Abstract
Objective
To define the relationship between autoantigen citrullination and different peptidylarginine deiminase (PAD) enzymes in rheumatoid arthritis (RA).
Methods
Citrullinated autoantigens were identified by immunoblotting control and ionomycin-activated human primary neutrophil lysate with RA sera. Autoantigen identity and citrullination sites were defined by mass spectrometry. PAD isoenzyme expression in human neutrophils was determined by immunoblotting. PAD substrate specificity was addressed in HL-60 cell lysates co-incubated with human recombinant PAD2, PAD3 and PAD4.
Results
Although prominent protein citrullination is observed in ionomycin activated neutrophils, RA sera only recognized a limited number of these citrullinated molecules. Among these, we identified that beta and gamma actins are citrullinated on at least ten arginine residues, generating a novel 47kDa species that is frequently recognized by RA autoantibodies. Interestingly, we showed that the PAD enzymes expressed in human neutrophils (i.e. PAD2, PAD3 and PAD4) have unique substrate specificities, independent of their subcellular distribution. Thus, only PAD2 was able to citrullinate native beta/gamma-actin, while histone H3 was only citrullinated by PAD4.
Conclusion
These studies identified beta and gamma actins as novel citrullinated autoantigens in RA, allowing enzyme specificity against intracellular substrates to be addressed. The studies provide evidence that PAD enzymes have the intrinsic capacity to select unique protein targets. We propose that unique PAD specificity may play a role in autoantigen selection in RA.
Keywords: Citrullination, rheumatoid arthritis, anti-CCP, actin, peptidylarginine deiminase
Introduction
Accumulating evidence suggests that enzymatic deimination (or citrullination) of proteins, a process catalyzed by the PAD enzymes,1 plays an essential role in RA pathogenesis.2 However, although advances have been made to support the pathogenic role of antigen citrullination in RA, important questions remain about the nature of the protein targets toward which anti-citrullinated protein antibodies (APCAs) are directed, and about the interaction between these autoantigen substrates and the PAD enzymes.
Among the five PAD enzymes encoded by humans (PAD1-PAD4, and PAD6), PAD2 and PAD4 have gained attention as potential candidates that may drive citrullination of self-antigens in RA. Both enzymes have been found in rheumatoid synovial tissue and fluid3–5 and PAD4 polymorphisms are genetically associated with RA development.6 Beside their distinct tissue distribution (PAD2 is widely expressed while PAD4 is preferentially expressed by hematopoietic cells),1 the major difference among these enzymes is that PAD2 has a cytoplasmic distribution while PAD4 mainly resides in the cell nucleus.7,8 These observations raise the possibility that different PADs target different cellular substrates, defined by both the cell type and the subcellular localization of the substrate. Therefore, distinct autoantibodies recognizing such citrullinated substrates might indicate the activity of specific PADs in intracellular protein citrullination.
In RA, little attention has been paid to defining whether the selection of citrullinated autoantigens is driven by unique PAD enzymes. This can be explained in part because current evidence suggests that protein citrullination may occur extracellularly.5 In this case, substrate selection by the PADs would not be limited by their subcellular localization. Under such circumstances, target specificity among PADs would be mechanistically irrelevant. Despite this, it is intriguing that except for PAD49,10 and the far upstream element-binding proteins11 which are nuclear, the majority of cellular targets identified as potential citrullinated autoantigens in RA have extranuclear distribution [e.g. filaggrin12,13; vimentin14,15; α-enolase, elongation factor-1a and adenyl cyclase-associated protein-116; F-actin capping protein alpha-1 subunit, asporin, cathepsin D, beta-actin, histamine receptor, protein disulfide-isomerase, ER60 precursor, and mitochondrial aldehyde dehydrogenase17; collagen type-I18 and II19; eukaryotic translation initiation factor-4G120; aldolase, phosphoglycerate kinase-1, calreticulin, and HSP6011]. The skewing of potential autoantigens to the extranuclear compartment raises questions about the role PAD4 if autoantigen citrullination occurs intracellularly and about the random selection of targets if citrullination occurs extracellularly. It is possible that such substrate specificity reflects the activity of unique PADs either intra (i.e. cytoplasmic PADs) or extracellularly. These studies provide evidence to support that the PAD enzymes have the intrinsic capacity to select unique protein targets.
Methods
Subjects
Sera from 39 patients with established RA were obtained from a convenience IRB approved cohort followed at the Johns Hopkins Arthritis Center. Sera from 15 healthy adults were used as comparison group.
Human PAD2-4 cloning and recombinant protein purification
Cloning and expression of recombinant (r)PAD4 was described elsewhere.10 Total RNA purified from ATRA-differentiated HL-60 cells (to clone PAD2) or human bone marrow (Cambrex) (to clone PAD3) was reverse-transcribed to generate cDNA. cDNAs were further cloned into pET28a (Novagen) for prokaryotic expression to generate an N-terminal His6-tagged fusion protein that was purified using Ni-NTA agarose (Qiagen) as described by the manufacturer.
Neutrophil isolation and activation
After IRB approval and informed consent, neutrophils were isolated from healthy donors as described10 and resuspended in HBSS without calcium/magnesium. CaCl2 was added to reach 2mM and neutrophils were further incubated in the absence or presence of 1 µM ionomycin for 4 hrs at 37°C. After incubation, the cells were lysed and boiled in SDS-sample buffer for further analysis by immunoblotting. Alternatively, the cells were lysed with IEF buffer (8M urea, 2M thiourea, 4% CHAPS, 1% DTT).
