Abstract
As a consumer of 95% of the oxygen we breathe, cytochrome c oxidase plays a major role in the energy balance of the cell. Regulation of its oxygen reduction and proton pumping activity is therefore critical to physiological function in health and disease. The location and structure of pathways for protons that are required to support cytochrome c oxidase activity are still under debate, with respect to their requirements for key residues and fixed waters, and how they are gated to prevent (or allow) proton backflow. Recent high resolution structures of bacterial and mammalian forms reveal conserved lipid and steroid binding sites as well as redox-linked conformational changes that provide new insights into potential regulatory ligands and gating modes. Mechanistic interpretation of these findings is discussed and their significance for understanding energy regulation.
Introduction
The proton pumping function of cytochrome c oxidase (CcO) has been the subject of intense study and controversy for many years, beginning with the question of whether CcO pumps protons at all[1]. While the pumping question is no longer in dispute and much progress has been made in defining the underlying mechanism of oxygen reduction, it remains unclear how this exergonic process drives proton translocation across a membrane. Part of the difficulty in addressing the issue relates to the apparently critical role of water in facilitating and gating the movement of protons [2-5]. Mutagenesis and crystallography are powerful tools for determining the structural features important for protein function, but they are less incisive when it comes to detecting the positions and roles of water molecules. Residues can be mutated but water cannot, and very high resolution crystal structures are necessary to reliably track the whereabouts of water. Obtaining the necessary resolution (usually better than 2.5 Å) with a membrane protein is a challenge. In this regard, some success has been achieved with CcO isolated from bovine and bacterial sources, clarifying some aspects of the pumping process but leading to conflicting conclusions regarding others.
A major physiological question that drives the quest to understand the pumping mechanism is: how is the efficiency of the process controlled? As the consumer of 95% of the oxygen we breathe and the terminal member of the mitochondrial electron transfer chain, cytochrome c oxidase is a major player in the energy equilibrium of organisms, contributing to the balance between ATP and heat production, exerting control over the level of aerobic metabolism, and affecting upstream production of reactive oxygen species [6, 7]. The intrinsic efficiency of the CcO proton pump has been proposed to be regulated by the rate of proton backflow through the protein [8-12] possibly via reversal of the normal exit pathway for pumped protons. However, neither the exit nor the backflow pathway has been located, although various routes are postulated [5, 13, 14]. It is clear that defining the proton pathways and understanding what structural features determine the rates of flow, during uptake, exit and backflow, is of fundamental importance to understanding energy balance and metabolic control in health and disease.
This review will summarize the insights gained from studies on the Rhodobacter enzyme (RsCcO) focusing on evidence for the importance of conserved lipid and steroid binding sites in structure and regulation, and for a role of conformational change and water positioning in the gating of proton pathways.
Conserved Lipid Binding Sites
The original RsCcO crystal structure ([15]; PDB ID: 1M56) revealed six phospholipids embedded in the structure, two buried in a cleft in subunit III and four surrounding the single transmembrane helix of subunit IV, in an arrangement that almost completely separated the latter subunit from interaction with the rest of the protein (FIGURE 1). The structure was unprecedented in its demonstration of major involvement of lipid in a membrane protein’s structural integrity.
Figure 1.

Rhodobacter sphaeroides cytochrome c oxidase (RsCcO) showing associated lipid. Four subunits: I,green; II,yellow; III, orange; IV, red. Lipid, phosphatidylethanolamine(PE), indicated as dark gray spheres. Drawing created in Chimera (UCSF) from PDB ID: 1M56.
Also striking were the images of lipids associated with the bovine enzyme: when structures were obtained at 1.8 Å resolution, a total of 13 different lipids per 13-subunit monomer were resolved and each binding site was specific for a particular lipid [16] (PDB ID: 2DYR). (FIGURE 2)
Figure 2.

Bovine cytochrome c oxidase dimer at 1.8 Å showing associated lipids (dark gray spheres). Drawing created in Pymol from PDB ID: 2DYR.
The importance of specific lipid associations was further emphasized when a second structure of the Rhodobacter enzyme [17] (PDB ID: 2GSM) was obtained at higher resolution (2.0 Å). This structure contained the two core subunits I and II, but was missing subunits III and IV that were initially seen as having the major lipid interactions. In the new structure, alkyl chains are seen embedded in the membrane domain of the protein surface, some with defined head groups of the sugar-containing detergents used in the purification and others without a resolved head group. Interestingly, the sites occupied by lipids and detergents in the RsCcO structure were found to overlay precisely with positions occupied by lipid or detergent in both Paracoccus and bovine CcO. Analysis of the residues that created the grooves holding the alkyl chains also showed a high level of conservation in all three structures [18], emphasizing the significance of the lipid binding sites. Such conserved sites have also been noted in other membrane proteins [19-22]. What these specifically-bound lipids contribute to the structural and functional properties of intrinsic membrane proteins remains to be clarified. A possibility is that they could act as flexible caulking between subunits and helices, shielding internal water channels while allowing conformational change. In fact, both proton uptake pathways, D and K, when inhibited by removal of a carboxyl at their entrance, can be chemically rescued by addition of μmolar levels of lipidic molecules with a carboxyl group, indicating a lipid binding site in close proximity [8, 23, 24].
