Abstract
Post-transplantation lymphoproliferative disorders (PTLD) arise in the immunosuppressed and are frequently Epstein-Barr virus (EBV) associated. The most common PTLD histological sub-type is diffuse large B-cell lymphoma (EBV+DLBCL-PTLD). Restoration of EBV-specific T-cell immunity can induce EBV+DLBCL-PTLD regression. The most frequent B-cell lymphoma in the immunocompetent is also DLBCL. ‘EBV-positive DLBCL of the elderly’ (EBV+DLBCL) is a rare but well-recognized DLBCL entity that occurs in the overtly immunocompetent, that has an adverse outcome relative to EBV-negative DLBCL. Unlike PTLD (which is classified as viral latency III), literature suggests EBV+DLBCL is typically latency II, i.e. expression is limited to the immuno-subdominant EBNA1, LMP1 and LMP2 EBV-proteins. If correct, this would be a major impediment for T-cell immunotherapeutic strategies. Unexpectedly we observed EBV+DLBCL-PTLD and EBV+DLBCL both shared features consistent with type III EBV-latency, including expression of the immuno-dominant EBNA3A protein. Extensive analysis showed frequent polymorphisms in EB-NA1 and LMP1 functionally defined CD8+ T-cell epitope encoding regions, whereas EBNA3A polymorphisms were very rare making this an attractive immunotherapy target. As with EBV+DLBCL-PTLD, the antigen presenting machinery within lymphomatous nodes was intact. EBV+DLBCL express EBNA3A suggesting it is amenable to immunotherapeutic strategies.
Keywords: Epstein-Barr virus, diffuse large B-cell lymphoma, EBNA3A, T-cell, epitope, immunotherapy, posttransplantation lymphoproliferative disorder
Introduction
Immunosuppression is the most frequently recognized predisposing factor to lymphoma. The importance of defective immunosurveillance in malignancy is most marked in cells with strong antigenic potential including those that have undergone viral induction. Consistent with this observation, is the archetypal immunosuppression associated lymphoma: post-transplantation lymphoproliferative disorder (PTLD). Relative to the immunocompetent patient, the risk of lymphoma after transplantation is increased by the order of 30-times [1, 2]. PTLD is frequently associated with Epstein-Barr virus (EBV) positivity within the malignant B-cells. PTLD is sub-divided into poly and monomorphic histological subtypes. It is believed to have a latency gene expression program (latency III) analogous to in-vitro EBV-transformed lymphoblastoid cell-lines (LCL), in which all EBV-latent antigens are expressed including the immuno-dominant EB-NA3A[3].
The most frequent histological sub-type of PTLD is diffuse large B-cell lymphoma (EBV+DLBCL-PTLD). Similarly, the most frequent B-cell lymphoma histology in the immunocompetent is DLBCL, representing 30% of all non-Hodgkin lymphoma. DLBCL is a biologically and clinically heterogeneous aggressive lymphoma. The World Health Organization (WHO) recently classified a provisional new sub-type of DLBCL arising in immunocompetent patients that is EBV-associated, termed ‘EBV-positive DLBCL of the elderly’ (EBV+DLBCL) [4, 5]. It is defined as occurring in those over 50 years of age without a predisposing immune deficiency [4]. Cases are consistently c-myc rearrangement negative [6]. In Korea, Turkey, Spain and Japan the rates of EBV+DLBCL are approximately 5-10% [7-10]. The proportion rises with age (∼30% in the >90 years) [8]. With the numbers of elderly predicted to markedly rise in many developed countries, EBV+DLBCL is likely to be an increasing health burden. EBV-encoded RNA in situ hybridization (EBER-ISH) on diagnostic DLBCL biopsies is not routine in many centres and data on frequency is limited and conflicting [9, 11, 12]. However a consistent finding from large series is that EBV+DLBCL is associated with reduced response to front-line chemotherapy therapy and reduced survival relative to EBV-negative DLBCL [7]. PTLD arises in patients given iatrogenic immunosuppression to prevent rejection of their transplanted organ. Antigen presentation in PTLD remains intact and defective immuno-surveillance is strongly implicated in pathogenesis [2]. Furthermore restoration of EBV-specific T-cell immunity by cellular immunotherapy can result in long-term regression [13]. The WHO postulate that EBV+DLBCL is related to deterioration in immunity as part of the ageing process [4]. If correct, this would also implicate defective immuno-surveillance and suggests that EBV+DLBCL could potentially be treated by novel immune-based strategies including adoptive immunotherapy. There is a well-defined hierarchy of EBV-specific T-cell immunity against EBV-latent antigens in which EBV-nuclear antigen (EBNA)-1 and Latent Membrane Proteins (LMP)-1 and 2 elicit sub-dominant responses relative to EBNA2/3A/3B and 3C. Furthermore, variations in functionally defined EBV T-cell epi-topes will influence whether EBV-specific T-cells generated by immunotherapy can recognize the viral strains expressed in the lymphoma. Therefore the prerequisite data necessary before such approaches can be considered includes EBV+DLBCL's viral protein expression, the lymphomas antigen presentation capability and the polymorphisms present within antigenic determinants. This study aims to demonstrate whether this new and highly aggressive lymphoma entity has characteristics amenable to T-cell based immunotherapies.
Materials and methods
Patient samples
All patients with EBV+DLBCL were aged over 50 years (mean 67 years, range 59-90). For EBV+DLBCL-PTLD, average age was 39 years (range 17-58). Patients were chosen solely on the availability of tissue. The majority of tissues were formalin-fixed paraffin-embedded (FFPE). DLBCL and EBV+DLBCL-PTLD samples were obtained from the Princess Alexandra Hospital, Brisbane, the Austin Hospital, Melbourne and the Department of Hematology and Oncology, Campus Virchow-Klinikum, Charité Universitäts-medizin Berlin. Three frozen tissues (two EBV+DLBCL and one EBV+DLBCL-PTLD) were supplied by the Australasian Leukaemia Lymphoma Group Tissue Bank. Cases were evaluated by a lymphoma pathologist (O.M.) as positive for EBV-encoded-RNA in-situ-hybridization (EBER-ISH) with a positive EBER-ISH reaction defined as ≥20% of nuclei positivity of examined cells [7]. EBV-tissue positivity was in all cases confirmed by real-time PCR for EBER-DNA (Table 1A). All patients were Human Immunodeficiency Virus (HIV) negative and (for EBV+DLBCL) not on immunosuppression medication. Transformed lymphoma and follicular lymphoma grade 3B were excluded. The study was approved by the relevant Human Research Ethics Committees and conducted in accordance with the Declaration of Helsinki. The patients reflected the ethnicity of the respective Hospital catchment areas. One was of Pacific Islander extraction (DL67); the others were Caucasian.
Table 1A.