Two-dimensional electrophoresis and mass spectrometry analysis
Samples in IEF buffer containing 0.2% Bio-lyte 3/10 ampholyte (BioRad), 1.5% 2-hydroxyethyl disulfide and 0.05% bromophenol blue were resolved using 3–10 IPG strips (Bio-Rad) and a BioRad IEF Cell system. The IPG strips were then incubated in equilibration buffer (6M urea, 30% w/v glycerol, 2% SDS, 0.05M tris pH 8.8) containing 1% DTT, followed by incubation in equilibration buffer containing 2.5% IAA. For the second dimension, the samples were resolved by electrophoresis on 8% SDS-polyacrylamide gels and transferred into nitrocellulose membrane. The protein pattern was visualized by Ponceau-S prior to immunoblotting using sera from RA patients. Once the antigen of interest was mapped, the spot was sliced from a two-dimensional gel stained with GelCode blue (Thermo Scientific) and analyzed by mass spectrometry to define the protein identity and to identify citrullination sites (Proteomics Core/Mass Spectrometry Facility, Johns Hopkins School of Medicine).
In vitro citrullination assays
Using siliconized tubes (Sigma), 1 µM human recombinant beta-actin, gamma-actin (GenWay), or 700 nM purified actin from human platelets (Cytoskeleton, Inc) plus 700 nM human recombinant histone H3.1 (New England Biolabs) were incubated alone or co-incubated with 700 nM rabbit PAD2 (Sigma), human rPAD2, rPAD3 or rPAD4 in buffer A (100 mM Tris pH 7.6, 5 mM DTT, 10 mM CaCl2). After 0–60 min at 37°C, reactions were stopped by adding SDS-sample buffer and boiling. Non-citrullinated and citrullinated recombinant beta-actin were also used for mass spectrometry analysis to identify citrullination sites, and to screen for anti-citrullinated beta-actin antibodies by immunoblotting using control and RA sera.
Cell lysates from PAD-negative undifferentiated HL-60 cells (3×106 cells/ml) were generated in buffer B (20 mM Tris pH 7.6, 1% NP40 and protease inhibitors) sonicated, cleared by centrifugation, and further incubated alone or co-incubated with 700 nM human rPAD2, rPAD3 or rPAD4 in the presence of 5 mM DTT and 10 mM CaCl2. After 60 min at 37°C, reactions were stopped by adding SDS-sample buffer and boiling. Protein citrullination was determined by anti–modified citrulline (AMC) immunoblotting, according to the manufacturer’s recommendations (Millipore).
Results
RA autoantibodies recognize a limited number of citrullinated antigens in activated primary neutrophils
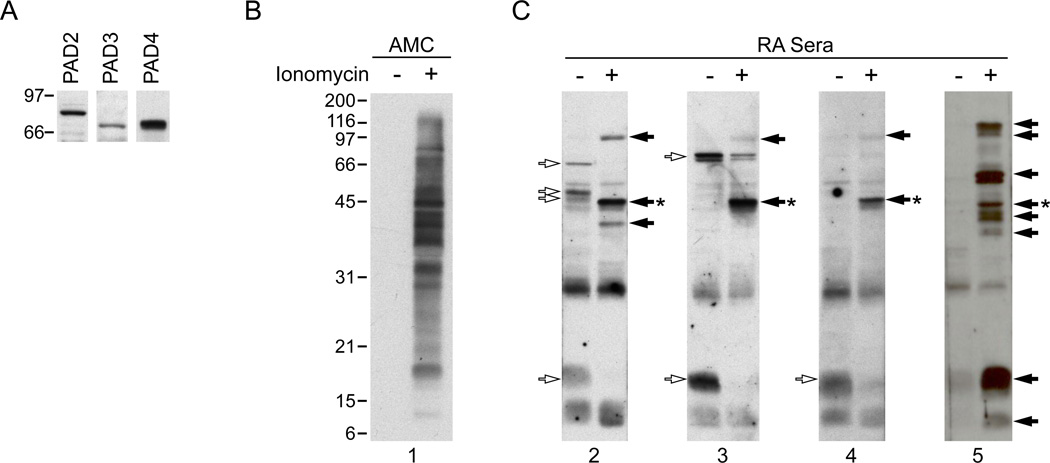
To better understand the independent role of the PAD enzymes in autoantigen citrullination in cells expressing multiple PADs, we initially focused on the study of human neutrophils. This cell type represents one of the most abundant inflammatory cells in the rheumatoid joint and has been widely used as a model for the study of protein citrullination. The cells constitutively expresses PAD47,21 and protein citrullination can be induced upon cell activation with different stimuli.22 In initial studies, we demonstrated that neutrophils express PADs 2 and 3 in addition to PAD4 protein (Figure 1A), making them a suitable system to study autoantigen citrullination by multiple PADs. To identify the patterns of citrullinated autoantigens generated in activated neutrophils, neutrophils were activated with ionomycin, and lysates of control and ionomycin-activated cells were analyzed for protein citrullination (Figure 1B) and recognition by RA sera (Figure 1C). While protein citrullination was absent in control neutrophils, ionomycin treatment induced massive citrullination, modifying molecules across the entire range of molecular weights (MW) detected by SDS-PAGE. Two different patterns of reactivity with RA sera were noted: i) molecules that were only detected in activated neutrophils (the focus of this study), and ii) antigens found in non-stimulated neutrophils, which either remained unchanged or disappeared upon cell activation. Interestingly, despite the large number of citrullinated proteins found in activated neutrophils (Figure 1B), sera from RA patients only detected a few of these molecules (Figure 1C and data not shown), confirming that RA autoantibodies recognize only a very small subset of the proteins citrullinated during PAD activation. Moreover, except for a few antigens that were co-detected by different sera, patterns of autoantigen recognition among RA sera were quite distinct.
Figure 1.
