Abstract
In recent years, the prospective isolation of hematopoietic stem and progenitor cells has identified the hierarchical structure of hematopoietic development and lineage-commitment. Moreover, these isolated cell populations allowed the elucitation of the molecular mechansims associated with lineage choice and revealed the indispensable functions of transcription factors as lineage determinants. This review summarizes current concepts regarding adult murine granulopoiesis and illustrates the importance of the transcription factors C/EBPα, PU.1 and GATA-2 for the development of neutrophil, eosinophil and basophil granulocytes.
Keywords: Granulopoiesis, transcription factors, C/EBPα
Introduction
Granulocytes are the most abundant type of myeloid cells in the blood stream and are characterized by two morphological features: the multilobulated shape of the nucleus and the eponymous, excessive enrichment of storage vesicles, named granules, in the cytoplasm. Based on the staining properties of these granules the granulocytes can be further subdivided into neutrophils, eosinophils and basophils. The most frequent granulocytes are neutrophils that are constantly generated in a high number in the bone marrow and circulate with the blood stream until activation in response to infections. After the conquest of the physical barriers, provided by skin and mucus membranes and infiltration of tissue by pathogens, neutrophils become activated by signals that are provoked by resident macrophages at the site of infection. The activated macrophages release cytokines that in turn activate locally endothelial cells and enable the capture of bypassing neutrophils to guide them into the infected tissue. Following the arrival at the site of infection, neutrophils combat microorganism via phagocytosis, the release of microbicidal proteins via degranulation as well as by neutrophil extracellular trap formation. Furthermore, neutrophils are able to regulate the immune response via recruitment and activation of additional neutrophils, macrophages and T cells. In contrast, eosinophils are resident in various organs such as the gastrointestinal tract, mammary glands as well as bone marrow and may contribute to tissue and immune homeostasis. Only a minor part of the eosinophils circulates with the blood stream and is recruited mainly upon T-helper 2-type responses (TH2) into sites of inflammation, where they produce several cytokines and lipid mediators and release toxic granule proteins. Additionally, eosinophils are able to initiate and amplify antigen-specific immune responses via antigen presentation to naïve and memory T cells. The least common granulocytes in the circulation are basophils. These cells are not only one of the main sources of histamine in allergic reactions, but also associated with the differentiation of naïve T cells towards TH2 cells during allergic and anti-parasitic immune responses via antigen presentation and IL-4 production.
Development of committed progenitors
Although the different types of granulocytes have quite diverse immune functions, the differentiation pathways of neutrophils, eosinophils as well as basophils show huge similarities. Like all other blood leukocytes, granulocytes derive from hematopoietic stem cells (HSC) in the bone marrow. These HSC possess multi-potentiality as well as the ability for self-renewal and develop into multipotent progenitors (MPP), which are characterized by a more limited proliferative potential, but retained ability to differentiate into various hematopoietic lineages.
At the moment a defined model for the hierarchy of multipotent progenitors is not available, because several studies have demonstrated different types of multipotent progenitors with myelo-lymphoid or myelo-erythroid potential, such as the lymphoid-primed multipotent progenitor (LMPP) [1] or a putative granulocyte-monocyte- lymphoid progenitor (GMLP) [2]. Downstream of the HSC and MPP populations starts the hematopoietic differentiation process in hematopoiesis leading to oligopotent and later on to lineage-committed progenitors with a diminished proliferation but increased differentiation. The contemporary model of hematopoiesis assumes that the decision for differentiation into the lymphoid/myeloid or megakaryocyte/ erythrocyte lineages probably occurs very early in hematopoiesis. Several studies have demonstrated that multipotent progenitors like the LMPP retain only minor megakaryocyte/ erythrocyte lineage potential, whereas the vast majority of progenitors appears to be committed to the granulocyte/monocyte as well as the lymphoid lineage [1,3].
In the next step of ongoing differentiation oligopotent progenitors with differentiation capacity for severala hematopoietic lineages develop from an ancestor, the common lymphoid progenitor (CLP) [4] and the common myeloid progenitor (CMP) [5]. The CLP carries differentiation potential for all types of lymphoid cells including B cells, T cells and NK cells and is the earliest population of oligopotent progenitors that upregulates the receptor for interleukin 7 (IL-7), an essential cytokine for T and B cell development. The CMP give rise to all types of myeloid colonies in clonogenic assays, while the granulocyte-monocyte progenitor (GMP) is restricted to granulocytes and macrophages and the megakaryocyte-erythrocyte progenitor (MEP) is delimitated to megakaryocytes and erythrocytes [5]. From the GMP derives the eosinophil lineage-committed progenitor (EoP) [6] and the basophil/mast cell progenitor (BMCP), which in turn gives rise to the mast cell progenitor (MCP) and the basophil progenitor (BaP) [7]. With regard to neutrophils a committed progenitor is not yet described (Figure 1).
Figure 1.
