Abstract
Natural Killer (NK) cells are important effector cells in both the innate and adaptive immune responses. Although they were identified almost 40 years ago, our understanding of how and where NK cells develop is rudimentary. In particular, we have only a limited understanding of the signaling pathways that need to be activated to cause NK cell commitment and maturation. Knowledge of this process is important as disruptions can lead to the development of highly aggressive NK cell malignancies. In this review, we discuss the known molecular mechanisms that trigger NK cell commitment, prompt them to mature and finally allow them to become functional killers. Known disruptions in this developmental process, and how they may contribute to malignancy, are also addressed.
Keywords: Animal, human, Natural Killer cell, transcription factors, cytokines, cell differentiation, lymphopoiesis, gene expression regulation, lymphoma
Introduction
Natural killer (NK) cells play a multitude of roles in host innate and adaptive immunity [1]. They possess the unique ability to recognize and lyse tumor and pathogen infected cells without any prior stimulation. This is achieved via direct lysis, and indirectly by antibody dependent cellular cytotoxicity. They also promote B and T cell differentiation and dendritic cell (DC) maturation [1]. In vivo, NK cells have a limited life span and therefore must be continually replenished [2]. It is well known that NK cells are derived from CD34+ hematopoietic progenitor cells (HPCs) and that IL-15 is crucial for their development and homeostasis [3-5]. Despite this our understanding of the molecular mechanisms controlling NK cell development remains rudimentary.
Mouse knockout models have proved to be a very useful tool for studying the molecular mechanisms controlling murine NK cell development. Most of this research has focused primarily on the later stages of development with only a few studies identifying transcription factors involved in NK cell commitment and early development. Although murine models are an invaluable tool for studying NK cell development, there are key differences between mouse and human NK cells. Therefore, it is essential that further research is conducted to confirm these results in humans.
A thorough understanding of how NK cells develop is important as disruptions in this developmental process are implicated in NK cell malignancies. Furthermore, an understanding of NK cell developmental mechanisms may enable future therapeutic manipulation. This review will discuss the different stages of NK cell development with an emphasis on the molecular mechanisms involved at each stage and disruptions leadingto malignancies.
NK cell developmental stages in mice
The major focus of NK cell developmental research has used the murine system. Here NK cell development occurs primarily in the bone marrow (BM), although immature NK cells are also present in the liver and thymus suggesting development may also occur at these sites [6]. Like other blood cells, NK cells develop from CD34+ HPCs that undergo a sequential developmental process where they gradually become more lineage restricted and lose their ability to self-renew. The acquisition of the β chain of the IL-2/15 receptor (CD122) marks one of the earliest steps in NK cell commitment [6]. These NK precursors (NKPs) lack expression of T cell associated proteins and are unable to differentiate into B , T , myeloid and erythroid cells, but can be stimulated to form mature NK cells in vitro [7]. NKPs also lack several NK cell receptors with the exception of NKG2D and 2B4, receptors normally involved in activation or inhibition of killing activity in mature NK cells [7, 8].
As NKPs mature they gradually acquire more of the receptors seen on mature NK cells. The development of immature NK (iNK) cells is associated with acquisition of NK1.1 and CD94-NKG2 [9]. These iNK cells then acquire Ly49 and DX5 and undergo a major expansion phase [9]. As these cells mature further, their proliferation rate slows and an increased expression of CD43 and the gaining cytotoxicity and IFN-γ production distinguishes mature NK (mNK) cells [6, 9].
NK cell developmental stages in humans
For several years it was believed that human NK cell development occurred primarily in the bone marrow. However, attempts to characterize NK cell developmental stages within the bone marrow have not been successful. The search for an alternative site of development led to the identification of a unique population of CD34+ CD45RA+ HPCs in secondary lymphoid tissues (SLTs) that was able to give rise to NK cells, T cells and myeloid DCs [4, 6]. These cells are now accepted as the first stage in NK cell development and are termed pro-NK cells (CD34+ CD45RA+CD117- CD94- CD122-) [3, 4].
Pro-NK cells are not IL-15 responsive however, following in vitro culture with FMS-like tyrosine kinase 3 ligand (FLT3L), IL-3 and IL-7, at least a fraction are able to give rise to IL-15 responsive, stage 2 pre-NK cells [10, 11]. This transition is marked by the acquisition of CD122 (IL-2/15Rβ) and c-Kit (a cytokine receptor also known as CD117) [11]. Following stimulation with IL-2 or IL-15, a proportion of stage 2 pre-NK cells are able to form cells that are committed to the NK cell lineage. These cells represent the third stage of NK cell development and are known as immature NK cells (iNK cells) [11].
Although iNK cells no longer express CD34 and are restricted to form mature NK cells, they themselves lack features of mature NK cells. In particular, iNK cells show an inability to produce interferon γ (IFN-γ) and lack cytotoxicity [11]. Furthermore, iNK cells show variable expression of CD56 and do not express CD94.
Functional maturation begins at stage 4 of NK cell development. Transition to this stage is associated with expression of the NK receptor, CD94-NKG2A [11, 12]. Stage 4 NK cells have a CD56bright CD16- phenotype and are the major population of mature NK (mNK) cells seen in SLTs (∼90%). However, they represent only 10% of NK cells seen in the circulation. Here they are believed to be the main producer of NK cell-derived IFN-γ. Although blood CD56bright CD16-NK cells are fully functional, within SLTs not all CD56brightCD16- NK cells express intracellular the cytolytic proteins perforin and granzyme B or produce IFN-γ. Therefore, it is likely that functional maturation occurs within this stage of development.