PAD expression in human primary neutrophils and autoantigen recognition by RA sera in control and ionomycin-activated neutrophils. A. PAD2, 3 and 4 are expressed in human neutrophils. Samples from freshly isolated neutrophils were analyzed by immunoblotting with antibodies against human PAD2, PAD3 and PAD4. B, C. Primary human neutrophils in HBSS containing 2 mM CaCl2 were incubated in the absence (−) or presence (+) of 1 µM ionomycin for 4 hrs at 37°C. Samples were analyzed by electrophoresis on 13% SDS-polyacrylamide gels and immunoblotted using an AMC antibody (B) or anti-CCP positive sera from RA patients (C). Data from 4 representative sera are shown in C. The unfilled arrows denote antigens detected in non-stimulated neutrophils, filled arrows mark antigens generated upon neutrophil activation and the asterisk denotes the 47kDa species that was further analyzed by mass spectrometry.
Beta/gamma actins are citrullinated in ionomycin-activated neutrophils and targeted by RA autoantibodies
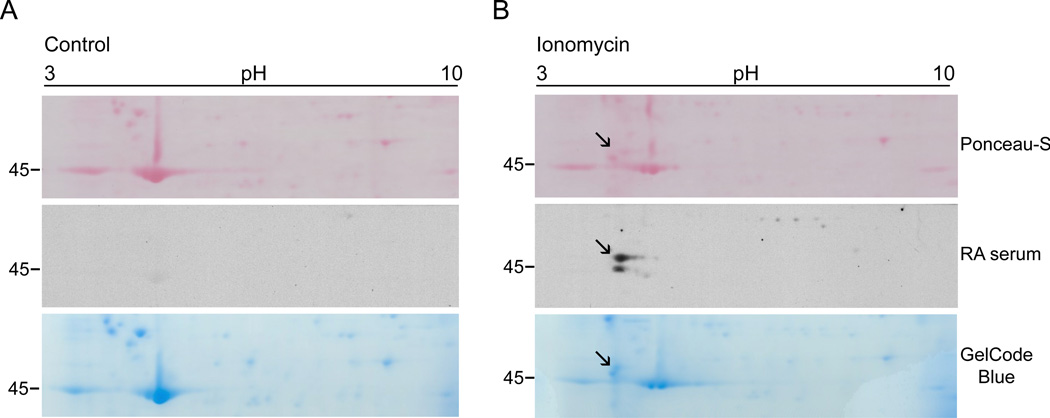
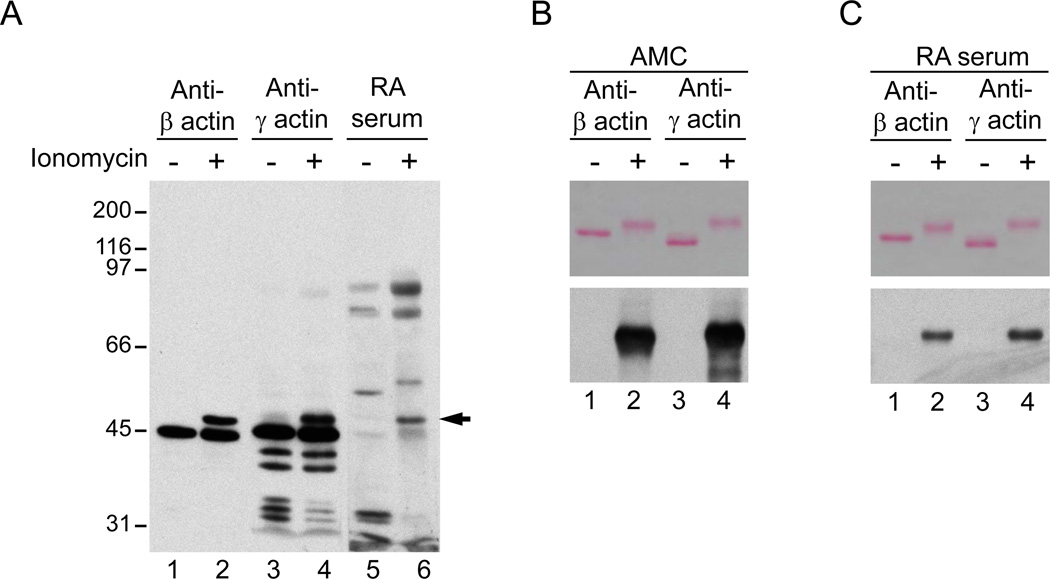
To define the identity of autoantigens recognized by autoantibodies in activated neutrophils, we used two-dimensional electrophoresis analysis and protein sequencing. We sought to identify a ~47kDa species which was among the few autoantigens detected by multiple RA sera (i.e. a similar band was recognized by ~30% of RA patients serum) (Figure 1B and data not shown). We mapped the isoelectric point and MW of the autoantigen by immunoblotting (Figure 2B), and then identified, harvested and sequenced the corresponding protein spot from a companion gelcode blue-stained gel. The protein was identified as cytoplasmic actin, a molecule that was previously described as a citrullinated autoantigen among many others detected in a proteomic analysis of RA synovial tissue.17 An immunoblot using standard antibodies recognizing the two major cytoplasmic actins beta- and gamma-actin (which only differ by 4 amino acids at the amino terminus)23,24 showed the detection of higher MW forms of these molecules only in ionomycin-activated neutrophils. Interestingly, these higher forms co-migrated exactly with the 47 kDa species detected by RA sera (Figure 3A). To further define beta- and gamma-actins as targets of citrullination, recombinant beta- or gamma-actin was incubated in the absence or presence of rabbit PAD2, and protein citrullination was determined by AMC immunoblotting. Interestingly, in the presence of rabbit PAD2, beta- and gamma-actins migrated as higher MW species (Figure 3B and 3C, upper panels), similar to endogenous actin in ionomycin-activated neutrophils. Moreover, we confirmed that the higher MW species of beta/gamma-actin generated by rabbit PAD2 were indeed citrullinated (Figure 3B, lower panel). Strikingly, when sera from RA patients were used to immunoblot beta/gamma-actin, only the citrullinated forms were detected (Figure 3C, lower panel). Indeed, antibodies against purified citrullinated beta-actin were identified by immunoblotting in 51% of RA sera (Table 1), but only in 1 of 15 controls (data not shown). Among the RA patients, anti-citrullinated actin antibodies tended to be more frequent in males and anti-CCP positive individuals, although these differences were not quite statistically significant in our sample.