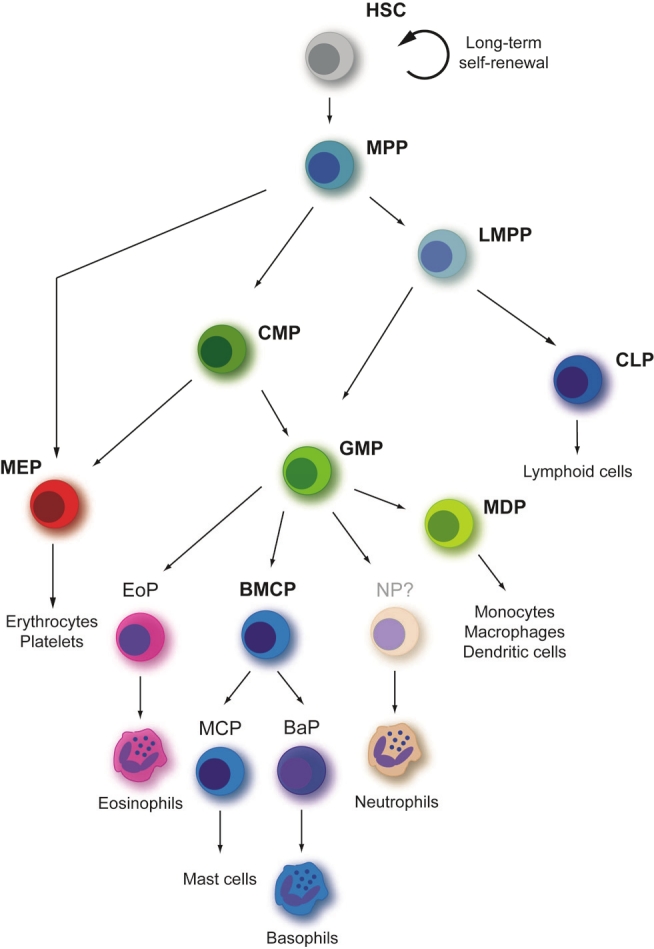
Hierarchy of hematopoietic progenitors. The developmental course shown in the scheme is proposed using results generated by prospective isolation and characterization of different progenitors in mouse. HSC, hematopoietic stem cell; MPP, multipotent progenitor; LMPP, lymphoid-primed multipotent progenitor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor; GMP, granulocyte- macrophage progenitor; MDP, monocyte-dendritic cell progenitor; BMCP, basophil-mast cell progenitor; EP, erythroid progenitor; MCP, mast cell progenitor; EoP, eosinophil progenitor; BaP, basophil progenitor. Figure adapted from [2,53,54].
Transcriptional regulation of myeloid progenitor commitment
The differentiation process from HSC into lineage- committed hematopoietic cells involves the selective activation of lineage-specific genes as well as the silencing of lineage-foreign genes and developmental regulators in a defined order. The orchestration of such complex lineage-determining programs is dependent on several factors, but extensive research has emphasized the essential role of gene regulatory networks in directing cell fate choice and lineage restriction. These gene regulatory networks are composed of several master transcription factors that join special features, such as mutual regulation of transcriptional activity by antagonism as well as lineage-determining functions via activation of lineage-specific genes and repression of lineage -foreign genes. One example for such gene regulatory networks is the choice for erythroid versus myeloid-lymphoid lineage restriction at the transition from MPP to LMPP or MEP that is regulated by the E-twenty six (Ets) family transcription factor PU.1 and the transcription factor GATA-binding protein 1 (GATA-1). GATA-1 is expressed in erythroid, megakaryocyte, mast as well as eosinophil lineages and contains zinc fingers, which mediate DNA binding to the WGA-TAR DNA sequence as well as protein-protein interaction [8,9]. In contrast to GATA-1, PU.1 is restricted to monocyte as well as B lymphoid lineages and consists of a N-terminal transactivation domain, a PEST-domain, and the eponymous Ets-domain at the C-terminus that mediates DNA binding to an 11 bp sequence with a central GGAA motif [10,11]. Additionally, both transcription factors are detectable in MPP and gene disruption studies have demonstrated the indispensable functions of GATA-1 and PU.1 for megakaryocyte/erythrocyte and myeloid/ lymphoid development, respectively. Analyses of systemic PU.1-deficient mice revealed a complete loss of CMP, GMP and CLP populations but normal numbers of MEP causing impaired lymphoid as well as myeloid cell development without influencing megakaryocyte/erythrocyte development [12]. In contrast, GATA-1-deficient mice die between embryonic day 10.5 and 11.5 due to severe anemia resulting from a maturation arrest of erythroid cells [13]. Further support for the lineage instructive role of GATA-1 originated from forced expression of GATA-1 in lineage-committed progenitors like GMP and CLP exclusively leading to megakaryocyte/ erythrocyte development [14]. Several other studies dealing with certain aspects of the molecular interaction of PU.1 and GATA-1 as well as their gene regulatory capacity revealed the cross-antagonism between these proteins involving direct physical interaction of both factors that results in an inhibition of the transactivation potential of the counterpart [15]. Based on these findings, GATA-1 is prospected as the erythroid/megakaryocyte lineage determinant, whereas PU.1 is regarded as the myeloid/ lymphoid lineage determinant.