Another interesting facet of crystallographically-defined, specific lipid binding sites is the observation that certain detergents, particularly sugar-based detergents, are capable of substituting for lipid molecules in these sites. The positioning of the sugar head groups indicate that they are particularly good substitutes because they stack and hydrogen-bond effectively with the aromatic residues that are concentrated in the region of the protein at the membrane interface[17]. This stable interaction may account for their unusual success in purifying [25, 26] and crystallizing membrane proteins [27].
A conserved steroid binding site
Of even greater significance with respect to the function and regulation of CcO is the discovery that one of the steroid / bile salt binding sites first recognized in the bovine crystal structure [28] and attributed to possible nucleotide binding, is also conserved in the bacterial enzyme [24]. In both cases, bile salts bound in this site appear to influence the activity of CcO by interacting with a carboxyl residue (E101II in Rs, E62 II in bovine) at the entrance to a key proton uptake route, the K pathway (FIGURE 3).
Figure 3.
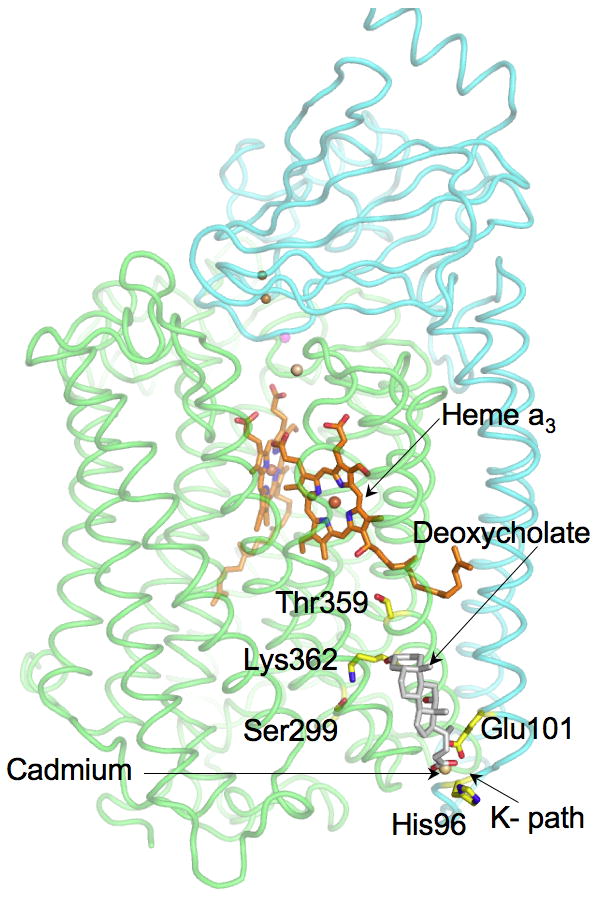
RsCcO two-subunit form (PDB ID: 3DTU)) showing the position of deoxycholate (light gray) in proximity to the K proton pathway. Subunit I, green; Subunit II, cyan. Labeled residues are those with a role in K path. Cadmium is located at the entrance of the path, bound to E101, His96, deoxycholate and water (not shown). Drawing was created in Pymol by Dr.Ling Qin.
Initial clues about the existence of this site in RsCcO came from studies of a mutant in which the conserved carboxyl, E101II, is replaced with an alanine, resulting in a strongly inhibited form (5% WT activity). Certain bile salts at μM levels were found to rescue the mutant, increasing its activity by more than 10 fold, to 50% of WT [23]. The most powerful chemical rescue was provided by deoxycholate, and a crystal structure of RsCcO determined in its presence resolved a single deoxycholate with its carboxyl group in close proximity to that of E101II (PDB ID: 3DTU), as predicted from its activating ability.
An unexpected finding, not predicted from the activation effects on the K-path, was that deoxycholate in RsCcO occupied a site that closely aligned with that of a cholate molecule in the bovine CcO structure[24]. This cholate site was previously noted to be associated with subunit 6a of bovine CcO at the interface of the dimer, rather than with subunits I and II of the alternate monomer, but in fact both steroids show a strong interaction with the homologous carboxyls at the mouth of the K channel (FIGURE 4). While it has long been known that certain detergents, including bile salts and Triton X100, inhibit bovine CcO [29], this new finding suggested that the inhibitory effect of cholate on the bovine enzyme is due to its ability to compromise the K path; a similar conclusion was reached by other investigators studying inhibition of bovine CcO by Triton X100 [30]. In contrast, the stimulatory effect on the E101II mutant is apparently due to the bile acid’s ability to bind and replace the needed carboxyl thus restoring some of the proton uptake capacity of the K-path. The E101IIA mutant therefore provides as a sensitive assay for ligand binding at the site, allowing the measurement of a compound’s ability to activate or inhibit the mutant form, or compete with a known activator or inhibitor.