Figure 2.
Two-dimentional mapping of the 47kDa RA autoantigen generated in ionomycin-activated neutrophils. Primary human neutrophils in HBSS containing 2 mM CaCl2 were incubated in the absence (A) or presence (B) of 1 µM ionomycin for 4 hrs at 37°C. Samples were resolved in the first dimension using 3–10 IPG strips and then by electrophoresis on 8% SDS-polyacrylamide gels. Proteins were visualized by Ponceau S staining (upper panel) prior to immunoblotting with RA patient sera (middle panel). Data from one representative serum is shown. Once the antigen was mapped, the spot was sliced from a GelCode blue-stained gel (lower panel) and used for protein sequencing. Note that the 47kDa species detected by RA sera (marked with the black arrow) is absent in control cells.
Figure 3.
Beta and gamma actin are citrullinated autoantigens in RA. A. Beta- and gamma-actin are modified in ionomycin activated neutrophils and the novel species co-migrates with the 47kDa RA autoantigen. Samples from control (lanes 1, 3, and 5) and ionomycin activated neutrophils (lanes 2, 4 and 6) were analyzed by immunoblotting using monoclonal antibodies against human beta- and gamma-actin (Sigma) or RA serum. The filled arrow denotes the 47kDa species detected by anti-actin antibodies and RA sera. B, C. Beta- and gamma-actin are citrullinated and recognized by RA autoantibodies. Recombinant human beta-actin (lanes 1 and 2) or gamma-actin (lanes 3 and 4) was incubated in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of rabbit PAD2. After 1 hr at 37°C, samples were divided in two and analyzed by electrophoresis on 8% SDS-polyacrylamide gels. After electrophoresis, proteins were visualized by Ponceau S staining (upper panels) prior to AMC immunoblotting (B, lower panel) or immunoblotting with RA sera (C, lower panel). Data from 1 representative serum containing anti-citrullinated actin antibodies is shown in C.
Table 1.
Characteristics of 39 RA patients according to anti-citrullinated beta-actin antibodies
| Parameter | Total (n = 39) |
Antibody (+) (n = 20) |
Antibody (−) (n = 19) |
P |
|---|---|---|---|---|
| Age, years | 61 ± 9 | 61 ± 8 | 61 ± 10 | 0.87* |
| Male, n (%) | 20 (52) | 13 (65) | 7 (37) | 0.079** |
| Caucasian, n (%) | 33 (85) | 16 (80) | 17 (89) | 0.66*** |
| RA duration, years; median (IQR) | 7 (4 – 18) | 9 (4 – 28) | 7 (3 – 10) | 0.25† |
| RF > 40 units, n (%) | 24 (62) | 12 (60) | 12 (63) | 0.84** |
| Anti-CCP > 20 units, n (%) | 30 (77) | 18 (90) | 12 (63) | 0.065*** |
| Anti-CCP > 60 units, n (%) | 29 (74) | 17 (85) | 12 (63) | 0.16*** |
Results are mean ± SD unless otherwise noted. IQR, interquartile range; RF, rheumatoid factor; CCP, cyclic citrullinated peptide. Comparison:
student’s t-test,
Chi-square goodness-of-fit test,
Fisher’s exact test,
Kruskal-Wallis test.
PAD2, PAD3 and PAD4 have distinct macromolecular substrate specificity
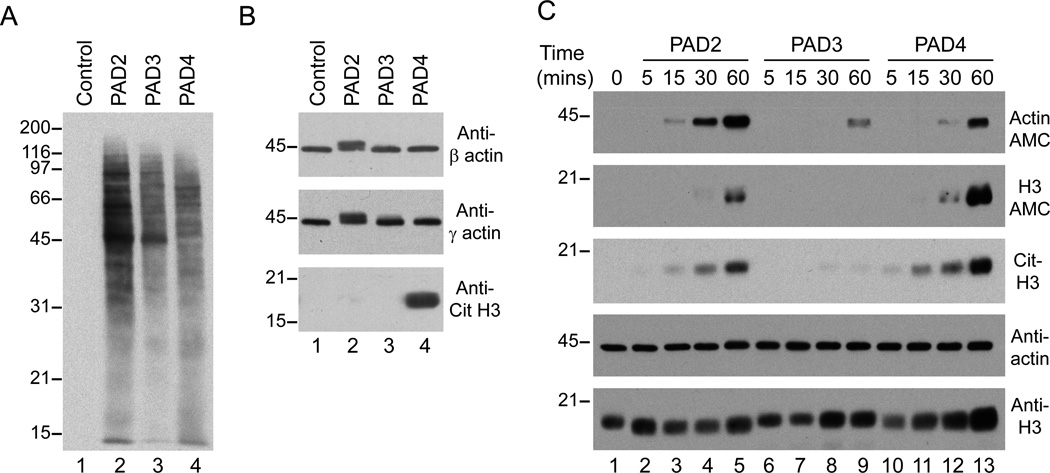
It is presently unknown whether citrullination of intracellular substrates occurs intra- or extracellularly in vivo in RA. Additionally, if citrullination occurs extracellularly, it is not known whether all PADs similarly modify all substrates. To gain insight into potential substrate specificity among PADs, we directly addressed the citrullination activity of human rPAD2, rPAD3 and rPAD4 against cellular substrates generated from HL-60 cell lysates that were sonicated and cleared by centrifugation. Using this approach, the PAD enzymes would have similar access to cellular substrates independent of the subcellular localization of the substrate or the enzyme. Interestingly, when HL-60 cell lysates were incubated in the absence or presence of human rPAD2, rPAD3 or rPAD4, different patterns of protein citrullination were generated (Figure 4A). These differences were particularly striking when specific substrates were analyzed (Figure 4B). Thus, only rPAD2 was able to citrullinate beta/gamma actin, as evidenced by a characteristic change in its MW, while histone H3 was only citrullinated by rPAD4. Furthermore, PAD3 (a cytoplasmic enzyme like PAD2) was unable to citrullinate actin, as well as histone H3. Further analysis using purified components confirmed the substrate preferences of these enzymes (Figure 4C). Thus, although rPAD2 and rPAD4 were able to modify purified human platelet actin (comprised of both beta and gamma actin) and recombinant histone H3, citrullination of actin and histone H3 was more efficient by rPAD2 and rPAD4, respectively. Moreover, rPAD3 was the least efficient in citrullinating these substrates. In regard to histone H3, it is interesting that differences between PAD2, PAD3 and PAD4 were more evident when H3 citrullination was determined using the AMC method (which detects all modified arginines), compared to the detection of citrullines specifically at residues 2, 8 and 17. Thus, it appears that in addition to substrate preference, PADs have different access to arginines within the same substrate.