Downstream of LMPP or GMLP, lineage choice embraces myeloid, as well as B or T lymphoid lineage and mainly depends on the gene regulatory network of the transcription factors PU.1, early B cell factor (EBF) and Notch. For myeloid lineage restriction, a high expression level of PU.1 is necessary, whereas low levels of PU.1 plus EBF expression establish the B lymphoid lineage restriction and Notch instructs the T lymphoid lineage choice. The B cell fate determinant EBF has been demonstrated to initiate the early B cell lineage program of gene expression (B29, VpreB and Pax5) and to antagonize expression of myeloid lineage genes (Cebpa, Sfpi1 and Id2) [16]. Additionally, the same regulatory properties have been shown for the T cell fate determinant Notch that inhibits B cell development via repression of EBF function as well as Pax5 expression, a secondary B cell lineage determinant [15].
The granulocyte versus monocyte lineage choice
Regarding granulopoiesis the transcription factor CCAAT-enhancer binding protein α (C/EBPα) has to be enumerated, since studies have demonstrated that conditional deletion of C/EBPα in bone marrow cells of mice using the Mx1-Cre system leads to a total lack of mature granulocytes and a partial lack of monocytes due to a differentiation block at the transition from CMP to GMP [17]. C/EBPα is the prototype of the C/ EBP family and displays all characteristic features of the transcription factor family, such as the N-terminal transactivation domain as well as the C-terminal DNA-binding domain consisting of a highly conserved basic region and a leucine zipper dimerization domain. Prerequisite for binding of C/EBPα to the cognate DNA-site is the homo- or heterodimerization with another transcription factor via the leucine zipper domain that in turn allows the basic region to bind to the CCAAT motif [18,19]. The need for C/ EBPα during the transition from CMP to GMP is possibly due to the transcriptional upregulation of PU.1, since forced C/EBPα expression in hematopoietic progenitors favors monopoiesis and not granulopoiesis, whereas exogenous C/EBPα in myeloid cell lines directs granulopoiesis [20]. Moreover, lineage choice between monocytes and granulocytes depends on the expression level of PU.1 and C/EBPα, which has been shown by studies using different mouse as well as in vitro models for diminished PU.1 expression in the hematopoietic system. In all experimental setups, reduced expression of PU.1 is followed by an augmented granulopoiesis to the disadvantage of monocyte development. In line with these findings, we have demonstrated that loss-of-function mutations of Btk in myeloid cells diminshed the C/EBPα as well as PU.1 expression resulting consequently also in an increased granulopoiesis at the expanse of monopoiesis [21]. Additionally, gene expression analyses of PU.1-deficient progenitors revealed a decreased or even absent expression of several monocyte-specific genes, like the macrophage scavenger receptor or the M-CSF receptor.
The indispensable functions of C/EBPα for granulocyte development are additionally pointed out by the transcriptional upregulation of several granulocyte-specific factors. One of these factors is the transcriptional repressor growth factor independent 1 (Gfi1), which is necessary for the repression of proliferation and of monocyte lineage-promoting factors such as M-CSF [22]. Another lineage-determining mechanism displays the repression of the microRNAs miR-21 and miR-196b Gfi-1 during granulopoiesis, since ectopic expression of both miRNAs in myeloid progenitors results in a complete block of G-CSF induced granulopoiesis [23]. Besides the upregulation of other transcriptional regulators, C/EBPα forces granulocyte development additionally by transactivation of various genes, such as genes coding for the G-CSF receptor [24,25] or for myeloperoxidase (MPO) [26], and downregulation of proliferation by direct interaction with the cell cycle regulator E2F [27,28]. In line with these experimental results is the association of inactivating C/EBPα mutations with hematopoietic malignancies like acute myeloid leukemia and high-risk myelodysplastic syndrome proposing that C/EBPα possesses the ability to arrest cell proliferation and to drive terminal differentiation [29]. Moreover, the transcription of miR-223 is activated by C/ EBPα that replaces the transcriptional repressor NFI-A upon activation of granulocytic differentiation [30] and results in the upregulation of miR- 223 during granulopoiesis [30,31]. Targets of miR-223 are the cell cycle regulator E2F1 and the monocyte-lineage promoting gene Mef2c leading to suppression of proliferation and induction of granulocyte differentiation [31]. A similar mechanism has been recently described for miR-34a that is also increased expressed during granulopoiesis by C/EBPα-mediated transcription and targets the cell cycle regulator E2F3 [32].
