Abstract
The complexes of the electron transport chain associate into large macromolecular assemblies, which are believed to facilitate efficient electron flow. We have identified a conserved mitochondrial protein, named Respiratory superComplex Factor 1 (Rcf1—Yml030w), that is required for the normal assembly of respiratory supercomplexes. We demonstrate that Rcf1 stably and independently associates with both Complex III and Complex IV of the electron transport chain. Deletion of the RCF1 gene caused impaired respiration, probably as a result of destabilization of respiratory supercomplexes. Consistent with the hypothetical function of these respiratory assemblies, loss of RCF1 caused elevated mitochondrial oxidative stress and damage. Finally, we show that knockdown of HIG2A, a mammalian homolog of RCF1, causes impaired supercomplex formation. We suggest that Rcf1 is a member of an evolutionarily conserved protein family that acts to promote respiratory supercomplex assembly and activity.
INTRODUCTION
Mitochondria are unique and complex organelles that perform essential functions in many aspects of cell biology. It is not surprising, therefore, that mitochondrial dysfunction is associated with many forms of human disease. This includes relatively common disorders, like cancer (Kroemer and Pouyssegur, 2008), diabetes (Patti and Corvera, 2010), and neurodegenerative disease (Lessing and Bonini, 2009). Beyond these common disorders, which are all age-related, mitochondrial dysfunction has been associated with the aging process itself. Manipulation of mitochondrial function, including by modulation of mitochondrial oxidative stress and mitochondrial DNA (mtDNA) mutability, has striking effects on longevity (Schriner et al., 2005; Trifunovic et al., 2004).
The intimate connection between oxidative stress, mitochondrial dysfunction and age-related disorders is thought to derive from the physical proximity of the electron transport chain (ETC) and the mtDNA. The ETC is the major source of reactive oxygen species (ROS) in most cells. This ROS load is then in immediate proximity to the mtDNA, which is highly sensitive to mutagenic insult. Mutated mtDNA is then postulated to be a causal factor in further ETC dysfunction, leading to a vicious cycle of ROS production (Wallace, 2005). The challenge for the ETC is to conduct reactive electrons via soluble carriers, through three distinct multi-protein complexes, to oxygen. When this conduction process is disturbed, the risk of ROS production increases. As this oxidative stress/ETC/aging axis is of such relevance to human disease, it is of great importance to understand the mechanisms that have evolved to prevent inappropriate ROS production. One biochemical phenomenon that has been postulated to control ETC malfunction and prevent mitochondrial ROS production is the assembly of the individual ETC complexes into massive conduction machines called respiratory supercomplexes (Cruciat et al., 2000; Lenaz and Genova, 2009).
The clear importance of mitochondria has led to extensive efforts to define the mitochondrial proteome in many species. The best current inventory of mammalian mitochondrial resident proteins consists of 1098 proteins (Pagliarini et al., 2008). Surprisingly, nearly 300 of these proteins are largely or completely unstudied (Pagliarini et al., 2008). Included among this number are many that are conserved throughout the eukaryotic kingdom. Such conservation implies that they perform a function that is of fundamental importance for cell viability. As a result, we initiated a project to identify the functions of a subset of these highly evolutionarily conserved, but uncharacterized, mitochondrial proteins. We previously described the function of SDH5/SDHAF2, which is required for assembly of ETC Complex II and is mutated in multiple families afflicted with the paraganglioma tumor syndrome (Bayley et al., 2010; Hao et al., 2009). We also described Vms1, which plays an important role in stress-responsive recruitment of the ubiquitin-proteasome machinery to mitochondria for protein degradation (Heo et al., 2010).
Herein, we describe the function of Rcf1, which we show is a stable component of ETC supercomplexes and is required for their normal stability. In Saccharomyces cerevisiae, ETC supercomplexes are assemblies in the mitochondrial inner membrane that contain, among other proteins, both Complex III (cytochrome bc1 complex) and Complex IV (cytochrome c oxidase complex) and perhaps Complex II (Stuart, 2008). In mammals, supercomplexes often also contain Complex I (Acin-Perez et al., 2008; Wittig and Schagger, 2009). These huge macromolecular assemblies are believed to enable more efficient electron flow between complexes and to promote complex stability (Suthammarak et al., 2010). This efficiency is believed to reduce electron loss from the ETC and ROS generation (Genova et al., 2008). Not surprisingly, we show that the rcf1Δ mutant, which exhibits decreased supercomplex assembly, has increased mitochondrial oxidative damage. We also show that knockdown of HIG2A, one of the mammalian orthologs of RCF1, impairs supercomplex assembly in mouse cells. We, therefore, speculate that the RCF1 gene family plays an evolutionarily conserved role in maintaining mitochondrial function through the optimal assembly of electron transport chain complexes. Thereby, it might be a component of the cellular system to delay ROS-induced and age-associated degeneration.
RESULTS
Rcf1 is an integral mitochondrial inner membrane protein required for normal respiration
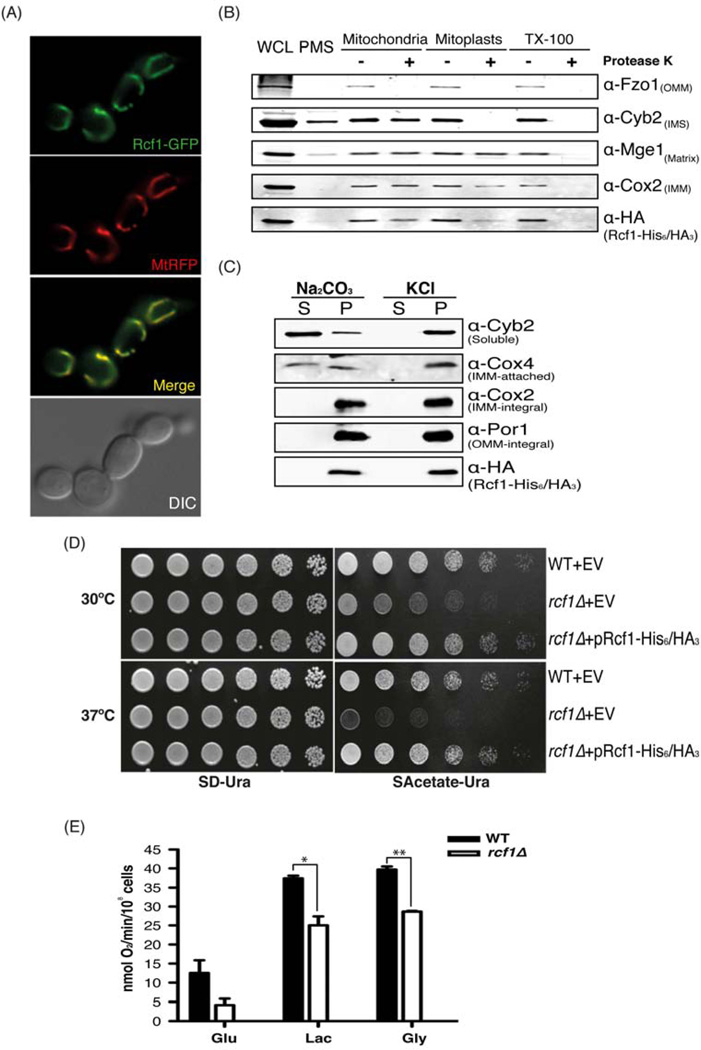
Using the yeast S. cerevisiae as the primary model system, we first verified mitochondrial localization of the Rcf1 (Yml030w/Aim31) protein using two complementary methods. First, we generated a strain expressing an Rcf1-GFP fusion protein from the native promoter, which is fully functional as assessed by suppression of the rcf1Δ phenotype (data not shown). This strain was co-transformed with a plasmid expressing a mitochondria-targeted red fluorescent protein (MtRFP). As seen in Figure 1A, we observed complete overlap between the mitochondrial RFP signal and the Rcf1-GFP signal. Under no conditions did we find extra-mitochondrial GFP fluorescence.
Figure 1. Rcf1 is a mitochondrial inner membrane protein important for respiration. (Also see Figure S1).
(A) The rcf1Δ mutant expressing Rcf1-GFP and MtRFP grown to log phase in SD medium was imaged by fluorescence microscopy. (B) Intact mitochondria, hypotonic-swollen mitoplasts, and TritonX-100–solubilized mitochondria of a strain expressing Rcf1-His6/HA3 were treated either with (+) or without (−) Proteinase K and analyzed by immunoblot with the whole cell lysate (WCL) and post mitochondrial supernatant (PMS). Cox2, Mge1, Cyb2, and Fzo1 are inner membrane, matrix, intermembrane space, and outer membrane proteins, respectively. (C) Soluble (S) and pellet (P) fractions from alkali and high salt extracted mitochondria expressing Rcf1-His6/HA3 were analyzed by immunoblot. Cyb2 is a soluble intermembrane space protein; Cox4 is an inner membrane-associated protein; Cox2 is an integral inner membrane protein; and Por1 is an integral outer membrane protein. (D) Five-fold serial dilutions of the indicated strains harboring an empty vector (EV) or a plasmid expressing Rcf1-His6/HA3 were spotted on plates and incubated at 30°C and 37°C. (E) Wild-type and rcf1Δ mutant strains of the BY4741 background were grown in glucose, lactate or glycerol to log phase and the rate of oxygen consumption was measured. Data shown is representative of three independent experiments and mean ± SD is shown. *, P<0.05; **, P<0.01
In parallel, we also biochemically examined the localization of an Rcf1 fusion protein with a dual His6/HA3-tag at its C-terminus. This fusion protein, expressed from the native promoter was also demonstrated to be fully functional (see Figure 1D). Upon biochemical fractionation, Rcf1-His6/HA3 was detected in the whole cell lysate, but was essentially absent from the post-mitochondrial supernatant (Figure 1B). The purified mitochondria fraction was subjected to Proteinase K digestion either with or without swelling to rupture the outer membrane (mitoplasts) or Triton X-100 to rupture all membranes. Rcf1 was partially degraded in mitoplasts and was completely degraded upon Triton X-100 treatment. This pattern is similar to that observed for Cox2, which is an integral protein of the mitochondrial inner membrane (Figure 1B). Independently, we found that this same Rcf1 fusion protein was stably associated with mitochondrial membranes even following high salt and bicarbonate extraction (Figure 1C). This is consistent with the bioinformatic prediction that Rcf1 and its relatives have two transmembrane domains, and are therefore likely to be integral membrane proteins (Figure S1A). Taken together, these results suggest that Rcf1 is an integral protein of the mitochondrial inner membrane as was found for one of the mammalian orthologs of RCF1 (Wang et al., 2006).