Figure 4.
PAD2, 3 and 4 have distinct substrate specificities. A, B. Sonicated HL-60 cell lysates were incubated in the absence (lane 1) or presence of human rPAD2, rPAD3 or rPAD4 (lanes 2–4, respectively) for 1 hr at 37°C. After terminating the reactions, samples were analyzed by electrophoresis on 13% SDS-polyacrylamide gels and immunoblotted with AMC antibodies (A) or antibodies against human beta-actin, gamma-actin (Sigma) and citrullinated histone H3 (citruline 2 + 8 + 17) (Abcam) (B). C. A mixture of purified human actin plus recombinant histone H3 was incubated in the absence (lane 1) or presence of human rPAD2 (lanes 2–5), rPAD3 (lanes 6–9) or rPAD4 (lanes 10–13) for 5–60 mins at 37°C. After terminating the reactions, samples were analyzed in duplicated (actin) or triplicated (histone H3) by electrophoresis on 13% SDS-polyacrylamide gels. After electrophoresis, proteins were visualized by immunoblotting with antibodies against AMC, citrullinated histone H3 (citrulline 2 + 8 + 17) (Cit-H3), human beta/gamma-actin and histone H3 (Abcam).
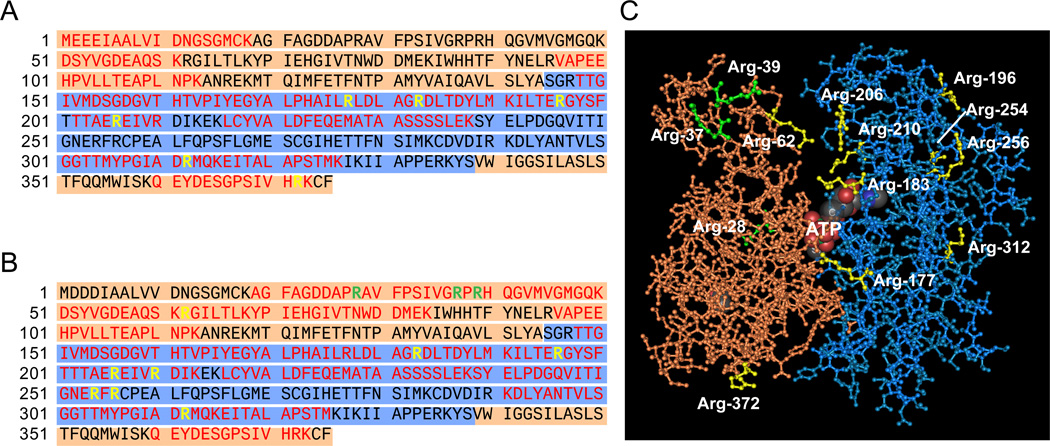
Citrullination sites in cytoplasmic actin form clusters in the three-dimensional structure
In order to define the distribution of peptidylcitrullines in actin, citrullination sites were identified using two different approaches: (i) endogenous citrullinated actin from activated neutrophils was isolated from a two-dimensional gel and subjected to protein sequencing (Figure 5A) and (ii) recombinant beta-actin was citrullinated in vitro with human rPAD2 and analyzed by mass spectrometry (Figure 5B). Since the amino acids surrounding all arginine residues are identical in the cytoplasmic actins, we did not include gamma-actin for this analysis. Peptides identified using the two approaches covered 69% of the actin sequence (which included 72.2% of the arginine content), leaving the citrullination status of five arginines (i.e. Arg-95, -116, -147, -290, and -335) undetermined. Five citrullination sites overlapped between endogenous and recombinant citrullinated actin (i.e. positions 183, 196, 206, 312, and 372) and 2 divergences were found (i.e. Cit-177 and -210, respectively). Peptides containing Cit-62, -354 and -356 were only isolated from citrullinated recombinant beta-actin. Three arginines were confirmed not to be modified by PAD2 (Arg-28, -37 and -39). Although we found no consensus sequences that may explain the targeting of distinct arginines by PAD2, it is noteworthy that non-modified arginines are flanked by proline residues at positions −1 or +1, supporting previous observations that this sequence (i.e. Pro-Arg or Arg-Pro) disfavors arginine deimination by PADs 25,26. The three-dimensional structure of beta-actin has been resolved and consists of two domains (called the large and small domains) with a cleft containing the bound nucleotide (usually ATP) (Figure 5C).28 Interestingly, the large domain of beta-actin contains the majority of the citrulline residues (i.e. 8 of the 10 peptidylarginines) identified in actin. Although these residues are far apart in the primary sequence, they form clusters in the three-dimensional protein structure. One cluster is located inside the actin cleft and includes residues 62, 183, 206 and 210. The second cluster is located in the surface of the large domain (residues 196, 254 and 256) opposite to the cleft region.