Taken together, the plethora of studies implicates the following model for monocyte versus granulocyte lineage choice: First, C/EBPα is needed for the transition from CMP to GMP by induction of PU.1 expression (Figure 2). High protein levels of PU.1 induce monopoiesis via interaction with other transcription factors like interferon regulatory factor 8 (IRF8) or activating protein-1 family transcription factors (AP-1/Jun proteins) and the transcriptional activation of monocyte-specific genes [20]. However, AP-1 family transcription factors are also able to heterodimerize with C/EBPα [33] implicating an inhibition mechanism of PU.1 for granulocyte development by sequestering the binding partners of C/EBPα. In contrast to the high protein levels of PU.1 that favor monopoiesis, insufficient activation of PU.1 transcription allows C/ EBPα to induce the granulopoiesis program accompanied by suppression of monopoiesis.
Figure 2.
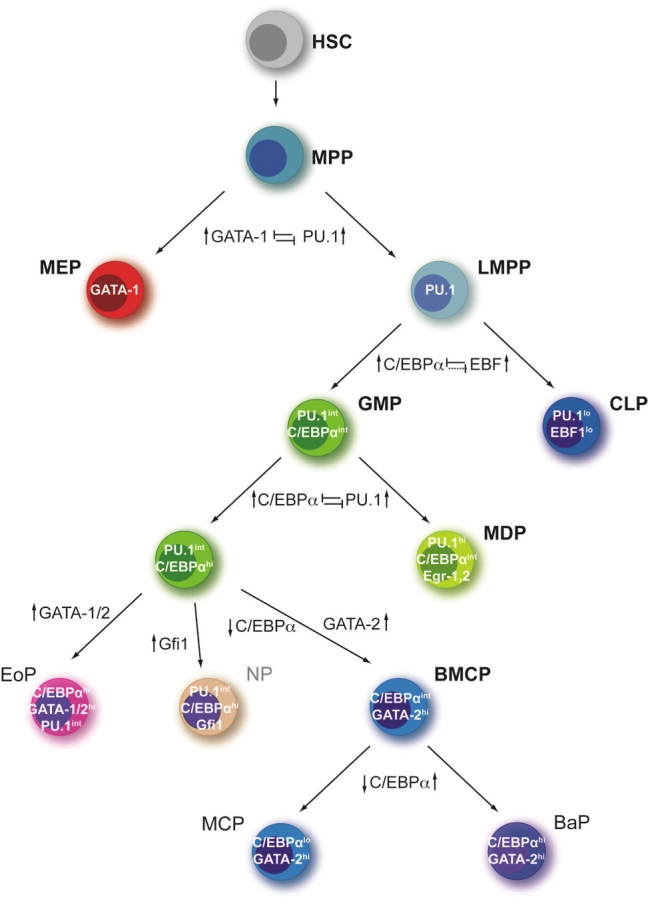
Transcriptional regulation of granulocyte lineage commitment. The scheme displays a simplified overview of gene regulatory networks, which have a major influence on hematopoietic lineage choice during hematopoiesis. Supposed (dashed lines) and proved (continuous lines) cross-antagonisms between key transcription factors which function to regulate binary cell fate choices are noted in the scheme. Additionally, transcription factors that are important for the generation of particular intermediates are noted in white. HSC, hematopoietic stem cell; MPP, multipotent progenitor; LMPP, lymphoid-primed multipotent progenitor; CLP, common lymphoid progenitor; CMP, common myeloid progenitor; MEP, megakaryocyte-erythrocyte progenitor; GMP, granulocyte-macrophage progenitor; EoP, eosinophil progenitor; MDP, monocyte-dendritic cell progenitor; BMCP, basophil-mast cell progenitor; MCP, mast cell progenitor; BaP, basophil progenitor. Figure adapted from [15].
The eosinophil versus basophil lineage choice
The lineage commitment towards the eosinophil or basophil lineage downstream of the GMP mainly depends on the expression of GATA transcription factors. First hints for the importance of GATA transcription factors revealed from gene expression analyses of eosinophils and basophils demonstrating that GATA-1 as well as GATA-2 are elusively expressed in EoP, BMCP and BaP, but not in GMP [34,35]. In addition the targeted deletion of the high-affinity GATA-binding site in the GATA-1 promoter resulted in the selective loss of eosinophils [36], whereas ectopic expression of GATA-2 in GMP introduced exclusively the development of eosinophils [35]. Nevertheless, the GATA transcription factors are not the solely transcription factors necessary for the development of eosinophils and basophils, since C/EBPα and PU.1 are also involved in lineage commitment. The importance of C/EBPα for eosinophil development has been shown by the loss of the eosinophil lineage after the deletion of C/EBPα in mice [37] and the disruption of C/EBPα expression in BMCP leading exclusively to mast cell development at the expanse of basophils [7]. Furthermore, Du and colleagues have demonstrated the synergistic transactivation of the promoter of the eosinophil-specific gene MBP (major basic protein) by GATA-1 and PU-1.
In summary, the gene regulatory network causing eosinophil versus basophil lineage choice is supposed to be dependent on the level of expression as well as on the order of expression of the transcription factor GATA-1 or GATA-2, C/EBPα and PU.1. For example, the upregulation of GATA-1 or GATA-2 along with a sustained C/EBPα expression at the GMP stage results in the development of eosinophils progenitors. However, for lineage commitment of basophils and mast cells the upregulation of GATA-2 expression accompanied by the downregulation of C/EBPα expression is needed. Downstream of the BMCP basophils as well as mast cells can develop depending on the expression level of C/EBPα: reintroduction of C/EBPα expression leads to the development of basophils, whereas further downregulation of C/EBPα expression results in mast cell development (Figure 2).