To begin to understand the role of Rcf1 in mitochondrial function, we generated an rcf1Δ mutant and subjected it to growth assays in comparison to a wild-type strain. The rcf1Δ mutant exhibited impaired growth on all non-fermentable carbon sources tested, including acetate (Figure 1D). This phenotype was exacerbated when the experiment was conducted at 37°C (Figure 1D, bottom), consistent with a destabilization of proteins or protein complexes in the rcf1Δ mutant. This slow growth phenotype on non-fermentable carbon sources is consistent with a partial failure in respiration in the rcf1Δ mutant. To directly assess respiration, we measured oxygen consumption in the wild-type and rcf1Δ mutant strains. The rcf1Δ mutant had roughly 30% of wild-type oxygen consumption on glucose medium and had less severe, but substantial, respiration defects on lactate and glycerol medium (Figure 1E and S1B). Together, these results demonstrate that Rcf1 is an integral protein of the mitochondrial inner membrane that is required for normal respiratory activity.
Rcf1 stably associates with Complex III and Complex IV of the electron transport chain
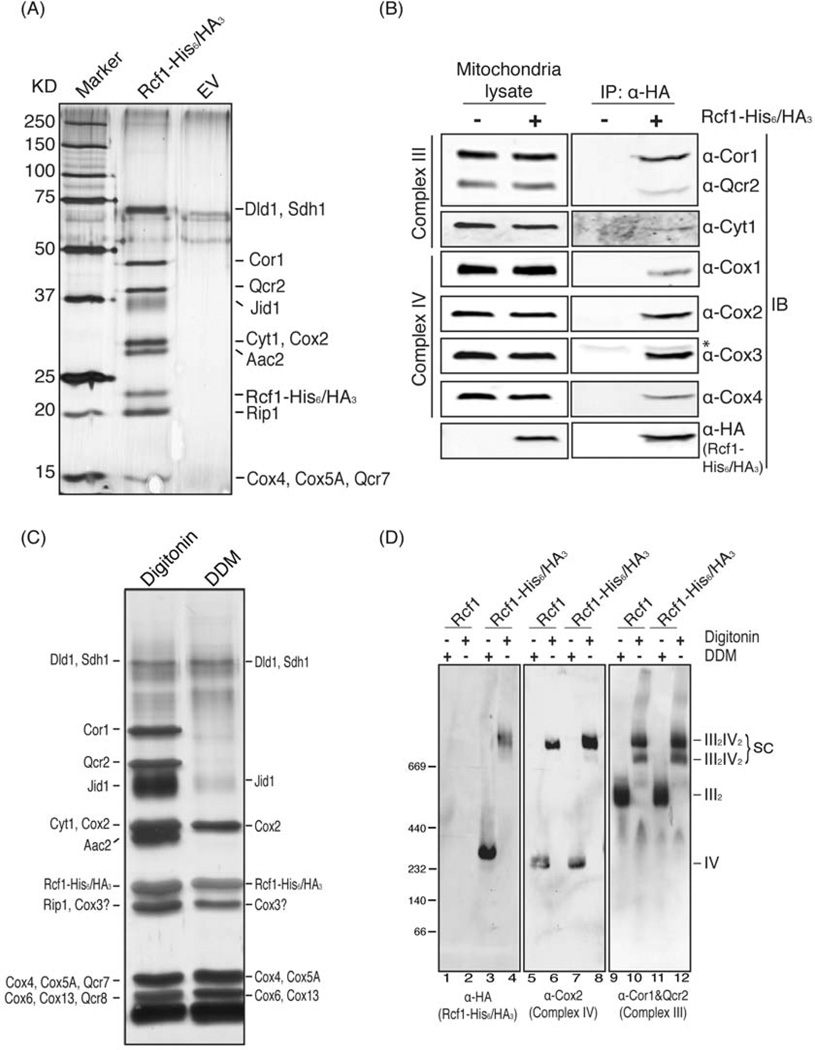
To determine the biochemical mechanism that underlies the role of Rcf1 in mitochondrial respiration, we identified proteins associated with Rcf1-His6/HA3 expressed at endogenous levels. After two-step purification, at least 9 discrete bands, which were absent in the empty vector negative control, were observed in the sample from the Rcf1-His6/HA3 strain (Figure 2A). These bands primarily contained subunits of either Complex III or Complex IV from the electron transport chain (Figure 2A and Table S1). We also observed the ATP/ADP carrier, Aac2, which was previously shown to associate with electron transport chain complexes (Dienhart and Stuart, 2008). Bands containing Dld1, Sdh1 and Jid1 were also observed. Co-purification of multiple components of both Complex III and Complex IV with Rcf1 was confirmed using immunoprecipitation of the Rcf1-His6/HA3 fusion protein (Figure 2B). Note that we observed a clear interaction with Cox3. Perhaps due to its sequence and chemical properties, Cox3 was not detected by mass spectrometry of the Rcf1 eluate (Table S1).
Figure 2. Rcf1 physically interacts with respiratory Complex III and IV.
(A) Mitochondria from an EV-containing wild-type strain or an rcf1Δ mutant strain expressing Rcf1-His6/HA3 were subjected to two-step purification. The final eluate was analyzed by silver staining. Discrete bands were excised and the protein identity was determined by mass spectrometry as indicated in the text. (B) Digitonin-solubilized mitochondria extracted from yeast strains either expressing Rcf1-His6/HA3 (+) or not (−) were immunoprecipitated using anti-HA antibody. One percent of the mitochondrial lysate and the final eluate were immunoblotted using anti-Cor1&Qcr2, Cyt1, Cox1, Cox2, Cox3, Cox4 and HA antibodies. * indicates IgG light chain. (C) Rcf1-His6/HA3 was purified from digitonin and DDM-solubilized mitochondria, respectively. The final eluate was visualized by silver stain and protein identities were determined by mass spectrometry. Cox3 is indicated based on SDS-PAGE/Western blot as it was not detected by mass spectrometry. (D) Mitochondria extracted from a strain harboring a plasmid expressing Rcf1 or Rcf1-His6/HA3 were solubilized by digitonin or DDM and subjected to BN-PAGE/Western blot. Rcf1, Complex III and Complex IV were immunoblotted by anti-HA, Cor1&Qcr 2 and Cox2 antibodies, respectively.
It is well known that Complex III and Complex IV physically associate with one another, forming respiratory supercomplexes (Schagger and Pfeiffer, 2000; Wittig and Schagger, 2009). Therefore, those two complexes simultaneously co-purifying with Rcf1 was not surprising. To further explore the inter-relationships between Rcf1, Complex III and Complex IV, we repeated the Rcf1-His6/HA3 purification experiment using both digitonin-solubilized and dodecylmaltoside (DDM)-solubilized mitochondria. While we again observed the same set of Complex III and Complex IV subunits in the digitonin sample, the Complex III subunits did not co-purify with Rcf1-His6/HA3 in the DDM-solubilized sample (Figure 2C). Digitonin has been shown to be mild enough to maintain Complex III/Complex IV associations in III2/IV and III2/IV2 supercomplexes, while DDM disrupts these interactions. This pattern of associations in the two detergents was confirmed by mass spectrometry of the entire eluate from the purifications. All complex IV subunits were observed in both digitonin and DDM, but Complex III subunits were only associated with Rcf1 in digitonin (Table S1). This implies that the interaction of Rcf1 with Complex IV is likely to be more stable than the interaction with Complex III. Interestingly, Complex II subunits also were detected in the Rcf1 eluate, albeit at apparently lower stoichiometry (Table S1). These observations were also confirmed using blue native-PAGE (BN-PAGE) electrophoresis. In DDM-solubilized mitochondrial lysates, Rcf1-His6/HA3 migrated in a complex of similar mobility to monomeric Complex IV, as exemplified by Cox2 (Figure 2D, lane 3). In digitonin, however, Rcf1-His6/HA3 primarily migrated at the position of the III2/IV2 heterotetrameric supercomplex (Figure 2D, lane 4). These data indicate that Rcf1 maintains strong association with Complex IV in either detergent condition, but loses the association with Complex III in DDM.