Figure 5.
Cirullination sites in actin. A, B and C. Amino acid sequence (gamma-actin in A and beta-actin in B) and tertiary structure (C) of actin. The structure of actin consists of two domains called the large and small domains (shown in blue and orange, respectively), with a cleft containing the bound nucleotide (marked as ATP in C). The amino acid sequence covered by mass spectrometry analysis of endogenous (A) and recombinant (B) citrullinated actin is shown in red, the arginines that are not citrullinated are shown in green and the potential arginine targets for citrullination are shown in yellow. The tertiary structure of actin (C) reveals clustering of citrullination sites around the cleft and the surface of the large domain opposite to the cleft region. The model shown in C was generated from the Molecular Modeling DataBase (NCBI) using the software Cn3D according to coordinates generated by Schutt et al.28
Discussion
Citrullinated autoantigens have emerged as key targets of the immune response in RA. Understanding the mechanisms that generate these antigens may identify unique pathways that regulate antigen drive in this disease, which might be relevant to the development of novel therapies. In this study, we identified several unique features of protein citrullination of potential relevance to RA pathogenesis: (i) Although a large number of proteins are citrullinated in activated neutrophils, ACPAs only recognize a limited number of these molecules; (ii) cytoplasmic actins represent a novel target for citrullination and a novel autoantigen in RA; (iii) primary human neutrophils express three different PAD isoenzymes (i.e. PAD2, PAD3 and PAD4); and (iv) the citrullination activity of each PAD appears to be specific and directed preferentially against distinct substrates, independent of their cellular localization.
Accumulating evidence from this and other studies strongly supports the notion that although citrullination plays a critical role in antigen recognition by ACPAs, the modification per se is not the only determinant that confers antibody binding.2 Although the structural context in which citrulline occurs may be shared among autoantigens, the striking observation that different RA sera have unique patterns of reactivity (see Figure 1) strongly supports the hypothesis that different ACPAs recognize unique sequences.2,29 Whether these unique patterns of antigen recognition among RA patients reflect citrullination of distinct autoantigens in different microenvironments is not yet known.
Citrullination sites have been confirmed in only few RA autoantigens generated in vitro or isolated from cells or tissues.10,16,30 Ten potential citrullination sites were identified in peptides generated from endogenous and recombinant citrullinated actin. Among these sites, two distribution patterns of peptidylcitrullines appear relevant. In the linear structure of actin, citrulline residues are found in pairs separated by 2–5 amino acid residues, which resemble the peptidylcitrulline distribution found in short peptides used for ACPAs detection.12,19,31,32 However, in the tertiary structure of the molecule, peptidylcitrullines formed clusters, in which citrulline residues far from each other in primary sequence converge to form citrulline-enriched regions within the molecule. Interestingly, a similar pattern of peptidylcitrulline distribution is observed in the tertiary structure of the autoantigen PAD4.10 Since the number and/or the conformation of citrulline residues appear to have important role in peptide recognition by ACPAs,2,12,31 it is possible that clusters of citrullines may allow more efficient antibody binding to citrullinated molecules. Considering that citrullinated autoantigens in RA are unlikely unified by common immunogenic sequences (since ACPAs do not cross-react among citrullinated autoantigens), but likely unified by common structural features, it would be important to address whether the clustering of peptidylcitrullines within the tertiary structure of molecules plays a role in autoantigen selection.
The sites and circumstances of citrullination of autoantigens in vivo in RA remain unclear. While fibrinogen is citrullinated extracellularly, likely by PADs which have leaked out of damaged cells,4,33 it is unknown where and under what circumstances citrullination of intracellular autoantigens occurs. The studies in this paper highlight the striking variation in the specificity of ACPA autoantibodies in different RA patients and raise important questions about the mechanisms underlying such variation. Importantly, these studies question the assumptions that all PADs share similar specificities and that the proposed extracellular presence of PADs in RA obliterate potentially important subcellular barriers. In spite of the cytoplasmic co-localization of actins, PAD2, and PAD3, it was surprising that these enzymes generated distinct patterns of citrullinated proteins. Moreover, the finding that in cell lysates, actin and histone-H3 are only citrullinated by PAD2 and PAD4, respectively, strongly supports the existence of substrate specificity among PADs. In this regard, studies using purified substrates demonstrate that although purified actin or histone H3 can be citrullinated by PAD2, PAD3 and/or PAD4, the enzymes have a clear substrate preference, which becomes more evident when the enzymes are exposed to a larger number of substrates (e.g. the full cellular content). In addition, this data strongly supports that target selection by PADs is independent of any other cellular component and therefore, is an intrinsic property of the enzyme. Interestingly, previous observations by Nakayama-Hamada et al34 demonstrated that human rPAD2 citrullinates purified fibrinogen and filaggrin several orders of magnitude more efficiently than human rPAD4, supporting the notion that PAD enzymes have either unique and/or preferential activity against macromolecular substrates.
