |
| =3 | 7/68 (10%) | 5/20 (25%) | |
| <3 | 4/68 (6%) | 10/20 (50%) | |
| CD5 positive | 69/73 (94%) | 27/29 (93%) | 0.99 |
| CD38 negative | 45/64 (70%) | 1/5 (20%) | <0.04 |
| Karyotype/ FISH | |||
| Variant translocation | 13/75* (17%) | 0/29 | <0.02 |
| Complex (≥3 abnormalities) | 15/74 (20%) | 13/28 (46%) | <0.02 |
| Single translocation | 25/74 (34%) | 1/28 (4%) | <0.002 |
| Trisomy 12 | 33/75 (44%) | 20/29 (69%) | <0.03 |
| 13q14 deletion | 32/68(47%) | 4/27 (15%) | <0.005 |
| TP53 deletion | 1/72 (1%) | 4/23 (17%) | <0.02 |
| ATM deletion | 0/72 (0%) | 2/24 (8%) | 0.06 |
| 6q deletion | 0/59 (0%) | 5/25 (20%) | <0.002 |
| Mutated cases | 33/41 (80%) | 2/20 (10%) | <0.0001 |
| IGHV4-39 | 1/41 (2%) | 5/20 (25%) | <0.02 |
| Treated cases | 35/64 (55%) | 21/28 (75%) | 0.1 |
| TFS interval (median +/-SEB) | 48 +/-12 months | 1.2 +/-1 months | <0.0001 |
CLL: chronic Iymphocytic leukemia, standard error. TFS: treatment-free survival.
: 11t(18;22), 2 t(2;18). M: male. F: female. ¤SE: standed error.
Figure 1.
Treatment-free survival (TFS) time from diagnosis for 92 CLLs : 64 SCL2-CLLs and 28 SCL3-CLLs. The median (+/- standard error) TFS for the SCL2-CLLS and SCL3-CLLs were 48 (+/−12) and 1.2 (+/−1) months respectively. X: censored case.
We also compared our FISH and molecular data with common CLL. Compared to the referent series of 325 CLLs published by Dohner et al., BCL2-CLLs displayed significantly more trisomy 12 (33/75 [44%] vs. 53/325 [16%], p<0.00001), and less ATM deletions (0/72 [0%] vs. 58/325 [22%], p<0.0001) and 6q deletion (0/59 [0%] vs. 21/325 [6%], p<0.05) [11]. Regarding IGHV status, BCL2-CLLS were mutated more frequently than a series of 2662 common CLLs (33/41 [80%] vs. 1426/2662 [54%], p<5.10-4) [12]. Moreover, BCL2-CLLs used the VH5-51 gene (3/41, 7%) more frequently than common CLLs (3/41 [7%] vs. 53/2662 [2%], p=0.05).
Finally, regarding the five patients with lymphoma (four MZLs, one unclassified lymphoma), all were CD10 negative, and four were CD5 positive (Table 1 and Supplementary Table 1). The Matutes score was 3 for one MZL, and <3 for the remaining lymphoma cases. Four of these five cases were CD38 positive. We observed four t (14;18) translocations and one variant t(2;18). In two cases [one t(14;18), one t(2;18)] an atypical signal pattern was observed, consistent with a 5’ side breakpoint of the BCL2 gene. These were analyzed with dual-color IG probes (IGH and IGK), and displayed atypical pattern, with a breakpoint in the constant region of IG. (Of note, one t(14;18)-CLL case harbored the same atypical signal with the BCL2 probe.) All lymphoma cases were simple karyotypes, and trisomy 12 was always associated with the BCL2-translocation.