Oligonucleotide primers and probes. Oligonucleotide primer/probe combinations for EBV gDNA real-time PCR
| Gene | Primer name | Sequence 5'-3' | Co-ordinate (AJ507799.2) |
|---|---|---|---|
| Alb* | Alb-F1 | GCTGTCATCTCTTGTGGGCTGT | 19583-19604 |
| Alb-R1 | AAACTCATGGGAGCTGCTGGTT | 19723-19702 | |
| Probe | CCTGTCATGCCCACACAAATCTCTCC | 19640-19665 | |
| EBER | EBER-F1 | AAACCTCAGGACCTACGCTGC | 6622-6642 |
| EBER-R1 | AGACACCGTCCTCACCAC | 6726-6709 | |
| Probe | TAGAGGTTTTGCTAGGGAGGAGACGTG | 6645-6671 |
Genbank accession number: NG_009291.1
Cell-lines
The following were used: Burkitt's lymphoma (BL) cell-lines without EBV: BJAB and Ramos; lymphoblastoid cell-line (LCL) DeMo (EBV+ B95-8); and T2 (transporter-associated protein [TAP1 and TAP2] deficient lymphoid cells).
In situ hybridization and immunohistochemistry
EBER -1 and -2 oligonucleotide probes (Leica Microsystems, UK) for EBER-ISH and immunohistochemistry for LMP1 (Dako, Germany), were performed as previously described [14]. EB-NA3A (ab16126, Abcam, UK) was used at a dilution of 1/1000, with a ready-to-use Goat Horse Radish Peroxidase secondary antibody. Human Leucocyte Antigen (HLA) class I, TAP1 and TAP2 (Abeam, UK ab70328, ab60112, ab60113 respectively) were performed as per manufacturer's instructions.
Nucleic acid extraction
RecoverAll Total Nucleic Acid Isolation Kit (Applied Biosystems, TX, USA) were employed for simultaneous DNA and total RNA extractions from FFPE tissue sections as manufacturer's instructions. For cell-lines and frozen tissues, DNA and RNA was extracted by QIAamp DNA blood mini-kit (QIAGEN, Hilden, Germany) and mirVana RNA isolation kit (Applied Biosystems, TX, USA) respectively as manufacturer's instructions. To remove contaminated DNA in total RNA, the TURBO DNA-free Kit (Applied Biosystems, USA) was used for all RNA samples. Both the quality and quantity of purified DNA and RNA were measured by a NanoDrop ND-100 spectrophotometer (Wilmington, DE, USA). All genomic DNA and total RNA were store at -20°C and -80°C, respectively.
Real-time comparative quantification PCR
Primers to identify EBV gene / expression (Sigma Aldrich, Victoria, Australia) were designed for PCR / real-time reverse-transcription (RT) PCR respectively, as shown in Table 1B. Primers were designed to pick up the transcripts in a spliced adjunction to eliminate the amplification of DNA remaining in the total RNA. Product sizes were between 60-100bp to ensure suitability for the amplification of RNA extracted from FFPE sections. β2M was used as the internal control for each sample. The DeMo (B95-8 EBV) LCL is used as a calibrator for all studied genes. Real-time RT-PCR was performed in 20 μl of reaction using SYBR Green PCR Master Mix (Applied Biosystems, UK) with 5μl diluted cDNA and the thermal cycle was as: 37°C × 10 minutes; 95°C × 10 minutes; 40 cycles: 95°C × 15 seconds, 60°C × 45 seconds running by the Rotor-Gene 3000. The data were analysed using Rotor-Gene 6.0 and Microsoft Excel 2007 and the amplified products were also checked by the separation in 2.5% agarose gel with ethidium bromide staining and visualized under UV light. The heat-map was generated by Genesis software v1.0. Primers for HLA-A, TAP1 and TAP2 for real-time RT-PCR are shown in Table 1B.
Table 1B.
Oligonucleotide primers for EBV gene expression profile and HLA class I, TAP1, TAP2 expressions by real-time RT-PCR
| Transcript | Primer name | Sequence 5'-3' | Co-ordinate (AJ507799.2) |
| EBNA2 | Y2/YH-T1F1 | GCTTAGCCAGTAACCCAGCACT | 35702-35711/36098-36109 |
| YH-R2 | TGCTTAGAAGGTTGTTGGCATG | 36181-36160 | |
| EBNA3A | BLRF3/BERF1-F1 | GGCTACGCGCATCGACACA | 80287-80293/80382-80393 |
| BERF1-R2 | TGTACATCTCGGTATTTGAAATCTGGG | 80445-80419 | |
| LMP1 | LMP1-F2 | TTGTATACTCCTACTGATGATCACCCTCC | 168768-168749/168670-168678 |
| LMP1-R2 | ACAATGCCTGTCCGTGCAAATTCC | 168623-168646 | |
| LMP2A | LMP2A-F1 | ATACGAAGAAGCGGGCAGAGG | 166435-166455 |
| LMP2A/B-R1 | GAGGTAGGGCGCAACAATTACAGG | 100-77 | |
| LMP2B | LMP2B-F1 | GGGAGGCCGTGCTTTAGGGG | 169422-169441 |
| LMP2A/B-R1 | GAGGTAGGGCGCAACAATTACAGG | 100-77 | |
| BART | BART-F1 | AGATGCCCTCCAGGTCAAAGA | 156884-156904 |
| BART-R1 | ATCCAGTGTCCCTCGTTTGG | 158669-158650 | |
| HLA class I* | HLA-A-F1 | AGGAGGAAGAGCTCAGATAGAAAA | 997-1020 |
| HLA-A-R1 | AAGCTGTGAGGGACACATCA | 1087-1068 | |
| TAP1** | TAP1-F1 | TCTCACCATAGCCAGTGCAG | 1034-1053 |
| TAP1-R1 | GTGGCCCATGGTGTTGTTAT | 1100-1080 | |
| TAP2*** | TAP2-F1 | TTCTCCTTTGGCAGCTCACT | 717-736 |
| TAP2-R1 | ATCCGCAAGTTGATTCGAGA | 796-777 | |
| B2M**** | B2M-F1 | TCACCCCCACTGAAAAAGATG | 329-349 |
| B2M-R1 | ATGATGCTGCTTACATGTCTCG | 430-409 |
CD8+ T-cell epitope sequencing for EBNA1, LMP1 and EBNA3A and phylogenetic analysis
To enable CD8+ T-cell epitope sequencing and phylogenetic analysis, selected DNA regions were chosen for PCR amplification and sequence analysis [15, 16]. The primers (Table 1C) were designed using Primer3 software and were synthesized by Sigma-Aldrich, Victoria, Australia. A nested PCR was done with the same primers in PCR1 or using inner primers for the samples that were negative in the first round PCR. All PCR products were purified using QIAquick Gel Extraction Kit (QIAGEN, Germany). The sequence analysis was performed using the BigDye 3.1 sequencing protocol (Applied Biosystems, USA). Briefly, the BigDye reaction was performed containing 1.2μl of BigDye terminator v3.1, 3.6 μl of 5x sequencing buffer, Ing template DNA per lOObp and 3.2 pmol primer in a total 12μl amplification reaction. The thermal cycles included an initial denature 94°C × 2 minutes and then 35 cycles: 94°C × 10 seconds, 50°C × 5 seconds and 60°C × 2 minutes. DNA was precipitated with 72μl of 70% isopropanol at room temperature × 15 minutes and centrifuged at 13,000rpm × 30 minutes. The DNA was dried for one hour at room temperature and diluted in 20μl formamide. The sequence analysis was run by ABI 3100 automated DNA sequencer (Applied Biosystems, CA, USA). Data was analysed with the FinchTV 1.4 DNA sequence analysis program (http://en.bio-soft.net/dna/FinchTV.html). The phylogenetic trees were drawn as previously outlined [16] from amino acid sequences using ClustalW2 (www.ebi.ac.uk/Tools/clustalw2) for EBV+DLBCL and EBV+DLBCL-PTLD and for three EBV strains from GenBank: B95-8 (V01555), AG876 (DQ279927.1) and GD1 (AY961628.3).