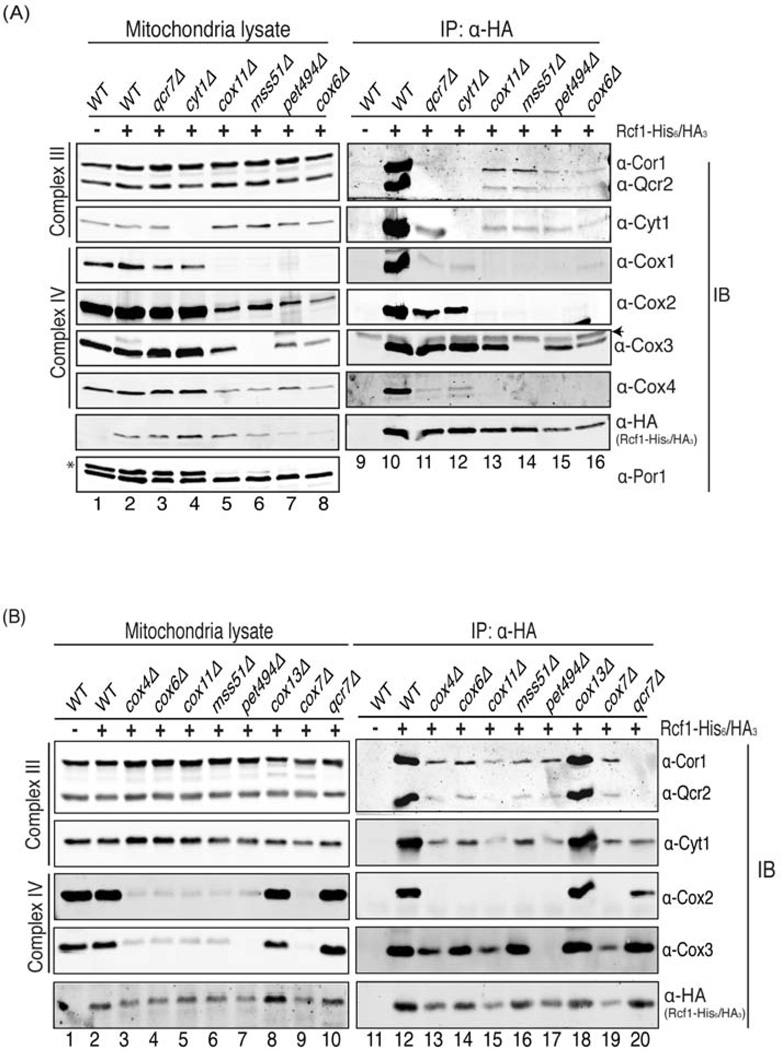
This pattern of associations between Rcf1 and the ETC complexes suggested two possibilities. One possible interpretation is that Rcf1 associates directly with Complex IV and the co-purification with Complex III is indirect, occurring via the Complex III/Complex IV interaction. The other possibility is that Rcf1 interacts with both complexes independently, perhaps even serving as a contact point between the two complexes. To systematically address this question, we monitored the association between Rcf1 and Complex III subunits (Cor1, Qcr2 and Cyt1) and between Rcf1 and Complex IV subunits (Cox1, Cox2, Cox3 and Cox4) in mutants that lead to the loss of either Complex III or Complex IV (Figure 3A). The co-purification of Complex IV with Rcf1 in DDM conditions was recapitulated in this assay. A qcr7Δ mutant causes severe failure of Complex III assembly and Rcf1 shows no association with Cor1 or Qcr2 in this mutant (Figure 3A-lane 11, 3B-lane 20), even though both Cor1 and Qcr2 are present in the mitochondrial lysate (Figure 3A-lane 3, 3B-lane 10). Interestingly, deletion of QCR7 does not destroy the interaction between Rcf1 and Cyt1, raising the possibility that Rcf1 might be more closely associated with Cyt1 than Cor1 or Qcr2. As expected, deletion of CYT1 also leads to the loss of Rcf1 interaction with Cor1 and Qcr2 (Figure 3A-lane 12). In both of these mutants, however, the Rcf1 interactions with Cox1, Cox2, Cox3 and Cox4 were maintained. For Cox1, Cox2 and Cox4, Rcf1 interactions were compromised, but remained significantly above background. Interestingly, the Rcf1/Cox3 interaction was not negatively affected by either the qcr7Δ or cyt1Δ mutations, suggesting that it might be more proximal to the Rcf1 interaction site than Cox1, 2 or 4. These data demonstrate that Rcf1 interacts stably with Complex IV even in the absence of association with Complex III.
Figure 3. Rcf1 interacts with Complex III and Complex IV independently. (Also see Figure S2).
(A) Rcf1 was immunoprecipitated using anti-HA antibodies from the digitonin-solubilized mitochondria of the indicated strains harboring either with (+) or without (−) a plasmid expressing Rcf1-His6/HA3. Both 1% crude lysate and final eluate were immunoblotted using anti-Cor1&Qcr2, Cyt1, Cox1, Cox2, Cox3, Cox4, HA and Por1 antibodies, respectively. Asterisk on the anti-Por1 blot indicates the Cox2 signal from a previous exposure and arrowhead on the anti-Cox3 blot indicates IgG light chain. (B) Mitochondria from the indicated strains were analyzed as in (A).
The more interesting question, however, is whether Rcf1 maintains an association with Complex III in the absence of Complex IV. To address this question, we monitored association between Rcf1 and Complex III subunits in four different mutants lacking properly assembled Complex IV. They each lead to distinct defects in the expression, translation or assembly of different Complex IV subunits. Deletion of COX11, which encodes a Cox1 assembly factor, causes impaired synthesis of Cox1 (Barrientos et al., 2004; Horng et al., 2004). Pet494 and Mss51 are translational activators of COX3 and COX1, respectively, and their deletion causes loss of their respective target (Costanzo and Fox, 1986; Perez-Martinez et al., 2003). Cox6 is a Complex IV subunit that might play a role in oxygen sensing and is indispensible for the stability and activity of Complex IV (Wright et al., 1995). None of these mutants has intact, properly assembled Complex IV. In all cases, however, Cor1, Qcr2 and Cyt1 co-purified with Rcf1, albeit at a reduced level compared to the wild-type strain (Figure 3A-lanes 13–16). As observed before, we again saw that Cox3 maintained an association with Rcf1 when other Complex IV subunits did not. In spite of this, depletion of Cox3, by mutation of PET494, did not prevent association of Rcf1 with Complex III subunits (Figure 3A-lane 15). Even after a stringent two-step purification, the same results were obtained for the wild-type, qcr7Δ and cox11Δ strains (Figure S2A and S2B).
The recent publication of higher resolution structures of respiratory supercomplexes (Althoff et al., 2011; Dudkina et al., 2011) has provided some insight into the more intimate relationship between Rcf1 and Cox3. These structures show that Cox3, Cox7 and Cox13 comprise the majority of the interface with Complex III. As shown above, loss of Cox3 failed to abolish the ability of Rcf1 to co-precipitate Complex III subunits (Figure 3B-lane 17). Additionally, neither loss of Cox7 or Cox13 abrogated the Rcf1/Complex III interaction (Figure 3B-lane 18, 19). Unlike the other Complex IV mutants, the cox13Δ mutant had no deleterious effect on the Rcf1 interaction with either Complex III subunits (Cor1, Qcr2 and Cyt1) or Complex IV subunits (Cox2 and Cox3). Therefore, even loss of those proteins that are likely to be most intimately associated with Rcf1 within Complex IV (see accompanying Vukotik, et al manuscript) doesn’t abolish the Rcf1 interaction with Cor1, Qcr2 and Cyt1. While it is possible that sub-complexes of Complex IV remain in any one of these mutants and could mediate the interaction between Rcf1 and Complex III, it is unlikely that the same sub-complex bearing that bridging activity is found in each of these many mutants. We, therefore, conclude that Rcf1 has the rather unusual property of interacting directly with components of Complex III and Complex IV, independent of association with the other.