Transcriptional regulation of terminal neutrophil differentiation
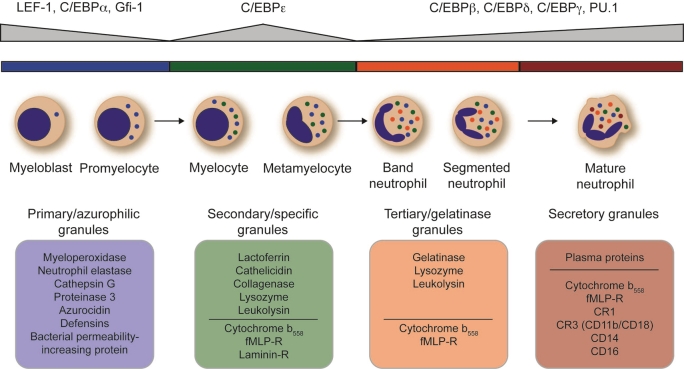
Terminal neutrophil granulopoiesis starts with the myeloblast and promyelocyte state, where the switch from proliferation to differentiation takes place, displayed by the loss of ability for cell division after the promyelocyte state. Moreover, the formation of the first granules starts, which are named primary or azurophilic granules. Primary granules are defined by a high content of myeloperoxidase (MPO), bactericidal permeability-increasing protein, defensins, and a family of structurally related serine proteases, namely cathepsin G, neutrophil elastase (ELANE) and proteinase 3 [38]. The most important transcription factors at myeloblast/ promyelocyte stage are C/EBPα and Gfi1, which are necessary for the suppression of monocyte development and proliferation as well as for the transcriptional activation of granulocyte-specific genes like MPO, ELANE or CEBPE [22,28,29]. The importance of Gfi1 and ELANE has been demonstrated by studies analyzing the genetic background of severe congenital neutropenia (SCN) and other forms of neutropenia. These studies revealed that one of the major causes for loss of neutrophil differentiation beyond promyelocyte state are mutations in the ELANE gene [39,40], but in rare cases of SCN also mutations of the GFI1 gene have been described [41]. Detailed analyses of Gfi1 in mice further supported the function of Gfi1 as molecular switch towards granulocyte development by suppression of monocyte-specific genes, like Csf1 (M-CSF) and Csf1r (M-CSFR) [42]. Recently, Skokowa and colleagues described another transcription factor important for terminal neutrophil differentiation - the lympohid enhancer-binding factor 1 (LEF-1) - that is highly expressed in promyelocytes and associated with severe congenital neutropenia (CN). Analyses of bone marrow samples of CN patients revealed a differentiation block at the promyelocytic stage and a greatly reduced or even absent expression of LEF-1 in comparison to bone marrow samples of healthy individuals. Moreover, reconstitution of LEF-1 in early hematopoietic progenitors of patients with CN corrected the defective granulopoiesis and resulted in mature neutrophils due to direct regulation of C/EBPα expression [43].
Ongoing differentiation beyond promyelocytes leads to the development of myelocytes and metamyelocytes, which are defined by the beginning of nuclear segmentation and the appearance of secondary (also called specific) granules as well as the exit from cell cycle. Secondary granules are characterized by high amounts of lactoferrin, cathelicidin, collagenase as well as leukolysin and lysozyme in the matrix [38,44]. The regulation of secondary granule protein expression and the exit from cell cycle mainly depends on the transcription factor C/ EBPε, whose expression peaks in myelocytes and metamyelocytes [28,45] (Figure 3). Based on studies using C/EBPε-deficient mice, which displayed neutrophil-specific defects including bilobed nuclei, abnormal respiratory burst, compromised bactericidal activity as well as impaired chemotaxis [46,47], the genetic cause of a very rare congenital disorder named neutrophil specific granule deficiency (SGD) has been delineated to the CEBPE locus [48]. Additional studies revealed essential functions of C/EBPε for the expression of secondary and tertiary granule proteins [46,49] and demonstrated the direct interaction of C/EBPε with E2F1 and Rb protein, finally leading to cell cycle exit [50].
Figure 3.
Transcription factors involved in terminal granulopoiesis. The terminal granulopoiesis that is characterized by sequential formation of different granule types and segmentation of the nucleus starts at the myeloblast/ promyelocyte stage and ends with mature neutrophils. Granule types not only differ in the time point at which they are formed, but also in their specific content, which is described at the bottom of the figure. Above the line in the boxes matrix content is depicted and beneath the proteins that are located to the vesicle membrane. At different stages of terminal granulopoiesis several transcription factors, which are indicated on top of the figure, are important for the regulation of maturation and timed expression of granule proteins. Figure adapted from [22,38,44].