Figure 4.
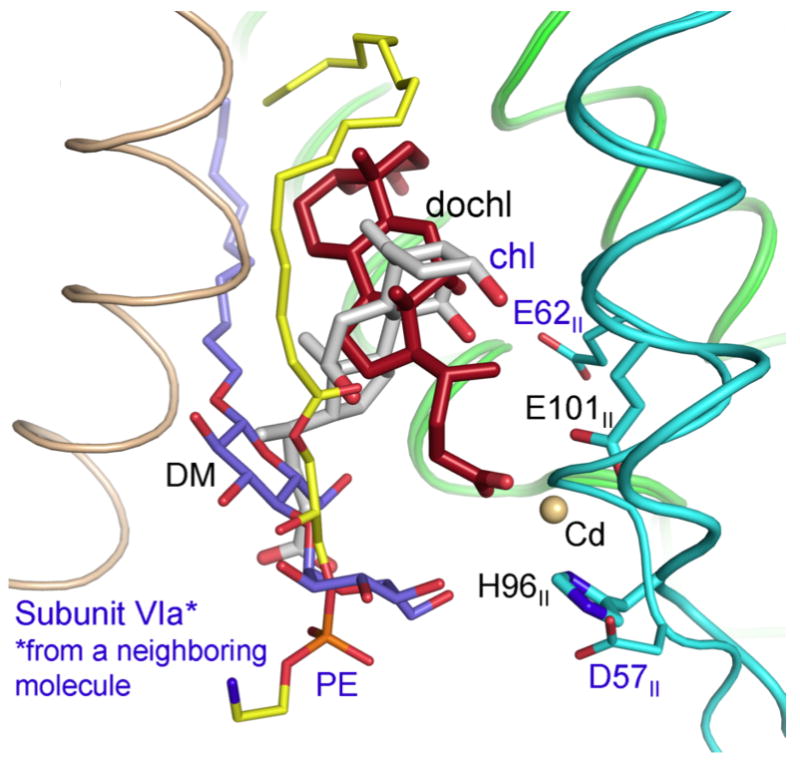
Overlay of Bovine CcO (PDB 2DYR) and RsCcO (PDB 3DTU) showing proximity of binding sites for cholate (red sticks) and deoxycholate (gray sticks). Associated lipids are shown: PE in bovine (yellow) and decylmaltoside in Rhodobacter (purple). Subunit VIa from the other monomer in the bovine dimer is shown (wheat). Subunit I (green). Subunit II (cyan). Drawing created in Pymol by Dr Ling Qin.
In the course of studies utilizing this system, it was found that there is a complex competition between ligands at the site. Notably, the level of activity of the E101IIA mutant is influenced not only by carboxyl containing compounds but also by certain detergents used for routine assay, even at the low concentrations that support maximal activity of wild type (e.g. 0.06% dodecylmaltoside). The results indicate that the binding pocket in the E101IIA mutant has high affinity for several detergents that, when bound, increase the blockage of the K path beyond the mutation itself; when these non-ionic detergents are displaced by bile salts or other molecules that provide a carboxyl, activity is partially restored. This detergent/ligand competition does not appear to affect so markedly the wildtype RsCcO, presumably because the pocket is less hydrophobic in the wildtype due to the strategic placement of the conserved carboxyl. The findings raise questions about which properties of the K path entrance region are most important in controlling proton uptake, the carboxyl itself or the ability of the carboxyl to create a more hydrophilic environment that allows water to be organized in the entrance region and prevents hydrophobic ligands from occupying it. The specificity of the K path entrance pocket for binding certain lipidic ligands may also explain conflicting experimental findings regarding the effects of mutations in this region [19, 31], likely due to different experimental or purification conditions, or different structural properties of various CcO forms.
A number of compounds appear to compete at the steroid binding site, as evidenced by their effects on the E101IIA mutant. Affinities in the micromolar concentration range have been determined for several bile salts, cholesterol-hemisuccinate, protoporphyrin IX, bilirubin, phytanic acid, arachidonic acid and some detergents [32]. It is important to note that the cholate binding sites in the bovine CcO have been postulated to represent regulatory nucleotide binding sites[33]; however, we were unable to demonstrate experimentally an effect of nucleotides in our system under a variety of conditions including long preincubation, even though computational modeling of the site supports that possibility.