Table 1C.
Oligonucleotide primers for PCR amplification and sequence analysis
| Gene | Primer name | Sequence 5'-3' | Co-ordinate (AJ507799.2) |
|---|---|---|---|
| EBNA1 | E1C-F1 | AAAAAGGAGGGTGGTTTGGA | 97040-97059 |
| E1C-R1 | CATTCCAAAGGGGAGACGAC | 97290-97271 | |
| LMP1 | LMP1C-F4 | TGACATGGTAATGCCTAGAAGTAAA | 167671-167695 |
| LMP1C-R3 | CTGGAGGTGGTCCTGACAAT | 168129-168110 | |
| EBNA2A | EBNA2A-F | AACTTCAACCCACACCATCA | 36925-36944 |
| EBNA2A-R | TTCTGGACTATCTGGATCAT | 37040-37021 | |
| EBNA2B* | EBNA2B-F | TACTCTTCCTCAACCCAGAA | 36749-36768 |
| EBNA2B-R | GGTGGTAGACTTAGTTGATG | 36868-36849 | |
| EBNA3A | E3A-F6 | ATGTATG CCATG GCCATTC | 92736-92754 |
| E3A-R6 | TCCTCCCAGATTTTCGTGAG | 93127-93108 | |
| E3A-F7 | TCGCCAGTGGTTGTATGTTC | 93008-93027 | |
| E3A-R7 | TTTCACCGGTAGCACCTTC | 93378-93360 | |
| E3A-F3 | ACGGCACAGGCTTGGAAT | 93279-93296 | |
| E3A-R8 | GTTGGGGGTTCTGGGACTT | 93673-93655 | |
| E3A-F8 | ACCAGAGGTCCCACAAAGC | 93605-93623 | |
| E3A-R3 | ACAGGGACGGGTTCTACTGG | 93949-93930 |
Note: GenBank accession number: DQ279927.1. EBNA2A and EBNA2B primers are for PCR amplification of EBV types 1 and 2, respectively.
Results
EBV+DLBCL has a latency profile similar to EBV+DLBCL-PTLD
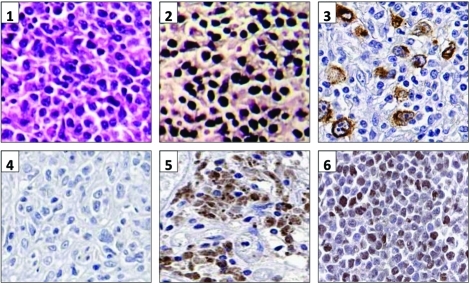
To further characterize the EBV latency profile, we performed real-time RT-PCR on a series of seven EBV+DLBCL and seven EBV+DLBCL-PTLD cases (Table 3). In agreement with prior work in EBV-positive cancers, expression was not identical in all samples [17-19]. There was no discernable differences between EBV+DLBCL and EBV+DLBCL-PTLD. In most cases the majority of EBV latent antigens were expressed, consistent with type III latency. EBNA3A was strongly expressed (defined as equivalent or above DeMo LCL) in 12/14 cases, whereas EBNA2 was strongly expressed in only 4/14, consistent with the low detection rate of EBNA2 by immunohistochemistry by others [4, 8]. The EBNA2 region we amplified (to distinguish EBV types 1 and 2) spanned this deleted region. Since DNA from this region could be amplified in all cases, the low EBNA2 / high EBNA3A gene expression pattern we observed in EBV+DLBCL was not due to the EBNA2 gene deletion. Protein expression was tested by immunohistochemistry in nine DLBCL cases with either high or low viral latent gene expression. LMP1 and EBNA3 protein was detectable in all cases with high gene expression, and not in those with low expression (Figure 1).
Table 3.
EBV gene expression profiles in EBV+DLBCL (DL4, 26, 29, 44, 67, 122, 82) and EBV+DLBCL-PTLD (PDL3-9), using DeMo (B95-8) LCL as a calibrator for all genes. The EBV-negative lines (Ramos and BJAB-) and EBV-negative DLBCL-PTLD primary samples (PDL1 and PDL2) were negative controls. P2M expression was used as an internal control for each gene and every duplicate experiment. Assays were repeated in two independent experiments.
| EBER | BART | LMP1 | LMP2A | LMP2B | EBNA2 | EBNA3A | |
|---|---|---|---|---|---|---|---|
| DEMO LCL | + | + | + | + | + | + | + |
| RAMO | - | - | - | - | - | - | - |
| BJAB- | - | - | - | - | - | - | - |
| PDL1 | - | - | - | - | - | - | - |
| PDL2 | - | - | - | - | - | - | - |
| PDL3 | + | + | + | + | - | - | + |
| PDL4 | + | + | + | - | + | + | + |
| PDL5 | + | + | + | + | + | + | + |
| PDL6 | + | + | - | - | + | - | - |
| PDL7 | + | + | + | + | + | - | - |
| PDL8 | + | + | + | - | - | - | - |
| PDL9 | + | + | + | - | - | - | - |
| DL4 | + | - | - | - | - | - | + |
| DL26 | + | + | + | - | - | - | + |
| DL29 | + | + | - | - | + | - | + |
| DL44 | + | + | - | - | - | + | + |
| DL67 | + | + | - | + | - | - | + |
| DL122 | + | + | - | + | - | - | + |
| DL82 | + | - | + | - | - | - | - |
Figure 1.
EBV latent gene and protein expression. 1. Panels 1 and 2: Haematoxylin and Eosin stain, and EBER-ISH respectively on DL67. Panel 3: LMP1 immunohistochemistry on DL26. Panels 4-6: EBNA3A immunohistochemistry on an EBV-negative DLBCL, an EBV+DLBCL-PTLD (PDL6) and on DeMo LCL respectively.
EBV+DLBCL has similar antigen processing machinery expression to EBV+DLBCL-PTLD
EBV+DLBCL-PTLD and LCL are known to be capable of presenting endogenous antigens for recognition by T-cells. Using immunohistochemistry (IHC), we compared EBV+DLBCL, EBV+DLBCL-PTLD and LCL for expression of the components of antigen processing machinery: MHC class I, TAP-1 and TAP-2. All were consistently present, with similar results obtained on both frozen and paraffin-wax sections. Control TAP deficient cell lines were not stainable for TAP-1 and TAP-2. To accurately quantify and compare the components required for antigen presentation, we performed real-time RT-PCR for MHC class I, TAP1 and TAP2 gene expression on EBV+DLBCL, EBV+DLBCL-PTLD, and EBV-negative DLBCL tissues, each relative to the LCL line DeMo. This line was chosen as it is known to be capable of presenting endogenous EBV antigens and is regularly used as a positive target cell control in functional T-cell assays. Levels were equivalent between samples for each of six EBV+DLBCL, seven EBV+DLBCL-PTLD, and seven EBV-negative DLBCL (P= NS) (Figure 2).