Discussion
The t(14;18)(q32;q21) translocation occurs in more than 80% of follicular lymphoma (FL) cases, in about 20% of diffuse large B-cell lymphoma cases, and, more rarely (<5%), in CLL. Whereas IG heavy-chain genes (IGH on 14q32) are often analyzed in CLL, the IG light-chain genes (IGkappa on 2p11 and IGlambda on 22q11) loci are not commonly investigated using FISH. Consequently, the frequency of variant translocations could be under estimated. These translocations result in the juxtaposition of BCL2 to IG. About 75% of BCL2 breakpoints are clustered in the major breakpoint region (MBR) and the minor cluster region (mcr), whereas the remaining breakpoints are scattered between these clusters, or at the 5’ side (variant cluster region or vcr) of the BCL2 gene. The breakpoints in the BCL2 gene of variant translocations, t(2;18) and t(18;22), map consistently to vcr. The vcr rearrangement implies that translocation must be associated with inversion, with a juxtaposition of the BCL2 and IG genes in a head-to-head configuration, and suggests a different pathogenic mechanism [13]. Actually, in three patients [2 t(14;18), 1 t(2;18)], FISH signals were consistent with a 5’ breakpoint at the cytogenetic level. Variant translocations are significantly more frequent in BCL2-CLLs than in BCL3-CLLs, this could be because, in these cases, BCL2 is activated at a mature B-cell stage, when IGL or IGK is rearranged, whereas BCL3 is activated at a more immature stage when IGH is rearranged. In a few cases, in both BCL2- and BCL3-CLLs, translocation presents as a subclonal change, suggesting that at least in some cases, it may represent a secondary aberration. It is interesting to note that both BCL2-translocations and BCL3-translocations are frequently associated with trisomy 12, suggesting oncogenic cooperation. Although 13q14 deletion is more frequent in BCL2-translocations, BCL3-translocations are more frequently associated with a complex karyotype, an ATM deletion, and a TP53 deletion, which are all poor cytogenetic prognostic factors. We and others have already observed that the IGHV4-39/D6-13/J5 rearrangement is frequent in BCL3-CLLS [7, 9, 14]. Although BCL2-CLLs use the VH5-51 gene more frequently than common CLLs, we have not observed any recurrent combination. Finally, BCL2-CLLs are more frequently mutated, and have longer TFS than BCL3-CLLs: this supports the hypothesis that BCL2-CLLs and BCL3-CLLs do not have the same prognosis, and need to be identified and distinguished.
Surprisingly, comparison with common CLL showed that BCL2-CLLs have significant differences, and are associated with good prognostic markers. Even if BCL2-CLLs are frequently associated with trisomy 12, they lack ATM and 6q deletions, ATM deletion being a poor prognostic factor. It is intriguing that none of the 72 cases tested actually showed a deletion of ATM, whereas the deletion is not as rare in CLL. This could suggest that both BCL2-translocation and ATM deletion could be exclusive oncogenic events. Accordingly, Put et al. observed no 11q deletions in a series of 40 cases with CLL and BCL2 rearrangements [15]. Among cell-surface markers, CD38 expression is associated with a poor clinical outcome [16, 17]. The majority of BCL2-CLLs (70%) in our series were CD38 negative. Finally, the majority of BCL2-CLLs (80%) were mutated, without any VH3-21 usage: these two last criteria are associated with a good prognosis.[18] The median TFS of BCL2-CLLs is 48 months, which is very close to the median TFS (49 months) of the Dohner subgroup with a “normal karyotype” [11]. Our result contrasts with a previously published series of eight t(14;18)-CLLs, with a median progression-free survival of only 20.6 months [19]. However, another study showed a TFS of 48 months in a series of 40 BCL2-CLLs [15]. Regarding the patients with a BCL2-translocation and trisomy 12 (n=33) in our series, the presence of the BCL2-translocation remains a favorable marker, even in patients who also exhibit markers of intermediate prognosis, such as trisomy 12. Actually, the median TFS of this group is longer than the median TFS of the reference group with trisomy 12 as found by Dohner et al. (48 vs. 33 months). Of note, there is no significant difference between BCL2-CLLs with trisomy 12 and BCL2-CLLs without trisomy 12 with regard to median TFS in our series. Thus, we extend in our large series of independent data, a recently published series which concluded that t(14;18) was not associated with an inferior outcome in CLL [15]. However, our results contrast with previous data which reported CLL patients with translocations or 14q32 rearrangements. Mayr et al. found a median TFS of 24 months, and a median OS of 94 months in CLL with translocations [1]. Juliusson et al. and Cavazzini et al. reported short TFS and/or OS in CLL with 14q32 abnormalities [5, 6]. Haferlach et al. found a significant correlation between CD38 expression and IGH rearrangement [2]. Also, Cavazzini et al. observed 8/14 (57%) unmutated CLLs with a 14q32 rearrangement [6]. Our data underline the fact that these chromosomal abnormalities need further characterization and may not be considered as a whole group. In the same way, Yin et al. have recently reported six cases of CLL/SLL, with a t(2;14)(p16;q32) translocation involving BCL11A and IGH genes, which have been characterized with atypical morphological features and unmutated IGHV genes [20].