The major population of NK cells seen in peripheral blood are CD56dim CD16+. There is considerable controversy regarding their development. It has been proposed that these cells may represent stage 5 of human NK cell development and differentiate from stage 4 CD56brightCD16- NK cells [12, 13]. There are several lines of evidence which support this hypothesis. For example, CD56bright CD16- NK cells have longer telomeres than CD56dim CD16+ NK cells, suggesting that the latter may be more terminally differentiated [14]. Furthermore, only some CD56bright CD16- NK cells are granular whilst all CD56dim CD16+ NK cells have a granular morphology, with the presence of granules conferring functional aptitude and therefore reflecting maturity [12]. Finally, it has been demonstrated that upon IL-2 stimulation in vitro, stage 4 CD56bright CD16- NK cells undergo phenotypic and functional changes to form cells similar to stage 5 CD56dim CD16+ NK cells [15]. Alternatively, CD56bright CD16- and CD56dim CD16+ NK cells may represent two terminally differentiated, functionally mature subsets. In this case, CD56dim CD16+ NK cells may develop from unique precursors that are not yet identified or from a common CD56bright/CD56dim NK cell pro-genitor. This hypothesis is supported by the observation that both CD56bright CD16- and CD56dim CD16+ NK cells play important roles within the innate immune response. Here CD56bright CD16- NK cells are the major mediators of NK cell-derived cytotoxicity while CD56bright CD16- NK cells produce copious amounts of NK cell-derived IFN-γ and other inflammatory cytokines. The importance of both these functions suggests that they represent two terminally differentiated, functionally mature subsets [16].
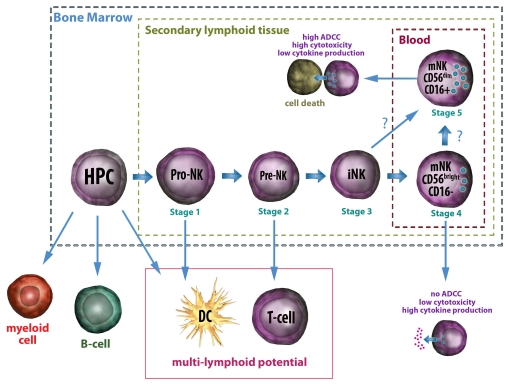
Despite the evidence supporting a role for SLTs in the development of NK cells, this does not reject a contribution from the BM to generate mature CD56+NK cells. One theory is that the initial stages of NK cell development occur in BM and, under the influence of stromal cell factors, CD34+ HPC differentiate to form CD34+ CD45RA+ pro-NK cells. These cells then travel to the lymph nodes and tonsils where, under the influence of IL-2, they further differentiate and eventually form CD56bright CD16- NK cells and then CD56dim CD16+ NK cells. This is summarized in Figure 1.
Figure 1.
Proposed Natural Killer (NK) Cell developmental pathway in Humans. In the bone marrow, HPCs differentiate to form pro-NK cells which are able to differentiate into dendritic cells (DC), T cells and NK cells. It is proposed that some of these then migrate to the lymph nodes and tonsils, where they undergo further differentiation, whilst others remain and mature in the bone marrow. As pro-NK cells differentiate they gradually lose their ability to self renew and become more restricted in lineage potential. Immature NK (iNK) cells are restricted to form mature NK (mNK) cells but are themselves not functional. iNK cells differentiate into CD56bright CD16- mNK cells in vitro; however the precursors of CD56dim CD16+ remain uncertain. Following maturation, both CD16+ and CD16- mNK cells migrate to the blood. Here they play important roles in the immune response with CD16+ mNK cells being the main mediators of cytotoxicity and ADCC, and showing modest secretion of cytokines; whilst CD16- mNK cells appear to have a more immunoregulatory role, producing copious amounts of interferon-γ(IFN-γ) and other cytokines, but showing limited cytotoxic capabilities and an inability to perform ADCC.
Alternatively, the presence of pre-NK cells and mNK cells within the BM contributes to the hypothesis that the complete developmental process may also occur at this site [17]. It remains unclear whether the mNK cells originate from BM or represent venous blood contamination. It is speculated that mNK cells developing at different sites may have specialized functions.
Forming committed NK precursors
Detailed knowledge regarding the signals required for commitment of HPCs to the NK cell lineage is lacking, but is likely mediated by cell to cell interactions within the microenvironment. This is evident by the enhanced number of NKPs seen when HSCs are cultured on relevant stromal cell lines compared to those cultured alone [18]. The molecular signals delivered by these cells are only just beginning to be elucidated, but appear to involve release of several cytokines including FLT3L and Stem Cell Factor (SCF). In vitro, addition of FLT3L or SCF to CD34+ HPCs greatly increased the frequency of NK cell precursor development and IL-15Ra expression [19]. Furthermore, mice deficient in FLT3L show reduced numbers of NK cells, DCs and HPCs [20]. Interestingly, mice that lack c-Kit, the receptor for SCF, do not have any NK cell deficiencies, suggesting that it is not essential for in vivo development of NK cells [21].
The Ax1/Gas6 signaling pathway has also been implicated in the commitment and differentiation of NK cells [22]. Ax1 is a tyrosine receptor kinase that is expressed by some CD34+ HPCs. Its ligand, Gas6, is also expressed by these cells, suggesting that Ax1 binds Gas6 in an autocrine fashion [22]. Interruption of this pathway greatly diminishes NK cell precursor frequency in human CD34+ HPCs cultured with SCF by impeding phosphorylation of c-Kit [22].
Using knockout models several transcriptional factors that appear to be involved in NK cell development have been identified and are summarized in Table 1. While there are a number of transcription factors that influence maturation of NK cells, a transcription factor that is essential for the generation of NKPs has yet to be identified. This is likely a result of the redundancy seen amongst transcription family members, making it hard to ascribe them to specific functions. One such family is the Ets family of transcription factors. Within this family two factors, Ets-1 and PU.1 have been demonstrated to affect NKP generation.
Table 1.