Figure 2.
HLA class I, TAP1 and TAP2 gene expression in EBV+DLBCL is equivalent to EBV+DLBCL-PTLD. The expression of HLA-A, TAP1 and TAP2 was quantified by real-time RT-PCR. DeMo B95-8 LCL was used as a reference. All experiments were run in duplicate and repeated once. No significant differences were observed between groups.
Polymorphisms in EBV regions encoding for CD8+ T-cell epitopes are infrequent for EBNA3A but common for EBNA1 and LMP1
There are 2 major types of EBV (types 1 and 2) distinguished by polymorphisms in EBNA2 [20]. All fourteen EBV+DLBCL and EBV+DLBCL-PTLD cases were type 1 (data not shown). Using direct sequence analysis, the gene variations of the EBNA1- and LMP1-C terminus functional domains were used to further type EBV into sub-strains [16]. Consistent with previous studies of EBV-positive lymphomas [21, 22] in 13/14 cases only one viral sub-strain could be identified. In polymorphic analysis of wild-type sub-strains in biopsy samples, the EBNA1 P-ala and LMP1 B95-8 sub-strains type the same as the type 1 laboratory strain B95-8. In EBV+DLBCL and EBV+DLBCL-PTLD tissues respectively only 1/7 and 3/7 samples were EBNA1 P-ala, versus 0/6 and 3/6 with LMP1 B95-8 sub-strains (Table 2). By EBNA1 typing, the most frequent sub-type was P-thr (7/14). For LMP1, the most common was Ch1 (4/12). PBMC from PDL7, DL4 and DL82 was sequenced and the detected EBNA1 and LMP1 sub-types were the same with that in the tumor (data not shown). Put together these data indicate considerable genetic diversity within the viral strains present in the lymphoma.
Table 2.
Summary of EBNA1 sub-types and LMP1 variants detected in EBV+DLBCL-PTLD and EBV+DLBCL
| EBNA1 SUB-TYPES | LMP1 VARIANTS | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No | Samp. | P-ala | P-ala-v2 | P-thr | V-val-v3 | B95-8 | Ch1 | Ch 2 | AL | NC | Med- | Med+ | |
| 1 | PDL3 | EBV + DLBCL - PTLD | +# | - | - | - | + | - | - | - | - | - | - |
| 2 | PDL4 | - | - | + | - | - | - | - | - | - | + | - | |
| 3 | PDL5 | - | - | + | - | ND | |||||||
| 4 | PDL6 | - | - | + | - | - | + | - | - | - | - | - | |
| 5 | PDL7 | +# | - | - | - | + | - | - | - | - | - | - | |
| 6 | PDL8 | - | + | + | - | - | - | - | - | + | + | - | |
| 7 | PDL9 | + | - | - | - | + | - | - | - | - | - | - | |
| 8 | DL4 | EBV + DLBCL | - | - | + | - | - | + | - | - | - | - | - |
| 9 | DL14 | +# | - | - | - | ND | |||||||
| 10 | DL26 | - | - | + | - | - | - | - | - | - | + | - | |
| 11 | DL29 | - | - | + | - | - | + | - | - | - | - | - | |
| 12 | DL44 | - | + | - | - | - | - | - | - | + | - | - | |
| 13 | DL67 | - | - | - | + | - | - | + | - | - | - | - | |
| 14 | DL82 | - | - | + | - | - | + | - | - | - | - | - | |
Note: EBNA1 and LMP1 were sub-typed as previously described.[15, 16] “+” the sub-type detected, and “-” is undetected. V-val-v3 is a sub-type of V-val (P529Q), due to two sub-types V-val-v1 and V-val-v2. ND: not done. PDL: EBV+DLBCL-PTLD; DL: EBV+DLBCL.
Some minor changes compared with P-ala: PDL3 and PLD7: E483D; DL14: E500K.
We performed EBNA3A sequencing in five EBV+DLBCL and four EBV+DLBCL-PTLD tissues in regions associated with ten functionally identified CD8+ T-cell epitopes presented by seven different (plus one currently unidentified) HLA class I alleles. This showed conservation of HLA class I epitopes in 85/90 epitopes tested (Table 4). Exceptions include DL67, a Pacific Islander who showed a type 1 EBNA2 by PCR but an HLA B*08 epitope (LLRGRAYGQ as opposed to FLRGRAYGL) identical to the type 2 prototype viral strain AG876. Extensive LMP1 and EB-NA3A sequencing (data not shown) in this case showed the patient to be compatible to a previously reported viral sub-strain in which both type 1 and type 2 features are known to coexist, that was originally identified in Papua New Guinea [23]. In all nine lymphoma cases the same HLA A*29 epitope (VSSDRGVAC as opposed to VFSDRGVAC) was identified. VSSDRGVAC is the most frequent variant observed in Australian Caucasians [24]. The HLA B*35.01 epitope YPLHEQHGMA was present in 5/9 cases. YPLHKQHGMA was observed in one EBV+DLBCL-PTLD case, and YTLHEQHGMA in three EBV+DLBCL cases.
Table 4.
EBNA3A HLA class 1 epitope polymorphisms compared to B95-8, GDI and AG876 strains.
| HLA Class I allele restriction | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Samples | A*24 | A*29 | A*30.02 | B*07 | B*07 | B*08 | *B*08 | B*35.01 | B*62 | Unknown |
| B95-8 | RYSIFFDY | VFSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGI | YPLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| GD1 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YPLHEQRGMA | LEKARGSTY | HLAAQGMAY |
| PDL3 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YPLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| PDL4 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YPLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| PDL7 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YPLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| PDL8 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YPLHKQHGMA | LEKARGSTY | HLAAQGMAY |
| DL26 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YPLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| DL82 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YPLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| DL29 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YTLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| DL44 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | FLRGRAYGL | YTLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| DL67 | RYSIFFDY | VSSDGRVAC | AYSSWMYSY | RPPIFIRRL | VPAPAGPIV | QAKWRLQTL | **LLRGRAYGQ | YTLHEQHGMA | LEKARGSTY | HLAAQGMAY |
| AG876 | CYSIFFDY | VPKDGRGAC | AYSSWMYSY | RPPIFLRRL | VPALAGPIV | QVKWRMTTL | LLRGRAYGQ | YPLHQQHSMA | LAKAPRRTY | ***H––– |
Underlined amino acids are different as compared to B95-8. Gray highlighted epitopes indicate differences among studied samples. There were 5/90 differences identified.
FLRGRAYGL is the functionally defined HLA B8 epitope. FLRGRAYGI is observed only in B95-8.