Rcf1 promotes the stability of Complex III/Complex IV supercomplexes
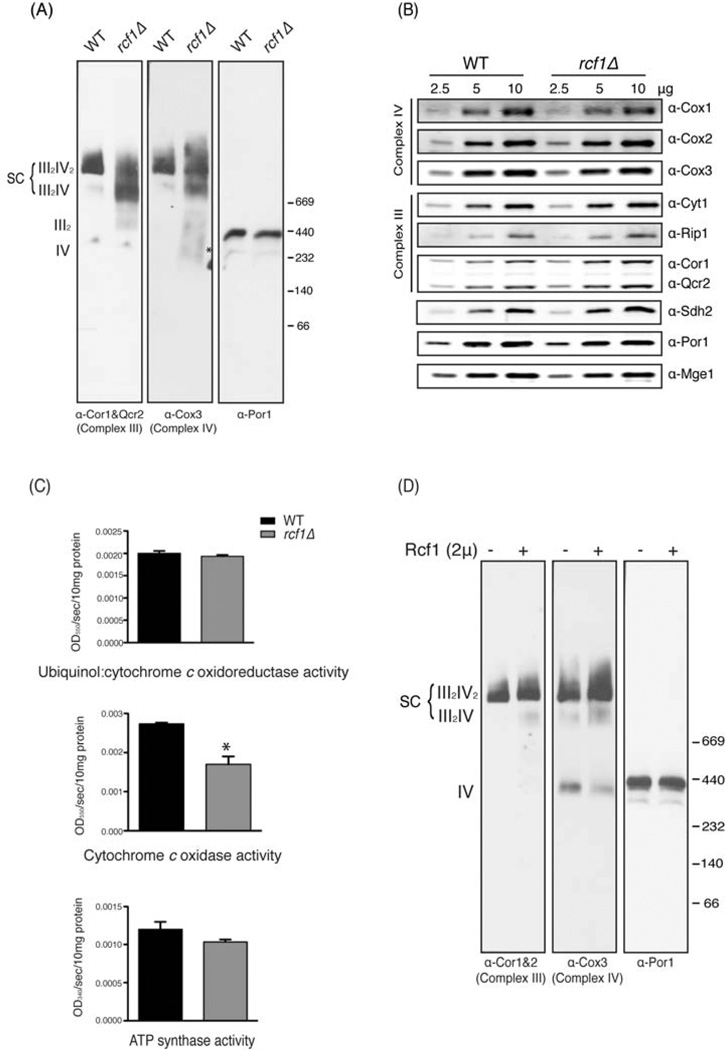
If Rcf1 interacts directly with both Complex III and Complex IV, and therefore serves as a point of contact between the two complexes, it is possible that an rcf1Δ mutant might exhibit defects in their association. BN-PAGE analysis was conducted on the wild-type and rcf1Δ mutant grown in raffinose medium to induce mitochondrial biogenesis. Complex III migration, as assessed by α-Cor1/Qcr2 immunoblot, in the wild-type strain was almost completely as the Complex III2/Complex IV2 tetramer (Figure 4A-left panel). A small amount of the III2/IV complex was observed, but there was no detectable free Complex III. Complex III in the rcf1Δ mutant, however, showed a loss of association with Complex IV. There was a shift from the fully assembled III2/IV2 complex toward the III2/IV complex and some free III2 dimer was detected. Examination of Complex IV (Cox3 immunoblot) showed a similar loss of the III2/IV2 complex and increase in the III2/IV complex and free Complex IV in the rcf1Δ mutant (Figure 4A-middle panel). A similar phenotype was observed for the rcf1Δ mutant in stationary phase cultures grown in glucose (Figure S3A). Again, we observed a shift from the fully assembled III2/IV2 complex toward the III2/IV and III2 complexes. We detected free Complex IV in the wild-type strain that was decreased in the rcf1Δ mutant, raising the possibility that Complex IV might be less stable to BN-PAGE analysis under some conditions in the absence of Rcf1. This appears to not be general, however, as we observed increased free Complex IV in the rcf1Δ mutant grown in raffinose medium (Figure 4A). The migration of Complex V was unaffected by loss of Rcf1 (Figure S3A). In both raffinose and glucose cultures, these effects on supercomplex organization occurred in the absence of any substantial effect on the steady-state levels of Complex III or Complex IV subunits (Figure 4B and S3B). While the rcf1Δ mutant may exhibit a defect in Complex IV assembly or stability, these data strongly suggest that there is a specific defect in the Complex III/Complex IV association.
Figure 4. Respiratory supercomplexes are impaired in the absence of RCF1. (Also see Figure S3).
(A) Mitochondria from wild-type and the rcf1Δ mutant grown to log phase in synthetic raffinose medium were analyzed by BN-PAGE/Western blot. Complex III and Complex IV were immunoblotted by anti-Cor1&Qcr2 and Cox3 antibodies, respectively. Porin serves as a control. (B) The mitochondria from (A) were subjected to SDS-PAGE and immunoblotted using Cox1, Cox2, Cox3, Cor1&Qcr2, Cyt1, Rip1,and Sdh2 antibodies. Por1 and Mge1 are loading controls. (C) Enzymatic activities of Complex III, Complex IV and Complex V were measured in lysates of the wild-type and rcf1Δ mutant grown in synthetic raffinose media to log phase. Each was measured in three independent cultures and data shown is mean + SD. *, P<0.05 (D) Mitochondria from the wild-type strain with or without a 2µ-based plasmid overexpressing Rcf1 grown in raffinose medium to log phase were analyzed by BN-PAGE/Western blot. Complex III, Complex IV and porin complex were immunoblotted by anti-Cor1&Qcr2, Cox3 and Por1 antibodies, respectively.
Measurement of complex activity following BN-PAGE was consistent with these results. As expected, Complex V/ATP synthase exhibited normal activity in the rcf1Δ mutant (Figure S3C, 4C). Total complex IV activity was decreased in the rcf1Δ mutant, and this impairment was due primarily to a loss of activity in the III2/IV2 complex (Figure S3C, 4C). The loss of Complex IV activity could be due to an intrinsic defect in Complex IV or due to loss of association with Complex III, which has been shown to result in Complex IV defects in mammalian systems (Acin-Perez et al., 2008; Gil Borlado et al., 2010). Quantitative assays confirmed these results and also showed that Complex III activity is unaffected by deletion of RCF1 (Figure 4C). We also noticed a slight but reproducible defect in the activity of Complex II in the rcf1Δ mutant (Figure S3C), possibly indicative that the interactions between Rcf1 and Complex II subunits (Table S1) have functional importance.
Rcf1 doesn’t appear to be a stoichiometric constituent of the Complex III/Complex IV supercomplex. When we immunoprecipitate Rcf1, we co-precipitate a smaller amount of both Complex III subunits (Cor1, Qcr2 and Cyt1) and Complex IV subunits (Cox1, Cox2, Cox3 and Cox4) than when we immunoprecipitate a typical Complex IV subunit, Cox4 (Figure S3D). Based on that and its apparent function, we reasoned that overexpression of Rcf1 might increase the amount of respiratory supercomplexes. As seen in Figure 4D, expression of Rcf1 from a multi-copy 2µ plasmid caused a subtle shift in the Complex IV migration pattern in cells grown in raffinose. Free Complex IV was lost and both the III2IV and the III2IV2 complexes were increased in abundance, which indicates that increased Rcf1 dosage can promote supercomplex formation. Although subtle, this effect is reproducible under distinct growth conditions— stationary phase in glucose medium (Figure S3E).
RCF1 genetically interacts with AAC2 and COX13 to stabilize respiratory supercomplexes
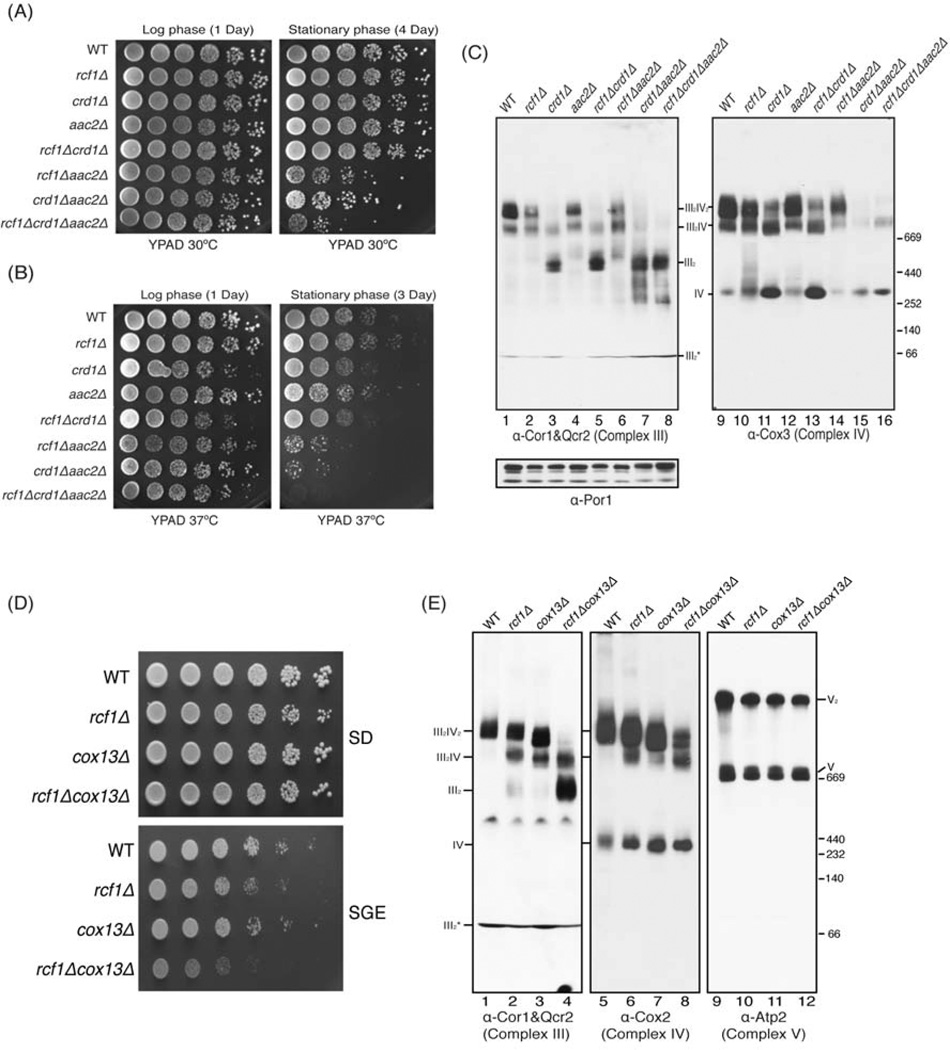
Two other molecules have been previously shown to be important for assembly of respiratory supercomplexes: the lipid cardiolipin and the ADP/ATP translocase Aac2. Loss of either of these molecules was found to destabilize supercomplexes (Dienhart and Stuart, 2008; Zhang et al., 2002). We wanted to examine the genetic relationship between RCF1, AAC2 and the gene encoding cardiolipin synthase CRD1, which is required for cardiolipin synthesis. Therefore, we constructed strains lacking these three genes in all possible combinations and examined their growth under different conditions. The aac2Δ strain exhibited a complete inability to grow on non-fermentable carbon sources, making it impossible to use such medium to examine respiratory growth (Figure S4A). As an alternative, we examined survival of these strains in stationary phase, which is also an indicator of respiratory capability (Gray et al., 2004). When grown at 30°C to log phase, all mutants exhibited normal survival (Figure 5A, left panel). Even at day 4 of stationary phase, the three single mutants showed no loss of viability, nor did the crd1Δ rcf1Δ double mutant. Both of the other double mutants, however, showed a substantial defect in survival and the triple mutant was even more severely affected (Figure 5A, right panel). When grown at 37°C, the same general pattern was evident, but the synergistic growth defects were more pronounced. These data demonstrated that loss of the RCF1 and CRD1 genes does not result in a synergistic or even additive phenotype. One possible explanation is that these two genes act in the same pathway to promote respiration, a conclusion that is supported by biochemical data described below. On the other hand, loss of AAC2 causes a synergistic survival defect with both the rcf1Δ and crd1Δ mutants. Such a result is consistent with Aac2 acting in parallel with both Rcf1 and Crd1 to promote supercomplex assembly or stability. Both Aac2 and cardiolipin have significant roles outside of supercomplex stabilization, which makes it difficult to draw mechanistic conclusions from genetic interactions.