Transcription factors involved in NK cell development as determined by mouse knockout models
| Transcription Factor | Phenotype of Knockout Mouse | References |
|---|---|---|
| Commitment | ||
| Ets-1 | Decrease in NK cell numbers | [23] |
| Impaired NKcell mediated cytotoxicity and IFN-γ secretion | ||
| PU.1* | Reduced production of NKPs and mNKs | [26] |
| Altered expression of receptors involved in development | ||
| Defective proliferation in response to cytokines | ||
| Maturation | ||
| STAT5 | Developmental block at the NKP-iNK stage resulting in diminished | [31-33] |
| E4BP4 | NK cell numbers | |
| Decrease in iNK cell numbers | [35] | |
| Almost undetectable mNK cells | ||
| Impaired NK cell mediated cytotoxicity | ||
| Id2 | Decrease in mNK cell numbers | [36] |
| Functional Maturation | ||
| MEF | Decrease in NK cell numbers | [39] |
| Impaired NK cell mediated cytotoxicity and IFN-γ secretion | ||
| Defective perforin expression | ||
| T-bet | Decrease in NK cell numbers | [40] |
| NK cells appear immature | ||
| Impaired NK cell mediated cytotoxicity and IFN-γ secretion | ||
| BLIMP1 | Decrease in peripheral mNK cell numbers | [42] |
| Decrease in Granzyme B expression | ||
PU.1-/- mice die embryonically, therefore the role of PU.1 in NK development was determined by transfer of PU.1-/- fetal liver hematopoietic stem cells to Rag1-/-γc-/- mice. NK: Natural Killer; NKP: NK progenitors; iNK: immature NK; mNK: mature NK; IFN-γ: interferon-γ STAT: signal transducer and activator of transcription; MEF: myeloid ELF1 (E74-like factor 1)-like factor
As NKPs develop from common lymphoid progenitor cells (CLPs), an increase in Ets-1 is seen [7]. This transcription factor is thought to be essential for NK cell development as mice deficient in Ets-1 have severe defects in NK cell lineages, showing reduced NK cell numbers and impaired NK cell cytolytic activity and IFN-γ secretion [23]. Ets-1 may influence NK cell commitment by increasing expression of CD122 [7, 24]. This increase in CD122 may lead to a further increase in Ets-1 expression as IL-15 has recently been shown to regulate Ets-1 post-transcriptionally via the MEK > ERK1/2 > MNK1 > elF4E pathway [25]. Interestingly, the few mNK cells that do develop in Ets-1 deficient mice express CD122 suggesting CD122 is not exclusively controlled by Ets-1 [23].
PU.1 is well known for the important role it plays in the development of multipotent lymphoid progenitors. Since PU.1 deficient mice die embryonically, the effect of PU.1 on NK cells has been demonstrated by transfer of PU.1-/- fetal liver hematopoietic stem cells (FL-HSCs) to Rag1-/-γc-/- mice [26]. These mice display reduced generation of NKP and mNK cells, suggesting that PU.1 plays a role in early NK cell differentiation [26]. Furthermore, PU.1-/- mNK cells have alterations in Ly49, IL-7Rα and c-Kit expression and have defective responses to IL-2 and IL-12 [26]. Interestingly though, when PU.1 expression is decreased, but not abolished, an increase in NK and T cell genes is seen in myeloid and B cells [27]. This suggests that PU.1 may enforce myeloid and B cell gene expression after lineage commitment by suppressing expression of NK and T cell genes [27].
Maturation of Precursor NK cells
Acquisition of the high-affinity β-chain of the IL-15 receptor, CD122, is an essential step in the commitment of HPCs to the NK cell lineage in both humans and mice. Therefore, it is no surprise that the ligand for this receptor, IL-15, would play a vital role in the maturation of NK cells. IL-15 is produced by accessory cells and presented to CD122 expressing NK cells in trans via the high-affinity IL-15Ra [28]. Mice that are genetically deficient in IL-15 or IL-15Ra lack NK cells [29]. Despite IL-15 being essential for development of mature NK cells, generation of NKPs from HPCs is IL-15 independent [8, 29].
One way in which IL-15 influences NK cell maturation is through the JAK3 > STAT5 signaling pathway. Binding of IL-15 or IL-2 to the IL-2/15Rβγ chain results in phosphorylation of STAT5 by JAK3. STAT5 then dimerises and translocates to the nucleus where it serves as a transcription factor [30]. The essential role for STAT5 in NK cell development can be seen in mice deficient in STAT5a or b which show diminished NK cell numbers and combined deletion results in an absence of NK cells [31, 32]. NK-specific deletion of STAT5 further confirmed this result, with deletion of STAT5 resulting in a developmental block at the NKP-iNK transition [33].
Beyond the role for Ax1/Gas6 in the development of NKPs, this pathway is also required for maturation of human NK cells. Interruption of the Ax1/Gas6 pathway results in fewer mature CD56bright NK cells with impaired IFN-γ production [22]. Interestingly, blockade of the Ax1/ Gas6 pathway does not affect natural cytotoxicity. This requirement is at least partially influenced by interactions between the Ax1/Gas6 pathway and the IL-15 signaling pathway, as impairment of the Ax1/Gas6 pathways impedes phosphorylation of STAT5 [22]. Similar results have also been demonstrated in mice, with knockout of Ax1 family members (Tryo3 and Mer) resulting in NK cells that were defective in both IFN-γ production and natural cytotoxicity [34].
The basic leucine zipper (bZIP) transcription factor, E4BP4, is essential for progression through the NKP-iNK and iNK-mNK transitions [35]. Mice lacking E4BP4 have considerably fewer iNK cells, almost no mNK cells and exhibit severely impaired NK cell-mediated cytotoxicity [35]. Importantly, they display normal numbers of NKPs, indicating that E4BP4 is not required for NK cell lineage specification. Like many NK cell transcription factors, E4BP4 expression appears to be IL-15 dependent [35]. One way in which E4BP4 may influence NK cell development is by inducing expression of Id2 [35]. The Id proteins, specifically Id2 and Id3, are thought to function by inhibiting E protein activity and preventing B or T cell development and allowing NK cell lymphopoiesis. Surprisingly, Id2 appears to be dispensable for the development of NKPs and iNK cells; however, it is essential for development of mNK cells [36]. This requirement for Id2 is concomitant with a decrease in Id3 expression, suggesting that NKPs may develop in Id2 deficient mice because Id3 is sufficient to compensate for loss of Id2. Consistent with this hypothesis, Id2-/- mice showed increased expression of Id3 in CLPs and NKPs [36].