Based on differences in the sequence alignment between PAD2, PAD3 and PAD4,35 there are two major regions in PADs that may participate in substrate selection, the N-terminal domain and the active site cleft. With regard to the active site cleft, it is interesting to note that Arg-374 in PAD4 is not conserved in PAD2 or PAD3 (which contain Gly at position 374). This Arg is directly involved in the recognition of histone N-terminal peptides36 and is absolutely required for H3 citrullination and for PAD4 enzyme activity.10,36 Interestingly, recent studies have shown that PAD3 has poor reactivity against small synthetic benzoylated arginine derivatives, known PAD4 substrates.27 Mutation of PAD3 Gly-374 to Arg was unable to restore the ability of PAD3 to citrullinated these molecules, suggesting that PAD3 and perhaps PAD2 have very different substrate specificities driven by other amino acid contacts outside the catalytic site. The N-terminal domain is not well conserved among the PAD enzymes and therefore may contribute to substrate specificity, potentially through extended macromolecular interactions outside the active site (i.e. through exosites). Defining these additional interactions may have important implications in the designing of inhibitors against specific PADs.
The discovery that different PAD isoforms have distinct substrate specificities has significant implications for RA. It is possible that only one or a few PADs may account for autoantigen citrullination in vivo. In this regard, future therapies targeting PAD enzymes might therefore be validly focused on PAD-selective inhibitors, instead of pan-PAD inhibitors which may have higher toxicity by affecting PADs with no relevance to RA. Further studies which elucidate PAD activation, specificity and functional consequences as they relate to specific ACPA antigens are of high priority.
Acknowledgments
We thank Xiaoming Zhu for her valuable technical assistance and Dr. Joan Bathon for providing valuable RA serum samples. F.A. is a Lowe Family Scholar in the Johns Hopkins Bayview Center for Innovative Medicine and is supported a Dana Foundation Scholars Program in Human Immunology, The Donald B. and Dorothy L. Stabler Foundation and NIH grant P30 AR053503. A.R. is supported by NIH Grant R37 DE-12354 and ACR-REF Within our Reach grant. E.D. is supported by NIH grant T32 AR048522. J.T.G is supported by NIH grant 1K23AR054112-01.
Footnotes
Statement
The Corresponding Author has the right to grant on behalf of all authors and does grant on behalf of all authors, an exclusive licence (or non exclusive for government employees) on a worldwide basis to the BMJ Publishing Group Ltd to permit this article (if accepted) to be published in ARD and any other BMJPGL products and sublicences such use and exploit all subsidiary rights, as set out in our licence).
References
- 1.Vossenaar ER, Zendman AJ, van Venrooij WJ, et al. Pad, a growing family of citrullinating enzymes: genes, features and involvement in disease. BioEssays. 2003;25:1106–1118. doi: 10.1002/bies.10357. [DOI] [PubMed] [Google Scholar]
- 2.Wegner N, Lundberg K, Kinloch A, et al. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunol Rev. 2010;233:34–54. doi: 10.1111/j.0105-2896.2009.00850.x. [DOI] [PubMed] [Google Scholar]
- 3.Foulquier C, Sebbag M, Clavel C, et al. Peptidyl arginine deiminase type 2 (PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in rheumatoid arthritis synovium in close association with tissue inflammation. Arthritis Rheum. 2007;56:3541–3553. doi: 10.1002/art.22983. [DOI] [PubMed] [Google Scholar]
- 4.Chang X, Yamada R, Suzuki A, et al. Localization of peptidylarginine deiminase 4 (PADI4) and citrullinated protein in synovial tissue of rheumatoid arthritis. Rheumatology (Oxford) 2005;44:40–50. doi: 10.1093/rheumatology/keh414. [DOI] [PubMed] [Google Scholar]
- 5.Kinloch A, Lundberg K, Wait R, et al. Synovial fluid is a site of citrullination of autoantigens in inflammatory arthritis. Arthritis Rheum. 2008;58:2287–2295. doi: 10.1002/art.23618. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki A, Yamada R, Chang X, et al. Functional haplotypes of PADI4, encoding citrullinating enzyme peptidylarginine deiminase 4, are associated with rheumatoid arthritis. Nat Genet. 2003;34:395–402. doi: 10.1038/ng1206. [DOI] [PubMed] [Google Scholar]
- 7.Nakashima K, Hagiwara T, Yamada M. Nuclear localization of peptidylarginine deiminase V and histone deimination in granulocytes. J Biol Chem. 2002;277:49562–49568. doi: 10.1074/jbc.M208795200. [DOI] [PubMed] [Google Scholar]
- 8.Mastronardi FG, Wood DD, Mei J, et al. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harris ML, Darrah E, Lam GK, et al. Association of autoimmunity to peptidyl arginine deiminase type 4 with genotype and disease severity in rheumatoid arthritis. Arthritis Rheum. 2008;58:1958–1967. doi: 10.1002/art.23596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrade F, Darrah E, Gucek M, et al. Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum. 2010;62:1630–1640. doi: 10.1002/art.27439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goeb V, Thomas-L'Otellier M, Daveau R, et al. Candidate autoantigens identified by mass spectrometry in early rheumatoid arthritis are chaperones and citrullinated glycolytic enzymes. Arthritis Res Ther. 2009;11:R38. doi: 10.1186/ar2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schellekens GA, de Jong BA, van den Hoogen FH, et al. Citrulline is an essential constituent of antigenic determinants recognized by rheumatoid arthritis-specific autoantibodies. J Clin Invest. 1998;101:273–281. doi: 10.1172/JCI1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Girbal-Neuhauser E, Durieux JJ, Arnaud M, et al. The epitopes targeted by the rheumatoid arthritis-associated antifilaggrin autoantibodies are posttranslationally generated on various sites of (pro)filaggrin by deimination of arginine residues. J Immunol. 1999;162:585–594. [PubMed] [Google Scholar]