Figure 5. RCF1 genetically interacts with AAC2 and COX13 to stabilize respiratory supercomplexes. (Also see Figure S4).
The indicated strains were grown in YPAD media to log or stationary phase as indicated were spotted on YPAD plates and incubated at 30°C (A) or 37°C (B). (C) Mitochondria from the indicated strains grown in raffinose medium to log phase were analyzed by BN-PAGE/Western blot. Complex III, Complex IV and porin complex were immunoblotted by anti-Cor1&Qcr2, Cox3 and Por1 antibodies, respectively. III2* indicates a Complex III intermediate. (D) The indicated strains were spotted on SD and SGlycerol/ethanol plates and incubated at 30°C. (E) Mitochondria from the indicated strains grown in 1% glucose medium for 1 day were analyzed by BN-PAGE/Western blot. Complex III, Complex IV and Complex V were immunoblotted by anti-Cor1&Qcr2, Cox2 and Atp2 antibodies, respectively. III2* indicates a Complex III intermediate.
To directly assess the assembly and stability of respiratory supercomplexes in these mutants, we subjected the same strains to BN-PAGE analysis after growth in raffinose. As observed before, the rcf1Δ mutant showed depletion of the III2/IV2 complex as determined both by Complex III and Complex IV immunoblot (Figure 5C-lanes 2, 10). The aac2Δ mutant exhibited a very similar phenotype (Lanes 4, 12). The crd1Δ single mutant showed a stronger defect, with an almost complete absence of the III2/IV2 complex and a significant amount of the III2 and IV species (Lanes 3, 11). As seen in the growth assays, the crd1Δ rcf1Δ double mutant showed no additive phenotype and was nearly identical to the crd1Δ single mutant (Lanes 5, 13). The aac2Δ rcf1Δ double mutant on the other hand showed a more substantial loss of the III2/IV2 complex than either single mutant (Lanes 6, 14). There was a clear synthetic phenotype in the aac2Δ crd1Δ double mutant, which had a nearly complete loss of all supercomplexes (Lanes 7, 15)). While deletion of AAC2 and CRD1 caused a decrease in the steady-state levels of some Complex IV subunits, deletion of RCF1 caused had no effect on any Complex III or Complex IV subunits even in the context of double mutants that had a synthetic effect on supercomplex organization (Figure S4B). Similar phenotypes were also observed in BN-PAGE analysis of these strains grown to stationary phase in glucose (Figure S4C). Another factor that occupies the predicted interface between Complex III and Complex IV is Cox13. In isolation, the cox13Δ mutant exhibits wild-type growth on both glucose and glycerol/ethanol plates (Figure 5D). The cox13Δ rcf1Δ double mutant, however, has a more dramatic growth phenotype than the rcf1Δ single mutant when grown under respiratory conditions (Figure 5D). This growth phenotype is accompanied by a severe loss of higher-order respiratory supercomplexes as determined by BN-PAGE analysis on the strain grown in different conditions. The cox13Δ single mutant shows depletion of the III2/IV2 complex similar to that observed in the rcf1Δ mutant (Figure 5E). The cox13Δ rcf1Δ double mutant, however, shows almost a complete loss of the intact III2/IV2 supercomplex and a marked shift to lower order complexes.
Increased mitochondrial oxidative stress in the rcf1Δ mutant
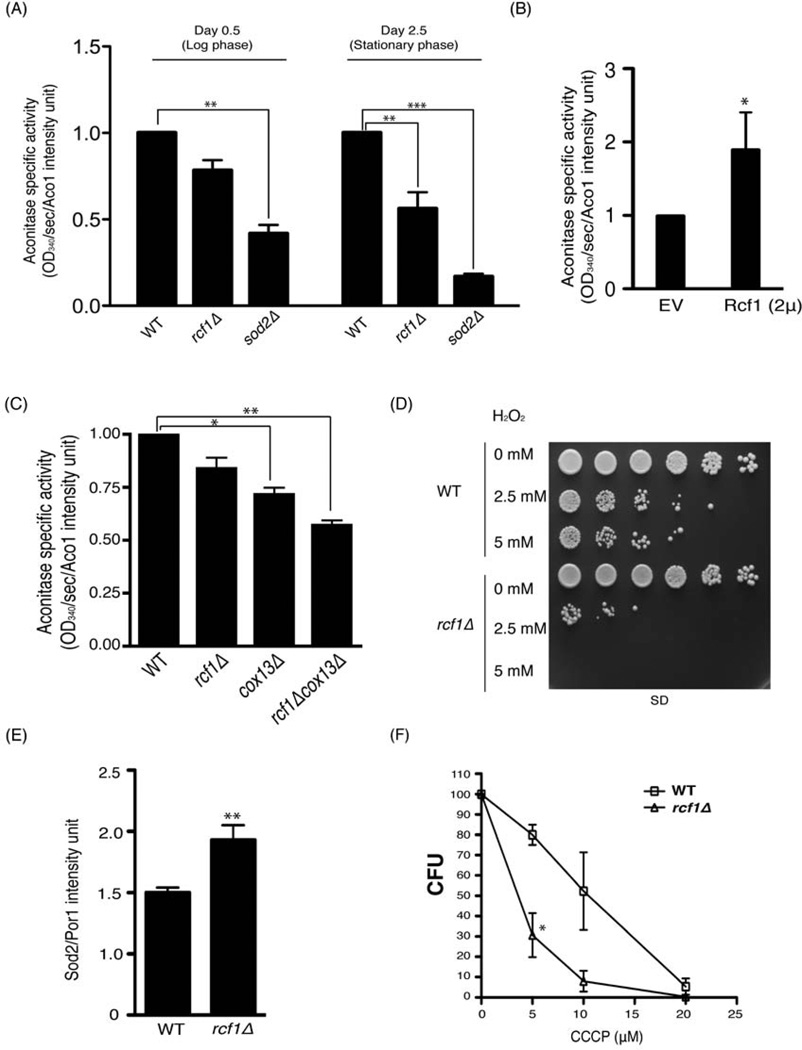
The physiological importance of mitochondrial ETC supercomplexes has not been precisely defined. It has been speculated, however, that the assembly of individual complexes into supercomplexes enables more efficient electron flow and decreases the risk of electron stalling. One predicted physiological manifestation of reduced electron stalling would be decreased generation of reactive oxygen species (ROS) by the ETC. As a means of assessing the level of oxidative damage in the mitochondrial matrix of the rcf1Δ mutant, we measured the activity of aconitase. This mitochondrial enzyme is highly susceptible to inactivation by ROS due to oxidation of an exposed Fe/S cluster. Therefore, aconitase activity is a measure of the in vivo burden of oxidative stress and damage in the mitochondrial matrix (Criscuolo et al., 2005; Gardner et al., 1995). In log phase cultures, we observed a ~20% decrease in aconitase activity in the rcf1Δ mutant, compared to a ~60% decrease in activity in a mutant lacking the mitochondrial Mn2+-dependent superoxide dismutase, Sod2 (Figure 6A). In stationary phase cultures, which are subject to enhanced respiratory activity and elevated oxidative stress, the aconitase activity was more severely affected in both mutants. We previously showed that overexpression of RCF1 caused a modest stabilization of higher-order supercomplex structures. RCF1 overexpression also caused a nearly 2-fold increase in aconitase activity (Figure 6B). As described previously, additional loss of COX13 confers a synergistic growth and supercomplex assembly phenotype upon the rcf1Δ mutant strain. The cox13Δ rcf1Δ double mutant also exhibited a more profound loss of aconitase activity relative to either of the two single mutants (Figure 6C).
Figure 6. Increased mitochondrial oxidative stress in the absence of RCF1. (Also see Figure S5).