DL67 (a Pacific Islander) showed a type 1 EBNA2 by PCR but an HLA class I epitope identical to the type 2 EBV strain AG876. Extensive LMP1 and EBNA3A sequencing showed this case to be compatible to a previously reported viral sub-strain originally identified in Papua New Guinea [23]
Amino acid deletion.
No phylogenetic distinction between EBV strains present in EBV+DLBCL versus EBV+DLBCL-PTLD
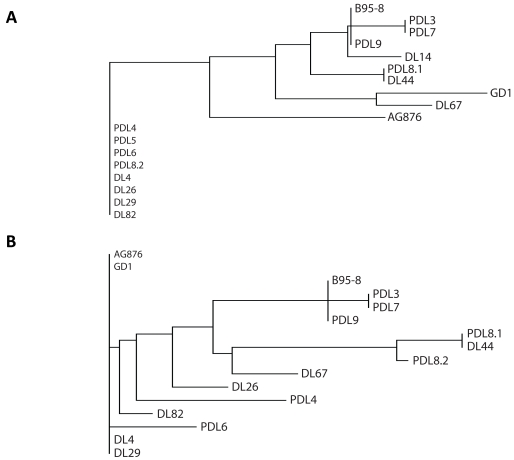
Phylogenetic analysis was performed to assess for relationships between viral strains present in patient biopsy samples. We compared results between EBV+DLBCL and EBV+DLBCL-PTLD. Previous work has shown that the phylogeny of EBV viral strains can be inferred from EBNA1 and LMP1 strain variation analysis, relative to three strains that have had full-length sequencing: B95-8, AG876 and GDI. With EBNA1, the tree had two main branches. The first branch had EBV+DLBCL-PTLD's PDL4, PDL5, PDL6 and EBV+DLBCL's DL26, DL29 (Figure 3A). The other branch then sub-divided into two, one of which contained AG876 and the other containing B95-8 and (in a sub-branch GDI). The EBV+DLBCL-PTLD samples PDL3, PDL7 and PDL9 were closely clustered to B95-8. For LMP1, a three branch phylogenetic tree was made (Figure 3B). One branch contained AG876 and GDI, another DL4 and DL29, and the last the remaining samples and B95-8, which again was closely clustered to PDL3, PDL7 and PDL9. There was broad overlap with the EBNA1 tree. However, there were some differences due to the presence or absence of a 30bp deletion present in the LMP1 C-terminus (this deletion is not present in B95-8 but is present in AG876 and GDI). This resulted in samples that formed the first EBNA1 branch, not clustering together in the LMP1 phylogram. PDL4 and DL26 sample had no 30bp deletion, whereas PDL6, DL4 and DL82, DL29 contained the 30bp deletion. The presence of clusters containing both EBV+DLBCL and PTLD-DLBCL biopsy samples suggests no phylogenetic distinction between viral strains implicated in DLBCL arising in the immunosuppressed versus the overtly immuno-competent.
Figure 3.
Phylogenetic Analysis of EBNA1 and LMP1 protein sequences from EBV+DLBCL and EBV+DLBCL-PTLD. The phylogeny of EBV strains can be inferred from EBNA1 and LMP1 strain variation analysis, relative to the prototype type 1 strain B95 -8, the prototype type 2 strain AG876, and the type 1 strain GDI. With EB-NA1 (A), the tree had 4 main branches. For LMP1 (B), a five branch pylogenetic tree was made. The first large cluster in EBNA1 is separated into 3 clusters in the LMP1 phylogram because of the 30bp deletion in the C -terminus (not present in B95-8). The first (PDL4 and DL26) samples has no 30bp deletion, the second (PDL6, DL4, DL29 and DL82) has the 30bp deletion and the third (AG876, GDI) contains the deletion. DL: EBV+DLBCL and PDL: EBV+DLBCL-PTLD.
Discussion
EBV+DLBCL has a poor outcome relative to EBV -negative DLBCL, indicating that new approaches need to be investigated [7-9]. Results from cellular immunotherapy in other EBV-associated malignancies are sufficiently encouraging for this modality to be explored in the setting of EBV+DLBCL [13, 25, 26]. However, before these strategies are undertaken, there are a number of fundamental questions that should be addressed. These include EBV+DLBCL's viral latency profile, the polymorphisms present within relevant EBVT-cell epitopes, and its antigen presentation capability.
Previous studies suggest that EBV+DLBCL generally have a viral latency type II pattern, i.e. expression is limited to EBNA1, LMP1 and LMP2. Only a minority of cases showed the EBV-latency type III pattern seen in LCL in which a broad range of EBV-latent antigens including EBNA3Aare expressed. The EBV-latency pattern is of critical importance for the rational design of EBV-specific cellular immunotherapies that target relevant tumor associated viral antigens. The reason for this is that in the hierarchy of EBV-latent antigen specific CD8+ T-cell immunity, the dominant response is against EB-NA2/3A/3B/3C, whereas EBNA1, LMP1 and LMP2 CD8+ T-cells are usually sub-dominant [27, 28]. Thus if EBV+DLBCL has a restricted viral latency profile it will be a major impediment for cellular immunotherapy.
Studies of EBV-positive cell-lines have broadly identified several forms of virus latency. Although classifying EBV-latency types based on cell-line data is useful, data from primary tumor biopsies indicate that such categorization is overly simplistic. Firstly, real-time RT-PCR indicates that patterns of virus gene expression are variable between primary tissues of the same histology (inter-patient variability) [17-19]. Secondly, tissue immunohistochemistry demonstrates differences in expression between malignant cells within the same biopsy (intra-patient variability) [29]. Furthermore, inferences regarding viral latency are frequently based on the use of antibodies to LMP1 (expressed in >90% of EBV+DLBCL) and EBNA2 (expressed in 25-30% of EBV+DLBCL) [4, 8]. The rationale is that EBNA2 acts as a transcriptional regulator of EBV nuclear antigens, and hence positivity can be regarded as a surrogate marker of EB-NA3A/3B/3C, and to indicate latency type III [8]. Prior work in EBV+DLBCL based on antibodies to LMP1 and EBNA2 suggest that EBV+DLBCL generally have a viral latency type II pattern, i.e. expression is limited to EBNA1, LMP1 and LMP2. Only a minority of cases showed the EBV-latency type III pattern seen in LCL in which a broad range of EBV-latent antigens including EBNA3A are expressed. However, data from primary tissues in other EBV-positive malignancies indicates that patterns of virus gene expression are more accurately identified by realtime RT-PCR [17]. However our gene expression data shows that the inferences from EBNA2 protein expression should be re-visited. Instead, EBV+DLBCL lymphomas have a variant type III pattern with EBNA2 down-regulated but the immuno-dominant EBNA3A expressed. The same expression profile was present in EBV+DLBCL-PTLD. To our knowledge our study is the first attempt to compare latency profiles in immuno-suppressed / non-immunosuppressed EBV-positive DLBCL by real-time RT-PCR. Our data in indicate that the presence or absence of iatrogenic immunosuppression does not influence the EBV-viral latency pattern.