- 14.Vossenaar ER, Radstake TR, van der HA, et al. Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann Rheum Dis. 2004;63:373–381. doi: 10.1136/ard.2003.012211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vossenaar ER, Despres N, Lapointe E, et al. Rheumatoid arthritis specific anti-Sa antibodies target citrullinated vimentin. Arthritis Res Ther. 2004;6:R142–R150. doi: 10.1186/ar1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinloch A, Tatzer V, Wait R, et al. Identification of citrullinated alpha-enolase as a candidate autoantigen in rheumatoid arthritis. Arthritis Res Ther. 2005;7:R1421–R1429. doi: 10.1186/ar1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matsuo K, Xiang Y, Nakamura H, et al. Identification of novel citrullinated autoantigens of synovium in rheumatoid arthritis using a proteomic approach. Arthritis Res Ther. 2006;8:R175. doi: 10.1186/ar2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suzuki A, Yamada R, Ohtake-Yamanaka M, et al. Anti-citrullinated collagen type I antibody is a target of autoimmunity in rheumatoid arthritis. Biochem Biophys Res Commun. 2005;333:418–426. doi: 10.1016/j.bbrc.2005.05.137. [DOI] [PubMed] [Google Scholar]
- 19.Burkhardt H, Sehnert B, Bockermann R, et al. Humoral immune response to citrullinated collagen type II determinants in early rheumatoid arthritis. Eur J Immunol. 2005;35:1643–1652. doi: 10.1002/eji.200526000. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki Y, Suzuki A, Sawada T, et al. Identification of citrullinated eukaryotic translation initiation factor 4G1 as novel autoantigen in rheumatoid arthritis. Biochem Biophys Res Commun. 2006;341:94–100. doi: 10.1016/j.bbrc.2005.12.160. [DOI] [PubMed] [Google Scholar]
- 21.Asaga H, Nakashima K, Senshu T, et al. Immunocytochemical localization of peptidylarginine deiminase in human eosinophils and neutrophils. J Leukoc Biol. 2001;70:46–51. [PubMed] [Google Scholar]
- 22.Neeli I, Khan SN, Radic M. Histone deimination as a response to inflammatory stimuli in neutrophils. J Immunol. 2008;180:1895–1902. doi: 10.4049/jimmunol.180.3.1895. [DOI] [PubMed] [Google Scholar]
- 23.Kabsch W, Vandekerckhove J. Structure and function of actin. Annu Rev Biophys Biomol Struct. 1992;21:49–76. doi: 10.1146/annurev.bb.21.060192.000405. [DOI] [PubMed] [Google Scholar]
- 24.Khaitlina SY. Functional specificity of actin isoforms. Int Rev Cytol. 2001;202:35–98. doi: 10.1016/s0074-7696(01)02003-4. [DOI] [PubMed] [Google Scholar]
- 25.Nomura K. Specificity and mode of action of the muscle-type protein-arginine deiminase. Arch Biochem Biophys. 1992;293:362–369. doi: 10.1016/0003-9861(92)90407-n. [DOI] [PubMed] [Google Scholar]
- 26.Stensland ME, Pollmann S, Molberg O, et al. Primary sequence, together with other factors, influence peptide deimination by peptidylarginine deiminase-4. Biol Chem. 2009;390:99–107. doi: 10.1515/BC.2009.019. [DOI] [PubMed] [Google Scholar]
- 27.Knuckley B, Causey CP, Jones JE, et al. Substrate specificity and kinetic studies of PADs 1, 3, and 4 identify potent and selective inhibitors of protein arginine deiminase 3. Biochemistry. 2010;49:4852–4863. doi: 10.1021/bi100363t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schutt CE, Myslik JC, Rozycki MD, et al. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 29.Snir O, Widhe M, von SC, et al. Multiple antibody reactivities to citrullinated antigens in sera from patients with rheumatoid arthritis: association with HLA-DRB1 alleles. Ann Rheum Dis. 2009;68:736–743. doi: 10.1136/ard.2008.091355. [DOI] [PubMed] [Google Scholar]
- 30.Hermansson M, Artemenko K, Ossipova E, et al. MS analysis of rheumatoid arthritic synovial tissue identifies specific citrullination sites on fibrinogen. Proteomics Clin Appl. 2010;4:511–518. doi: 10.1002/prca.200900088. [DOI] [PubMed] [Google Scholar]
- 31.Perez ML, Gomara MJ, Ercilla G, et al. Antibodies to citrullinated human fibrinogen synthetic peptides in diagnosing rheumatoid arthritis. J Med Chem. 2007;50:3573–3584. doi: 10.1021/jm0701932. [DOI] [PubMed] [Google Scholar]
- 32.Lundberg K, Kinloch A, Fisher BA, et al. Antibodies to citrullinated alpha-enolase peptide 1 are specific for rheumatoid arthritis and cross-react with bacterial enolase. Arthritis Rheum. 2008;58:3009–3019. doi: 10.1002/art.23936. [DOI] [PubMed] [Google Scholar]
- 33.Takizawa Y, Suzuki A, Sawada T, et al. Citrullinated fibrinogen detected as a soluble citrullinated autoantigen in rheumatoid arthritis synovial fluids. Ann Rheum Dis. 2006;65:1013–1020. doi: 10.1136/ard.2005.044743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama-Hamada M, Suzuki A, Kubota K, et al. Comparison of enzymatic properties between hPADI2 and hPADI4. Biochem Biophys Res Commun. 2005;327:192–200. doi: 10.1016/j.bbrc.2004.11.152. [DOI] [PubMed] [Google Scholar]
- 35.Arita K, Hashimoto H, Shimizu T, et al. Structural basis for Ca(2+)-induced activation of human PAD4. Nat Struct Mol Biol. 2004;11:777–783. doi: 10.1038/nsmb799. [DOI] [PubMed] [Google Scholar]
- 36.Arita K, Shimizu T, Hashimoto H, et al. Structural basis for histone N-terminal recognition by human peptidylarginine deiminase 4. Proc Natl Acad Sci U S A. 2006;103:5291–5296. doi: 10.1073/pnas.0509639103. [DOI] [PMC free article] [PubMed] [Google Scholar]