Functional maturation and homeostasis of mNK cells
The development of functionally mature NK cells is associated with the expression of cytotoxic granules (namely perforin and granzyme B) and the ability to secrete IFN-γ. Concurrent with its important role in NK cell development, IL-15 is also thought to influence functional maturation by regulating perforin expression via the STAT5 pathway [37, 38]. Perforin expression has also been shown to be directly regulated by MEF, a member of the Ets family of transcription factors, with MEF-/- mice having reduced NK cell numbers with severally impaired perforin expression and cytotoxic ability. In addition, MEF-/- NK cells also show a reduced ability to secrete IFN-γ [39]
The T-box family of transcription factors, specifically T-bet (Tbx21) and Eomesodermin (Eomes), have also been shown to influence NK cell functional maturation. T-bet is thought to trigger functional maturation in NK cells by regulating perforin and granzyme B expression. Mice deficient in T-bet show reduced numbers of NK cells which appear immature and have impaired function [40]. Interestingly combined deletion of T-bet and heterozygous deletion of Eomesodermin exacerbates this effect, with NK cell numbers being further diminished and expressing significantly less perforin [41]. One way in which T-bet may lead to functional maturation of NK cells is through the up-regulation of BLIMP1. It has recently been shown that T-bet/- NK cells have significantly reduced expression of BLIMP1, an essential transcription factor for high granzyme B expression by NK cells. Interestingly, BLIMP1 is not required for most cyto-kine production and cytotoxicity but is required for NK cell homeostasis [42].
In addition to influencing development of NKPs, FLT3L is also important for mature NK cell homeostasis in vivo. It has recently been shown that the effect of FLT3L on NK cells may be influenced, at least in part, by the tight link in DC and NK cell homeostasis. When mice were administered recombinant FLT3L expansion of both CD11chi DCs and NK cells was observed. However, ablation of CD11chi DCs greatly reduced expansion of NK cells following FLT3L administration [43]. It is thought that FLT3L therapy induces expansion of IL-15 producing CD11chi DCs, this in turn expands NK cells [43].
MicroRNAs (miRNAs) have recently been shown to be indispensable for NK cell homeostasis. miRNAs are short (∼22nt), non-coding RNA molecules that bind to target mRNAs and cause transcriptional repression or degradation. Deletion of Dicer or DiGeorge syndrome critical region 8 (Dgcr8), two enzymes involved in miRNA biogenesis, results in a decrease in NK cells in the blood, spleen and liver as a result of decreased turnover rate and survival [44]. The remaining NK cells show defects in ITAM-containing activating NK cell receptors and retarded expansion during mouse cytomegalovirus infection
miRNAs also appear to be vital for NK cell activation. When human and mouse mNK cells are expanded and stimulated with IL-15 or IL-2 in vitro they show a dynamic change in their miRNA expression profiles [45, 46]. Of these miRNAs, miR223 shows a significant decrease in mouse NK cells following activation and has been shown to target murine granzyme B in vitro [45].
NK cell malignancies
NK cells are the cause of several aggressive lymphomas and leukemia. Although these diseases are rare in western countries, they are relatively common in Asian populations [47, 48]. The World Health Organization (WHO) divides NK cell neoplasms into four main groups: Extranodal NK/T cell lymphoma, Nasal Type (ENKTL); Aggressive NK cell Leukemia/Lymphoma; Chronic Lymphoproliferative Disorders of NK cells (CLPD-NK), and NK cell lymphoblastic leukemia/lymphoma [49]. These are summarized in Table 2. The treatment of NK cell malignancies is beyond the scope of this review, for which readers are directed elsewhere [48, 50, 51].
Table 2.
NK cell Malignancies
| Natural killer (NK)-cell lymphoblastic leukemia/lymphoma | Chronic lymphoproliferative disorders of NK-cells | Aggressive NK-cell leukemia | Extranodal NK/T-cell lymphoma, nasal type | |
|---|---|---|---|---|
| Definition | Provisional entity WHO 2008 Leukemia due to NK-cell precursors | Provisional entity WHO 2008 Persistent (> 6 months) increase in peripheral blood NK cells (≥ 2.0 ×109/L) with no other identifiable cause | Rare neoplastic proliferation of NK-cells almost always associated with EBV causing an aggressive systemic disease | Predominantly extranodal lymphoma causing vascular damage, destruction and prominent necrosis in association with EBV |
| Presenting features | Constitutional symptoms Leukemic blood picture Cytopenias Not associated with EBV infection | Median age of onset 60 years No sex predominance No racial or genetic predisposition Presentation ranges from asymptomatic to systemic symptoms and cytopenias Lymphadenopathy and organomegaly rare Not associated with EBV infection | More prevalent among Asian populations Median age of onset 42 years Slight male predominance Fever, constitutional symptoms, leukemic blood picture, cytopenias are common Hypersensitivity to mosquito bites, chronic active EBV infection | More prevalent in Asians, native American populations of Mexico, Central and South America Median age of onset 50-60 years Male predominance 3:1 Nasal and non-nasal mass lesions Disease activity can be monitored by measuring circulating EBV DNA |
| Natural history | Aggressive disease process | Chronic indolent disease process | Very aggressive disease process Median survival less than 2 months | Aggressive disease process with variable prognosis Median survival 4-12 months |
| Sites involved | Peripheral blood and bone marrow | Peripheral blood and bone marrow predominantly | Peripheral blood, bone marrow, liver and spleen are commonest sites Skin involvement is uncommon | Upper aerodigestive tract (nasal cavity, nasopharynx, paranasal sinuses, palate) commonly Other sites include skin, soft tissue, gastrointestinal tract and testis, rarely bone marrow |