Finally, MZL could be associated with t(14;18), this is in contrast to a recent publication that reported 29/239 MZL cases that had abnormal karyotypes that carried a translocation involving IGH, but not the BCL2 gene [21].
In conclusion, different IG-translocations have different prognoses. Therefore, screening and identification of partner genes may be warranted in CLL. Variant translocations involving IGL and IGK are not as rare, but could be missed by classical FISH analyses using only IGH probes. The majority of BCL2-CLLs are characterized by CD5+, the CD38- immunophenotype, a Matutes score ≥4, no deletion of ATM, and a mutated IGHV status. Although frequently associated with trisomy 12, BCL2-translocations may be associated with a good prognosis.
Authorship and disclosures
F. N-K wrote the manuscript, designed and performed research, and analyzed the data. CL performed the statistical analyses. EC, AG performed the research. IL, IR-W, CL, SF-F, EC-B, EL, VR, LM, CB, MAC-R, FM, VE, ST, ND, SR, SS, PT, LB, NG, CG, BQ performed the cytogenetic analyses. CS, KM performed the morphological review, H.M-B performed the immunological review, FD performed the molecular review and contributed to the writing of this manuscript. All authors approved the final version of the manuscript. The authors report no potential conflicts of interest.
Acknowledgments
We thank A. Grelier, J. Ong, L Merlin, for technical assistance, N. Put for critically reading the manuscript, and E. Floc’h and M. Simon (Newmed Publishing Services and Amgen France) for the English revision of the manuscript.
Supplementary Table 1.
Clinical data: Binet stages and lymphocyte counts at diagnosis. Survival was calculated from the diagnosis.
| CLL | Gender/Age | Binet Stage | Lymphocyte count × 109/l | CD5/ CD23 | Time to first treatment (months) | Treatments | Follow up* (months) |
|---|---|---|---|---|---|---|---|
| 1 | F/73 | C | un | +/− | 108 | CVP | A (108) |
| 2 | M/75 | A | 14.6 | +/+ | 4 | CHOP/R | A (20) |
| 3 | F/68 | A | 57.1 | +/− | un | un | un |
| 4 | M/66 | A | 13.7 | +/nd | no | A (84) | |
| 5 | M/72 | A | 14 | +/+ | No | A (36) | |
| 6 | M/69 | A | 13.3 | -/+ | 12 | Cb | A(72) |
| 7 | F/68 | A | 20.7 | +/+ | 36 | FR;R | A (48) |
| 8 | M/54 | A | 8.4 | +/− | un | un | un |
| 9 | F/70 | A | un | +/+ | No | A (48) | |
| 10 | M/69 | A | un | +/− | No | A (108) | |
| 11 | M/74 | A | 7.8 | +/+ | 36 | Cb;FR | A (132) |
| 12 | M/82 | un | 24 | +/+ | un | un | un |
| 13 | M/80 | A | 6.1 | +/+ | un | un | un |
| 14 | M/60 | B | 92.4 | -/+ | 0 | FCR | un |
| 15 | M/70 | A | 9.1 | +/+ | No | A (24) | |
| 16 | M/66 | A | 10.2 | +/+ | No | A (12) | |
| 17 | M/48 | A | un | +/+ | 12 | Cb;CHOP;F;FC;Alemtuzumab | D (204) |
| 18 | M/57 | A | 15.2 | +/+ | No | A (60) | |
| 19 | M/62 | A | 6.2 | +/+ | No | A (48) | |
| 20 | M/72 | un | 10.9 | nd/nd | No | A (40) | |