The last step of terminal granulopoiesis, the differentiation into band and segmented neutrophils leads to mature neutrophils with finally segmented nuclei and tertiary as well as secretory granules. Tertiary granules are mainly defined by a high content of gelatinase as well as leukolysin and lysozyme, whereas secretory granules only contain plasma proteins in their matrix. The most important feature of a secretory vesicle is its membrane that is highly enriched for receptors such as Mac-1/CD11b, CD14, FcγRII (CD16) or formyl-peptide receptor (fMLP-R) [44]. In the course of neutrophil terminal differentiation, C/EBPα expression gradually diminishes during the myeloblast stage. C/EBPε peaks at the myelocyte/metamyelocyte stage, whereas the expression level of the transcription factors PU.1, C/EBPβ, C/EBPδ and C/EBPγ increases continously during development to the metamyelocyte stage [45]. However, gene deletion studies using C/EBPβ- or C/EBPδ- deficient mice revealed no hematopoietic abnormalities with regard to terminal granulopoiesis. Still, Hirai and colleagues have demonstrated the indispensable role of C/EBPβ during emergency granulopoiesis in response to cytokine treatment or fungal infection. In contrast, C/EBPα and C/EBPε were not upregulated under these conditions [51]. In the case of the transcription factor PU.1, a conditional gene deletion model revealed a PU.1-dependent transcriptional activation of gp91phox, a component of the cytochrome b558 of the NADPH oxidase, as well as of Mac-1/CD11b [52].
Concluding remarks
The process of differentiation and lineage commitment during granulopoiesis depends strongly on the defined activation of lineage-determining gene programs as well as the repression of lineage- foreign gene programs. This concerted regulation of genetic programs can be achieved by the activation of lineage-specific transcription factors that are often involved in several lineage -decisions were they induce quite different cell fates depending on the presence of other transcription factors co-expressed in the cell or on the level and order of protein expression. Therefore, the granulocyte differentiation process clearly demonstrates the importance of regulatory networks that integrate several intrinsic and extrinsic signals for the outcome of differentiation and cell fate decisions.
References
- 1.Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, Anderson K, Sitnicka E, Sasaki Y, Sigvardsson M, Jacobsen SE. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Iwasaki H, Akashi K. Myeloid lineage commitment from the hematopoietic stem cell. Immunity. 2007;26:726–740. doi: 10.1016/j.immuni.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Iwasaki H, Akashi K. Hematopoietic developmental pathways: on cellular basis. Oncogene. 2007;26:6687–6696. doi: 10.1038/sj.onc.1210754. [DOI] [PubMed] [Google Scholar]
- 4.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 5.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki H, Mizuno S, Mayfield R, Shigematsu H, Arinobu Y, Seed B, Gurish MF, Takatsu K, Akashi K. Identification of eosinophil lineage- committed progenitors in the murine bone marrow. J Exp Med. 2005;201:1891–1897. doi: 10.1084/jem.20050548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arinobu Y, Iwasaki H, Gurish MF, Mizuno S, Shigematsu H, Ozawa H, Tenen DG, Austen KF, Akashi K. Developmental checkpoints of the basophil/mast cell lineages in adult murine hematopoiesis. Proc Natl Acad Sci USA. 2005;102:18105–18110. doi: 10.1073/pnas.0509148102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. GATA switches as developmental drivers. J Biol Chem. 2010;285:31087–31093. doi: 10.1074/jbc.R110.159079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morceau F, Schnekenburger M, Dicato M, Diederich M. GATA-1: friends, brothers, and coworkers. Ann N Y Acad Sci. 2004;1030:537–554. doi: 10.1196/annals.1329.064. [DOI] [PubMed] [Google Scholar]
- 10.Gangenahalli GU, Gupta P, Saluja D, Verma YK, Kishore V, Chandra R, Sharma RK, Ravindranath T. Stem cell fate specification: role of master regulatory switch transcription factor PU. 1 in differential hematopoiesis. Stem Cells Dev. 2005;14:140–152. doi: 10.1089/scd.2005.14.140. [DOI] [PubMed] [Google Scholar]
- 11.Sharrocks AD. The ETS-domain transcription factor family. Nat Rev Mol Cell Biol. 2001;2:827–837. doi: 10.1038/35099076. [DOI] [PubMed] [Google Scholar]