In general, stimulation of the E101IIA mutant occurs when the effector molecule has hydrophobic character and certain structural features that allow it to bind, as well as a carboxyl group or some ability to facilitate proton uptake into the K path, such as organizing water. Inhibition occurs when the compound can bind but interferes with proton uptake. Striking in this regard is the fact that, in all the crystal structures obtained of the native CcO from bacteria or bovine sources, neither detergent nor lipid is resolved in the steroid site, only water or cholate or deoxycholate when present. This may be an indication that the site is designed to be open to allow proton access via water chains. Competition between activators and inhibitors of E101IIA suggest that they are binding at the same site as the bile salts [32], but the physiological significance of any of these molecules and the precise nature of their influence on protein conformation or water organization in or near the K-path remains unclear. One clue to possible clinical importance comes from a report [34] that bilirubin, at levels found in neonatal jaundice, has a direct inhibitory effect on neuronal CcO, an effect that is counteracted by bile salts. Since these are both competing ligands at the K-path steroid site, it is tempting to think that this might be the site of interaction responsible for the observed cellular response.
Computational analysis and crystallographic studies are currently aimed at understanding the molecular details of the interactions involved and identifying other ligands, including nucleotides, that may be able to bind at the site and exert physiological regulatory effects.
Conformational change and conserved water
A prevalent view of the mechanism of CcO is that proton pumping does not depend on any significant conformational changes beyond rearrangements of side chain positions and water molecules [35-37] In contrast, in the case of the bovine CcO, structural changes in the vicinity of heme a have been observed in reduced crystals and interpreted to be involved in a pumping mechanism associated with a proton pathway designated the H path [38], but neither the pathway [39] nor the conformational changes were apparent in the bacterial enzyme [22, 40]. However, recent crystal structures of the Rhodobacter CcO in dithionite-reduced compared to oxidized states, have revealed conformational changes that differ in some respects from those seen in bovine CcO, involving a shift in position of heme a3 and of closely associated regions of the structure [2, 41].
These new structures provide evidence for involvement of conformational change in a gating mechanism that allows the opening of the K path for substrate protons to be correlated with closing of the D path that conveys pumped protons. In the oxidized structure, entrance of protons from the K path into the heme a3-CuB active site appears to be blocked by a tight hydrogen bond (2.6 Å) between the OH of the CuB His-Tyr-ligand and the OH of the farnesyl group of heme a3 (FIGURE 5). In the reduced RsCcO structure, movement of the heme a3 porphyrin ring away from CuB breaks the bond and creates a larger opening (4.1Å) at the bottom of the active site. Formation of a chain of waters leading into the heme a3 – CuB crevice is seen in the reduced crystal, suggesting a path for proton uptake from the K pathway below. A shift in helix VIII and the K-path residue T359 appears to be associated with the water rearrangement. In addition, the reduced crystal shows the loss of a water from a position close to CuB ([2]; W301) that has been invoked as part of a connection between the top of the D path and the active site[4]. This change suggests a mechanism to allow alternating access of protons from K and D paths, a necessary feature of the CcO pumping mechanism.
Figure 5.
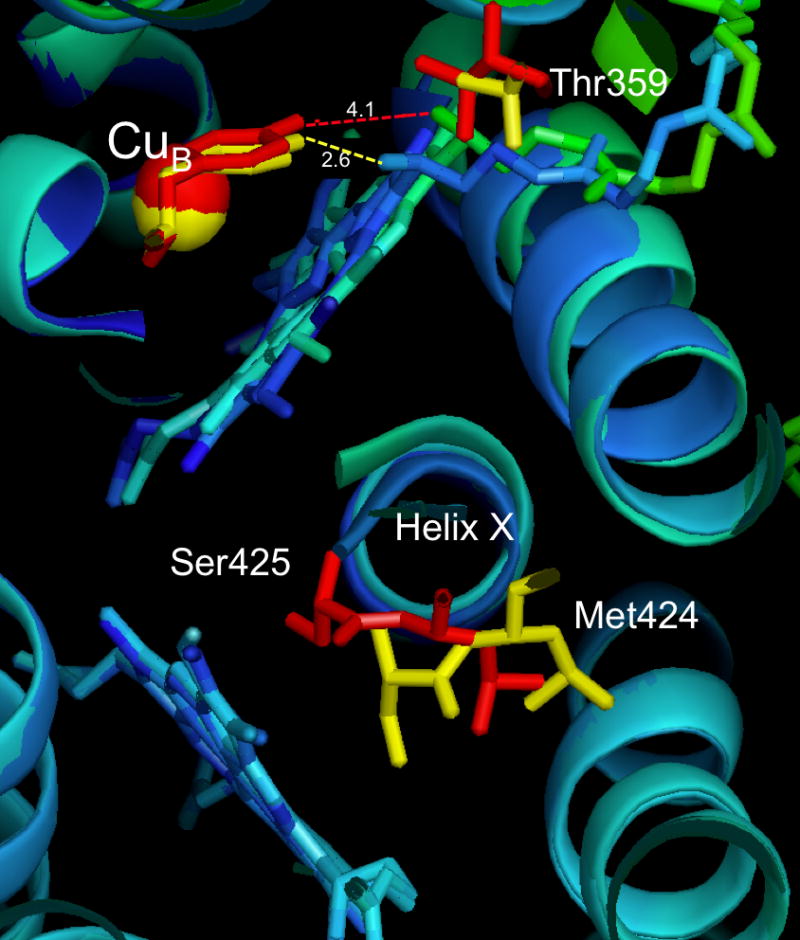
Overlay of crystal structures of oxidized (PDB ID: 2GSM) and reduced (PDB ID: 3FYE) forms of two-subunit RsCcO. Red indicates reduced and yellow indicates oxidized, for CuB, Tyr288, Thr359, Met424 and Ser425 where significant movement is observed. Hydrogen bond lengths between the Tyr288-OH and the heme a3 farnesyl-OH are indicated by dotted lines, 2.6 Å (yellow, oxidized) and 4.1Å (red, reduced). Heme a3 is cyan and green (reduced), blue (oxidized). The central helix X is rendered in turquoise (oxidized) dark blue (reduced). The drawing is a cross-section viewed from the inside of the membrane, created in Pymol.