| 21 | M/52 | un | un | nd/nd | No | A (36) | |
| 22 | M/54 | A | un | +/+ | 36 | Cb;FCR | A (96) |
| 23 | M/69 | A | un | +/+ | 84 | Cb | A (84) |
| 24 | F/62 | B | un | -/+ | 0 | Cb;FC;CHOP/R;Alemtuzumab | D(48) |
| 25 | M/45 | A | 66.2 | +/+ | 4 | Cb;FC;Cb | A (216) |
| 26 | F/56 | A | un | -/- | 120 | Cb | A (264) |
| 27 | F/74 | A | 6.6 | +/+ | 120 | Cb | A (144) |
| 28 | M/75 | A | 5.6 | +/+ | No | A (15) | |
| 29 | F/64 | A | 22.4 | +/− | 84 | Cb | A (84) |
| 30 | M/70 | A | 73.6 | +/+ | No | A (6) | |
| 31 | M/64 | B | 9.38 | +/+ | 0 | Cb | un |
| 32 | M/54 | A | 5.4 | +/+ | No | A (12) | |
| 33 | M/68 | A | 16.5 | +/+ | No | A (20) | |
| 34 | M/65 | A | 15.6 | +/+ | 5 | RC | A (12) |
| 35 | M/57 | A | 15.4 | +/− | 12 | Cb | A(72) |
| 36 | M/73 | A | un | +/+ | No | A (24) | |
| 37 | M/82 | A | 15 | +/+ | No | A (12) | |
| 38 | F/66 | A | un | +/+ | No | A (40) | |
| 39 | M/32 | A | 100 | +/+ | 48 | FCR | A(48) |
| 40 | M/57 | A | 68.6 | +/+ | No | A (48) | |
| 41 | M/72 | A | 5.6 | +/+ | No | A (4) | |
| 42 | F/67 | A | 18 | +/+ | No | A (60) | |
| 43 | M/75 | A | un | +/− | 48 | Cb | A(72) |
| 44 | M/60 | A | 25 | +/+ | 48 | FC | A (48) |
| 45 | M/61 | A | 9.2 | +/+ | un | un | un |
| 46 | M/50 | A | 15.6 | +/+ | un | un | un |
| 47 | M/74 | A | 8.8 | +/+ | un | un | un |
| 48 | M/61 | A | 13.8 | +/+ | 24 | Cb | A (96) |
| 49 | F/78 | A | 5.7 | +/+ | No | A (84) | |
| 50 | M/51 | A | 7.38 | +/+ | No | A (12) | |
| 51 | M/68 | un | un | +/+ | 36 | FC | A (48) |
| 52 | M/63 | A | 19.4 | +/+ | 72 | CHOP;R/Pentostatine/C | A (168) |
| 53 | F/71 | A | un | +/+ | 12 | RC | A (36) |
| 54 | M/63 | un | un | +/+ | 36 | FC | A (48) |
| 55 | M/72 | un | 17.8 | +/+ | un | un | un |
| 56 | F/49 | A | un | +/− | No | A (36) | |
| 57 | M/70 | A | 9.3 | +/+ | No | A (108) | |
| 58 | M/43 | A | 106.2 | +/+ | No | A (24) | |
| 59 | M/36 | A | 11.9 | +/+ | 36 | Cb | A(72) |
| 60 | F/75 | A | un | +/+ | No | A (36) | |
| 61 | F/66 | A | 11.1 | +/+ | 0 | Cb | A (24) |
| 62 | M/55 | A | 11 | +/+ | 44 | CHOP/F;FCR;ASCT | A (96) |
| 63 | M/74 | A | un | +/nd | 24 | Cb;CHOPR | A (26) |
| 64 | M/69 | un | un | +/nd | un | un | un |
| 65 | M/31 | A | 9.97 | +/+ | un | un | un |
| 66 | M/57 | A | 9 | +/+ | No | A (84) | |
| 67 | M/54 | B | un | +/+ | No | A (98) | |
| 68 | M/53 | A | 19.2 | +/+ | No | A (26) | |
| 69 | F/64 | A | 16.1 | +/+ | No | A (12) | |
| 70 | M/56 | A | 24.8 | +/+ | 48 | CHOP/RR/DHAP | A (96) |
| 71 | M/71 | A | 24.6 | +/+ | un | un | un |
| 72 | M/75 | A | un | +/+ | 24 | Cb | A (36) |
| 73 | F/55 | A | 82.7 | +/+ | 0 | FRC | A (24) |
| 74 | M/56 | A | un | +/+ | 156 | Cb;FR | A (192) |
| 75 | F/57 | A | 21.6 | +/+ | 12 | Cb/P | A (12) |
| MZL | |||||||
| 76 | F/76 | 12.8 | +/− | 24 | CHOP/R | A(24) | |
| 77 | M/57 | 8 | +/+ | un | un | un | |
| 78 | M/76 | 8.8 | +/− | No | A (12) | ||
| 79 | M/57 | 33 | +/+ | 5 | FCR | A (12) | |
| 80 | M/65 | 5.7 | -/- | No | A (36) | ||