| Immunophenotype | No specific markers May be CD2+, CD5+, CD7+, and cCD3s+ T cell markers with CD16 and CD56 expression in the absence of B-cell or myeloid markers CD94 or CD161 may be more specific but are not commonly tested | Diminished or lost CD2, CD7, and CD57 expression sCD3-, cCD3s+, CD16+, CD56 weak Cytotoxic molecules* + Aberrant coexpression of CD5 or uniform CD8 Diminished CD161 | CD2+, sCD3-, cCD3s+, CD56+ Cytotoxic molecules + Frequent CD16 and FAS ligand expression CD11b may be expressed CD57 usually negative EBER-ISH positive | CD2+, sCD3-, cCD3s+, CD56+ Cytotoxic molecules+ Negative: CD4, CD5, CD8, TCR5, CD16, CD57 Commonly positive: CD43, CD45RO, HLA-DR, CD25, FAS Occasionally positive: CD7, CD30 EBER-ISH positive |
| Morphology | PB: Intermediate to large monocytoid cells, immature nuclear features with high nuclearcytoplasmic ratio | PB: Intermediate sized lymphoid cells with round nuclei, condensed chromatin Moderate amounts of slightly basophilic cytoplasm containing fine or coarse azurophilic granules BM: Intrasinusoidal and interstitial infiltration of lymphoid cells with minimally irregular nuclei and modest amounts of pale cytoplasm | PB: Ranges from the appearance of normal LGLs$ to lymphoid cells with atypical enlarged nuclei with irregular foldings, open chromatin or distinct nucleoli Ample amount of pale or lightly basophilic cytoplasm containing azurophilic granules BM: Range of disease infiltration of usually monotonous cells with intermingled reactive histiocytes and sometimes haemophagocytosis Associated apoptotic bodies and necrosis is common | Soft tissue: Angiocentric or angiodestructive growth pattern with fibrinoid changes in blood vessels, there may be associated inflammatory infiltrate Broad cytological spectrum with variation in size, irregularly folded and sometimes elongated nuclei, granular chromatin, inconspicuous or small nucleoli Moderate amount of pale or clear cytoplasm |
| Current treatment strategies | Intensive combination chemotherapy | Monitor for progression /transformation | Combination chemotherapy including L-asparaginase e.g. SMILE# Consider allogeneic hematopoietic stem cell transplantation (HSCT) | Chemoradiotherapy or radiotherapy alone for localized disease Combination chemotherapy including L-Asparaginase e.g. SMILE for disseminated disease Consider high-dose chemotherapy and autologous or allogeneic (HSCT) |
The most common NK malignancy is ENKTL and therefore the majority of research is focused on this disease. ENKTL is characterized by a diffuse proliferation of mature NK cells and is generally localized to the nasal cavity, although disease can occur at other sites [48, 52]. The mechanisms underlying ENKTL development are poorly understood, however its strong association with the Epstein-Barr virus (EBV) suggests the virus may have an etiological role [48, 52]. EBV is a gamma-herpes virus with known oncogenic properties that is implicated in a range of B, T and NK cell lymphoid malignancies [53]. Although its principal reservoir is B cells, large expansions of circulating EBV infected NK cells are apparent in EBV associated hemophagocytic syndrome and chronic active EBV syndrome [54].
Patients with ENKTL show elevated serum levels of IL-15 compared to healthy controls [55]. This increase in IL-15 allows ENKTL tumor survival, likely through phosphorylation of AKT, a serine/threonine kinase with anti-apoptotic activity [55]. In addition, over expression of IL-15 in mice causes development of fatal lymphocytic leukemia with a T/NK phenotype [56]. It has also been suggested that over-expression of T-bet may be a transforming event in the development of ENKTL. ENKTL cells show amplification and over-expression of T-bet which may have lead to tumorgenesis through loss of growth control and over-function [57] . Several other transcription factors associated with NK cell development including Eomes, MEF and Ets1 have also been recently shown to be expressed in ENKTL [7, 39, 58].
Similar to ENKTL, Aggressive NK cell Leukemia/Lymphoma is also believed to be triggered by infection of NK cells with the EBV [48]. As aggressive NK cell Leukernia/Lymphoma is such a rare disease, our understanding of it is limited. Based on the expression of CD56 and CD94 and the lack of CD3, this disease appears to have a mature NK cell origin [59]. However, morphologically, malignant cells appear immature with a fine chromatin pattern and large, pale cytoplasm [48, 50]. As such, it is proposed that transformation may occur at an early stage of development, possibly during transition from stage 3 to stage 4, and trigger up regulation of these surface receptors.
CLPD-NK is a rare entity that is characterized by expansion of mature looking NK cells in the peripheral blood. Based on the expression of CD16 and low levels of CD56, this disease appears to have a stage 5 mature NK cell origin [48, 50]. CLPD-NK is speculated to develop in response to a stimulus, likely of viral origin, activating mNK cells and selecting for clones [50]. The origin of this stimulus, however, remains undefined but is likely to reside in the bone marrow where it is presented to mNK cells by infected DCs [50].
Unlike the other NK cell malignancies, NK cell Lymphoblastic Leukemia/Lymphoma appears to have an immature NK cell origin. Neoplastic cells express CD56 but have a blastic appearance, suggesting they likely arise from stage 3 iNK cells [48, 60]. Likely as a result of its rarity, little is known about what causes NK cell Lymphoblastic Leukemia/Lymphoma. As with CLPD-NK, EBV is not expressed within the malignant NK cell.
Concluding Remarks
An understanding of normal human NK cells development will enable the pathogenesis of NK cell malignancies to be elucidated. Perturbations in cytokines, transcription factors, miRNAs and transcription factors are all likely to be implicated, as well as the transforming role of EBV. However, the majority of research remains in the mouse. Future research into pathogenesis is needed, that specifically integrates observations from patient material with in vivo and in vitro models of human NK cell development. Insights into how disruptions in normal NK cell developmental pathways contribute to tumorgenesis are critical for the development of new targeted therapies.
Acknowledgments
S.A.M. and M.K.G. are supported by the NHMRC (Australia). Work in the laboratory relevant to this area is supported by NHMRC, the Australian Centre for Vaccine Development and the Queensland State Government.