Our data indicates considerable genetic diversity within the viral strains detected between lymphoma biopsies. The strains were associated with known polymorphisms in functionally identified EBNA1 and LMP1 CD8+ T-cell epitopes. For example the HLA A*02 B95-8 defined 9mer LMP1 epitope (YLLEMLWRL) has between one and three amino acid differences from Ch1 (YFLEILWRL), Ch2 (YFLEILWRL), NC (YFLEILWRL) and Med- (YLLEILWRL) sub-strains, that were identified in 83% of biopsies tested. The Med- found in 3/12 cases does not appear to influence HLA peptide binding or recognition by YLLEMLWRL-specific CTL clones [30]. However Ch1, Ch2 and NC have changes in the anchoring position within the epitopes that do interfere with MHC-I binding and fail to elicit a CTL response in chromium release assays [30]. However these strains have no differences with another well-defined functionally confirmed HLA A*02 restricted epitope (YLQQNWWTL) [30, 31]. Outside of BL, EBNA1 sequence analysis in EBV-positive lymphoma samples is limited. We demonstrate considerable genetic diversity from the B95-8 strain, with 79% of biopsies harbouring a non-P-ala sub-strain. Previous work has shown that this sequence polymorphism greatly influences T-cell recognition [32]. Of note, sub-typing covered the amino acid residue 524 located in the EBNA1 epitope described by Bell et al. 2008 [32]. P-ala codes for Threonine (ACT) at the 524 position (T-cell epitope YNLRRGTAL) whereas sub-types P-thr or V-val contain Isoleucine (ATT) (coding for YNLRRGIAL) and the P-ala-v2 subtype encodes for Valine (GTT) (resulting in YNLRRGVAL). In our study, only 3/7 (DLBCL-PTLD) and 1/7 (EBV+DLBCL) biopsies had the P -ala sub-type. Polymorphism at this site has previously been shown to influence CD8+ T-cell recognition of an endogenously processed HLA B*08 binding EBNA1 epitope [32]. Polymorphism in the EBNA1 amino acid residue 524 site also influences the frequency of EBNA1-specific CD8+ T-cells that can be generated in-vitro in pre-therapy blood from patients PTLD [33]. In Australia 24.7% are HLA B*08 and 46.5% type as HLA A*02 [34]. The high incidence of EBNA1 non-P-ala, LMP1 non-B95.8, and HLA B*08, HLA A*02 alleles in our population has implications for future vaccines, which will need to be designed to cater for EBNA1 and LMP1 strain variation in epitope encoding regions. Any future EBV vaccine that exclusively delivers the B95-8 sequence is likely to stimulate CD8+ T-cells which in a significant proportion of individuals may have a limited capacity in recognizing the EBNA1 and LMP1 epitopes encoded by the EBV sub-strain within the tumor.
Our data in indicate that the presence or absence of iatrogenic immunosuppression does not influence the EBV-viral latency pattern. To further examine the relationship between EBV+DLBCL and EBV+DLBCL-PTLD, we typed viral strains so as to perform a phylogenetic analysis. The lack of phylogenetic distinction that we observed between the viral strains identified in EBV+DLBCL and EBV+DLBCL-PTLD biopsy samples further emphasize the similarities in viral biology between the two entities. Further work is required to establish the pathogenic links between EBV+DBLCL and EBV+DLBCL-PTLD.
Latency III is the expression profile used during infectious mononucleosis [35]. Normally, the cytotoxic T-lymphocyte (CTL) response targets the immuno-dominant EBNA2,3A,3B,3C latent proteins [27]. The CTL response eliminates EBV-latently infected cells before they develop into a pathogenic lymphoproliferation. This is exemplified in individuals who develop PTLD due to iatrogenic immunosuppression following organ transplantation. Selective immune impairment in EBV-positive HL suggests a similar role of immunopathogenesis in HL arising in immuno-competentvs immunodeficient [36, 37]. Recon-stitution of EBV-specific CTL has been associated with objective clinical response in EBV-positive lymphomas arising in both overtly immunocompetent and immunosuppressed patients, and may be a potential strategy for EBV+DLBCL [13, 26, 38, 39]. Results to date suggest cellular immunotherapy is relatively low-risk and might be an attractive option for the older patient. Antigen processing and presentation is known to be intact in EBV-positive PTLD and in LCL. We observed that HLA class I, TAP1 and TAP2 in EBV+DLBCL expression were equivalent to EBV+DLBCL-PTLD (and EBV-negative DLBCL), and was greater than a control LCL in the majority of cases. In contrast to EB-NA1 and LMP1, we observed extensive conservation of functionally defined CD8+ T-cell epitopes (85/90 or 94% of epitopes sequenced) encoded by the immuno-dominant EBNA3A, making this an attractive immunotherapy target. All nine patients exhibited the HLA A*29 epitope VSSDRGVAC (which is identical to GD1, but distinct to B95-8's VFSDRGVAC). This is the most frequent form of the epitope seen in Australian Caucasians and does not appear to influence immunogenicity [24]. The 5/90 epitope variations included an EBV+DLBCL case in which the HLA B*08 epitope LLRGRAYGQ (identical to the type 2 prototype viral strain AG876), and an EBV+DLBCL-PTLD case where the HLA B*35.01 epitope YPLHKQHGMA was found. These variants are known to be essentially unrecognized by FLRGRAYGL and YPLHEQHGMA specific CTL respectively [40]. There were also three EBV+DLBCL cases of YTLHEQHGMA. This epitope shows reduced recognition by YPLHEQHGMA specific CTL [41].
EBV+DLBCL have a highly adverse outcome to chemotherapy relative to EBV-negative DLBCL [7-10]. We demonstrate that EBV+DLBCL has many similarities to EBV+DLBCL-PTLD, including expression of EBNA3A, indicating that such an approach may be feasible in EBV-positive DLBCL arising in the overtly immunocompetent as well as in the context of PTLD. Further studies are required to assess the nature of EBV specific T-cell immunity in these patients. Strategies such as immunotherapies specifically targeting the virus within the malignant cell should be actively explored.
Acknowledgments
Work in the Clinical Immunohaematology Laboratory relevant to this project was supported by the NHMRC (Australia), Atlantic Philanthropies, Cancer Council Queensland, the Australian Centre for Vaccine Development and the Leukaemia Foundation. We thank the Australasian Leukaemia Lymphoma Group Tissue Bank for assistance with samples.