This finding is particularly interesting because it fits with a previously proposed mechanism for proton pumping [13] that predicts that reduction of CuB will trigger the loss of the tight hydrogen bond at the bottom of the heme a3 crevice, as actually observed in the reduced crystal structure. In the model, reduction of CuB followed by movement of a proton from the D-path to the vicinity of its His334 ligand, weakened the bonding of CuB to the His-Tyr ligand, resulting in internal electronic rearrangements within this unique cross-linked pair. The internal rearrangement in turn caused the His-Tyr-OH hydrogen bond with the farnesyl–OH to be broken, opening access to water molecules and K-path protons[13]. Aside from giving a new perspective on the role of this unusual CuB ligand as an important gate for the K path, the model and the reduced structure emphasize the critical function of water positioning in forming and breaking proton paths.
It is important to note that these conformational changes are also observed in crystals of the four-subunit RsCcO, and so are not dependent on the crystal type [2]. In addition, the effects of reduction are reversible: when a crystal is reduced and then allowed to reoxidize, it returns to the oxidize conformation. However there are other concerns regarding the significance of the structural changes observed in Rs and bovine CcO, since they differ in several respects.
A concern regarding all crystal structures of redox active metalloproteins is the known ability of high intensity x-rays to release electrons from the protein and solvent, so that the metal centers in an oxidized crystal may become reduced during data collection [2, 22, 41-45] . To address this question we have examined the spectra of frozen RsCcO crystals during x-ray exposure [41] and found the noteworthy result that, although heme a appears to be rapidly reduced even at 70 °K, the observed spectrum is not that of the native reduced CcO, but shows a split peak at 589 and 610 nm in the alpha-band region. When the crystal is briefly annealed (warmed for a few seconds) the spectrum then becomes that of a normal reduced heme a at 606 nm (Figure 6). This finding suggests that the frozen crystal retains the protein structure of the oxidized form even when the metal center is reduced, leading to a “strained” configuration at the metal center giving rise to the altered spectrum. This finding explains why conformational differences can be seen between reduced and oxidized forms, but could imply that we are not seeing the full extent of change. An even more rigorous test was applied by Yoshikawa and colleagues working with the bovine CcO [46]. Using 400 crystals and a few seconds exposure each, they showed that the same oxidized structure was obtained as when they collected data for much longer periods on one crystal.
Figure 6.

Spectra of crystals of wildtype RsCcO during X-ray irradiation at 100 degree K. Time of exposure and peak wavelengths are indicated on graph. A double peak at 589 and 610 nm is shown to develop during 10 minutes radiation. A peak at 606 nm is observed after annealing. Similar results were obtained with wildtype four-subunit crystals. Drawing created in PLOT.
It is important to note that there are some commonalities between the Rs and bovine reduced structures: the bovine CcO shows the same significant movement of residues in helix X between the two hemes as RsCcO (FIGURE 5) and a similar, though smaller, shift in the heme a3 porphyrin ring in the CN/reduced form[47]. But the structures differ in the degree of heme a3 movement and the resultant opening of the bottom of the active site crevice, as well as the movement of helix VIII. The RsCcO also shows no changes in the vicinity of the equivalent of the H pathway (close to the position of Asp 51, bovine numbering). These inconsistencies and the lack of change found in Paracoccus CcO [40] and the thermophilic ba3 oxidase [22] remain to be resolved, but could well be the result of differences in completeness of reduction and deoxygenation, differences in water content, and/or restrictions imposed by dissimilar crystal contacts.