References
- 1.Moretta A, Marcenaro E, Parolini S, Ferlazzo G, Moretta L. NK cells at the interface between innate and adaptive immunity. Cell Death Differ. 2008;15:226–233. doi: 10.1038/sj.cdd.4402170. [DOI] [PubMed] [Google Scholar]
- 2.Yokoyama WM, Kim S, French AR. The dynamic life of natural killer cells. Annu Rev Immunol. 2004;22:405–429. doi: 10.1146/annurev.immunol.22.012703.104711. [DOI] [PubMed] [Google Scholar]
- 3.Galy A, Travis M, Cen D, Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 4.Miller J, Alley K, McGlave P. Differentiation of natural killer (NK) cells from human primitive marrow progenitors in a stroma-based long-term culture system: identification of a CD34+7+ NK progenitor. Blood. 1994;83:2594–2601. [PubMed] [Google Scholar]
- 5.Colucci F, Caligiuri MA, Di Santo JP. What does it take to make a natural killer? Nat Rev Immunol. 2003;3:413–425. doi: 10.1038/nri1088. [DOI] [PubMed] [Google Scholar]
- 6.Boos MD, Ramirez K, Kee BL. Extrinsic and intrinsic regulation of early natural killer cell development. Immunol Res. 2008;40:193–207. doi: 10.1007/s12026-007-8006-9. [DOI] [PubMed] [Google Scholar]
- 7.Rosmaraki EE, Douagi I, Roth C, Colucci F, Cumano A, Santo JPD. Identification of committed NK cell progenitors in adult murine bone marrow. European Journal of Immunology. 2001;31:1900–1909. doi: 10.1002/1521-4141(200106)31:6<1900::aid-immu1900>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 8.Vosshenrich CAJ, Ranson T, Samson SI, Corcuff E, Colucci F, Rosmaraki EE, Di Santo JP. Roles for Common Cytokine Receptor y-Chain-Dependent Cytokines in the Generation, Differentiation, and Maturation of NK Cell Precursors and Peripheral NK Cells in Vivo. The Journal of Immunology. 2005;174:1213–1221. doi: 10.4049/jimmunol.174.3.1213. [DOI] [PubMed] [Google Scholar]
- 9.Kim S, lizuka K, Kang HS, Dokun A, French AR, Greco S, Yokoyama WM. In vivo developmental stages in murine natural killer cell maturation. Nat Immunol. 2002;3:523–528. doi: 10.1038/ni796. [DOI] [PubMed] [Google Scholar]
- 10.Freud AG, Becknell B, Roychowdhury S, Mao HC, Ferketich AK, Nuovo GJ, Hughes TL, Marburger TB, Sung J, Baiocchi RA, Guimond M, Caligiuri MA. A human CD34(+) subset resides in lymph nodes and differentiates into CD56bright natural killer cells. Immunity. 2005;22:295–304. doi: 10.1016/j.immuni.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Freud AG, Yokohama A, Becknell B, Lee MT, Mao HC, Ferketich AK, Caligiuri MA. Evidence for discrete stages of human natural killer cell differentiation in vivo. J Exp Med. 2006;203:1033–1043. doi: 10.1084/jem.20052507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freud AG, Caligiuri MA. Human natural killer cell development. Immunol Rev. 2006;214:56–72. doi: 10.1111/j.1600-065X.2006.00451.x. [DOI] [PubMed] [Google Scholar]
- 13.Cooper MA, Fehniger TA, Caligiuri MA. The biology of human natural killer-cell subsets. Trends Immunol. 2001;22:633–640. doi: 10.1016/s1471-4906(01)02060-9. [DOI] [PubMed] [Google Scholar]
- 14.Ouyang Q, BAERLOCHER G, VULTO I, LANSDORP PM. Telomere Length in Human Natural Killer Cell Subsets. Annals of the New York Academy of Sciences. 2007;1106:240–252. doi: 10.1196/annals.1392.001. [DOI] [PubMed] [Google Scholar]
- 15.Ferlazzo G, Thomas D, Lin SL, Goodman K, Morandi B, Muller WA, Moretta A, Munz C. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell Ig-like receptors and become cytolytic. J Immunol. 2004;172:1455–1462. doi: 10.4049/jimmunol.172.3.1455. [DOI] [PubMed] [Google Scholar]
- 16.Cooper MA, Fehniger TA, Turner SC, Chen KS, Ghaheri BA, Ghayur T, Carson WE, Caligiuri MA. Human natural killer cells: a unique innate immunoregulatory role for the CD56bright subset. Blood. 2001;97:3146–3151. doi: 10.1182/blood.v97.10.3146. [DOI] [PubMed] [Google Scholar]
- 17.Caligiuri MA. Human natural killer cells. Blood. 2008;112:461–469. doi: 10.1182/blood-2007-09-077438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Santo JP. Natural killer cell developmental pathways: a question of balance. Annu Rev Immunol. 2006;24:257–286. doi: 10.1146/annurev.immunol.24.021605.090700. [DOI] [PubMed] [Google Scholar]
- 19.Yu H, Fehniger TA, Fuchshuber P, Thiel KS, Vivier E, Carson WE, Caligiuri MA. Flt3 Ligand Promotes the Generation of a Distinct CD34+ Human Natural Killer Cell Progenitor That Responds to lnterleukin-15. Blood. 1998;92:3647–3657. [PubMed] [Google Scholar]
- 20.McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, Roux ER, Teepe M, Lyman SD, Peschon JJ. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- 21.Waskow C, Rodewald HR. Lymphocyte development in neonatal and adult c-Kit-deficient (c-KitW/W) mice. Adv Exp Med Biol. 2002;512:1–10. doi: 10.1007/978-1-4615-0757-4_1. [DOI] [PubMed] [Google Scholar]
- 22.Park IK, Giovenzana C, Hughes TL, Yu JH, Trotta R, Caligiuri MA. The Axl/Gas6 pathway is required for optimal cytokine signaling during human natural killer cell development. Blood. 2009;113:2470–2477. doi: 10.1182/blood-2008-05-157073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton K, Muthusamy N, Fischer C, Ting C-N, Walunas TL, Lanier LL, Leiden JM. The Ets-1 Transcription Factor Is Required for the Development of Natural Killer Cells in Mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 24.Ye S-K, Kim TJ, Won SS, Yoon TJ, Park TK, Yoo YC, Kim Y-N, Lee HC, Ikuta K, Chung M-H, Lee KH. Transcriptional regulation of the mouse interleukin-2 receptor [beta] chain gene by Ets and Egr-1. Biochemical and Biophysical Research Communications. 2005;329:1094–1101. doi: 10.1016/j.bbrc.2005.02.073. [DOI] [PubMed] [Google Scholar]