un: unknown, nd: not done, CLL: chronic lymphocytic leukemia, MZL: marginal zone lymphoma,Cb: chlorambucil, P: prednisone, CHOP: cyclophasphamide/doxorubicine/vincristine/prednisone, F: fludarabine, R: rituximab, C: cyclophosphamide, DHAP: dexamethasone, cytarabine, cisplatine, ASCT: autologous stem cell transplantation
: defined as the time from diagnosis to last follow up; A: alive, D: dead.
References
- 1.Mayr C, Speicher MR, Kofler DM, Buhmann R, Strehl J, Busch R, et al. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107(2):742–51. doi: 10.1182/blood-2005-05-2093. [DOI] [PubMed] [Google Scholar]
- 2.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21(12):2442–51. doi: 10.1038/sj.leu.2404935. [DOI] [PubMed] [Google Scholar]
- 3.Van Den Neste E, Robin V, Francart J, Hagemeijer A, Stul M, Vandenberghe P, et al. Chromosomal translocations independently predict treatment failure, treatment-free survival and overall survival in B-cell chronic lymphocytic leukemia patients treated with cladribine. Leukemia. 2007;21(8):1715–22. doi: 10.1038/sj.leu.2404764. [DOI] [PubMed] [Google Scholar]
- 4.Mitelman F, Johansson B, Mertens F, editors. 2010. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. http://cgap.nci.nih.gov/Chromosomes/ Mitelman. [Google Scholar]
- 5.Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323(11):720–4. doi: 10.1056/NEJM199009133231105. [DOI] [PubMed] [Google Scholar]
- 6.Cavazzini F, Hernandez JA, Gozzetti A, Russo Rossi A, De Angeli C, Tiseo R, et al. Chromosome 14q32 translocations involving the immunoglobulin heavy chain locus in chronic lymphocytic leukaemia identify a disease subset with poor prognosis. Br J Haematol. 2008;142(4):529–37. doi: 10.1111/j.1365-2141.2008.07227.x. [DOI] [PubMed] [Google Scholar]
- 7.Chapiro E, Radford-Weiss I, Bastard C, Luquet I, Lefebvre C, Callet-Bauchu E, et al. The most frequent t(14;19)(q32;q13)-positive B-cell malignancy corresponds to an aggressive subgroup of atypical chronic lymphocytic leukemia. Leukemia. 2008;22(11):2123–7. doi: 10.1038/leu.2008.102. [DOI] [PubMed] [Google Scholar]
- 8.Michaux L, Dierlamm J, Wlodarska I, Bours V, Van den Berghe H, Hagemeijer A. t(14;19)/ BCL3 rearrangements in lymphoproliferative disorders: a review of 23 cases. Cancer Genet Cytogenet. 1997;94(1):36–43. doi: 10.1016/s0165-4608(96)00247-6. [DOI] [PubMed] [Google Scholar]
- 9.Martin-Subero JI, Ibbotson R, Klapper W, Michaux L, Callet-Bauchu E, Berger F, et al. A comprehensive genetic and histopathologic analysis identifies two subgroups of B-cell malignancies carrying a t(14;19)(q32;q13) or variant BCL3-translocation. Leukemia. 2007;21(7):1532–44. doi: 10.1038/sj.leu.2404695. [DOI] [PubMed] [Google Scholar]
- 10.Huh YO, Abruzzo LV, Rassidakis GZ, Parry-Jones N, Schlette E, Brito-Bapabulle V, et al. The t (14;19)(q32;q13)-positive small B-cell leukaemia: a clinicopathologic and cytogenetic study of seven cases. Br J Haematol. 2007;136(2):220–8. doi: 10.1111/j.1365-2141.2006.06416.x. [DOI] [PubMed] [Google Scholar]