References
- 1.Curtis RE, Travis LB, Rowlings PA, Socie G, Kingma DW, Banks PM, Jaffe ES, Sale GE, Horowitz MM, Witherspoon RP, Shriner DA, Weisdorf DJ, Kolb HJ, Sullivan KM, Sobocinski KA, Gale RP, Hoover RN, Fraumeni JF, Jr, Deeg HJ. Risk of lymphoproliferative disorders after bone marrow transplantation: a multi-institutional study. Blood. 1999;94:2208–2216. [PubMed] [Google Scholar]
- 2.Tran H, Nourse J, Hall S, Green M, Griffiths L, Gandhi MK. Immunodeficiency-associated lymphomas. Blood Rev. 2008;22:261–281. doi: 10.1016/j.blre.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 3.Young LS, Rickinson AB. Epstein-Barr virus: 40 years on. Nat Rev Cancer. 2004;4:757–768. doi: 10.1038/nrc1452. [DOI] [PubMed] [Google Scholar]
- 4.Nakamura S, Jaffe ES, Swerdlow SH. EBV positive diffuse large B-cell lymphoma of the elderly. In: Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri S, Stein H, Thiel J, Vardiman JW, editors. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer Press; 2008. pp. 243–244. [Google Scholar]
- 5.Dojcinov SD, Venkataraman G, Pittaluga S, Wlodarska I, Schrager JA, Raffeld M, Hills RK, Jaffe ES. Age-related EBV-associated lymphoproliferative disorders in the Western population: a spectrum of reactive lymphoid hyperplasia and lymphoma. Blood. 2011;117:4726–4735. doi: 10.1182/blood-2010-12-323238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura N, Nakamine H, Tamaru J, Nakamura S, Yoshino T, Ohshima K, Abe M. The distinction between Burkitt lymphoma and diffuse large B-Cell lymphoma with c-myc rearrangement. Mod Pathol. 2002;15:771–776. doi: 10.1097/01.MP.0000019577.73786.64. [DOI] [PubMed] [Google Scholar]
- 7.Park S, Lee J, Ko YH, Han A, Jun HJ, Lee SC, Hwang IG, Park YH, Ahn JS, Jung CW, Kim K, Ann YC, Kang WK, Park K, Kim WS. The impact of Epstein-Barr virus status on clinical outcome in diffuse large B-cell lymphoma. Blood. 2007;110:972–978. doi: 10.1182/blood-2007-01-067769. [DOI] [PubMed] [Google Scholar]
- 8.Oyama T, Yamamoto K, Asano N, Oshiro A, Suzuki R, Kagami Y, Morishima Y, Takeuchi K, Izumo T, Mori S, Ohshima K, Suzumiya J, Nakamura N, Abe M, Ichimura K, Sato Y, Yoshino T, Naoe T, Shimoyama Y, Kamiya Y, Kinoshita T, Nakamura S. Age-related EBV-associated B -cell lymphoproliferative disorders constitute a distinct clinicopathologic group: a study of 96 patients. Clin Cancer Res. 2007;13:5124–5132. doi: 10.1158/1078-0432.CCR-06-2823. [DOI] [PubMed] [Google Scholar]
- 9.Saez AI, Saez AJ, Artiga MJ, Perez-Rosado A, Camacho Fl, Diez A, Garcia JF, Fraga M, Bosch R, Rodriguez-Pinilla SM, Mollejo M, Romero C, Sanchez-Verde L, Pollan M, Piris MA. Building an outcome predictor model for diffuse large B-cell lymphoma. Am J Pathol. 2004;164:613–622. doi: 10.1016/S0002-9440(10)63150-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uner A, Akyurek N, Saglam A, Abdullazade S, Uzum N, Onder S, Barista I, Benekli M. The presence of Epstein-Barr virus (EBV) in diffuse large B-cell lymphomas (DLBCLs) in Turkey: special emphasis on ‘EBV-positive DLBCL of the elderly’. APMIS. 2011;119:309–316. doi: 10.1111/j.1600-0463.2011.02736.x. [DOI] [PubMed] [Google Scholar]
- 11.Gibson SE, Hsi ED. Epstein-Barr virus-positive B-cell lymphoma of the elderly at a United States tertiary medical center: an uncommon aggressive lymphoma with a nonger-minal center B-cell phenotype. Hum Pathol. 2009;40:653–661. doi: 10.1016/j.humpath.2008.10.007. [DOI] [PubMed] [Google Scholar]
- 12.Morales D, Beltran B, De Mendoza FH, Riva L, Yabar A, Quinones P, Butera JN, Castillo J. Epstein-Barr virus as a prognostic factor in de novo nodal diffuse large B-cell lymphoma. Leuk Lymphoma. 2010;51:66–72. doi: 10.3109/10428190903308015. [DOI] [PubMed] [Google Scholar]
- 13.Heslop HE, Slobod KS, Pule MA, Hale GA, Rousseau A, Smith CA, Bollard CM, Liu H, Wu MF, Rochester RJ, Amrolia PJ, Hurwitz JL, Brenner MK, Rooney CM. Long-term outcome of EBV -specific T-cell infusions to prevent or treat EBV-related lymphoproliferative disease in transplant recipients. Blood. 2010;115:925–935. doi: 10.1182/blood-2009-08-239186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gandhi MK, Lambley E, Burrows J, Dua U, Elliott S, Shaw PJ, Prince HM, Wolf M, Clarke K, Underhill C, Mills T, Mollee P, Gill D, Marlton P, Seymour JF, Khanna R. Plasma Epstein-Barr Virus (EBV) DNA Is a Biomarker for EBV-Positive Hodgkin's Lymphoma. Clin Cancer Res. 2006;12:460–464. doi: 10.1158/1078-0432.CCR-05-2008. [DOI] [PubMed] [Google Scholar]
- 15.Bhatia K, Raj A, Guitierrez MI, Judde JG, Spangler G, Venkatesh H, Magrath IT. Variation in the sequence of Epstein Barr virus nuclear antigen 1 in normal peripheral blood lymphocytes and in Burkitt's lymphomas. Onco-gene. 1996;13:177–181. [PubMed] [Google Scholar]
- 16.Nguyen-Van D, Ernberg I, Phan-Thi Phi P, Tran-Thi C, Hu L. Epstein-Barr virus genetic variation in Vietnamese patients with nasopharyngeal carcinoma: full-length analysis of LMP1. Virus Genes. 2008;37:273–281. doi: 10.1007/s11262-008-0262-9. [DOI] [PubMed] [Google Scholar]
- 17.Bell AI, Groves K, Kelly GL, Croom-Carter D, Hui E, Chan AT, Rickinson AB. Analysis of Epstein-Barr virus latent gene expression in endemic Burkitt's lymphoma and nasopharyngeal carcinoma tumour cells by using quantitative real-time PCR assays. J Gen Virol. 2006;87:2885–2890. doi: 10.1099/vir.0.81906-0. [DOI] [PubMed] [Google Scholar]
- 18.Deacon EM, Pallesen G, Niedobitek G, Crocker J, Brooks L, Rickinson AB, Young LS. Epstein-Barr virus and Hodgkin's disease: transcriptional analysis of virus latency in the malignant cells. J Exp Med. 1993;177:339–349. doi: 10.1084/jem.177.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rea D, Fourcade C, Leblond V, Rowe M, Joab I, Edelman L, Bitker MO, Gandjbakhch I, Suberbielle C, Farcet JP, et al. Patterns of Epstein -Barr virus latent and replicative gene expression in Epstein-Barr virus B cell lymphoproliferative disorders after organ transplantation. Transplantation. 1994;58:317–324. [PubMed] [Google Scholar]