(A) Cell lysates of the indicated strains grown in SD media for 0.5 and 2.5 days were subjected to aconitase activity assay and immunoblotted with anti-Aco1 antibody. Aconitase activity was determined specifically by normalizing NADP reduction rate to Aco1 protein band intensity. sod2Δ was used as a control. Mean ± SD of three independent cultures is shown. **, P<0.01; ***, P<0.0005 (B) The wild-type strain harboring either EV or a 2µ-based plasmid overexpressing Rcf1 were grown for 1 day in 1% glucose and analyzed as in (A). Mean ± SD of three independent cultures is shown. *, P<0.05 (C) The indicated strains were harvested after being grown in SD media for 0.5 day and analyzed as in (A). Mean ± SD of three independent cultures is shown. *, P<0.05; **, P<0.01 (D) Log phase cultures (0.5 day) of the wild-type and rcf1Δ mutant were pre-treated with H2O2 for 2 hours, spotted on synthetic glucose medium and incubated at 30°C. (E) Log phase cultures (0.5 day) of the indicated strains were harvested and immunoblotted for Sod2, which was normalized to Por1 intensity. Mean ± SD of six independent cultures is shown. **, P<0.01 (F) Wild-type and the rcf1Δ mutant strains of the BY4741 background were grown to log phase in glucose media and equal number of cells were plated on media containing the indicated concentration of CCCP. Number of colonies was counted after 48 hours. Mean ± SD of three independent cultures is shown. *, P<0.05
We also monitored the susceptibility of wild-type and mutant strains to exogenous hydrogen peroxide. Mutants with higher in vivo production of ROS are typically more sensitive to this exogenous stress. Compared to wild-type, the dose-dependent lethality in response to hydrogen peroxide is exacerbated in the rcf1Δ mutant (Figure 6D). In addition, we also measured the expression of the endogenous ROS defense system, as exemplified by the mitochondria-specific superoxide dismutase Sod2. The rcf1Δ mutant has elevated Sod2 protein levels relative to the wild-type control (Figure 6E)., which is consistent with this mutant having elevated endogenous oxidative stress, particularly in mitochondria. We hypothesize that this increased oxidative damage and sensitivity are results of accelerated ROS production from the electron transport chain. This ETC dysfunction is a predictable result of impaired assembly of individual complexes into supercomplexes, which promotes reliable, solid-state electron transmission. As another manifestation of ETC dysfunction, the rcf1Δ mutant exhibited impaired maximal respiration in the presence of CCCP, which causes dissipation of the mitochondrial membrane potential and a compensatory acceleration of respiration in wild-type cells (Figure S5-compare Figure 1E). The impaired stimulated respiration in the rcf1Δ mutant was accompanied by markedly increased sensitivity to CCCP-induced cell death (Figure 6F).
The HIG2A mammalian homolog of RCF1 plays a role in supercomplex stability
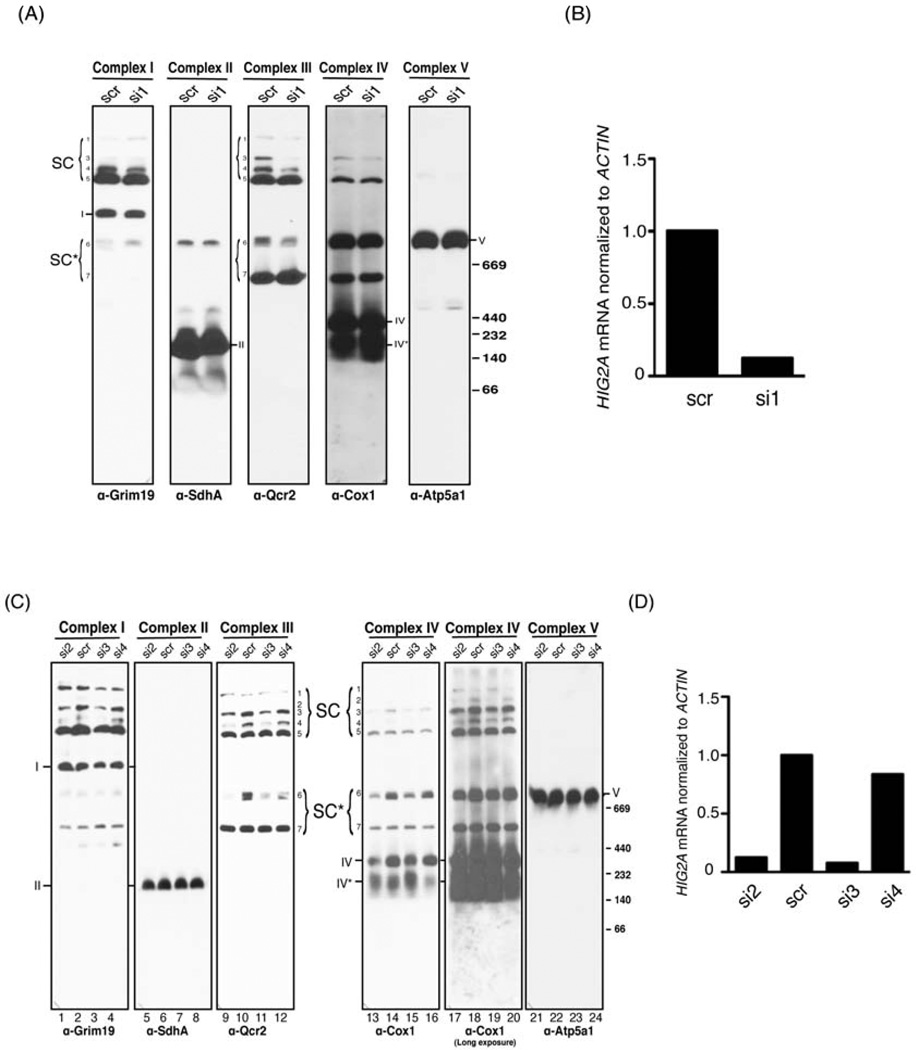
As previously described, our selection of RCF1 for detailed study was based partially upon evolutionary conservation amongst eukaryotes. We were interested to determine whether the role that we discovered for Rcf1 in supercomplex organization might also extend to RCF1 orthologs in other species. The mouse and human genomes contain five homologs of RCF1, which are sub-divided into two classes defined by the two genes with the broadest expression pattern, HIG1A and HIG2A (Figure S1A). HIG1A was originally described as being strongly inducible by hypoxia in a HIF-1-dependent manner (Denko et al., 2000; Kasper and Brindle, 2006). To determine whether either HIG1A or HIG2A were required for normal supercomplex assembly or stability in mammalian cells, we transfected cells with siRNAs targeted to these two genes. Knockdown of HIG1A (Figure S6B) had no effect on the pattern of supercomplex assembly (Figure S6A), which is more complicated in mammalian cells due to the presence of Complex I and the stable incorporation of both Complex II and Complex V (Acin-Perez et al., 2008). Knockdown of HIG2A (Figure 7B), on the other hand, caused a depletion of all higher order supercomplexes that contain Complex IV, particularly the I+II+III+IV (#3), I+III+IV (#4) and III+IV (#6) species (Figure 7A) (Acin-Perez et al., 2008). Supercomplex species that do not contain Complex IV, like the two I+III+V complexes (#1 and #5), were either unaffected or slightly increased upon HIG2A depletion (Figure 7A). To more rigorously evaluate a role for HIG2A and its encoded protein in supercomplex stability. We generated three additional siRNAs targeted to HIG2A, two of which caused significant depletion of the HIG2A mRNA and the other did not (Figure 7D). The two additional efficacious siRNAs (Si2 and Si3) caused an identical phenotype to that observed for Si1 (Figure 7C). Specifically, we again observed a loss of supercomplex species containing Complex IV and either unaffected or increased levels of Complex IV-independent species. At longer exposure of the anti-Cox1 immunoblot, we also saw a profound depletion of the #2 I+II+II+IV species in Si2 and Si3-treated cells (Figure 7C-5th panel). We observed a slight reduction in the amount of free, fully assembled Complex IV and a slight increase in an incomplete Complex IV (IV*). While these results demonstrate a role for HIG2A in C2C12 myoblast cells, it is possible that the four additional mammalian HIG family members serve a related function in different cell types or different conditions. Determining their precise roles and contributions in Complex IV and supercomplex assembly and stability requires additional study.
Figure 7. Mammalian HIG2A is critical for normal respiratory supercomplex organization. (Also see Figure S6).
(A) Mitochondria extracted from C2C12 cells transfected with either scrambled (scr) or HIG2A-targeted siRNA (si1) were solubilized in 2% digitonin and resolved by BN-PAGE, followed by immunoblotting with the indicated antibodies. Respiratory complex species are indicated following the convention of (Acin-Perez et al., 2008), specifically: 1 (I+III+V); 2 (I+II+III+IV); 3 (I+II+III+IV); 4 (I+III+IV); 5 (I+III+V); 6 (III+IV); 7 (III+IV)*. (B) HIG2A mRNA level normalized to ACTIN was measured from cells in (A) using quantitative RT-PCR. (C) Mitochondria extracted from C2C12 cells transfected with scrambled (scr) or one of three unique HIG2A-targeted siRNA (si2, 3, 4) were treated as in (A). A longer exposure of the anti-Cox1 immunoblot is shown to enable visualization of the supercomplexes #1, 2 and 4. Note that si4 is not efficacious in HIG2A knockdown. (D) HIG2A mRNA level normalized to ACTIN was measured from cells in (C) using quantitative RT-PCR.
DISCUSSION
Based on the data presented herein, we conclude that Rcf1 is a component of respiratory supercomplexes and is required for their normal stability. We also conclude that Rcf1 is not essential for basal mitochondrial function, but is essential for optimally efficient ETC function and respiration. Evidence that supports these conclusions will be detailed below.
First, Rcf1 directly interacts with respiratory supercomplexes. When we purified Rcf1, we co-purified stoichiometric amounts of both Complex III and Complex IV, the major components of S. cerevisiae ETC supercomplexes. We considered the alternative explanation that Rcf1 is a previously unidentified component of Complex IV. The association with Complex III could be indirect, occurring only as a consequence of the interaction between Complex III and Complex IV. The fact that extraction of mitochondria with the relatively harsh detergent DDM causes the Rcf1/Complex III association to be lost could be seen as supporting this view. However, none of the seven distinct COX mutants that we have analyzed, six of which exhibit failed assembly of Complex IV and the destruction of many of its subunits, have lost the association of Rcf1 with Complex III subunits. Interactions of Rcf1 with Cor1, Qcr2 and Cyc1 are maintained in spite of the fact that Rcf1 has lost all detectable interaction with Cox1, Cox2, Cox3, Cox4 and presumably other Complex IV subunits for which we don’t have usable antibodies. Similarly, loss of the Complex III subunit Cyt1 leads to a complete failure of Rcf1 to interact with central Complex III subunits Cor1 and Qcr2, but interactions with Complex IV subunits are maintained.