Conclusions
The mechanism and regulation of coupling between electron transfer and proton pumping in cytochrome c oxidase remains a complex unsolved problem of major physiological importance, given the central role of this enzyme in aerobic energy metabolism. The ability to obtain high-resolution structures in which lipid and water positions can be observed and conformational changes dependent on redox state detected, is providing new insight into possible proton gating modes and regulatory ligands. The crystallographic capture of new catalytic intermediates and the discovery of new crystal forms, with better resolved water and lipidic ligands, will be key to further illuminating this subtle and intricate process.
Highlights.
Cytochrome c oxidase is a critical player in regulation of aerobic metabolism
Lipid binding sites are conserved, indicating structural and functional roles
Conformational change is involved in regulation of proton uptake in K path
Frozen crystals maintain structure even when reduced by x-‐rays
Acknowledgments
We thank Dr. Ling Qin for several original figures used in this review.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wikstrom M, Krab K. Proton-pumping cytochrome c oxidase. Biochimica Et Biophysica Acta-Bioenergetics. 1979;549:177–222. doi: 10.1016/0304-4173(79)90014-4. [DOI] [PubMed] [Google Scholar]
- 2.Qin L, Liu J, Mills DA, Proshlyakov DA, Hiser C, Ferguson-Miller S. Redox-dependent conformational changes in cytochrome C oxidase suggest a gating mechanism for proton uptake. Biochemistry. 2009;48:5121–5130. doi: 10.1021/bi9001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu J, Sharpe MA, Qin L, Ferguson-Miller S, Voth GA. Storage of an excess proton in the hydrogen-bonded network of the d-pathway of cytochrome C oxidase: identification of a protonated water cluster. J Am Chem Soc. 2007;129:2910–2913. doi: 10.1021/ja067360s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wikstrom M, Verkhovsky MI, Hummer G. Water-gated mechanism of proton translocation by cytochrome c oxidase. Biochimica Et Biophysica Acta-Bioenergetics. 2003;1604:61–65. doi: 10.1016/s0005-2728(03)00041-0. [DOI] [PubMed] [Google Scholar]
- 5.Sharpe MA, Krzyaniak MD, Xu S, McCracken J, Ferguson-Miller S. EPR evidence of cyanide binding to the Mn(Mg) center of cytochrome c oxidase: support for Cu(A)-Mg involvement in proton pumping. Biochemistry. 2009;48:328–335. doi: 10.1021/bi801391r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kadenbach B, Arnold S, Lee I, Huttemann M. The possible role of cytochrome c oxidase in stress-induced apoptosis and degenerative diseases. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 2004;1655:400–408. doi: 10.1016/j.bbabio.2003.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Huttemann M, Lee I, Pecinova A, Pecina P, Przyklenk K, Doan J. Regulation of oxidative phosphorylation, the mitochondrial membrane potential, and their role in human disease. Journal of Bioenergetics and Biomembranes. 2008;40:445–456. doi: 10.1007/s10863-008-9169-3. [DOI] [PubMed] [Google Scholar]
- 8.Fetter J, Sharpe M, Qian J, Mills D, Ferguson-Miller S, Nicholls P. Fatty acids stimulate activity and restore respiratory control in a proton channel mutant of cytochrome c oxidase. FEBS Lett. 1996;393:155–160. doi: 10.1016/0014-5793(96)00874-5. [DOI] [PubMed] [Google Scholar]
- 9.Ferguson-Miller S, Babcock GT. Heme/Copper Terminal Oxidases. Chemical Reviews. 1996;96:2889–2908. doi: 10.1021/cr950051s. [DOI] [PubMed] [Google Scholar]
- 10.Mills DA, Ferguson-Miller S. Influence of structure, pH and membrane potential on proton movement in cytochrome oxidase. Biochim Biophys Acta. 2002;1555:96–100. doi: 10.1016/s0005-2728(02)00261-x. [DOI] [PubMed] [Google Scholar]
- 11.Mills DA, Schmidt B, Hiser C, Westley E, Ferguson-Miller S. Membrane potential-controlled inhibition of cytochrome c oxidase by zinc. J Biol Chem. 2002;277:14894–14901. doi: 10.1074/jbc.M111922200. [DOI] [PubMed] [Google Scholar]
- 12.Mills DA, Tan Z, Ferguson-Miller S, Hosler J. A role for subunit III in proton uptake into the D pathway and a possible proton exit pathway in Rhodobacter sphaeroides cytochrome c oxidase. Biochemistry. 2003;42:7410–7417. doi: 10.1021/bi0341307. [DOI] [PubMed] [Google Scholar]