- 11.Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 12.Darzentas N, Hadzidimitriou A, Murray F, Hatzi K, Josefsson P, Laoutaris N, et al. A different ontogenesis for chronic lymphocytic leukemia cases carrying stereotyped antigen receptors: molecular and computational evidence. Leukemia. 2010;24(1):125–32. doi: 10.1038/leu.2009.186. [DOI] [PubMed] [Google Scholar]
- 13.Willis TG, Dyer MJ. The role of immunoglobulin translocations in the pathogenesis of B-cell malignancies. Blood. 2000;96(3):808–22. [PubMed] [Google Scholar]
- 14.Schweighofer CD, Huh YO, Luthra R, Sargent RL, Ketterling RP, Knudson RA, et al. The B cell antigen receptor in atypical chronic lymphocytic leukemia with t(14;19)(q32;q13) demonstrates remarkable stereotypy. Int J Cancer. 2010 doi: 10.1002/ijc.25605. [DOI] [PubMed] [Google Scholar]
- 15.Put N, Meeus P, Chatelain B, Rack K, Boeckx N, Nollet F, et al. Translocation t(14;18) is not associated with inferior outcome in chronic lymphocytic leukemia. Leukemia. 2009;23(6):1201–4. doi: 10.1038/leu.2009.44. [DOI] [PubMed] [Google Scholar]
- 16.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94(6):1840–7. [PubMed] [Google Scholar]
- 17.Shanafelt TD, Rabe KG, Kay NE, Zent CS, Jelinek DF, Reinalda MS, et al. Age at diagnosis and the utility of prognostic testing in patients with chronic lymphocytic leukemia. Cancer. 116(20):4777–87. doi: 10.1002/cncr.25292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tobin G, Thunberg U, Johnson A, Thorn I, Soderberg O, Hultdin M, et al. Somatically mutated Ig V(H)3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood. 2002;99(6):2262–4. doi: 10.1182/blood.v99.6.2262. [DOI] [PubMed] [Google Scholar]
- 19.Nowakowski GS, Dewald GW, Hoyer JD, Paternoster SF, Stockero KJ, Fink SR, et al. Inter-phase fluorescence in situ hybridization with an IGH probe is important in the evaluation of patients with a clinical diagnosis of chronic lymphocytic leukaemia. Br J Haematol. 2005;130(1):36–42. doi: 10.1111/j.1365-2141.2005.05548.x. [DOI] [PubMed] [Google Scholar]
- 20.Yin CC, Lin KI, Ketterling RP, Knudson RA, Medeiros LJ, Barron LL, et al. Chronic lymphocytic leukemia With t(2;14)(p16;q32) involves the BCL11A and IgH genes and is associated with atypical morphologic features and unmutated IgVH genes. Am J Clin Pathol. 2009;131(5):663–70. doi: 10.1309/AJCPXLY46UPFLISC. [DOI] [PubMed] [Google Scholar]
- 21.Salido M, Baro C, Oscier D, Stamatopoulos K, Dierlamm J, Matutes E, et al. Cytogenetic aberrations and their prognostic value in a series of 330 splenic marginal zone B-cell lymphomas: a multicenter study of the Splenic B-Cell Lymphoma Group. Blood. 2010;116(9):1479–88. doi: 10.1182/blood-2010-02-267476. [DOI] [PubMed] [Google Scholar]