In general, while mutations impairing either Complex III or IV assembly don’t eliminate the interaction of Rcf1 with the other Complex, the interactions are significantly weakened. One exception to this is the Complex IV subunit, Cox3. The qcr7Δ and cyt1Δ (Complex III) mutations eliminate the interaction of Rcf1 with the core Cor1 and Qcr2 subunits of Complex III. They also greatly impair the Rcf1 interaction with Complex IV subunits Cox1 and Cox4, and to a lesser extent, Cox2. The Rcf1 interaction with Cox3, however, is essentially unaffected. The Rcf1/Cox3 interaction also seems to be unusually persistent in the face of Complex IV destruction. The cox11Δ, mss51Δ and cox6Δ (all Complex IV) mutations each eliminate the interaction of Rcf1 with Cox1, Cox2 and Cox4, but the interaction with Cox3 is maintained, albeit at a reduced level. In spite of this seemingly special relationship between Cox3 and Rcf1, loss of Pet494, which completely blocks the expression of Cox3, does not destroy the interaction of Rcf1 with Complex III.
The data demonstrating an intimate association between Rcf1 and Cox3 has been confusing until recently. If Rcf1 interacts independently with both Complex III and Complex IV, it must occupy a position close to the interface between the two in the respiratory supercomplex. The structure of the III2/IV2 supercomplex from S. cerevisiae, however, suggested that Cox3 was situated on the face of Complex IV that was opposite to the Complex III interaction site (Heinemeyer et al., 2007). This model was based on low-resolution data and the rotational orientation of Complex IV was quite speculative. Recently, much higher resolution electron microscopy reconstructions of the bovine I/III2/IV supercomplex were completed and published (Althoff et al., 2011; Dudkina et al., 2011). These data clearly show the Complex IV orientation to be rotated relative to Complex III, placing Cox3 at the site of the Complex III/Complex IV interface. Interestingly, the docking of the crystal structures of Complex III and Complex IV into the EM model demonstrated a clear gap between the two complexes, particularly in the juxta-membrane region (Dudkina et al., 2011). Based on its small size and two transmembrane domains, this is the region that should be occupied by Rcf1. It is possible that in addition to cardiolipin, which was postulated to occupy this position, Rcf1 fills this space and stabilizes the Complex III/Complex IV interaction.
Second, RCF1 has a robust genetic interaction with AAC2, which has been previously implicated in stabilizing ETC supercomplexes (Dienhart and Stuart, 2008). In assays of stationary phase survival, RCF1 and AAC2 exhibited a profound synthetic phenotype, particularly at elevated temperature. Such a growth phenotype is potentially due to destabilization of supercomplexes as we showed using blue native-PAGE. AAC2 has a nearly identical relationship with CRD1, which encodes cardiolipin synthase and is necessary for the synthesis of this lipid. Like Aac2, cardiolipin has also been implicated in the stability of respiratory supercomplexes. There was no additivity, on the other hand, between RCF1 and CRD1. While we don’t yet know how these molecules inter-relate in their stabilization of supercomplexes, these varied genetic interactions imply that they don’t all function identically or independently.
We also observed a strong genetic interaction between RCF1 and COX13. The Cox13 protein is predicted to occupy the interface between Complex III and Complex IV. Our data shows a modest loss of the III2/IV2 supercomplexes in the cox13Δ single mutant, which is dramatically exacerbated in the cox13Δ rcf1Δ double mutant. This synergistic loss of supercomplexes is accompanied by a synthetic growth defect and accelerated generation of oxidative stress, as determined by loss of aconitase activity. These phenotypes are consistent with the proposed role of Rcf1 in supercomplex stabilization and with the proposed role of supercomplexes in prevention of oxidant production.
Finally, loss of Rcf1 leads to a loss of the fully assembled III2/IV2 supercomplexes and an increase in lower order structures. The majority of Complex III and IV appears to assume the III2/IV assemblage, but a significant amount of both complexes dissociate completely from the other. It is important to note that this effect seems to be specific as assembly of the other OXPHOS complexes is not impaired. HIG2A, one of the mammalian homologs of RCF1, appears to play a similar role in mouse cells. Treatment with any of three siRNAs that deplete the HIG2A mRNA causes a reduction of all Complex IV-containing respiratory supercomplexes.
While we have not explored this possibility to date, it is likely that the extent and nature of respiratory supercomplexes is tightly regulated and responsive to environmental conditions. For example, we predict that nutrient availability, oxygen concentration and ATP demand are all likely stimuli for adjustment of supercomplex status. This could be accomplished through the transcriptional or post-transcriptional regulation of RCF1 or its homologs in other species. The mouse ortholog of RCF1 whose depletion had no effect on supercomplex organization in C2C12 cells, HIG1A, is robustly induced by hypoxia (Denko et al., 2000). Perhaps HIG1A plays a more prominent role under hypoxia or in different cells. Indeed it has been shown to protect pancreatic β-cells from apoptosis in response to a variety of stimuli (Wang et al., 2006). Due to its relatively simple supercomplex content, S. cerevisiae has only one interaction that can be targeted for regulation, between Complex III and Complex IV. In mammals, however, a much more complicated pattern exists. Complexes I, II, III, IV and V are all components of respiratory supercomplexes and their inclusion and stoichiometry is variable (Acin-Perez et al., 2008). The makeup of these complexes is a likely target of metabolic regulation.
We certainly don’t fully understand the consequences of the loss of supercomplex formation. We do observe, however, that there is a substantial reduction in respiratory activity as measured by total oxygen consumption in the rcf1Δ mutant. Perhaps of more interest, it appears that the rcf1Δ mutant also has elevated oxidative stress and damage in the mitochondrial matrix. This was determined by the loss of aconitase activity, hypersensitivity to exogenous hydrogen peroxide and an upregulation of the endogenous oxidative stress defenses. These are common phenotypes amongst strains that are combatting an elevated level of endogenous oxidative stress. One of the factors that have kept us from an understanding of the effects of supercomplex assembly and disassembly is an inability to genetically manipulate supercomplex formation without disrupting other mitochondrial functions. To the best that we can determine, RCF1 deletion causes a specific defect in supercomplex formation in yeast and HIG2A silencing does the same in mouse cells. These will be valuable tools to understand the role and importance of the assembly of these intricate machines.
EXPERIMENTAL PROCEDURES
Assessment of sub-mitochondrial localization
These experiments were performed following a protocol adapted from (Boldogh and Pon, 2007). Mitochondria were incubated in the isotonic SH buffer or hypertonic H buffer (20mM Hepes-KOH) with and without 1% Triton X-100. Protease K was added and incubated on ice for 20–30 min and digestion was stopped by adding PMSF. Samples were denatured in 6X Laemmli buffer and resolved by 12% SDS-PAGE, followed by immunoblot. To assess the solubility of Rcf1, intact mitochondria were extracted by 100 mM Na2CO3 and 1 M KCl for 20 min on ice. Supernatant was isolated by centrifuging samples at 100,000 × g for 20 min (MLA-130, Beckman Coulter) and precipitated in 15% TCA. The precipitated supernatant fraction and insoluble membrane fraction were solubilized in 1X Laemmli buffer, and resolved by 12% SDS-PAGE followed by immunoblot.
Two-step Rcf1-His6/HA3 purification
Crude mitochondria isolated from strains grown in synthetic glycerol/ethanol medium were solubilized in 0.8% digitonin or dodecylmaltoside (DDM) for 1 hour at 4°C. For the first step of the purification, cleared mitochondria lysate was incubated with equilibrated Ni-NTA beads for 1 hour at 4°C. Beads were washed 5 times with buffer containing 20 mM imidazole. Protein was eluted by 250 mM imidazole. For the second step of the purification, final eluates of the first purification were mixed with anti-HA antibody conjugated agarose (Sigma) for 1 hour at 4°C. Agarose was washed 5 times and eluted by 1 mg/ml HA peptide. Eighty percent of the final precipitate was dissolved in Laemmli buffer and resolved by SDS-PAGE followed by silver stain. Protein bands excised from the gel and the rest of 20% of the final precipitate were analyzed by mass spectrometry.
Blue-Native polyacrylamide gel electrophoresis (BN-PAGE)
BN-PAGE was performed as described previously (Wittig et al., 2006). Yeast mitochondria were solubilized in 1% digitonin or DDM and mammalian mitochondria were solubilized in 2% digitonin. Lysate was resolved on a 3–13% gradient native gel using a PROTEAN® II xi Cell gel running system (Bio-rad). Western blot was performed as the regular procedure using a Trans-Blot® transfer cell (Bio-rad). Membrane was blocked in 5% non-fat milk/TBS and probed with antibodies as indicated.
Solution ETC complex activity assay
These experiments were performed following a protocol adapted from (Franco et al., 2010). Ubiqinol: cytochrome c oxidoreductase (Complex III) activity was determined by measuring the rate of cytochrome c reduction by ubiquinol at 550nm. Complex III specific activity was calculated by deducing the rate of cytochrome c reduction of a parallel reaction with antimycin A. Cytochrome c oxidase (Complex IV) activity was determined by measuring the rate of cytochrome c oxidation. Complex IV specific activity was verified by adding KCN into the reaction mixture. ATP synthase (Complex V) activity was determined by measuring the rate of NADH oxidation. ATP synthase specific activity was verified by adding oligomycin to stop the reaction. The specific Complex V activity was calculated by deducing the rate of NADH oxidation in the presence of oligomycin.