- 13.Sharpe MA, Ferguson-Miller S. A chemically explicit model for the mechanism of proton pumping in heme-copper oxidases. J Bioenerg Biomembr. 2008;40:541–549. doi: 10.1007/s10863-008-9182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugitani R, Stuchebrukhov AA. Molecular dynamics simulation of water in cytochrome c oxidase reveals two water exit pathways and the mechanism of transport. Biochim Biophys Acta. 2009;1787:1140–1150. doi: 10.1016/j.bbabio.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Svensson-Ek M, Abramson J, Larsson G, Tornroth S, Brzezinski P, Iwata S. The X-ray crystal structures of wild-type and EQ(I-286) mutant cytochrome c oxidases from Rhodobacter sphaeroides. J Mol Biol. 2002;321:329–339. doi: 10.1016/s0022-2836(02)00619-8. [DOI] [PubMed] [Google Scholar]
- 16.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci U S A. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin L, Sharpe MA, Garavito RM, Ferguson-Miller S. Conserved lipid-binding sites in membrane proteins: a focus on cytochrome c oxidase. Curr Opin Struct Biol. 2007;17:444–450. doi: 10.1016/j.sbi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koepke J, Olkhova E, Angerer H, Muller H, Peng G, Michel H. High resolution crystal structure of Paracoccus denitrificans cytochrome c oxidase: new insights into the active site and the proton transfer pathways. Biochim Biophys Acta. 2009;1787:635–645. doi: 10.1016/j.bbabio.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Hasan SS, Yamashita E, Ryan CM, Whitelegge JP, Cramer WA. Conservation of Lipid Functions in Cytochrome bc Complexes. J Mol Biol. 2011;413 doi: 10.1016/j.jmb.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roszak AW, Gardiner AT, Isaacs NW, Cogdell RJ. Brominated lipids identify lipid binding sites on the surface of the reaction center from Rhodobacter sphaeroides. Biochemistry. 2007;46:2909–2916. doi: 10.1021/bi062154i. [DOI] [PubMed] [Google Scholar]
- 22.Liu B, Chen Y, Doukov T, Soltis SM, Stout CD, Fee JA. Combined microspectrophotometric and crystallographic examination of chemically reduced and X-ray radiation-reduced forms of cytochrome ba3 oxidase from Thermus thermophilus: structure of the reduced form of the enzyme. Biochemistry. 2009;48:820–826. doi: 10.1021/bi801759a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin L, Mills DA, Hiser C, Murphree A, Garavito RM, Ferguson-Miller S, Hosler J. Crystallographic location and mutational analysis of Zn and Cd inhibitory sites and role of lipidic carboxylates in rescuing proton path mutants in cytochrome c oxidase. Biochemistry. 2007;46:6239–6248. doi: 10.1021/bi700173w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin L, Mills DA, Buhrow L, Hiser C, Ferguson-Miller S. A conserved steroid binding site in cytochrome C oxidase. Biochemistry. 2008;47:9931–9933. doi: 10.1021/bi8013483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosevear P, VanAken T, Baxter J, Ferguson-Miller S. Alkyl glycoside detergents: a simpler synthesis and their effects on kinetic and physical properties of cytochrome c oxidase. Biochemistry. 1980;19:4108–4115. doi: 10.1021/bi00558a032. [DOI] [PubMed] [Google Scholar]
- 26.Suarez MD, Revzin A, Narlock R, Kempner ES, Thompson DA, Ferguson-Miller S. The functional and physical form of mammalian cytochrome c oxidase determined by gel filtration, radiation inactivation, and sedimentation equilibrium analysis. J Biol Chem. 1984;259:13791–13799. [PubMed] [Google Scholar]
- 27.Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 29.Yu C-A, Yu L, King TE. Studies on Cytochrome Oxidase: Interactions of cytochrome oxidase protein with phospholipids and cytochrome c. Journal of Biological Chemistry. 1975;250:1383–1392. [PubMed] [Google Scholar]
- 30.Antalik M, Jancura D, Palmer G, Fabian M. A role for the protein in internal electron transfer to the catalytic center of cytochrome c oxidase. Biochemistry. 2005;44:14881–14889. doi: 10.1021/bi050824z. [DOI] [PubMed] [Google Scholar]
- 31.Richter O-MH, Dürr KL, Kannt A, Ludwig B, Scandurra FM, Giuffrè A, Sarti P, Hellwig P. Probing the access of protons to the K pathway in the Paracoccus denitrificans cytochrome c oxidase. FEBS Journal. 2005;272:404–412. doi: 10.1111/j.1742-4658.2004.04480.x. [DOI] [PubMed] [Google Scholar]