- 12.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU. 1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki H, Mizuno S, Wells RA, Cantor AB, Watanabe S, Akashi K. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462. doi: 10.1016/s1074-7613(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 15.Laslo P, Pongubala JM, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin Immunol. 2008;20:228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 16.Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- 17.Zhang P, Iwasaki-Arai J, Iwasaki H, Fenyus ML, Dayaram T, Owens BM, Shigematsu H, Levantini E, Huettner CS, Lekstrom-Himes JA, Akashi K, Tenen DG. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP α. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Lekstrom-Himes J, Xanthopoulos KG. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J Biol Chem. 1998;273:28545–28548. doi: 10.1074/jbc.273.44.28545. [DOI] [PubMed] [Google Scholar]
- 19.Johnson PF. Molecular stop signs: regulation of cell-cycle arrest by C/EBP transcription factors. J Cell Sci. 2005;118:2545–2555. doi: 10.1242/jcs.02459. [DOI] [PubMed] [Google Scholar]
- 20.Friedman AD. Transcriptional control of granulocyte and monocyte development. Oncogene. 2007;26:6816–6828. doi: 10.1038/sj.onc.1210764. [DOI] [PubMed] [Google Scholar]
- 21.Fiedler K, Sindrilaru A, Terszowski G, Kokai E, Feyerabend TB, Bullinger L, Rodewald HR, Brunner C. Neutrophil development and function critically depend on Bruton tyrosine kinase in a mouse model of X-linked agammaglobulinemia. Blood. 2011;117:1329–1339. doi: 10.1182/blood-2010-04-281170. [DOI] [PubMed] [Google Scholar]
- 22.Borregaard N. Neutrophils, from marrow to microbes. Immunity. 2010;33:657–670. doi: 10.1016/j.immuni.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 23.Velu CS, Baktula AM, Grimes HL. Gfi1 regulates miR-21 and miR-196b to control myelopoiesis. Blood. 2009;113:4720–4728. doi: 10.1182/blood-2008-11-190215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohaus S, Petrovick MS, Voso MT, Sun Z, Zhang DE, Tenen DG. PU. 1 (Spi-1) and C/ EBP α regulate expression of the granulocyte-macrophage colony-stimulating factor receptor α gene. Mol Cell Biol. 1995;15:5830–5845. doi: 10.1128/mcb.15.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith LT, Hohaus S, Gonzalez DA, Dziennis SE, Tenen DG. PU. 1 (Spi-1) and C/EBP α regulate the granulocyte colony-stimulating factor receptor promoter in myeloid cells. Blood. 1996;88:1234–1247. [PubMed] [Google Scholar]
- 26.Wang W, Wang X, Ward AC, Touw IP, Friedman AD. C/EBPα and G-CSF receptor signals cooperate to induce the myeloperoxidase and neutrophil elastase genes. Leukemia. 2001;15:779–786. doi: 10.1038/sj.leu.2402094. [DOI] [PubMed] [Google Scholar]
- 27.D'Alo F, Johansen LM, Nelson EA, Radomska HS, Evans EK, Zhang P, Nerlov C, Tenen DG. The amino terminal and E2F interaction domains are critical for C/EBP α-mediated induction of granulopoietic development of hematopoietic cells. Blood. 2003;102:3163–3171. doi: 10.1182/blood-2003-02-0479. [DOI] [PubMed] [Google Scholar]
- 28.Theilgaard-Monch K, Jacobsen LC, Borup R, Rasmussen T, Bjerregaard MD, Nielsen FC, Cowland JB, Borregaard N. The transcriptional program of terminal granulocytic differentiation. Blood. 2005;105:1785–1796. doi: 10.1182/blood-2004-08-3346. [DOI] [PubMed] [Google Scholar]
- 29.Koschmieder S, Halmos B, Levantini E, Tenen DG. Dysregulation of the C/EBPα differentiation pathway in human cancer. J. Clin. Oncol. 2009;27:619–628. doi: 10.1200/JCO.2008.17.9812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fazi F, Rosa A, Fatica A, Gelmetti V, De Marchis ML, Nervi C, Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Johnnidis JB, Harris MH, Wheeler RT, Stehling- Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 32.Pulikkan JA, Peramangalam PS, Dengler V, Ho PA, Preudhomme C, Meshinchi S, Christopeit M, Nibourel O, Muller-Tidow C, Bohlander SK, Tenen DG, Behre G. C/EBPα regulated microRNA-34a targets E2F3 during granulopoiesis and is down-regulated in AML with CEBPA mutations. Blood. 2010;116:5638–5649. doi: 10.1182/blood-2010-04-281600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cai DH, Wang D, Keefer J, Yeamans C, Hensley K, Friedman AD. C/EBP α: AP-1 leucine zipper heterodimers bind novel DNA elements, activate the PU. 1 promoter and direct monocyte lineage commitment more potently than C/EBP α homodimers or AP-1. Oncogene. 2008;27:2772–2779. doi: 10.1038/sj.onc.1210940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zon LI, Yamaguchi Y, Yee K, Albee EA, Kimura A, Bennett JC, Orkin SH, Ackerman SJ. Expression of mRNA for the GATA-binding proteins in human eosinophils and basophils: potential role in gene transcription. Blood. 1993;81:3234–3241. [PubMed] [Google Scholar]