Isolation of mammalian mitochondria
The procedure was adapted from (Bozidis et al., 2007). C2C12 cells were washed twice with PBS, scraped from dishes and pelleted. Roughly 150 mg cell pellet was resuspended in 2 ml of ice-cold MTE buffer. Cells were lysed by sonication (continuous pulse on power setting of 3.5) 3 times for 10 sec each. Cell debris and nuclei were pelleted at 1,400 × g for 10 min. Crude mitochondria were recovered from the supernatant fraction by centrifuging at 10,000 × g for 10 min. Mitochondrial pellet was rinsed with MTE and resuspended in it. Protein concentration was determined by Bradford assay.
Statistics
Data are presented as the means ±standard deviation (SD). Statistical significance was evaluated by the Student t test. *, P<0.05; **, P<0.01; ***, P<0.0005
Electron transport chain complexes associate into respiratory supercomplexes
Rcf1, a pan-eukaryotic inner membrane protein, binds Complexes III and IV independently
Rcf1 is required for normal supercomplex organization in yeast and mammals
Loss of Rcf1 causes increased oxidative damage.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the Rutter laboratory as well as the laboratories of Dennis Winge, Tim Formosa, David Stillman, Janet Shaw, Jindrich Kopecek, Jerry Kaplan and Vincenzo Zara for technical support and helpful discussions. We especially thank the Winge lab and Oleh Khalimonchuk for many ETC antibodies, protocols and instruction on BN-PAGE and oxygen consumption assays. We thank the Shaw lab for the anti-Fzo1, Mge1 antibodies and mito-RFP constructs and fluorescence microscopy and the Kopecek lab for the preparation of DBH2. We also thank Kaplan lab for the anti-Sod2 antibody and Dr. Vincenzo Zara for the anti-Cor1&Qcr2, Rip1 antibodies. This work was supported by NIH grants RO1GM087346 (to JR) and R24DK092784 (to JR).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yu-Chan Chen, Department of Biochemistry, University of Utah School of Medicine.
Eric B. Taylor, Department of Biochemistry, University of Utah School of Medicine
Noah Dephoure, Department of Cell Biology, Harvard Medical School.
Jin-Mi Heo, Department of Biochemistry, University of Utah School of Medicine.
Aline Tonhato, Department of Biochemistry, University of Utah School of Medicine.
Ioanna Papandreou, Division of Radiation and Cancer Biology, Department of Radiation Oncology at Stanford University School of Medicine.
Nandita Nath, Division of Radiation and Cancer Biology, Department of Radiation Oncology at Stanford University School of Medicine.
Nicolas C. Denko, Division of Radiation and Cancer Biology, Department of Radiation Oncology at Stanford University School of Medicine
Steven P. Gygi, Department of Cell Biology, Harvard Medical School
Jared Rutter, Department of Biochemistry, University of Utah School of Medicine.
LITERATURE CITED
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I(1)III(2)IV(1) EMBO J. 2011 doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. Embo J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayley JP, Kunst HP, Cascon A, Sampietro ML, Gaal J, Korpershoek E, Hinojar-Gutierrez A, Timmers HJ, Hoefsloot LH, Hermsen MA, et al. SDHAF2 mutations in familial and sporadic paraganglioma and phaeochromocytoma. Lancet Oncol. 2010 doi: 10.1016/S1470-2045(10)70007-3. [DOI] [PubMed] [Google Scholar]
- Boldogh IR, Pon LA. Purification and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2007;80:45–64. doi: 10.1016/S0091-679X(06)80002-6. [DOI] [PubMed] [Google Scholar]
- Bozidis P, Williamson CD, Colberg-Poley AM. Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Curr Protoc Cell Biol. 2007;Chapter 3(Unit 3):27. doi: 10.1002/0471143030.cb0327s37. [DOI] [PubMed] [Google Scholar]
- Costanzo MC, Fox TD. Product of Saccharomyces cerevisiae nuclear gene PET494 activates translation of a specific mitochondrial mRNA. Mol Cell Biol. 1986;6:3694–3703. doi: 10.1128/mcb.6.11.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criscuolo F, Gonzalez-Barroso Mdel M, Le Maho Y, Ricquier D, Bouillaud F. Avian uncoupling protein expressed in yeast mitochondria prevents endogenous free radical damage. Proc Biol Sci. 2005;272:803–810. doi: 10.1098/rspb.2004.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA. The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem. 2000;275:18093–18098. doi: 10.1074/jbc.M001901200. [DOI] [PubMed] [Google Scholar]
- Denko N, Schindler C, Koong A, Laderoute K, Green C, Giaccia A. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin Cancer Res. 2000;6:480–487. [PubMed] [Google Scholar]
- Dienhart MK, Stuart RA. The yeast Aac2 protein exists in physical association with the cytochrome bc1-COX supercomplex and the TIM23 machinery. Mol Biol Cell. 2008;19:3934–3943. doi: 10.1091/mbc.E08-04-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci U S A. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco T, Marco R, Stefania M, Valentina F. Preparation of yeast mitochondria and <em>in vitro</em> assay of respiratory chain complex activities. 2010 [Google Scholar]
- Gardner PR, Raineri I, Epstein LB, White CW. Superoxide radical and iron modulate aconitase activity in mammalian cells. J Biol Chem. 1995;270:13399–13405. doi: 10.1074/jbc.270.22.13399. [DOI] [PubMed] [Google Scholar]
- Genova ML, Baracca A, Biondi A, Casalena G, Faccioli M, Falasca AI, Formiggini G, Sgarbi G, Solaini G, Lenaz G. Is supercomplex organization of the respiratory chain required for optimal electron transfer activity? Biochim Biophys Acta. 2008;1777:740–746. doi: 10.1016/j.bbabio.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Gil Borlado MC, Moreno Lastres D, Gonzalez Hoyuela M, Moran M, Blazquez A, Pello R, Marin Buera L, Gabaldon T, Garcia Penas JJ, Martin MA, et al. Impact of the mitochondrial genetic background in complex III deficiency. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. "Sleeping beauty": quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2004;68:187–206. doi: 10.1128/MMBR.68.2.187-206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao HX, Khalimonchuk O, Schraders M, Dephoure N, Bayley JP, Kunst H, Devilee P, Cremers CW, Schiffman JD, Bentz BG, et al. SDH5, a gene required for flavination of succinate dehydrogenase, is mutated in paraganglioma. Science. 2009;325:1139–1142. doi: 10.1126/science.1175689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemeyer J, Braun HP, Boekema EJ, Kouril R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, et al. A stress-responsive system for mitochondrial protein degradation. Mol Cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng YC, Cobine PA, Maxfield AB, Carr HS, Winge DR. Specific copper transfer from the Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J Biol Chem. 2004;279:35334–35340. doi: 10.1074/jbc.M404747200. [DOI] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK. Mammalian gene expression program resiliency: the roles of multiple coactivator mechanisms in hypoxia-responsive transcription. Cell Cycle. 2006;5:142–146. doi: 10.4161/cc.5.2.2353. [DOI] [PubMed] [Google Scholar]
- Kroemer G, Pouyssegur J. Tumor cell metabolism: cancer's Achilles' heel. Cancer Cell. 2008;13:472–482. doi: 10.1016/j.ccr.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML. Structural and functional organization of the mitochondrial respiratory chain: a dynamic super-assembly. Int J Biochem Cell Biol. 2009;41:1750–1772. doi: 10.1016/j.biocel.2009.04.003. [DOI] [PubMed] [Google Scholar]
- Lessing D, Bonini NM. Maintaining the brain: insight into human neurodegeneration from Drosophila melanogaster mutants. Nat Rev Genet. 2009;10:359–370. doi: 10.1038/nrg2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Corvera S. The role of mitochondria in the pathogenesis of type 2 diabetes. Endocr Rev. 2010;31:364–395. doi: 10.1210/er.2009-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schriner SE, Linford NJ, Martin GM, Treuting P, Ogburn CE, Emond M, Coskun PE, Ladiges W, Wolf N, Van Remmen H, et al. Extension of murine life span by overexpression of catalase targeted to mitochondria. Science. 2005;308:1909–1911. doi: 10.1126/science.1106653. Epub 2005 May 1905. [DOI] [PubMed] [Google Scholar]
- Stuart RA. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J Bioenerg Biomembr. 2008;40:411–417. doi: 10.1007/s10863-008-9168-4. [DOI] [PubMed] [Google Scholar]
- Suthammarak W, Morgan PG, Sedensky MM. Mutations in mitochondrial complex III uniquely affect complex I in Caenorhabditis elegans. J Biol Chem. 2010;285:40724–40731. doi: 10.1074/jbc.M110.159608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cao Y, Chen Y, Gardner P, Steiner DF. Pancreatic beta cells lack a low glucose and O2-inducible mitochondrial protein that augments cell survival. Proc Natl Acad Sci U S A. 2006;103:10636–10641. doi: 10.1073/pnas.0604194103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- Wittig I, Schagger H. Supramolecular organization of ATP synthase and respiratory chain in mitochondrial membranes. Biochim Biophys Acta. 2009;1787:672–680. doi: 10.1016/j.bbabio.2008.12.016. [DOI] [PubMed] [Google Scholar]
- Wright RM, Simpson SL, Lanoil BD. Oxygen regulation of the cytochrome c oxidase subunit VI gene, COX6, in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;216:676–685. doi: 10.1006/bbrc.1995.2675. [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.