- 25.Grund EM, Spyropoulos DD, Watson DK, Muise-Helmericks RC. Interleukins 2 and 15 Regulate Ets1 Expression via ERK1/2 and MNK1 in Human Natural Killer Cells. Journal of Biological Chemistry. 2005;280:4772–4778. doi: 10.1074/jbc.M408356200. [DOI] [PubMed] [Google Scholar]
- 26.Colucci F, Samson SI, DeKoter RP, Lantz 0, Singh H, Di Santo JP. Differential requirement for the transcription factor PU.1 in the generation of natural killer cells versus B and T cells. Blood. 2001;97:2625–2632. doi: 10.1182/blood.v97.9.2625. [DOI] [PubMed] [Google Scholar]
- 27.Kamath MB, Houston IB, Janovski AJ, Zhu X, Gowrisankar S, Jegga AG, DeKoter RP. Dose -dependent repression of T-cell and natural killer cell genes by PU.1 enforces myeloid and B -cell identity. Leukemia. 2008;22:1214–1225. doi: 10.1038/leu.2008.67. [DOI] [PubMed] [Google Scholar]
- 28.Burkett PR, Koka R, Chien M, Chai S, Boone DL, Ma A. Coordinate Expression and Trans Presentation of Interleukin (IL)-15Ra and IL-15 Supports Natural Killer Cell and Memory CD8+ T Cell Homeostasis. The Journal of Experimental Medicine. 2004;200:825–834. doi: 10.1084/jem.20041389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy MK, Glaccum M, Brown SN, Butz EA, Viney JL, Embers M, Matsuki N, Charrier K, Sedger L, Willis CR, Brasel K, Morrissey PJ, Stocking K, Schuh JCL, Joyce S, Peschon JJ. Reversible Defects in Natural Killer and Memory Cd8 T Cell Lineages in Interleukin 15-Deficient Mice. The Journal of Experimental Medicine. 2000;191:771–780. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becknell B, Caligiuri MA. lnterleukin-2, Inter-leukin-15, and Their Roles in Human Natural Killer Cells. In: Frderick WA, editor. Advances in Immunology. Academic Press; 2005. pp. 209–239. [DOI] [PubMed] [Google Scholar]
- 31.Imada K, Bloom ET, Nakajima H, Horvath-Arcidiacono JA, Udy GB, Davey HW, Leonard WJ. Stat5b Is Essential for Natural Killer Cell-mediated Proliferation and Cytolytic Activity. The Journal of Experimental Medicine. 1998;188:2067–2074. doi: 10.1084/jem.188.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moriggl R, Topham DJ, Teglund S, Sexl V, McKay C, Wang D, Hoffmeyer A, van Deursen J, Sangster MY, Bunting KD, Grosveld GC, Ihle JN. Stat5 Is Required for IL-2-lnduced Cell Cycle Progression of Peripheral T Cells. Immunity. 1999;10:249–259. doi: 10.1016/s1074-7613(00)80025-4. [DOI] [PubMed] [Google Scholar]
- 33.Eckelhart E, Warsch W, Zebedin E, Simma 0, Stoiber D, Kolbe T, Rülicke T, Mueller M, Casanova E, Sexl V. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 2011;117:1565–1573. doi: 10.1182/blood-2010-06-291633. [DOI] [PubMed] [Google Scholar]
- 34.Caraux A, Lu Q, Fernandez N, Riou S, Di Santo JP, Raulet DH, Lemke G, Roth C. Natural killer cell differentiation driven by Tyro3 receptor tyrosine kinases. Nat Immunol. 2006;7:747–754. doi: 10.1038/ni1353. [DOI] [PubMed] [Google Scholar]
- 35.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJM. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 36.Boos MD, Yokota Y, Eberl G, Kee BL. Mature natural killer cell and lymphoid tissue-inducing cell development requires Id2-mediated suppression of E protein activity. Journal of Experimental Medicine. 2007;204:1119–1130. doi: 10.1084/jem.20061959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu C-R, Ortaldo JR, Curiel RE, Young HA, Anderson SK, Gosselin P. Role of a STAT Binding Site in the Regulation of the Human Perforin Promoter. The Journal of Immunology. 1999;162:2785–2790. [PubMed] [Google Scholar]
- 38.Zhang J, Scordi I, Smyth MJ, Lichtenheld MG. Interleukin 2 Receptor Signaling Regulates the Perforin Gene through Signal Transducer and Activator of Transcription (Stat)5 Activation of Two Enhancers. The Journal of Experimental Medicine. 1999;190:1297–1308. doi: 10.1084/jem.190.9.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lacorazza HD, Miyazaki Y, Di Cristofano A, Deblasio A, Hedvat C, Zhang J, Cordon-Cardo C, Mao S, Pandolfi PP, Nimer SD. The ETS Protein MEF Plays a Critical Role in Perforin Gene Expression and the Development of Natural Killer and NK-T Cells. Immunity. 2002;17:437–449. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 40.Townsend MJ, Weinmann AS, Matsuda JL, Salomon R, Farnham PJ, Biron CA, Gapin L, Glimcher LH. T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity. 2004;20:477–494. doi: 10.1016/s1074-7613(04)00076-7. [DOI] [PubMed] [Google Scholar]
- 41.Intlekofer AM, Takemoto N, Wherry EJ, Long-worth SA, Northrup JT, Palanivel VR, Mullen AC, Gasink CR, Kaech SM, Miller JD, Gapin L, Ryan K, Russ AP, Lindsten T, Orange JS, Goldrath AW, Ahmed R, Reiner SL. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat Immunol. 2005;6:1236–1244. doi: 10.1038/ni1268. [DOI] [PubMed] [Google Scholar]