- 35.Iwasaki H, Mizuno S, Arinobu Y, Ozawa H, Mori Y, Shigematsu H, Takatsu K, Tenen DG, Akashi K. The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 2006;20:3010–3021. doi: 10.1101/gad.1493506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu C, Cantor AB, Yang H, Browne C, Wells RA, Fujiwara Y, Orkin SH. Targeted deletion of a high-affinity GATA-binding site in the GATA-1 promoter leads to selective loss of the eosinophil lineage in vivo. J Exp Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein α-deficient mice. Proc Natl Acad Sci USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham CT. Neutrophil serine proteases: specific regulators of inflammation. Nat Rev Immunol. 2006;6:541–550. doi: 10.1038/nri1841. [DOI] [PubMed] [Google Scholar]
- 39.Horwitz M, Benson KF, Person RE, Aprikyan AG, Dale DC. Mutations in ELA2, encoding neutrophil elastase, define a 21-day biological clock in cyclic haematopoiesis. Nat Genet. 1999;23:433–436. doi: 10.1038/70544. [DOI] [PubMed] [Google Scholar]
- 40.Dale DC, Person RE, Bolyard AA, Aprikyan AG, Bos C, Bonilla MA, Boxer LA, Kannourakis G, Zeidler C, Welte K, Benson KF, Horwitz M. Mutations in the gene encoding neutrophil elastase in congenital and cyclic neutropenia. Blood. 2000;96:2317–2322. [PubMed] [Google Scholar]
- 41.Person RE, Li FQ, Duan Z, Benson KF, Wechsler J, Papadaki HA, Eliopoulos G, Kaufman C, Bertolone SJ, Nakamoto B, Papayannopoulou T, Grimes HL, Horwitz M. Mutations in proto-oncogene GFI1 cause human neutropenia and target ELA2. Nat Genet. 2003;34:308–312. doi: 10.1038/ng1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zarebski A, Velu CS, Baktula AM, Bourdeau T, Horman SR, Basu S, Bertolone SJ, Horwitz M, Hildeman DA, Trent JO, Grimes HL. Mutations in growth factor independent-1 associated with human neutropenia block murine granulopoiesis through colony stimulating factor-1. Immunity. 2008;28:370–380. doi: 10.1016/j.immuni.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skokowa J, Cario G, Uenalan M, Schambach A, Germeshausen M, Battmer K, Zeidler C, Lehmann U, Eder M, Baum C, Grosschedl R, Stanulla M, Scherr M, Welte K. LEF-1 is crucial for neutrophil granulocytopoiesis and its expression is severely reduced in congenital neutropenia. Nat Med. 2006;12:1191–1197. doi: 10.1038/nm1474. [DOI] [PubMed] [Google Scholar]
- 44.Borregaard N, Cowland JB. Granules of the human neutrophilic polymorphonuclear leukocyte. Blood. 1997;89:3503–3521. [PubMed] [Google Scholar]
- 45.Bjerregaard MD, Jurlander J, Klausen P, Borregaard N, Cowland JB. The in vivo profile of transcription factors during neutrophil differentiation in human bone marrow. Blood. 2003;101:4322–4332. doi: 10.1182/blood-2002-03-0835. [DOI] [PubMed] [Google Scholar]
- 46.Yamanaka R, Barlow C, Lekstrom-Himes J, Castilla LH, Liu PP, Eckhaus M, Decker T, Wynshaw- Boris A, Xanthopoulos KG. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc Natl Acad Sci USA. 1997;94:13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lekstrom-Himes J, Xanthopoulos KG. CCAAT/ enhancer binding protein epsilon is critical for effective neutrophil-mediated response to inflammatory challenge. Blood. 1999;93:3096–3105. [PubMed] [Google Scholar]
- 48.Lekstrom-Himes JA, Dorman SE, Kopar P, Holland SM, Gallin JI. Neutrophil-specific granule deficiency results from a novel mutation with loss of function of the transcription factor CCAAT/enhancer binding protein epsilon. J Exp Med. 1999;189:1847–1852. doi: 10.1084/jem.189.11.1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verbeek W, Lekstrom-Himes J, Park DJ, Dang PM, Vuong PT, Kawano S, Babior BM, Xanthopoulos K, Koeffler HP. Myeloid transcription factor C/EBPepsilon is involved in the positive regulation of lactoferrin gene expression in neutrophils. Blood. 1999;94:3141–3150. [PubMed] [Google Scholar]
- 50.Gery S, Gombart AF, Fung YK, Koeffler HP. C/EBPepsilon interacts with retinoblastoma and E2F1 during granulopoiesis. Blood. 2004;103:828–835. doi: 10.1182/blood-2003-01-0159. [DOI] [PubMed] [Google Scholar]
- 51.Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG. C/EBPβ is required for "emergency"; granulopoiesis. Nat Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- 52.Dakic A, Metcalf D, Di Rago L, Mifsud S, Wu L, Nutt SL. PU. 1 regulates the commitment of adult hematopoietic progenitors and restricts granulopoiesis. J Exp Med. 2005;201:1487–1502. doi: 10.1084/jem.20050075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissman IL, Shizuru JA. The origins of the identification and isolation of hematopoietic stem cells, and their capability to induce donor- specific transplantation tolerance and treat autoimmune diseases. Blood. 2008;112:3543–3553. doi: 10.1182/blood-2008-08-078220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–661. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]