- 20.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKenzie J, Gray D, Pinto-Paes R, Barrezueta LF, Armstrong AA, Alexander FA, McGeoch DJ, Jarrett RF. Analysis of Epstein-Barr virus (EBV) nuclear antigen 1 subtypes in EBV-associated lymphomas from Brazil and the United Kingdom. J Gen Virol. 1999;80(Pt 10):2741–2745. doi: 10.1099/0022-1317-80-10-2741. [DOI] [PubMed] [Google Scholar]
- 22.Jones K, Nourse JP, Morrison L, Nguyen-Van D, Moss DJ, Burrows SR, Gandhi MK. Expansion of EBNAl-specific effector T cells in post-transplant lymphoproliferative disorders. Blood. doi: 10.1182/blood-2010-03-274076. [DOI] [PubMed] [Google Scholar]
- 23.Burrows JM, Khanna R, Sculley TB, Alpers MP, Moss DJ, Burrows SR. Identification of a naturally occurring recombinant Epstein-Barr virus isolate from New Guinea that encodes both type 1 and type 2 nuclear antigen sequences. J Virol. 1996;70:4829–4833. doi: 10.1128/jvi.70.7.4829-4833.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khanna R, Slade RW, Poulsen L, Moss DJ, Burrows SR, Nicholls J, Burrows JM. Evolutionary dynamics of genetic variation in Epstein-Barr virus isolates of diverse geographical origins: evidence for immune pressure-independent genetic drift. J Virol. 1997;71:8340–8346. doi: 10.1128/jvi.71.11.8340-8346.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bollard CM, Aguilar L, Straathof KC, Gahn B, Huls MH, Rousseau A, Sixbey J, Gresik MV, Carrum G, Hudson M, Dilloo D, Gee A, Brenner MK, Rooney CM, Heslop HE. Cytotoxic T Lymphocyte Therapy for Epstein-Barr Virus+ Hodgkin's Disease. J Exp Med. 2004;200:1623–1633. doi: 10.1084/jem.20040890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haque T, Wilkie GM, Jones MM, Higgins CD, Urquhart G, Wingate P, Burns D, McAulay K, Turner M, Bellamy C, Amlot PL, Kelly D, MacGilchrist A, Gandhi MK, Swerdlow AJ, Crawford DH. Allogeneic cytotoxic T-cell therapy for EBV-positive posttransplantation lymphoproliferative disease: results of a phase 2 multicenter clinical trial. Blood. 2007;110:1123–1131. doi: 10.1182/blood-2006-12-063008. [DOI] [PubMed] [Google Scholar]
- 27.Hislop AD, Taylor GS, Sauce D, Rickinson AB. Cellular responses to viral infection in humans: lessons from Epstein-Barr virus. Annu Rev Immunol. 2007;25:587–617. doi: 10.1146/annurev.immunol.25.022106.141553. [DOI] [PubMed] [Google Scholar]
- 28.Rickinson AB, Moss DJ. Human cytotoxic T lymphocyte responses to Epstein-Barr virus infection. Annu Rev Immunol. 1997;15:405–431. doi: 10.1146/annurev.immunol.15.1.405. [DOI] [PubMed] [Google Scholar]
- 29.Rowe M, Niedobitek G, Young LS. Epstein-Barr Virus gene expression in Post-Transplantation Lymphoproliferative Disorders. Springer Semin Immunopathol. 1998;20:389–403. doi: 10.1007/BF00838051. [DOI] [PubMed] [Google Scholar]
- 30.Duraiswamy J, Burrows JM, Bharadwaj M, Burrows SR, Cooper L, Pimtanothai N, Khanna R. Ex vivo analysis of T-cell responses to Epstein-Barr virus-encoded oncogene latent membrane protein 1 reveals highly conserved epitope sequences in virus isolates from diverse geographic regions. J Virol. 2003;77:7401–7410. doi: 10.1128/JVI.77.13.7401-7410.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards RH, Sitki-Green D, Moore DT, Raab-Traub N. Potential selection of LMP1 variants in nasopharyngeal carcinoma. J Virol. 2004;78:868–881. doi: 10.1128/JVI.78.2.868-881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bell MJ, Brennan R, Miles JJ, Moss DJ, Burrows JM, Burrows SR. Widespread sequence variation in Epstein-Barr virus nuclear antigen 1 influences the antiviral T cell response. J Infect Dis. 2008;197:1594–1597. doi: 10.1086/587848. [DOI] [PubMed] [Google Scholar]
- 33.Jones K, Nourse JP, Morrison L, Nguyen-Van D, Moss DJ, Burrows SR, Gandhi MK. Expansion of EBNA1-specific effector T cells in posttransplantation lymphoproliferative disorders. Blood. 2010;116:2245–2252. doi: 10.1182/blood-2010-03-274076. [DOI] [PubMed] [Google Scholar]
- 34.Witt C, Sayer D, Christiansen F. HLA-A, -B and -DRB1 Allele Frequencies in a Population from Western Australia. Human Immunology. 2004;65:861–862. [Google Scholar]
- 35.Thorley-Lawson DA, Gross A. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N Engl J Med. 2004;350:1328–1337. doi: 10.1056/NEJMra032015. [DOI] [PubMed] [Google Scholar]
- 36.Gandhi MK, Lambley E, Duraiswamy J, Dua U, Smith C, Elliott S, Gill D, Marlton P, Seymour J, Khanna R. Expression of LAG-3 by tumorinfiltrating lymphocytes is coincident with the suppression of latent membrane antigenspecific CD8+ T-cell function in Hodgkin lymphoma patients. Blood. 2006;108:2280–2289. doi: 10.1182/blood-2006-04-015164. [DOI] [PubMed] [Google Scholar]
- 37.Gandhi MK, Moll G, Smith C, Dua U, Lambley E, Ramuz O, Gill D, Marlton P, Seymour JF, Khanna R. Galectin-1 mediated suppression of Epstein-Barr virus specific T-cell immunity in classic Hodgkin lymphoma. Blood. 2007;110:1326–1329. doi: 10.1182/blood-2007-01-066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khanna R, Moss D, Gandhi MK. Technology insight: Applications of emerging immunotherapeutic strategies for Epstein-Barr virusassociated malignancies. Nat Clin Pract Oncol. 2005;2:138–149. doi: 10.1038/ncponc0107. [DOI] [PubMed] [Google Scholar]
- 39.Merlo A, Turrini R, Dolcetti R, Martorelli D, Muraro E, Comoli P, Rosato A. The interplay between Epstein-Barr virus and the immune system: a rationale for adoptive cell therapy of EBV-related disorders. Haematologica. 2010;95:1769–1777. doi: 10.3324/haematol.2010.023689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Burrows SR, Kienzle N, Winterhalter A, Bharadwaj M, Altman JD, Brooks A. Peptide-MHC class I tetrameric complexes display exquisite ligand specificity. J Immunol. 2000;165:6229–6234. doi: 10.4049/jimmunol.165.11.6229. [DOI] [PubMed] [Google Scholar]
- 41.Burrows JM, Burrows SR, Poulsen LM, Sculley TB, Moss DJ, Khanna R. Unusually high frequency of Epstein-Barr virus genetic variants in Papua New Guinea that can escape cytotoxic T-cell recognition: implications for virus evolution. J Virol. 1996;70:2490–2496. doi: 10.1128/jvi.70.4.2490-2496.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]