- 42.Kallies A, Carotta S, Huntington ND, Bernard NJ, Tarlinton DM, Smyth MJ, Nutt SL. A role for Blimp1 in the transcriptional network controlling natural killer cell maturation. Blood. 2011;117:1869–1879. doi: 10.1182/blood-2010-08-303123. [DOI] [PubMed] [Google Scholar]
- 43.Guimond M, Freud AG, Mao HC, Yu JH, Blaser BW, Leong JW, Vandeusen JB, Dorrance A, Zhang JY, Mackall CL, Caligiuri MA. In Vivo Role of Flt3 Ligand and Dendritic Cells in NK Cell Homeostasis. Journal of Immunology. 2010;184:2769–2775. doi: 10.4049/jimmunol.0900685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bezman NA, Cedars E, Steiner DF, Blelloch R, Hesslein DGT, Lanier LL. Distinct Requirements of MicroRNAs in NK Cell Activation, Survival, and Function. J. Immunol. 185:3835–3846. doi: 10.4049/jimmunol.1000980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fehniger TA, Wylie T, Germino E, Leong JW, Magrini VJ, Koul S, Keppel CR, Schneider SE, Koboldt DC, Sullivan RP, Heinz ME, Crosby SD, Nagarajan R, Ramsingh G, Link DC, Ley TJ, Mardis ER. Next-generation sequencing identifies the natural killer cell microRNA transcrip-tome. Genome Research. doi: 10.1101/gr.107995.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park KU, Jin P, Sabatino M, Feng J, Civini S, Khuu H, Berg M, Childs R, Stroncek D. Gene expression analysis of ex vivo expanded and freshly isolated NK cells from cancer patients. J Immunother. 2010;33:945–955. doi: 10.1097/CJI.0b013e3181f71b81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwong YL. Natural killer-cell malignancies: diagnosis and treatment. Leukemia. 2005;19:2186–2194. doi: 10.1038/sj.leu.2403955. [DOI] [PubMed] [Google Scholar]
- 48.Oshimi K. Progress in understanding and managing natural killer-cell malignancies. Br J Haematol. 2007;139:532–544. doi: 10.1111/j.1365-2141.2007.06835.x. [DOI] [PubMed] [Google Scholar]
- 49.WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues. Lyon: WHO; 2008. IARC. [Google Scholar]
- 50.Semenzato G, Zambello R. Natural Killer Cell Disorders. Hematology Education:The Education Program for the Annual Congress of the European Hematology Association. 2009;3:294–301. [Google Scholar]
- 51.Ishida F, Kwong YL. Diagnosis and management of natural killer-cell malignancies. Expert Rev Hematol. 2010;3:593–602. doi: 10.1586/ehm.10.51. [DOI] [PubMed] [Google Scholar]
- 52.Chan JKC, Quintanilla-Martinez L, Ferry JA, Peh SC. Extranodal NK/T-cell Lymphoma, Nasal Type. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW, editors. WHO Classification of Tumors of Hematopoietic and Lymphoid Tissues. 4. Lyon: IARC; 2008. pp. 285–288. [Google Scholar]
- 53.Tran H, Nourse J, Hall S, Green M, Griffiths L, Gandhi MK. Immunodeficiency-associated lymphomas. Blood Reviews. 2008;22:261–281. doi: 10.1016/j.blre.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular Responses to Viral Infection in Humans: Lessons from Epstein-Barr Virus. Annual Review of Immunology. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 55.Chow C, Liu A, Chan W, Lei K, Chan W, Lo A. AKT plays a role in the survival of the tumor cells of extranodal NK/T-cell lymphoma, nasal type. Haematologica. 2005;90:274–275. [PubMed] [Google Scholar]
- 56.Fehniger TA, Suzuki K, Ponnappan A, Van-Deusen JB, Cooper MA, Florea SM, Freud AG, Robinson ML, Durbin J, Caligiuri MA. Fatal Leukemia in Interleukin 15 Transgenic Mice Follows Early Expansions in Natural Killer and Memory Phenotype Cd8+ T Cells. The Journal of Experimental Medicine. 2001;193:219–232. doi: 10.1084/jem.193.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye Y, Li T, Zhang B, Guo Z. Amplification and specific expression of T-bet gene in nasal NK/T-cell lymphoma. Leukemia & Lymphoma. 2007;48:168–173. doi: 10.1080/10428190600955902. [DOI] [PubMed] [Google Scholar]
- 58.Zhang S, Li T, Zhang B, Nong L, Aozasa K. Transcription factors engaged in development of NK cells are commonly expressed in nasal NK/T-cell lymphomas. Human Pathology In Press. Corrected Proof. [DOI] [PubMed]
- 59.Mori KL, Egashira M, Oshimi K. Differentiation stage of natural killer cell-lineage lym-phoproliferative disorders based on phenotypic analysis. British Journal of Haematology. 2001;115:225–228. doi: 10.1046/j.1365-2141.2001.03038.x. [DOI] [PubMed] [Google Scholar]
- 60.Liang X, Graham DK. Natural killer cell neoplasms. Cancer. 2008;112:1425–1436. doi: 10.1002/cncr.23316. [DOI] [PubMed] [Google Scholar]
- 61.Hasserjian RP, Harris NL. NK-cell lymphomas and leukemias: a spectrum of tumors with variable manifestations and immunopheno-type. Am J Clin Pathol. 2007;127:860–868. doi: 10.1309/2F39NX1AL3L54WU8. [DOI] [PubMed] [Google Scholar]
- 62.Jaffe ES. Classification of natural killer (NK) cell and NK-like T-cell malignancies. Blood. 1996;87:1207–1210. [PubMed] [Google Scholar]
- 63.Cheung MMC, Chan JKC, Wong KF. Natural killer cell neoplasms: A distinctive group of highly aggressive lymphomas/leukemias. Seminars in Hematology. 2003;40:221–232. doi: 10.1016/s0037-1963(03)00136-7. [DOI] [PubMed] [Google Scholar]
- 64.Gill H, Liang RH, Tse E. Extranodal natural killer/t-cell lymphoma, nasal type. Adv Hematol. 2010 doi: 10.1155/2010/627401. 2010: 627401. [DOI] [PMC free article] [PubMed] [Google Scholar]