- 32.Hiser C, Buhrow L, Liu J, Ferguson-Miller S. New Ligands of the Conserved Steroid Binding Site of Cytochrome c Oxidase. Biophysical Journal. 2012 in press. [Google Scholar]
- 33.Napiwotzki J, Shinzawa-Itoh K, Yoshikawa S, Kadenbach B. ATP and ADP bind to cytochrome c oxidase and regulate its activity. Biol Chem. 1997;378:1013–1021. doi: 10.1515/bchm.1997.378.9.1013. [DOI] [PubMed] [Google Scholar]
- 34.Vaz AR, Delgado-Esteban M, Brito MA, Bolanos JP, Brites D, Almeida A. Bilirubin selectively inhibits cytochrome c oxidase activity and induces apoptosis in immature cortical neurons: assessment of the protective effects of glycoursodeoxycholic acid. J Neurochem. 2010;112:56–65. doi: 10.1111/j.1471-4159.2009.06429.x. [DOI] [PubMed] [Google Scholar]
- 35.Wikstrom M, Ribacka C, Molin M, Laakkonen L, Verkhovsky M, Puustinen A. Gating of proton and water transfer in the respiratory enzyme cytochrome c oxidase. Proc Natl Acad Sci U S A. 2005;102:10478–10481. doi: 10.1073/pnas.0502873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovic DM, Stuchebrukhov AA. Proton pumping mechanism and catalytic cycle of cytochrome c oxidase: Coulomb pump model with kinetic gating. FEBS Lett. 2004;566:126–130. doi: 10.1016/j.febslet.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 37.Ghosh N, Prat-Resina X, Gunner MR, Cui Q. Microscopic pKa analysis of Glu286 in cytochrome c oxidase (Rhodobacter sphaeroides): toward a calibrated molecular model. Biochemistry. 2009;48:2468–2485. doi: 10.1021/bi8021284. [DOI] [PubMed] [Google Scholar]
- 38.Yoshikawa S, Muramoto K, Shinzawa-Itoh K. Proton-pumping mechanism of cytochrome C oxidase. Annu Rev Biophys. 2011;40:205–223. doi: 10.1146/annurev-biophys-042910-155341. [DOI] [PubMed] [Google Scholar]
- 39.Lee HM, Das TK, Rousseau DL, Mills D, Ferguson-Miller S, Gennis RB. Mutations in the putative H-channel in the cytochrome c oxidase from Rhodobacter sphaeroides show that this channel is not important for proton conduction but reveal modulation of the properties of heme a. Biochemistry. 2000;39:2989–2996. doi: 10.1021/bi9924821. [DOI] [PubMed] [Google Scholar]
- 40.Harrenga A, Michel H. The cytochrome c oxidase from Paracoccus denitrificans does not change the metal center ligation upon reduction. Journal of Biological Chemistry. 1999;274:33296–33299. doi: 10.1074/jbc.274.47.33296. [DOI] [PubMed] [Google Scholar]
- 41.Liu J, Qin L, Ferguson-Miller S. Crystallographic and online spectral evidence for role of conformational change and conserved water in cytochrome oxidase proton pump. Proc Natl Acad Sci U S A. 2011;108:1284–1289. doi: 10.1073/pnas.1012846108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshikawa S, Shinzawa-Itoh K, Nakashima R, Yaono R, Yamashita E, Inoue N, Yao M, Fei MJ, Libeu CP, Mizushima T, Yamaguchi H, Tomizaki T, Tsukihara T. Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science. 1998;280:1723–1729. doi: 10.1126/science.280.5370.1723. [DOI] [PubMed] [Google Scholar]
- 43.Macedo S, Pechlaner M, Schmid W, Weik M, Sato K, Dennison C, Djinovic-Carugo K. Can soaked-in scavengers protect metalloprotein active sites from reduction during data collection? Journal of Synchrotron Radiation. 2009;16:191–204. doi: 10.1107/S0909049509003331. [DOI] [PubMed] [Google Scholar]
- 44.Sommerhalter M, Lieberman RL, Rosenzweig AC. X-ray crystallography and biological metal centers: is seeing believing? Inorg Chem. 2005;44:770–778. doi: 10.1021/ic0485256. [DOI] [PubMed] [Google Scholar]
- 45.Lee HJ, Svahn E, Swanson JM, Lepp H, Voth GA, Brzezinski P, Gennis RB. Intricate role of water in proton transport through cytochrome c oxidase. J Am Chem Soc. 2010;132:16225–16239. doi: 10.1021/ja107244g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aoyama H, Muramoto K, Shinzawa-Itoh K, Hirata K, Yamashita E, Tsukihara T, Ogura T, Yoshikawa S. A peroxide bridge between Fe and Cu ions in the O2 reduction site of fully oxidized cytochrome c oxidase could suppress the proton pump. Proc Natl Acad Sci U S A. 2009;106:2165–2169. doi: 10.1073/pnas.0806391106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muramoto K, Ohta K, Shinzawa-Itoh K, Kanda K, Taniguchi M, Nabekura H, Yamashita E, Tsukihara T, Yoshikawa S. Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate. Proc Natl Acad Sci U S A. 2010;107:7740–7745. doi: 10.1073/pnas.0910410107. [DOI] [PMC free article] [PubMed] [Google Scholar]


